ABSTRACT
A double ampC (AmpCG183D) and ampD (AmpDH157Y) genes mutations have been identified by whole genome sequencing in a Pseudomonas aeruginosa (PaS) that became resistant (PaR) in a patient treated by ceftolozane/tazobactam (C/T). To precisely characterize the respective contributions of these mutations on the decreased susceptibility to C/T and on the parallel increased susceptibility to imipenem (IMI), mutants were generated by homologous recombination in PAO1 reference strain (PAO1- AmpCG183D, PAO1-AmpDH157Y, PAO1-AmpCG183D/AmpDH157Y) and in PaR (PaR-AmpCPaS/AmpDPaS). Sequential time-kill curve experiments were conducted on all strains and analyzed by semi-mechanistic PKPD modeling. A PKPD model with adaptation successfully described the data, allowing discrimination between initial and time-related (adaptive resistance) effects of mutations. With PAO1 and mutant-derived strains, initial EC50 values increased by 1.4, 4.1, and 29-fold after AmpCG183D , AmpDH157Y and AmpCG183D/AmpDH157Y mutations, respectively. EC50 values were increased by 320, 12.4, and 55-fold at the end of the 2 nd experiment. EC50 of PAO1-AmpCG183D/AmpDH157Y was higher than that of single mutants at any time of the experiments. Within the PaR clinical background, reversal of AmpCG183D, and AmpDH157Y mutations led to an important decrease of EC50 value, from 80.5 mg/L to 6.77 mg/L for PaR and PaR-AmpCPaS/AmpDPaS, respectively. The effect of mutations on IMI susceptibility mainly showed that the AmpCG183D mutation prevented the emergence of adaptive resistance. The model successfully described the separate and combined effect of AmpCG183D and AmpDH157Y mutations against C/T and IMI, allowing discrimination and quantification of the initial and time-related effects of mutations. This method could be reproduced in clinical strains to decipher complex resistance mechanisms.
KEYWORDS: Pseudomonas aeruginosa, ceftolozane/tazobactam, adaptive resistance, ampC, betalactamase, ampD, antibiotic resistance
INTRODUCTION
Multi-drug resistant (MDR) Pseudomonas aeruginosa resistant to carbapenems are among the critical priority pathogens list of WHO (1). β-lactams (BL) are the main therapeutic class used against these MDR bacteria, mostly associated with a β-lactamase inhibitor (BLI). Ceftolozane/tazobactam (C/T) is a BL/BLI with preserved efficacy on MDR P. aeruginosa (2, 3). Yet P. aeruginosa resistance to C/T has been observed in patients on several occasions (4 – 11), mainly due to mutations in ampC gene associated with mutations in ampC regulators, such as ampR and/or ampD (4, 8, 11). However, the identification of a particular mutation hardly predicts the precise reduction of antibiotic susceptibility especially since the same ampC mutation can induce increased resistance to C/T but restore the susceptibility to imipenem (IMI) (12). Furthermore P. aeruginosa resistance development may arise from multiple factors [mutations in chromosomal resistance gene(s), single nucleotide polymorphisms or deletions, horizontal acquisitions of resistance gene(s)] (13), and take several forms (acquired or adaptive resistance) that cannot be discriminated by minimum inhibitory concentration (MIC) determinations (14). To cope with that issue, semi-mechanistic PKPD modeling approaches have been developed to consider the full time-course of bacterial growth, killing, and emergence of resistance in response to different antibiotic exposure profiles (15).
We have detected a resistance to C/T that appeared during treatment in a clinical isolate of P. aeruginosa, after mutations in ampC and ampD. Thus, the objective of this study was to characterize the decreased susceptibility to C/T induced by those mutations, using P. aeruginosa PAO1 reference and mutant-derived strains as well as semi-mechanistic PKPD modeling, to capture the effect of each mutation on acquired and adaptive resistance. In parallel, the developed approach has been used to characterize the effect of these mutations on IMI susceptibility recovery.
RESULTS
Characterization of clinical isolates and mutant-derived strains
The whole genome sequencing (WGS) analysis confirmed that the MDR IMI-resistant P. aeruginosa, susceptible to C/T (PaS) and the C/T resistant but IMI susceptible P. aeruginosa (PaR) clinical isolates belonged to the same sequence type ST252 and were isogenic. Analysis of Single Nucleotide Polymorphisms (SNPs) has shown around 300 SNPs in PaR compared to PaS (Table S1). This led to the identification of two mutations with possible implication in resistance to C/T: a C548A nucleotide substitution, leading to a G183D substitution in the chromosomal cephalosporinase (AmpC), and a C469T nucleotide mutation, leading to a H157Y substitution in AmpD, an N-acetyl-anhydromuramyl-L-alanine-amidase that negatively controls expression of ampC gene.
To decipher the role of each mutation on C/T resistance, two PAO1 strains with single mutation (PAO1-AmpCG183D and PAO1-AmpDH157Y) and one PAO1 strain with double mutations (PAO1-AmpCG183D/AmpDH157Y) were obtained. An increase in basal ampC expression was observed in the isogenic PAO1 mutant strain carrying the AmpDH157Y mutation (Fig. S1).
C/T MIC values of PAO1-AmpCG183D and PAO1-AmpDH157Y were identical to that of PAO1 (MIC = 0.25 mg/L), while that of the PAO1 double mutant (PAO1-AmpCG183D/AmpDH157Y) was increased eightfold (MIC = 2 mg/L). A double reverted PaR isolate (PaR-AmpCPaS/AmpDPaS) was obtained with a MIC close to that of PaS (MIC = 1 mg/L) and considerably reduced compared with that of PaR (MIC = 2 mg/L versus 32 mg/L) (Table 1).
TABLE 1.
Minimal inhibitory concentrations (mg/L) and EC50 of C/T at the start (EC50,off) of the first TKC and at the end (EC50,on) of the second TKC (mg/L) of PAO1 and mutant-derived strains and of clinical pair of P. aeruginosa (PaS and PaR), and isogenic mutant-derived PaR a
| MIC | EC50,off [CI 95%] c | EC50,on [CI 95%] | EC50,on/EC50,off | |
|---|---|---|---|---|
| (mg/L) | mg/L | mg/L | ||
| Mutant-derived reference strains | ||||
| PAO1 | 0.25 | 0.0444 [0.00818–0.104] |
0.551 [0.408–0.743] |
12.4 |
| PAO1- AmpCG183D | 0.25 | 0.0606 [0.0294–0.0988] (1.4 b ) |
19.4 [15.8–23.4] |
320 |
| PAO1- AmpDH157Y | 0.25 | 0.181 [0.121–0.263] (4.1 b ) |
2.20 [1.83–2.59] |
12.2 |
| PAO1- AmpCG183D/AmpDH157Y | 2 | 1.29 [0.499–2.55] (29 b ) |
70.8 [60.5–83.1] |
55 |
| Mutant-derived clinical isolates | ||||
| PaS | 1 | 1.36 [1.04–1.73] |
155 [124–92] |
114 |
| PaR | 32 d |
80.5
[66.4–92] (59 b ) |
420 [371–485] |
5.2 |
| PaR-AmpCPaS/AmpDPaS | 2 |
6.77
[5.37–8.17] (5.0 b ) |
269 [220–345] |
40 |
Results in italics should be interpreted with caution, as the PaR strain was previously exposed to C/T, thus could be already adapted at the start of the first TKC.
fold-change compared to initial strain (PAO1 or PaS).
CI 95%, Confidence intervals 95%.
In Bold: resistant considering EUCAST Breakpoint of 4 mg/L.
Concerning IMI, the clinical isolate PaS was resistant to IMI with a MIC of 4 mg/L, whereas the clinical isolate PaR was susceptible with a MIC value of 1 mg/L. MICs of PAO1 and PAO1-AmpDH157Y to IMI were 0.5 mg/L. The values of MIC for strains carrying the AmpCG183D mutation (PAO1-AmpCG183D and PAO1-AmpCG183D/AmpDH157Y) were decreased to 0.125 mg/L (Table 2).
TABLE 2.
Minimal inhibitory concentrations (mg/L) and EC50 of IMI at the start (EC50,off) of the first TKC and at the end (EC50,on) of the second TKC (mg/L) of PAO1 and mutant-derived strains
| MIC | EC50,off [CI 95%] b | EC50,on [CI 95%] | EC50,on/EC50,off | |
|---|---|---|---|---|
| (mg/L) | mg/L | mg/L | ||
| Mutant-derived reference strains | ||||
| PAO1 | 0.5 | 0.411 [0.250–0.638] |
24.8 [21.3–29.3] |
60.3 |
| PAO1- AmpCG183D | 0.125 | 1.86 [1.49–2.26] (4.5 a ) |
2.25 [1.91–2.57] |
1.2 |
| PAO1- AmpDH157Y | 0.5 | 2.03 [1.58–2.64] (4.9 a ) |
7.68 [4.62–11.1] |
3.8 |
| PAO1- AmpCG183D/AmpDH157Y | 0.125 | 0.495 [0.397–0.603] (1.2 a ) |
3.29 [2.84–3.80] |
6.6 |
fold-change compared to initial strain PAO1.
CI 95%, Confidence intervals 95%.
PKPD modeling
No degradation of C/T was observed in MHB. IMI degradation followed a mono-exponential decay with degradation half-life t1/2= 16.4 h (data not shown) comparable with 17 hr which was found by Yadav et al. in the same experimental conditions (16).
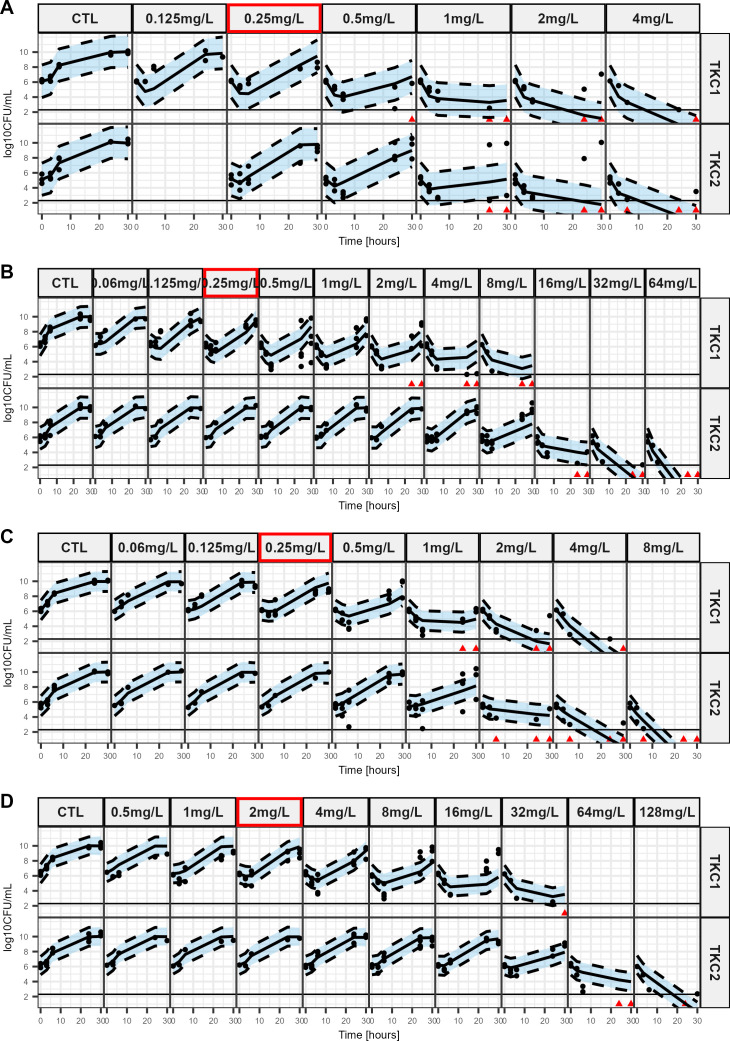
C/T experimental data are represented as dots in Fig. 1 for PAO1 mutant-derived strains and on Fig. S2 for clinical isolates. For PAO1 mutant-derived strains and clinical isolates, the first time-kill curves (TKCs) showed bactericidal effect of C/T for concentrations over their corresponding MIC. Initial CFU decreased followed by a regrowth occurred for all mutant strains until the concentration corresponding to MIC in the first time-kill but for higher concentration in the second time-kill, particularly for PAO1-AmpCG183D (Fig. 1B) and the double mutant (Fig. 1D).
Fig 1.
Visual predictive checks of the final model based on in vitro sequential time kill data of ceftolozane/tazobactam for PAO1 and mutant-derived strains. The plots show the observed time-kill data at different C/T concentrations with model predictions as medians and 80% confidence intervals around the median (shaded area). The red frame represents the MIC value of the corresponding strain. Dots represent countable plates; red triangles represent data below the limit of quantification (BLQ: 2.3 log10 CFU/mL). Data below the limit of detection are plotted as 0.1 CFU/mL. (A). PAO1 (B). PAO1-AmpCG183D, (C). PAO1- AmpDH157Y, (D). PAO1-AmpCG183D/AmpDH157Y.
For all strains, killing and regrowth were well described by the model (Fig. 1; Fig. S2), and susceptibilities to C/T are reflected by EC50 values before (EC50,off, 2nd column) and after (EC50,on, 3rd column) exposure to C/T (Table 1).
For PAO1 mutant-derived strains, AmpDH157Y mutation induced acquired resistance, with a 4.1-fold increase in EC50,off compared to PAO1 (Table 1), while the AmpCG183D mutation had a similar EC50,off than PAO1 with a 1.4-fold increase and an overlapping 95% confidence interval (CI). But AmpCG183D/AmpDH157Y double mutation had the greatest effect with an EC50,off value increased by 29-fold. In terms of adaptive resistance, PAO1 EC50,on was 12.4-fold higher than its corresponding EC50,off. This ratio between EC50,on and EC50,off was virtually unchanged (12.2) after AmpDH157Y mutation and tremendously increased (320-fold) after AmpCG183D mutation, but intermediate (55) after AmpCG183D/AmpDH157Y double mutation.
For clinical isolates, EC50,off value increased from 1.36 mg/L for PaS to 80.5 mg/L for PaR corresponding to a 59-fold difference and decreased to 6.77 mg/L for the double reverted PaR mutant. PaS EC50,on was estimated at 155 mg/L, that is 114-fold higher than EC50,off.
Other modeling parameters values for C/T are provided in Table S2 and S3.
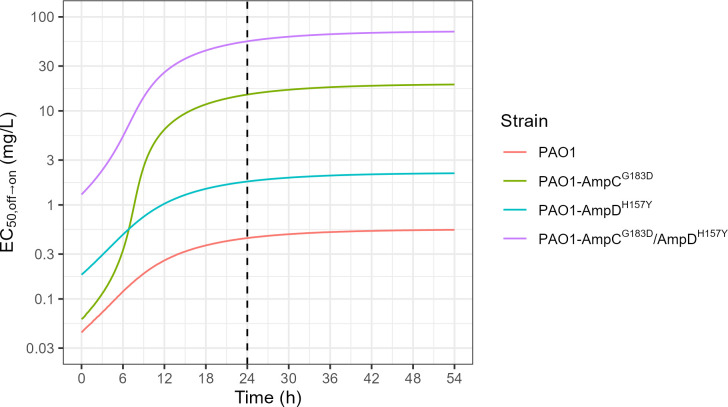
The effect of time on PAO1 and mutant-derived strains susceptibility to C/T (EC50, off→on), reflecting adaptive resistance development, is illustrated in Fig. 2. Simulations showed that at any time, for C/T, EC50, off→on of PAO1 is always the lowest, whereas that of PAO1-AmpCG183D/AmpDH157Y is always the highest. The PAO1-AmpCG183D curve starts at almost the same value than that of PAO1 and increase rapidly from 6 hours until to reach a plateau. The PAO1-AmpDH157Y curve evolves parallel to that of double mutant strain.
Fig 2.
EC50,off→on over time simulated from the final model based on the C/T in vitro sequential time-kill data. Time is defined as the time since the beginning of the first time-kill. Each colored line shows the empirical EC50 for a different strain. Dashed vertical line shows time = 24 hours which is the time at which samples from the first time-kill are taken to prepare the inoculum of the second time-kill.
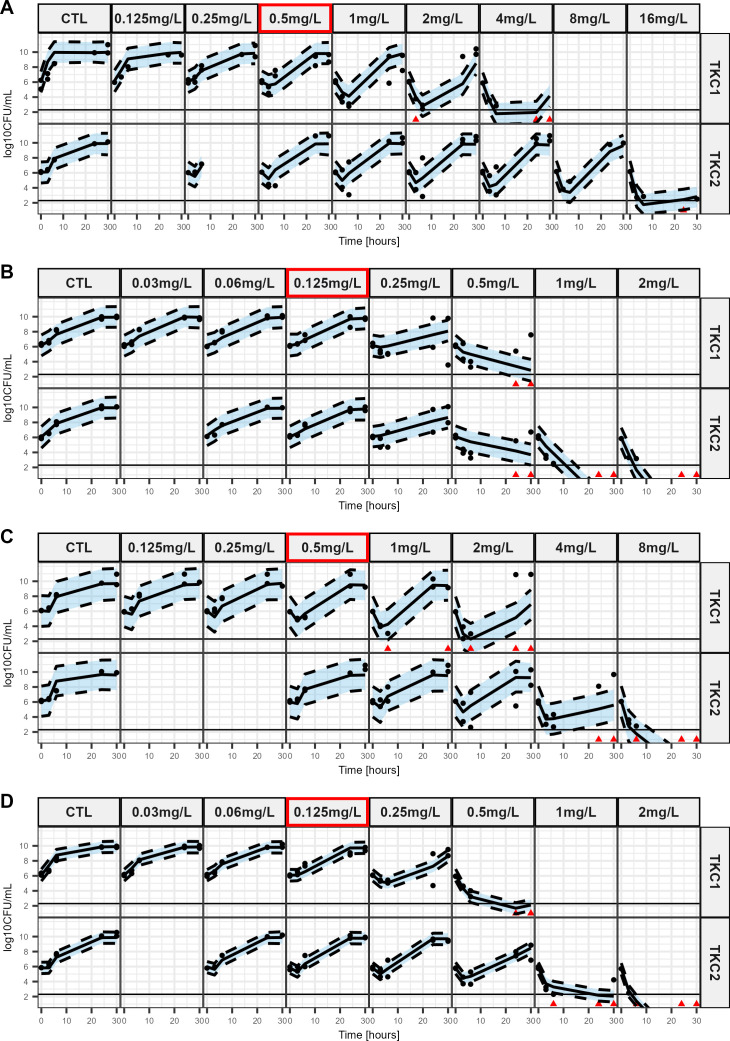
IMI experimental data for PAO1 and mutant-derived strains are represented as dots in Fig. 3. For all strains, initial CFU decrease followed by a regrowth occurred in the first time-kill for concentration close to (PAO1-AmpCG183D, Fig. 3B and the double mutant, Fig. 3D) or slightly higher (PAO1, Fig. 3A and PAO1-AmpDH157Y, Fig. 3C) than the corresponding MIC. In the second time-kill, regrowths at higher concentrations are mainly observed for PAO1. Killing and regrowth were well described by the model (Fig. 3), and susceptibilities to IMI are reflected by EC50 values before (EC50,off, 2nd column) and after (EC50,on, 3rd column) exposure to IMI (Table 2).
Fig 3.
Visual predictive checks of the final model based on in vitro sequential time-kill data of imipenem for PAO1 and mutant-derived strains. The plots show the observed time-kill data at different IMI concentrations with model predictions as medians and 80% confidence intervals around the median (shaded area). The red frame represents the MIC value of the corresponding strain. Dots represent countable plates; red triangles represent data below the limit of quantification (BLQ: 2.3 log10 CFU/mL). Data below the limit of detection are plotted as 0.1 CFU/mL. (A) PAO1 (B) PAO1-AmpCG183D, (C) PAO1- AmpDH157Y, (D) PAO1-AmpCG183D/AmpDH157Y.
For PAO1 and mutant-derived strains, AmpCG183D and AmpDH157Y mutations alone induced greater acquired resistance than AmpCG183D/AmpDH157Y double mutation, with respectively a 4.5, 4.9, and 1.2-fold (with an overlap in 95% CI) increase in EC50,off compared to PAO1 (Table 2). In terms of adaptive resistance, PAO1 EC50,on was 60.3-fold higher than its corresponding EC50,off while it increased a lot less for the mutant-derived strains. Notably, strains with the AmpCG183D mutation have markedly lower EC50,on than PAO1 (2.25 and 3.29 mg/L vs 24.8 mg/L) and slightly lower than PAO1-AmpDH157Y (2.25 and 3.29 mg/L vs 7.68 mg/L). Other model parameters values for IMI are provided in Table S4.
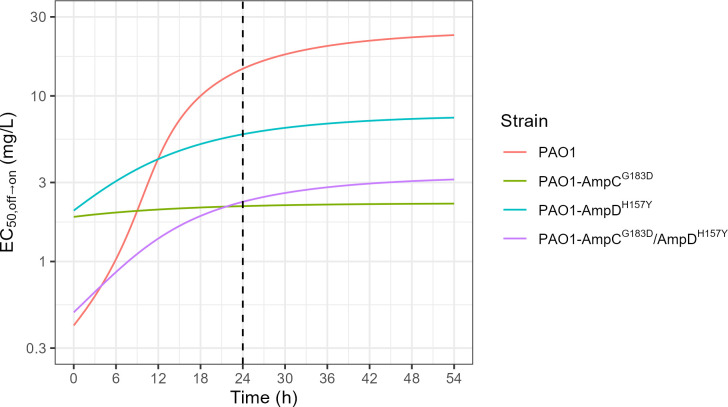
The effect of time on PAO1 and mutant-derived strains susceptibility to IMI, reflecting adaptive resistance development, is illustrated in Fig. 4. Simulations showed that the PAO1 curve increased with time to reach the highest EC50,off→on value at 54 hours. PAO1-AmpCG183D curve is almost flat and the PAO1-AmpDH157Y and the PAO1-AmpCG183D/AmpDH157Y double mutant curves are almost parallel.
Fig 4.
EC50,off→on over time simulated from the final model based on the imipenem in vitro sequential time-kill data. Time is defined as the time since the beginning of the first time-kill. Each colored line shows the empirical EC50 for a different strain. Dashed vertical line shows time = 24 hours which is the time at which samples from the first time-kill are taken to prepare the inoculum of the second time-kill.
Moreover, the EC50, off→on at 24 hours of PAO1 and mutant-derived strains were consistent with MIC values (Fig. 4 and Table 2) with higher EC50, off→on for PAO1 and PAO1-AmpDH157Y than for PAO1-AmpCG183D and the PAO1-AmpCG183D/AmpDH157Y double mutant strain.
DISCUSSION
AmpCG183D and AmpDH157Y mutations detected in PaR, and presumably responsible for C/T resistance associated with recovery of IMI susceptibility, were reproduced in a reference PAO1 strain to investigate specifically their consequences on C/T and IMI resistance variability. Mutants were generated using homologous recombination, which has the advantages over other commonly used methods, of being scarless and reflecting the gene function in its native context (17). Semi-mechanistic PKPD modeling was conducted from sequential TKC data to facilitate the selection of a model (18), and allow precise quantitative discrimination between acquired resistance due to mutation and consecutive adaptive resistance (19, 20).
AmpCG183D mutation induced limited, if any, acquired resistance of PAO1 to C/T (EC50,off increased by only 1.4-fold). This mutation was first described in vitro (4) and later in vivo after treatment by C/T (8, 21, 22). Its effect on C/T resistance has been investigated by using an expression plasmid harboring AmpCG183D in a PAO1 reference strain, knocked-out for ampC (4), and reporting high-level resistance to C/T (MIC = 32 mg/L). Yet pUC plasmids are high-copy number plasmids, leading to an overexpression of the plasmid genes (23), whereas homologous recombination, which has been used in this study, allows the determination of the effect of AmpCG183D in the native context. Other authors have cloned the ampC gene with its promoter into Escherichia coli to investigate the effect of the AmpCG183D substitution, confirming its role in C/T resistance (8). A recent study (24) showing that this mutation allows more flexibility to AmpC leading to an enhanced enzyme affinity toward ceftolozane could explain the adaptive resistance to C/T observed in the PAO1-AmpCG183D strain (Table 1).
Mutations in AmpD are expected to increase ampC expression, leading to high β-lactamase production (25, 26). Indeed, AmpDH157Y mutation in this study induced a basal overexpression of ampC, confirmed by qPCR experiments on PAO1 mutant-derived strains (Fig. S1). AmpDH157Y mutation was responsible for acquired resistance of PAO1 to C/T, characterized by a 4.1-fold increase of EC50,off. In the present study, initial higher-level expression of ampC may be responsible for a limited initial decrease in susceptibility to C/T. However, this ampC overexpression had no effect on adaptive resistance, as EC50,on/EC50,off ratios were similar between PAO1 and PAO1-AmpDH157Y (Table 1).
More interestingly, the double mutations led to a “synergistic type” of effect on the acquired resistance to C/T, with an EC50,off value for PAO1-AmpCG183D/AmpDH157Y, 29-fold higher than that of PAO1 (1.29 mg/L vs 0.0444 mg/L, respectively). The observation was different for adaptive resistance, which was moderate for the AmpDH157Y mutant (EC50,on/EC50,off = 12) and much higher for the AmpCG183D (EC50,on/EC50,off = 320), but intermediate for PAO1-AmpCG183D/AmpDH157Y (EC50,on/EC50,off = 55). However, the difference in susceptibility to C/T between the PAO1-AmpCG183D/AmpDH157Y and the wild-type PAO1 increased with adaptation as shown by a 128-fold increase of EC50,on (70.8 mg/L and 0.551 mg/L, respectively) compared with 29-fold increase for the EC50,off values. These results highlight previous observations suggesting that high-level C/T resistance was achieved when mutations occurred on both ampC gene and ampC regulators (4).
The combined effects of acquired and adaptive resistance on C/T with time, induced by the various mutations are illustrated in Fig. 2. The PKPD model with adaptation used to describe these experimental data has been initially proposed by Mouton et al. and since that, used on a number of occasions (15, 27, 28). It assumes that adaptation starts at time zero (when 100% of the bacteria are in the “off” state), and keeps developing until the bacterial population becomes fully adapted (100% “on”). Yet, the adaptation development rate constitutes an interesting but neglected information. This issue is complexified here by the presence of two mutations. During adaptation development, when only a continuously changing fraction of the bacterial population is adapted, the bactericidal effect is no more characterized by an Emax model, but by an equation combining two Emax models, one for the “on” and the other for the “off” bacteria (Eq 1). For any given fraction of the bacterial population that is adapted, we could determine the C/T concentration at which 50% of this maximum effect is reached, defined as EC50,off→on. However, as opposed to the EC50 value of an Emax model, this parameter value is not fixed but varies with the fraction of the bacterial population that is adapted (Fig. S3a), which itself varies with time (Fig. S3b). Because of these two driving effects, hence EC50,off→on is time-dependent (Fig. S3c).
In simulations (Fig. 2), The PAO1 and PAO1-AmpDH157Y curves are parallel, consistent with the fact that AmpDH157Y mutation confers initial resistance but then similar adaptive resistance to PAO1. The PAO1-AmpCG183D curve starts at almost the same value than that of PAO1 due to the virtual lack of acquired resistance, but raises very sharply to cross the PAO1-AmpDH157Y curve after about 6 hours. Figure 2 also shows that adaptive resistance develops rapidly in these in vitro conditions, with about 90% of adaptation reached after 24 hours.
In the clinical isolates, only the double mutant was identified after treatment. Yet, results obtained with PaS and PaR are more difficult to interpret than those obtained with PAO1 and mutant-derived strains, since the two types of resistance cannot be differentiated in PaR. Acquired resistance was characterized genotypically, whereas adaptive resistance, likely due to multiple and complex variations of gene expression, could only be characterized indirectly and unspecifically from reduction of antimicrobial efficacy assessed by EC50 determinations. Indeed, PaS had never been exposed to C/T, whereas PaR was recovered after a 2 weeks C/T treatment. Therefore, adaptive resistance may have developed in vivo in PaR. Then, comparing PaR and PaS, EC50,off values (80.5 and 1.36 mg/L) may reflect a mixed effect due to acquired and adaptive resistance. Yet, the relatively low EC50,off value of PaR-AmpCPaS/AmpDPaS (6.77 mg/L), almost back to that of PaS (1.36 mg/L), suggests that this double mutation explains most but not all of PaR resistance to C/T.
In this clinical case, as PaS became PaR during treatment, C/T resistance increased (32-fold MIC increase), while IMI resistance reverted (eightfold MIC reduction). This reversion could be linked with AmpCG183D mutation that has been found in PaR, since it has been previously described to decrease the MIC of IMI (4, 8, 12, 22, 24). This observation was confirmed in our study by a fourfold IMI MIC decrease of PAO1 mutant-derived strains containing the AmpCG183D mutation (Table 2). Simulations (Fig. 4 and Table 2) showed that strains carrying the AmpCG183D mutation are less prone to adaptive resistance with flattened curves and lower EC50,off→on from 24 hours (Fig. 4) compared to strains with a wild-type AmpC. This is consistent with previous studies that have shown AmpC mutations (including AmpCG183D) decrease the affinity to imipenem (4, 8, 12, 22, 24). Regarding the AmpDH157Y mutation, the effect appeared to be less important than AmpCG183D mutations but slows adaptive resistance over time compared to PAO1 illustrated by lower EC50,off→on from the 12th hour (Fig. 4). However, EC50,off→on at time 0 is more difficult to interpret since the strains with single mutations presented a higher EC50,off→on value than PAO1 wild type and PAO1 carrying the double mutation.
Several limitations of the study should be noted. Most of the information provided by this study come from PAO1 and derived strains, which constitutes an optimal tool for precise mechanistic characterizations, but do not represent clinical isolates. Furthermore, data obtained in vitro should always be very carefully extrapolated in clinics. In this study, we focused on the two clinically observed mutations that could be directly related with known mechanisms of resistance to ceftolozane, AmpCG183D and AmpDH157Y. However, while the double reverted mutant PaR-AmpCPaS/AmpDPaS has showed decreased C/T MIC and EC50,off, it was still less susceptible to C/T than PaS. Thus, even if their roles are probably minor, the effect of other mutations among the SNPs observed in PaR compared to PaS cannot be excluded. Likewise, the other mutations present in the genes involved in the regulation or in ampC of the clinical isolates that appeared before C/T treatment were not reproduced in PAO1. For example, both clinical isolates (PaS and PaR) had two other mutations in ampC gene (AmpCT105A and AmpCR233D) and one mutation in the AmpC-repressor AmpR (AmpRG85S) in comparison with PAO1. These mutations could have a cumulative effect with the AmpCG183D and AmpDH157Y mutations identified in the PaR strain after C/T treatment.
PAO1 strains and their mutants exhibited different growth patterns in the C/T and the IMI experiments. Indeed, even in absence of drug a lag-time could be present in one set of experiments but not in the other as evidenced by the Tlag parameter estimates in Table S2 and S4 and the growth curves in Fig. 1 and 3. We could not find and experimental or biological reason for this intriguing phenomenon. However, the goal of the study was to quantify the in vitro difference of effects between the different studied strains rather than mechanistically describe to bacterial growth. Thus, we decided to estimate growth parameters specific to each experiment set, leading to a proper yet overfitted description of the growth data. This enabled us to have a correct quantitative description of drug effect while having the adverse effect of making the model unsuitable for predictions of future experiments before any external validation is conducted.
Another limitation of the study is that due to the limited number (n = 3) of replicates for each drug-bug pair, the observed variability in bacterial response could not be attributed to a specific model parameter but only quantified through the residual unexplained variability term (σ2, Table S2and S4). Notably, this variability parameter is high for C/T against PAO1 (Table S2, 1st column) and C/T against PAO1-AmpDH157Y (Table S4, 3rd column). These high variabilities, reflect the presence of concentrations for which regrowth was observed in some replicates but not in others, e.g., 1 and 2 mg/L for the second TKC of C/T against PAO1 (Fig. 1A). Again, this limits the predictive ability of the model while not invalidating the quantitative description of the experimental data.
In this study, we showed that AmpCG183D AmpDH157Y double mutation in P. aeruginosa was responsible for acquisition of resistance to C/T in a clinical and a laboratory strain. In the same strains, we also showed that AmpCG183D was mainly responsible for reversal of resistance to IMI. Using newly created PAO1 mutants, sequential time-kill curves and PKPD modeling, we were able to precisely quantify the impact of each mutation on acquired and adaptive resistances to C/T and IMI.
MATERIALS AND METHODS
Patient characteristics and clinical isolates
A 58-year-old female patient, under chemotherapy for breast cancer, developed a catheter-related bacteremia. Empirical treatment (IMI 1 g tid plus amikacin 30 mg/kg qd) was started, then replaced 2 days later by C/T (Zerbaxa, MSD, Puteaux, France), 1.5 g tid, after an MDR IMI-resistant P. aeruginosa (MIC of 8 mg/L), susceptible to C/T (PaS) with an MIC equal to 1 mg/L, was isolated. After 2 weeks of treatment, a C/T resistant but IMI susceptible P. aeruginosa (PaR) was isolated from blood cultures with MIC equal to 32 mg/L and 1 mg/L for C/T and IMI, respectively. Patient was eventually treated successfully with imipenem/cilastatin 0.5 g qid for 14 days.
Whole genome sequence analysis of PaS and PaR was conducted by Ion Proton technology (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Quality check of raw reads was done using FastQC v.0.11.5 and MultiQC v.0.9, and no trimming was performed. Mean coverage of 40× was determined after alignment against the P. aeruginosa PAO1 reference genome (GenBank GCA_000006765.1). Reads were then assembled using Mimicking Intelligent Read Assembly software. Contigs were analyzed using Geneious software (Biomatters Ltd., Auckland, New Zealand). Antimicrobial resistance genes were identified using ResFinder 4.0 (29). Genes with a 60% minimum length and a percentage of identity >98% were considered. Sequence types (STs) were determined using the Pseudomonas multilocus sequence typing database (https://cge.cbs.dtu.dk/services/MLST/). SNPs were called against PAO1 reference sequence (GenBank GCA_000006765.1) using freebayes (v.1.3.1) (30).
Construction of ampC/ampD mutant strains by homologous recombination and characterization
All the strains and primers used in the study are presented in Tables S5 and S6. P aeruginosa PAO1 (ATCC 15692) was used as a reference strain for mutant production. Construction of plasmids and induction of AmpC and AmpD mutations was accomplished as previously described (31), with slight modifications. Briefly, 700 base pair sequences of the flanking regions of the targeted mutation were PCR-amplified with primers containing the mutation. All primers were designed with Snapgene software (from Insightful Science; available at snapgene.com). The PCR fragments were gel-purified and inserted into pEXG2-linearized plasmid by Gibson assembly (New England Biolabs, Evry, France). The assembled plasmid was transformed into competent E. coli JKe201 and then plated on Luria Bertani (LB) agar containing 10 mg/L of gentamicin and 100 µM of diaminopimelic acid. Sequenced-verified clones were mated for 4 hours with the PAO1 reference strain at 37°C and then plated on LB agar containing 30 mg/L of gentamicin allowing to select PAO1 that have integrated the plasmid. Colonies were picked and grown in LB for 4 hours and streaked on 20% sucrose plates overnight at 30°C. Targeted mutations in clones were confirmed by Sanger sequencing.
For clinical strains, AmpCG183D and AmpDH157Y mutations found in PaR isolate were reverted by cloning part of the whole ampC and ampD genes from PaS into PaR, using a similar protocol. Seven hundred base pair sequences upstream and downstream of the targeted mutation in ampC or ampD genes were PCR-amplified from the susceptible PaS clinical isolate and then assembled by Gibson assembly into pFOG plasmid (17). Then, the mating step was done with E. coli JKE201 containing pFOG plasmid and PaR resistant isolate. Sanger sequencing was used to confirm the presence of the mutation in each mutant strain.
ampC gene expression analysis
Bacterial suspensions were collected to obtain a maximum inoculum of 109 CFU. Samples were centrifuged (12,000 g, 5 minutes), and the pellets were extracted with the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Bacteria were lysed using 0.1 mm Glass Beads (Scientific industries) for 15 minutes under horizontal shaking. Samples were treated with the TURBO DNA-Free Kit using the rigorous protocol (Thermo Fischer Scientific, Illkirch-Graffenstaden, France) to eliminate residual DNA. The concentration of total RNA was estimated using the Nanodrop One system (Thermo Fischer Scientific, Illkirch-Graffenstaden, France).
Extracted total RNA was used to synthetize cDNA with the FIREScript RT cDNA synthesis kit (Solis Biodyne, Tartu, Estonia) according to the manufacturer’s instructions, using an iCycler PCR system (Bio-Rad, Marnes-la-Coquette, France).
Primers were designed with Geneious Prime software (Biomatters Ltd., Auckland, New Zealand). Quantitative PCR was performed from cDNA with the ONEGreen FAST qPCR Premix (Ozyme, Saint-Cyr-l'École, France) using the CFX96 Touch Real-Time PCR Detection System. All samples were analyzed in duplicate, and the relative expression of genes was normalized by the expression of a reference gene (rpsl). The relative differences in mRNA expression levels were determined using comparative cycle threshold (Ct) method (2-ΔΔCt). The results were analyzed with the CFX Maestro Software (Bio-rad).
Minimum Inhibitory Concentration
Fresh bacteria grown overnight on Muller-Hinton Agar plate were resuspended in cation-adjusted Muller-Hinton broth II (MHBII) (Fluka Biochemika, Sigma-Aldrich, France) to an optical density (600 nm) of 0.1 (corresponding to 1 × 108 CFU/mL). This bacterial suspension was adjusted to 1 × 106 CFU/mL in MHBII. Twofolds geometric dilution of each tested antibiotic were performed in 96-well plates using a pipetting robot (ASSIST Plus, INTEGRA Biosciences AG, Zizers, Switzerland). Bacterial inoculates were added to obtain a final concentration of 5 × 105 CFU/mL. Plates were then incubated 16–20 hours at 35°C +/- 1°C in an ambient air incubator. MIC was determined as the lowest drug concentration inhibiting visible bacterial growth. Results were confirmed by an absorbance measurement at 600 nm with an automatic plate-reader (Infinite 200 Pro, Tecan Trading AG, Männedorf, Switzerland). MICs were performed in triplicate.
Sequential time-kill curve experiment
Bacterial inoculum was prepared by suspension of the bacteria from a 2 hours logarithmic-growth-phase culture in cation-adjusted MHBII, adjusted to a final concentration of 1 × 106 CFU/mL. The experiments were performed in 24-well plates (Greiner Bio-One, Courtaboeuf, France), with an Assist Plus pipetting robot (Integra Biosciences, Zizers, Switzerland). Antibiotics were added to obtain concentrations corresponding to 0.5 × to 16 × MIC for C/T and 0.25 × to 8 × MIC for IMI. To compensate for IMI degradation, (16) doses were added at 9 and 24 hours post inoculation (44% and 35% of the initial dose at 9 and 24 hours, respectively). The 24-well plates were incubated at 35°C for 30 hours under agitation (130 rpm). Bacteria were counted at 0, 3, 6, 24, and 30 hours by agar plating using an automatic plater (easySpiral pro, Interscience, Saint Nom la Bretèche, France). At the start of the experiment, the wells contained 2.2 mL of bacterial suspension, and no less than 1.88 mL after the last sampling at 30 hours. As previously described (18), at 24 hours, a second TKC was started, using bacteria from wells where regrowth was observed in presence of antibiotics. Protocol was identical as the first TKC, except for antibiotics concentration that were increased (up to 64× MIC). The number of CFU was counted after incubation at 35°C for 16 to 20 hours on MH agar plate. Semi-automatic plate reader was used for bacterial count on agar plates (Scan300, Interscience, Saint Nom la Bretèche, France). The limit of quantification was fixed at 200 CFU/mL. At least one growth control, performed without addition of antibiotic, was included in each experiment. All experiments were performed at least three times.
Antibiotic stability assays
Antibiotic assays (HPLC-MSMS) were performed during TKC experiments to ensure stability of C/T and IMI in MHBII. Procedures are detailed in the Text S2.
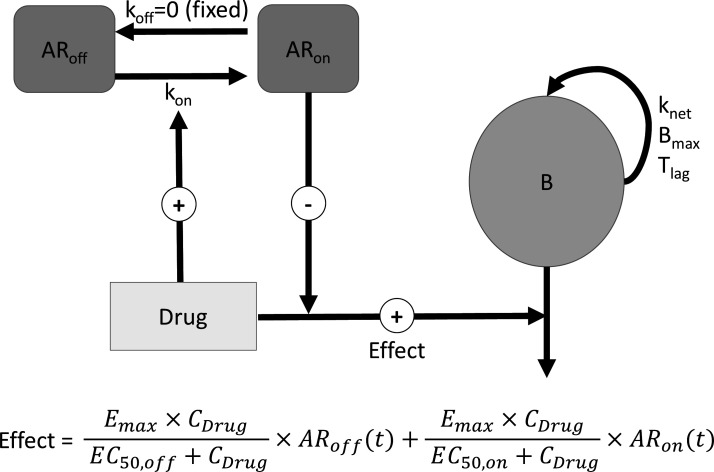
Pharmacodynamic modeling
Data were analyzed using a PKPD model with adaptation (Fig. 5). Drug (C/T or IMI) effect follows Equation 1:
Fig 5.
Schematic representation of the final PKPD model. B: Bacterial; ARon and ARoff: virtual compartments representing the fraction of adapted and non-adapted bacteria, respectively; koff: rate constant for reversal of adaptive resistance; kon: rate constant for development of adaptive resistance; Tlag: growth lag time; Bmax: maximal bacterial density in the system; knet: difference between growth and death of bacteria in the absence of antibiotic; Emax: maximum rate constant for drug effect; EC50,off: drug concentration necessary to reach 50% of Emax when 0% of bacteria are adapted; EC50,on: drug concentration necessary to reach 50% of Emax when 100% of bacteria are adapted. Parameters are defined in Tables S4 to S6 and model equations.
| [1] |
where Emax corresponds to the maximal effect rate constant, CDrug the drug (C/T or IMI) concentration, EC50,off the drug concentration necessary to reach 50% of Emax in non-adapted bacteria, ARoff the proportion of non-adapted bacteria, EC50,on the drug concentration necessary to reach 50% of Emax in adapted bacteria and ARon the proportion of adapted bacteria.
For each strain, the bactericidal effect of both drugs can be described by an Emax model for both the adapted and non-adapted bacteria, with a common Emax but distinct EC50 values (respectively, EC50,off at time zero, and EC50,on at time infinity)
Comparisons of EC50,off values of the various mutants with that of the reference PAO1 characterize the acquired resistance, whereas comparisons between EC50,off and EC50,on for a particular strain reflects adaptive resistance. The fractions of non-adapted ARoff and adapted ARon bacteria as a function of time were computed using the equations 2 and 3,
| [2] |
| [3] |
where kon represents the adaptation rate. To study the change in EC50 value from EC50,off before adaptation starts, to EC50,on when 100% of bacteria are adapted the following procedure was followed: the drug effect at any given time t (effect) was computed for a wide range of concentrations [i.e., CDrug∈ (0,EC50,on)] according to equation (1). The concentration for which Effect = 50% Emax at t was found and called EC50,off→on. The procedure was reproduced for all times between the beginning and the end of TKCs [i.e., t ∈(0,54)] to yield EC50,off→on at all time-points. Details and full differential equations are provided in Text S1.
ACKNOWLEDGMENTS
We would like to thank Lucie Laval, Agnès Audurier and Margaux Thiry for technical assistance in microbiology as well as Christophe Adier for technical assistance in analytics.
Luc Deroche’s PhD was supported by a grant from Institut National de la Santé et de la Recherche Médicale (Inserm) and Conseil Régional Nouvelle-Aquitaine (n°2019-1R30222).
Contributor Information
Julien M. Buyck, Email: julien.buyck@univ-poitiers.fr.
Boudewijn L. de Jonge, Shionogi Inc., Florham Park, New Jersey, USA
DATA AVAILABILITY
The whole-genome sequences of the strains analyzed in this study are available under BioProject accession number PRJNA998736 (strains PaS and PaR).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aac.00480-23.
contains supplement table S1 to S6, supplemental Figure S1 to s3 and supplemental text S1 and S2.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- 2. Juan C, Zamorano L, Pérez JL, Ge Y, Oliver A, Spanish Group for the Study of Pseudomonas, Spanish Network for Research in Infectious Diseases . 2010. Activity of a new antipseudomonal cephalosporin, CXA-101 (FR264205), against carbapenem-resistant and multidrug-resistant Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 54:846–851. doi: 10.1128/AAC.00834-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karlowsky JA, Lob SH, DeRyke CA, Hilbert DW, Wong MT, Young K, Siddiqui F, Motyl MR, Sahm DF. 2022. In vitro activity of ceftolozane-tazobactam, imipenem-relebactam, ceftazidime-avibactam, and comparators against Pseudomonas aeruginosa isolates collected in United States hospitals according to results from the SMART surveillance program, 2018 to 2020. Antimicrob Agents Chemother 66:e0018922. doi: 10.1128/aac.00189-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cabot G, Bruchmann S, Mulet X, Zamorano L, Moyà B, Juan C, Haussler S, Oliver A. 2014. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 58:3091–3099. doi: 10.1128/AAC.02462-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fraile-Ribot PA, Cabot G, Mulet X, Periañez L, Martín-Pena ML, Juan C, Pérez JL, Oliver A. 2018. Mechanisms leading to in vivo ceftolozane/tazobactam resistance development during the treatment of infections caused by MDR Pseudomonas aeruginosa. J Antimicrob Chemother 73:658–663. doi: 10.1093/jac/dkx424 [DOI] [PubMed] [Google Scholar]
- 6. Fraile-Ribot PA, Mulet X, Cabot G, Del Barrio-Tofiño E, Juan C, Pérez JL, Oliver A. 2017. In vivo emergence of resistance to novel cephalosporin-β-lactamase inhibitor combinations through the duplication of amino acid D149 from OXA-2 β-Lactamase (OXA-539) in sequence type 235 Pseudomonas aeruginosa. Antimicrob Agents Chemother 61:e01117-17. doi: 10.1128/AAC.01117-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vanscoy B, Mendes RE, Castanheira M, McCauley J, Bhavnani SM, Forrest A, Jones RN, Okusanya OO, Friedrich LV, Steenbergen J, Ambrose PG. 2013. Relationship between ceftolozane-tazobactam exposure and drug resistance amplification in a hollow-fiber infection model. Antimicrob Agents Chemother 57:4134–4138. doi: 10.1128/AAC.00461-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. MacVane SH, Pandey R, Steed LL, Kreiswirth BN, Chen L. 2017. Emergence of ceftolozane-tazobactam-resistant Pseudomonas aeruginosa during treatment is mediated by a single AmpC structural mutation. Antimicrob Agents Chemother 61:e01183-17. doi: 10.1128/AAC.01183-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gomis-Font MA, Pitart C, Del Barrio-Tofiño E, Zboromyrska Y, Cortes-Lara S, Mulet X, Marco F, Vila J, López-Causapé C, Oliver A. 2021. Emergence of resistance to novel cephalosporin-β-lactamase inhibitor combinations through the modification of the Pseudomonas aeruginosa MexCD-OprJ efflux pump. Antimicrob Agents Chemother 65:e0008921. doi: 10.1128/AAC.00089-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ruedas-López A, Alonso-García I, Lasarte-Monterrubio C, Guijarro-Sánchez P, Gato E, Vázquez-Ucha JC, Vallejo JA, Fraile-Ribot PA, Fernández-Pérez B, Velasco D, Gutiérrez-Urbón JM, Oviaño M, Beceiro A, González-Bello C, Oliver A, Arca-Suárez J, Bou G. 2022. Selection of AmpC β-lactamase variants and metallo-β-lactamases leading to ceftolozane/tazobactam and ceftazidime/avibactam resistance during treatment of MDR/XDR Pseudomonas aeruginosa infections. Antimicrob Agents Chemother 66:e0206721. doi: 10.1128/AAC.02067-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. VanScoy BD, Mendes RE, Castanheira M, McCauley J, Bhavnani SM, Jones RN, Friedrich LV, Steenbergen JN, Ambrose PG. 2014. Relationship between ceftolozane-tazobactam exposure and selection for Pseudomonas aeruginosa resistance in a hollow-fiber infection model. Antimicrob Agents Chemother 58:6024–6031. doi: 10.1128/AAC.02310-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cabot G, Kim K, Mark BL, Oliver A, Khajehpour M. 2023. Biochemical insights into imipenem collateral susceptibility driven by ampC mutations conferring ceftolozane/tazobactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 67:e0140922. doi: 10.1128/aac.01409-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cortes-Lara S, Barrio-Tofiño ED, López-Causapé C, Oliver A, GEMARA-SEIMC/REIPI Pseudomonas study Group . 2021. Predicting Pseudomonas aeruginosa susceptibility phenotypes from whole genome sequence resistome analysis. Clin Microbiol Infect 27:1631–1637. doi: 10.1016/j.cmi.2021.05.011 [DOI] [PubMed] [Google Scholar]
- 14. Gollan B, Grabe G, Michaux C, Helaine S. 2019. Bacterial persisters and infection: past, present, and progressing. Annu Rev Microbiol 73:359–385. doi: 10.1146/annurev-micro-020518-115650 [DOI] [PubMed] [Google Scholar]
- 15. Nielsen EI, Friberg LE. 2013. Pharmacokinetic-pharmacodynamic modeling of antibacterial drugs. Pharmacol Rev 65:1053–1090. doi: 10.1124/pr.111.005769 [DOI] [PubMed] [Google Scholar]
- 16. Yadav R, Landersdorfer CB, Nation RL, Boyce JD, Bulitta JB. 2015. Novel approach to optimize synergistic carbapenem-aminoglycoside combinations against carbapenem-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 59:2286–2298. doi: 10.1128/AAC.04379-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cianfanelli FR, Cunrath O, Bumann D. 2020. Efficient dual-negative selection for bacterial genome editing. BMC Microbiol 20:129. doi: 10.1186/s12866-020-01819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Landersdorfer CB, Nation RL. 2021. Limitations of antibiotic MIC-based PK-PD metrics: looking back to move forward. Front Pharmacol 12:770518. doi: 10.3389/fphar.2021.770518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chauzy A, Ih H, Jacobs M, Marchand S, Grégoire N, Couet W, Buyck JM. 2020. Sequential time-kill, a simple experimental trick to discriminate between pharmacokinetics/pharmacodynamics models with distinct heterogeneous subpopulations versus homogenous population with adaptive resistance. Antimicrob Agents Chemother 64:e00788-20. doi: 10.1128/AAC.00788-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jacobs M, Grégoire N, Couet W, Bulitta JB. 2016. Distinguishing antimicrobial models with different resistance mechanisms via population pharmacodynamic modeling. PLoS Comput Biol 12:e1004782. doi: 10.1371/journal.pcbi.1004782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khil PP, Dulanto Chiang A, Ho J, Youn J-H, Lemon JK, Gea-Banacloche J, Frank KM, Parta M, Bonomo RA, Dekker JP. 2019. Dynamic emergence of mismatch repair deficiency facilitates rapid evolution of ceftazidime-avibactam resistance in Pseudomonas aeruginosa acute infection. mBio 10:e01822-19. doi: 10.1128/mBio.01822-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boulant T, Jousset AB, Bonnin RA, Barrail-Tran A, Borgel A, Oueslati S, Naas T, Dortet L. 2019. A 2.5-year within-patient evolution of Pseudomonas aeruginosa isolates with in vivo acquisition of ceftolozane-tazobactam and ceftazidime-avibactam resistance upon treatment. Antimicrob Agents Chemother 63:e01637-19. doi: 10.1128/AAC.01637-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin-Chao S, Chen WT, Wong TT. 1992. High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II. Mol Microbiol 6:3385–3393. doi: 10.1111/j.1365-2958.1992.tb02206.x [DOI] [PubMed] [Google Scholar]
- 24. Slater CL, Winogrodzki J, Fraile-Ribot PA, Oliver A, Khajehpour M, Mark BL. 2020. Adding insult to injury: mechanistic basis for how AmpC mutations allow Pseudomonas aeruginosa to accelerate cephalosporin hydrolysis and evade avibactam. Antimicrob Agents Chemother 64:e00894-20. doi: 10.1128/AAC.00894-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jacobs C, Huang LJ, Bartowsky E, Normark S, Park JT. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J 13:4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Torrens G, Hernández SB, Ayala JA, Moya B, Juan C, Cava F, Oliver A. 2019. Regulation of AmpC-driven β-lactam resistance in Pseudomonas aeruginosa: different pathways, different signaling. mSystems 4:e00524-19. doi: 10.1128/mSystems.00524-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mouton JW, Vinks AA, Punt NC. 1997. Pharmacokinetic-pharmacodynamic modeling of activity of ceftazidime during continuous and intermittent infusion. Antimicrob Agents Chemother 41:733–738. doi: 10.1128/AAC.41.4.733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Minichmayr IK, Aranzana-Climent V, Friberg LE. 2022. Pharmacokinetic/pharmacodynamic models for time courses of antibiotic effects. Int J Antimicrob Agents 60:106616. doi: 10.1016/j.ijantimicag.2022.106616 [DOI] [PubMed] [Google Scholar]
- 29. Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, Fagelhauer L, Chakraborty T, Neumann B, Werner G, Bender JK, Stingl K, Nguyen M, Coppens J, Xavier BB, Malhotra-Kumar S, Westh H, Pinholt M, Anjum MF, Duggett NA, Kempf I, Nykäsenoja S, Olkkola S, Wieczorek K, Amaro A, Clemente L, Mossong J, Losch S, Ragimbeau C, Lund O, Aarestrup FM. 2020. Resfinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75:3491–3500. doi: 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv. doi: 10.48550/arXiv.1207.3907 [DOI] [Google Scholar]
- 31. Ude J, Tripathi V, Buyck JM, Söderholm S, Cunrath O, Fanous J, Claudi B, Egli A, Schleberger C, Hiller S, Bumann D. 2021. Outer membrane permeability: antimicrobials and diverse nutrients bypass porins in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 118:e2107644118. doi: 10.1073/pnas.2107644118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains supplement table S1 to S6, supplemental Figure S1 to s3 and supplemental text S1 and S2.
Data Availability Statement
The whole-genome sequences of the strains analyzed in this study are available under BioProject accession number PRJNA998736 (strains PaS and PaR).