Abstract
ErCas12a is a class 2 type V CRISPR–Cas nuclease isolated from Eubacterium rectale with attractive fundamental characteristics, such as RNA self-processing capability, and lacks reach-through royalties typical for Cas nucleases. This study aims to develop a ErCas12a-mediated genome editing tool applicable in the model yeast Saccharomyces cerevisiae. The optimal design parameters for ErCas12a editing in S. cerevisiae were defined as a 21-nt spacer flanked by 19 nt direct repeats expressed from either RNApolII or III promoters, achieving near 100% editing efficiencies in commonly targeted genomic locations. To be able to transfer the ErCas12a genome editing tool to different strain lineages, a transportable platform plasmid was constructed and evaluated for its genome editing efficiency. Using an identical crRNA expression design, the transportable ErCas12a genome editing tool showed lower efficiency when targeting the ADE2 gene. In contrast to genomic Ercas12a expression, episomal expression of Ercas12a decreases maximum specific growth rate on glucose, indicating ErCas12a toxicity at high expression levels. Moreover, ErCas12a processed a multispacer crRNA array using the RNA self-processing capability, which allowed for simultaneous editing of multiple chromosomal locations. ErCas12a is established as a valuable addition to the genetic toolbox for S. cerevisiae.
Keywords: CRISPR–Cas, ErCas12a (MAD7), genome engineering, multiplexing, Saccharomyces cerevisiae
This study established ErCas12a, CRISPR-endonuclease that possesses RNA self-processing capability and the advantage of lacking reach-through royalties typical of Cas-nucleases, as a valuable addition to the S. cerevisiae genetic toolbox.
Introduction
Originally studied for their role in prokaryotic adaptive immunity (Ishino et al. 1987, Mojica et al. 1993, 2005, Barrangou et al. 2007, Brouns et al. 2008, Marraffini and Sontheimer 2008, Garneau et al. 2010), clustered regularly interspaced short palindromic repeats (CRISPR)‐associated (Cas) systems have since been studied for their use in genome editing (Sapranauskas et al. 2011, Jinek et al. 2012, Cong et al. 2013, Mali et al. 2013). In this application, precisely and efficiently introducing double stranded breaks (DSBs) is critical to activate and recruit the DNA repair machinery to the targeted location for precise genetic modifications. The freedom to design spacers at will makes the sequence guided endonuclease function of CRISPR–Cas systems attractive for this purpose.
Genome editing strategies rely on a single effector Cas protein, consisting of a single crRNA-programmable multidomain, belonging to one of the class 2 CRISPR–systems. In contrast, class 1 CRISPR-systems rely on hetero multimer complexes, making them less attractive as genome editing tool (Makarova et al. 2020). Applying CRISPR–Cas systems for gene editing is, therefore, based on the introduction of a class 2 Cas protein loaded with a CRISPR–RNA (crRNA). In Archaeal and Prokaryotic hosts, the endonuclease programming RNA can be composed of an RNA duplex resulting from the combination of a crRNA and a tracrRNA (Jinek et al. 2012) or composed of a single RNA molecule (Zetsche et al. 2015) like for Cas9 (class 2 type II) and Cas12a (class 2 type V-A), respectively. Upon complementarity of the spacer and protospacer and presence of the essential protospacer adjacent motif (PAM) (Garneau et al. 2010), this ribonucleoprotein complex cleaves the DNA. For efficient application in heterologous hosts, the CRISPR class 2 type II system (Cas9) has been simplified by connecting the crRNA to the tracrRNA in a chimeric single guide RNA (sgRNA) (Jinek et al. 2012). Targeted DSBs can be exploited to direct DNA repair mechanisms to the target locus. In cells with a functional homologous recombination (HR) machinery, adding a DNA repair fragment containing flanking homology sequences to the target site resolves the otherwise lethal dsDNA break, thereby incorporating the desired genetic modification (Capecchi 1989).
Within the class 2 Cas proteins, the type II, which includes the Cas9 signature protein, was the first to be harnessed for gene editing applications (Jinek et al. 2012) and has remained the dominant endonuclease for editing applications. While first applications were demonstrated in human cell lines, the Cas9 system has shown efficacy in a wide range of hosts (DiCarlo et al. 2013, Feng et al. 2013, Jiang et al. 2013, Mizuno et al. 2014, Juergens et al. 2018). For effective endonuclease activity, the spacer sequence must be adjacent to a CRISPR-system dependent PAM. The popular Streptococcus pyogenes Cas9 (SpyCas9) recognizes the 5′-NGG-3′ PAM, a sequence that occurs frequently enough to target editing events in most genes within a genome. However, the occurrence and location of PAM sequences may limit in vivo site-directed mutagenesis approaches designed to alter a single nucleotide. For instance, targeting a chromosomal region with low GC content could be more difficult using SpyCas9. Therefore, increased flexibility in PAM recognition sequences is desired, and this has been one of the driving forces both for studies aiming at altering Cas9 PAM specificity (Hu et al. 2018) and the search for alternative CRISPR–Cas systems (Zetsche et al. 2015, Li et al. 2016, Edraki et al. 2019).
Class 2 type V CRISPR–Cas systems are one such alternative. The first type V Cas protein characterized in yeast was Cas12a (originally Cpf1) and was shown to differ from type II proteins in significant ways (Zetsche et al. 2015, Swiat et al. 2017, Verwaal et al. 2018). Cas12a proteins recognize T-rich PAM sequences (5′-TTTV-3′ or more general 5′-YTTN-3′), conversely to Cas9. Cas12a cleavage occurs distal from the PAM sequence and leaves staggered end cuts, while Cas9 cuts proximal to the PAM and generates blunt ends. Interestingly, its effector module requires neither RNAse nor tracrRNA to process its crRNA. The Cas12a effector protein recognizes the direct repeats (DR) preceding spacers in the CRISPR array and self-processes them to produce single molecule programming crRNAs.
One interesting Cas12a protein is ErCas12a (also referred to as MAD7), isolated from an Eubacterium rectale specimen found in Madagascar (Inscripta Inc., Boulder, CO; Gill et al. 2018). In contrast to previously characterized Cas12a nucleases from Acidaminococcus, and Francisella species that share high amino acid sequence identity (>95%), ErCas12 exhibits less than 45% of identity with the above-mentioned nucleases while keeping all features of class 2 type V-A endonucleases. Interestingly, next to its technological advantages, ErCas12a is currently the only Cas nuclease with a free commercial research license, stimulating the development of this endonuclease as a method for genome editing. ErCas12a editing has been evaluated in several organisms (Liu et al. 2019, 2020, Wierson et al. 2019, Price et al. 2020, Jarczynska et al. 2021, Lin et al. 2021, Rojek et al. 2021, Zhang et al. 2021, Vanegas et al. 2022) (Table 1), but fundamental questions remain over the optimal method of expression, critical design elements, and multiplex editing. These design elements, including expression system, DR length, spacer length and PAM, can have considerable impact on editing efficiency (Swiat et al. 2017) and a consensus design has not yet been reached. Thus, there is a need to establish the optimal design features for implementing ErCas12a editing in Saccharomyces cerevisiae.
Table 1.
Overview of reported ErCas12a genome editing.
| Organism | ErCas12a expression | crRNA | PAM (5′→3′) | DR length (bp) | Spacer length (bp) | Editing efficiency (%) | Reference |
|---|---|---|---|---|---|---|---|
| E. coli | Unreported | Unreported | YTTN | 21 | 21 | 100% | Inscripta Inc. |
| S. cerevisiae | Unreported | Unreported | YTTN | 36 | 21 | 66% | Inscripta Inc. |
| Aspergillus nidulans, Aspergillus niger, Aspergillus aculeatus, Aspergillus oryzae | AMA1 plasmid pAnTEF1a | AMA1 plasmid RNApolIII (pAoU6b) | YTTN | 19 | 21 | 50% | Jarczynska et al. (2021) |
| Aspergillus species: Aspergillus nidulans, A. niger, A. oryzae, and A. campestris | AMA1 plasmid pAnTEF1a | AMA1 plasmid RNApolIII (U3), glycine tRNA-based splicing | TTTN | 35 | 21 | Unreported | Vanegas et al. (2022) |
| Rice and wheat protoplasts | Agrobacterium transferred T-DNA pZmUbi-1c | Episomal expression, RNApolII pUbi-1, HH&HDV | TTTN | 35 or 21 | 20–24 | 66% | Lin et al. (2021) |
| Rice | Episomal expression, pUbi-1 | Episomal expression, RNApolII pUbi-1, HH&HDV | TTTV | Unreported | 19–23 | 89% | Zhang et al. (2021) |
| Mouse | Nucleofection (protein/RNA) | Nucleofection (RNA) | YTTN | 35 | 21 | 81% | Liu et al. (2020) |
| Rat | Nucleofection (protein/RNA) | Nucleofection (RNA) | YTTN | 35 | 21 | 25% | Liu et al. (2020) |
| Human cells | Nucleofection (protein/RNA) | Nucleofection (RNA) | YTTN | 35 | 21 | 23% | Liu et al. (2020) |
| Bacillus subtilis | Episomal expression, pGRAC | Episomal expression, pVEG | YTTN | 35 | 21 | 100% | Price et al. (2020) |
| CHO | Transfection (protein, plasmid) | Transfection (RNA; plasmid, RNApolIII (pU6)) | Unreported | Unreported | Unreported | 32% | Rojek et al. (2021) |
| Zebrafish | Injection (RNA) | Injection (RNA) | YTTN | 35 | 21 | 90% | Wierson et al. (2019) |
| S. cerevisiae | Genomic expression, pScPGK1d | pUDP240 plasmid, RNApolII (pScTDH3e), HH&HDV, panARS origin of replication | TTTV | 19 | 21 | 100% | This study |
| S. cerevisiae | pUDP293 plasmid (panARS origin of replication), pScPGK1d | pUDP293 plasmid, RNApolII (pScTDH3e), HH&HDV, panARS origin of replication | TTTV | 19 | 21 | 31.5% | This study |
TEF1 promoter from Aspergillus nidulans.
U6 promoter from Aspergillus oryzae.
Ubi-1 promoter from Zea mays.
PGK1 promoter from S. cerevisiae.
TDH3 promoter from S. cerevisiae.
The goal of the present study was to optimize the ErCas12a-based genome editing in S. cerevisiae. To this end, we explored ways to improve the efficiency of genome editing by tuning the length of the DRs and of the spacers. We also evaluated approaches to express the crRNA array by testing different designs and consequently proposed design principles for application of ErCas12a in S. cerevisiae. Furthermore, we applied these principles to investigate applicability of ErCas12a to multiplexing strategies.
Materials and method
Strains and cultivation conditions
The yeast strains used in this study are shown in Table 2. Strains were grown on complex yeast extract–peptone–dextrose (YPD) medium, consisting of 20.0 g l−1 glucose, 20.0 g l−1 bacto peptone, and 10.0 g l−1 bacto yeast extract or on synthetic medium (SMD) containing 5 g l−1 (NH4)2SO4, 3 g l−1 KH2PO4, 0.5 g l−1 MgSO4·7H2O, 1.0 ml l−1 trace element solution, and was supplemented with 20 g l−1 glucose and 1.0 ml l−1 vitamin solution (Verduyn et al. 1992). SMD with urea as nitrogen source (SMD-urea) contained 6.6 g l−1 K2SO4, 3 g l−1 KH2PO4, 0.5 g l−1 MgSO4·7H2O, 1.0 ml l−1 trace element solution, and was supplemented with 20 g l−1 glucose, 1.0 ml l−1 vitamin solution and 2.3 g l−1 CO(NH2)2. As required, medium was supplemented with G418 (200 mg l−1) or hygromycin (200 mg l−1). Solid medium was obtained by addition of 20.0 g l−1 bacto agar. Saccharomyces cerevisiae strains were grown in an Innova 44R shaker (Eppendorf, Hamburg, Germany) at 30°C and 200 rpm in 500 ml shake flasks containing 100 ml medium or on stationary plates containing solid medium. For plasmid propagation, Escherichia coli XL 1-Blue cells (New England Biolabs, Ipswich, MA) were grown in 15 ml Greiner tubes containing 5 ml lysogeny broth (LB) medium at 37°C and 200 rpm in an Innova 4000 incubator shaker (Eppendorf) or on stationary plates containing solid LB medium supplemented with 20.0 g l−1 bacto agar. When required, LB medium was supplemented with ampicillin (100 mg l−1), kanamycin (50 mg l−1), spectinomycin (100 mg l−1), or chloramphenicol (25 mg l−1). Plates used for selection of E. coli transformants were grown stationary overnight at 37°C and S. cerevisiae cultures were grown at 30°C for 3 days in a stationary incubator. S. cerevisiae and E. coli cultures were stocked as 1 ml aliquots in a −80°C freezer after addition of 30% (v/v) glycerol.
Table 2.
Strains used in this study.
| Strain | Relevant genotype | Reference |
|---|---|---|
| CEN.PK113-7D | MATa MAL2-8c SUC2 | Entian and Kotter (2007) |
| IMX2600 | MATa MAL2-8c SUC2 Δcan1::Spycas9-natNT2 | van den Broek et al. (2023) |
| IMX2713 | MATa MAL2-8c SUC2 Δcan1::Spycas9-natNT2 ΔX-2*::pPGK1-Ercas12a-tPHO5 | This study |
| IME795 | MATa MAL2-8c SUC2 pGGKd018 | This study |
| IME796 | MATa MAL2-8c SUC2 pUDE1093 | This study |
| IMK1049, colony 1 | MATa MAL2-8c SUC2 Δcan1::Spycas9-natNT2 ΔX-2*::pPGK1-Ercas12a-tPHO5 Δade2 | This study |
| IMK1050, colony 2 | MATa MAL2-8c SUC2 Δcan1::Spycas9-natNT2 ΔX-2*::pPGK1-Ercas12a-tPHO5 Δade2 | This study |
| IMX2898, colony 1 | MATa MAL2-8c SUC2 Δcan1::Spycas9-natNT2 ΔX-2*::pPGK1-Ercas12a-tPHO5 ΔXI-3*::XdcrtE ΔYPRCτ3*::XdcrtI ΔII-1*::XdcrtYB | This study |
| IMX2899, colony 2 | MATa MAL2-8c SUC2 Δcan1::Spycas9-natNT2 ΔX-2*::pPGK1-Ercas12a-tPHO5 ΔXI-3*::XdcrtE ΔYPRCτ3*::XdcrtI ΔII-1*::XdcrtYB | This study |
Molecular biology techniques
The Ercas12a gene was de novo synthesized and cloned in plasmid by GeneArt (Thermo Scientific, Walthman, MA). Gibson assembly was performed using Gibson Assembly Master Mix (New England Biolabs) according to the supplier’s instructions. Golden Gate assembly was done according to Lee et al. (2015) with 20 fmol of each fragment with BsaI or BsmBI enzymes (New England Biolabs) and T7 DNA ligase (New England Biolabs). PCR for diagnostic purposes was performed using DreamTaq PCR Master Mix (Thermo Fisher Scientific) according to supplier’s instructions. Phusion® High-Fidelity DNA polymerase (Thermo Fisher Scientific) was used for fragment amplification for cloning purposes according to supplier’s instructions. All primers (Table S1, Supporting Information) were ordered at Sigma-Aldrich (St Louis, MO). Isolation of yeast genomic material for PCR verification was done with the LiAc-SDS protocol according to Lóoke et al. (2011). Plasmid isolation was performed using GeneJET Miniprep kit (Thermo Fisher Scientific) and PCR products were purified using GeneJET PCR Miniprep Kit (Thermo Fisher Scientific), both according to the supplier's instructions. When required, PCR products were isolated from gel using ZymocleanTM Gel DNA Recovery Kits (Zymo Research, Irvine, CA) according to the supplier’s instructions. Unless specified otherwise, plasmid sequencing was done with Sanger sequencing by Macrogen Europe (Amsterdam, the Netherlands).
Plasmid construction (supplemental protocol)
Construction of ErCas12a-expressing plasmid
The Ercas12a gene sequence was retrieved from Inscripta Inc. (https://www.inscripta.com/products/mad7-nuclease, consulted April 2020, WP_055225123.1). To optimize protein expression and nuclear localization, the sequence was codon optimized for S. cerevisiae and the nuclear localization signal (NLS) SV40 (Kalderon et al. 1984) was added to the C-terminus. The plasmid pUDE1093 (Addgene #204227) expressing the ErCas12a CRISPR nuclease from the strong constitutive pPGK1 promoter and tPHO5 terminator was constructed by Golden Gate cloning with BsaI of pYTK011, pGGKp177, and pUD1171 (coSc-Ercas12a) into the backbone of pGGKd018, replacing the BsaI-flanked GFP dropout with the Ercas12a expression cassette. Correct assembly of pUDE1093 was verified by colony PCR using the primers 10320 and 10325 and sequence verified by next generation sequencing at Plasmidsaurus (Eugene, OR; https://www.plasmidsaurus.com/).
Construction of ErCas12a genome editing platform plasmids
The platform plasmids pUDP239–pUDP242 consisting of four parts were assembled via Gibson Assembly using unique 60 bp synthetic HR sequences (SHR-sequences A, F, C, and I) incorporated during PCR amplification (Kuijpers et al. 2013). The yeast marker cassette, A-pAgTEF1-KanMXR-tAgTEF1-B, was amplified from plasmid pMEL13 (Mans et al. 2015) using primers 3748 and 3749. The yeast origin B-panARSopt-C was amplified from pUD530 (Gorter de Vries et al. 2017) using primers 4672 & 3856. The E. coli origin and marker, I-ori blaR-A, was amplified from pUD532 (Gorter de Vries et al. 2017) with primers 3274 & 3275. The gRNA insertion fragment was designed in four different combinations of promoter/terminator pair and DR sequences. The two DR sequences differ in length: a long DR of 35 nt (GTCAAAAGACCTTTTTAATTTCTACTCTTGTAGAT) and a short DR of 19 nt (AATTTCTACTCTTGTAGAT). Two types of promoter/terminator pairs were used: transcription by RNA polymerase III (pSNR52 and tSUP4) or II in combination with ribozymes (pTDH3 and tTDH3; hammerhead (HH) and hepatitis delta virus (HDV) ribozymes). Therefore, four different crRNA expression cassettes were ordered (pUD1190, pUD1191, pUD1194, and pUD1195) from Gene Art (ThermoFisher Scientific), each containing a GFP dropout site flanked with BsaI recognition sites for replacement by the crRNA for easy cloning. The plasmids were PCR amplified with primers 3283 & 4068 to obtain the four different SHR-flanked crRNA insertion sites fragments with either a long or short DR and either a RNA polymerase II (pTDH3) or III (pSNR52) promotor. The PCR-amplified fragments were purified, and different platform plasmids were assembled by combining the appropriate fragments in a Gibson Assembly reaction. Correct assembly was verified by colony PCR of green colonies over the A, B, C, and I SHR-sequences of pUDP239–pUDP242 using the primers 16503 & 8401, 9719 & 10345, 10344 & 4369, and 16501 & 6818, respectively.
The episomal plasmid enabling the expression of Ercas12a together with the crRNA was constructed by Gibson assembly of five fragments flanked with compatible SHR sequences (Kuijpers et al. 2013). The yeast marker cassette A-pAgTEF1-hphNT1R-tAgTEF1-B was amplified from plasmid pMEL12 (Mans et al. 2015) using primers 3748 & 3749. The yeast origin F-panARSopt-C was amplified from pUD530 (Gorter de Vries et al. 2017) using primers 4672 & 3856. The E. coli origin and marker, I-ori blaR-A, was amplified from pUD532 (Gorter de Vries et al. 2017) with primers 3274 & 3275. The crRNA insertion expression cassette (short DR and RNApolII design) was amplified from pUD1190 with primers 3283 & 4068. The Ercas12a expression cassette was amplified from pUDE1093 with primers 17934 & 9393. Gibson assembly of these five fragments resulted in pUDP293 (Addgene #204228). Correct construction was PCR-verified with primers 9719, 7487, 4377, 10345, 10344, 4369, 16501, 6818, 16503, and 8401. Also, the plasmid was Sanger sequenced using primers 3847, 3276, 4672, 7487, 7488, 19481, 19482, 19483, 19484, 19485, 19486, 3288, 3274, and 3275 and sequence verified by next generation sequencing at Plasmidsaurus (Eugene, OR).
General construction of crRNA expressing plasmids
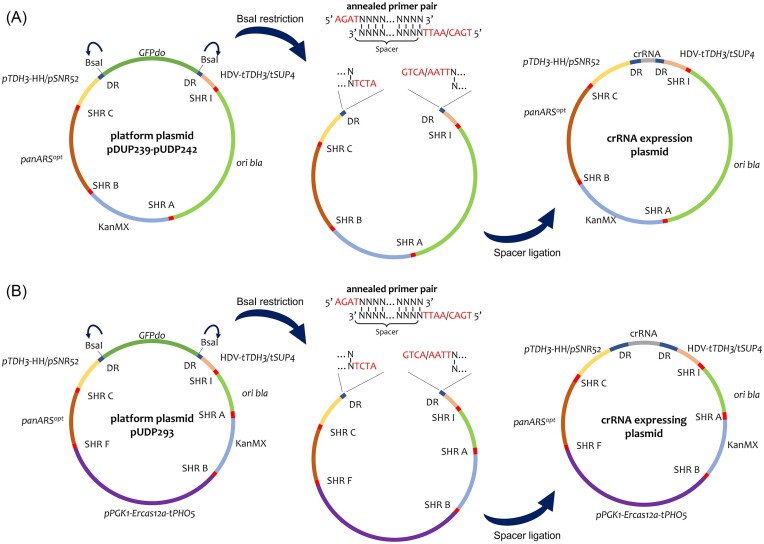
The plasmids used in this study for crRNA expression were constructed according to a generalized Golden Gate cloning scheme (Fig. 1). Platform plasmids contained a GFP dropout (GFPdo) within a crRNA expression cassette flanked by BsaI restriction sites, DRs, ribozymes in case of RNA polymerase II promoter/terminator regulation, and finally a promoter and terminator. Spacers were ordered as primer pairs with each primer containing four nucleotide overhangs complementary to the 5′ or 3′ four nucleotide overhangs generated after BsaI restriction of the platform plasmid. The annealing of the primer pair thus creates a DNA fragment with four nucleotide overhangs complementary to the restricted platform plasmid (Fig. 1). Plasmids expressing crRNAs were constructed by Golden Gate assembly of platform plasmid and annealed oligonucleotide followed by green/white screening. Correct assembly of the crRNA into the platform plasmids was verified by diagnostic PCR and Sanger sequencing.
Figure 1.
Construction of crRNA expression plasmids. Platform plasmids (A) pUDP239–pUDP242 and (B) pUDP293 carrying crRNA expression cassette (promoter: yellow; DR: dark blue; terminator: pink) with GFPdo (dark green), ColE1 ori bla (light green), G418 yeast resistance marker cassette (KanMX) (blue), pPGK1-Ercas12a-tPHO5 expression cassette (purple; B), and panARSopt yeast origin of replication (Liachko and Dunham 2014) (brown), separated by synthetic homologous recombination (SHR) sequences (red) (Kuijpers et al. 2013). BsaI restriction is used to cut the plasmid at BsaI restriction sites. Sticky ends left from BsaI restriction are complementary to four nucleotide overhangs of annealed spacer primer pair, which can also constitute a spacer-[DR-spacer]N array for multiplex editing at N + 1 genomic targets. The 3′ primer overhang sequence is dependent of choice of DR. The oligo pair is annealed and ligated of into the crRNA expression cassette generates crRNA expression plasmid.
Construction of ADE2 targeting plasmids and repair fragments
To construct crRNA expressing plasmids targeting the ADE2 gene in S. cerevisiae, complementary primers (18327, 18328, 18329, 19010, 19011, and 19012) containing a crRNA sequence targeting ADE2 were annealed and cloned into plasmids pUDP239, pUDP240, pUDP241, and pUDP242 via Golden Gate assembly with BsaI, resulting in plasmids pUDP285, pUDP286, pUDP287, pUDP288, pUDP289, pUDP290, pUDP291, and pUDP292, respectively. Inserts were PCR-verified with primers 10344 & 6097 and Sanger sequenced using primers 3283 & 11551. Repair fragments for ADE2 deletion were constructed by annealing the primers 10155 & 10156.
Construction of XI-3, YPRCτ3, and II-1 crRNA expression plasmids and repair fragments
Plasmids expressing crRNA targeting the XI-3 (Mikkelsen et al. 2012), YPRCτ3 (Flagfeldt et al. 2009), and II-1 (Babaei et al. 2021) loci individually, a duplex editing plasmid targeting XI-3 and YPRCτ3 and a plasmid targeting all three loci simultaneously as a multiplex crRNA array were constructed with platform plasmid pUDP240 as backbone. These three chromosomal locations were chosen as they are nonessential, thereby not interfering with any cellular activities, and promote high transcription. To construct crRNA expressing plasmids pUDP295 (XI-3), pUDP296 (YPRCτ3), pUDP311 (II-1), pUDP312 (XI-3 and YPRCτ3), and pUDP313 (XI-3, YPRCτ3, and II-1), annealed primer pairs (primers 19489 & 19490 for XI-3, 19 870 & 19871 for II-1, 19491 & 19492 for YPRCτ3, 19771 & 19772 for XI-3 and YPRCτ3 duplex editing, and 19881 & 19882 for XI-3, YPRCτ3, and II-1 triplex editing) were cloned into pUDP240 via BsaI Golden Gate assembly. Correct construction was PCR-verified and Sanger sequenced using primers 2655 & 6097.
To construct repair fragments for single editing, pUDE1111 (ori bla hphNT1 2 µm ConLS-pTDH3-ymNeonGreen-tADH1-ConR1) was PCR amplified with primers containing 60 bp overhangs homologous to the target: 15396 & 15397 for XI-3; 14022 & 14023 for YPRCτ3; 19773 & 19774 for II-1. For duplex editing, YPRCτ3 was repaired with the ymNeongreen fragment amplified from pUDE1111 with primers 14022 & 14023 and XI-3 was repaired with the ymScarletI fragment amplified from pUDE1112 (ori bla hphNT1 2 µm ConLS-pTDH3-ymScarletI-tADH1-ConR1) with primers 15396 & 15397. For multiplex editing, the transcriptional units including the Xanthophyllomyces dendrorhous genes XdcrtYB, XdcrtE, and XdcrtI (Verwaal et al. 2007) were PCR amplified from pUD1248, pUD1249, and pUD1250 with primers 19883 & 19884, 19547 & 19548, and 19549 & 19550, respectively, and were used as repair fragments.
Construction of Ercas12a-expressing strains
A S. cerevisiae strain expressing the CRISPR nucleases Ercas12a and Spycas9, was constructed from the Spycas9-expressing strain IMX2600 (CEN.PK113-7D Δcan1::Spycas9-natNT2) (van den Broek et al. 2023) (Table 2) using the CRISPR–Cas9-system (Mans et al. 2015). The Ercas12a expression cassette was amplified from pUDE1093 using primers 11074 & 13597 incorporating flanks with homology to the X-2 genomic location (Mikkelsen et al. 2012) required for in vivo HDR. IMX2600 was cotransformed with 500 ng of the plasmid pUDR547 expressing the gRNA targeting the X-2 intergenic region and 1000 ng of the pPGK1-Ercas12a-tADH1 repair fragment. Transformants were selected on YPD plates supplemented with hygromycin B. Correct integration was verified by diagnostic PCR using the primers 13662 & 13663. After plasmid recycling, the Ercas12a-expressing strain was named as IMX2713 (CEN.PK113-7D Δcan1::Spycas9-natNT2, ΔX-2::Ercas12a).
Genome editing procedure
crRNA design
All oligonucleotides targeting genomic sites were designed as two annealing primer sequences containing four nucleotide overhangs complementary to the BsaI restriction overhangs of the plasmids. To design the spacer sequence, the CHOPCHOP tool (Labun et al. 2019) was used to find a list of candidate spacers ranked by predicted functionality, searching for 5′-TTTV-3′ PAM. This list was checked for RNA secondary structure with the RNAfold online tool (Lorenz et al. 2011). Spacer sequences with no predicted secondary structure interfering with the DR sequence were checked for off-target effects with an in-house BLAST search tool against the IMX2600 strain genome and the top result with no off-targets was chosen.
Transformation procedure
For genomic editing, the recipient yeast strain was transformed in triplicate with 500 ng crRNA expression plasmid DNA and 1000 ng repair fragment DNA. In all cases, a control culture of the same strain was transformed with 500 ng of the crRNA expression plasmid without repair fragment. Saccharomyces cerevisiae strains were transformed according to the LiAc/ssDNA/PEG transformation protocol (Gietz et al. 1992). After transformation, 1 ml YPD was added to the cells for a 2-h recovery at 30°C prior to plating on selective medium. All transformations for determining editing efficiencies were performed in triplicate.
Genomic editing and screening
Transformants were genotyped by diagnostic PCR with primers specific for the targeted genomic location. In cases where ADE2 was targeted, visual red/white screening was performed and up to ten red colonies and two white colonies were chosen for genotyping. In cases the repair fragment contained an ymNeonGreen expression cassette, transformants were visually screened under blue light for fluorescence before genotyping. Visual orange/white screening was used before PCR analysis for multiplex editing as the repair fragments each carried a different gene in the X. dendrorhous β-carotene pathway. Correct integration of all three genes leads to orange phenotype due to β-carotene accumulation (Verwaal et al. 2018). Editing efficiency was calculated as colonies showing appropriate reporter phenotype (red/fluorescent/orange) over total transformants.
Growth rate determination
To determine maximum specific growth rates on glucose, the yeast strains were cultivated on SMD. When selective pressure for plasmid maintenance was required, IME795 (CEN.PK113-7D pGGKd018) and IME796 (CEN.PK113-7D pUDE1093) were grown on SMD-Urea supplemented with G418. Cultures were inoculated from stocked aliquots, grown overnight at 30°C and transferred to fresh medium. From this preculture, the experimental cultures were inoculated in triplicate at an OD660 of 0.2 on their corresponding medium. Optical density (OD) was measured at set time intervals with a Jenway 7200 scanning spectrophotometer (Cole-Parmer Inc, Chicago, MI) at 660 nm. To remove plasmids from IME795 and IME796, strains were restreaked on nonselective medium, until restreaking on selective medium (G418) yielded no colonies. Maximum specific growth rates were determined by plotting the natural logarithm of the growth curve and performing linear regression on the exponential portion of the growth curve (at least five data points).
Analytical techniques
Culture supernatants were collected as previously described in Hassing et al. (2019), glucose and ethanol concentrations were analysed on an Agilent 1260 Infinity HPLC (Agilent Technologies Inc, Santa Clara, CA) equipped with an Animex HPX-87H ion exchange column (Bio-Rad, Hercules, CA) at 60°C and 5 mM H2SO4 solution as mobile phase a flow rate of 0.6 ml min−1.
Flow cytometry and cell sorting
After the transformation procedure, the cell suspension was plated onto selective medium (one-fifth of the cells) and transferred to 20 ml liquid selective medium in 100 ml shake flasks (four-fifth of the cells) and cultivated for 2 days at 30°C while shaking at 200 rpm. Then, 1 ml of the cell suspension was transferred to 20 ml fresh YPD medium supplemented with 200 mg l−1 of hygromycin in 100 ml shake flasks and grown for 2 days. Finally, 1 ml of cell suspension was transferred to 100 ml nonselective YPD medium in 500 ml shake flasks for optimal expression of the gene coding a fluorescent protein and grown for 1 day at 30°C while shaking at 200 rpm. In vivo assembly efficiency of ymNeongreen and ymScarletI genes in IMX2713 was analyzed by measuring fluorescence levels in the BD FACSAria™ II SORP Cell Sorter (BD Biosciences, Erembodegem-Dorp, Belgium) equipped with 355, 445, 488, 561, and 640 nm lasers and a 70-m nozzle and operated with filtered FACSFlow™ software (BD Biosciences). The fluorophore ymScarletI was excited by the 561-nm laser and emission was detected through a 582-nm bandpass filter with a bandwidth of 15 nm. The fluorophore ymNeongreen was excited by the 488 nm laser and emission was detected through a 545-nm bandpass filter with a bandwidth of 30 nm. The cytometer performance was evaluated prior to each experiment by running a CST cycle with CS&T Beads (BD Biosciences) and the drop delay for sorting was determined by running an Auto Drop Delay cycle with Accudrop Beads (BD Biosciences). For each sample, 100 000 events were analyzed. Cell morphology was analyzed by plotting forward scatter (FSC) against side scatter (SSC) and the appropriate cell size was gated. Gated cells were used to determine the fluorescence intensity of the cells. Gating windows for fluorescence intensity were based on the fluorescence of the cells transformed with solely ymNeongreen or ymScarletI as repair fragment for sole integrations. Cells in the gate ymNeongreen+ymScarletI+ were sorted separately on nonselective YPD plates and grown for 2 days. FACS data was analyzed using the Flowing Software version 2.5.1 (Turku Centre for Biotechnology, Finland).
Whole genome sequencing
Yeast genomic DNA of transformants IMX2713, IMK1050, IMK1051, IMX2898, and IMX2899 was isolated using QIAGEN Genomic DNA isolation buffer set in combination with the Genomic-tip 100/G columns (Qiagen, Hilden, Germany), following manufacturer’s instructions. Genomic DNA concentrations were measured with the BR ds DNA kit (Invitrogen, Carlsbad, CA) using a Qubit 2.0 Fluorometer (Thermo Fisher Scientific). Whole genome sequencing using TruSeq DNA PCR-Free Library Preparation (150 bp paired ends, 350 bp insert) on a NovaSeq 6000 S4 sequencer was performed by Macrogen Europe.
All Illumina sequencing data (Table S5, Supporting Information) are available at NCBI (https://www.ncbi.nlm.nih.gov/) under the bioproject accession number PRJNA977855 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA977855). The raw Illumina reads were mapped using the Burrows–Wheeler Alignment tool (BWA) (Li 2013) against a chromosome-level reference genome of IMX2600 (NCBI bioproject accession number PRJNA976676 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA976676) (van den Broek et al. 2023) to which four extra contigs containing the ErCas12a and Xdcrt integration cassettes were added (supplementary sequences). The alignments were further processed with SAMtools (Danecek et al. 2021) and visualized using the Integrative Genomics Viewer (IGV) (Robinson et al. 2011). Sequence variants were called using Pilon (Walker et al. 2014), ReduceVCF (https://github.com/AbeelLab/genometools/blob/master/scala/abeel/genometools/reducevcf/ReduceVCF.scala) was used to extract the variants and VCFannotator (http://vcfannotator.sourceforge.net/) was used to annotate the variants.
Results
Construction of a ErCas12a nuclease expressing S. cerevisiae strain
To evaluate ErCas12a-mediated genome editing in S. cerevisiae, two strategies for nuclease expression were explored. Expression from a chromosomal location ensures stable gene expression, allows recurrent use of the endonuclease and a constant selective pressure is not required, whereas plasmid-based expression allows for quick removal of the nuclease encoding DNA after the editing event is completed and enables easy transfer into different yeast lineages. Plasmid-free nuclease expression requires integration of the respective expression cassette into the genome of S. cerevisiae. First, the cas12a gene from E. rectale (Ercas12a) was codon optimized for S. cerevisiae and the SV40 nuclear localization sequence was added to the C-terminus of the protein to ensure nuclear localization. The Ercas12a gene was cloned under control of the constitutive PGK1 promoter and the PHO5 terminator. The Ercas12a expression cassette was integrated into the X-2 site (Mikkelsen et al. 2012), regarded as safe integration site, using the Spycas9-system (Mans et al. 2015), guided by a specific gRNA expressed from pUDR547, in the S. cerevisiae strain IMX2600 (CEN.PK113-7D Δcan1::Spycas9-natNT2) resulting in IMX2713 (CEN.PK113-7D Δcan1::cas9-natNT2 ΔX-2::Ercas12a). SpyCas9 and ErCas12a endonucleases rely on distinct crRNA/tracrRNA configurations and dissimilar PAM sequences. Nevertheless, this disparity does not pose an obstacle to their simultaneous expression.
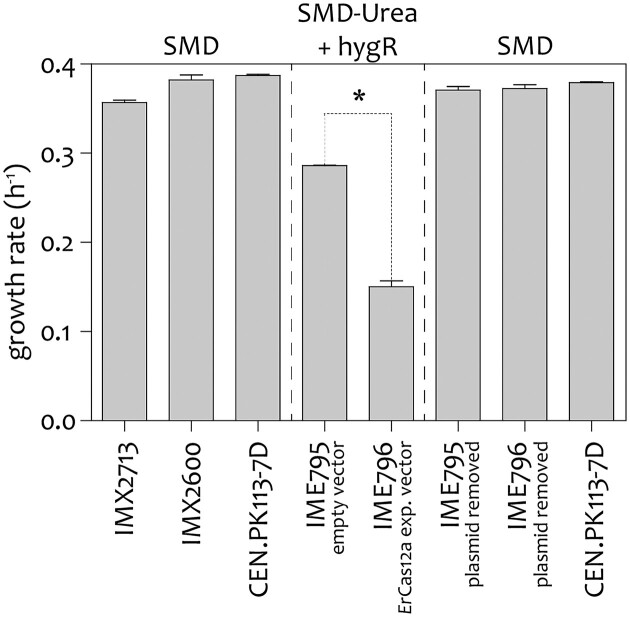
To study the impact of Ercas12a expression in IMX2713, the specific growth rate on SMD was determined. The strain IMX2713 (CEN.PK113-7D Δcan1::cas9-natNT2 ΔX-2::Ercas12a) exhibited a nonsignificant 10% decrease in growth rate (0.36 ± 0.004 h−1) in comparison to its parental strain IMX2600 (0.39 ± 0.010 h−1) and laboratory strain CEN.PK113–7D (0.40 ± 0.001 h−1), demonstrating that genomic integration of Ercas12a had no significant impact on growth (Fig. 2).
Figure 2.
Maximum specific growth rate of strains expressing ErCas12a and their control strains. IMX2713 expressing ErCas12a from its genomic DNA, control strain IMX2600 and CEN.PK113-7D were cultivated in shake flasks on SMD with glucose as sole carbon source. IME796 expressing ErCas12a from the multicopy plasmid pUDE1093 (2 µm KanMX pTDH3-gfpdo-tADH1 pPGK1-Ercas12a-tPHO5 and its reference strain IME795 containing the empty vector pGGKd018 (2 µm KanMX) were grown on SMD with glucose as carbon source and urea as nitrogen source supplemented with G418 for selective pressure for plasmid maintenance. After plasmid removal, the strains were grown on SMD with glucose as carbon source with CEN.PK113-7D as control strain. The measurements were performed in two (IMX2713 and IMX2600) or three (IME795 & IME796) biological replicates and two technical replicates per sample. Significant differences in specific growth rate relative to the control strain based on a two-tailed homoscedastic paired t-test are indicated with a * (P < .05), ** (P < .01), or *** (P < .001).
To evaluate whether increased expression negatively impacts growth of S. cerevisiae, Ercas12a was expressed from the 2-µm (high copy) plasmid pUDE1093 under control of the same constitutive promoter used in IMX2713. The strain IME796 [pUDE1093 (pPGK1-Ercas12a-tPHO5) grew significantly slower with a 52% reduction of the maximum specific growth rate relative to the control strain IME795 that harboured the pGGKd018 empty vector (IME796 µmax = 0.14 ± 0.00 h−1, IME795 µmax = 29 ± 0.00 h−1)]. We hypothesized that an increased gene copy number of the plasmid-borne expression system might result in enhanced gene expression of ErCas12a that in return could be toxic to the cell (Fig. 2). Curing the plasmids fully restored specific growth rate in strains IME796−pUDE1093 and IME795−pGGKd018 to that of the CEN.PK113-7D reference strain. On SMD with glucose as carbon source the reference strain CEN.PK113-7D grew with a growth rate of 0.379 ± 0.00 h−1, IME796−pUDE1093 and IME795−pGGKd018 grew with a growth rate of 0.371 ± 0.01 h−1 and 0.376 ± 0.00 h−1, respectively (Fig. 2). These results were in line with earlier characterization of FnCas12a and SpyCas9, where plasmid borne expression of CRISPR endonucleases led to similar toxic effect and reduction of growth rates (Generoso et al. 2016, Swiat et al. 2017).
Defining parameters for optimal ErCas12a-mediated genome editing in S. cerevisiae
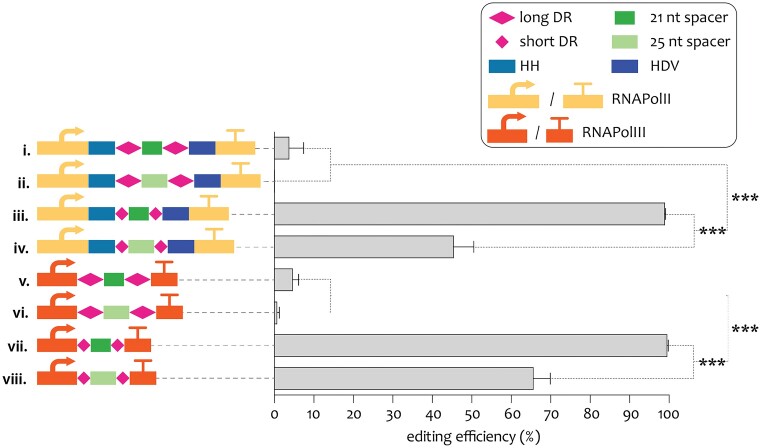
To optimize the editing procedure with ErCas12a in S. cerevisiae, a series of constructs with variable designs for the crRNA expression cassette were made to evaluate the effect of different promoter systems, length of DRs and spacer length on editing efficiency. Two expression cassettes of the crRNA were assessed, the first was based on the RNA polymerase III (RNApolIII) regulatory sequences pSNR52 and tSUP4 classically used to express crRNA; and the second was based on the RNA polymerase II (RNApolII) dependent TDH3 regulatory sequences pTDH3/tTDH3. Since RNA polymerase II expression concomitantly results in 5′ RNA capping and 3′ poly-A tailing, the pTDH3/tTDH3 constructs incorporated both HammerHead (HH) and Hepatitis Delta Virus (HDV) ribozymes enabling maturation of the crRNA after self-cleavage (Gao and Zhao 2014). Two DR sequences were tested with either the short 19 nt 5′-AATTTCTACTCTTGTAGAT-3′ or the longer 35 nt 5′-GTCAAAAGACCTTTTTAATTTCTACTCTTGTAGAT-3′ DR. The long DR is corresponding to the DR found in the E. rectale CRISPR array and was used in most of the studies implicating ErCas12a (Table 1). A previous study established that editing in S. cerevisiae using the Cas12a nuclease from Francisella novicida (FnCas12a) was more efficient when using shorter DRs corresponding to the highly conserved core region of the Cas12a DR among prokaryotes, than when using the native long DR (Swiat et al. 2017, Randazzo et al. 2021). Additionally, two spacers of 21 or 25 nt were evaluated. All eight combinations (23) were constructed and combined using spacer sequences that targets the S. cerevisiae ADE2 locus; deletion of ADE2 causes an accumulation of 5-amino-imidazole ribonucleotide that upon oxidation produces a red/pink colour (Dorfman 1969), facilitating the visual detection of the edited transformants (Fig. 3). The spacers CCGGTTGTGGTATATTTGGTGTGGA (25 nt) and CCGGTTGTGGTATATTTGGTG (21 nt) were selected based on the presence of a 5′-TTTV-3′ PAM sequence located at T738 within the ADE2 open reading frame. Separately, the eight plasmids targeting ADE2 (pUDP285–pUDP292), were cotransformed with a 120 bp repair oligo containing 60 bp homology upstream and downstream the ADE2 gene for proper chromosomal repair using the native yeast HDR machinery into the S. cerevisiae strain IMX2713 (can1Δ::Spycas9-natNT2 X-2Δ::pPGK1-Ercas12a-tPHO5).
Figure 3.
Efficiency of crRNA expression designs transformed into IMX2713 targeting ADE2. Different combinations of expression system, spacer length and DR length were investigated: i–iv RNApolII (pTDH3 and tADH1) expression system and ribozymes flanking the DR-spacer-DR sequences; v–viii RNApolIII (pSNR52 and tSUP4) expression system; i, ii, v, and vi long (35 nt) DR sequences; ii, iv, vi, and viii short (19 nt) DR sequences; i, iii, v, and vii short (21 nt) spacer; ii, iv, vi, and viii long (25 nt) spacer. Efficiency was calculated as the ratio of red transformants over the total number of transformants of three biological replicates. Significant differences in editing efficiencies based on a two-tailed homoscedastic paired t-test are indicated with a * (P < .05), ** (P < .01), or *** (P < .001).
For each transformation, 10 red and two white colonies were genotyped by diagnostic PCR to confirm the correct ADE2 deletion (Figure S1, Supporting Information). Since all genotyped red colonies harboured the expected deletion and the white colonies still showed presence of the wild-type ADE2 allele, the targeting efficiencies (%) were estimated by the ratio of red colonies over all transformants. The most impactful design element was the length of the DR, as constructs carrying the long DR (35 nt) did not exceed a targeting efficiency of 5%. This is remarkable, since the long DR sequence for ErCas12a has been widely used in previous literature (Table 1). In contrast, all designs that included the short DR version (19 nt) exhibited at least 45% targeting efficiencies regardless of the spacer length and the promoter systems used (Fig. 3; Table S2, Supporting Information). Next, from the four configurations with short DRs, the two constructs with shorter spacers (21 nt) were significantly better, reaching over 99% efficiency irrespective of the expression system used. As the strain IMX2713 expressed both the ErCas12a and SpyCas9 endonucleases, the strain IMX2600 solely expressing SpyCas9 was transformed with the pUDP287 plasmid targeting ADE2 with the highest efficiency and its repair fragment. The absence of red colonies and high number of white colonies shows that Cas9 and crRNAs designed for Cas12a are incompatible with each other and the observed editing is purely performed by ErCas12a (Table S3, Supporting Information).
Overall, these data indicated that a combination of 21 nt spacer and short flanking DR represented an optimized design for highly efficient ErCas12a-mediated genome editing in S. cerevisiae. Due to the applicability of RNApolII expression systems across difference Saccharomycotina yeast species, it was decided to continue with the RNApolII design. Based on these considerations, pUDP240 (pTDH3-HH-S.DR-GFPdo-S.DR-HDV-tTDH3) containing a versatile GFP dropout construct for easy cloning of crRNAs was chosen as basic plasmid architecture for further experiments (Fig. 1A).
ErCas12a can edit commonly used integration sites with high efficiency in S. cerevisiae
For metabolic engineering applications, it is important that the engineered function (e.g. heterologous metabolic pathway) remains mitotically stable, which is usually achieved through chromosomal integration. To further assess the applicability of ErCas12a to engineer the S. cerevisiae genome, we have selected three nonessential chromosomal regions that promote high transcriptional activities: XI-3 (Mikkelsen et al. 2012), YPRCτ3 (Flagfeldt et al. 2009), and II-1 (Babaei et al. 2021). Spacers were designed for each genomic location and assembled into pUDP240 resulting in pUDP295 (crRNA XI-3), pUDP296 (crRNA YPRCτ3), and pUDP311 (crRNA II-1). Conversely to ADE2 editing, the successfully targeted transformants showed no distinctive phenotype, therefore, to determine the targeting efficiency a repair DNA fragment including the ymNeonGreen fluorescent reporter (Botman et al. 2019) flanked with corresponding homology arms was used. To determine the editing efficiency, the strain IMX2713 (can1Δ::Spycas9-natNT2 X-2Δ::pPGK1-Ercas12a-tPHO5) was transformed with pUDP296 (crRNA XI-3), pUDP300 (crRNA YPRCτ3), and pUDP311 (crRNA II-1) and their respective repair fragments (Table S4, Supporting Information). The transformations targeting the XI-3 and YPRCτ3 loci yielded a 100% editing efficiency as all transformants exhibited a fluorescent phenotype resulting from the integration of the ymNeonGreen cassette. The transformation targeting II-1 reached a near 100% editing efficiency of 99.1 ± 0.8%. Similarly to ADE2, transforming IMX2600 with the ErCas12a-designed crRNAs targeting XI-3, YPRCτ3, and II-1 did not result in editing (Table S3, Supporting Information). As precedingly performed for ADE2, for each transformation ten green fluorescent colonies were subjected to genotyping and all fluorescent transformants were shown to have a correct insertion of ymNeonGreen repair at the targeted genomic site (Figure S2, Supporting Information).
ErCas12a can process multispacer crRNA arrays for multiplex editing in S. cerevisiae
The most attractive feature of class 2 type V-A nucleases over class 2 type II is their ribonuclease activity enabling self-processing of the pre-crRNA array. This property becomes pertinent when several crRNA have to be expressed simultaneously to target different chromosomal locations in a so-called multiplexing editing approach. By combining multiple spacers interspaced by DR sequences in a single array, the Cas12a nuclease can cleave the polycistronic pre-crRNA array into single mature crRNAs (Fig. 4A). To assess the multiplexing ability of ErCas12a in S. cerevisiae, the previously tested spacers targeting XI-3 and YPRCτ3 were combined in a single DR-spacer-DR-spacer-DR array and cloned into pUDP240 yielding pUDP312 (Table 3). The use of repair fragments expressing the fluorescent proteins ymScarlet and ymNeongreen for resolving the ErCas12a-induced DSB at XI-3 and YPRCτ3, respectively enabled analysis of the transformed population by flow cytometry (Fig. 4B). A 96.5 ± 4.9% editing efficiency was obtained for duplex editing of the XI-3 and YPRCτ3 genomic integration sites (Fig. 4C; Figure S3 and Table S4, Supporting Information). These results showed that the ErCas12a nuclease can process a polycistronic crRNA array for multiplex editing.
Figure 4.
(A) Overview of multiplex genome editing in S. cerevisiae with ErCas12a. (1) crRNAs and repair DNA are designed and ordered as oligos. (2) crRNAs are cloned into the crRNA expression plasmid using the predesigned BsaI compatible overhangs. (3) The crRNA expressing plasmid is cotransformed with the DNA repair fragments into the ErCas12a expressing strain IMX2713. (4) The pre-crRNA multispacer array is transcribed and processed by the flanking ribozymes. (5) ErCas12a self-processes the pre-crRNA array into mature crRNAs, recognizes the pseudoknot and is guided to the target location. (6) ErCas12a induces precise DSB and the native homology directed repair machinery resolves the DSB using the supplied repair DNA fragment as template, thereby incorporating the intended genomic modification. (B) Screening strategy for the evaluation of duplex editing in IMX2713. IMX2713 was cotransformed with pUDP312 and repair DNA fragments ymScarletI and ymNeongreen for targeting and integration in XI-3 and YPRCτ3, respectively. After transformation, the cells were cultivated on YPD G418 medium, transferred to fresh selective medium, transferred to nonselective YPD, and screened by FACS. (C) FACS screening of the transformed cell population. Cells were plotted for ymNeongreen (y-axis) and ymScarletI (x-axis) fluorescence levels and gated for single (ymNeongreen upper left gate, ymScarletI, bottom right gate) or double (ymNeongreen and ymScarletI, upper right gate) fluorescence. (D) Sorted cell populations grown on YPD. (E) Schematic representation of multiplex editing strategy for integration of XdcrtE, XdcrtI, and XdcrtYB in XI-3, YPRCτ3, and II-1, respectively. (F) Plate of grown cells after cotransforming of IMX2713 with pUDP311 targeting XI-3, YPRCτ3, and II-1 and the respective DNA repair fragments encoding XdcrtE,XdcrtI, and XdcrtYB. Orange colonies indicate correct integration of all three Xdcrt genes due to accumulation of the orange pigment β-carotene.
Table 3.
Plasmids used in this study.
| Plasmid | Genotype | Reference |
|---|---|---|
| pUDR547 | ori bla hph 2 µm gRNA X-2 gRNA X-2 | Postma et al. (2021) |
| pYTK011 | ori cat pPGK1 | Lee et al. (2015) |
| pYTK053 | ori cat tADH1 | Lee et al. (2015) |
| pUD1171 | ori cat coSc-NLSSV40-ErCas12a | GeneArt™ |
| pGGKd015 | ori bla ConLS-GFPdo-ConR1 | Hassing et al. (2019) |
| pGGKd018 | ori aadA1 kanMXR 2 µm GFPdo | Randazzo et al. (2021) |
| pUD530 | ori aph3’ SHRB panARSopt SHRC | GeneArt™ |
| pUD531 | ori aph3’ SHRC pTDH3BsaI BsaICYC1t SHRI | GeneArt™ |
| pUD532 | ori aph3’ SHRI ori bla SHRA | GeneArt™ |
| pMEL12 | ori bla 2 μm hphNT1 gRNA-CAN1.Y | Mans et al. (2015) |
| pMEL13 | ori bla 2 μm kanMX gRNA-CAN1.Y | Mans et al. (2015) |
| pUD1190 | ori aph3’ pTDH3-HH-S.DR-GFPdo-S.DR-HDV-tTDH3 | GeneArt™ |
| pUD1191 | ori aph3’ pTDH3-HH-L.DR-GFPdo-L.DR-HDV-tTDH3 | GeneArt™ |
| pUD1195 | ori aph3’ pSNR52-S.DR-GFPdo-S.DR-tSUP4’ | GeneArt™ |
| pUD1196 | ori aph3’ pSNR52-L.DR-GFPdo-L.DR-tSUP4’ | GeneArt™ |
| pUDE1093 | ori aadA1 kanMX 2 µm pPGK1-Ercas12a-tPHO5 | This study, addgene #204227 |
| pUDE1111 | ori bla hph 2 µm ConLS-pTDH3-ymNeonGreen-tADH1-ConR1 | This study |
| pUDE1112 | ori bla hph 2 µm ConLS-pTDH3-ymScarletI-tADH1-ConR1 | This study |
| pUD1248 | ori cat pPGK1-XdcrtYB-tPGK1 | This study |
| pUD1249 | ori cat pHHF2-XdcrtE-tADH1 | This study |
| pUD1250 | ori cat pTDH3-XdcrtI-tTDH1 | This study |
| pUDP239 | ori bla kanMX panARSoptpTDH3-HH-L.DR-GFPdo-L.DR-HDV- tTDH3 | This study |
| pUDP240 | ori bla kanMX panARSoptpTDH3-HH-S.DR-GFPdo-S.DR-HDV- tTDH3 | This study |
| pUDP241 | ori bla kanMX panARSoptpSNR52-L.DR-GFPdo-L.DR-tSUP4’ | This study |
| pUDP242 | ori bla kanMX panARSoptpSNR52-S.DR-GFPdo-S.DR-tSUP4’ | This study |
| pUDP285 | ori bla kanMX panARSoptpTDH3-HH-L.DR-21 nt crRNA ADE2-L.DR-HDV-tTDH3 | This study |
| pUDP286 | ori bla kanMX panARSoptpTDH3-HH-L.DR-25 nt crRNA ADE2-L.DR-HDV-tTDH3 | This study |
| pUDP287 | ori bla kanMX panARSoptpTDH3-HH-S.DR-21 nt crRNA ADE2-S.DR-HDV-tTDH3 | This study |
| pUDP288 | ori bla kanMX panARSoptpTDH3-HH-S.DR-25 nt crRNA ADE2-S.DR-HDV-tTDH3 | This study |
| pUDP289 | ori bla kanMX panARSoptpSNR52-L.DR-21 nt crRNA ADE2-L.DR-tSUP4’ | This study |
| pUDP290 | ori bla kanMX panARSoptpSNR52-L.DR-25 nt crRNA ADE2-L.DR-tSUP4’ | This study |
| pUDP291 | ori bla kanMX panARSoptpSNR52-S.DR-21 nt crRNA ADE2-S.DR-tSUP4’ | This study |
| pUDP292 | ori bla kanMX panARSoptpSNR52-S.DR-25 nt crRNA ADE2-S.DR-tSUP4’ | This study |
| pUDP295 | ori blaR kanMX panARSoptpTDH3-HH-S.DR-crRNA XI-3-S.DR-HDV-tTDH3 | This study |
| pUDP296 | ori bla kanMX panARSoptpTDH3-HH-S.DR-crRNA YPRCτ3-S.DR-HDV-tTDH3 | This study |
| pUDP311 | ori bla kanMX panARSoptpTDH3-HH-S.DR-crRNA II-1 -S.DR-HDV-tTDH3 | This study |
| pUDP312 | ori bla kanMX panARSoptpTDH3-HH-S.DR-crRNA XI-3-S.DR-crRNA YPRCτ3-S.DR-HDV-tTDH3 | This study |
| pUDP313 | ori bla kanMX panARSoptpTDH3-HH-S.DR-crRNA XI-3-S.DR-crRNA YPRCτ3-S.DR-crRNA II-1-S.DR-HDV-tTDH3 | This study |
| pUDP293 | ori blaR hph panARSoptpTDH3-HH-S.DR-GFPdo-S.DR-HDV- tTDH3, pPGK1-Ercas12a-tPHO5 | This study addgene #204228 |
| pUDP299 | ori bla hph panARSoptpTDH3-HH-S.DR-crRNA ADE2-S.DR-HDV-tTDH3, pPGK1-Ercas12a-tPHO5 | This study |
Similarly, a polycistronic crRNA array comprising the previously tested spacers targeting XI-3, YPRCτ3, and II-1 were combined in a single array and cloned into the pUDP240 yielding pUDP313 (Table 3). The repair of the three DSB introduced at XI-3, YPRCτ3, and II-1 was achieved by cotransforming three repair DNA fragments containing respectively the X. dendrorhous carotenogenic genes crtE, crtI, and crtYB (Fig. 4D). Simultaneous expression of the three Xdcrt genes in S. cerevisiae leads to accumulation of β-carotene, which colour the yeast colonies orange (Verwaal et al. 2007). Cotransformation of the plasmid encoding the multispacer crRNA array and the β-carotene genes into IMX2713 resulted in an editing efficiency of 31.3 ± 6.6% (Fig. 4E; Table S4, Supporting Information). Whole genome sequencing of two orange colonies verified the integration of the three β-carotene genes into their respective genomic locations (Figure S4, Supporting Information). Genotyping of 10 white colonies revealed that in all cases the integration of the XdcrtYB gene into the II-1 genomic location was missing and in two colonies the XdcrtI gene in YPRCτ3 as well, resulting in absence of β-carotene accumulation (Figure S5, Supporting Information).
Whole genome sequencing of ErCas12a-edited strains strongly suggest absence of off-targeting
Although the programmable specificity of Cas9 and Cas12a primarily depends on the spacer sequence of the gRNA and the presence of a PAM sequence at the genomic target, off-target cleaving activity could still take place at sequences with up to five mismatches in the gRNA sequence (Zhang et al. 2015). Therefore, potential off-target DSBs in CRISPR-mediated editing may pose a major concern in phenotypic analysis. To investigate the extent of off-targeting by ErCas12a, two ∆ade2 strains (IMK1050 and IMK1051) and two β-carotene producing strains (IMX2898 and IMX2899) obtained by ErCas12a genome editing, were subjected to whole genome sequence analysis (Table S5, Supporting Information). Compared to the parental strain IMX2713 (Spycas9 Ercas12a), IMK1050 (Spycas9 Ercas12a Δade2) had one nucleotide variation (CHRXIII in gene TRI1 C654A, resulting in Asn218Lys), and IMK1051 (Spycas9 Ercas12a Δade2) had four nucleotide variations in intergenic regions (CHRI A408C, CHRXI A465625T, CHRXIV A478C, and CHRXV G637531C). Similarly, the analysis of the genome sequence of the carotenogenic S. cerevisiae strains IMX2898 and IMX2899 (Spycas9 Ercas12a XdcrtE XdcrtI XdcrtYB) did not reveal many single nucleotide variations (SNVs). IMX2898 harbored one SNV (CHRXI in gene SSH4 G370C resulting in Glu124Gln) and IMX2899 four (CHRIII in YEL074W C122A causing Pro41His and the synonymous mutation A120C, CHRIV in NUM1 T4188G causing Asn-1396-Lys and CHRXIV A478C in an intergenic region) of which one SNV located in telomeric region of CHRXIV was also observed in IMK1051 (Table S6, Supporting Information). The detection of mutations A408C and A478C on CHRI and CHRXIV, which are both located in the telomeres of their respective chromosomes, stresses that these SNVs occur in rather redundant regions of the genome. Moreover, nucleotide BLAST analysis of the region of the SNV (± 500 bp upstream–downstream of the SNV location) did not identify potential a PAM (TTTV) and spacer seed sequence susceptible to result in off-target restriction. Thus, there was no indication that the mutations in these strains were located near sequences recognizable by ErCas12a based on the supplied crRNAs.
Moreover, comparison of genome sequence of IMX2713 (Ercas12a Spycas9) and IMX2600 (Spycas9) revealed only one mutation in MSH5, a gene located on CHRIV and that encodes for a protein involved in meiosis. Thus solely expressing Ercas12a without presence of a crRNA spacer sequence was not mutagenic.
Since connection between the mutations in the different strains was absent, these mutations were likely to have been introduced as a result of regular DNA replication during cell division rather than a consequence of genome editing with ErCas12a.
Designing and testing a transportable ErCas12a-mediated editing platform
To foster the use of this CRISPR tool, a plasmid combining the Ercas12a gene and the crRNA expression cassettes was constructed that can easily be transferred to other S. cerevisiae lineages without investing time in constructing a strain expressing the endonuclease from a chromosomal location. As demonstrated earlier, such overexpression of Ercas12a from a high copy number plasmid significantly reduced the growth rate. The transportable editing system pUDP293 (Fig. 1B) was designed to express Ercas12a and contained the easy to clone crRNA insertion part with the previously determined optimal configurations for crRNA expression. The highly efficient ADE2 spacer was cloned into the transportable plasmid to test the functionality of the transportable ErCas12a plasmid and to determine the editing efficiency with episomally expressed Ercas12a. Therefore, the S. cerevisiae reference strain CEN.PK113-7D was co-transformed with the transportable plasmid along with repair fragments for ADE2 deletion. Based on the ratio of red colonies over the total number of transformants an editing efficiency of 31.5 ± 11.5% was determined (Table S4, Supporting Information). A total of 10 red colonies were subjected to genotyping for the ADE2 deletion and confirmed the deletion of ADE2 (Figure S6, Supporting Information). The editing efficiency obtained with this plasmid borne Ercas12a expression was 3-fold lower than with Ercas12a expression from a chromosomal locus. Although reduced, the efficacy of this plasmid system will facilitate the application of ErCas12a allowing for genome editing in multiple Saccharomyces lineages and this editing system has also the potential to be transferred in other yeast species (Liachko and Dunham 2014, Juergens et al. 2018).
Discussion
The CRISPR–Cas genome editing technology has greatly accelerated strain engineering in the last decade. However, the use of the hallmark endonuclease SpyCas9 is impeded by uncertainties such as a complex patent landscape (Ledford 2022, Shaffer 2022). In addition, although already widespread, the SpyCas9 imposes design principles that sometimes might restrict its field of application, such as its GC-rich PAM sequence, a gRNA that includes complex structural elements that prevent implementation of simple multiplex editing design, limit the applicability of this endonuclease (Swarts and Jinek 2018).
Hence, facilitating access to CRISPR systems with more transparent and less restrictive intellectual property (IP) rights, as well as a more adaptable multiplexing framework, would further advance the democratization of CRISPR editing technologies. An alternative CRISPR–Cas system relies on the class 2 type V endonuclease Cas12a (formerly known as Cpf1). Notably, the patent landscape for Cas12a appears clearer, given that its discovery was initially only attributed to the Zhang group at the Broad Institute (Zetsche et al. 2015, Zhang et al. 2019). Subsequently, Inscripta Inc. (https://www.inscripta.com/) identified a novel class of Cas12a enzyme distinct from the original Cpf1 (Gill et al. 2018). This class 2 type V endonuclease, derived from E. rectale (also known as MAD7), was made available for research, development, and academic purposes through a permissive licensing agreement. Although ErCas12a has been studied in various species (Liu et al. 2019, 2020, Wierson et al. 2019, Price et al. 2020, Jarczynska et al. 2021, Lin et al. 2021, Rojek et al. 2021, Zhang et al. 2021, Vanegas et al. 2022) (Table 1) including S. cerevisiae, the targeting efficiency per organism varied significantly and greatly depended on the parameters used. Especially, application of ErCas12a in the model microorganism S. cerevisiae only reached an editing efficiency of 66%, and the parameters were unfortunately not reported (Inscripta Inc.; Table 1). Here, we demonstrated that efficiency of genome editing using the ErCas12a system could be improved to reach nearly 100%, when the endonuclease is integrated in the genome. In addition, to go beyond the canonical CRISPR–Cas9, the ErCas12a further expanded genome accessibility by facilitating access of AT-rich regions.
Our work confirmed and expanded design guiding principles for the construction of gRNA expression cassette of Cas12a enzymes. Of all tested parameters, the length of the CRISPR DR sequence was the most critical. Out of the two configurations tested, the use of the shorter DR (19 nt) systematically resulted in a better targeting efficiency (Fig. 3). Similar results were reported for the application of the F. novicida Cas12a in S. cerevisiae (Swiat et al. 2017), demonstrating that this was not specific to a single Cas12a enzyme, but that this principle could be potentially extended to all class 2 type V endonucleases. One attractive feature of Cas12a endonucleases is their RNAse activity that allows the nuclease to self-process the crRNA. The result of this ribonuclease activity is a mature crRNA with a 19 nt DR (Zetsche et al. 2017). The size reduction of the DR from 35 to 19 nt (Table 1) might allow skipping of the pre-crRNA self-processing steps into the mature crRNA, which would enhance the synthesis of the mature crRNA, and therefore improve targeting efficiency. While the use of short DR for FnCas12a has already been performed successfully in other living systems (Zetsche et al. 2017), whether this holds true for ErCas12a editing in other yeast species beyond S. cerevisiae (Juergens et al. 2018) requires further investigation. Next to the design of the DR structural part of the crRNA, the design of a spacer specific for the editing event is crucial and, based on the presented results, guidelines for crRNA design for ErCas12a can be summarized as: (i) find a 5′-TTTV-3′ PAM sequence at the genomic targeted site of interest, (ii) the GC-content of the spacer sequence can be between 30% and 70%, (iii) add the 19-nt DR sequences flanking the spacer, (iv) the secondary structure (determined using RNAfold online tool; Lorenz et al. 2011) of the DR sequences should remain intact and not interfere with the spacer (Creutzburg et al. 2020), and (v) off-targets at other nonintended genomic locations should be avoided. The efficiency of the crRNA targeting can be affected by other parameters such as local chromatin structures, but the contribution of these factors is still poorly understood and were not considered in the design process.
The S. cerevisiae strain IMX2713, which expressed both SpyCas9 and ErCas12a endonucleases, revealed that the concurrent expression of these two enzymes was achievable without any adverse effects on the cell’s physiological functions. Consequently, this engineered strain could be considered a novel platform strain for genetic engineering. This innovation allows for the exploration of a broader genomic landscape without the necessity for multiple cycles of introducing and eliminating plasmids carrying the endonucleases. If needed, the removal of the sequences encoding the CRISPR enzymes could be performed after all desired modifications have been incorporated. Such a step is particularly essential for ErCas12a, as the provision of strains for royalty-free use hinges on the absence of ErCas12a in the final strain and product.
While plasmid borne expression of Ercas12a was sufficient to edit ADE2 with 31.5% efficiency, this constitutes a significant reduction from the 100% editing efficiency achieved with genome integrated Ercas12a. Aside from ErCas12a toxicity at high expression levels, another difference between plasmid borne and genomic expression is the timing of the concurrent expression of ErCas12a (transcription and translation), the editing event and the repair of the double strand break with a linear repair DNA. While constitutive genomic expression ensures active ErCas12a presence upon transformation of the crRNA expressing plasmid and the double stranded DNA repair fragment, the expression of Ercas12a and of the gRNA from the same plasmid delays the time at which all three essential elements to perform the editing event are concomitantly present. To be optimal, the rate of the enzyme synthesis and gRNA expression must be fast and synchronized, while in the meantime the degradation of the linear repair DNA has to remain low. A too fast degradation of the linear repair DNA might explain this reduced efficiency. It is worth mentioning that similar construction using SpyCas9 instead of ErCas12a resulted in 100% efficiency (Generoso et al. 2016), which might indicate a difference between Cas9 and Cas12a enzymes. Mitigation strategies to compensate this premature degradation could be beneficial such as the elongation of the repair (> 120 bp used as default in this study), protection of the linear DNA by specific short sequences (Biswas et al. 1995) or protein binding protection (Norouzi et al. 2021). Despite a reduced editing efficiency, the transportable plasmid pUDP293 could be valuable for accessing multiple strains issued from different genetic background.
In conclusion, ErCas12a was established as part of the S. cerevisiae genetic toolbox, able to edit several commonly used integration sites with (near) 100% efficiency in a single editing event as well as in multiplex editing. The single crRNAs or multispacer crRNA arrays could be ordered as two short annealing oligonucleotides and could easily and rapidly be cloned in purposed plasmids. The presented crRNA design criteria in combination with the developed genome editing strategy for ErCas12a was shown to expand the genetic toolbox for editing of the yeast genome.
Supplementary Material
Acknowledgements
We gratefully acknowledge Dr Anna Wronska and Dr Jasmijn Hassing for constructing strain IMX2600, Laura Sierra Herras for constructing the Xdcrt expressing plasmids pUDE1248, pUD1249, and pUD1250, and Marcel van den Broek for assisting the genome analysis of the strains sequenced in the study. We are thankful to Prof Dr J.T. Pronk (Delft University of Technology), Dr V.M. Boer, and Mr T. Elink Schuurman (HEINEKEN Supply Chain B.V.) for their support during this project.
Contributor Information
Nicole X Bennis, Department of Biotechnology, Delft University of Technology, van der Maasweg 9, 2627 HZ Delft, The Netherlands.
Jonah P Anderson, Department of Biotechnology, Delft University of Technology, van der Maasweg 9, 2627 HZ Delft, The Netherlands.
Siebe M C Kok, Department of Biotechnology, Delft University of Technology, van der Maasweg 9, 2627 HZ Delft, The Netherlands.
Jean-Marc G Daran, Department of Biotechnology, Delft University of Technology, van der Maasweg 9, 2627 HZ Delft, The Netherlands.
Authors’ contributions
J.M.G.D. and N.X.B. designed experiments. N.X.B., J.A., and S.K. executed the experimental work. N.X.B. and J.M.G.D. supervised the study. N.X.B., J.A., and J.M.G.D. wrote the manuscript. All authors read and approved the final manuscript.
Conflict of interest
None declared.
Funding
This work was performed within the Top consortia for Knowledge and Innovation (TKIs) AgriFood which was granted a PPP allowance from the Ministry of Economic Affairs and Climate Policy (Project Habitats #PPPS1701).
References
- Babaei M, Sartori L, Karpukhin Aet al. Expansion of EasyClone-MarkerFree toolkit for Saccharomyces cerevisiae genome with new integration sites. FEMS Yeast Res. 2021;21:foab027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau Het al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. [DOI] [PubMed] [Google Scholar]
- Biswas I, Maguin E, Ehrlich SDet al. A 7-base-pair sequence protects DNA from exonucleolytic degradation in Lactococcus lactis. Proc Natl Acad Sci USA. 1995;92:2244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botman D, de Groot DH, Schmidt Pet al. In vivo characterisation of fluorescent proteins in budding yeast. Sci Rep. 2019;9:2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren Met al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–92. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox Det al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutzburg SCA, Wu WY, Mohanraju Pet al. Good guide, bad guide: spacer sequence-dependent cleavage efficiency of Cas12a. Nucleic Acids Res. 2020;48:3228–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle Jet al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali Pet al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman BZ. The isolation of adenylosuccinate synthetase mutants in yeast by selection for constitutive behavior in pigmented strains. Genetics. 1969;61:377–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edraki A, Mir A, Ibraheim Ret al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol Cell. 2019;73:714–26.e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian KD, Kotter P. Yeast genetic strain and plasmid collections. Method Microbiol. 2007;36:629–66. [Google Scholar]
- Feng Z, Zhang B, Ding Wet al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23:1229–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagfeldt DB, Siewers V, Huang Let al. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast. 2009;26:545–51. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. JIPB. 2014;56:343–9. [DOI] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion Met al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. [DOI] [PubMed] [Google Scholar]
- Generoso WC, Gottardi M, Oreb Met al. Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J Microbiol Methods. 2016;127:203–5. [DOI] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RAet al. Improved method for high efficiency transformation of intact yeast cells. Nucl Acids Res. 1992;20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill RT, Garst A, Lipscomb TEW. Inscripta Inc. Nucleid acid-guided nucleases. US patent 9,982,279 B981, 2018. [Google Scholar]
- Gorter de Vries AR, de Groot PA, van den Broek Met al. CRISPR-Cas9 mediated gene deletions in lager yeast Saccharomyces pastorianus. Microb Cell Fact. 2017;16:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassing EJ, de Groot PA, Marquenie VRet al. Connecting central carbon and aromatic amino acid metabolisms to improve de novo 2-phenylethanol production in Saccharomyces cerevisiae. Metab Eng. 2019;56:165–80. [DOI] [PubMed] [Google Scholar]
- Hu JH, Miller SM, Geurts MHet al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino Ket al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarczynska ZD, Rendsvig JKH, Pagels Net al. DIVERSIFY: a fungal multispecies gene expression platform. ACS Synth Biol. 2021;10:579–88. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox Det al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara Iet al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergens H, Varela JA, Gorter de Vries ARet al. Genome editing in Kluyveromyces and Ogataea yeasts using a broad-host-range Cas9/gRNA co-expression plasmid. FEMS Yeast Res. 2018;18:foy012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WDet al. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. [DOI] [PubMed] [Google Scholar]
- Kuijpers NG, Solis-Escalante D, Bosman Let al. A versatile, efficient strategy for assembly of multi-fragment expression vectors in Saccharomyces cerevisiae using 60 bp synthetic recombination sequences. Microb Cell Fact. 2013;12:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Krause Met al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H. Major CRISPR patent decision won't end tangled dispute. Nature. 2022;603:373–4. [DOI] [PubMed] [Google Scholar]
- Lee ME, DeLoache WC, Cervantes Bet al. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth Biol. 2015;4:975–86. [DOI] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Genomics. 2013;1303. 10.48550/arXiv.41303.43997. [DOI] [Google Scholar]
- Li Y, Pan S, Zhang Yet al. Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016;44:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I, Dunham MJ. An autonomously replicating sequence for use in a wide range of budding yeasts. FEMS Yeast Res. 2014;14:364–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Zhu Z, Liu Get al. Genome editing in plants with MAD7 nuclease. J Genet Genomics. 2021;48:444–51. [DOI] [PubMed] [Google Scholar]
- Liu RM, Liang LL, Freed Eet al. Synthetic chimeric nucleases function for efficient genome editing. Nat Commun. 2019;10:5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Schiel JA, Maksimova Eet al. ErCas12a CRISPR-MAD7 for model generation in human cells, mice, and rats. CRISPR J. 2020;3:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looke M, Kristjuhan K, Kristjuhan A. Extraction of genomic DNA from yeasts for PCR-based applications. BioTechniques. 2011;50:325–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Honer Zu Siederdissen Cet al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Iranzo Jet al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Micro. 2020;18:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KMet al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R, van Rossum HM, Wijsman Met al. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015;15:fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science. 2008;322:1843–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Buron LD, Salomonsen Bet al. Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab Eng. 2012;14:104–11. [DOI] [PubMed] [Google Scholar]
- Mizuno S, Dinh TT, Kato Ket al. Simple generation of albino C57BL/6 J mice with G291T mutation in the tyrosinase gene by the CRISPR/Cas9 system. Mamm Genome. 2014;25:327–34. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez Jet al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–82. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Juez G, Rodriguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol Microbiol. 1993;9:613–21. [DOI] [PubMed] [Google Scholar]
- Norouzi M, Panfilov S, Pardee K. High-efficiency protection of linear DNA in cell-free extracts from Escherichia coli and Vibrio natriegens. ACS Synth Biol. 2021;10:1615–24. [DOI] [PubMed] [Google Scholar]
- Postma ED, Dashko S, van Breemen Let al. A supernumerary designer chromosome for modular in vivo pathway assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2021;49:1769–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MA, Cruz R, Bryson Jet al. Expanding and understanding the CRISPR toolbox for Bacillus subtilis with MAD7 and dMAD7. Biotech Bioeng. 2020;117:1805–16. [DOI] [PubMed] [Google Scholar]
- Randazzo P, Bennis NX, Daran JMet al. gEL DNA: a cloning- and polymerase chain reaction-free method for CRISPR-based multiplexed genome editing. CRISPR J. 2021;4:896–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler Wet al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek J, Basavaraju Y, Nallapareddy Set al. Mad7: an IP friendly CRISPR enzyme. Res Sq. 2021. (pre-print). 10.21203/rs.21203.rs-1004025/v1004021. [DOI] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux Cet al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer C. Broad defeats Berkeley CRISPR patent. Nat Biotechnol. 2022;40:445. [DOI] [PubMed] [Google Scholar]
- Swarts DC, Jinek M. Cas9 versus Cas12a/Cpf1: structure-function comparisons and implications for genome editing. WIREs RNA. 2018;9:e1481. [DOI] [PubMed] [Google Scholar]
- Swiat MA, Dashko S, den Ridder Met al. FnCpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae. Nucleic Acids Res. 2017;45:12585–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek M, Ortiz-Merino RA, Bennis NXet al. , Draft genome sequence of the Saccharomyces cerevisiae SpyCas9 expressing strain IMX2600, a laboratory and platform strain from the CEN.PK lineage for cell-factory research. Microbiol Res Announ. 2023. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanegas KG, Rendsvig JKH, Jarczynska ZDet al. A Mad7 system for genetic engineering of filamentous fungi. J Fungi. 2022;9:16. 10.3390/jof9010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WAet al. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–17. [DOI] [PubMed] [Google Scholar]
- Verwaal R, Buiting-Wiessenhaan N, Dalhuijsen Set al. CRISPR/Cpf1 enables fast and simple genome editing of Saccharomyces cerevisiae. Yeast. 2018;35:201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwaal R, Wang J, Meijnen JPet al. High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Appl Environ Microb. 2007;73:4342–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea Tet al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierson WA, Simone BW, WareJoncas Zet al. Expanding the CRISPR toolbox with ErCas12a in zebrafish and human cells. CRISPR J. 2019;2:417–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OOet al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju Pet al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35:31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Zetsche B, Ran FAet al. Novel CRISPR enzymes and systems. 2019;patent US 2019/0233814 A1. [Google Scholar]
- Zhang XH, Tee LY, Wang XGet al. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4:e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ren Q, Tang Xet al. Expanding the scope of plant genome engineering with Cas12a orthologs and highly multiplexable editing systems. Nat Commun. 2021;12:1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.