Abstract
The phylum Nucleocytoviricota includes the largest and most complex viruses known. These “giant viruses” have a long evolutionary history that dates back to the early diversification of eukaryotes, and over time they have evolved elaborate strategies for manipulating the physiology of their hosts during infection. One of the most captivating of these mechanisms involves the use of genes acquired from the host—referred to here as viral homologs or “virologs”—as a means of promoting viral propagation. The best-known examples of these are involved in mimicry, in which viral machinery “imitates” immunomodulatory elements in the vertebrate defense system. But recent findings have highlighted a vast and rapidly expanding array of other virologs that include many genes not typically found in viruses, such as those involved in translation, central carbon metabolism, cytoskeletal structure, nutrient transport, vesicular trafficking, and light harvesting. Unraveling the roles of virologs during infection as well as the evolutionary pathways through which complex functional repertoires are acquired by viruses are important frontiers at the forefront of giant virus research.
Keywords: auxiliary metabolic genes, giant viruses, mimicry, nucleocytoviricota, viral diversity, viral evolution
The authors discuss recent research into the complex functions encoded in the genomes of giant viruses.
Main text
Large DNA viruses in the phylum Nucleocytoviricota comprise a diverse group of pathogens that infect eukaryotic hosts that range from unicellular protists to vertebrates (Aylward et al. 2021). This phylum encompasses several families that include metazoan viruses that are well-studied due to their role in deadly outbreaks in humans, livestock, and other animals, such as the Poxviridae, Asfarviridae, and Ascoviridae/Iridoviridae. In addition, this phylum includes the families Mimiviridae, Phycodnaviridae, and Marseilleviridae that comprise mostly viruses of algae, amoebae, and other protists. Although the term “giant virus” has historically been used inconsistently and usually only with reference to those viruses that possess particularly large genome lengths or capsid sizes, we will use the term here to broadly refer to all members of the Nucleocytoviricota owing to their shared evolutionary history.
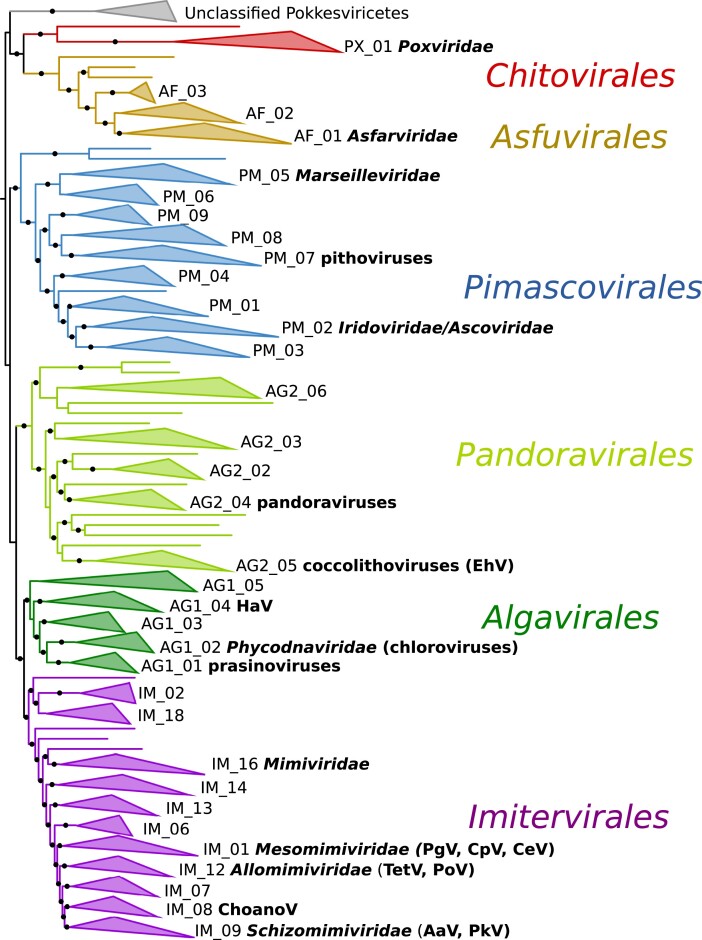
Although only 11 families are officially classified within the Nucleocytoviricota according to the International Committee for the Taxonomy of Viruses (ICTV), recent cultivation-independent viral diversity surveys have discovered a vast diversity of giant viruses in the biosphere that far exceeds those in these established groups (Fig. 1). Early approaches using viral marker genes were the first to show that diverse giant viruses were present in environmental samples (Chen et al. 1996, Short and Suttle 2002), and subsequent metagenomic surveys have shown that they are abundant in habitats ranging from ocean waters, permafrost and other soil systems, wastewater, and hot springs (Yau et al. 2011, Schulz et al. 2018, 2020, Bäckström et al. 2019, Moniruzzaman et al. 2020a, Rigou et al. 2022, Kavagutti et al. 2023, reviewed in Schulz et al. 2022). The hosts of these uncultivated viruses remain largely unclear, but co-occurrence analysis and examination of recent host–virus gene transfer has been used to link several uncultivated viruses to potential eukaryotic hosts (Schulz et al. 2020, Meng et al. 2021, Bhattacharjee et al. 2023). Indeed, the known hosts of isolated giant viruses have grown markedly in the last decade, and it now seems likely that most eukaryotic lineages are infected by these viruses in nature. Recent phylogenomic analysis of both cultivated and uncultivated viral genomes has estimated that at least 32 families exist, and this still likely represents a lower bound that will increase as more lineages continue to be discovered through a combination of cultivation-based efforts and further metagenomic surveys (Aylward et al. 2021).
Figure 1.
Phylogeny of the Nucleocytovoricota, adapted from Aylward et al. (2021). Collapsed branches depict families or family-level lineages. Families accepted by the ICTV are in bold italic, and commonly used family names that have not yet been officially accepted are in bold. Notable viruses mentioned in the text are listed in bold. Aav, Aureococcus anophagefferens virus; ChoanoV1, Choanoflagellate virus; CpV, Chrysochromulina parva virus; HaV, Heterosigma akashiwo virus; IC, Internode Certainty; PgV, Phaeocystis globosa virus; TetV, Tetraselmis virus.
Members of the Nucleocytoviricota are known for their large genome sizes, reaching up to 2.7 million base-pairs in the case of some pandoraviruses (Philippe et al. 2013, Legendre et al. 2019), and routinely achieving lengths >500 kb in many other lineages (Aylward et al. 2021). As an increasing number of giant virus genomes have been sequenced over the last decade, a resonant theme has been the presence of many “cellular like” genes that are otherwise rare or absent in other viral lineages. Early examples that were identified in some of the first poxvirus genomes to be sequenced include immunomodulatory genes involved in manipulating host defenses, such as interleukin homologs and cytokines (Hughes 2002, Hughes and Friedman 2005). More recently, many metagenome-derived giant virus genomes have been shown to encode homologs of cytoskeletal components such as actin and myosin, and components of central carbon metabolic pathways, such as glycolysis and the TCA cycle (Moniruzzaman et al. 2020a, Ha et al. 2021, Kijima et al. 2021, Da Cunha et al. 2022). Historically, the term “auxiliary metabolic gene” has been used to refer to bacteriophage-encoded metabolic genes, whereas immunomodulatory genes in poxviruses have often been referred to as “mimics” due to their similarity to host counterparts. In light of the growing breadth of cellular-derived genes encoded by viruses, which encompass a wide range of pathways and often lack known functions during infection, we will hereafter refer to these genes as simply virologs (viral homologs). This term, used previously in this context (Sorouri et al. 2022), simply refers to viral genes with clear sequence homology to cellular counterparts, and does not imply any specific function or evolutionary history.
Virologs are hardly unique to members of the Nucleocytoviricota. Indeed, a wide range of DNA and RNA viruses encode host-derived virologs that play key roles in infection. Early studies of marine bacteriophages that infect cyanobacteria in the ocean revealed that many encode photosynthesis genes in their genomes (Mann et al. 2003), and the list of virologs collectively encoded by members of the Caudoviricetes has grown to encompass processes involved in nitrogen, phosphorus, sulfur, and central carbon metabolism (reviewed in Hurwitz and U’Ren 2016, Warwick-Dugdale et al. 2019). Some recently described “jumbo bacteriophages” have genomes >200 kb in length and encode a particularly impressive complement of predicted metabolic genes (Fridman et al. 2017, Michniewski et al. 2021, Weinheimer and Aylward 2022). In addition, herpesviruses have been known to encode host-derived immunomodulatory genes for decades (Schönrich et al. 2017), and members of a recently discovered phylum of herpesvirus relatives, the Mirusviricota, also possess large genomes that encode numerous virologs (Gaïa et al. 2023). Even the small genomes of RNA viruses can encode several virologs that play roles in infection (Lasso et al. 2021). Despite the propensity of viruses from all realms to acquire and co-opt host genes, we will focus this review on members of the Nucleocytoviricota owing to their particularly large genome sizes and rich complement of virologs, as well as the large number of studies that have recently been published on this group. Moreover, it is likely that different viral groups employ virologs in a similar manner, and an in-depth examination of the Nucleocytoviricota will, therefore, provide useful insights into viral infection strategies more broadly.
Recent work has underscored the richness of virologs found in giant virus genomes, raising questions regarding their role during infection and the extent to which they may be involved in manipulating host physiology. Indeed, the impressive functional repertoires encoded in the genomes of giant viruses have led some to describe these viruses as “quasi-autonomous” from their hosts (Claverie and Abergel 2010, 2016). A key organizing principle for elucidating the physiological shifts that take place during infection is the virocell concept (Box 1). The virocell concept emphasizes the intracellular activities of viruses during infection over their extracellular phase, and thereby promotes the view of viruses as dynamic biological agents with their own form of metabolism (i.e. cellular metabolism during infection). Viral manipulation of host physiology is hardly limited to giant viruses; in fact numerous studies have elucidated how viruses of plants and animals hijack critical host processes to elevate virus production (Thaker et al. 2019, Walsh and Naghavi 2019). The virocell concept takes on a new dimension in the context of giant viruses, however, due to the particularly complex strategies for cellular manipulation that are encoded in the genomes of their genomes. For example, the manipulation of central carbon metabolism and cytoskeletal networks during infection have been widely reported in a diverse array of both RNA and DNA viruses, but giant viruses stand out in that they encode glycolysis, TCA cycle, actin, and myosin virologs in their genomes.
Box 1. Virocells and the concept of a virus.
Several authors have reformulated the concept for a virus by emphasizing the intracellular phase of the viral life cycle (i.e. the virus factory, or, more generally, the virocell) over that of extracellular virions (Claverie 2006, Forterre 2010). Traditionally, viruses have largely been defined in terms of the biochemical properties of their virions—inert particles packages with nucleic acid—but viewing viruses as virocells promotes the view that viruses should instead be defined as infected cells. In this way, viruses can be viewed as “cellular” organisms with their own distinct metabolism, in the sense that viral infection drastically alters the cellular metabolic state. The virocell concept, therefore, takes center stage in our understanding of virologs and mimicry in viruses, especially in large DNA viruses that deploy broad functional repertoires to transform cellular physiology during infection. Although the perspective that viruses can be viewed as a kind of infected cellular life form is controversial, the virocell concept has nonetheless proven itself to be a useful reframing of traditional views, i.e. important for understanding host–virus dynamics during infection. Recent work has also extended these concepts to explicitly examine the impact of virologs on host metabolism during infection (Rosenwasser et al. 2016). Moreover, some authors have gone farther and suggested that viruses should instead be defined as processes rather than entities (Dupré and Guttinger 2016), a view which puts even more emphasis on cellular dynamics that take place during infection.
One further consideration of virocell metabolism is that hosts will not necessarily encode homologs of the viral genes deployed by viruses during infection. Indeed, many virologs are the product of ancient acquisitions from cellular lineages, and as viral host range changes over time, viruses may infect hosts that no longer encode homologs of these genes. Moreover, even if cellular homologs are present, it is possible that virologs have evolved novel functions that are distinct from their cellular counterparts. These are critical considerations because they raise the possibility that virocell metabolism can reach beyond the manipulation of existing cellular machinery and confer new physiological capabilities onto their infected hosts. One excellent recent example is the wide range of giant viruses that have been found to encode rhodopsins (Needham et al. 2019, Rozenberg et al. 2020, Hososhima et al. 2022); this virolog may manipulate phototaxis during infection in a way that facilitates virus propagation.
Box 2. Diverse mechanisms of mimicry.
Although the most obvious cases of viral mimicry are those in which genes in viral and cellular lineages have clear sequence homology, there are many cases of mimicry that involve structural similarities where the shared evolutionary history between viral and host counterparts cannot be demonstrated. This may be because sequences have diverged too far for sequence homology to be detected, or because viruses convergently evolved motifs that resemble cellular structures from unrelated genetic material. One recent study performed a large-scale structural comparison between viral and host proteins and concluded that only 30% of putative viral mimics could be identified through examination of sequence homology (Lasso et al. 2021), indicating that the scale of viral mimicry is far larger than commonly thought. In addition to the direct mimicry of cellular structures, many viruses also employ alternative strategies to hijack host cellular processes. One example is phosphomimicry, in which viruses can manipulate the phosphorylation of host proteins and thereby disrupt normal cellular processes. Viral phosphomimetics have been shown to manipulate a variety of processes, including spliceosomal activity and translation (Huang et al. 2002, Jha et al. 2017). Given the diverse and unexpected mechanisms of viral mimicry it is tantalizing to consider the large number of uncharacterized proteins encoded in giant virus genomes that may play an as-yet unknown role in host manipulation. This is especially true in lineages such as the pandoraviruses, which contain genomes > 2.5 Mbp in size in which genes potentially evolve de novo from intergenic regions (Legendre et al. 2018).
Many virologs have been identified in viruses recently, and in many cases it is still unclear what role they play during infection. One clear possibility is that viral genes have the same function as their eukaryotic counterparts, and are needed by viruses to maintain or extend host cellular processes during infection (i.e. exaptation while retaining biochemical activity, as reviewed in Koonin et al. 2022). Shutoff of host metabolism is widely observed during infection and may be triggered either as a byproduct of viral disruption of host physiology or as a host defense mechanism that reduces available energy for viral production. Regardless of the cause, in this scenario virologs can be viewed as a means of maintaining host physiology sufficient to support virion production. Alternatively, virologs may have evolved distinct functions compared to their cellular counterparts, either due to distinct regulation or alternative enzymatic or structural activities. For some virologs both of these scenarios may be correct depending on the virus in question; e.g. members of both the Mimiviridae and Phycodnaviridae encode active superoxide dismutase (SOD) enzymes that likely play a role in the detoxification of reactive oxygen species (ROS) during the early stages of infection, whereas a SOD homolog in some poxviruses has been shown to be an inactive decoy enzyme that inhibits the activity of cellular homologs (Teoh et al. 2003, Kang et al. 2014, Lartigue et al. 2015). On a similar theme, various metabolic enzymes such as glycoside hydrolases and oxidoreductases appear to have evolved novel structural roles in various giant virus lineages (Klose et al. 2015, Krupovic et al. 2020, Villalta et al. 2022, Alempic et al. 2023). Even if the enzymatic activity of virologs is the same as their cellular homologs, their kinetics or context of expression may have vastly different effects on cellular physiology during infection. For example, viral genes involved in central carbon metabolism may play roles in mitigating the buildup of ROS, which may occur if host enzymes begin to degrade at different rates. Given these complications, functional predictions based on genomic data alone must be treated with a degree of caution when inferring the role of various virologs during infection.
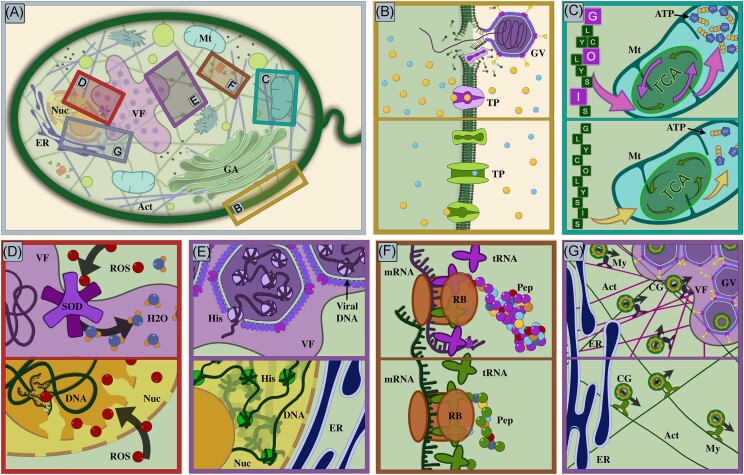
Here, we outline current knowledge of the diversity of virologs known to be encoded in members of the Nucleocytoviricota together with recent advances in our knowledge of how these viral genes are used during infection. A summary of notable virologs is provided in Table 1, and select processes are illustrated in Fig. 2. Many of these processes are enormously complex and we cannot hope to provide a complete overview of their functionality within cells. We, therefore, focus these sections on providing brief overviews of the processes themselves, a summary of notable viruses known to encode these virologs, and any experimental evidence that can link virologs to specific roles during infection.
Table 1.
Overview of functions encoded in giant virus genomes.
| Process | Notable enzymes | Prevalence | Relevant citations |
|---|---|---|---|
| Central carbon metabolism | Glycolysis, TCA cycle, and fermentation enzymes | Broadly distributed, but most prevalent in Imitervirales | Schvarcz and Steward (2018), Rodrigues et al. (2019), Moniruzzaman et al. (2020a), Blanc-Mathieu et al. (2021) |
| Histones | All four primary histones | Broadly distributed, but most prevalent in Pimascovirales and medusavirus | Boyer et al. (2009), Yoshikawa et al. (2019), Liu et al. (2021) |
| Translation | tRNA synthetases, translation termination factors | Widespread but particularly prevalent in members of the Imitervirales | Raoult et al. (2004), Schulz et al. (2017), Abrahão et al. (2018) |
| Sphingolipid synthesis | Entire pathway | Coccolithoviruses | Monier et al. (2009), Vardi et al. (2009) |
| Cytoskeleton | Actin, myosin, dynein, and kinesin | Widespread but particularly prevalent in members of the Imitervirales | Ha et al. (2021), Kijima et al. (2021), Da Cunha et al. (2022) |
| Ubiquitin signaling | E1, E2, and E3 Ub-associated enzymes, ubiquitin homologs | Widespread | Iyer et al. (2006), poxvirus virologs reviewed in Lant and de Motes (2021) |
| Nutrient transporters and ion channels | Ammonium, phosphate transporters, and K + transporters | Prevalent in Algavirales and many members of the Imitervirales | Plugge et al. (2000), Frohns et al. (2006), Monier et al. (2012, 2017) |
| Vesicular trafficking | SNAREs, Rab- and Ran-like GTPases, and Sec1/Munc18-like proteins | Imitervirales, Algavirales, and Pimascovirales | Neveu et al. (2022) |
| Immunoregulatory genes | Virokines and virocepters | Poxviruses, irodoviruses, and asfarviruses | Reviewed in McFadden and Murphy (2000), Elde and Malik (2009) |
| Caspases | Caspase and Metacaspase-like homologs | Poxviruses, irodoviruses, and asfarviruses | Bideshi et al. (2005), Wilson et al. (2017) |
| Polysaccharide metabolism | Glycoside hydrolases | Widespread | Reviewed in Speciale et al. (2022) |
| Rhodopsins | Channelrhodopsins and heliorhodopsins | Imitervirales and coccolithoviruses | Rozenberg et al. (2020), Zabelskii et al. (2020) |
| Cell cycle | Cyclins and components of the anaphase promoting complex | Poxviruses, Medusavirus, and Clandestinovirus | Mo et al. (2009), Yoshikawa et al. (2019), Rolland et al. (2021) |
Figure 2.
Illustration of possible roles of virologs in diverse processes within virocells. Panel (A): overview of a eukaryotic cell featuring key organelles and a virus factory. Each area highlighted using rectangles of different colors is shown in detail in panels (B)–(G). The top halves of panels (B)–(G) represent processes in infected cells carried out in part by virus-encoded virologs, while the bottom halves represent processes as they occur in healthy cells. Virus-encoded proteins and related processes or organelles are shown in shades of purple. Panel (B): virus infection of a cell and subsequent injection of viral DNA initiate reprogramming of the healthy cell metabolism into a virocell. Viral-encoded potassium channels cause depolarization of the host membrane and thereby facilitate viral entry. In the virocell, viral-encoded transporters help import nutrients, facilitating the infection process. Panel (C): virologs involved in central carbon metabolism (glycolysis and TCA cycle—purple letters and arrows) potentially augment cellular energy metabolism, leading to increased production of ATP. Panel (D): ROS cause damage to cellular components. During viral replication, viral-encoded SODs potentially prevent damage to viral proteins and nucleic acids within the virocell. Panel (E): While histones in a regular cell contribute to the structural organization of cellular DNA, within a virocell, viral-encoded histones can help package and organize viral DNA within the capsid. Panel (F): many GVs encode tRNAs in their genomes, which can potentially augment the cellular tRNA pool to sustain viral protein production. Panel (G): although the functions of viral-encoded actin and myosin genes have yet to be characterized, they could potentially assist in the localization and structure of the virus factory. Viral myosin might help transport molecular cargoes related to virus replication and assembly processes along the actin filaments. Abbreviations: Nuc—nucleus, ER—endoplasmic reticulum, VF—virus factory, Mt—mitochondria, GA—Golgi apparatus, Act—actin filaments, GV—giant virus, TP—transporters, ROS—reactive oxygen species, SOD—superoxide dismutase, H2O—water, His—histone, RB—ribosome, Pep—peptide chain, tRNA—transfer RNA, My—myosin, and CG—molecular cargo.
Translation
Viruses are known to depend primarily on the host translation machinery for carrying out synthesis of virus-specific structural and nonstructural proteins. A key feature of many members of the Nucleocytoviricota is the presence of diverse genes involved in translation, thereby potentially making them partially independent of the host machinery for successful propagation. Components of translation are particularly common across many families in the Imitervirales and Pimascovirales, but they can also be found in a wide range of genomes in other orders. Early examination of chloroviruses found that tRNAs and translation elongation factors are found in this group (Li et al. 1997, Duncan et al. 2020), and subsequently tRNAs have been found in a wide range of giant viruses (Wilson et al. 2005, Fischer et al. 2010, Philippe et al. 2013). The description of Acanthamoeba polyphaga mimivirus, now classified in the species Mimivirus bradfordmassiliense, dramatically expanded this set to include additional tRNAs, translation factors (initiation, elongation, and termination) and, most strikingly, four aminoacyl-tRNA synthetases genes (aaRS: Arginyl-RS, Cysteinyl-RS, Methionyl-RS, and Tyrosyl-RS) (Raoult et al. 2004). These genes encode for enzymes that attach cognate amino acids to specific tRNAs, and are essential for the translation process that up until that time had only been found in the genomes of cellular organisms.
Subsequent discoveries have continued to expand the repertoire of translation-associated genes in giant viruses. Notably, a metagenomic study recovered the genome of klosneuvirus (Schulz et al. 2017), which encodes 19 aaRS genes, and culture-based studies identified these genes in a wide range of other isolates, including pandoraviruses and members of the Pimascovirales and Asfuvirales orders (Philippe et al. 2013, Andreani et al. 2017). Tupanviruses, members of the Mimiviridae family that have been isolated from soda lakes in Brazil and the deep Atlantic Ocean, represent the most remarkable example so far, with 20 aaRSs, 70 tRNAs, and 11 other translation factors encoded in their genomes (Abrahão et al. 2018). Excluding ribosomal proteins, tupanviruses have even more translation-related elements than some cellular organisms, such as Encephalitozoon cuniculi (eukaryota), Nanoarchaeum equitans (archaea), and Candidatus Carsonella ruddii (bacteria). The broad host-range described for tupanviruses in comparison to other amoeba-infecting viruses might be related to its expanded set of translation-related genes, which may allow tupanviruses to infect hosts with a broader range of codon usage profiles.
Like many other virologs in giant viruses, the functional role of translation-related genes remains to be largely determined, although important insights have been gained through characterization of some translation genes in pandoraviruses and members of the Imitervirales and Algavirales. One bioinformatic study examining tRNAs encoded in chloroviruses noted that 17 different tRNAs for 14 different amino acids clustered in the middle of the viral genomes (Duncan et al. 2020). The authors suggested that these tRNAs are primarily involved in overcoming the codon usage differences between chloroviruses and their host. tRNA genes are also found in coccolithoviruses, prasinoviruses, and rhaphidoviruses, but to a lesser extent compared to chloroviruses (Wilson et al. 2009, Ogura et al. 2016, Fromm et al. 2022). In addition, an elongation factor (EF3) encoded by chloroviruses is expressed within the first hour of infection and appears to be important for the synthesis of viral proteins throughout the replication cycle (Yamada et al. 1993). In mimivirus, the role of methyonyl- and tyrosyl-aaRSs were experimentally validated in activating tyrosine and methionine, respectively (Abergel et al. 2007). The silencing of the mimivirus R458 gene, a homolog of the initiation factor 4A, causes deregulation in translation of proteins associated with transcription, particle structure, DNA repair/topology, and others (Bekliz et al. 2018). Although these results collectively suggest that translation-associated genes can play key roles in the translation of viral proteins, there is reason to doubt that all of these genes are critical for viral propagation. For example, the genome of Bodo saltans virus encodes five total aaRS genes, but three appear to be nonfunctional pseudogenes that show signs of recent nonsense mutations or ORF disruptions (Deeg et al. 2018). Moreover, a recent study developed extensive new tools for the genetic manipulation of pandoraviruses and revealed that most proteins involved in translation were not essential (Bisio et al. 2023).
Replication, transcription, and mRNA maturation
Several genes involved in DNA replication, transcription, and mRNA maturation are highly conserved in the Nucleocytoviricota and are encoded in almost all members of this phylum. DNA replication is achieved using a family B DNA polymerase (PolB), which is one of the best phylogenetic markers for giant viruses, and it has been used extensively in PCR-based or marker gene surveys of giant virus diversity (Short and Suttle 2002, Li et al. 2018, Endo et al. 2020). Several helicases and topoisomerases are also highly conserved and broadly represented in these viruses. A superfamily II helicase (SFII) is often used as a marker gene in phylogenetic reconstruction, whereas both family II and IB topoisomerases are also found in a range of giant viruses but have somewhat complementary distributions and may play similar functional roles (Iyer et al. 2006, Aylward et al. 2021). Some giant viruses, in particular members of the Imitervirales, also encode a MutS protein likely involved in mismatch repair (Ogata et al. 2011, Priet et al. 2015, Gallot-Lavallée et al. 2017, Claverie and Abergel 2018). Lastly, most giant viruses appear to encode a multisubunit ribonucleotide reductase (RNR), which facilitates dNTP production during replication (Gammon et al. 2010). The phylogeny of RNR subunits is often inconsistent with other viral genes involved in DNA replication, however, suggesting that they have been acquired multiple times independently from different eukaryotic lineages (Filée et al. 2008, Yutin and Koonin 2012a).
Almost all giant viruses also encode a multi-subunit RNA polymerase (RNAP) that they use for gene expression. Prasinoviruses and chloroviruses are the primary exceptions to this, and they are therefore dependent on host enzymes for transcription during the nuclear stages of their infection (Moreau et al. 2010, Van Etten et al. 2019). The RNAP encoded by most giant viruses is homologous to the same enzyme used by bacteria, archaea, and eukaryotes for gene expression, and it therefore represents a sophisticated transcriptional apparatus that is essentially unknown elsewhere in the virosphere except in some large bacteriophages (Ceyssens et al. 2014, Sokolova et al. 2020, Weinheimer and Aylward 2020). Viral RNAP allows many giant viruses to complete their infection cycle in the cytoplasm, but some of these viruses can still use host enzymes for gene expression by transiently recruiting nuclear transcriptional machinery to virus factories (Fabre et al. 2017). In addition to RNAP, several giant viruses also typically encode a wide range of transcription factors, including several that are highly conserved and are often used in phylogenetic reconstruction (Iyer et al. 2006, Yutin et al. 2013, Guglielmini et al. 2019). Giant viruses also often encode several genes involved in mRNA maturation, including an mRNA capping enzyme and polyA polymerase (Priet et al. 2015), providing some striking similarities to eukaryotic machinery. Phylogenetic analysis of viral genes involved in DNA replication and transcription has revealed signatures of ancient coevolution and gene exchange with early eukaryotes (Box 3).
Box 3. Deeply intertwined coevolution of eukaryotes and giant viruses.
The highly conserved nature of virologs involved in DNA replication, transcription, and mRNA maturation suggests that these are the most ancient enzymes in the Nucleocytoviricota and that they were present in the last common ancestor of this phylum. Hence, several phylogenetic studies have examined the evolution of these enzymes in depth to shed light on their origins. These viral proteins were most likely acquired from cellular groups at some stage, but examining the evolutionary relationships between these proteins in giant viruses and eukaryotes is complicated by the long stem branch leading to modern eukaryotes (Betts et al. 2018). This long stem branch, together with recent geochemical findings (Brocks et al. 2023), implies that proto-eukaryotic lineages thrived on earth for hundreds of millions of years prior to the emergence of the last eukaryotic common ancestor (LECA). During this period, proto-eukaryotes presumably coevolved with their viruses, and the ancestor of the Nucleocytoviricota likely emerged at this time (Forterre and Gaïa 2016). Phylogenetic analysis of RNA polymerase has suggested that viruses acquired this gene from proto-eukaryotes prior to the emergence of LECA, and then subsequently transferred a copy back (Guglielmini et al. 2019). Modern eukaryotes have three copies of RNAP, and this reciprocal transfer may have given rise to the RNAP II we see today. Likewise, modern eukaryotes have four copies of family B DNA polymerases, and the delta polymerase (PolDelta) likely arose from a transfer from giant viruses that took place prior to the emergence of LECA (Takemura et al. 2015, Karki and Aylward 2023). Evidence for eukaryotic acquisition of viral genes has also been reported for topoisomerase II and actin (Da Cunha et al. 2022, Guglielmini et al. 2022). These studies have demonstrated that modern eukaryotic and viral genomes are a product of deeply intertwined coevolution. Although the details of eukaryogenesis remain enigmatic, several scenarios in which giant viruses play central roles have been postulated (Takemura 2020, Bell 2022).
Central carbon metabolism
Genes involved in sugar and amino acid metabolism were initially observed in prasinoviruses that infect members of the the prasinophyte genera Ostreococcus, Bathycoccus, and Micromonas (Weynberg et al. 2009, 2011, Moreau et al. 2010), and subsequent work identified the TCA cycle enzyme citrate synthase in tupanviruses (Rodrigues et al. 2019). A broad metagenomic survey of giant virus diversity identified the widespread presence of glycolysis and TCA cycle enzymes in a wide range of giant viruses, especially members of the Imitervirales and Algavirales (Moniruzzaman et al. 2020a). One recently isolated member of the Imitervirales that infects Prymnesium kappa encodes a particularly large complement of energy metabolism genes, including all four succinate dehydrogenase subunits (Blanc-Mathieu et al. 2021). Interestingly, a virus that encodes a complete glycolysis or TCA cycle pathway has not been identified, indicating that viruses do not reconstitute complete metabolic pathways using their own enzymes. It is possible that these enzymes are involved in boosting flux in existing metabolic pathways and that the viruses are still reliant on some host enzymes during infection. Although the activity of most of these enzymes has not been experimentally verified, one study examined a putative isocitrate dehydrogenase encoded in a pandoravirus and reported enzymatic activity (Aherfi et al. 2022). The activity of other TCA cycle genes in pandoraviruses is unclear and many appear to be highly divergent from any cellular homologs, however.
Increased glucose metabolism is a common aspect of virocell metabolism in a wide variety of viruses (Sánchez-García et al. 2021, Sumbria et al. 2021), including norovirus, herpesviruses, and some relatively small RNA viruses such as Dengue and Zika virus (Vastag et al. 2011, Allonso et al. 2015, Fontaine et al. 2015, Passalacqua et al. 2019, Pang et al. 2021). It is, therefore, plausible that giant virus-encoded enzymes play a role in hijacking host metabolism that is similar to that observed in other viruses. For example, if a cell is in a quiescent state at the time of infection, increasing flux in central carbon metabolism may lead to increased energy availability for viral replication. Although the reason giant viruses encode their own central metabolic enzymes rather than simply manipulating cellular homologs remains unknown, it is possible that this allows the viruses to more precisely control the process of metabolic hijacking, potentially instigate this process earlier in the infection cycle, or increase the amount of substrate used for processes such as post-translational modifications that are necessary during infection. Moreover, by encoding these enzymes, viruses may obviate possible red queen dynamics that could occur if viruses were solely reliant on cellular genes, which could mutate and thereby evade viral control. Some of these possibilities have been discussed in a recent review (Brahim Belhaouari et al. 2022).
Aside from genes involved in glycolysis and the TCA cycle, other central metabolic enzymes have also been found in giant viruses. One study examined the genome of Oceanusvirus kaneohense (Tetraselmis virus 1) and identified several genes involved in fermentation, including pyruvate–formate lyase and mannitol-1-phosphate dehydrogenase (Schvarcz and Steward 2018). It was hypothesized that the host may use fermentation for energy generation during low oxygen conditions, which can occur during periods of high respiration in eutrophic ocean waters. Importantly, many algal blooms lead to anoxia due to the high respiration of both algae and heterotrophic bacteria, suggesting that viral-mediated fermentation could play an important role in viral infection during these periods of high host cell density.
Sphingolipid metabolism
Sphingolipids are essential structural components of all eukaryotic membranes and are important signaling lipids in diverse cellular pathways. Viruses exploit membrane lipids such as sphingolipids in all steps of their replication cycle, including during cell entry and egress. Diverse viruses remodel their host’s sphingolipid metabolism during infection, including human immunodeficiency virus (HIV), hepatitis C virus, and rhinoviruses (Schneider-Schaulies and Schneider-Schaulies 2015). Moreover, sphingolipids are known to be enriched in lipid rafts that are involved in viral entry in a receptor-mediated manner. One of the most extensively researched phenomena of mimicry in the Nucleocytoviricota is the coordination of sphingolipid biosynthesis in the context of viral infection of the coccolithophore Emiliania huxleyi by its eponymous virus EhV. Emiliania huxleyi often forms large scale algal blooms that are routinely infected by EhV, which is thought to lead to demise of the bloom. A near-complete pathway for sphingolipid biosynthesis is encoded by the EhV genome (Wilson et al. 2005), and to-date this is the only virus that encodes for such enzymes, although diverse viruses rewire their host sphingolipids as part of their infection cycle (Schneider-Schaulies and Schneider-Schaulies 2015). Phylogenomic investigation has predicted that several genes in this pathway have been horizontally transferred between E. huxleyi and its virus, or vice versa (Monier et al. 2009). Experimental work on the viral-encoded rate limiting enzyme serine palmitoyltransferase of this pathway (Ziv et al. 2016), has confirmed its functional activity and demonstrated it has different substrate specificity than its algal homolog. This metabolic shift is part of the major lipid rewiring induced by viral infection that leads to production of odd-based fatty acids (Schleyer et al. 2019).
Interestingly, the EhV-encoded sphingolipid biosynthesis pathway is upregulated during infection, concomitant with the downregulation of the host counterpart (Rosenwasser et al. 2014), resulting in both the production of virus-specific glycosphingolipids (vGSLs) that are highly enriched in the virus membranes as well as the induction of programmed cell death (PCD) during lytic phase of infection (Vardi et al. 2009, Schleyer et al. 2019). Since these lipids are integrated into the virion, they can affect virion structural integrity and virion decay rate under UV stress in the upper ocean. Furthermore, since vGSLs are produced exclusively during viral infection of EhV, it has been used successfully as an effective metabolic biomarker to detect active viral infection during E. huxleyi blooms (Vardi et al. 2009, Fulton et al. 2014, Laber et al. 2018). High vGSL-producing strains of EhV, which are extremely virulent and harbor a greater infectivity at high host densities, provide a selective advantage under laboratory conditions. However, field data obtained from natural environments suggest a better survival rate of slow glycosphingolipid producing EhVs, where lower host densities are encountered (Nissimov et al. 2019). The metabolic arms race between the virus and its algal host around the sphingolipid biosynthetic pathways is reflected by the production of variety of GSL molecules with diverse chemical composition between susceptible, resistant (Hunter et al. 2015, Schleyer et al. 2023) and virocells of E. huxleyi cells (Vardi et al. 2009, Schleyer et al. 2019). Lastly, virus-derived sphingolipids can be detected in extracellular vesicles released by infected cells, which suggests that they may act as intercellular communicating signals during viral infection in algal blooms (Schatz et al. 2017, 2021). These vesicles can lead to faster viral infection dynamics and prolong half-life of viral infectivity.
Cytoskeletal dynamics
Some of the most recently discovered virologs include the cytoskeletal components actin, kinesin, and myosin. Kinesin homologs were the first to be identified in the small genomes of viruses that infect crustacea (the “mininucleoviridae” within the Pimascovirales) (Subramaniam et al. 2020), and subsequent studies of metagenome-derived viruses identified multiple actin and myosin homologs (so-called viractins and virmyosins) in other viruses (Ha et al. 2021, Kijima et al. 2021, Da Cunha et al. 2022). Most of the actin and myosin homologs appear to be encoded by members of the Imitervirales. A metatranscriptomic analysis of giant viruses in surface waters near the coast of California identified expression of several of these cytoskeletal components, confirming that they are expressed in natural conditions (Ha et al. 2021), but the precise role they play during infection remains unknown.
Similar to the hijacking of host central carbon metabolism, the subversion of the host cytoskeleton during infection is also widespread in different lineages of eukaryotic viruses (Taylor et al. 2011). Actin plays an important role in poxvirus egress and intercellular transmission (Rottner and Stradal 2009, Taylor et al. 2011), and similar dynamics have been observed for other giant viruses (Murti et al. 1985, Jouvenet et al. 2006). Viral interactions with the cytoskeleton can occur at several stages of infection, such as initial contact with cells, cell entry, and egress (Taylor et al. 2011). The precise role of viral-encoded actin, myosin, and kinesin homologs, therefore, remains unclear due to the complexity of cellular reorganization that can occur during viral infections as well as the numerous processes that involve cytoskeletal dynamics.
DNA packaging
Histones represent some of the most intriguing examples of virologs identified in giant virus genomes. These genes were initially identified in viruses of the Marseilleviridae (Boyer et al. 2009, Thomas et al. 2011), which encode fused genes with homology to H3–H4 and H2B–H2A core histones. Subsequent analysis of Acanthamoeba castellanii medusavirus revealed individual genes with homology to all four core histones (H2A, H2B, H3, and H4) as well as the linker histone (H1) (Yoshikawa et al. 2019). Another study cultivated a medusavirus relative, clandestinovirus, that also encodes four core histones (Rolland et al. 2021). Viral-encoded histones have low amino acid identity to eukaryotic homologs (< 30%) and phylogenetic analysis of the different core histones have placed them at the root of their respective clades, indicating they have an ancient evolutionary origin. In addition to those found in marseilleviruses and their relatives, a recent examination of a broad diversity of metagenome-derived giant viruses genomes revealed that histone subunits are also encoded by a wide range of viruses within the Imitervirales and Algavirales (Moniruzzaman et al. 2020a).
In marseilleviruses, core histones are packaged in the virion, and it has been hypothesized that they play a role in viral genome organization (Boyer et al. 2009, Fabre et al. 2017, Okamoto et al. 2018). Recent studies have reported crystal structures of marseillevirus histones and demonstrated that they indeed form nucleosome structures similar to eukaryotic homologs (Liu et al. 2021, Valencia-Sánchez et al. 2021). One study also demonstrated that the histones are essential for viral infectivity (Liu et al. 2021). Histones, therefore, appear to be a unique case among virologs in that they are not necessarily involved in manipulation of the host during infection, but rather are used to preserve the integrity of viral DNA in virions. A recent review has comprehensively discussed the presence of histone subunits in both members of the Nucleocytoviricota as well as other viral lineages (Talbert et al. 2022).
Nutrient and ion transport
Several transporters and ion channels have been identified in giant viruses, and some of the most in-depth experimental work has been performed on these virologs. The first example of a potassium channel encoded by a virus was found in chloroviruses, and extensive molecular characterization demonstrated its ion selectivity (Plugge et al. 2000). This ion channel is packaged in virions of Paramecium bursaria chlorella virus (PBCV-1) and is responsible for rapid depolarization of the cell membrane that facilitation of DNA release into the cell in the initial stages of infection (Kang et al. 2005, Frohns et al. 2006). Subsequent studies have found that potassium channels are common in a wide range of giant viruses (Siotto et al. 2014, Moniruzzaman et al. 2020a), and confirmation of ion selectivity has also been reported in a protein encoded by a member the Imitervirales (Kukovetz et al. 2020). Hence, channel-mediated membrane depolarization is potentially a common aspect for cell entry among viruses that do not exploit host phagocytosis or macropinocytosis for uptake. A wide range of other ion channels have been reported in giant viruses, such as mechanosensitive ion channels, but their role during infection remains largely unknown (Greiner et al. 2018).
Other studies have shown that viral-encoded transporters can influence nutrient acquisition during infection. One notable study examined an ammonium transporter encoded in a prasinovirus that infected Ostreococcus tauri, a small algae that is globally abundant in coastal waters (Monier et al. 2017). This study showed that the ammonium transporter was expressed and altered cellular nitrogen uptake during infection. This was an important finding not just for clarifying the role of this viral transporter, but also for demonstrating that virologs can have important implications for global biogeochemical cycling. Indeed, a diverse array of nutrient transporters have also been identified in giant virus genomes, including several high affinity phosphate transporters that are potentially involved in nutrient acquisition during infection (Weynberg et al. 2011, Monier et al. 2012, Moniruzzaman et al. 2020a). Collectively, these findings suggest that giant viruses manipulate the nutrient transport dynamics of their hosts substantially during infection, which may be critical in environments such as the ocean where the lack of particular macronutrients may limit virion production.
Vesicular trafficking
Numerous giant virus genomes encode components of vesicular trafficking machinery that potentially play important roles in cell entry, virion morphogenesis, and egress. Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), typically involved in vesicle fusion, were first identified in mimiviruses and coccolithoviruses (Wilson et al. 2005, Kloepper et al. 2007). Subsequent analysis identified a Rab GTPase encoded in mimivirus (Zade et al. 2015), which was found to have structural similarities to mammalian Rab5 proteins involved in regulating endosomal trafficking (Ku et al. 2017). Work focusing on medusavirus has also identified Ran-like GTPases that are potentially involved in nuclear import/export (Yoshikawa et al. 2019). Most recently, a genomic survey identified a broad diversity of SNAREs, Ras superfamily GTPases, N-ethylmaleimide-sensitive factors, and Sec1/Munc18-like proteins across a wide range of viruses within the Algavirales, Imitervirales, and Pimascovirales, demonstrating that these genes are commonly encoded in numerous giant virus lineages (Neveu et al. 2022). Moreover, this study also expressed several giant virus SNAREs and showed that they can form stable complexes with neuronal SNARE proteins, indicating that they are functional.
Vesicular trafficking is likely a critical component of virion morphogenesis that recruits membranes derived from the endoplasmic reticulum (ER) to the virus factory, where they ultimately give rise to the inner membrane of new virions (Mutsafi et al. 2013, Suárez et al. 2013). Treatment with vesicular trafficking inhibitors leads to defective virion morphogenesis in marseilleviruses (Arantes et al. 2016), cedratviruses (Silva et al. 2018), orpheoviruses (Souza et al. 2019), and pandoraviruses (Andrade et al. 2019), suggesting that vesicular trafficking from the ER is a key component of successful infection. Transcriptomic evidence suggests that some of the SNARE and Rab proteins are expressed only a few hours post infection, consistent with the hypothesis that they are inactive in the early stages of infection and primarily play a role during virion assembly (Wilson et al. 2005, Legendre et al. 2010). It remains unclear whether these virologs are directly involved in virion morphogenesis, however; given their patchy distribution across giant viruses it has been suggested that they are more likely involved in interfering with host vesicular trafficking rather than participating in core virion morphogenesis processes themselves (Neveu et al. 2022). In addition, one recent study on a dynamin virolog provided experimental evidence that it was involved in mitochondrial remodeling (Sheikh et al. 2023), suggesting that the scope of viral manipulation of host membranes extends well beyond the nucleus and ER.
Interestingly, membrane vesicles also play key roles in cell entry and egress in giant viruses (Rodrigues et al. 2016). Phagocytosis is the preferred mechanism of cell entry for the largest viruses, but for hosts in the genus Acanthamoeba this process is stimulated only for virions > 500 nm in size (Korn and Weisman 1967). Smaller marseilleviruses are only 250 nm in diameter, but during infection they induce the formation of large membrane vesicles that can encase dozens to thousands of virions as they are released from cells (Arantes et al. 2016). These membrane-bound groups are large enough to stimulate phagocytosis and can, therefore, go on to infect new hosts. The molecular mechanisms that underpin the formation of giant vesicles during marseillevirus infection are unclear, but it underscores the dramatic impact that viruses have on vesicular trafficking and membrane dynamics during infection.
Ubiquitin signaling
Ubiquitin signaling is a critical regulator of a wide range of cellular processes, and it is not surprising that viruses have evolved strategies to manipulate this system during infection (Lant and de Motes 2021). The ubiquitination process involves a cascade of three enzymes: E1 Ub-activation enzymes, E2 Ub-conjugating enzymes, and E3 Ub-ligating enzymes (Komander and Rape 2012). Homologs of all of these components have been reported in various members of the Nucleocytoviricota. For example, many poxviruses encode E3 homologs with ubiquitin ligase activity (Mansouri et al. 2003, Nerenberg et al. 2005) and homologs of E1 and E2 enzymes have been reported in members of the Mimiviridae and Phycodnaviridae (Iyer et al. 2006). Members of the Imitervirales appear to encode a particularly large complement of ubiquitin signaling genes; Rheavirus sinusmexicani (Cafeteria roenbergensis virus) encodes a E1 Ub-activating enzyme and six E2 Ub-conjugating enzymes (Fischer et al. 2010), while O. kaneohense encodes 14 E3 and one E2 enzyme (Schvarcz and Steward 2018). Both R. sinusmexicani and O. kaneohense also encode several deubiquitination enzymes and one ubiquitin homolog each, and these genes are also commonly found in poxviruses (Afonso et al. 1999, Bawden et al. 2000, Tulman et al. 2004, Fischer et al. 2010), indicating that widespread manipulation of ubiquitination during infection is widespread in the Nucleocytoviricota.
Experimental analysis of the roles of ubiquitin signaling virologs has been done in both poxviruses and African Swine Fever Virus (ASFV). ASFV encodes a homolog of a E2 Ub-conjugating enzyme, which is expressed throughout infection and packaged into the virion. Inhibition of the proteasome during infection by ASFV severely limited viral production (Barrado-Gil et al. 2017). The E2 virolog is recruited to virus factories during infection, where it may play a role in modulating several different key aspects of virion biogenesis (Freitas et al. 2018). Other studies have provided evidence that this virolog is also involved in suppressing the host immune response by antagonizing interferon signaling (Barrado-Gil et al. 2021, Riera et al. 2022), highlighting the various potential benefits of hijacking the host proteasome during infection. Proteasome inhibition experiments also showed that a functional ubiquitin–proteasome complex was required for viral gene expression, formation of virus factories, and viral DNA replication in vaccinia virus (Satheshkumar et al. 2009, Teale et al. 2009). Proteasome activity appears to be particularly important early in infection where it is necessary for viral genome release (Mercer et al. 2012). Poxviruses also encode several E3 Ub-ligases that have been shown to be important for virulence, though their precise roles remain unclear (reviewed in Lant and de Motes 2021).
Many members of the Nucleocytoviricota also encode numerous repeat domains that are linked to ubiquitin signaling. Ankyrin repeats are some of the most common functional annotations that arise in computational analysis of giant virus genomes, and many proteins with these motifs often contain F-box-like domains that interact with host ubiquitin ligases (Mercer et al. 2005, Sperling et al. 2008). Kelch repeats that likely mediate interactions with the cullin family of E3 ubiquitin ligases are also common in giant viruses, and in poxviruses these repeats have been linked to virulence (Kochneva et al. 2005, Beard et al. 2006, Balinsky et al. 2007). Protein repeat content has been linked to genome size in giant viruses, suggesting that larger viruses encode a higher capacity for complex protein–protein interactions (Shukla et al. 2018). Some of these protein–protein interaction domains are likely involved other functions unrelated to the proteasome, however, and may not necessarily be involved in ubiquitin signaling.
Control of ROS
In cellular organisms, SOD is a key protein in regulating redox homeostasis and mitigating oxidative stress (Miao and St. Clair 2009). When ROS in the cell accumulate to a toxic level, SODs catalyze the reduction of superoxide radicals (O2•–) to molecular oxygen and hydrogen peroxide (Sheng et al. 2014). All four cellular SOD families contain redox-active metal ions at their active sites: nickel in Ni-SOD, manganese in Mn-SOD, iron in Fe-SOD, and copper–zinc in Cu,Zn-SOD. Homologs of Cu, Zn-SOD have been found in several phylogenetically divergent giant viruses, including mimiviruses, chloroviruses, and poxviruses. During viral infection and replication, high ROS levels inside the host cell could damage viral DNA and replication machinery, and viral-encoded SOD may mitigate this damage. The chlorovirus PBCV-1 was found to produce an enzymatically active Cu,Zn-SOD and package it in the virion, and it was suggested that the SOD protein was involved in mitigating oxidative stress early in viral infection, potentially leading to a shorter replication cycle and larger burst size (Kang et al. 2014). Megavirus chilense of the Mimiviridae family also encodes and expresses a fully active Cu,Zn-SOD. Interestingly, the activation of this viral SOD protein seems to be independent of copper chaperon proteins, unlike its eukaryotic counterpart (Lartigue et al. 2015). Similar to the chloroviruses enzyme, the Megavirus-encoded SOD presumably decreases the concentration of ROS in the early stages of viral infection.
The Amsacta moorei entomopoxvirus AMV255 was also reported to express a catalytically active Cu,Zn-SOD (Becker et al. 2004), although the protein was not essential for viral infection. Most known SOD homologs encoded by other poxviruses are not enzymatically functional and lack both the zinc and the copper ion binding domain required for catalytic activity (Smith et al. 1991, Almazán et al. 2001). Intriguingly, the SOD-like proteins encoded by two leporipoxviruses, shope fibroma virus (SFV) and myxoma virus (MYX) do not have copper binding residues, but retain the capacity to bind zinc and cellular copper chaperones, which is typically required for the activation of eukaryotic SODs (Cao et al. 2002, Teoh et al. 2003, 2005). It was hypothesized that the SOD homologs in these viruses did not have antioxidant function, but instead act as decoys to sequester cellular copper chaperones, and thereby downregulate host SOD activity. This may increase ROS levels in the host intracellular environment, and in turn inhibit apoptosis and stimulate cell proliferation (Teoh et al. 2005). The contrasting roles of SODs encoded by different members of the Nucleocytoviricota underscore the difficulty in assigning definitive functional roles to viral enzymes based on bioinformatic predictions alone.
Cell cycle
The eukaryotic cell cycle is strictly controlled by various cellular proteins such as cyclins, cyclin-dependent kinases (Cdk), cell division cycle proteins (Cdc), and various other kinases. A wide range of mammalian viruses have been found to encode proteins that manipulate the host cell cycle and control replication (Mo et al. 2012, Fan et al. 2018), and these include proteins with clear homologs in eukaryotes (i.e. virologs). Herpesviruses, baculoviruses, and even retroviruses have been shown to encode cyclins used to subvert the cell cycle (Nicholas et al. 1992, Belyavskyi et al. 1998, LaPierre et al. 1998), demonstrating that many viruses have independently acquired these genes and converged on a common strategy for manipulating host growth dynamics. Here, we will focus on poxviruses and other members of the Nucleocytoviricota where these genes have recently been described.
An important factor in cell cycle control in the mammalian cell is the Anaphase Promoting Complex or Cyclosome (APC/C) composed of various proteins including APC11 and APC2. Virologs of APC11 have been found in orf virus (ORFV), a poxvirus, which can bind to APC2 in the cyclosome but lacks ubiquitin ligase activity. Binding of this virolog to APC/C prevents cell cycle progression and leads to accumulation of cells in G2/M phase. PACR promotes viral propagation but is not an essential protein in ORFV (Mo et al. 2009). Homologs of PACR can be found in a range of other poxviruses, indicating this is a common strategy in this family (Mo et al. 2009, Fan et al. 2018).
Numerous virologs of cell cycle regulators have recently been found in the genomes of medusavirus, cultivated from hot spring water in Japan using A. castellanii, and clandestinovirus, cultivated from wastewater using Vermamoeba vermiformis (Yoshikawa et al. 2019, Rolland et al. 2021). Both of these giant viruses likely belong to the same broad lineage, though their loss of some hallmark giant virus genes and orthologous displacement of others have obscured their overall evolutionary relationship to other giant viruses (Zhang et al. 2023). Clandestinovirus in particular was found to encode a plethora of cell cycle manipulating proteins in its genome, including virologs of Cyclin A2 and Cdc123. Cyclin A2 can bind to either CDK1 and CDK2 to regulate G2-M phase and S phase, respectively. Cdc123 interacts with eukaryotic initiation factor 2 (eIF2) and is involved in promotion of G1 to S phase transition. Medusavirus encodes a Cyclin B homolog, which is predicted to be associated with the M-phase promoting factor (MPF) that is involved in regulating G2-M phase transition (Fan et al. 2018, Yoshikawa et al. 2019). The presence of these putative cell cycle regulators may endow clandestinovirus and medusavirus with substantial control over the host growth phase. The infection cycles of clandestinovirus and medusavirus both contain a nuclear phase and are rather long, with viral release beginning only 16 and 14 hours postinfection, respectively, which may make control of cell division particularly important for these viruses.
Light sensing
Rhodopsins are light energy transducers that play important roles in energy production and phototaxis in a wide range of microbial lineages (for a recent review; see Rozenberg et al. 2021). Multiple studies have identified homologs of rhodopsins in the genomes of giant viruses, including several members of the Imitervirales (Needham et al. 2019, Moniruzzaman et al. 2020a, Farzad et al. 2022). Moreover, several environmental surveys have found that viral rhodopsins are widespread in marine environments (Yutin and Koonin 2012b, Philosof and Béjà 2013, López et al. 2017, Olson et al. 2018, Ha et al. 2023). Viral rhodopsins fall into several distinct groups and have been acquired from cellular lineages multiple times independently. The first study to examine these genes in giant viruses identified two distinct but evolutionarily related clades of viral rhodopsins, termed groups I and II, that encompassed a broad phylogenetic diversity (Yutin and Koonin 2012b). It was suggested that these rhodopsins arose from an ancient viral acquisition due to their divergence compared to extant cellular homologs. One study examining a group I rhodopsin encoded in a choanoflagellate virus suggested that these enzymes act as proton pumps and are potentially involved in energy metabolism during infection (Needham et al. 2019), but a subsequent study provided evidence that rhodopsins in this group are cation channels (i.e. channelrhodopsins) (Zabelskii et al. 2020). Structural analysis of a member of the viral group II rhodopsin clade also suggested that these are channelrhodopsins (Bratanov et al. 2019).
A separate group of channelrhodpsins are also prevalent in marine giant viruses (Rozenberg et al. 2020). These rhodopsins have been shown to act as light-driven anion pumps, and they are widespread in a wide range of both green algae and giant viruses. Phylogenetic analysis has revealed that they form a distinct clade compared with the group I and II viral rhodopsins, and that viruses most likely acquired them recently from green algal hosts. The exact role that channelrhodpsins play during viral infection remains unclear, but it has been hypothesized that they allow viruses to manipulate cellular signaling pathways during infection. For example, channelrhodopsins may allow viruses to influence phototaxis and swimming behavior in their hosts, possibly driving cells toward nutrient conditions more permissive for viral production (Gallot-Lavallée and Archibald 2020, Rozenberg et al. 2020).
Lastly, heliorhodopsins, a new group of rhodopsins that is widespread in bacteria, archaea, and protists, have also been found in giant viruses (Pushkarev et al. 2018). The function of viral heliorhodopsins is somewhat enigmatic due to their inverted membrane topology and divergent evolutionary history compared to other rhodopsins. One recent study working with a coccolithovirus heliorhodopsin has provided in vitro evidence that they act as proton pumps that depolarize the host membrane during infection (Hososhima et al. 2022). This could possibly interfere with host defenses or as a mechanism for superinfection exclusion.
Polysaccharide metabolism
Many viral proteins are glycosylated by host-encoded glycosyltransferases (GTs) located at the ER and Golgi apparatus (Olofsson and Hansen 1998). Viral-encoded GTs have been reported for some bacteriophages, poxviruses, herpesviruses, and baculoviruses, but rather than modifying viral structural glycoproteins, these enzymes are mainly expressed as virulence factors to modify host molecules, or to protect viral DNA from host restriction enzymes (Markine-Goriaynoff et al. 2004). In 1993, studies of chloroviruses suggested that they encode enzymes to glycosylate their structural proteins (Wang et al. 1993). Subsequent work showed that PBCV-1 encodes for at least seven putative GTs involved in the glycosylation of the Vp54 major capsid protein (Van Etten 2003). The Vp54 glycan repertoire is complex and atypical (De Castro et al. 2013), synthesized by a set of GTs with multiple functional domains (Speciale et al. 2019). It has been hypothesized that these rare glycans could extend the range of potential hosts (Piacente et al. 2014b), maintain the structural integrity of virions (Van Etten et al. 2017), or mediate adhesion to the host (Piacente et al. 2015).
Chlorovirus genomes also encode enzymes involved in the synthesis of hyaluronan and chitin, two polysaccharides that accumulate on the surface of the infected cells early during infection (DeAngelis et al. 1997, Graves et al. 1999, Kawasaki et al. 2002). It is unclear what role these cell surface polysaccharides play during infection, but a number of hypotheses have been proposed, including that they impair the attachment of additional chloroviruses (i.e. superinfection exclusion; Graves et al. 1999), protect the infected cell from the uptake and digestion by grazing protists, or stimulate cell clumping cells (thereby increasing the probability of future infections) (Van Etten et al. 2017). Chloroviruses also encode for numerous polysaccharide-degrading enzymes that are involved in degrading the cell wall of the host both during viral entry and exit (Kawasaki et al. 2002, Van Etten et al. 2017).
Mimivirus and its close relatives also possess a versatile glycosylation machinery for the biosynthesis of rare glycans previously thought to be restricted to bacteria (Piacente et al. 2012, Notaro et al. 2022). These glycans have been found in association with the layer of fibers surrounding the viral capsids, which vary in sugar composition according to the clade (Notaro et al. 2022). It is suggested that such complex machinery comes with a fitness cost after it was shown in mimivirus that several GT genes are lost after repeated passaging (Boyer et al. 2011). The resulting emergence of a bald version of the virus (M4) highlights how glycosylated fibers might be important in natural settings where competition between viruses for the same amoeba host is high (Boyer et al. 2011, Notaro et al. 2022). Glycosylated fibers may also shape interactions between giant viruses and their viral parasites, the so-called “virophages.” For example, the inability of the M4 strain to successfully propagate a parasitic virophage illustrates the potential role of fibers and their glycosylation for virophage attachment (Boyer et al. 2011). Overall, fibril glycans are important for interactions with the host cell (Luther et al. 2011, Piacente et al. 2014a), and may play a role in protecting the virions from the environment or the conditions found inside the phagocytic vacuole (Luther et al. 2011, Piacente et al. 2014a, 2015).
Interestingly, many members of the Nucleocytoviricota also encode collagen homologs in their genomes, which are thought to also play a role in virion stability. Collagens are a class of proteins found most notably in animals, where they provide structure for tissues. To date collagen-like have been identified in most pandoraviruses, pithoviruses, and members of the Mimiviridae (Baumann 2016). It was originally hypothesized that collagen is part of the dense network of fibers surrounding giant viruses such as mimivirus (Klose et al. 2010), but the resistance of the fibers to treatment with lysozyme and collagenase cast doubt on this idea (Luther et al. 2011). Nevertheless, biotinylation has shown these collagen-like proteins to be present on the surface of mimivirus particles (Shah et al. 2014) indicating they play a structural role. Interestingly, viral collagen has been shown to be modified after translation. Lysine residues are hydroxylated, and then glucosylated, as opposed to the galactosylation conserved across animals (Luther et al. 2011).
As new giant virus genomes become available, more putative glycogenes are being found for which their functions have yet to be elucidated. Klosneuviruses, e.g. have one of the most complex glycosylation machineries observed in viruses, with 30 GTs encoded by the recently sequenced fadolivirus (de Oliveira et al. 2022, Speciale et al. 2022). Another member of the Imitervirales that infects a marine haptophyte, Biavirus raunefjordenense, encodes for 48 putative GTs genes, the highest number of GTs found for a giant virus to date (Van Etten et al. 2017, Speciale et al. 2022). In general, although GTs have been characterized most in chloroviruses and members of the Mimiviridae, these genes have also been found in the genomes of marselleiviruses, medusaviruses, molliviruses, pandoraviruses, and pithoviruses (Piacente et al. 2015, Van Etten et al. 2017, Speciale et al. 2022), suggesting that viral-encoded glycosylation is a widespread phenomenon in giant viruses.
Immunoregulation
The immunomodulatory genes encoded by poxviruses and herpesviruses were the earliest virologs to be discovered, and they remain the most thoroughly investigated. Although herpesviruses belong to the phylum Peploviricota and are unrelated to the Nucleocytoviricota, historically their immunomodulatory virologs have been studied together with those of poxviruses, and so we will include some discussion of these viruses where appropriate. Other reviews have discussed viral-encoded immunomodulatory genes in more detail (McFadden and Murphy 2000, Elde and Malik 2009, Haller et al. 2014).
The examination of immunomodulatory virologs has also been instrumental in providing insights related to the interplay between viruses and both the innate and adaptive arms of the vertebrate immune system. For example, to limit cross-talk between infected and neighboring cells, viruses deploy decoy receptors and cytokines referred to as viroceptors and virokines, respectively. Textbook examples of viroceptors are the soluble IFNα (Colamonici et al. 1995, Symons et al. 1995) and IFNγ receptors (Upton et al. 1992, Alcamí and Smith 1995) encoded by poxviruses of which vaccinia B8R and B18R are the best studied instances. Uniquely, these receptors are not membrane bound, like their cellular counterparts, but secreted to sequester extracellular interferon. Several essential cytokines such as like IL-10 (Moore et al. 1990, Ouyang et al. 2014) and IL-6 (Moore et al. 1996, Russo et al. 1996) are also encoded by large DNA viruses. Notably, IL-10 is encoded by several unrelated viruses and has been acquired independently by poxviruses and herpesviruses that infect hosts ranging from mammals to fish (Ouyang et al. 2014). The independent acquisition of the same cellular gene by different viruses is rare overall, but it has been documented for cellular factors that are largely viewed as master regulators.
Cell death is a common infection outcome that can either enhance or diminish viral propagation (Danthi 2016), and it comes to no surprise that some immunomodulatory virologs manipulate cell death pathways. BCL2 family proteins, which display pro- and antiapoptotic activities, are encoded by several members of the Nucleocytoviricota (Afonso et al. 1996, Brun et al. 1996, Cuconati and White 2002). Other studies have illustrated the fundamental roles for effectors of emerging cell death pathways like necroptosis in tipping infection outcomes. The executioner of necroptosis, MLKL, has been acquired independently by both poxviruses and RNA viruses. Poxviruses encode the kinase-like domain of MLKL (Palmer et al. 2021), which functions as a pseudosubstrate of RIPK3 to suppress necroptosis (Petrie et al. 2019). Repeated duplications and losses of poxvirus encoded MLKL indicate a volatile history of this virolog that is suggestive of a molecular arms race (Palmer et al. 2021). In contrast, caliciviruses (e.g. noroviruses), which are single-stranded RNA viruses, have hijacked and repurposed the MLKL 4HB domain to drive viral egress by cell lysis. Surprisingly, this virolog appears to catalyze cell death independent of established cellular effectors of necroptosis, apoptosis, and pyroptosis (Wang et al. 2023). The poxvirus and calicivirus MLKL virologs demonstrate that viruses may capture different domains to drive distinct activities.
New insights into the activation of immune responses and counteraction by viruses can be gleaned from recently discovered virologs. For instance, a family of ultraconserved micropeptides that comprise the mitochondrial response (MISTR) circuit (Sorouri et al. 2020, 2022) have been acquired by three distinct members of the Nucleocytoviricota that collectively infect mammals, fish, and algae. The independent acquisition by viruses that infect divergent hosts implies that these factors are linked to core cellular processes. Indeed, MISTR factors interface with electron transport chain complexes. MISTR consists of MISTR1, MISTR AntiViral, and MISTR Hypoxia encoded by the NDUFA4, C15orf48, and NDUFA4L2 genes, respectively. Cellular and viral MISTR factors impact immune responses including virus-induced apoptosis (Sorouri et al. 2022) in a manner reflecting their patterns of gene regulation. Specifically, host MISTRAV, which is an interferon-stimulated gene, is proapoptotic, while the virolog encoded by squirrelpox is antiapoptotic. Likewise, downregulation of cellular MISTR1 by immune cues is antiapoptotic. The MISTR virologs point to essential roles for poorly understood higher-order configurations of the electron transport complexes—known as respiratory chain supercomplexes (Sorouri et al. 2020)—in the playbook of infection. Future functional characterization of virologs of other interferon-stimulated genes will provide increased resolution of conflict occurring in the early stages of infection. An exciting example is the gene encoded by squirrelpox (Darby et al. 2014) that is similar to OAS—a fundamental antiviral interferon stimulated gene that senses double-stranded RNA (Wickenhagen et al. 2021, Zhou et al. 2021, Lee et al. 2023).
Programmed cell death
Caspases (Cysteine-dependent aspartate-directed proteases) are a family of endoproteases mostly known as orchestrators of PCD in animals during development, tissue homeostasis, and pathology. Caspases belong to a broader family of C14 proteases that includes a wide range of metacaspases and paracaspases encoded by plants, fungi, and even bacteria, where they have been implicated in regulatory roles (Minina et al. 2017). Although many animal viruses encode a wide range of caspase inhibitors that suppress cell death and provide the viruses with enough time undergo their replication cycle (Best 2008, Connolly and Fearnhead 2017), research over the last few decades has shown that many viruses often co-opt host caspases in several unexpected ways during infection (Connolly and Fearnhead 2017).
Several members of the Nucleocytoviricota also encode their own caspase-like C14 proteases. Caspases are common in ascoviruses, where they have been implicated in the induction of a modified form of apoptosis that promotes viral transmission (Bideshi et al. 2005). In some ascoviruses the viral caspases do not seem to induce apoptosis, but they are still necessary for infection and they may play a role in the remodeling of actin fibers that is similar to that seen during apoptosis (Bideshi et al. 2005, Asgari 2007, Connolly and Fearnhead 2017). Other studies have found a broad diversity of metacaspases encoded by other members of the Nucleocytoviricota, in particular members of the Imitervirales and Algavirales that reside in marine environments, but it remains unclear what role they may play during infection (Wilson et al. 2017, Moniruzzaman et al. 2020a).
Insights into the role of viral caspases can be gleaned from studies of viruses that co-opt host metacaspases. This has been studied in depth in the coccolithovirus E. huxleyi virus (EhV), where it has been shown that host metacaspases are upregulated and activated during successful infection, and that reduction in caspase activity leads to decreased viral production (Bidle et al. 2007). Predicted metacaspase cleavage regions can be found in several EhV-encoded proteins, suggesting that caspase activity is needed for the maturation of viral proteins. This has also been suggested for chloroviruses and other members of the Algavirales, where key enzymes such as the DNA-packaging ATPase (chloroviruses) and DNA polymerase (Ectocarpus siliculosus virus) contain predicted caspase cleavage sites (Bidle et al. 2007, Van Etten et al. 2019). Indeed, caspase cleavage of viral proteins has also been implicated in the proliferation of a wide range of different RNA and DNA viruses (Connolly and Fearnhead 2017), demonstrating that co-option of host caspases is widespread in viruses.
Conclusions and perspectives
The prevalence of viruses within the phylum Nucleocytoviricota in the biosphere raises many important questions. Perhaps most obviously, why do these viruses encode so many unusual genes that are not often found in other viral lineages? The evolutionary forces that drive the accumulation of diverse virologs in the Nucleocytovirocota—and genome gigantism in general—have remained largely unclear. Several hypotheses have been put forward to explain how a complex set of encoded proteins may benefit giant viruses (for a broader review of tradeoffs in viral life history traits, see Edwards et al. 2021). First, the complex structural components encoded in many members of the Nucleocytoviricota—which often encode multiple capsid proteins and a wide range of other structural elements integrated into virions—may enhance capsid stability and prevent decay in the environment. Rates of virion decay are quite high in marine viruses (Suttle and Chen 1992), and enhanced virion stability may be a major benefit that giant viruses have compared to smaller viruses. High virion stability may be particularly important for viruses that infect low-abundance hosts in which the time in between infections is long.
Second, a broad range of virologs may allow giant viruses to broaden their tropism and infect hosts spanning a broader phylogenetic breadth. Indeed, the virus factories of many giant viruses appear capable of viral production in a range of different hosts—mimivirus has even been reported to replicate in human macrophages, albeit in specific laboratory conditions (Ghigo et al. 2008). Third, viruses may be able to partially circumvent red queen dynamics by removing their reliance on some cellular proteins that could otherwise mutate or evade viral manipulation during infection. Encoding a range of virologs thereby provides enhanced control over cellular dynamics. Lastly, it is likely that many virologs increase infection efficiency, potentially by decreasing the latent period of infection. One rare study that directly compared the infection dynamics of a giant virus and an RNA virus capable of infecting the same protist host found that the burst size was much larger for the RNA virus, but the latent period was shorter for the giant virus (Lawrence et al. 2006). Although more studies directly comparing the infection programs of giant viruses to smaller viruses infecting the same host are necessary to make any generalizations, this study hints that giant viruses may have greater infection efficiency at the cost of total progeny produced compared to smaller viruses. These tradeoffs are generally consistent with those observed when comparing the life history strategies of different bacteriophages (De Paepe and Taddei 2006).
Another important future frontier will be understanding the evolutionary origins of individual virologs and the extent to which virus–host gene exchange has shaped the evolution of both giant viruses and eukaryotes. Most virologs likely represent cases of host-to-virus gene transfer that have taken place over varying time-scales. While genes involved in DNA replication or transcription likely represent ancient gene transfers that were acquired by members of the Nucleocytoviricota early in their evolution, other virologs, such as some rhodopsins and ammonium transporters, represent clear cases of recent acquisition from distinct cellular lineages (see Fig. 3 for examples; broader discussion in Box 3). Although the mechanisms of gene transfer from eukaryotes to viruses are often unclear, recent studies suggest that selfish genetic elements may be a major conduit of host-to-virus gene transfer (Fixsen et al. 2022, Rahman et al. 2022). Moreover, other recent studies have begun to highlight how viruses can also transfer genes back into their host lineages, demonstrating that widespread bidirectional gene exchange is a major factor that shapes both viral and eukaryotic genomes (Irwin et al. 2018, 2022). Many giant viruses can endogenize into the genomes of their hosts, representing the largest endogenous viral elements recognized to date (Moniruzzaman et al. 2020b, 2022). Although giant virus endogenization is best characterized in the green algae, it appears to be quite common across a wide range of other protist groups (Filée 2014, Aylward and Moniruzzaman 2021, Hannat et al. 2021), and has been studied for decades in the brown algae Ectocarpus siliculosus (Müller et al. 1990).
Figure 3.
Phylogenetic trees of select virologs mentioned in the text. Some virologs have deep placement indicative of ancient gene exchange (DNA primase and mRNA capping enzyme), while others have been acquired more recently (K+ transporter and myosin). Moreover, some virologs appear to have been acquired by viruses multiple times independently (rhodopsin, synaptobrevin, Bax1 apoptosis inhibitor and K+ transporter).
Our understanding of giant viruses has advanced dramatically in the last two decades. After the discovery of mimivirus, it remained a possibility that giant viruses were an obscure lineage that infected primarily amoeba. But in-depth phylogenomic studies quickly demonstrated that mimivirus was part of a broader lineage that included poxviruses, iridoviruses, asfarviruses, and chloroviruses—all groups that had largely been studied individually prior to that point (Iyer et al. 2001, 2006). Subsequent culture-based and metagenomic approaches demonstrated that many of these groups are widespread in a broad range of ecosystems around the globe, and belong to the same phylum. We now know that eukaryotic viruses with large dsDNA genomes and a complex array of encoded proteins encompass an astounding diversity that includes a broad range of protist viruses and many emerging animal pathogens. Future work that continues to expand our understanding of the diversity of these viruses in the biosphere, the roles of encoded virologs during infection, and the impact of host–virus gene exchange on cellular and viral evolution will undoubtedly yield many more surprises in the years to come.
Acknowledgement
We are grateful to Oded Beja and Keiichi Inoue for helpful discussion of viral rhodopsins. We apologize to any colleagues whose important work was not discussed. J.S.A. is a CNPq/CAPES/FAPEMIG researcher.
Contributor Information
Mohammad Moniruzzaman, Rosenstiel School of Marine Atmospheric, and Earth Science, University of Miami, Coral Gables, FL 33149, United States.
Maria Paula Erazo Garcia, Department of Biological Sciences, Virginia Tech, 926 West Campus Drive, Blacksburg, VA 24061, United States.
Roxanna Farzad, Department of Biological Sciences, Virginia Tech, 926 West Campus Drive, Blacksburg, VA 24061, United States.
Anh D Ha, Department of Biological Sciences, Virginia Tech, 926 West Campus Drive, Blacksburg, VA 24061, United States.
Abdeali Jivaji, Department of Biological Sciences, Virginia Tech, 926 West Campus Drive, Blacksburg, VA 24061, United States.
Sangita Karki, Department of Biological Sciences, Virginia Tech, 926 West Campus Drive, Blacksburg, VA 24061, United States.
Uri Sheyn, Department of Biological Sciences, Virginia Tech, 926 West Campus Drive, Blacksburg, VA 24061, United States.
Joshua Stanton, Department of Biological Sciences, Virginia Tech, 926 West Campus Drive, Blacksburg, VA 24061, United States.
Benjamin Minch, Rosenstiel School of Marine Atmospheric, and Earth Science, University of Miami, Coral Gables, FL 33149, United States.
Danae Stephens, Rosenstiel School of Marine Atmospheric, and Earth Science, University of Miami, Coral Gables, FL 33149, United States.
Dustin C Hancks, Department of Immunology, University of Texas Southwestern Medical Center, 6000 Harry Hines Blvd, Dallas, TX, United States.
Rodrigo A L Rodrigues, Laboratório de Vírus, Departamento de Microbiologia, Universidade Federal de Minas Gerais, Belo Horizonte 31270-901, MG, Brazil.
Jonatas S Abrahao, Laboratório de Vírus, Departamento de Microbiologia, Universidade Federal de Minas Gerais, Belo Horizonte 31270-901, MG, Brazil.
Assaf Vardi, Department of Plant and Environmental Sciences, Weizmann Institute of Science, 7610001 Rehovot, Israel.
Frank O Aylward, Department of Biological Sciences, Virginia Tech, 926 West Campus Drive, Blacksburg, VA 24061, United States; Center for Emerging, Zoonotic, and Arthropod-Borne Infectious Disease, Virginia Tech, Blacksburg, VA 24061, United States.
Conflict of interest
None declared.
References
- Abergel C, Rudinger-Thirion J, Giegé Ret al. Virus-encoded aminoacyl-tRNA synthetases: structural and functional characterization of mimivirus TyrRS and MetRS. J Virol. 2007;81:12406–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahão J, Silva L, Silva LSet al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat Commun. 2018;9:749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso CL, Neilan JG, Kutish GFet al. An African swine fever virus Bc1-2 homolog, 5-HL, suppresses apoptotic cell death. J Virol. 1996;70:4858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso CL, Tulman ER, Lu Zet al. The genome of Melanoplus sanguinipes Entomopoxvirus. J Virol. 1999;73:533–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aherfi S, Brahim Belhaouari D, Pinault Let al. Incomplete tricarboxylic acid cycle and proton gradient in Pandoravirus massiliensis: is it still a virus?. ISME J. 2022;16:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcamí A, Smith GL. Vaccinia, cowpox, and camelpox viruses encode soluble gamma interferon receptors with novel broad species specificity. J Virol. 1995;69:4633–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alempic J-M, Bisio H, Villalta Aet al. No fitness impact of the knockout of the two main components of mimivirus genomic fiber and fibril layer. bioRxiv. 2023. https://www.biorxiv.org/content/10.1101/2023.04.28.538727v3. [Google Scholar]
- Allonso D, Andrade IS, Conde JNet al. Dengue virus NS1 protein modulates cellular energy metabolism by increasing glyceraldehyde-3-phosphate dehydrogenase activity. J Virol. 2015;89:11871–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán F, Tscharke DC, Smith GL. The vaccinia virus superoxide dismutase-like protein (A45R) is a virion component that is nonessential for virus replication. J Virol. 2001;75:7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade ACdSP, dos Santos Pereira Andrade AC, de Miranda Boratto PVet al. New isolates of Pandoraviruses: contribution to the study of replication cycle steps. J Virol. 2019;93. 10.1128/jvi.01942-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreani J, Khalil JYB, Baptiste Eet al. Orpheovirus IHUMI-LCC2: a new virus among the giant viruses. Front Microbiol. 2017;8:2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes TS, Rodrigues RAL, dos Santos Silva LKet al. The large Marseillevirus explores different entry pathways by forming giant infectious vesicles. J Virol. 2016;90:5246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari S. A caspase-like gene from Heliothis virescens ascovirus (HvAV-3e) is not involved in apoptosis but is essential for virus replication. Virus Res. 2007;128:99–105. [DOI] [PubMed] [Google Scholar]
- Aylward FO, Moniruzzaman M, Ha ADet al. A phylogenomic framework for charting the diversity and evolution of giant viruses. PLoS Biol. 2021;19:e3001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylward FO, Moniruzzaman M. ViralRecall - a flexible command-line tool for the detection of giant virus signatures in ’Omic Data. Viruses. 2021;13. 10.3390/v13020150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström D, Yutin N, Jørgensen SLet al. Virus genomes from deep sea sediments expand the ocean megavirome and support independent origins of viral gigantism. mBio. 2019;10. 10.1128/mBio.02497-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balinsky CA, Delhon G, Afonso CLet al. Sheeppox virus kelch-like gene SPPV-019 affects virus virulence. J Virol. 2007;81:11392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrado-Gil L, Del Puerto A, Galindo Iet al. African swine fever virus ubiquitin-conjugating enzyme is an immunomodulator targeting NF-κB activation. Viruses. 2021;13. 10.3390/v13061160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrado-Gil L, Galindo I, Martínez-Alonso Det al. The ubiquitin-proteasome system is required for African swine fever replication. PLoS ONE. 2017;12:e0189741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann S. Functional analysis of collagen galactosyltransferases and the identification of collagens in giant viruses. Zürich: University of Zurich, 2016. [Google Scholar]
- Bawden AL, Glassberg KJ, Diggans Jet al. Complete genomic sequence of the Amsacta moorei Entomopoxvirus: analysis and comparison with other poxviruses. Virology. 2000;274:120–39. [DOI] [PubMed] [Google Scholar]
- Beard PM, Froggatt GC, Smith GL. Vaccinia virus kelch protein A55 is a 64 kDa intracellular factor that affects virus-induced cytopathic effect and the outcome of infection in a murine intradermal model. J Gen Virol. 2006;87:1521–9. [DOI] [PubMed] [Google Scholar]
- Becker MN, Greenleaf WB, Ostrov DAet al. Amsacta moorei Entomopoxvirus expresses an active superoxide dismutase. J Virol. 2004;78:10265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekliz M, Azza S, Seligmann Het al. Experimental analysis of Mimivirus translation initiation factor 4a reveals its importance in viral protein translation during infection of Acanthamoeba polyphaga. J Virol. 2018;92. 10.1128/JVI.00337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell PJL. Eukaryogenesis: the rise of an emergent superorganism. Front Microbiol. 2022;13:858064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyavskyi M, Braunagel SC, Summers MD. The structural protein ODV-EC27 of Autographa californica nucleopolyhedrovirus is a multifunctional viral cyclin. Proc Natl Acad Sci. 1998;95:11205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best SM. Viral subversion of apoptotic enzymes: escape from death row. Annu Rev Microbiol. 2008;62:171–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts HC, Puttick MN, Clark JWet al. Integrated genomic and fossil evidence illuminates life's early evolution and eukaryote origin. Nat Ecol Evol. 2018;2:1556–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee AS, Schulz F, Woyke Tet al. Genomics discovery of giant fungal viruses from subsurface oceanic crustal fluids. ISME Commun. 2023;3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bideshi DK, Tan Y, Bigot Yet al. A viral caspase contributes to modified apoptosis for virus transmission. Genes Dev. 2005;19:1416–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidle KD, Haramaty L, Barcelos E Ramos Jet al. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc Natl Acad Sci USA. 2007;104:6049–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisio H, Legendre M, Giry Cet al. Evolution of giant pandoravirus revealed by CRISPR/Cas9. Nat Commun. 2023;14:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc-Mathieu R, Dahle H, Hofgaard Aet al. A persistent giant algal virus, with a unique morphology, encodes an unprecedented number of genes involved in energy metabolism. J Virol. 2021;95. 10.1128/JVI.02446-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Azza S, Barrassi Let al. Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc Natl Acad Sci USA. 2011;108:10296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Yutin N, Pagnier Iet al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci USA. 2009;106:21848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahim Belhaouari D, Pires De Souza GA, Lamb DCet al. Metabolic arsenal of giant viruses: host hijack or self-use?. eLife. 2022;11. 10.7554/eLife.78674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratanov D, Kovalev K, Machtens J-Pet al. Unique structure and function of viral rhodopsins. Nat Commun. 2019;10:4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocks JJ, Nettersheim BJ, Adam Pet al. Lost world of complex life and the late rise of the eukaryotic crown. Nature. 2023;618:767–73. [DOI] [PubMed] [Google Scholar]
- Brun A, Rivas C, Esteban Met al. African swine fever virus gene A179L, a viral homologue of bcl-2, protects cells from programmed cell death. Virology. 1996;225:227–30. [DOI] [PubMed] [Google Scholar]
- Cao JX, Teoh MLT, Moon Met al. Leporipoxvirus Cu-Zn superoxide dismutase homologs inhibit cellular superoxide dismutase, but are not essential for virus replication or virulence. Virology. 2002;296:125–35. [DOI] [PubMed] [Google Scholar]
- Ceyssens P-J, Minakhin L, Van den Bossche Aet al. Development of giant bacteriophage ϕKZ is independent of the host transcription apparatus. J Virol. 2014;88:10501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Suttle CA, Short SM. Genetic diversity in marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl Environ Microb. 1996;62:2869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J-M, Abergel C. An expanding family of highly diverse large dsDNA viruses infecting a wide phylogenetic range of aquatic eukaryotes. Viruses. 2018;10. 10.3390/v10090506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J-M, Abergel C. Giant viruses: the difficult breaking of multiple epistemological barriers. Stud Hist Philos Biol Biomed Sci. 2016;59:89–99. [DOI] [PubMed] [Google Scholar]
- Claverie J-M, Abergel C. Mimivirus: the emerging paradox of quasi-autonomous viruses. Trends Genet. 2010;26:431–7. [DOI] [PubMed] [Google Scholar]
- Claverie J-M. Viruses take center stage in cellular evolution. Genome Biol. 2006;7:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colamonici OR, Domanski P, Sweitzer SMet al. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J Biol Chem. 1995;270:15974–8. [DOI] [PubMed] [Google Scholar]
- Connolly PF, Fearnhead HO. Viral hijacking of host caspases: an emerging category of pathogen-host interactions. Cell Death Differ. 2017;24:1401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuconati A, White E. Viral homologs of BCL-2: role of apoptosis in the regulation of virus infection. Genes Dev. 2002;16:2465–78. [DOI] [PubMed] [Google Scholar]
- Da Cunha V, Gaia M, Ogata Het al. Giant viruses encode actin-related proteins. Mol Biol Evol. 2022;39. 10.1093/molbev/msac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danthi P. Viruses and the diversity of cell death. Annu Rev Virol. 2016;3:533–53. [DOI] [PubMed] [Google Scholar]
- Darby AC, McInnes CJ, Kjær KHet al. Novel host-related virulence factors are encoded by squirrelpox virus, the main causative agent of epidemic disease in red squirrels in the UK. PLoS ONE. 2014;9:e96439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro C, Molinaro A, Piacente Fet al. Structure of N-linked oligosaccharides attached to chlorovirus PBCV-1 major capsid protein reveals unusual class of complex N-glycans. Proc Natl Acad Sci USA. 2013;110:13956–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira EG, Carvalho JVRP, Botelho BBet al. Giant viruses as a source of novel enzymes for biotechnological application. Pathogens. 2022;11. 10.3390/pathogens11121453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe M, Taddei F. Viruses’ life history: towards a mechanistic basis of a trade-off between survival and reproduction among phages. PLoS Biol. 2006;4:e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis PL, Jing W, Graves MVet al. Hyaluronan synthase of chlorella virus PBCV-1. Science. 1997;278:1800–3. [DOI] [PubMed] [Google Scholar]
- Deeg CM, Chow C-ET, Suttle CA. The kinetoplastid-infecting Bodo saltans virus (BsV), a window into the most abundant giant viruses in the sea. eLife. 2018;7:e33014. 10.1101/214536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan GA, Dunigan DD, Van Van Etten JL. Diversity of tRNA clusters in the Chloroviruses. Viruses. 2020;12. 10.3390/v12101173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré J, Guttinger S. Viruses as living processes. Stud Hist Philos Sci Part C Stud Hist Philos Biol Biomed Sci. 2016;59:109–16. [DOI] [PubMed] [Google Scholar]
- Edwards KF, Steward GF, Schvarcz CR. Making sense of virus size and the tradeoffs shaping viral fitness. Ecol Lett. 2021;24:363–73. [DOI] [PubMed] [Google Scholar]
- Elde NC, Malik HS. The evolutionary conundrum of pathogen mimicry. Nat Rev Micro. 2009;7:787–97. [DOI] [PubMed] [Google Scholar]
- Endo H, Blanc-Mathieu R, Li Yet al. Biogeography of marine giant viruses reveals their interplay with eukaryotes and ecological functions. Nat Ecol Evol. 2020;4:1639–49. [DOI] [PubMed] [Google Scholar]
- Fabre E, Jeudy S, Santini Set al. Noumeavirus replication relies on a transient remote control of the host nucleus. Nat Commun. 2017;8:15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Sanyal S, Bruzzone R. Breaking bad: how viruses subvert the cell cycle. Front Cell Infect Microbiol. 2018;8:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzad R, Ha AD, Aylward FO. Diversity and genomics of giant viruses in the North Pacific Subtropical Gyre. Front Microbiol. 2022;13:1021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filée J, Pouget N, Chandler M. Phylogenetic evidence for extensive lateral acquisition of cellular genes by nucleocytoplasmic large DNA viruses. BMC Evol Biol. 2008;8:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filée J. Multiple occurrences of giant virus core genes acquired by eukaryotic genomes: the visible part of the iceberg?. Virology. 2014;466–467:53–9. [DOI] [PubMed] [Google Scholar]
- Fischer MG, Allen MJ, Wilson WHet al. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci USA. 2010;107:19508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fixsen SM, Cone KR, Goldstein SAet al. Poxviruses capture host genes by LINE-1 retrotransposition. eLife. 2022;11. 10.7554/eLife.63332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine KA, Sanchez EL, Camarda Ret al. Dengue virus induces and requires glycolysis for optimal replication. J Virol. 2015;89:2358–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Gaïa M. Giant viruses and the origin of modern eukaryotes. Curr Opin Microbiol. 2016;31:44–9. [DOI] [PubMed] [Google Scholar]
- Forterre P. Giant viruses: conflicts in revisiting the virus concept. Intervirology. 2010;53:362–78. [DOI] [PubMed] [Google Scholar]
- Freitas FB, Frouco G, Martins Cet al. African swine fever virus encodes for an E2-ubiquitin conjugating enzyme that is mono- and di-ubiquitinated and required for viral replication cycle. Sci Rep. 2018;8:3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman S, Flores-Uribe J, Larom Set al. A myovirus encoding both photosystem I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells. Nat Microbiol. 2017;2:1350–7. [DOI] [PubMed] [Google Scholar]
- Frohns F, Käsmann A, Kramer Det al. Potassium ion channels of Chlorella viruses cause rapid depolarization of host cells during infection. J Virol. 2006;80:2437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm A, Schatz D, Ben-Dor Set al. Complete genome sequence of virus strain M1, isolated from an induced bloom in Bergen, Norway. Microbiol Resour Announc. 2022;11:e0007122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton JM, Fredricks HF, Bidle KDet al. Novel molecular determinants of viral susceptibility and resistance in the lipidome of Emiliania huxleyi. Environ Microbiol. 2014;16:1137–49. [DOI] [PubMed] [Google Scholar]
- Gaïa M, Meng L, Pelletier Eet al. Mirusviruses link herpesviruses to giant viruses. Nature. 2023;616:783–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallot-Lavallée L, Archibald JM. Evolutionary biology: viral rhodopsins illuminate algal evolution. Curr Biol. 2020;30:R1469–71. [DOI] [PubMed] [Google Scholar]
- Gallot-Lavallée L, Blanc G, Claverie J-M. Comparative genomics of Chrysochromulina ericina virus and other microalga-infecting large dna viruses highlights their intricate evolutionary relationship with the established Mimiviridae family. J Virol. 2017;91. 10.1128/JVI.00230-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammon DB, Gowrishankar B, Duraffour Set al. Vaccinia virus-encoded ribonucleotide reductase subunits are differentially required for replication and pathogenesis. PLoS Pathog. 2010;6:e1000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo E, Kartenbeck J, Lien Pet al. Ameobal pathogen mimivirus infects macrophages through phagocytosis. PLoS Pathog. 2008;4:e1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves MV, Burbank DE, Roth Ret al. Hyaluronan synthesis in virus PBCV-1-infected chlorella-like green algae. Virology. 1999;257:15–23. [DOI] [PubMed] [Google Scholar]
- Greiner T, Moroni A, Van Etten JLet al. Genes for membrane transport proteins: not so rare in viruses. Viruses. 2018;10. 10.3390/v10090456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J, Gaia M, Da Cunha Vet al. Viral origin of eukaryotic type IIA DNA topoisomerases. Virus Evol. 2022;8:veac097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J, Woo AC, Krupovic Met al. Diversification of giant and large eukaryotic dsDNA viruses predated the origin of modern eukaryotes. Proc Natl Acad Sci USA. 2019;116:19585–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha AD, Moniruzzaman M, Aylward FO. Assessing the biogeography of marine giant viruses in four oceanic transects. ISME Commun. 2023;3:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha AD, Moniruzzaman M, Aylward FO. High transcriptional activity and diverse functional repertoires of hundreds of giant viruses in a coastal marine system. mSystems. 2021;6:e0029321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller SL, Peng C, McFadden Get al. Poxviruses and the evolution of host range and virulence. Infect Genet Evol. 2014;21:15–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannat S, Pontarotti P, Colson Pet al. Diverse trajectories drive the expression of a giant virus in the oomycete plant pathogen. Front Microbiol. 2021;12:662762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hososhima S, Mizutori R, Abe-Yoshizumi Ret al. Proton-transporting heliorhodopsins from marine giant viruses. eLife. 2022;11. 10.7554/eLife.78416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T-S, Nilsson CE, Punga Tet al. Functional inactivation of the SR family of splicing factors during a vaccinia virus infection. EMBO Rep. 2002;3:1088–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Poxvirus genome evolution by gene gain and loss. Mol Phylogenet Evol. 2005;35:186–95. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Origin and evolution of viral interleukin-10 and other DNA virus genes with vertebrate homologues. J Mol Evol. 2002;54:90–101. [DOI] [PubMed] [Google Scholar]
- Hunter JE, Frada MJ, Fredricks HFet al. Targeted and untargeted lipidomics of Emiliania huxleyi viral infection and life cycle phases highlights molecular biomarkers of infection, susceptibility, and ploidy. Front Mar Sci. 2015;2. 10.3389/fmars.2015.00081. [DOI] [Google Scholar]
- Hurwitz BL, U'Ren JM. Viral metabolic reprogramming in marine ecosystems. Curr Opin Microbiol. 2016;31:161–8. [DOI] [PubMed] [Google Scholar]
- Irwin NAT, Martin BJE, Young BPet al. Viral proteins as a potential driver of histone depletion in dinoflagellates. Nat Commun. 2018;9:1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin NAT, Pittis AA, Richards TAet al. Systematic evaluation of horizontal gene transfer between eukaryotes and viruses. Nat Microbiol. 2022;7:327–36. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Balaji S, Koonin EVet al. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–84. [DOI] [PubMed] [Google Scholar]
- Jha S, Rollins MG, Fuchs Get al. Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature. 2017;546:651–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Windsor M, Rietdorf Jet al. African swine fever virus induces filopodia-like projections at the plasma membrane. Cell Microbiol. 2006;8:1803–11. [DOI] [PubMed] [Google Scholar]
- Kang M, Duncan GA, Kuszynski Cet al. Chlorovirus PBCV-1 encodes an active copper-zinc superoxide dismutase. J Virol. 2014;88:12541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Dunigan DD, VAN Etten JL. Chlorovirus: a genus of Phycodnaviridae that infects certain chlorella-like green algae. Mol Plant Pathol. 2005;6:213–24. [DOI] [PubMed] [Google Scholar]
- Karki S, Aylward FO. Resolving ancient gene transfers clarifies the early co-evolution of eukaryotes and giant viruses. bioRxiv. 2023. 10.1101/2023.07.11.548585. [DOI]
- Kavagutti VS, Bulzu P-A, Chiriac CMet al. High-resolution metagenomic reconstruction of the freshwater spring bloom. Microbiome. 2023;11:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Tanaka M, Fujie Met al. Chitin synthesis in chlorovirus CVK2-infected chlorella cells. Virology. 2002;302:123–31. [DOI] [PubMed] [Google Scholar]
- Kijima S, Delmont TO, Miyazaki Uet al. Discovery of viral myosin genes with complex evolutionary history within plankton. Front Microbiol. 2021;12:683294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepper TH, Kienle CN, Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol Biol Cell. 2007;18:3463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose T, Herbst DA, Zhu Het al. A mimivirus enzyme that participates in viral entry. Structure. 2015;23:1058–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose T, Kuznetsov YG, Xiao Cet al. The three-dimensional structure of mimivirus. Intervirology. 2010;53:268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva G, Kolosova I, Maksyutova Tet al. Effects of deletions of kelch-like genes on cowpox virus biological properties. Arch Virol. 2005;150:1857–70. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The Ubiquitin Code. Annu Rev Biochem. 2012;81:203–29. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Dolja VV, Krupovic M. The logic of virus evolution. Cell Host Microbe. 2022;30:917–29. [DOI] [PubMed] [Google Scholar]
- Korn ED, Weisman RA. Phagocytosis of latex beads by Acanthamoeba. II. Electron microscopic study of the initial events. J Cell Biol. 1967;34:219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Yutin N, Koonin E. Evolution of a major virion protein of the giant pandoraviruses from an inactivated bacterial glycoside hydrolase. Virus Evol. 2020;6:veaa059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B, You JA, Oh K-Jet al. Crystal structures of two forms of the Acanthamoeba polyphaga mimivirus Rab GTPase. Arch Virol. 2017;162:3407–16. [DOI] [PubMed] [Google Scholar]
- Kukovetz K, Hertel B, Schvarcz CRet al. A functional K channel from Tetraselmis Virus 1, a member of the Mimiviridae. Viruses. 2020;12. 10.3390/v12101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laber CP, Hunter JE, Carvalho Fet al. Coccolithovirus facilitation of carbon export in the North Atlantic. Nat Microbiol. 2018;3:537–47. [DOI] [PubMed] [Google Scholar]
- Lant S, de Motes CM. Poxvirus interactions with the host Ubiquitin system. Pathogens. 2021;10:1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPierre LA, Casey JW, Holzschu DL. Walleye retroviruses associated with skin tumors and hyperplasias encode cyclin D homologs. J Virol. 1998;72:8765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue A, Burlat B, Coutard Bet al. The megavirus chilensis Cu,Zn-superoxide dismutase: the first viral structure of a typical cellular copper chaperone-independent hyperstable dimeric enzyme. J Virol. 2015;89:824–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasso G, Honig B, Shapira SD. A sweep of Earth’s virome reveals host-guided viral protein structural mimicry and points to determinants of human disease. Cell Syst. 2021;12:82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JE, Brussaard CPD, Suttle CA. Virus-specific responses of Heterosigma akashiwo to infection. Appl Environ Microb. 2006;72:7829–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Le Pen J, Yatim Aet al. Inborn errors of OAS-RNase L in SARS-CoV-2-related multisystem inflammatory syndrome in children. Science. 2023;379:eabo3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Alempic J-M, Philippe Net al. Pandoravirus celtis illustrates the microevolution processes at work in the giant genomes. Front Microbiol. 2019;10:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Audic S, Poirot Oet al. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in mimivirus. Genome Res. 2010;20:664–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Fabre E, Poirot Oet al. Diversity and evolution of the emerging Pandoraviridae family. Nat Commun. 2018;9:2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hingamp P, Watai Het al. Degenerate PCR primers to reveal the diversity of giant viruses in coastal waters. Viruses. 2018;10. 10.3390/v10090496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lu Z, Sun Let al. Analysis of 74 kb of DNA located at the right end of the 330-kb Chlorella virus PBCV-1 genome. Virology. 1997;237:360–77. [DOI] [PubMed] [Google Scholar]
- Liu Y, Bisio H, Toner CMet al. Virus-encoded histone doublets are essential and form nucleosome-like structures. Cell. 2021;184. 10.1016/j.cell.2021.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López JL, Golemba M, Hernández Eet al. Microbial and viral-like rhodopsins present in coastal marine sediments from four polar and subpolar regions. FEMS Microbiol Ecol. 2017;93. 10.1093/femsec/fiw216. [DOI] [PubMed] [Google Scholar]
- Luther KB, Hülsmeier AJ, Schegg Bet al. Mimivirus collagen is modified by bifunctional lysyl hydroxylase and glycosyltransferase enzyme. J Biol Chem. 2011;286:43701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann NH, Cook A, Millard Aet al. Bacterial photosynthesis genes in a virus. Nature. 2003;424:741. [DOI] [PubMed] [Google Scholar]
- Mansouri M, Bartee E, Gouveia Ket al. The PHD/LAP-domain protein M153R of Myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J Virol. 2003;77:1427–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markine-Goriaynoff N, Gillet L, Van Etten JLet al. Glycosyltransferases encoded by viruses. J Gen Virol. 2004;85:2741–54. [DOI] [PubMed] [Google Scholar]
- McFadden G, Murphy PM. Host-related immunomodulators encoded by poxviruses and herpesviruses. Curr Opin Microbiol. 2000;3:371–8. [DOI] [PubMed] [Google Scholar]
- Meng L, Endo H, Blanc-Mathieu Ret al. Quantitative assessment of nucleocytoplasmic large DNA virus and host interactions predicted by co-occurrence analyses. mSphere. 2021;6. 10.1128/mSphere.01298-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer AA, Fleming SB, Ueda N. F-Box-like domains are present in most Poxvirus ankyrin repeat proteins. Virus Genes. 2005;31:127–33. [DOI] [PubMed] [Google Scholar]
- Mercer J, Snijder B, Sacher Ret al. RNAi screening reveals proteasome- and Cullin3-dependent stages in vaccinia virus infection. Cell Rep. 2012;2:1036–47. [DOI] [PubMed] [Google Scholar]
- Miao L, St. Clair DK. Regulation of superoxide dismutase genes: implications in diseases. Free Radic Biol Med. 2009;47:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewski S, Rihtman B, Cook Ret al. A new family of “megaphages” abundant in the marine environment. ISME Commun. 2021;1:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina EA, Coll NS, Tuominen Het al. Metacaspases versus caspases in development and cell fate regulation. Cell Death Differ. 2017;24:1314–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo M, Fleming SB, Mercer AA. Cell cycle deregulation by a poxvirus partial mimic of anaphase-promoting complex subunit 11. Proc Natl Acad Sci USA. 2009;106:19527–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo M, Shahar S, Fleming SBet al. How viruses affect the cell cycle through manipulation of the APC/C. Trends Microbiol. 2012;20:440–8. [DOI] [PubMed] [Google Scholar]
- Monier A, Chambouvet A, Milner DSet al. Host-derived viral transporter protein for nitrogen uptake in infected marine phytoplankton. Proc Natl Acad Sci USA. 2017;114:E7489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A, Pagarete A, de Vargas Cet al. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Res. 2009;19:1441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A, Welsh RM, Gentemann Cet al. Phosphate transporters in marine phytoplankton and their viruses: cross-domain commonalities in viral-host gene exchanges. Environ Microbiol. 2012;14:162–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniruzzaman M, Erazo-Garcia MP, Aylward FO. Endogenous giant viruses contribute to intraspecies genomic variability in the model green alga. Virus Evol. 2022;8:veac102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniruzzaman M, Martinez-Gutierrez CA, Weinheimer ARet al. Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses. Nat Commun. 2020a;11:1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniruzzaman M, Weinheimer AR, Martinez-Gutierrez CAet al. Widespread endogenization of giant viruses shapes genomes of green algae. Nature. 2020b;588:141–5. [DOI] [PubMed] [Google Scholar]
- Moore KW, Vieira P, Fiorentino DFet al. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science. 1990;248:1230–4. [DOI] [PubMed] [Google Scholar]
- Moore PS, Boshoff C, Weiss RAet al. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–44. [DOI] [PubMed] [Google Scholar]
- Moreau H, Piganeau G, Desdevises Yet al. Marine prasinovirus genomes show low evolutionary divergence and acquisition of protein metabolism genes by horizontal gene transfer. J Virol. 2010;84:12555–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller DG, Kawai H, Stache Bet al. A virus infection in the marine brown alga Ectocarpus siliculosus (Phaeophyceae). Bot Acta. 1990;103:72–82. [Google Scholar]
- Murti KG, Chen M, Goorha R. Interaction of frog virus 3 with the cytomatrix. III. Role of microfilaments in virus release. Virology. 1985;142:317–25. [DOI] [PubMed] [Google Scholar]
- Mutsafi Y, Shimoni E, Shimon Aet al. Membrane assembly during the infection cycle of the giant Mimivirus. PLoS Pathog. 2013;9:e1003367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham DM, Yoshizawa S, Hosaka Tet al. A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators. Proc Natl Acad Sci USA. 2019;116:20574–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerenberg BTH, Hovey Nerenberg BT, Taylor Jet al. The Poxviral RING protein p28 is a ubiquitin ligase that targets ubiquitin to viral replication factories. J Virol. 2005;79:597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu E, Khalifeh D, Fasshauer D. Megaviruses contain various genes encoding for eukaryotic vesicle trafficking factors. Traffic. 2022;23:414–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas J, Cameron KR, Honess RW. Herpesvirus saimiri encodes homologues of G protein-coupled receptors and cyclins. Nature. 1992;355:362–5. [DOI] [PubMed] [Google Scholar]
- Nissimov JI, Talmy D, Haramaty Let al. Biochemical diversity of glycosphingolipid biosynthesis as a driver of Coccolithovirus competitive ecology. Environ Microbiol. 2019;21:2182–97. [DOI] [PubMed] [Google Scholar]
- Notaro A, Poirot O, Garcin EDet al. Giant viruses of the Megavirinae subfamily possess biosynthetic pathways to produce rare bacterial-like sugars in a clade-specific manner. Microlife. 2022;3. 10.1093/femsml/uqac002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Ray J, Toyoda Ket al. Two new subfamilies of DNA mismatch repair proteins (MutS) specifically abundant in the marine environment. ISME J. 2011;5:1143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Hayashi T, Ueki S. Complete genome sequence of a Phycodnavirus, Heterosigma akashiwo virus strain 53. Genome Announc. 2016;4. 10.1128/genomeA.01279-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Miyazaki N, Reddy HKNet al. Cryo-EM structure of a Marseilleviridae virus particle reveals a large internal microassembly. Virology. 2018;516:239–45. [DOI] [PubMed] [Google Scholar]
- Olofsson S, Hansen JE. Host cell glycosylation of viral glycoproteins–a battlefield for host defence and viral resistance. Scand J Infect Dis. 1998;30:435–40. [DOI] [PubMed] [Google Scholar]
- Olson DK, Yoshizawa S, Boeuf Det al. Proteorhodopsin variability and distribution in the North Pacific Subtropical Gyre. ISME J. 2018;12:1047–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang P, Rakus K, van Beurden SJet al. IL-10 encoded by viruses: a remarkable example of independent acquisition of a cellular gene by viruses and its subsequent evolution in the viral genome. J Gen Virol. 2014;95:245–62. [DOI] [PubMed] [Google Scholar]
- Palmer SN, Chappidi S, Pinkham Cet al. Evolutionary profile for (host and viral) MLKL indicates its activities as a battlefront for extensive counteradaptation. Mol Biol Evol. 2021;38:5405–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang H, Jiang Y, Li Jet al. Aberrant NAD metabolism underlies Zika virus-induced microcephaly. Nat Metab. 2021;3:1109–24. [DOI] [PubMed] [Google Scholar]
- Passalacqua KD, Lu J, Goodfellow Iet al. Glycolysis is an intrinsic factor for optimal replication of a norovirus. mBio. 2019;10. 10.1128/mBio.02175-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie EJ, Sandow JJ, Lehmann WILet al. Viral MLKL homologs subvert necroptotic cell death by sequestering cellular RIPK3. Cell Rep. 2019;28:3309–19. [DOI] [PubMed] [Google Scholar]
- Philippe N, Legendre M, Doutre Get al. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341:281–6. [DOI] [PubMed] [Google Scholar]
- Philosof A, Béjà O. Bacterial, archaeal and viral-like rhodopsins from the Red Sea. Environ Microbiol Rep. 2013;5:475–82. [DOI] [PubMed] [Google Scholar]
- Piacente F, Bernardi C, Marin Met al. Characterization of a UDP-N-acetylglucosamine biosynthetic pathway encoded by the giant DNA virus Mimivirus. Glycobiology. 2014a;24:51–61. [DOI] [PubMed] [Google Scholar]
- Piacente F, De Castro C, Jeudy Set al. Giant virus Megavirus chilensis encodes the biosynthetic pathway for uncommon acetamido sugars. J Biol Chem. 2014b;289:24428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacente F, Gaglianone M, Laugieri MEet al. The autonomous glycosylation of large DNA viruses. Int J Mol Sci. 2015;16:29315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacente F, Marin M, Molinaro Aet al. Giant DNA virus mimivirus encodes pathway for biosynthesis of unusual sugar 4-amino-4,6-dideoxy-D-glucose (Viosamine). J Biol Chem. 2012;287:3009–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plugge B, Gazzarrini S, Nelson Met al. A potassium channel protein encoded by chlorella virus PBCV-1. Science. 2000;287:1641–4. [DOI] [PubMed] [Google Scholar]
- Priet S, Lartigue A, Debart Fet al. mRNA maturation in giant viruses: variation on a theme. Nucleic Acids Res. 2015;43:3776–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkarev A, Inoue K, Larom Set al. A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature. 2018;558:595–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MJ, Haller SL, Stoian AMMet al. LINE-1 retrotransposons facilitate horizontal gene transfer into poxviruses. eLife. 2022;11. 10.7554/eLife.63327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert Cet al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–50. [DOI] [PubMed] [Google Scholar]
- Riera E, García-Belmonte R, Madrid Ret al. African swine fever virus ubiquitin-conjugating enzyme pI215L inhibits IFN-I signaling pathway through STAT2 degradation. Front Microbiol. 2022;13:1081035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigou S, Santini S, Abergel Cet al. Past and present giant viruses diversity explored through permafrost metagenomics. Nat Commun. 2022;13:5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues RAL, Abrahão JS, Drumond BPet al. Giants among larges: how gigantism impacts giant virus entry into amoebae. Curr Opin Microbiol. 2016;31:88–93. [DOI] [PubMed] [Google Scholar]
- Rodrigues RAL, Arantes TS, Oliveira GPet al. The complex nature of Tupanviruses. Adv Virus Res. 2019;103:135–66. [DOI] [PubMed] [Google Scholar]
- Rolland C, Andreani J, Sahmi-Bounsiar Det al. Clandestinovirus: a giant virus with chromatin proteins and a potential to manipulate the cell cycle of its host Vermamoeba vermiformis. Front Microbiol. 2021;12:715608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwasser S, Mausz MA, Schatz Det al. Rewiring host lipid metabolism by large viruses determines the fate of Emiliania huxleyi, a bloom-forming alga in the ocean. Plant Cell. 2014;26:2689–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwasser S, Ziv C, Creveld SGVet al. Virocell metabolism: metabolic innovations during host-virus interactions in the ocean. Trends Microbiol. 2016;24:821–32. 10.1016/j.tim.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Rottner K, Stradal TEB. Poxviruses taking a ride on actin: new users of known hardware. Cell Host Microbe. 2009;6:497–9. [DOI] [PubMed] [Google Scholar]
- Rozenberg A, Inoue K, Kandori Het al. Microbial rhodopsins: the last two decades. Annu Rev Microbiol. 2021;75:427–47. [DOI] [PubMed] [Google Scholar]
- Rozenberg A, Oppermann J, Wietek Jet al. Lateral gene transfer of anion-conducting channelrhodopsins between green algae and giant viruses. Curr Biol. 2020;30:4910–20. [DOI] [PubMed] [Google Scholar]
- Russo JJ, Bohenzky RA, Chien MCet al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci USA. 1996;93:14862–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-García FJ, Pérez-Hernández CA, Rodríguez-Murillo Met al. The role of tricarboxylic acid cycle metabolites in viral infections. Front Cell Infect Microbiol. 2021;11:725043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satheshkumar PS, Anton LC, Sanz Pet al. Inhibition of the ubiquitin-proteasome system prevents vaccinia virus DNA replication and expression of intermediate and late genes. J Virol. 2009;83:2469–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D, Rosenwasser S, Malitsky Set al. Communication via extracellular vesicles enhances viral infection of a cosmopolitan alga. Nat Microbiol. 2017;2:1485–92. [DOI] [PubMed] [Google Scholar]
- Schatz D, Schleyer G, Saltvedt MRet al. Ecological significance of extracellular vesicles in modulating host-virus interactions during algal blooms. ISME J. 2021;15:3714–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleyer G, Kuhlisch C, Ziv Cet al. Lipid biomarkers for algal resistance to viral infection in the ocean. Proc Natl Acad Sci USA. 2023;120:e2217121120. 10.1073/pnas.2217121120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleyer G, Shahaf N, Ziv Cet al. In plaque-mass spectrometry imaging of a bloom-forming alga during viral infection reveals a metabolic shift towards odd-chain fatty acid lipids. Nat Microbiol. 2019;4:527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Schaulies J, Schneider-Schaulies S. Sphingolipids in viral infection. Biol Chem. 2015;396:585–95. [DOI] [PubMed] [Google Scholar]
- Schönrich G, Abdelaziz MO, Raftery MJ. Herpesviral capture of immunomodulatory host genes. Virus Genes. 2017;53:762–73. [DOI] [PubMed] [Google Scholar]
- Schulz F, Abergel C, Woyke T. Giant virus biology and diversity in the era of genome-resolved metagenomics. Nat Rev Micro. 2022;20:721–36. [DOI] [PubMed] [Google Scholar]
- Schulz F, Alteio L, Goudeau Det al. Hidden diversity of soil giant viruses. Nat Commun. 2018;9:4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz F, Roux S, Paez-Espino Det al. Giant virus diversity and host interactions through global metagenomics. Nature. 2020;578:432–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz F, Yutin N, Ivanova NNet al. Giant viruses with an expanded complement of translation system components. Science. 2017;356:82–5. [DOI] [PubMed] [Google Scholar]
- Schvarcz CR, Steward GF. A giant virus infecting green algae encodes key fermentation genes. Virology. 2018;518:423–33. [DOI] [PubMed] [Google Scholar]
- Shah N, Hülsmeier AJ, Hochhold Net al. Exposure to mimivirus collagen promotes arthritis. J Virol. 2014;88:838–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh S, Pánek T, Gahura Oet al. A novel group of dynamin-related proteins shared by eukaryotes and giant viruses is able to remodel mitochondria from within the matrix. Mol Biol Evol. 2023;40:msad134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Abreu IA, Cabelli DEet al. Superoxide dismutases and superoxide reductases. Chem Rev. 2014;114:3854–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short SM, Suttle CA. Sequence analysis of marine virus communities reveals that groups of related algal viruses are widely distributed in nature. Appl Environ Microb. 2002;68:1290–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Chatterjee A, Kondabagil K. The number of genes encoding repeat domain-containing proteins positively correlates with genome size in amoebal giant viruses. Virus Evol. 2018;4:vex039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva LKDS, Andrade ACDSP, Dornas FPet al. Cedratvirus getuliensis replication cycle: an in-depth morphological analysis. Sci Rep. 2018;8:4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siotto F, Martin C, Rauh Oet al. Viruses infecting marine picoplancton encode functional potassium ion channels. Virology. 2014;466-467:103–11. [DOI] [PubMed] [Google Scholar]
- Smith GL, Chan YS, Howard ST. Nucleotide sequence of 42 kbp of vaccinia virus strain WR from near the right inverted terminal repeat. J Gen Virol. 1991;72. 10.1099/0022-1317-72-6-1349. [DOI] [PubMed] [Google Scholar]
- Sokolova ML, Misovetc I, Severinov KV. Multisubunit RNA polymerases of jumbo bacteriophages. Viruses. 2020;12. 10.3390/v12101064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorouri M, Chang T, Hancks DC. Mitochondria and viral infection: advances and emerging battlefronts. mBio. 2022;13:e0209621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorouri M, Chang T, Jesudhasan Pet al. Signatures of host-pathogen evolutionary conflict reveal MISTR-A conserved MItochondrial STress Response network. PLoS Biol. 2020;18:e3001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza F, Rodrigues R, Reis Eet al. In-depth analysis of the replication cycle of Orpheovirus. Virol J. 2019;16:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speciale I, Duncan GA, Unione Let al. The -glycan structures of the antigenic variants of chlorovirus PBCV-1 major capsid protein help to identify the virus-encoded glycosyltransferases. J Biol Chem. 2019;294:5688–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speciale I, Notaro A, Abergel Cet al. The astounding world of glycans from giant viruses. Chem Rev. 2022;122:15717–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling KM, Schwantes A, Schnierle BSet al. The highly conserved orthopoxvirus 68k ankyrin-like protein is part of a cellular SCF ubiquitin ligase complex. Virology. 2008;374:234–9. [DOI] [PubMed] [Google Scholar]
- Suárez C, Welsch S, Chlanda Pet al. Open membranes are the precursors for assembly of large DNA viruses. Cell Microbiol. 2013;15:1883–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K, Behringer DC, Bojko Jet al. A new family of DNA viruses causing disease in crustaceans from diverse aquatic biomes. mBio. 2020;11. 10.1128/mBio.02938-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumbria D, Berber E, Mathayan Met al. Virus infections and host metabolism—can we manage the interactions?. Front Immunol. 2021;11. 10.3389/fimmu.2020.594963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA, Chen F. Mechanisms and rates of decay of marine viruses in seawater. Appl Environ Microb. 1992;58:3721–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons JA, Alcamí A, Smith GL. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–60. [DOI] [PubMed] [Google Scholar]
- Takemura M, Yokobori S-I, Ogata H. Evolution of eukaryotic DNA polymerases via interaction between cells and large dna viruses. J Mol Evol. 2015;81:24–33. [DOI] [PubMed] [Google Scholar]
- Takemura M. Medusavirus ancestor in a proto-eukaryotic cell: updating the hypothesis for the viral origin of the nucleus. Front Microbiol. 2020;11:571831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Armache K-J, Henikoff S. Viral histones: pickpocket's prize or primordial progenitor?. Epigenetics Chromatin. 2022;15:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MP, Koyuncu OO, Enquist LW. Subversion of the actin cytoskeleton during viral infection. Nat Rev Micro. 2011;9:427–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale A, Campbell S, Van Buuren Net al. Orthopoxviruses require a functional ubiquitin-proteasome system for productive replication. J Virol. 2009;83:2099–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh MLT, Turner PV, Evans DH. Tumorigenic poxviruses up-regulate intracellular superoxide to inhibit apoptosis and promote cell proliferation. J Virol. 2005;79:5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh MLT, Walasek PJ, Evans DH. Leporipoxvirus Cu,Zn-superoxide dismutase (SOD) homologs are catalytically inert decoy proteins that bind copper chaperone for SOD. J Biol Chem. 2003;278:33175–84. [DOI] [PubMed] [Google Scholar]
- Thaker SK, Ch'ng J, Christofk HR. Viral hijacking of cellular metabolism. BMC Biol. 2019;17:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas V, Bertelli C, Collyn Fet al. Lausannevirus, a giant amoebal virus encoding histone doublets. Environ Microbiol. 2011;13:1454–66. [DOI] [PubMed] [Google Scholar]
- Tulman ER, Afonso CL, Lu Zet al. The genome of canarypox virus. J Virol. 2004;78:353–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton C, Mossman K, McFadden G. Encoding of a homolog of the IFN-gamma receptor by myxoma virus. Science. 1992;258:1369–72. [DOI] [PubMed] [Google Scholar]
- Valencia-Sánchez MI, Abini-Agbomson S, Wang Met al. The structure of a virus-encoded nucleosome. Nat Struct Mol Biol. 2021;28:413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten JL, Agarkova I, Dunigan DDet al. Chloroviruses have a sweet tooth. Viruses. 2017;9. 10.3390/v9040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten JL, Agarkova IV, Dunigan DD. Chloroviruses. Viruses. 2019;12. 10.3390/v12010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten JL. Unusual life style of giant chlorella viruses. Annu Rev Genet. 2003;37:153–95. [DOI] [PubMed] [Google Scholar]
- Vardi A, Van Mooy BAS, Fredricks HFet al. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science. 2009;326:861–5. [DOI] [PubMed] [Google Scholar]
- Vastag L, Koyuncu E, Grady SLet al. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 2011;7:e1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalta A, Schmitt A, Estrozi LFet al. The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield. eLife. 2022;11. 10.7554/eLife.77607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Naghavi MH. Exploitation of cytoskeletal networks during early viral infection. Trends Microbiol. 2019;27:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhang D, Orchard RCet al. Norovirus MLKL-like protein initiates cell death to induce viral egress. Nature. 2023;616:152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IN, Li Y, Que Qet al. Evidence for virus-encoded glycosylation specificity. Proc Natl Acad Sci USA. 1993;90:3840–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick-Dugdale J, Buchholz HH, Allen MJet al. Host-hijacking and planktonic piracy: how phages command the microbial high seas. Virol J. 2019;16:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinheimer AR, Aylward FO. A distinct lineage of Caudovirales that encodes a deeply branching multi-subunit RNA polymerase. Nat Commun. 2020;11:4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinheimer AR, Aylward FO. Infection strategy and biogeography distinguish cosmopolitan groups of marine jumbo bacteriophages. ISME J. 2022;16:1657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weynberg KD, Allen MJ, Ashelford Ket al. From small hosts come big viruses: the complete genome of a second Ostreococcus tauri virus, OtV-1. Environ Microbiol. 2009;11:2821–39. [DOI] [PubMed] [Google Scholar]
- Weynberg KD, Allen MJ, Gilg ICet al. Genome sequence of Ostreococcus tauri virus OtV-2 throws light on the role of picoeukaryote niche separation in the ocean. J Virol. 2011;85:4520–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickenhagen A, Sugrue E, Lytras Set al. A prenylated dsRNA sensor protects against severe COVID-19. Science. 2021;374:eabj3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WH, Gilg IC, Moniruzzaman Met al. Genomic exploration of individual giant ocean viruses. ISME J. 2017;11:1736–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WH, Schroeder DC, Allen MJet al. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309:1090–2. [DOI] [PubMed] [Google Scholar]
- Wilson WH, Van Etten JL, Allen MJ. The Phycodnaviridae: the story of how tiny giants rule the world. Curr Top Microbiol Immunol. 2009;328:1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Fukuda T, Tamura Ket al. Expression of the gene encoding a translational elongation factor 3 homolog of Chlorella virus CVK2. Virology. 1993;197:742–50. [DOI] [PubMed] [Google Scholar]
- Yau S, Lauro FM, DeMaere MZet al. Virophage control of antarctic algal host-virus dynamics. Proc Natl Acad Sci USA. 2011;108:6163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa G, Blanc-Mathieu R, Song Cet al. Medusavirus, a novel large DNA virus discovered from hot spring water. J Virol. 2019;93. 10.1128/JVI.02130-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Colson P, Raoult Det al. Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol J. 2013;10:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Koonin EV. Hidden evolutionary complexity of Nucleo-Cytoplasmic Large DNA viruses of eukaryotes. Virol J. 2012a;9:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Koonin EV. Proteorhodopsin genes in giant viruses. Biol Direct. 2012b;7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabelskii D, Alekseev A, Kovalev Ket al. Viral rhodopsins 1 are an unique family of light-gated cation channels. Nat Commun. 2020;11. 10.1038/s41467-020-19457-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zade A, Sengupta M, Kondabagil K. Extensive in silico analysis of Mimivirus coded Rab GTPase homolog suggests a possible role in virion membrane biogenesis. Front Microbiol. 2015;6:929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Takemura M, Murata Ket al. “Mamonoviridae”, a proposed new family of the phylum Nucleocytoviricota. Arch Virol. 2023;168:80. [DOI] [PubMed] [Google Scholar]
- Zhou S, Butler-Laporte G, Nakanishi Tet al. A Neanderthal OAS1 isoform protects individuals of European ancestry against COVID-19 susceptibility and severity. Nat Med. 2021;27:659–67. [DOI] [PubMed] [Google Scholar]
- Ziv C, Malitsky S, Othman Aet al. Viral serine palmitoyltransferase induces metabolic switch in sphingolipid biosynthesis and is required for infection of a marine alga. Proc Natl Acad Sci USA. 2016;113:E1907–16. [DOI] [PMC free article] [PubMed] [Google Scholar]