Summary
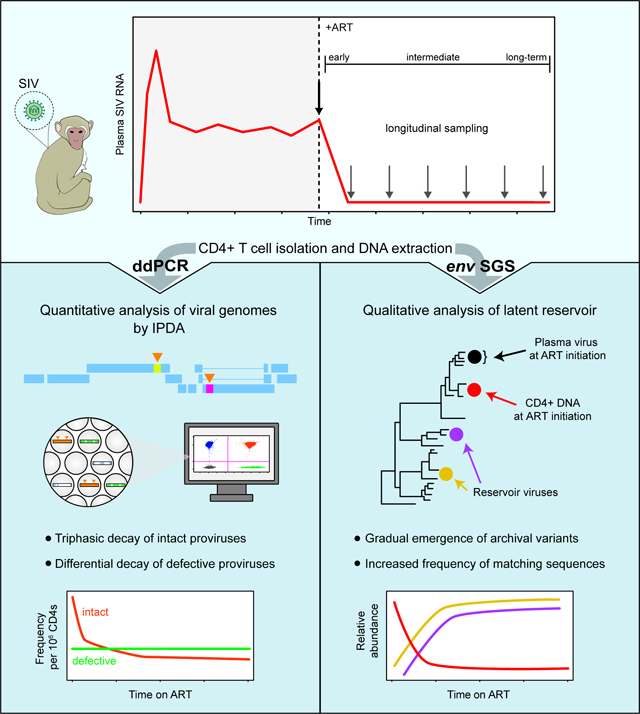
The decay kinetics of HIV-1-infected cells are critical to understand virus persistence. We evaluated the frequency of simian immunodeficiency (SIV)-infected cells for 4 years of antiretroviral therapy (ART). The intact proviral DNA assay (IPDA) and an assay for hypermutated proviruses revealed short- and long-term infected cell dynamics in macaques starting ART ~1 year after infection. Intact SIV genomes in circulating CD4+T cells showed triphasic decay with an initial phase slower than decay of plasma virus, a second phase faster than second phase decay of intact HIV-1, and a stable third phase reached after 1.6–2.9 years. Hypermutated proviruses showed bi- or mono-phasic decay, reflecting different selective pressures. Viruses replicating at ART initiation had mutations conferring antibody escape. With time on ART, viruses with fewer mutations became more prominent, reflecting decay of variants replicating at ART initiation. Collectively, these findings confirm ART efficacy and indicate cells enter the reservoir throughout untreated infection.
eTOC Blurb
SIV-infected macaques are an important animal model for people living with HIV (PLWH). Fray et al. measured the decay of SIV-infected cells after treatment with antiretroviral therapy. They found that SIV-infected cells decay more rapidly than in PLWH and provide a benchmark for evaluating future cure strategies in this model.
Graphical Abstract
Introduction
Antiretroviral therapy (ART) halts HIV-1 replication, resulting in a rapid biphasic drop in viremia to below the detection limit1. However, ART is not curative due to a pool of latently infected CD4+ T cells that harbor replication-competent transcriptionally silent proviruses2–4. This small population of cells has a half-life of 3.7 years, guaranteeing lifetime HIV-1 persistence even with optimal ART5,6. Regardless of the length of treatment, viremia rapidly rebounds if ART is stopped7,8.
The decay kinetics of plasma virus and infected CD4+ T cells provide insights into the establishment and persistence of the latent reservoir1,9,10. However, the measurement of infected cell populations is complicated because in people living with HIV-1 (PLWH) on ART, most infected cells harbor proviruses with fatal defects, including large deletions and APOBEC3G/F-mediated G-to-A hypermutation11–19. To address this problem, we developed a digital droplet PCR (ddPCR) assay in which individual proviruses suspended in nanoliter-sized droplets are interrogated with two strategically chosen amplicons that are more likely to detect intact viral genomes20,21. This intact proviral DNA assay (IPDA) provides a more accurate measurement of the reservoir size by excluding most defective proviruses.
The IPDA was used to confirm the long-term stability of the reservoir21–24 and more recently, to measure the decay of intact and defective HIV-1 genomes during the first 2 years of ART25. Some intact genomes detected at ART initiation may be linear unintegrated viral DNA molecules in resting cells26,27. However, these forms decay within days28,29, and in activated cells, integration occurs within hours of completion of reverse transcription30. Thus, most viral genomes detected with the IPDA are integrated proviruses. White et al. used the IPDA to show that after ART initiation intact genomes decayed with biphasic kinetics characterized by a rapid initial phase (t1/2 = 12.9 days) and a slower second phase (t1/2 = 19 months) that was still faster than the slow reservoir decay observed in PLWH on long-term ART5,24,25,31. In this same study, cells with defective proviruses showed monophasic decay, and 2LTR circles, which are also defective for replication showed biphasic decay and long-term stability25,32–35. These findings raise questions about the nature and location of the populations of infected cells that produce most of the plasma virus, the mechanisms by which they decay, and the relationship between these rapidly decaying populations and the latent reservoir.
These questions can be more easily addressed in macaques infected with simian immunodeficiency virus (SIV) because the sequence of the infecting virus and the timing of infection and ART can be controlled36–39. As in PLWH, there is viremia, progressive CD4+ T cell depletion, chronic immune activation, decay of viremia upon ART initiation, immune control in some instances, and a latent reservoir established in resting CD4+ T cells37,40–50. Measurement of the SIV latent reservoir is also complicated by defective viral genomes20,51. The decay of SIV-infected cells in macaques on ART has not been fully described.
Here, we evaluated changes in the frequency of SIV-infected cells for 4 years following ART initiation. We used the IPDA and an assay for hypermutated proviruses to compare the decay rates of intact and defective proviruses. We used single-genome sequencing (SGS) of plasma virus and proviral DNA to examine qualitative changes in infected cell populations during ART. The sequencing analyses provided strong evidence for the efficacy of ART and insights into whether the latent reservoir is established predominantly at ART initiation or continuously throughout untreated infection52–56. The changes in infected cell populations identified here should be considered in the design and evaluation of cure interventions.
Results
Cohort characteristics
We studied Indian-origin rhesus macaques (cohort 18–02, n=10) inoculated with SIVmac251 by repetitive low-dose intrarectal challenge (Fig. 1A, Fig. S1). After 48 weeks, animals were placed on a combination ART regimen of tenofovir disoproxil fumarate, emtricitabine, and dolutegravir (TDF/FTC/DTG)57,58. They were maintained on ART without additional interventions for >4 years (Fig. 1A). Cohort details are included in the supplement and the STAR Methods section (Fig. S1A).
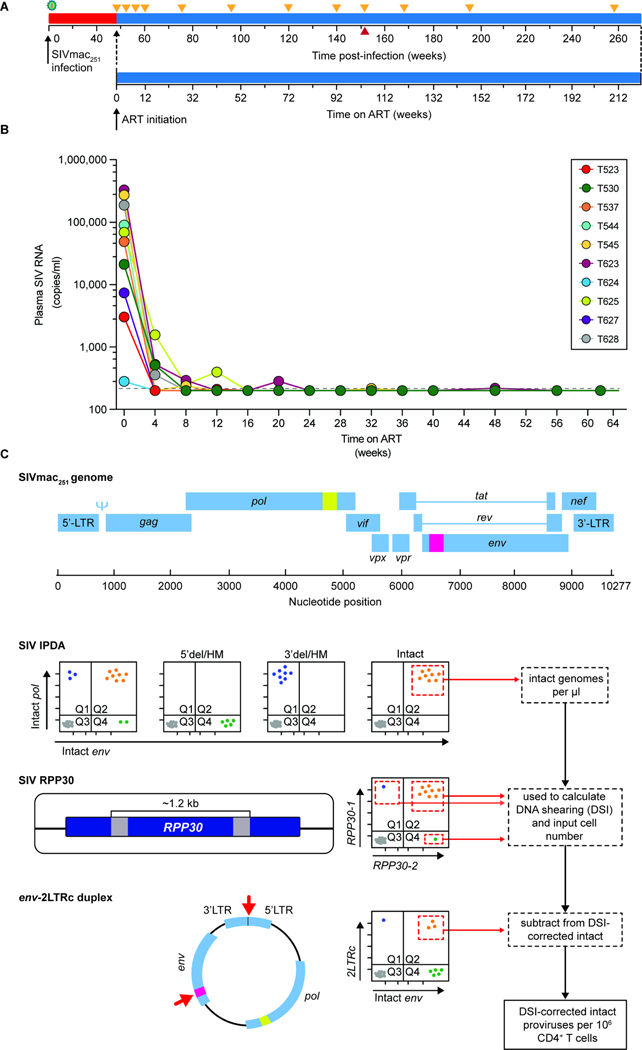
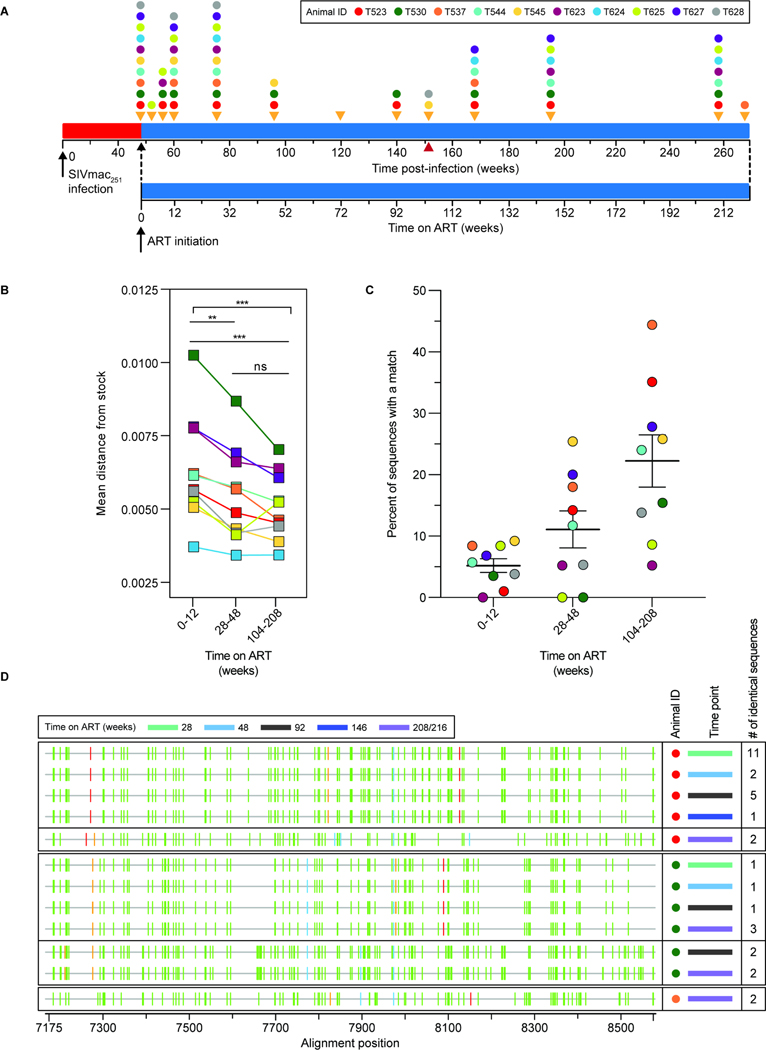
Figure 1. Study design.
(A) Timeline of infection, initiation of ART, and sampling. Animals were infected by repetitive intrarectal challenge with SIVmac251 and maintained untreated for 48 weeks after initial challenge (red) before initiation of ART (blue). Orange triangles indicate sampling times. The red triangle indicates the last time point before two animals (T545 and T628) were necropsied due to unrelated complications. (B) Plasma SIV RNA levels after ART initiation. The assay detection limit (200 copies/ml) is indicated by the dashed gray line. (C) Design of the SIV Intact Proviral DNA Assay (IPDA)20. Locations of two amplicons in the SIV genome are indicated in yellow (pol) and pink (env). Data from 3 individual assays are used to determine the frequency of intact proviruses per 106 CD4+ T cells. Events from quadrant 2 (Q2) of the IPDA measure the number of intact genomes. Proviruses with deletions or hypermutation affecting the pol amplicon (5’del/HM) appear in Q4 while proviruses with deletions or hypermutation affecting the env amplicon (3’del/HM) appear in Q1. Proviruses with defects affecting both amplicons appear in Q3 along with droplets lacking a provirus. The number of intact proviruses (Q2) is corrected for shearing using the DNA Shearing Index (DSI), and the input cell number is determined using ddPCR amplification of two similarly spaced amplicons in a cellular gene (RPP30) as described20,21. 2LTR circles are quantitated in a separate ddPCR assay using two amplicons, one spanning the LTR-LTR junction62 and one in env (red arrows)20. Events in Q2 of the env-2LTRc duplex assay are subtracted from the number of intact proviruses to correct for 2LTR circles.
Changes in plasma virus upon ART initiation
All animals were infected by SIVmac251 after one or more challenges, and showed peak viremia typical of SIV infection of rhesus macaques (geometric mean = 1.05 × 107 copies/ml) (Fig. S1A)41. ART rapidly suppressed viral replication as demonstrated by an exponential decrease in plasma SIV RNA (Fig. 1B, Fig. S1B). For 6 of 10 animals, plasma SIV RNA levels fell below the detection limit within 4 weeks (Fig. 1B, Fig. S1B). 4 of 10 animals experienced infrequent blips during the first 48 weeks of ART, after which plasma SIV RNA remained undetectable (Fig. 1B).
IPDA analysis of reservoir size and changes in intact proviruses on ART
According to classic viral dynamics models1,9,10,59, the rapid drop in viremia following ART initiation (Fig. 1B) reflects the loss of productively infected cells. However, this prediction is difficult to verify because cells that produce most of the plasma virus may not be in the circulation25,60, and because many cells carry defective proviruses20,21,61. Therefore, we used the SIV IPDA20 to quantitate cells with intact proviruses. This assay excludes most deleted or hypermutated proviruses20,21. Our analysis also included a duplex ddPCR assay for a cellular gene (RPP30) and a duplex ddPCR assay for 2LTR circles (Fig. 1C)20,62
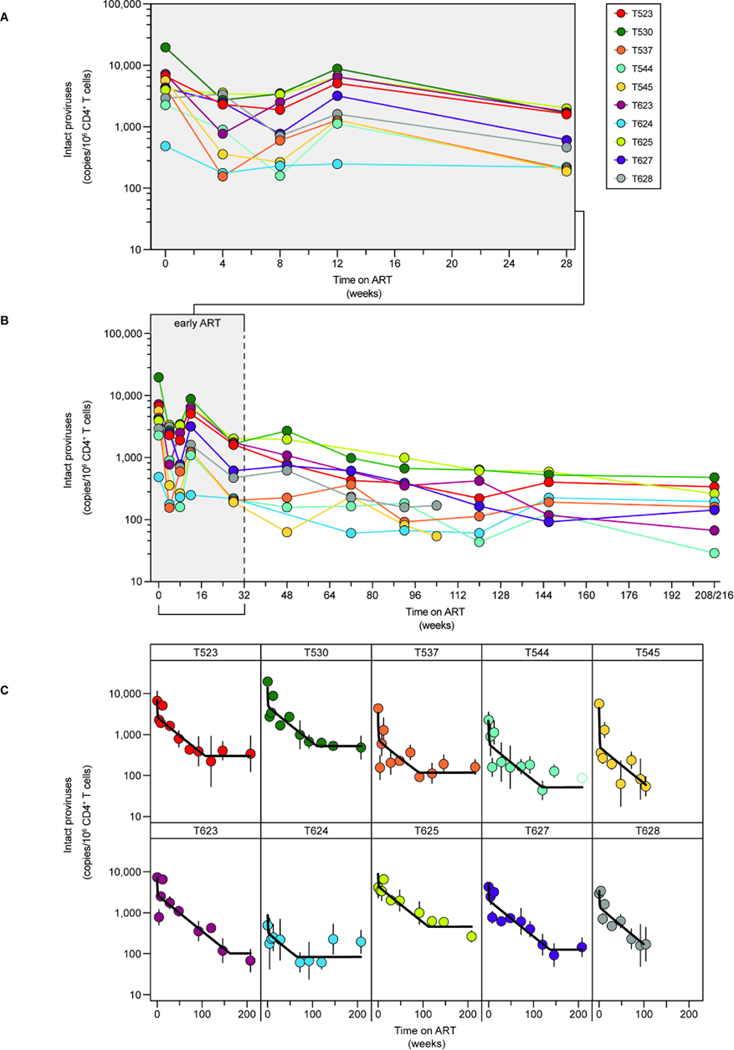
At ART initiation, intact proviral frequencies in circulating CD4+ T cells were similar to those in untreated PLWH (Fig. 2A–C, geometric mean frequency 3,642 copies per 106 CD4+ T cells, compared to 2,255 copies per 106 CD4+ T cells for HIV-125). We did not observe a relationship between viral load at week 0 and the frequency of intact proviruses at either week 0 or at a later time point (week 92) after which the more labile populations of infected cells have largely decayed (Fig. S1B, C). We observed rapid initial decay of intact proviruses over the first 4 weeks of ART, a small but consistent increase around 12 weeks (Fig. 2A), then slower decay for ~2 years until stabilization occurred (Fig. 2B). We used non-linear mixed-effects modeling to determine the decay rate of intact proviruses. Changes in the frequency of cells harboring intact genomes were best described by a three-phase model, which is most evident in plots for individual macaques (Fig. 2C, Table S1).
Figure 2. Decay of intact proviruses after ART initiation.
Frequency of intact proviruses during (A) the first 6 months of ART or (B) over the first 4 years of treatment. Datapoints represent the geometric mean from at least 3 replicates per animal. Open circles indicate that no positive events were detected by the IPDA; the y-axis value indicates the limit of detection based on the number of cell equivalents assayed. (C) Decay of intact proviruses in individual animals during ART. Data were fitted to a triphasic model (black line) as described in the Methods. Vertical lines indicate the standard deviations.
During the first 4 weeks of ART, intact SIV proviruses decayed with a half-life of 3.3 days (95% CI: 2.5–4.5 days), faster than the initial decay of HIV-1 proviruses in people initiating ART (12.9 days25). Although fast compared to the subsequent decay of the remaining intact SIV proviruses, it is nevertheless slower than the decay of plasma SIV RNA over the same period (Fig. S1D, E). Therefore, the very rapid initial decay of viremia may reflect the equally rapid death of infected cells located primarily in tissues, as seen for HIV-125.
Given the strong suppression of viremia upon ART initiation, the increase in intact proviruses at week 12 was unexpected. Single genome sequencing described below did not reveal any unique properties of variants detected at week 12, such as an increase in divergence. This increase at approximately the same time post-treatment has also been observed in an independent cohort of SIV-infected animals (unpublished data, Fray) and may reflect a redistribution of infected cells from tissues to blood63,64. In modeling the decay kinetics, we tested models that included (Table S1) or ignored the week 12 time point (Table S2). Whether the week 12 data were included or not, the best fit model was consistent with a triphasic decay pattern. All results discussed below are based on the full dataset (Table S1). It is unclear whether this increase in intact proviruses also occurs in animals that are started on ART after the relatively short periods of untreated infection characteristic of many cure studies.
The rapid initial decay of intact SIV proviruses was followed by a slower second phase (t1/2 = 8.1 months, 95% CI: 6.5–10.0 months). This decay is also faster than the second phase decay of intact HIV-1 proviruses (t1/2 = 19 months25). The more rapid first and second-phase decay of intact SIV genomes relative to HIV-1 has not been previously described. After a mean of 2.3 years (95% CI: 1.6–2.9 years), the slope changes to a stable third phase with no decay (Fig. 2C, Tables S1, S3). Most SIV cure experiments are conducted during the first 2 years of ART, when the more rapid second phase decay is still occurring. Thus, adequately powered control groups are needed to distinguish natural and intervention-induced decay.
Decay of 2LTR circles during ART
2LTR circles showed triphasic decay similar to that measured for intact proviruses (Fig. S2, Tables S1, S4). After 4 years of ART, 2LTR circles were rare. env+2LTR circles, which are subtracted from the Q2 IPDA result, were only detected in 2 of 8 animals at weeks 208–216 (Fig. S2). The low frequency of env+2LTR circles after 4 years of ART could reflect the death of cells harboring the circles and/or cellular proliferation causing dilution of the circles which are not copied during replication of host cells. Overall, total and env+ 2LTR circles showed decay kinetics that were very similar to those described for intact proviruses (Fig. S2, Tables S1, S3), as is the case with HIV-125.
An assay for hypermutated proviruses
In addition to detecting intact proviruses in Q2, the HIV-1 IPDA detects proviruses with hypermutation and/or deletions in Q1 and proviruses with 5’ deletions in Q421. Because of differences in the distribution of defects, the SIV IPDA uses different amplicons20, and many of the defective proviruses end up in Q3 (Fig. 1C) with the droplets that do not contain a provirus (estimated ~55% compared to ~3.8% for HIV based on in silico analyses of near-full-length sequencing data20,21). For this reason, the SIV IPDA does not provide quantitation of defective proviruses. To determine whether decay of intact proviruses is due to virus-dependent factors such as viral cytopathic effects and immune clearance or to virus-independent factors such as normal CD4+ T cell turnover, we developed a method to quantify one class of defective SIV genomes. APOBEC3G/F-mediated hypermutation introduces multiple missense and nonsense mutations in most open reading frames, likely precluding high-level viral gene expression20. We therefore developed a ddPCR assay specific for hypermutated SIV proviruses based on the same principles and amplicon positions as the SIV IPDA. We used fluorescently labeled pol and env probes with 2 G→A mutations at frequently hypermutated positions within each probe and unlabeled competitor probes with the non-mutated sequences (Fig. S3). Details of the Hypermutated Proviral DNA Assay (HPDA) are provided in the supplement (Fig. S3).
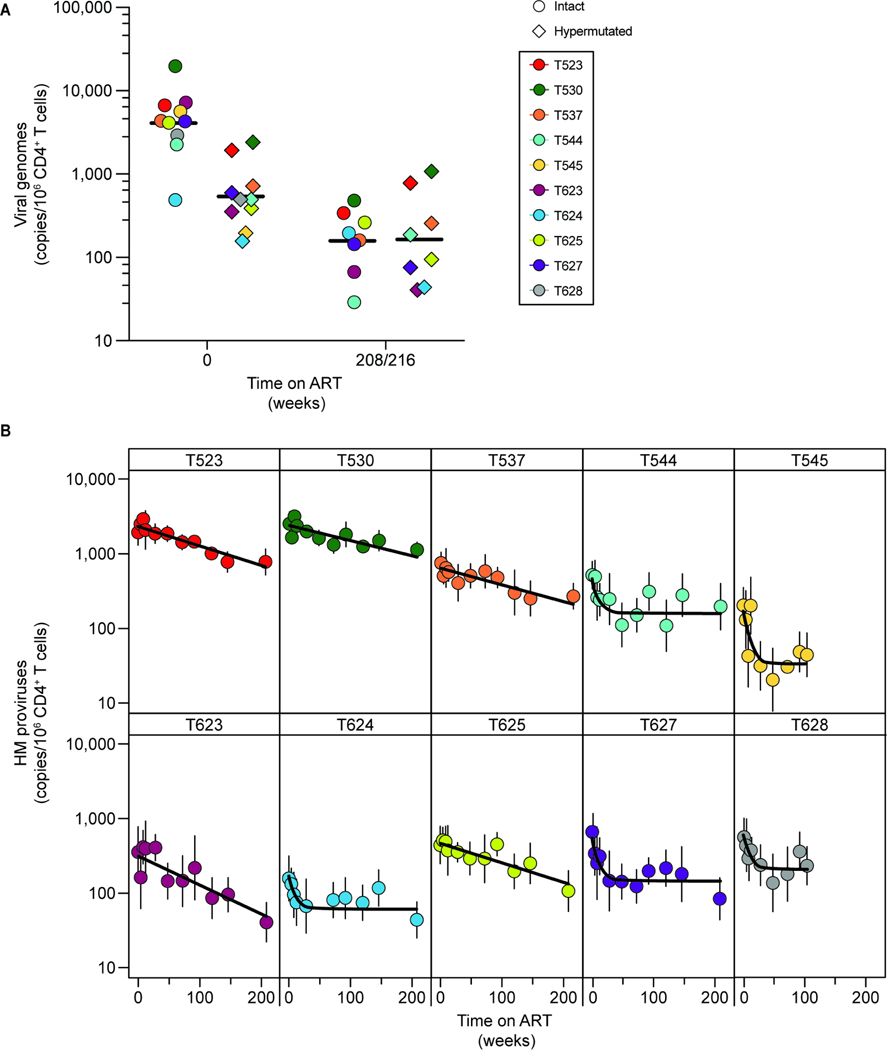
Hypermutated proviruses were less frequent than intact proviruses (geometric mean frequency = 539 copies per 106 CD4+ T cells, compared to 3,642 intact copies per 106 CD4+ T cells, Fig. 3A). Hypermutated proviruses were most abundant at the time of ART initiation (Fig. 3A) and decayed with kinetics different from those of intact proviruses (Fig. 3B) and variable between animals (Table S5). The best-fit model for hypermutated proviruses was not uniform. There was biphasic decay for 5 animals and monophasic decay for the other 5 (Fig. 3B, Table S5). For the animals with biphasic decay, hypermutated proviruses decayed initially with an average half-life of 42 days (95% CI: 31–55 days), followed by a stable second phase with no decay (Fig. 3B, Tables S1, S5). For the other 5 animals, we observed monophasic decay with an average half-life of 27.8 months (95% CI: 19.5–39.9 months) (Fig. 3B, Tables S1, S5). These kinetics are strikingly different from those described for cells harboring intact SIV proviruses. Overall, the decay of hypermutated proviruses is slower, such that by week 216 the average frequencies are similar (Fig. 3A).
Figure 3. Decay of hypermutated proviruses after ART initiation.
(A) Comparison of the frequency of intact and hypermutated proviruses prior to ART initiation (t=0) and at the latest time point analyzed (week 208 for all animals except T537, in which case it is week 216). Each datapoint represents the geometric mean for the IPDA or HPDA. A total of 3 replicates for each animal are included for the IPDA, and 6 replicates for the HPDA. IPDA data are corrected for env+2LTR circles and shearing. HPDA data are corrected for shearing. Black lines show the geometric means. (B) Decay of hypermutated proviruses on ART. Data were fitted to biphasic or monophasic decay models (black line) as described in the Methods section. Vertical lines represent the standard deviations.
Analysis of infected cell decay using gag PCR
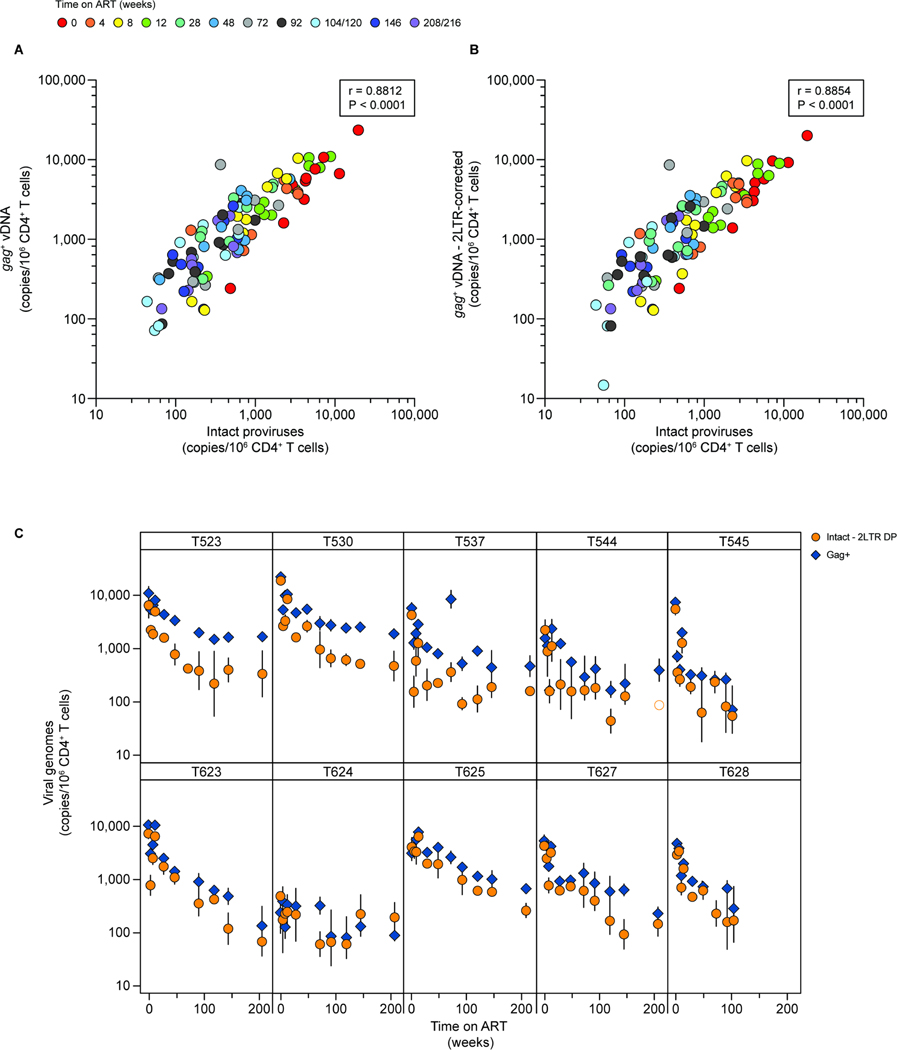
Many SIV reservoir studies use a single amplicon in the well-conserved gag gene to quantify SIV DNA65,66. We therefore analyzed viral decay using a ddPCR assay targeting gag67. This assay gave values similar to, or slightly higher than those measured by the IPDA (Fig. 4A). This reflects the fact that the ratio of intact to defective proviruses is higher for HIV-1 than for SIV20,21,51 and the fact that some defective SIV proviruses have hypermutation or deletions affecting the gag amplicon and thus are also excluded by the gag PCR assay. We found a strong positive correlation between the gag and IPDA assays (r=0.8812, R2=0.7765, 95% CI=0.8281–0.9186, p<0.0001, n=100; Pearson’s test). Even when the gag data were corrected by the same env+2LTR correction applied to the IPDA values (Fig. 4B; r=0.8854, R2=0.7840, 95% CI=0.8341–0.9216, p<0.0001, n=100; Pearson’s test), gag values were, in general, higher, likely reflecting the detection of some defective proviruses with the gag assay.
Figure 4. Comparison of gag ddPCR and IPDA quantification of SIV DNA.
(A) Correlation between gag+ SIV DNA copies and intact proviruses quantified using the IPDA. Individual datapoints represent the number of copies detected using either an amplicon in gag (y-axis)67 or the IPDA (x-axis)20. Each point is the geometric mean of 3 replicates. Intact proviruses are corrected for shearing (DSI) and env+2LTR circles. The correlation between the two variables was calculated using Pearson’s coefficient. Data from one animal, T624, were excluded from this analysis due to failure of the gag amplicon resulting from mutations or deletion. (B) Correlation between gag+ SIV DNA copies and intact proviruses, with gag values corrected using the same env+2LTR correction factor applied to the IPDA data, calculated using Pearson’s coefficient. (C) Comparison of the decay of SIV gag copies and intact proviruses for the animals in cohort 18–02. IPDA data are corrected for env+2LTR circles and DNA shearing. Vertical lines represent the standard deviations. See also Figure S4.
As with intact proviruses, the decay of gag+ DNA was best described by a triphasic model (Fig. 4C, Tables S1, S6). The average first phase half-life was 6.5 days (95% CI: 5.9–7.4 days), not significantly different from the initial decay of intact proviruses. However, at the later time points the half-lives became longer relative to intact proviruses (Table S6). We calculated an average second phase half-life of 13.0 months (95% CI: 11.8–14.1 months; Table S1). After a mean of 34.5 months (95% CI: 27.3–43.3 months) there was a third phase with essentially no decay (Tables S1, S6). These results are consistent with the fact that the gag assay is also capturing some defective proviruses, which generally have a longer half-life24,25. Indeed, the gag probe detected hypermutated proviruses with 1 to 3 G→A mutations in the probe binding site (Fig. S4A–C). Consistent with this observation, the greatest differential between gag and IPDA measurements was found in animals with a higher frequency of hypermutated proviruses detected by the HPDA (animals T523, T530 and T537; Fig. 4C, Fig. S4B, D), although as time on ART increased, there was a greater discrepancy between the two assays in most animals. Therefore, the gag assay is a good approximation of the total abundance of viral DNA, including defective proviruses, but overestimates the frequency of intact proviruses, especially after longterm ART.
Qualitative changes in infected cell populations on ART
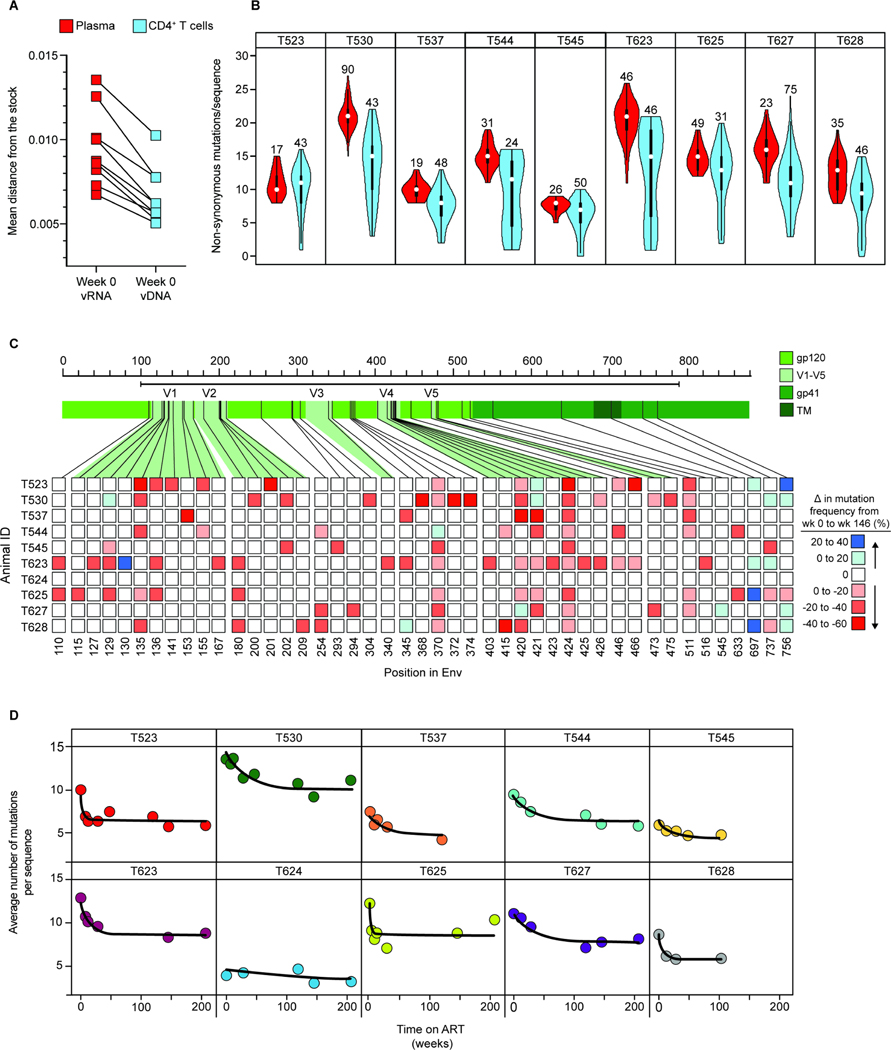
To understand qualitative changes in the pool of SIV-infected cells following ART initiation, we collected a large number of env sequences by single-genome sequencing of DNA from peripheral blood CD4+ T cells at weeks 0–216 of ART (Fig. 5A). After excluding sequences with deletions, frameshifts, or hypermutation, we were left with ~1700 sequences that were non-defective in the env gene. We determined the average pairwise distance (APD) from the consensus sequence of the inoculum57. We found no increase in APD over ~4 years of treatment (Fig. 5B, Table S7). Thus, despite the enormous diversification and divergence away from the stock sequence during untreated infection, there was no further evolution during ART. In fact, APD decreased with increasing time on ART (Fig. 5C). The mechanism underlying this apparent “reverse evolution” is described below.
Figure 5. Analysis of env sequences from CD4+ T cell DNA during ART.
(A) Timeline for sequence analysis. Colored dots indicate sampling times for each animal. (B) Average pairwise distance from the stock57 of env sequences collected at early (weeks 0–12), intermediate (weeks 28–48) or late (weeks 104–216) time points after ART initiation. The number of sequences analyzed for each animal is listed in Table S7. Statistical significance was determined using one-way ANOVA (GraphPad Prism). (C) Percent of non-defective env sequences that exactly match another independent sequence from the same sample at early, intermediate, or late time points on ART. Sequences with hypermutation (as determined using Los Alamos National Lab’s Hypermut Program99), or deletions were excluded from this analysis. Animal T624 was excluded due to a high frequency of identical sequences at all time points, reflecting early control of replication (Fig. 1B, Fig. S13). (D) Highlighter plots of identical hypermutated env sequences from 3 animals. The animal (colored dot, see key in panel A), time point (colored bar), and the number of times an identical sequence was recovered from a given sample are indicated. Vertical bars indicate a nucleotide difference from the consensus of the stock57. The color of the bar represents the mutant nucleotide (red – thymine, blue – cytosine, orange – guanine, green – adenine). Alignment numbering corresponds to nucleotide positions based on the SIVmac239 reference (accession no: M33262.1).
We also analyzed env sequences for evidence of proliferation of infected cells, as has been clearly demonstrated in PLWH68–72. In macaques infected with SIVmac251 for ~1.5 years and treated with ART for 60 weeks20, we previously found only 2 pairs of identical sequences among more than 500 independent env sequences. Another study used integration site analysis to recover matching integration sites from macaques treated during acute infection, although at low abundance73. The extended follow-up here allowed us to observe the appearance of identical sequences over time. Among ~1700 non-defective sequences from 9 animals, the fraction of identical sequences increased with time on ART (Fig. 5C) as seen in PLWH12,72. However, proviruses with identical sequences could contain polymorphisms elsewhere74,75. Therefore, we extended our analysis to include 226 defective, hypermutated sequences collected from 3 animals (Fig. 5D) and captured identical sequences. Proviruses with extensive hypermutation are unable to replicate20, and therefore must arise through proliferation or through infection of multiple cells by the same viral variant with APOBEC3G/F-mediated hypermutation subsequently occurring in exactly the same way in each cell. Analysis of the likelihood of G→A mutation at each G in env (see Methods) showed that the likelihood of the latter scenario is extremely low (p<10−7). Thus, infected cells with identical hypermutated env sequences arise through a proliferative process that also likely explains at least some of the identical intact env sequences observed at later time points.
env sequence analysis and evidence of antibody pressure
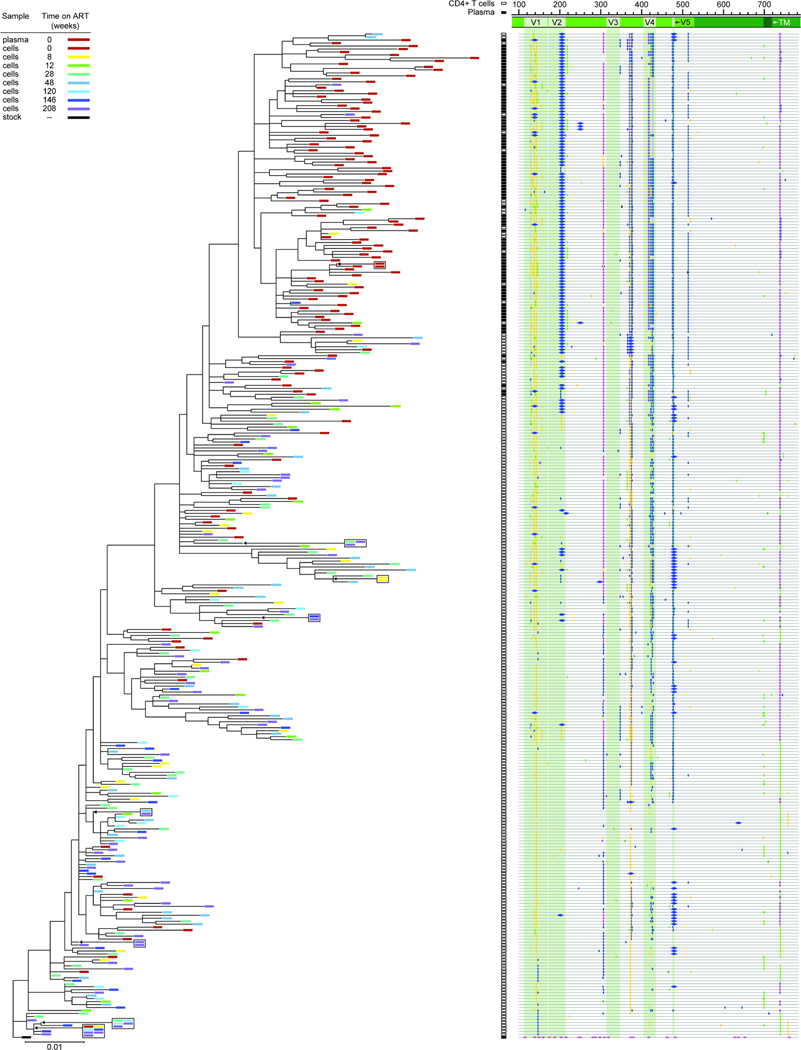
We also compared env sequences from plasma virus at ART initiation to non-defective proviral env sequences from circulating CD4+ T cells at various time points on ART (Fig. 5A). A representative maximum likelihood phylogenetic tree for animal T530 is shown in Figure 6. This tree is rooted on the consensus of the stock sequences57 and illustrates the enormous sequence divergence from the stock and intra-individual diversification arising during untreated infection. Similar patterns were observed for all other animals (Fig. S5–12). Although all animals were infected with the same stock, the divergence occurred differently in each animal as shown by a composite phylogenetic tree (Fig. S13A). The exception is T624 (Fig. S13B), which spontaneously controlled viral replication to low levels before ART (Fig. S1). However, for the other 9 animals, in all cases mutations accumulated in the gp120 variable regions. The V1/V2 and V4 regions of the SIV env gene are under strong selective pressure from the antibody response76–90, and key amino acid positions consistently accumulate non-synonymous mutations in MHC-disparate animals, coincident with the development of autologous neutralizing antibodies (aNabs). We found many previously reported mutations80,88, including at N202, W345, D415, R420, P421, and R424. In 9 animals, we found the R424Q mutation, an early indicator of escape from humoral immunity88. Other mutations alter potential N-linked glycosylation sites (PNGs), including D415N, which creates a new PNG, and non-synonymous changes at residue N202, which removes a PNG in V183,84,88. These mutations were observed in more than one, and sometimes as many as 9 of 10 animals. In 2 animals (T530 and T623), we also observed characteristic deletions in V4 (Fig. 6, Fig. S9) that mediate escape from aNabs80,88.
Figure 6. Sequencing analysis of env diversity in plasma virus and proviral DNA from CD4+ T cells for T530.
Phylogenetic tree and highlighter plot of env sequences collected from either plasma or CD4+ T cell DNA from animal T530. Highlighter plot was generated using Los Alamos National Lab’s Highlighter program. The tree is rooted on a consensus sequence of the stock57. Sequence analysis includes 2054 nt.
Plasma sequences at ART initiation show high divergence from the stock relative to proviral sequences in CD4+ T cells
The time 0 plasma sequences represent virus actively replicating at ART initiation. In addition to the ~1700 sequences collected from CD4+ T cell DNA, we collected over 300 plasma sequences from 9 animals at week 0. Compared to proviral env sequences in circulating CD4+ T cells at the same time, plasma sequences showed a high mean divergence from the stock in all 9 animals analyzed (Fig. 7A). Plasma sequences were consistently mutated at the positions associated with antibody pressure (Fig. 6, Fig. S5–12). No wild-type sequences that clustered closer to the root were recovered from plasma from any animal at week 0 (Fig. 6, Fig. 7B, Fig. S5–12). Importantly, the time 0 plasma sequences were found predominantly on distal branches (Fig. 6, Fig. 7B, Fig. S5–12). This can be illustrated by comparing the number of non-synonymous mutations away from the stock per sequence for the time 0 plasma and cellular sequences (Fig. 7B). Some circulating CD4+ T cells present at ART initiation contained SIV variants that had accumulated antibody escape mutations similar to those in plasma, while other CD4+ T cells present at the same time harbored variants that had fewer of these mutations and that were not seen in plasma at the time of ART initiation. These are likely ancestral sequences from earlier pre-ART time points preserved in the latent reservoir. This finding was consistent for all animals and not related to the depth of sampling (Fig. 7B).
Figure 7. Differences in mutation frequency between time 0 plasma virus and cellular sequences at time 0 and week 146.
(A) Comparison of average pairwise distance from the stock57 between sequences collected from plasma (red) or peripheral blood CD4+ T cell DNA (blue) at the time of ART initiation (t=0). The number of sequences analyzed and standard deviations are given in Table S7. (B) Violin plots comparing the number of non-synonymous amino acid mutations per env sequence in plasma (red) vs. CD4+ T cell DNA (blue) at the time of ART initiation. Numbers above the violin plots indicate the number of sequences included in the analysis. (C) Heat map depicting changes in mutation frequency at different positions in env between sequences collected from week 0 and week 146 for each animal. Week 146 was chosen as a late time point because slight increases in mutational frequency were observed between week 146 and 208, reflecting distortions introduced by the proliferation of particular infected cell clones. Red or blue squares indicate increases or decreases, respectively, in the frequency of sequences with a mutation at the indicate position. Analysis covers sequenced positions where there were mutations away from the stock consensus in at least 20% of sequences at that time point. (D) Decay of non-synonymous mutations in env sequences during ART. Data were fitted to a biphasic model (black line) as described in the Methods section. The number of sequences included in this analysis, mean APD, and standard deviation for each animal at the indicated timepoints are included in Table S7.
Sequences with large numbers of antibody escape mutations become less abundant at later time points
In contrast to the normal pattern of increasing divergence over time in untreated infection41,74,88,91,92, sequences with large numbers of antibody escape mutations became less abundant at the later time points on ART (Fig. 7C). This is evident in the phylogenetic trees (Fig. 6, Fig. S5–12). Relatively unmutated sequences close to the root were frequently isolated at later time points. At weeks 120–216 on ART, sequences with few or no mutations relative to the stock became more apparent. We analyzed the kinetics of the decay process by examining changes in the mean number of non-synonymous mutations per sequence (Fig. S5–12. 7D). Modeling revealed a biphasic decay pattern. The initial phase of decay has a half-life of 11.4 weeks (95% CI: 4.6–28.1 weeks). The second phase is so slow as to be statistically indistinguishable from “no decay.” The net result of these qualitative changes is summarized in Fig. 7C, which shows that at almost all positions in the env ectodomain mutations are less frequent after 3 years of ART than at ART initiation. This finding is consistent with the decay process described above. The infected cells that decay after ART initiation carry proviruses with multiple immune escape mutations. As these cells decay, less mutated ancestral sequences persisting in the latent reservoir become more apparent. Strikingly, many of these variants closest to the root of the trees are from later on-ART time points (Fig. 6, Fig. S5–12). These results provide strong evidence that ART arrests viral evolution; but more importantly, they show that viruses are deposited in the latent reservoir throughout untreated infection and not just at ART initiation.
Discussion
To inform HIV-1 cure strategies, we characterized quantitative and qualitative changes in SIV-infected CD4+ T cells following ART initiation during chronic infection and over the next 4 years. Our findings provide critical insights into the dynamics of infected cells over a time frame in which many intervention studies are conducted.
Overall, the decay kinetics of cells harboring intact SIV proviruses are similar to those reported for HIV-125. In both cases, there is complex, multiphasic decay, with the final phase representing the stable persistence of a latent reservoir in CD4+ T cells. For both viruses, there are different decay rates for intact and defective proviruses, likely reflecting selective pressures operating on cells capable of expression viral genes. In both cases, cells with 2LTR circles decay with kinetics similar to those of intact proviruses. Together, these findings support the use of the non-human primate (NHP) model in HIV-1 cure research.
Our study did uncover important differences in decay kinetics that must be considered in cure studies. Notably, although the frequency of intact genomes at the start of ART was similar for SIV and HIV-1 (geometric mean frequency = 3,642 vs. 2,255 per 106 CD4+ T cells25, the first and second phase half-lives for intact SIV genomes are significantly shorter than those reported for HIV-1 (3.3 days and 8.1 months for SIV vs. 12.9 days and 19 months for HIV-125). Differences in the second phase decay kinetics are particularly important given that most cure studies in NHP models involve interventions carried out during second phase decay. The relatively quasi-stable third phase is not reached until ~2.3 years of ART. Studies of cure interventions during this second phase must include robust untreated controls to distinguish natural vs. intervention-induced decay.
By following treated macaques for 4 years, we observed in the same animals both the early and late phase dynamics shaping the latent reservoir. Most of the plasma virus is produced by rapidly decaying cells that are not present in the circulation (Fig. S1D, E)60. However, most infected CD4+ T cells in the circulation at ART initiation are also labile and decay during the first 2.3 years without entering the stable latent reservoir. The stable reservoir becomes apparent as rapidly decaying cells are eliminated (Figs. 5–7). The proliferation of infected cells generates large clones, which after several years comprise a substantial fraction of the reservoir (Fig. 5C, D). This proliferation is particularly evident in HIV-1-infected individuals on long-term ART68–70, and may be responsible for the inflection in the decay of intact proviruses seen after 7 years of ART24.
We also developed an assay for hypermutated SIV proviruses. Most hypermutated SIV genomes have premature stop codons in most genes20,93 and are unlikely to give rise to high-level viral gene expression. We demonstrate slower decay of cells with hypermutated proviruses relative to intact proviruses, consistent with selective processes acting against cells that express viral genes21,23–25,94.
We also compared a single amplicon in gag to the IPDA. The gag assay provides a reasonable measure of intact proviruses at the beginning of ART. However, it becomes progressively less accurate with increasing time on ART as cells with intact proviruses decay and cells with defective proviruses, many of which are detected by the gag assay, persist.
Lastly, we used env SGS to determine whether the quantitative changes in infected cell populations were accompanied by qualitative changes. We show that infected cell populations change in composition during ART. The populations that decay initially are recently infected cells carrying proviruses with immune escape mutations. As these populations decay, ancestral variants with fewer mutations that are archived in the latent reservoir and that are not present in the plasma at the time of ART initiation become more prominent. Our results show that sequences arising throughout the period of untreated infection can be deposited in the latent reservoir. The reservoir includes both early sequences with few escape mutations as well as sequences that have accumulated more escape mutations and are deposited shortly before the time of ART initiation. All of these variants can persist in the reservoir without immune selection as long as viruses remain latent. What changes is the relative proportion of these early and late variants. The change is due to the decay processes defined here. At the time of ART initiation, the population of productively infected cells, as sampled by sequencing of plasma virus, consists exclusively of sequences with many escape mutations (red bars associated with black squares in Fig. 6 and Figs. S5–12). Some of these variants persist and are deposited in the reservoir after several years of ART (blue and purple bars near the top of Fig. 6, Figs. S5–12). Variants with fewer mutations are also preserved in the reservoir and become more prominent with increasing time on ART, as most of the variants that were actively replicating at the time of ART initiation decay. In the long-term, variants from both early and late in untreated infection persist in the reservoir.
This finding is important in the context of recent studies suggesting that the reservoir is predominantly comprised of cells infected at or around the time of ART initiation52,53,56,95. The observed decrease in APD from the stock, the observation that sequences with more mutations in the env variable regions become less apparent at later time points, and the emergence of archival variants during ART are more consistent with a model in which sequences enter the reservoir continually throughout untreated infection54. Latency explains how the cells harboring archival sequences persist during untreated infection in the context of high levels of immune activation, CD4+ T cell death and surveillance from the host immune system. Some of these factors are altered by ART, skewing the reservoir towards variants replicating around the time of treatment initiation. However, it is incorrect to conclude that the reservoir is composed exclusively of such variants.
There are limitations to our study. First, we analyzed only peripheral blood CD4+ T cells. Although we hypothesize that the increase in intact proviruses observed at week 12 is due to the redistribution of CD4+ T cells from tissues to the blood, we have not directly tested this due to limited tissue sample availability from this longitudinal cohort.
Furthermore, the discrepancy between the initial decay rates of plasma viremia and intact proviruses in the blood suggests that the cells producing plasma virus are found at another anatomical location. It will be important to test this hypothesis in future studies using contemporaneous blood and tissue samples. Another important consideration is that the animals in cohort 18-02 were treated during chronic infection. Many NHP studies initiate ART much earlier and may show different decay dynamics.
Together, these findings reveal that the population of SIV-infected CD4+ T cells is dynamic and changes quantitatively and qualitatively for several years following ART initiation. These data provide a framework for evaluating and interpreting intervention trials utilizing the SIV/NHP model.
STAR Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Robert F. Siliciano (rsiliciano@jhmi.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Sequences generated in this study have been deposited in Genbank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This study does not report original code and all computational resources used are publicly available and listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| SIVmac251 swarm57 | Dr. Dan Barouch, BIDMC | N/A |
| Biological samples | ||
| SIVmac251-infected Rhesus macaque PBMCs | Dr. Dan Barouch, BIDMC | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Platinum Taq DNA Polymerase High Fidelity | ThermoFisher Scientific | Cat # 11304011 |
| SuperScript III Reverse Transcriptase | ThermoFisher Scientific | Cat # 18-080-044 |
| dNTP Mix (10 mM each) | ThermoFisher Scientific | Cat # 18-427-088 |
| UltraPure 1M Tris-HCl Buffer (pH 8.0) | ThermoFisher Scientific | Cat # 15-568-025 |
| RNaseOUT Recombinant Ribonuclease Inhibitor | ThermoFisher Scientific | Cat # 10777019 |
| Critical commercial assays | ||
| QIAamp DNA Mini Kit | Qiagen | Cat # 51304 |
| CD4+ T Cell Isolation Kit, Non-Human Primate | Miltenyi Biotec | Cat # 130-092-144 |
| 2X ddPCR Supermix for Probes (no dUTPs) | Bio-Rad | Cat # 186-3024 |
| Deposited data | ||
| SIV - plasma RNA sequences | This paper | OQ168641-OQ168979 |
| SIV - non-defective proviral DNA sequences | This paper | OQ168980-OQ170751 |
| SIV - hypermutated (defective) proviral sequences | This paper | OQ170752-OQ170785 |
| Experimental models: Organisms/strains | ||
| Rhesus macaque (Macaca mulatta) infected with SIVmac251 | Indian origin | Animals T523, T530, T537, T544, T545, T623, T624, T625, T627 & T628 |
| Oligonucleotides | ||
| Oligonucleotides, standard desalting (Sanger sequencing & ddPCR) - see Methods and Table S8 | IDT | N/A |
| FAM/VIC labeled probes with MGB quencher, HPLC purified (see Table S8) | Applied Biosystems | N/A |
| Unlabeled competition probes w/MGB quencher, HPLC purified (see Table S8) | Applied Biosystems | N/A |
| FAM/HEX labeled probes w/MGB-NFQ quencher, HPLC purified (see Table S8) | IDT | N/A |
| Recombinant DNA | ||
| Synthetic double-stranded DNA controls (gBlocks) - see Methods | IDT | N/A |
| Software and algorithms | ||
| QuantaSoft Analysis-Pro | Bio-Rad | http://www.bio-rad.com/en-us/product/qx200-droplet-digital-pcr-system?ID=MPOQQE4VY |
| Geneious Prime | Dotmatics | https://www.geneious.com/prime/ |
| MEGA v7.0 & v11 | MEGA | https://www.megasoftware.net/ |
| Clustal W | EMBL-EBI | http://www.ebi.ac.uk/Tools/msa/clustalw2/ |
| GraphPad Prism | Dotmatics | https://www.graphpad.com/ |
| Monolix 2020R1 | Lixoft | https://lixoft.com/ |
| Los Alamos National Labs - Highlighter | Keele et al.91 | https://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html |
| Los Alamos National Labs - Hypermut 2.0 | Rose et al.99 | https://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html |
Experimental model and subject details
Animals
Animal work was approved by the Institutional Care & Use Committees of Bioqual and the NIH and determined to be in accordance with the guidelines outlined by the Animal Welfare Act and Regulation (USDA) and the Guide for the Care & Use of Laboratory Animals, 8th Edition (NIH).
Cohort 15–09: The details of this study have been described elsewhere20,93. Near-full-length sequencing data collected from SIV-infected Rhesus macaques (n=7, Bender, et al.20; or n=16, Liu, et al.93) were used for assay development and verification of assay targets, as detailed in the “HPDA sequence validation” subsection below.
Cohort 18–02: 10 outbred Indian-origin rhesus macaques (Macaca mulatta) were housed at Bioqual Inc. in Rockville, MD. Animals were infected via repetitive intrarectal challenge with the SIVmac251 swarm57 until infection was confirmed by detection of SIV RNA in the plasma via qPCR. After ~48 weeks, the animals were placed on a daily regimen of tenofovir disoproxil fumarate, emtricitabine, and dolutegravir (TDF/FTC/DTG, Gilead Sciences, Inc.58. The animals were MHC class I genotyped as previously described96. MHC genotypes, ages and sexes of the animals in this cohort are detailed below.
| Animal ID | Sex | Age at time of infection (years) | MHC-A haplotype 1 | MHC-A haplotype 2 | MHC-B haplotype 1 | MHC-B haplotype 2 |
|---|---|---|---|---|---|---|
| T523 | F | 7.6 | A*002 | A*701 | B*017 | B*017 |
| T530 | F | 6.6 | A*023 | A*002 | B*001 | B*024 |
| T537 | F | 5.8 | A*011 | A*012 | B*012 | B*012 |
| T544 | M | 3.6 | A*004 | A*019 | B*024 | B*015 |
| T545 | M | 4.8 | A*004 | A*002 | B*015 | B*055 |
| T623 | M | 4.9 | A*004 | A*018 | B*015 | B*076 |
| T624 | M | 5.1 | A*028 | A*002 | B*001 | B*055 |
| T625 | M | 4.8 | A*002 | A*002 | B*015 | B*043 |
| T627 | M | 3.3 | A*004 | A*025 | B*048 | B*069 |
| T628 | M | 4.4 | A*001 | A*004 | B*012 | B*012 |
Method details
SIV viral load quantification
Plasma SIV RNA was quantified using a qRT-PCR assay targeting gag as described97.
CD4+ T cell isolation & DNA extraction
PBMCs were collected from whole blood, viably frozen in DMSO, and stored in liquid nitrogen until sample processing. Cells were rapidly thawed at 37°C in RPMI + 20% heat-inactivated FBS and subjected to negative depletion using the Miltenyi NHP CD4 Isolation Kit according to the manufacturer’s protocol. After negative depletion, up to 5 × 106 cells were pelleted and resuspended in 200 μl PBS, then lysed for DNA extraction using the QiAamp DNA Mini Kit according to the manufacturer’s directions.
Droplet digital PCR (ddPCR)
Specific primer and probe sequences for each assay are described in Table S820,67. Details specific to individual assays are described below under sub-headings. Conditions common to all ddPCR experiments are described here:
Samples were assayed in triplicate unless indicated otherwise. Reactions were conducted in a total volume of 22 μl with 10 μl 2X Bio-Rad ddPCR Supermix (no dUTPs). Primers were included at a final concentration of 600 nM, and probes at a final concentration of 200 nM. Droplets were made using the Bio-Rad QX200 manual droplet generator and thermal-cycled using a Bio-Rad QX200 Thermalcycler and the following conditions: 95°C for 10 minutes, 50 cycles of (30 seconds at 94°C, 2 minutes at 56°C), and 10 minutes at 98°C, with a ramp rate of 2°C/s. Cycling conditions were modified from the original protocol described by Bender et al.20 to include more cycles, a lower extension temperature and a longer extension time. Droplets were read by the Bio-Rad QX200 Reader and analyzed with Quantasoft Studio Software. Wells with <10,000 droplets were not analyzed.
RPP30
Host genomes in each sample were quantified using a duplex ddPCR assay with 2 amplicons specific for the Rhesus macaque Ribonuclease P/MRP Subunit 30 (RPP30) gene and spaced at the same distance as the SIV IPDA amplicons20,21. The RPP30 primer/probe pairs are listed in Table S8. Primers were included at a final concentration of 500 nM and probes at a final concentration of 250 nM. RPP30 data were used to correct for DNA shearing and to determine the number of host cells per sample.
IPDA
The IPDA was performed as described20 using the primer/probe pairs indicated in Table S8.
env-2LTRc
The env-2LTRc duplex assay was performed as described20,62 using the primer/probe pairs indicated in Table S8.
Hypermutated Proviral DNA Assay (HPDA)
The HPDA was performed using the primer/probe pairs described in Table S8. 6 replicates were performed per sample to account for the overall lower frequency of hypermutated proviruses. Details of assay design, optimization and validation are given below.
HPDA – sensitivity and specificity
The HPDA is an assay designed to quantify hypermutated proviruses, based on the same principles as the IPDA (Fig. S3A)1. To test whether the HPDA correctly distinguishes between templates that are intact at both positions and those which are hypermutated at both positions, we designed synthetic DNA templates to match the pol and env sequence encompassing the region of the HPDA amplicons (Fig. S3B). We designed these templates to reflect the intact probe sequence (no G→A mutations) or hypermutated probe sequence (two G→A mutations) at one or both amplicons, and tested them using the HPDA, or, as a control, the IPDA (Fig. S3C). Both assays performed as predicted for proviruses matching hypermutated or intact sequences at one or both amplicons (Fig. S3C). These data are consistent with control experiments performed when validating the SIV IPDA1 and provide evidence that switching the labeled and unlabeled probes does not affect the assay’s ability to differentiate between intact and hypermutated templates. Using these controls, we determined that the HPDA is sensitive and specific for hypermutated proviruses and performs well using the same thermal-cycling conditions as the IPDA.
HPDA – sequence validation
Although the HPDA performed well when used to measure templates which exactly match one of two target sequences, in vivo, SIV proviruses may contain polymorphisms which could affect probe binding. Because the assay we describe here essentially relies on two bases within each amplicon to discriminate intact and hypermutated proviruses, it is important to determine whether additional sequence variation in the probe site could affect our ability to detect hypermutated genomes in biological samples from infected macaques. We therefore analyzed the same 266 sequences used to design the IPDA1 for sequence conservation within the probe binding site (Fig. S3D, E). By this analysis, approximately 30% of hypermutated proviruses would be missed by our assay due to deletions encompassing both probe sites. However, ~90% of proviruses with sequence information for at least one of two probe sites (i.e. proviruses without deletions at both amplicons) would be detected by the HPDA. In addition to these 266 sequences, we extended our analysis to a dataset of 259 near full-length (nFL) hypermutated sequences collected from 16 SIVmac251-infected macaques from that same cohort (Fig. S3F, G) (1509)3. This dataset was generated using PCR product size as an exclusion-criteria to specifically collect nFL sequences, therefore proviruses with deletions are not included. The majority (81% for pol and 66% for env) of hypermutated proviruses included in this analysis directly matched the HPDA probe sequences, emphasizing that the HPDA probe positions with G→A mutations are indeed good indicators of APOBEC3G/F-mediated hypermutation status. Interestingly, proviruses with additional G→A mutations (three or four) in the env probe binding site were relatively common in this dataset (~11%, compared to ~3% in the first dataset). Of the nFL hypermutated sequences with 3 or 4 G→A mutations in the region of the HPDA env probe, 93% had 2 G→A mutations at the pol probe binding site and would therefore be detected by the HPDA in Q1. Although approximately 16% of the hypermutated sequences from the second dataset had SNPs that could prevent the hypermutated env probe from binding, we found that 85% of these sequences were hypermutated at both positions in the pol probe binding site. Based on this analysis, we concluded that the most accurate way to estimate the frequency of hypermutated proviruses would be to sum the positive events detected in quadrants 1, 2 and 4 by the HPDA to give an estimate of “total hypermutated proviruses,” rather than attempt to categorize positive droplets based on the region of hypermutation (i.e. 5’HM (Q1), 3’HM (Q4) or 2XHM (Q2)). An important implication of this analysis is that single-positive events do not necessarily represent proviruses that are deleted at the other amplicon.
gag ddPCR
We designed synthetic double-stranded DNA templates (corresponding to nt: 1185–1620 in the SIVmac251 reference genome – accession no: M19499.1) to adapt the previously described SIV gag qPCR assay67 to the Bio-Rad ddPCR platform (Fig. S4A, C; Table S8). Primers were included at a final concentration of 500 nM and the probe at a final concentration of 250 nM.
gag mutation analysis
Sequences from SIV-infected macaques20,93 were analyzed for G→A sequence polymorphisms in the gag probe binding site (Fig. S4C). Sequences with 1-to-5 mutations were used to design synthetic DNA templates to determine the effect of mismatches on assay performance, including the amplitude of positive droplets (Fig. S4A, C). 250 copies of each template were loaded in a 22 μl reaction in triplicate and scored for fluorescence in the FAM channel.
Envelope sequencing & analysis
DNA used for ddPCR was also used to collect individual env sequences by limiting dilution nested PCR. Each sample was diluted for an outer PCR using the primers SOUTF – (5’ – GGC TAA GGC TAA TAC ATC TTC TGC ATC – 3’) and NOUTR (5’ – TTT AAG CAA GCA AGC GTG GAG – 3’)82 at a final concentration of 0.2 μM with Platinum Taq and the following cycling conditions: 5 minutes at 95°C, 35 cycles of (94°C for 1 minute, 58°C for 1 minute, 72°C for 3 minutes) then 10 minutes at 72°C82. PCR products were diluted 1 in 2 with 10 mM Tris-HCl and 2 μl were used as the template for a nested inner PCR with the primers EnvF2 (5’ – GGA ACA ACT CAG TGC CTA CCA GAT AAT GGT G – 3’) and EnvR2 (5’ – GTA GGT CAG TTC AGT CCT GAG GAC TTC TCG – 3’)98 at a final concentration of 0.20 μM and the following PCR conditions: 2 minutes at 94°C, 41 cycles of (94°C for 30 seconds, 55°C for 30 seconds, 72°C for 2 minutes) then 3 minutes at 72°C98. 3 μl of each inner PCR product was added to 17 μl loading dye, run on an agarose gel and visualized using UV illumination. Gels in which <30% of wells were positive were considered to be at limiting dilution and to have resulted from a single template. Positive wells were submitted for Sanger Sequencing (Genewiz). Chromatograms were assembled using the “de novo assembly” function in Geneious Prime. Chromatograms with one or more overlapping peaks – indicative of Taq polymerase-induced error or multiple proviral templates in a single well – were removed from analysis.
Phylogenetic analysis
Sequences were defined as “non-defective” based on the absence of premature stop codons, deletions, or evidence of APOBEC3G/F-mediated G-to-A hypermutation, assessed using Los Alamos National Lab’s Hypermut 2.0 program99. Only non-defective sequences were used for phylogenetic tree construction (Fig. 6, Figs. S5–13) and other analyses including: percent of sequences with a match (Fig. 5), average pairwise-distance calculations (Table S7, Fig. 5B and Fig. 7A), and frequency of non-synonymous mutations (Fig. 7B–D). The evolutionary history was inferred by using the Maximum Likelihood method implemented in MEGA11100 using the optimal model selected by the program. Trees were rooted using a consensus of the stock sequences57.
Average pairwise distances were calculated between the consensus sequence of the virus stock57 and all non-defective envelope nucleotide sequences retrieved from plasma and gDNA. Calculations were implemented in MEGA v.7 with a p-distance substitution model. Analyses were run for 1000 replicates.
To determine the likelihood of identical hypermutated sequences arising independently in different infected cells, we calculated the frequency of G→A mutation at each G in the sequenced region of env in 226 hypermutated sequences from 3 animals (Fig. 5D) and used per site mutations probabilities in simulations to determine how often identical hypermutated sequences would arise assuming each site behaved independently.
Isolation of RNA from plasma for sequencing
To allow for consistent amplification of plasma virus, plasma aliquots were thawed and ~10,000 copies of SIV RNA were used. Viral RNA was extracted as previously described101. Viral RNA pellets were resuspended in 20ul Buffer TDR (5 mM Tris-HCl, 1 mM DDT, 2.5 mM RNaseOUT) and immediately used for reverse transcription with SuperScript III. A mixture of 20 μl SIV RNA, 2.5 μl of 2 μM primer NOUTR (5’ – TTT AAG CAA GCA AGC GTG GAG – 3’)82 and 2.5 μl 10 mM dNTPs was heated to 65°C for 10 minutes then placed on ice for 1 minute. A reaction mixture of 10 μl 5X First Strand Buffer, 13.5 μl water, 0.5 μl 0.1 M DTT, 0.5 μl SuperScript III and 0.5 μl 40 U/μl RNaseOUT was then added to reaction volume of 50 μl. The mixture was heated at 50°C for 55 minutes then inactivated at 85°C for 10 minutes. The resulting cDNA was serial diluted with 10 mM Tris-HCl to reach limiting dilution as input for single-genome sequencing using the approach described above.
Modeling of decay processes
We used a non-linear mixed effect approach to fit multiple models for the decay of SIV DNA species in macaques infected with SIV on ART. Given the long time scale of the study, the following general model was used:
| (1) |
where is the variable of interest, is its baseline value, is the fraction of that decays in the first phase with decay rate , and is the fraction of which decays in the second phase with decay rate . After this potential biphasic decay lasting until time , there could be a third phase with decay rate , which would be smaller than the two initial phases.
With this general formula, multiple models can be tested. If we fix , then the third phase of decay starting at time is flat and remains at the value of attained at time . On the other hand, if we fix at a large value (say ) then we never reach and have the usual biphasic decay model. In this latter case, with , we can test for a single decay model, estimating only . Note that in the case of a single decay, is not estimated. Using this approach, we statistically compare the quality of the fits (using the corrected Bayesian Information Criteria, BIC) among these models102. In addition, we also tested a hybrid model, (particularly for the hypermutated DNA), where some macaques (specifically T544, T545, T624, T627 and T628) had a biphasic decay, and another group of macaques (T523, T530, T537, T623 and T625) had a single phase of decay. This was prompted by visual inspection of the decay patterns across the animals.
To fit the model of the decay processes – equations (1) above – to the data, we used a nonlinear mixed-effects model. This approach assumes that a model parameter is drawn from a distribution with a fixed part , which is the median value of the parameter in the population, and a random term , which is assumed to be normally distributed with zero mean and standard deviation . We assumed that parameters b1, b2, b3 follow a lognormal distribution (to guarantee that they are positive), and A is logit normal (between 0 and 1). We fit instances of the model to the data, with different parameters fixed as described above, and estimate the parameters using the software Monolix 2020R1 (Lixoft)103. We calculated 95% CI for the parameters based on the Fisher Information matrix using the Monolix function confintmlx.
For intact proviruses, gag+ DNA and 2LTR circles, the best fit models were triphasic, with the last phase after , essentially flat, . For the env+ and env− 2LTR species, the biphasic and triphasic models had similar statistical support, with biphasic slightly better, because was very high. We suspect that this is the case due to multiple censored values (i.e. no positive events were detected) for these species. We note that we couldn’t fit the 2LTR species for T624, because it showed almost no decay with many censored values. Finally, for hypermutated SIV proviruses, the hybrid model gave the best fits.
We used the same general modeling equations (1) to quantify the decay of non-synonymous mutations over time, which was well described by a biphasic decline.
Quantification and statistical analysis
Details regarding the statistical tests used and the represented data can be found in the figure legends, or, for the mathematical modeling in the “Modeling of decay processes” section. Statistical tests were considered significant when P values were less than 0.05. Statistical analyses were performed via GraphPad Prism (Prism Software) or, for the modeling of decay processes, Monolix2020R1 (Lixoft).
Supplementary Material
Highlights.
Intact SIV genomes decay with triphasic kinetics upon treatment with ART.
Defective (hypermutated) proviruses decay with mono or biphasic kinetics.
Early after treatment, labile populations of recently infected cells decay.
Long-term ART reveals persistence of archival variants & infected cell proliferation
Acknowledgements
This work was supported by the NIH Martin Delaney Collaboratory grant UM1AI164556 to D.H.B and by Howard Hughes Medical Institute to R.F.S. Animal studies were supported by AI124377, AI128751, AI149670, AI164556, AI169615 for D.H.B. F.R.S is supported by the Office of the NIH Director and National Institute of Dental & Craniofacial Research (DP5OD031834). Portions of this work were performed under the auspices of the U.S. Department of Energy through Los Alamos National Laboratory, which is operated by Triad National Security, LLC, for the National Nuclear Security Administration of the US Department of Energy (Contract 89233218CNA000001). Support was also provided by NIH Grants R01-AI028433, R01-OD011095, P01-AI131365 (to A.S.P.), R01-AI15270301; and UM1-AI164561 (to R.M.R.).
Footnotes
Declaration of Interests
Aspects of HIV-1 IPDA are subject of a patent application PCT/ US16/28822 filed by Johns Hopkins University. R.F.S. is an inventor on this application. Accelevir Diagnostics holds an exclusive license for this patent application. R.F.S. holds no equity interest in Accelevir Diagnostics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, and Ho DD (1997). Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 387, 188–191. [DOI] [PubMed] [Google Scholar]
- 2.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, and Fauci AS (1997). Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 94, 13193–13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. (1999). Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 5, 512–517. [DOI] [PubMed] [Google Scholar]
- 4.Wong JK, Hezareh M, Günthard HF, Havlir DV, Ignacio CC, Spina CA, and Richman DD (1997). Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 278, 1291–1295. [DOI] [PubMed] [Google Scholar]
- 5.Crooks AM, Bateson R, Cope AB, Dahl NP, Griggs MK, Kuruc JD, Gay CL, Eron JJ, Margolis DM, Bosch RJ, et al. (2015). Precise quantitation of the latent HIV-1 reservoir: Implications for eradication strategies. J. Infect. Dis 212, 1361–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, and Siliciano RF (2003). Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med 9, 727–728. [DOI] [PubMed] [Google Scholar]
- 7.Davey RT, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, et al. (1999). HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A. 96, 15109–15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, Khoruts A, Estes JD, Anderson J, Callisto SP, et al. (2015). Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A. 112, 1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, and Markowitz M. (1995). Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 373, 123–126. [DOI] [PubMed] [Google Scholar]
- 10.Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, and Hahn BH (1995). Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 373, 117–122. [DOI] [PubMed] [Google Scholar]
- 11.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, et al. (2016). Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 22, 1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohn LB, Silva IT, Oliveira TY, Rosales RA, Parrish EH, Learn GH, Hahn BH, Czartoski JL, McElrath MJ, Lehmann C, et al. (2015). HIV-1 integration landscape during latent and active infection. Cell. 160, 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, and Malim MH (2003). DNA deamination mediates innate immunity to retroviral infection. Cell. 113, 803–809. [DOI] [PubMed] [Google Scholar]
- 14.Hiener B, Horsburgh BA, Eden J, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, et al. (2017). Identification of genetically intact HIV-1 proviruses in specific CD4+ T cells from effectively treated participants. Cell Rep. 21, 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho Y, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, and Siliciano RF (2013). Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 155, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imamichi H, Dewar RL, Adelsberger JW, Rehm CA, O’Doherty U, Paxinos EE, Fauci AS, and Lane HC (2016). Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci U S A. 113, 8783–8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, Kuo H, Hua S, Chen H, Ouyang Z, et al. (2017). Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest. 127, 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez G, Xu X, Chermann JC, and Hirsch I. (1997). Accumulation of defective viral genomes in peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected individuals. J Virol. 71, 2233–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheehy AM, Gaddis NC, Choi JD, and Malim MH (2002). Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral vif protein. Nature. 418, 646–650. [DOI] [PubMed] [Google Scholar]
- 20.Bender AM, Simonetti FR, Kumar MR, Fray EJ, Bruner KM, Timmons AE, Tai KY, Jenike KM, Antar AAR, Liu P, et al. (2019). The landscape of persistent viral genomes in ART-treated SIV, SHIV, and HIV-2 infections. Cell Host Microbe. 26, 73–85.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, et al. (2019). A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature. 566, 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falcinelli SD, Kilpatrick KW, Read J, Murtagh R, Allard B, Ghofrani S, Kirchherr J, James KS, Stuelke E, Baker C, et al. (2021). Longitudinal dynamics of intact HIV proviral DNA and outgrowth virus frequencies in a cohort of individuals receiving antiretroviral therapy. J Infect Dis. 224, 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gandhi RT, Cyktor JC, Bosch RJ, Mar H, Laird GM, Martin A, Collier AC, Riddler SA, Macatangay BJ, Rinaldo CR, et al. (2021). Selective decay of intact HIV-1 proviral DNA on antiretroviral therapy. J Infect Dis. 223, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peluso MJ, Bacchetti P, Ritter KD, Beg S, Lai J, Martin JN, Hunt PW, Henrich TJ, Siliciano JD, Siliciano RF, et al. (2020). Differential decay of intact and defective proviral DNA in HIV-1-infected individuals on suppressive antiretroviral therapy. JCI Insight. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White JA, Simonetti FR, Beg S, McMyn NF, Dai W, Bachmann N, Lai J, Ford WC, Bunch C, Jones JL, et al. (2022). Complex decay dynamics of HIV virions, intact and defective proviruses, and 2LTR circles following initiation of antiretroviral therapy. Proc Natl Acad Sci U S A. 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bukrinsky MI, Stanwick TL, Dempsey MP, and Stevenson M. (1991). Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science. 254, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, and Chen IS (1990). HIV-1 entry into quiescent primary lymphocytes: Molecular analysis reveals a labile, latent viral structure. Cell. 61, 213–222. [DOI] [PubMed] [Google Scholar]
- 28.Pierson TC, Zhou Y, Kieffer TL, Ruff CT, Buck C, and Siliciano RF (2002). Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J Virol. 76, 8518–8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Zhang H, Siliciano JD, and Siliciano RF (2005). Kinetics of human immunodeficiency virus type 1 decay following entry into resting CD4+ T cells. J Virol. 79, 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray JM, Kelleher AD, and Cooper DA (2011). Timing of the components of the HIV life cycle in productively infected CD4+ T cells in a population of HIV-infected individuals. J Virol. 85, 10798–10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, and Siliciano RF (2003). Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 9, 727–728. [DOI] [PubMed] [Google Scholar]
- 32.Butler SL, Johnson EP, and Bushman FD (2002). Human immunodeficiency virus cDNA metabolism: Notable stability of two-long terminal repeat circles. J Virol. 76, 3739–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierson TC, Kieffer TL, Ruff CT, Buck C, Gange SJ, and Siliciano RF (2002). Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J Virol. 76, 4138–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharkey ME, Teo I, Greenough T, Sharova N, Luzuriaga K, Sullivan JL, Bucy RP, Kostrikis LG, Haase A, Veryard C, et al. (2000). Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat Med. 6, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharkey M, Triques K, Kuritzkes DR, and Stevenson M. (2005). In vivo evidence for instability of episomal human immunodeficiency virus type 1 cDNA. J Virol. 79, 5203–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Del Prete GQ, and Lifson JD (2018). Nonhuman primate models for studies of AIDS virus persistence during suppressive combination antiretroviral therapy. Curr Top Microbiol Immunol. 417, 69–109. [DOI] [PubMed] [Google Scholar]
- 37.Dinoso JB, Rabi SA, Blankson JN, Gama L, Mankowski JL, Siliciano RF, Zink MC, and Clements JE (2009). A simian immunodeficiency virus-infected macaque model to study viral reservoirs that persist during highly active antiretroviral therapy. J Virol. 83, 9247–9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar N, Chahroudi A, and Silvestri G. (2016). Animal models to achieve an HIV cure. Curr Opin HIV AIDS. 11, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z, Schuler T, Zupancic M, Wietgrefe S, Staskus KA, Reimann KA, Reinhart TA, Rogan M, Cavert W, Miller CJ, et al. (1999). Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science. 286, 1353–1357. [DOI] [PubMed] [Google Scholar]
- 40.Brandin E, Thorstensson R, Bonhoeffer S, and Albert J. (2006). Rapid viral decay in simian immunodeficiency virus-infected macaques receiving quadruple antiretroviral therapy. J Virol. 80, 9861–9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keele BF, Li H, Learn GH, Hraber P, Giorgi EE, Grayson T, Sun C, Chen Y, Yeh WW, Letvin NL, et al. (2009). Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J Exp Med. 206, 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q, Duan L, Estes JD, Ma Z, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, and Haase AT (2005). Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 434, 1148–1152. [DOI] [PubMed] [Google Scholar]
- 43.Ma Z, Abel K, Rourke T, Wang Y, and Miller CJ (2004). A period of transient viremia and occult infection precedes persistent viremia and antiviral immune responses during multiple low-dose intravaginal simian immunodeficiency virus inoculations. J Virol. 78, 14048–14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malouli D, Gilbride RM, Wu HL, Hwang JM, Maier N, Hughes CM, Newhouse D, Morrow D, Ventura AB, Law L, et al. (2022). Cytomegalovirus-vaccine-induced unconventional T cell priming and control of SIV replication is conserved between primate species. Cell Host Microbe. 30, 1207–1218.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nowak MA, Lloyd AL, Vasquez GM, Wiltrout TA, Wahl LM, Bischofberger N, Williams J, Kinter A, Fauci AS, Hirsch VM, et al. (1997). Viral dynamics of primary viremia and antiretroviral therapy in simian immunodeficiency virus infection. J Virol. 71, 7518–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okoye AA, Hansen SG, Vaidya M, Fukazawa Y, Park H, Duell DM, Lum R, Hughes CM, Ventura AB, Ainslie E, et al. (2018). Early antiretroviral therapy limits SIV reservoir establishment to delay or prevent post-treatment viral rebound. Nat Med. 24, 1430–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen A, Zink MC, Mankowski JL, Chadwick K, Margolick JB, Carruth LM, Li M, Clements JE, and Siliciano RF (2003). Resting CD4+ T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J. Virol 77, 4938–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanstrom AE, Del Prete GQ, Deleage C, Elser SE, Lackner AA, and Hoxie JA (2018). The SIV envelope glycoprotein, viral tropism, and pathogenesis: Novel insights from nonhuman primate models of AIDS. Curr HIV Res. 16, 29–40. [DOI] [PubMed] [Google Scholar]
- 49.Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, and Lackner AA (1998). Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 280, 427–431. [DOI] [PubMed] [Google Scholar]
- 50.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, et al. (2014). Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature. 512, 74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long S, Fennessey CM, Newman L, Reid C, O’Brien SP, Li Y, Del Prete GQ, Lifson JD, Gorelick RJ, and Keele BF (2019). Evaluating the intactness of persistent viral genomes in simian immunodeficiency virus-infected rhesus macaques after initiating antiretroviral therapy within one year of infection. J Virol. 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abrahams M, Joseph SB, Garrett N, Tyers L, Moeser M, Archin N, Council OD, Matten D, Zhou S, Doolabh D, et al. (2019). The replication-competent HIV-1 latent reservoir is primarily established near the time of therapy initiation. Sci Transl Med. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brodin J, Zanini F, Thebo L, Lanz C, Bratt G, Neher RA, and Albert J. (2016). Establishment and stability of the latent HIV-1 DNA reservoir. Elife. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brooks K, Jones BR, Dilernia DA, Wilkins DJ, Claiborne DT, McInally S, Gilmour J, Kilembe W, Joy JB, Allen SA, et al. (2020). HIV-1 variants are archived throughout infection and persist in the reservoir. PLoS Pathog. 16, e1008378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Böni J, Hirschel B, Weber R, Trkola A, and Günthard HF (2008). HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci U S A. 105, 16725–16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pankau MD, Reeves DB, Harkins E, Ronen K, Jaoko W, Mandaliya K, Graham SM, McClelland RS, Matsen Iv FA, Schiffer JT, et al. (2020). Dynamics of HIV DNA reservoir seeding in a cohort of superinfected kenyan women. PLoS Pathog. 16, e1008286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Del Prete GQ, Scarlotta M, Newman L, Reid C, Parodi LM, Roser JD, Oswald K, Marx PA, Miller CJ, Desrosiers RC, et al. (2013). Comparative characterization of transfection- and infection-derived simian immunodeficiency virus challenge stocks for in vivo nonhuman primate studies. J Virol. 87, 4584–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Del Prete GQ, Smedley J, Macallister R, Jones GS, Li B, Hattersley J, Zheng J, Piatak M, Keele BF, Hesselgesser J, et al. (2016). Short communication: Comparative evaluation of coformulated injectable combination antiretroviral therapy regimens in simian immunodeficiency virus-infected rhesus macaques. AIDS Res Hum Retroviruses. 32, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perelson AS, Neumann AU, Markowitz M, Leonard JM, and Ho DD (1996). HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science. 271, 1582–1586. [DOI] [PubMed] [Google Scholar]
- 60.He X, Aid M, Ventura JD, Borducchi E, Lifton M, Liu J, and Barouch DH (2022). Rapid loss of CD4 T cells by pyroptosis during acute SIV infection in rhesus macaques. J Virol., e0080822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, et al. (2016). Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med 22, 1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Policicchio BB, Cardozo EF, Sette P, Xu C, Haret-Richter G, Dunsmore T, Apetrei C, Pandrea I, and Ribeiro RM (2018). Dynamics of simian immunodeficiency virus two-long-terminal-repeat circles in the presence and absence of CD8+ cells. J Virol. 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bucy RP, Hockett RD, Derdeyn CA, Saag MS, Squires K, Sillers M, Mitsuyasu RT, and Kilby JM (1999). Initial increase in blood CD4(+) lymphocytes after HIV antiretroviral therapy reflects redistribution from lymphoid tissues. J Clin Invest. 103, 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pakker NG, Notermans DW, de Boer RJ, Roos MT, de Wolf F, Hill A, Leonard JM, Danner SA, Miedema F, and Schellekens PT (1998). Biphasic kinetics of peripheral blood T cells after triple combination therapy in HIV-1 infection: A composite of redistribution and proliferation. Nat Med. 4, 208–214. [DOI] [PubMed] [Google Scholar]
- 65.Lifson JD, Rossio JL, Piatak M, Parks T, Li L, Kiser R, Coalter V, Fisher B, Flynn BM, Czajak S, et al. (2001). Role of CD8(+) lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J Virol. 75, 10187–10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, and Roederer M. (2005). Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 434, 1093–1097. [DOI] [PubMed] [Google Scholar]
- 67.Cline AN, Bess JW, Piatak M, and Lifson JD (2005). Highly sensitive SIV plasma viral load assay: Practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol. 34, 303–312. [DOI] [PubMed] [Google Scholar]
- 68.Bui JK, Sobolewski MD, Keele BF, Spindler J, Musick A, Wiegand A, Luke BT, Shao W, Hughes SH, Coffin JM, et al. (2017). Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 13, e1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hosmane NN, Kwon KJ, Bruner KM, Capoferri AA, Beg S, Rosenbloom DI, Keele BF, Ho YC, Siliciano JD, and Siliciano RF (2017). Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J. Exp. Med 214, 959–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lorenzi JC, Cohen YZ, Cohn LB, Kreider EF, Barton JP, Learn GH, Oliveira T, Lavine CL, Horwitz JA, Settler A, et al. (2016). Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc. Natl. Acad. Sci. U. S. A 113, E7908-E7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, et al. (2014). HIV latency. specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 345, 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wagner TA, McLaughlin S, Garg K, Cheung CYK, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, and Frenkel LM (2014). HIV latency. proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 345, 570–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ferris AL, Wells DW, Guo S, Del Prete GQ, Swanstrom AE, Coffin JM, Wu X, Lifson JD, and Hughes SH (2019). Clonal expansion of SIV-infected cells in macaques on antiretroviral therapy is similar to that of HIV-infected cells in humans. PLoS Pathog. 15, e1007869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laskey SB, Pohlmeyer CW, Bruner KM, and Siliciano RF (2016). Evaluating clonal expansion of HIV-infected cells: Optimization of PCR strategies to predict clonality. PLoS Pathog. 12, e1005689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patro SC, Brandt LD, Bale MJ, Halvas EK, Joseph KW, Shao W, Wu X, Guo S, Murrell B, Wiegand A, et al. (2019). Combined HIV-1 sequence and integration site analysis informs viral dynamics and allows reconstruction of replicating viral ancestors. Proc Natl Acad Sci U S A. 116, 25891–25899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buckley KA, Li P, Khimani AH, Hofmann-Lehmann R, Liska V, Anderson DC, McClure HM, and Ruprecht RM (2003). Convergent evolution of SIV env after independent inoculation of rhesus macaques with infectious proviral DNA. Virology. 312, 470–480. [DOI] [PubMed] [Google Scholar]
- 77.Burns DP, and Desrosiers RC (1991). Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J Virol. 65, 1843–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Campbell BJ, and Hirsch VM (1994). Extensive envelope heterogeneity of simian immunodeficiency virus in tissues from infected macaques. J Virol. 68, 3129–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi WS, Collignon C, Thiriart C, Burns DP, Stott EJ, Kent KA, and Desrosiers RC (1994). Effects of natural sequence variation on recognition by monoclonal antibodies neutralize simian immunodeficiency virus infectivity. J Virol. 68, 5395–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ita S, Hill AK, Lam EC, Dufort FJ, Yang X, Newman R, Leviyang S, Fofana IB, and Johnson WE (2018). High-resolution sequencing of viral populations during early simian immunodeficiency virus infection reveals evolutionary strategies for rapid escape from emerging env-specific antibody responses. J Virol. 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kinsey NE, Anderson MG, Unangst TJ, Joag SV, Narayan O, Zink MC, and Clements JE (1996). Antigenic variation of SIV: Mutations in V4 alter the neutralization profile. Virology. 221, 14–21. [DOI] [PubMed] [Google Scholar]
- 82.Lamers SL, Nolan DJ, Rife BD, Fogel GB, McGrath MS, Burdo TH, Autissier P, Williams KC, Goodenow MM, and Salemi M. (2015). Tracking the emergence of host-specific simian immunodeficiency virus env and nef populations reveals nef early adaptation and convergent evolution in brain of naturally progressing rhesus macaques. J Virol. 89, 8484–8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Overbaugh J, Rudensey LM, Papenhausen MD, Benveniste RE, and Morton WR (1991). Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J Virol. 65, 7025–7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Overbaugh J, and Rudensey LM (1992). Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J Virol. 66, 5937–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sato S, Yuste E, Lauer WA, Chang EH, Morgan JS, Bixby JG, Lifson JD, Desrosiers RC, and Johnson WE (2008). Potent antibody-mediated neutralization and evolution of antigenic escape variants of simian immunodeficiency virus strain SIVmac239 in vivo. J Virol. 82, 9739–9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Torres JV, Anderson DE, Malley A, Banapour B, Axthelm MK, Benjamini E, and Gardner MB (1993). SIV envelope glycoprotein epitopes recognized by antibodies from infected or vaccinated rhesus macaques. J Med Primatol. 22, 129–137. [PubMed] [Google Scholar]
- 87.Wu F, Ourmanov I, Kuwata T, Goeken R, Brown CR, Buckler-White A, Iyengar R, Plishka R, Aoki ST, and Hirsch VM (2012). Sequential evolution and escape from neutralization of simian immunodeficiency virus SIVsmE660 clones in rhesus macaques. J Virol. 86, 8835–8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yeh WW, Rahman I, Hraber P, Coffey RT, Nevidomskyte D, Giri A, Asmal M, Miljkovic S, Daniels M, Whitney JB, et al. (2010). Autologous neutralizing antibodies to the transmitted/founder viruses emerge late after simian immunodeficiency virus SIVmac251 infection of rhesus monkeys. J Virol. 84, 6018–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yeh WW, Brassard LM, Miller CA, Basavapathruni A, Zhang J, Rao SS, Nabel GJ, Mascola JR, Letvin NL, and Seaman MS (2012). Envelope variable region 4 is the first target of neutralizing antibodies in early simian immunodeficiency virus mac251 infection of rhesus monkeys. J Virol. 86, 7052–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yuste E, Johnson W, Pavlakis GN, and Desrosiers RC (2005). Virion envelope content, infectivity, and neutralization sensitivity of simian immunodeficiency virus. J Virol. 79, 12455–12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. (2008). Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 105, 7552–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, et al. (1999). Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol 73, 10489–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu P, Keele BF, Abbink P, Mercado NB, Liu J, Bondzie EA, Chandrashekar A, Borducchi EN, Hesselgesser J, Mish M, et al. (2020). Origin of rebound virus in chronically SIV-infected rhesus monkeys following treatment discontinuation. Nat Commun. 11, 5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Einkauf KB, Osborn MR, Gao C, Sun W, Sun X, Lian X, Parsons EM, Gladkov GT, Seiger KW, Blackmer JE, et al. (2022). Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell. 185, 266–282.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jones BR, Kinloch NN, Horacsek J, Ganase B, Harris M, Harrigan PR, Jones RB, Brockman MA, Joy JB, Poon AFY, et al. (2018). Phylogenetic approach to recover integration dates of latent HIV sequences within-host. Proc Natl Acad Sci U S A. 115, E8958-E8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, et al. (2009). Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med. 15, 1322–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barouch DH, Ghneim K, Bosche WJ, Li Y, Berkemeier B, Hull M, Bhattacharyya S, Cameron M, Liu J, Smith K, et al. (2016). Rapid inflammasome activation following mucosal SIV infection of rhesus monkeys. Cell. 165, 656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stone M, Keele BF, Ma Z, Bailes E, Dutra J, Hahn BH, Shaw GM, and Miller CJ (2010). A limited number of simian immunodeficiency virus (SIV) env variants are transmitted to rhesus macaques vaginally inoculated with SIVmac251. J Virol. 84, 7083–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rose PP, and Korber BT (2000). Detecting hypermutations in viral sequences with an emphasis on G --> A hypermutation. Bioinformatics. 16, 400–401. [DOI] [PubMed] [Google Scholar]
- 100.Tamura K, Stecher G, and Kumar S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38, 3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tosiano MA, Jacobs JL, Shutt KA, Cyktor JC, and Mellors JW (2019). A simpler and more sensitive single-copy HIV-1 RNA assay for quantification of persistent HIV-1 viremia in individuals on suppressive antiretroviral therapy. J Clin Microbiol. 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burnham KP, Anderson DR (2002). Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach, 2nd Edition. (Springer; ). [Google Scholar]
- 103.Lavielle M. (2014) Mixed Effects Models for the Population Approach: Models, Tasks, Methods and Tools, (Chapman and Hall/CRC; ). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences generated in this study have been deposited in Genbank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This study does not report original code and all computational resources used are publicly available and listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| SIVmac251 swarm57 | Dr. Dan Barouch, BIDMC | N/A |
| Biological samples | ||
| SIVmac251-infected Rhesus macaque PBMCs | Dr. Dan Barouch, BIDMC | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Platinum Taq DNA Polymerase High Fidelity | ThermoFisher Scientific | Cat # 11304011 |
| SuperScript III Reverse Transcriptase | ThermoFisher Scientific | Cat # 18-080-044 |
| dNTP Mix (10 mM each) | ThermoFisher Scientific | Cat # 18-427-088 |
| UltraPure 1M Tris-HCl Buffer (pH 8.0) | ThermoFisher Scientific | Cat # 15-568-025 |
| RNaseOUT Recombinant Ribonuclease Inhibitor | ThermoFisher Scientific | Cat # 10777019 |
| Critical commercial assays | ||
| QIAamp DNA Mini Kit | Qiagen | Cat # 51304 |
| CD4+ T Cell Isolation Kit, Non-Human Primate | Miltenyi Biotec | Cat # 130-092-144 |
| 2X ddPCR Supermix for Probes (no dUTPs) | Bio-Rad | Cat # 186-3024 |
| Deposited data | ||
| SIV - plasma RNA sequences | This paper | OQ168641-OQ168979 |
| SIV - non-defective proviral DNA sequences | This paper | OQ168980-OQ170751 |
| SIV - hypermutated (defective) proviral sequences | This paper | OQ170752-OQ170785 |
| Experimental models: Organisms/strains | ||
| Rhesus macaque (Macaca mulatta) infected with SIVmac251 | Indian origin | Animals T523, T530, T537, T544, T545, T623, T624, T625, T627 & T628 |
| Oligonucleotides | ||
| Oligonucleotides, standard desalting (Sanger sequencing & ddPCR) - see Methods and Table S8 | IDT | N/A |
| FAM/VIC labeled probes with MGB quencher, HPLC purified (see Table S8) | Applied Biosystems | N/A |
| Unlabeled competition probes w/MGB quencher, HPLC purified (see Table S8) | Applied Biosystems | N/A |
| FAM/HEX labeled probes w/MGB-NFQ quencher, HPLC purified (see Table S8) | IDT | N/A |
| Recombinant DNA | ||
| Synthetic double-stranded DNA controls (gBlocks) - see Methods | IDT | N/A |
| Software and algorithms | ||
| QuantaSoft Analysis-Pro | Bio-Rad | http://www.bio-rad.com/en-us/product/qx200-droplet-digital-pcr-system?ID=MPOQQE4VY |
| Geneious Prime | Dotmatics | https://www.geneious.com/prime/ |
| MEGA v7.0 & v11 | MEGA | https://www.megasoftware.net/ |
| Clustal W | EMBL-EBI | http://www.ebi.ac.uk/Tools/msa/clustalw2/ |
| GraphPad Prism | Dotmatics | https://www.graphpad.com/ |
| Monolix 2020R1 | Lixoft | https://lixoft.com/ |
| Los Alamos National Labs - Highlighter | Keele et al.91 | https://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html |
| Los Alamos National Labs - Hypermut 2.0 | Rose et al.99 | https://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html |