Abstract
Existing technologies for site-specific integration of kilobase-sized DNA sequences in bacteria are limited by low efficiency, a reliance on recombination, the need for multiple vectors, and challenges in multiplexing. To address these shortcomings, we introduce a substantially improved version of our previously reported Tn7-like transposon from Vibrio cholerae, which uses a Type I-F CRISPR–Cas system for programmable, RNA-guided transposition. The optimized insertion of transposable elements by guide RNA–assisted targeting (INTEGRATE) system achieves highly accurate and marker-free DNA integration of up to 10 kilobases at ~100% efficiency in bacteria. Using multi-spacer CRISPR arrays, we achieved simultaneous multiplexed insertions in three genomic loci and facile, multi-loci deletions by combining orthogonal integrases and recombinases. Finally, we demonstrated robust function in biomedically and industrially relevant bacteria and achieved target- and species-specific integration in a complex bacterial community. This work establishes INTEGRATE as a versatile tool for multiplexed, kilobase-scale genome engineering.
DNA technologies to stably integrate genes and pathways into the genome enable the generation of engineered cells with entirely new functions. Applications of this approach have already yielded commercial products, with examples including chimeric antigen receptor T cell therapies1, genetically modified crops2 and cell factories producing diverse compounds and medicines3. In many of these applications, genomic integration is preferred over plasmid-based methods for maintaining heterologous genes in engineered cells, owing to improved stability in the genome, better control of copy numbers and regulatory concerns regarding biocontainment of recombinant DNA4,5. However, the generation of modified cells with kilobases of changes across the genome remains practically challenging, often requiring inefficient, multi-step processes that are time- and resource-intensive. A facile, efficient and versatile method that allows programmable genomic integrations in multiplex would accelerate advances in cellular programming.
In bacteria, DNA integration can be achieved through several approaches that use endogenous or foreign integrases4,5, transposases6,7, recombinases8,9 or homologous recombination (HR) machinery10-13, which can be further combined with CRISPR–Cas to improve efficiency14,15. Despite being widely used, these methods, nevertheless, have substantial drawbacks. For example, recombination-mediated genetic engineering (recombineering) using λ-red or RecET recombinase systems in Escherichia coli allows programmable genomic integrations, specified by the homology arms flanking the foreign DNA cassette13,16. However, recombineering efficiency is generally low (less than 1 in 103–104)17 without selection of a co-integrating selectable marker8 or CRISPR–Cas-mediated counter-selection of unedited alleles14 and, thus, cannot be easily multiplexed to make simultaneous insertions into the same cell. There is a limited number of robust selectable markers (for example, antibiotic resistance genes) that must be removed from the genome during a separate excision step for subsequent reuse, and expression of Cas9 for negative selection can cause unintended DNA double-strand breaks (DSBs) that lead to cytotoxicity18-20. Practically, recombineering has a payload size limit of only 3–4 kb in many cases, making it less useful for genomic integration of pathway-sized DNA cassettes. Finally, unknown requirements for host-specific factors or cross-species incompatibilities of phage recombination proteins make E. coli recombineering systems challenging to adapt to other bacterial species, requiring optimizations21 or screening of new recombinases22.
Integrases and transposases, such as ICEBs1 and Tn7, have also been used for genomic integration4,23. These systems recognize specific attachment sites and cannot be easily reprogrammed, thus requiring the prior presence of these sites or their separate introduction in the genome24. Other more portable transposons, such as Mariner and Tn5, generate non-specific integrations that have been used for genome-wide transposon mutagenesis libraries25-28. However, these transposons cannot be targeted to specific genomic loci, and large-scale screens are needed to isolate desired clones. More recently, a catalytically dead Cas9 has been fused to either a transposase or a recombinase to provide better site specificity, which showed success in mostly in vitro studies29,30. Autocatalytic Group II RNA introns, which are selfish genetic elements in bacteria, have also been used for genomic transpositions and insertions31,32. This system uses an RNA intermediate to guide insertions but suffers from inconsistent efficiencies ranging from 1% to 80%, depending on the target site and species33, and a limited cargo size of 1.8 kb34.
An ideal genome insertion technology should provide efficient single-step DNA integration for high-capacity cargos with high specificity and programmability, without relying on DSBs or HR. We recently described a new category of programmable integrases whose sequence specificity is governed exclusively by guide RNAs35. Motivated by the bioinformatic description of Tn7-like transposons encoding nuclease-deficient CRISPR–Cas systems36, we selected a candidate CRISPR-transposon from V. cholerae (Tn6677) and reconstituted RNA-guided transposition in an E. coli host. DNA integration occurs ~50 base pairs (bp) downstream of the genomic site targeted by the CRISPR RNA (crRNA) and requires transposition proteins TnsA, TnsB and TnsC, in conjunction with the RNA-guided DNA targeting complex TniQ-Cascade35,37 (Fig. 1a,b). Remarkably, bacterial transposons have hijacked at least three distinct CRISPR–Cas subtypes35,38,39, and work from Zhang and colleagues demonstrated that the Type V-K effector protein, Cas12k, also directs targeted DNA integration, albeit with lower fidelity40. These studies underscore the exaptation that allowed transposons to repurpose RNA-guided DNA targeting systems for selfish propagation, and they highlight the promise of programmable integrase systems, which we named INTEGRATE, for genome engineering41,42.
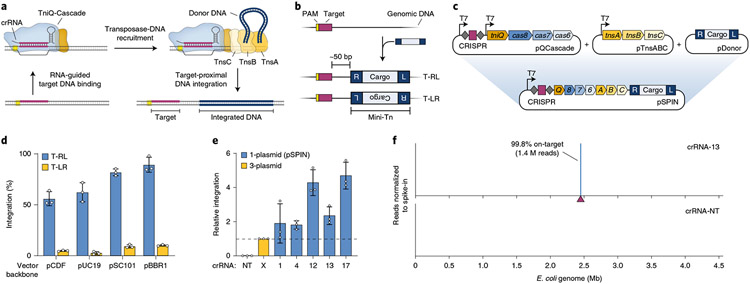
Fig. 1 ∣. Streamlined single-plasmid system for RNA-guided DNA integration.
a, Schematic of INTEGRATE using a Type I-F V. cholerae CRISPR-transposon. b, RNA-guided DNA integration occurs ~50 bp downstream of the target site, in one of two possible orientations (T-RL and T-LR); the mini-transposon (mini-Tn) comprises a genetic cargo flanked by left (L) and right (R) transposon ends. c, Top, a three-plasmid INTEGRATE system encodes protein–RNA components on pQCascade and pTnsABC and the mini-Tn on pDonor. Bottom, a single-plasmid INTEGRATE system (pSPIN) drives protein–RNA expression with a single promoter, on the same vector as the donor DNA. d, qPCR-based quantification of integration efficiency with crRNA-4 for pSPIN with distinct vector backbones and differing copy numbers. e, Relative integration efficiencies for the three-plasmid or single-plasmid (pSPIN) expression system across a non-targeting (NT) and five distinct targeting crRNAs. Data are normalized to the three-plasmid system; pSPIN contained the pBBR1 backbone. f, Normalized Tn-seq data with crRNA-13 and a non-targeting crRNA (crRNA-NT) for pSPIN containing the pBBR1 backbone. Genome-mapping reads are normalized to the reads from a spike-in control (Methods); the target site is denoted by a maroon triangle. Data in d and e are shown as mean ± s.d. for n = 3 biologically independent samples.
INTEGRATE combines the high-efficiency, seamless integrations of transposases with the programmability of CRISPR-mediated targeting. However, our previous system35, demonstrated in E. coli, required multiple cumbersome genetic components and displayed low efficiency for larger insertions in dual orientations. In this study, we developed a vastly improved INTEGRATE system that uses streamlined expression vectors to direct highly accurate insertions at ~100% efficiency effectively in a single orientation, independent of the cargo size, without requiring selection markers. Because INTEGRATE does not rely on homology arms specific to each target site, multiple simultaneous genomic insertions into the same cell could be rapidly generated using CRISPR arrays with multiple targeting spacers, and INTEGRATE paired with Cre-LoxP was used to achieve precise genomic deletions. Furthermore, we demonstrated the portability and high site specificity of INTEGRATE in other species, such as Klebsiella oxytoca and Pseudomonas putida, highlighting its broad utility for bacterial genome engineering. Finally, we showed that INTEGRATE functions as an effective genetic tool for engineering specific strains in a complex mammalian gut microbiome.
Results
An optimized, single-plasmid system for high-efficiency, RNA-guided DNA integration
We previously employed a three-plasmid expression system to reconstitute RNA-guided DNA integration in E. coli, whereby pQCascade and pTnsABC encoded the necessary protein–RNA components, and pDonor contained the mini-transposon (mini-Tn, also known as donor DNA)35 (Fig. 1c). To streamline our strategy and eliminate both antibiotic burden and the need for multiple transformation events, we serially reduced the number of independent promoters and plasmids and ultimately arrived at a single-plasmid INTEGRATE construct (pSPIN), in which one promoter drives expression of the crRNA and polycistronic messenger RNA, directly upstream of the mini-Tn (Fig. 1c, Supplementary Fig. 1 and Supplementary Table 1). This design allows for modular substitution of the promoter and/or genetic cargo for user-specific applications and for straightforward subcloning into distinct vector backbones.
After identifying an optimal arrangement of the CRISPR array and operons (Supplementary Fig. 1), we transformed E. coli BL21(DE3) with four pSPIN derivatives encoding a lacZ-specific crRNA on distinct vector backbones and monitored the efficiency of RNA-guided transposition by quantitative polymerase chain reaction (qPCR). Surprisingly, our streamlined plasmids exhibited enhanced integration activity, with efficiency exceeding 90% using the pBBR1 vector backbone (Fig. 1d), and showed substantially stronger bias for insertion events in which the transposon right end was proximal to the target site (T-RL), as compared to the original three-plasmid expression system (Supplementary Fig. 2). To determine whether increased efficiency would translate across multiple targets, we assessed integration at five target sites used in our previous study35 and found that our pSPIN vector was consistently 2–5 times more efficient (Fig. 1e). Our single-plasmid INTEGRATE system maintained high-fidelity activity and an absence of insertion events with a non-targeting crRNA, as reported by genome-wide transposon insertion sequencing (Tn-seq; Fig. 1f and Supplementary Fig. 3). This high degree of specificity was further verified by isolating clones and confirming the unique presence of a single insertion by whole-genome, single-molecule real-time (SMRT) sequencing and structural variant analysis (Methods and Supplementary Table 2).
We next assessed the role of expression level by modifying the promoter driving protein–RNA expression. Using a panel of constitutive promoters of varying expression strength, we found that higher expression drove higher rates of integration, without any deleterious effect on genome-wide specificity (Fig. 2a,b and Supplementary Fig. 4a). Efficient integration was also achieved with a natural broad-host promoter recently adopted for metagenomic microbiome engineering43 (Fig. 2a), and the use of constitutive promoters allowed us to demonstrate high-accuracy integration in additional E. coli strains, including MG1655 and BW25113, without any requirement for host recombination factors (Supplementary Fig. 4b,c). Interestingly, we also noticed that RNA-guided DNA integration readily proceeded when cells were grown at room temperature and reached ~100% efficiency (without selection for the integration event) while maintaining 99.7% on-target specificity, even for the low-strength J23114 promoter (Fig. 2c and Supplementary Fig. 4d).
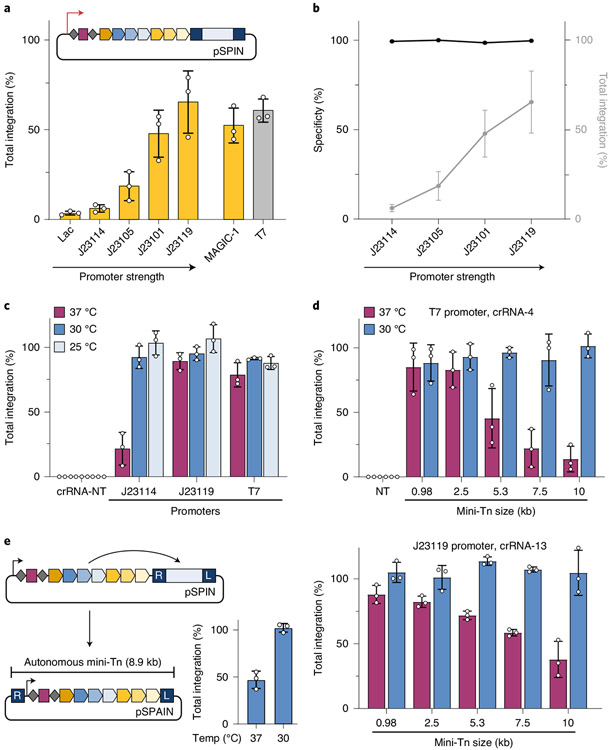
Fig. 2 ∣. INTEGRATE supports high-efficiency insertion of large (10-kb) genetic payloads.
a, qPCR-based quantification of integration efficiency with crRNA-4 as a function of pSPIN promoter identity; MAGiC-1 is taken from ref. 43. b, DNA integration specificity (black) for the promoters shown, as determined by Tn-seq, calculated as the percent of on-target reads relative to all genome-mapping reads (Methods); total integration efficiencies (qPCR) are plotted in gray. c, qPCR-based quantification of integration efficiency with crRNA-4 as a function of culture temperature and promoter strength. Integration reaches ~100% efficiency at lower growth temperatures for all constructs, including the weak J23114 promoter. d, qPCR-based quantification of integration efficiency with variable mini-Tn sizes, after culturing at either 30 °C or 37 °C. The promoter and crRNA used in each panel are shown at the top; experiments were performed with a two-plasmid system comprising pEffector-B (Supplementary Fig. 1c) and pDonor. Unless specified, transposition assays elsewhere in this study use a 0.98-kb mini-Tn. e, Schematic of a single-plasmid autonomous INTEGRATE system (pSPAIN; left) and qPCR-based quantification of integration efficiency with crRNA-4 after culturing at 30 °C and 37 °C (right). The inserted DNA encodes all the necessary machinery for further mobilization. Integration efficiency data in a–e are shown as mean ± s.d. for n = 3 biologically independent samples.
To better understand this temperature effect, we followed the kinetics of transposition in liquid culture experiments. For both strong and weak promoters, the integration efficiency plateaued as the cells approached stationary phase at 37 °C, suggesting that rapid growth of the bacterial population at higher temperatures can limit transposition (Supplementary Fig. 5a). This effect was most apparent for the low-strength J23114 promoter, where the slower onset of exponential growth at 30 °C allowed more time for integration to reach its maximum efficiency of ~90%. In addition, simple dilution of a culture grown at 37 °C into fresh media also boosted integration efficiencies (Supplementary Fig. 5b). We also found that integration products could be detected within 2 h after transformation (Supplementary Fig. 5c), suggesting that INTEGRATE can be deployed without conventional replicating plasmids. Indeed, when we delivered the donor DNA encoding chloramphenicol resistance to cells in the form of a linear PCR product, we readily isolated drug-resistant clones that uniformly contained the on-target insertion (Supplementary Fig. 5d and Methods).
Motivated by the enhanced integration efficiencies at lower temperature growth, we reexamined the effect of cargo size on transposition. We previously found that, whereas the V. cholerae machinery integrated a ~1-kb cargo with optimal efficiency, larger cargos were poorly mobilized35. Remarkably, when we expressed protein–RNA components from a single effector plasmid (pEffector-B; Supplementary Fig. 1c) and cultured cells at 30 °C, we could integrate mini-transposons spanning 1–10 kb with ~100% efficiency, with no observable size-dependent effects (Fig. 2d) and without the need for marker selection. The same pattern was observed across multiple target sites and promoters, and the specificity of 10-kb insertions was verified by Tn-seq and SMRT sequencing (SMRT-seq) (Fig. 2d and Supplementary Fig. 6). As native CRISPR-containing transposons are frequently several tens of kb in size35,36, we anticipate that INTEGRATE has the potential to mobilize payloads beyond 10 kb. To further leverage the large-cargo capability, we generated a single-plasmid autonomous INTEGRATE system (pSPAIN), in which the protein–RNA coding genes were cloned within the mini-Tn itself, and showed that this construct also directed targeted integration at ~100% efficiency (Fig. 2e). We envision that autonomous INTEGRATE systems, by virtue of mobilizing themselves according to the user-defined CRISPR array content, can serve as potent gene drive elements capable of programmed self-propagation in mixed community environments.
Development of orthogonal integrases for iterative DNA insertions
As strain engineering often requires multiple insertions and knockouts to be performed in distinct genomic regions, we next sought to evaluate the amenability of the V. cholerae system for iterative integration events. We first cloned a derivative of pSPIN using a temperature-sensitive plasmid backbone, isolated a clonal strain containing a lacZ-specific insertion (target-4) and cured the plasmid (Methods). Next, we reintroduced the machinery to generate a proximal insertion at variable distances upstream of target-4, but we used a mini-Tn whose distinct cargo could be selectively tracked by qPCR (Fig. 3a). Previous studies demonstrated that Tn7 and Tn7-like transposons exhibit target immunity40,44,45, whereby integration is prevented at target sites already containing another transposon copy. When we compared integration across a panel of crRNAs for strains with and without a pre-existing mini-Tn, we found that the V. cholerae transposon also exhibited target immunity, with ~20% relative efficiency at target sites ~5 kb away (Fig. 3a and Methods). This effect was ablated when we instead targeted a glmS-proximal site (target-1) that was >1 Mbp from the pre-existing insertion, demonstrating that iterative insertions are straightforward but more efficient when spaced far apart.
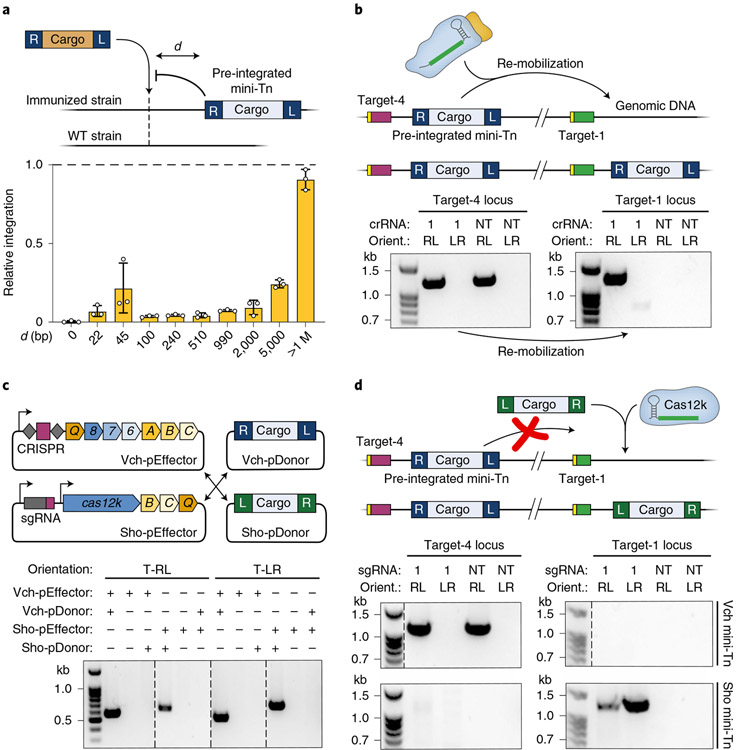
Fig. 3 ∣. Orthogonal INTEGRATE systems facilitate multiple, iterative insertions.
a, Effect of target immunity on RNA-guided DNA integration. An E. coli strain containing a single, genomically integrated mini-Tn was generated, and the efficiency of additional transposition events using crRNAs targeting d-bp upstream was determined by qPCR. The relative efficiency for each crRNA in the immunized versus WT strain is plotted. b, Top, schematic showing re-mobilization of a genomically integrated mini-Tn (target-4) to a new genomic site (target-1) with crRNA-1. Bottom, PCR products probing for the mini-Tn at target-4 (left) and target-1 (right), resolved by agarose gel electrophoresis; Orient., integration orientation. The mini-Tn is efficiently transposed to target-1 by crRNA-1, without apparent loss of the mini-Tn at target-4. c, Top, schematic of orthogonal INTEGRATE systems from V. cholerae (Vch; Type I-F) and S. hofmannii (Sho; Type V-K); the mini-Tn is encoded on pDonor, separately from pEffector. Bottom, PCR products probing for RNA-guided DNA integration at target-4 with both systems, resolved by gel electrophoresis. Integration proceeds only with a cognate pairing between the expression and donor plasmids. d, Top, schematic of strategy to make a second DNA insertion by leveraging the orthogonal ShoINT system, where the Vch mini-Tn remains inert. Bottom, PCR products probing for either the Vch mini-Tn (top) or Sho mini-Tn (bottom) at target-4 (left) and target-1 (right), resolved by agarose gel electrophoresis. The Sho mini-Tn is efficiently integrated at target-1 by sgRNA-1, without loss of the Vch mini-Tn at target-4. Data in a are shown as mean ± s.d. for n = 3 biologically independent samples. Gel source can be found in Source Data Fig. 3.
The simultaneous presence of a genomically integrated mini-Tn and distinct plasmid-borne mini-Tn produces an interesting scenario in which the transposase machinery can theoretically employ either DNA molecule as the donor substrate for integration (Supplementary Fig. 7a). Using cargo-specific primers, we showed that new insertions at target-1 were indeed a heterogeneous mixture of both mini-Tn donors, although the higher-copy plasmid source was heavily preferred (Supplementary Fig. 7a). To further investigate intramolecular transposition events, we transformed our clonally integrated strain with a plasmid encoding the protein–RNA machinery without donor DNA and monitored re-mobilization of the pre-existing mini-Tn from target-4 to target-1. Integration at target-1 was readily observed, but, surprisingly, we saw no PCR evidence of mini-Tn loss at target-4, despite the expectation that the transposon mobilizes through a cut-and-paste mechanism35 (Fig. 3b and Methods), suggesting that lesions resulting from donor DNA excision are rapidly resolved by HR, as has been observed with Tn7 (ref. 46).
To avoid any low-level contamination between donor DNA molecules, we explored the use of multiple RNA-guided transposases whose cognate transposon ends would be recognized orthogonally. Guided by prior bioinformatic description and experimental validation of transposons encoding Type V-K CRISPR–Cas systems35,38,40, we developed a new INTEGRATE system derived from Scytonema hofmannii strain PCC 7110 (hereafter called ShoINT; Supplementary Fig. 7b). We note that the protein components are 30–55% identical to the homologous system described by Strecker et al. (ShCAST)40, which derives from a distinct S. hofmannii strain (Supplementary Table 3). ShoINT catalyzes RNA-guided DNA integration with 20–40% efficiency and strongly favors integration in the T-LR orientation, albeit with detectable bi-directional integration at multiple target sites (Supplementary Fig. 7c-e). Next, we combined pEffector plasmids for the V. cholerae INTEGRATE system (VchINT) or ShoINT with either its own cognate pDonor or pDonor from the other system and found that each RNA-guided integrase was exclusively active on its respective mini-Tn substrate (Fig. 3c). With this knowledge, we were able to sequentially introduce a new cargo at a different locus (at target-1) using ShoINT, without any secondary mobilization of the pre-existing VchINT mini-Tn (at target-4) (Fig. 3d). We expect the same lack of cross-reactivity between VchINT and ShCAST, based on similarities between the Type V-K proteins and transposon ends. This approach of using systems with transposon ends that are sufficiently distinct will enable orthogonal and iterative integration events for distinct genetic payloads.
We were keen to carefully compare genome-wide integration specificity between Type I-F VchINT and Type V-K ShoINT systems, particularly in light of the significant off-target insertions previously observed for ShCAST40. After developing an alternative, unbiased next-generation sequencing (NGS) approach to query genome-wide integration events, which does not require the MmeI restriction enzyme used in Tn-seq, we first verified that this random fragmentation-based method returned similar specificity information for VchINT (Supplementary Fig. 8a). When the same method was applied to ShoINT and ShCAST, we found that only ~5–50% of integration events were on-target, with substantial numbers of insertions distributed randomly across the genome (Supplementary Fig. 8b,c). These experiments highlight the remarkable fidelity of Type I-F INTEGRATE systems and the need for further mechanistic studies to dissect the molecular basis of promiscuous integration by Cas12k-associated transposases. It will also be interesting to investigate the evolutionary forces that shaped I-F and V-K transposons, given the competing selective pressures to spread widely without restrictive targeting constraints, while retaining enough specificity to mitigate fitness costs on the host.
Single-step multiplexed DNA insertions using INTEGRATE
CRISPR–Cas systems are naturally capable of multiplexing because of the way that CRISPR arrays are transcribed and processed, and transposase-mediated DNA integration exhibits intrinsic compatibility with multiple different genomic target sites, as there is no requirement for target-specific homology arms. Thus, INTEGRATE provides a unique potential for multi-spacer CRISPR arrays to direct insertion of the same cargo at multiple genomic targets simultaneously (Fig. 4a), which significantly reduces time and complexity for strain engineering projects requiring multi-copy integration. We explored this by first cloning a series of multiple-spacer arrays into pSPIN and found that the integration efficiency of a lacZ-specific crRNA was unchanged for two spacers and reduced by <two-fold for three spacers, depending on relative spacer position, when cells were cultured at 37 °C (Fig. 4b). Tn-seq analyses with double- and triple-spacer arrays revealed >99% on-target transposition, with characteristics that were otherwise indistinguishable from single-plex insertions for each target site (Fig. 4c and Supplementary Fig. 9), and we further verified multiplexed insertions by whole-genome SMRT-seq of double- and triple-insertion clones (Supplementary Table 2).
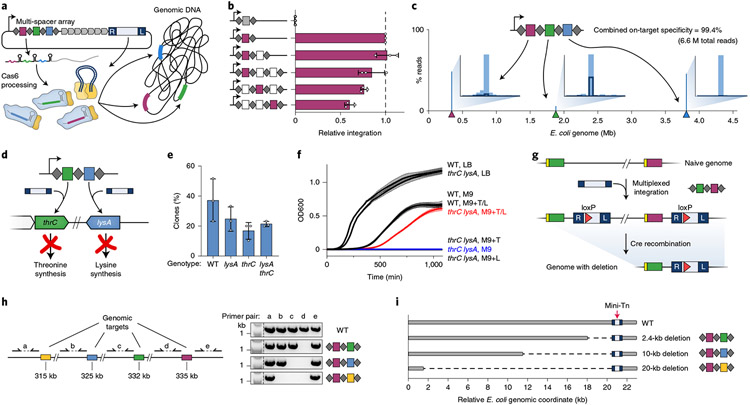
Fig. 4 ∣. Multi-spacer CRISPR arrays direct single-step multiplexed insertions.
a, Schematic of multiplexed integration with pSPIN encoding a multi-spacer CRISPR array. b, qPCR-based quantification of integration efficiency with non-targeting crRNA (crRNA-NT; gray) and crRNA-4 (maroon), encoded in a single-, double- or triple-spacer CRISPR array in the position indicated; white squares represent other genome-targeting spacers. Data are normalized to the single-spacer array efficiency. c, Tn-seq data for a triple-spacer CRISPR array, plotted as percent of total genome-mapping reads. Target sites are denoted by colored triangles, and insets show the distribution of integration events within a 42–58-bp window downstream of each target site. d, Schematic of multiplexed thrC- and lysA-specific spacers for single-step generation of threonine–lysine auxotrophic E. coli. e, Recovery percentage of the indicated clonal genotypes (WT, single-knockout or double-knockout) using multiplexed pSPIN. f, Growth curves for WT and double-knockout clones cultured in LB or M9 minimal media with or without supplemented threonine (T) and lysine (L). g, Experimental approach to generate programmed genomic deletions. A double-spacer array directs multiplexed insertion of two mini-Tn copies carrying LoxP sites; subsequent introduction of Cre recombinase leads to precise excision of the genomic fragment spanning the LoxP sites. h, Left, genomic locus targeted for deletion. Right, double-spacer arrays used to generate defined deletions and PCR products with indicated primer pairs showing the presence or absence of genomic fragments flanking each target site, resolved by agarose gel electrophoresis. i, Programmed genomic deletions generated in h (2.4-, 10- or 20-kb) were further verified by whole-genome SMRT-seq (Methods). The mini-transposon is indicated with a red arrow. Data in b and e are shown as mean ± s.d. for n = 3 biologically independent samples. Data in f are shown as mean ± s.d. for three technical replicates. Gel source can be found in Source Data Fig. 4.
To further confirm that simultaneous insertions were indeed occurring within each individual chromosome rather than population wide, we designed an experiment to generate auxotrophic E. coli strains requiring both threonine and lysine for viability by insertionally inactivating thrC and lysA47 (Fig. 4d and Supplementary Fig. 10a-c). Double-knockout clones could be rapidly isolated after a single transformation step (Fig. 4e) and exhibited selective growth in M9 minimal media only when both threonine and lysine were supplemented (Fig. 4f). To probe the stability of integration-based knockouts, we cultured clones in rich media for five serial overnight passages without removing the expression plasmid and observed no subsequent change in the media requirements (Supplementary Fig. 10d), thus confirming the potency of phenotypic outcomes driven by multiplexed INTEGRATE.
Finally, we explored the combined use of RNA-guided integrases with site-specific recombinases to mediate facile, programmable, one-step genomic deletions. Specifically, we inserted a LoxP site within the mini-Tn cargo and generated double-spacer CRISPR arrays to drive multiplexed integration at two target sites. We subsequently used Cre recombinase to excise the chromosomal region between the LoxP sites, thus resulting in a precise deletion with a single mini-Tn left behind (Fig. 4g and Methods). We designed CRISPR arrays to produce 2.4-, 10- and 20-kb deletions and confirmed the deletions via diagnostic PCR analysis and unbiased, whole-genome SMRT-seq (Fig. 4h,i and Supplementary Fig. 11). These experiments highlight the potential synergies of INTEGRATE with existing technologies and the ease and versatility with which RNA-guided integrases can be leveraged for diverse and programmable genome-scale genetic modifications.
Broad-host-range activity of RNA-guided integrases
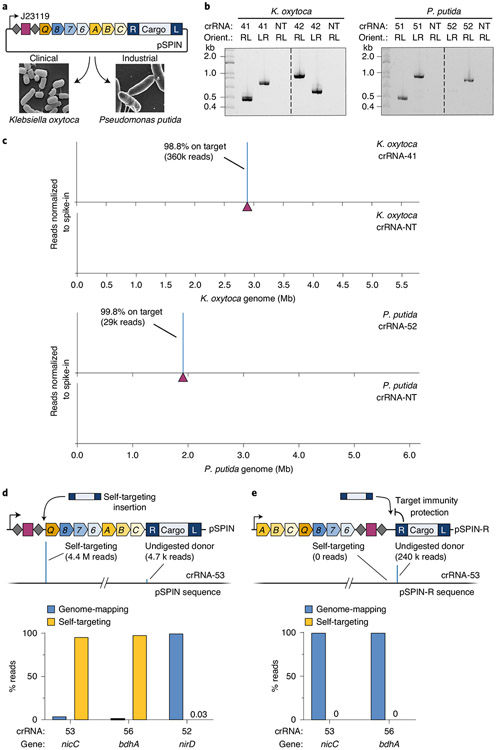
Mobile genetic elements, especially transposons, often ensure their evolutionary success by functioning robustly across a broad range of hosts, without a requirement for specific host factors48. Given this expectation, as well as the efficiency with which the V. cholerae machinery directs RNA-guided transposition in E. coli, we set out to evaluate INTEGRATE activity in other Gram-negative bacteria. We selected Klebsiella oxytoca, a clinically relevant pathogen implicated in drug-resistant infections49 and an emerging model organism for biorefinery50, and Pseudomonas putida, an important bacterial platform for biotechnological and industrial applications51,52 (Fig. 5a). Using a pSPIN derivative driven by the constitutive J23119 promoter, we targeted four non-essential metabolic genes (xylA, galK, lacZ and malK) and one antibiotic resistance gene (ampR) in K. oxytoca, as well as intergenic regions (upstream of PP_2928 and benR) or genes previously edited (nirC, nirD, bdhA and PP_3889) in P. putida53,54. For all ten targets, we observed highly accurate RNA-guided DNA integration by both PCR and Tn-seq, with similar integration distance and orientation bias profiles as seen in E. coli (Fig. 5b,c and Supplementary Fig. 12a-d). DNA insertions were virtually absent with a non-targeting crRNA, and on-target specificity was >95% on average, with the only outlier resulting from a prominent Cascade off-target binding site (Supplementary Fig. 12e). Given the potential for INTEGRATE to exhibit off-target activity similar to canonical CRISPR–Cas systems55, we developed a computational tool for guide RNA design and off-target prediction (Supplementary Fig. 13). We anticipate updating this algorithm as additional mechanistic insights for programmable, RNA-guided integrases are acquired.
Fig. 5 ∣. Robust and highly accurate INTEGRATE activity in additional Gram-negative bacteria.
a, Schematic showing the use of pSPIN constructs with constitutive J23119 promoter and broad-host pBBR1 backbone for RNA-guided DNA insertions in K. oxytoca and P. putida. Micrographs are taken from refs. 50,63. b, PCR products probing for integration at two different genomic loci in K. oxytoca (left) and P. putida (right), resolved by agarose gel electrophoresis. c, Normalized Tn-seq data with select targeting and non-targeting crRNAs for K. oxytoca (top) and P. putida (bottom). Genome-mapping reads are normalized to the reads from a spike-in control; the target site is denoted by a maroon triangle. d, Top, self-targeting of the spacer within the CRISPR array inactivates the pSPIN-encoded INTEGRATE system and was detected for select crRNAs by Tn-seq (middle); undigested donor reads are artifacts of NGS library preparation. Bottom, P. putida crRNAs targeting nicC and bdhA, but not nirD, show substantial plasmid self-targeting relative to genomic integration, as assessed by Tn-seq. e, Top, a modified vector (pSPIN-R) encodes the CRISPR array proximal to the mini-Tn, whereby self-targeting is blocked by target immunity. Bottom, P. putida crRNAs targeting nicC and bdhA no longer show any evidence of self-targeting with pSPIN-R, as assessed by Tn-seq. Gel source can be found in Source Data Fig. 5.
Interestingly, for two of the P. putida crRNAs, we observed a substantial enrichment of Tn-seq reads mapping to pSPIN, precisely 48–50 bp downstream of the spacer in the CRISPR array (Fig. 5d). We previously reported that Cascade-directed DNA integration exhibits a high degree of protospacer adjacent motif (PAM) promiscuity, including low activity with the mutant 5′-AC-3′ self-PAM present within CRISPR repeats flanking the spacer35. Indeed, we observed evidence of low-level self-targeting in all of our E. coli Tn-seq datasets (Supplementary Table 4), and we suspected that the apparent abundance of self-targeting insertions for P. putida crRNAs targeting nicC and bhdA genes resulted from a fitness cost of the intended knockout and concomitant selective pressure to inactivate the pSPIN expression vector. We hypothesized that this ‘escape’ outcome could be avoided by redesigning the expression vector such that the self-target CRISPR array would be in close proximity to the mini-Tn and, thus, become protected by transposon target immunity. When we transformed P. putida with modified pSPIN-R vectors encoding the exact same crRNAs but at the 3′ end of the fusion transcript, we found that self-targeting was completely abrogated (Fig. 5e and Supplementary Fig. 12f). This illustrates how mechanistic knowledge of the transposition pathway can directly inform technology development efforts.
Finally, we sought to harness INTEGRATE for specific manipulation of bacteria in a mixed community environment by taking advantage of our streamlined single-plasmid vector. We used bacterial conjugation to deliver pSPIN from a donor E. coli strain into a complex bacterial community derived from the mouse gut; the pSPIN construct was designed to specifically target the lacZ locus of a K. oxytoca strain added to the community. After isolating transconjugants, we observed robust and high-efficiency RNA-guided transposition to the target site across distinct microbiome community sources and with different donor-to-recipient ratios (Supplementary Fig. 14). Altogether, these experiments highlight the utility of RNA-guided integrases for programmable genetic modifications across diverse bacterial species and within complex microbiota.
Discussion
Through systematic engineering steps, we developed an optimized set of vectors to leverage INTEGRATE for targeted DNA integration applications in diverse bacterial species, without the need for DSBs, HR or cargo-specific marker selection. These streamlined constructs can be easily modified to include user-specific guide RNAs and genetic cargos, and they catalyze highly accurate, large DNA insertions at ~100% efficiency after a single transformation step. Moreover, by employing multi-spacer CRISPR arrays within the same seamless workflow, we demonstrated efficient multiplexing for simultaneous insertions, and we combined multiplexed INTEGRATE with Cre-LoxP to generate programmed genomic deletions. Notably, as target-specific homology arms are not required, the mini-Tn is compatible with any target site, thus significantly reducing the complexity of the donor DNA and accelerating the experiment compared to HR, particularly for large-scale multiplex applications and metabolic engineering56-58.
This genetic engineering toolkit can be harnessed to generate large guide RNA libraries, which will enable high-throughput screening of rationally designed DNA insertions that are not easily accessible with random transposase-based strategies. Libraries of multiplexed guide RNAs will also enable synthetic lethality screening and investigations of pairwise interactions at the genome scale in bacteria, approaches that are straightforward in eukaryotes using CRISPR–Cas9 (ref. 59) but less accessible in NHEJ-deficient bacteria. Furthermore, INTEGRATE can help advance existing technologies for engineering strains or complex communities, particularly those currently employing non-programmable23 or non-specific43 transposases that could benefit from programmable, site-specific insertions. Our observation of highly active integration at lower culturing temperatures provides a strategy for increasing the efficacy of genetic manipulations, and we anticipate that the broad temperature range of the system holds promise for general utility across diverse species. Finally, our finding that integration can be quickly established within a bacterial cell population and accessed with transient delivery of linear donor DNAs might enable future applications in microbial species that cannot be stably transformed with replicating plasmids.
Despite key advantages of RNA-guided DNA integration for bacterial engineering, we note some specific drawbacks that users should take into account. First, because transposon end sequences are essential for specific recognition by the transposase machinery, INTEGRATE is not suited for applications where precise, scarless insertions or point mutations are required; these applications will require gene editing- and/or recombination-based approaches, such as CRISPR–Cas9-coupled recombineering8,14,60. However, many other applications are not constrained by relatively short cargo-flanking sequences, including simple insertional gene knockouts, strain tagging or stable transgene integration into safe harbor regions. In addition, future transposon engineering might enable further reductions in the size of transposon ends or their conversion into functional parts, such as peptide linkers for in-frame gene tagging. Second, applications involving iterative DNA insertions will need to carefully consider transposon target immunity and/or the risk of pre-existing insertions being mobilized by their cognate transposition machinery. Although these effects might affect efficiencies, iterative insertions can still proceed using the same VchINT system, followed by routine validation of clones. However, homologous INTEGRATE systems provide avenues for circumventing these potential issues. The orthogonality of the V. cholerae and S. hofmannii INTEGRATE systems serves to illustrate the promise of combining multiple phylogenetically distinct transposases. As more CRISPR-transposon systems are discovered and functionally validated for both Type I and Type V, we envision the INTEGRATE toolkit expanding into a robust set of programmable, RNA-guided integrases that act orthogonally and are fully cross-compatible.
In addition to its utility for strain engineering, INTEGRATE systems might be particularly useful for species- and target-specific genetic manipulations in mixed bacterial communities and microbiome niches, via the ability to broadly deliver all the necessary machinery on a single vector by conjugation61. Furthermore, using our compact construct designs, we generated a fully autonomous CRISPR-transposon that was capable of high-efficiency integration. In future studies, we envision mobilizing similar constructs on broad-host-range conjugative plasmids, pre-programmed with multiple-spacer CRISPR arrays, to genetically modify desired bacterial species at user-defined target sites. Such self-driving systems would leverage the natural ability of transposons to propagate both within and between host genomes, while maintaining tight experimental control over specificity. These future gene drive applications can be used to inactivate antibiotic resistance genes or virulence factors62 or introduce genetic circuits and synthetic pathways in a targeted manner.
Methods
Plasmid construction
All V. cholerae INTEGRATE plasmid constructs were generated from pQCascade, pTnsABC and pDonor (described previously35) using a combination of Gibson assembly, restriction digestion, ligation or inverse (around-the-horn) PCR. All PCR fragments for cloning were generated using Q5 DNA Polymerase (NEB).
Different vector backbone versions of pSPIN were cloned by generating a PCR fragment of the INTEGRATE expression construct and mini-transposon and combining with digested vector backbone in a Gibson assembly reaction. pSPAIN was generated by Gibson assembly: a 0.98-kb mini-Tn was first cloned into pBBR1, followed by double digestion within the mini-Tn and insertion of the INTEGRATE expression construct.
Components of the ShoINT system were synthesized by GenScript, and the system was cloned in-house. Cas12k and the single guide RNA (sgRNA) were cloned as two separate cassettes on a pCDFDuet-1 (pCDF) plasmid; the native TnsB-TnsC-TniQ operon was cloned on a pCOLADuet-1 (pCOLA) plasmid; and the mini-Tn was cloned on a pUC19 plasmid. Sho-pEffector was generated from these plasmids using Gibson assembly. The ShCAST system was synthesized by GenScript according to the constructs described previously40, with pHelper on pUC19 and pDonor on pCDF backbones. Pairwise protein sequence similarities among the VchINT, ShoINT and ShCAST machinery can be found in Supplementary Table 3.
Each construct containing a spacer was first generated with a filler sequence containing tandem BsaI recognition sites in place of the spacer (for VchINT and ShoINT) or tandem BbsI sites in place of the spacer (for ShCAST). New spacers were then cloned into the array by phosphorylating complementary oligonucleotides with T4 PNK (NEB), hybridizing the oligonucleotides and ligating them into the BsaI- or BbsI-digested plasmid. Double- and triple-spacer arrays were cloned by combining two or three oligoduplexes with compatible sticky ends in a single ligation reaction. crRNAs for VchINT were designed with 32-nt spacers targeting genomic sites with a 5′ CC PAM. sgRNAs for ShoINT and ShCAST were designed with 23-nt spacers targeting genomic sites with a 5′ RGTN and 5′ NGTT PAM, respectively. All spacer sequences used for this study are available in Supplementary Table 5. We note that our guide RNA design algorithm (described in Supplementary Fig. 11) was not used to generate the spacers for this study.
Cloning reactions were used to transform NEB Turbo E. coli, and plasmids were extracted using Qiagen Miniprep columns and confirmed by Sanger sequencing (GENEWIZ). Transformed cells were cultured in liquid LB media or solid LB agar media, with the addition of 100 μg ml−1 of carbenicillin for pUC19 plasmids, 50 μg ml−1 of spectinomycin for pCDF and pSC101* plasmids, and 50 μg ml−1 of kanamycin for pCOLA, pSC101 and pBBR1 plasmids. All plasmid construct sequences are available in Supplementary Table 1, and a subset is available from Addgene.
E. coli culturing and general transposition assays
A full list of strains used for transposition experiments is provided in Supplementary Table 6.
All E. coli transformations were performed using homemade chemically competent cells and standard heat shock transformation, followed by recovery in LB at 37 °C and plating on LB agar media with the appropriate antibiotics at the concentrations described above. Typical transformation efficiencies were >103 colony-forming units per microgram of total DNA. All standard transposition assays in E. coli involved incubation at 37 °C for 24 h after recovery and plating. However, for experiments involving incubation at 30 °C or 25 °C, cells were grown for an extended total of 30 h to produce enough cell material for downstream analyses. To control for this extended incubation time, all incubations for Fig. 4c were performed uniformly for 30 h, including the 37 °C incubations. For similar reasons, a 30-h incubation was performed for the ΔrecA transposition assays (Supplementary Fig. 4c) owing to a significantly slower growth rate of the ΔrecA strain.
For most experiments involving an IPTG-inducible T7 promoter, transformed cells were plated directly on LB agar plates containing 0.1 mM IPTG for 24 h after recovery. Exceptions were made for the pUC19 pSPIN construct (Fig. 1d), transposition assays performed for Fig. 1e and all ShoINT and ShCAST experiments, for which transformed cells were first plated on LB agar without induction and incubated for 16 h and then scraped and replated on LB agar with 0.1 mM IPTG and incubated for an additional 24 h. This replating protocol was generally used when initial transformation efficiencies were low, potentially from IPTG-induced toxicity; separating the transformation and induction steps allowed enough cells to be generated for lysis and further analysis. To avoid any adverse effects of IPTG degradation on transposition efficiency, LB agar plates were made fresh with frozen IPTG stocks, stored at 4 °C and used within 7 d of preparation. All cell culturing after transformation and recovery was performed on solid media to avoid competitive growth effects causing enrichment of rare events, with the exception of kinetics experiments (Supplementary Fig. 5).
Experiments involving three plasmids (pDonor, pTnsABC and pQCascade or variants thereof) were performed by first transforming chemically competent cells with pTnsABC and pDonor, picking a single colony and growing overnight with double antibiotic selection, generating chemically competent cells using standard methods and then transforming these cells with the pQCascade plasmid. Experiments involving two plasmids were performed by co-transforming chemically competent cells with both plasmids simultaneously. We note that this generally resulted in lower transformation efficiencies and required more input DNA than single-plasmid transformations.
Transposition assays in K. oxytoca and P. putida
A full list of strains used for transposition experiments is provided in Supplementary Table 6.
Electrocompetent K. oxytoca cells were generated as follows. Cells were grown overnight to saturation and then diluted 1:100 and grown to OD600 of ~0.4–0.5. Cells were then placed on ice for 15–30 min, washed three times with ice-cold 10% glycerol and then concentrated 100-fold in ice-cold 10% glycerol. Next, 50 μl of cells were electroporated with 50 ng of plasmid, using 0.1-cm cuvettes at 1.8 kV. Cells were recovered in 1 ml of LB media for 2 h at 37 °C and were plated on LB agar with selection at 37 °C for 24 h.
For P. putida transformations, electrocompetent cells were generated following a previously described protocol64. Briefly, overnight cultures were washed three times with 300 mM sucrose and concentrated 50-fold. Cells were then distributed into 100-μl aliquots and separately electroporated with 100 ng of plasmid using 0.2-cm cuvettes at 2.5 kV. Cells were recovered in 1 ml of LB media for 2 h at 30 °C and were plated on LB agar with selection at 30 °C for 24 h.
All transposition assays for K. oxytoca and P. putida were performed by transforming with a pSPIN construct on a pBBR1 backbone, expressed from a constitutive J23119 promoter. Cells were incubated on LB agar for 24 h after recovery. Colonies were then scraped for genomic DNA (gDNA) extraction using the Wizard Genomic DNA Purification Kit (Promega).
PCR and qPCR analysis of transposition
E. coli cells transformed with INTEGRATE machinery were scraped from LB agar plates and resuspended in liquid LB, and the OD600 of the resulting suspensions was taken. From each resuspension, approximately 3.2 × 108 cells (equivalent of 200 μl of OD600 = 2.0 of resuspended cells) were retained for lysis and downstream analysis. In scenarios where less than this amount of cell resuspension was recovered, the entire resuspension was used for lysis. Cells were pelleted by centrifugation at 4,000g for 2 min, the LB supernatant was poured off and cells were resuspended in 80 μl of deionized (DI) water, followed by lysis at 95 °C for 10 min. The lysates were cooled to room temperature and pelleted by centrifugation at 4,000g for 2 min, and the supernatant was diluted 20-fold in DI water and used for subsequent analyses. Further lysate dilutions were sometimes used, as we have observed polymerase inhibition from raw lysates at higher concentrations than the 20-fold dilution, especially during qPCR.
PCR reactions for E. coli samples were performed using Q5 Polymerase (NEB) in a 12.5-μl reaction containing 200 μM dNTPs, 0.5 μM of each primer and 5 μl of diluted lysate supernatant. Primer pairs comprised one mini-Tn-specific primer and one genome-specific primer; each primer pair probes for integration in either the T-RL or T-LR orientation. PCR amplicons were generated over 30 PCR cycles and were resolved by electrophoresis on 1–1.5% agarose gels stained with SYBR Safe (Thermo Fisher Scientific). PCR reactions for K. oxytoca and P. putida were performed using a similar primer design strategy as for E. coli, with Q5 Polymerase in a standard 50-μl reaction mixture and with 20 ng of extracted gDNA as input instead of cell lysate.
qPCR reactions were performed on 2 μl of diluted lysates in 10-μl reactions, containing 5 μl of SsoAdvanced Universal SYBR Green 2× Supermix (BioRad), 2 μl of 2.5 μM mixed primer pair and 1 μl of water. Each lysate sample was analyzed with three separate qPCR reactions involving three primer pairs, as described previously35: two pairs comprise one mini-Tn-specific primer and one genomic-specific primer probing for either the T-RL or T-LR integration orientation, and one pair comprises two genome-specific reference primers at the rssA locus. Primer pairs were designed to amplify a product between 100 and 250 bp and were confirmed to have amplification efficiencies between 90% and 110% using serially diluted lysates. A full list of qPCR primers used in this study is provided in Supplementary Table 7. Integration efficiency (%) for each orientation is defined as 100 × (2^ΔCq), where ΔCq is the Cq(genomic reference pair) – Cq(T-RL pair OR T-LR pair); the total integration efficiency is the sum of both orientation efficiencies.
We note that our qPCR protocol was previously benchmarked using lysate samples simulating known integration efficiencies and orientation biases35. However, as efficiency is dependent on Cq measurement of both the genomic reference primer pair and the integration junction primer pair, variation in either measurement affects the final calculated efficiency value. This can lead to apparent measurements of >100% when the actual integration efficiency is close to 100%, particularly because variations in ΔCq values are amplified as the magnitude of raw Cq values increases. Thus, qPCR noise affects efficiency measurements more disproportionately at higher efficiencies.
Isolation of clonally integrated E. coli colonies
We previously reported that single colonies might be genetically heterogeneous (that is, non-clonal) when integration occurs contemporaneously with colony expansion35. Therefore, all clonal isolation steps were preceded by a ‘bottlenecking’ step, where all colonies from the first solid media growth were scraped and pooled together, resuspended in LB and plated at an appropriate dilution to obtain a fresh set of colonies. Colonies were then picked and resuspended in 100 μl of DI water, followed by lysis at 95 °C for 10 min. Then, 5 μl of lysate was used as an input template for PCR, as described above. Colonies were identified as clonal using three sets of PCRs per target site per lysate, as described previously35. Briefly, two PCR pairs probed for the presence of the T-RL and T-LR integration orientation, respectively, and a third pair amplified across the genomic region of the expected insertion junction. A colony was considered clonal when only one of the first two primer pairs resulted in successful amplification and when the third pair solely amplified a larger product corresponding to the genomic region with integrated mini-Tn. When crRNA-4 (targeting lacZ) was used for integration, blue/white screening was used to select for white colonies, which were then confirmed with the above PCR strategy.
Liquid culture time course experiments
While performing initial kinetics experiments, we noticed that pSPIN plasmids with constitutive promoters, which were extracted from NEB Turbo cloning cells, contained contaminating gDNA with targeted integration that was detectable at low levels with both endpoint PCR as well as qPCR, especially at early time points after a fresh transformation reaction using pSPIN. To avoid this low-level gDNA contamination generating an artifact during time course experiments, we instead used plasmids that were passaged in and extracted from E. coli strain BW25113, which does not have the corresponding genomic site targeted by crRNA-4.
For each sample in the time course experiment, three separate transformations were performed and pooled together after a 1-h recovery at 37 °C. The pooled recovery was then split into three equal volumes, each of which was used to inoculate a 25-ml liquid LB outgrowth culture. The cultures were incubated while shaking continuously for 24 h at either 37 °C or 30 °C. At each time point indicated in Supplementary Fig. 5a, a 1-ml sample was taken from each liquid culture for OD600 measurement (WPA Biowave, 2.0 max reading) and subsequent analysis of integration efficiencies by qPCR; samples were either lysed at 95 °C or frozen at −20 °C within 10 min of collection. For early time points with dilute cultures, 1-ml samples were pelleted entirely and resuspended in a sufficient volume to achieve a Cq value of 18–20 for the genomic qPCR primer pair. For later time points with significantly turbid cultures, dilution of the sample was performed based on the OD600 measurements, as described in the qPCR section above.
Transposition with linear mini-Tn
Linear donors were generated by PCR amplification of a 1,104-bp donor sequence containing a full chloramphenicol resistance cassette (Supplementary Table 1) from a non-replicative plasmid template. A subsequent DpnI digestion and gel extraction step ensured that no intact plasmid was present in the linear donor sample. Control transformations of an E. coli pir+ strain with the resulting amplicons were performed to confirm that there was no contaminating plasmid left in the linear DNA sample.
Chemically competent cells harboring a constitutive pEffector plasmid with either non-targeting crRNA or crRNA-4 were transformed with 500–600 ng of the linear donor using heat shock transformation, as described above. After a 1-h recovery at 37 °C, cells were plated directly onto LB agar containing 25 μg ml−1 of chloramphenicol and incubated an additional 16 h at 37 °C before the resulting colonies were counted. Colonies were then scraped and bottlenecked onto a fresh LB agar plate with chloramphenicol selection, followed by PCR analysis of colonies, as described above.
VchINT target immunity experiments
A pSPIN derivative with crRNA-4 (targeting lacZ) on a pSC101* temperature-sensitive backbone was used to integrate a 0.98-kb mini-Tn in BL21(DE3) cells at 30 °C for 30 h. A clonal insertion strain was isolated as described above, and the pSPIN plasmid was cured by isolating a colony from cells cultured at 37 °C overnight in liquid LB media. The resulting cells were made chemically competent and co-transformed with a separate pDonor containing a different cargo alongside a pEffector construct with crRNA targeting a site d-bp away from the original crRNA-4 target (as indicated in Fig. 3a). qPCR was then performed, with mini-Tn-specific primers designed to bind within the cargo to distinguish it from the original crRNA-4 insertion. For each target site, normalization was done by performing the same transposition and qPCR assay in wild-type (WT) BL21(DE3) cells and dividing the immunized qPCR efficiency by the WT efficiency. We note that, due to the presence of two identical repeats of the mini-Tn R end and L end (111 bp and 149 bp in length, respectively) from the original and new insertions, it is possible that the observed target immunity phenotype is affected by low-level recombination between these repetitive sequences, which is not taken into account in our analyses.
Mini-Tn remobilization experiments
BL21(DE3) cells with a clonal crRNA-4 (lacZ) insertion, isolated and cured of INTEGRATE plasmids as described above, were made chemically competent. These cells were transformed with a pEffector construct with crRNA-1 (targeting downstream of glmS), without any donor plasmid containing a new mini-Tn. Presence of the mini-Tn at both lacZ and glmS was probed for by PCR, as described above.
Mini-Tn competition experiments were performed similarly, where cells were transformed with a pEffector construct with crRNA-1 alongside a pDonor that carried the same mini-Tn as the lacZ insertion, except for a 5-bp mutation at the 3′ end of the R-end. This mutation site was used to design mini-Tn-specific primers to distinguish the genome-borne and plasmid-borne mini-Tn at both lacZ and glmS sites.
VchINT/ShoINT orthogonality experiments
For the orthogonality experiments in Fig. 3c, BL21(DE3) cells were co-transformed with a two-plasmid combination of either Vch-pEffector or Sho-pEffector and either Vch-pDonor or Sho-pDonor. The spacers for both systems were designed to target the same region of the lacZ locus. For PCR analysis of integration activity, transposon-specific primers were designed to bind in the R end or L end of the mini-Tn.
For data shown in Fig. 3d, BL21(DE3) cells containing a clonal lacZ insertion were co-transformed with Sho-pEffector and Sho-pDonor. The Sho-sgRNA was designed with a spacer targeting a similar region near the glmS locus that is targeted by Vch crRNA-1. PCR analysis was performed as described above.
Amino acid auxotrophy experiments
M9 minimal media were prepared with the following components: 1× M9 salts (Difco), 0.4% glucose, 2 mM MgSO4 and 0.1 mM CaCl2. M9 agar was prepared as above, with the addition of 15 g L−1 of dehydrated agar (BD). L-threonine and/or L-lysine was supplemented at 1 mM, as indicated.
For individual thrC or lysA targeting experiments, BL21(DE3) cells were transformed with a pSPIN construct with a crRNA targeting either gene. Transformed cells were incubated on LB agar at 37 °C for 24 h. Bottlenecking and clonal insertion identification by PCR were performed as described above, and cells were then evaluated for ability to grow in M9 minimal media with and without addition of the appropriate amino acid.
For multiplexed targeting of both thrC and lysA, BL21(DE3) cells were transformed with a pSPIN construct expressing a thrC–lysA-targeting double-spacer array. Cells were then incubated and bottlenecked on LB agar, as above, and bottlenecked colonies were then stamped onto M9 agar plates supplemented with no amino acids, only threonine or lysine or both amino acids, to identify a growth phenotype. For data presented in Fig. 4e, this screen was performed on 30 colonies for each of three independent experiments.
The OD600 growth curve analysis was performed by first inoculating WT BL21(DE3) cells or isolated auxotrophic strains from −80 °C glycerol stocks into LB media for overnight growth. Then, 1 ml of each culture was pelleted at 16,000g and resuspended in 1 ml of DI water and then used to inoculate a culture on a 96-well cell culture plate in the desired growth media at a 1:1,000 dilution. Growth assays were then performed with a Synergy H1 plate reader shaking at 37 °C for 18 h, with the OD at 600 nm taken every 5 min. Each sample was measured in three technical replicates in separate wells on the sample plate, and values were normalized to blank wells containing media only.
Cre-LoxP genomic deletion experiments
A modified pSPIN construct was generated, in which the mini-Tn was modified to include a 34-bp LoxP recognition sequence for Cre recombinase, and a double-spacer CRISPR array encoded crRNA-4 and a second spacer targeting the same strand 2.4, 10 or 20 kb away from crRNA-4. BL21(DE3) cells were transformed with pSPIN, incubated and bottlenecked, and colonies with clonal double T-RL insertions were isolated by a combination of blue/white screening and PCR, as described above. We note that, although the two targets for the 2.4-kb deletion were close enough to each other to elicit target immunity effects, we were still readily able to isolate the desired clone. Double-insertion clones were made chemically competent and then transformed with a plasmid expressing Cre recombinase from an IPTG-inducible T7 promoter (a gift from N. Geijsen; Addgene no. 62730). Cells were incubated at 37 °C for 16 h and bottlenecked, and colonies that underwent recombination were isolated by PCR. We observed small colonies and low transformation efficiencies when transformed cells were plated on LB agar containing 0.1 mM IPTG, whereas we could readily isolate recombined clones without IPTG induction, suggesting that low-level leaky Cre expression in the absence of induction was sufficient for recombination. Thus, all Cre-recombinase transformations were performed with no IPTG present.
Tn-seq library preparation and sequencing
Transformations for Tn-seq transposition assays were carried out as described above, using donor plasmids containing a mini-Tn where the 8-bp terminal repeat of the R end was mutated to generate an MmeI recognition sequence (from 5′-TGGTGATA-3′ to 5′-TGGTGGAA-3′). We previously showed that a mini-Tn with this mutation is still active, with a ~50% decrease in total integration efficiency35. Transformed cells were incubated on LB agar at 37 °C for 24 h, except for assays shown in Supplementary Figs. 4d and 6b, in which cells were incubated at 30 °C for 30 h. Colonies were then scraped and resuspended in liquid LB media, and 0.5 ml (approximately 2 × 109 cells) was used for gDNA extraction with the Wizard Genomic DNA Purification Kit (Promega), which typically yielded 50 μl of 0.5–1.5 μg μl−1 gDNA.
NGS libraries were prepared in parallel in PCR tubes. First, 1 μg of gDNA was digested with 4 U of MmeI (NEB) for 2 h at 37 °C in a 50-μl reaction containing 50 μM S-adenosyl methionine and 1× CutSmart buffer, followed by heat inactivation at 65 °C for 20 min. MmeI digestion results in the generation of 2-nt 3′ overhangs. Reactions were cleaned up with 1.4× Mag-Bind TotalPure NGS magnetic beads (Omega) according to the manufacturer’s instructions, and elutions were done using 30 μl of 10 mM Tris-Cl, pH 7.0. Double-stranded i5 universal adaptors containing a 3′-terminal NN overhang were ligated to the MmeI-digested gDNA in a 20-μl ligation reaction consisting of 16.86 μl of MmeI-digested gDNA, 5 nM adaptor, 400 U of T4 DNA ligase (NEB) and 1× T4 DNA ligase buffer. Reactions were carried out at room temperature for 30 min and then cleaned up with magnetic beads. Because the donor plasmid (pDonor, pSPIN or pSPIN-R) contains a copy of the mini-Tn that can also be digested with MmeI and ligated with i5 adaptor, we included a restriction enzyme recognition site (HindIII for pDonor or Bsu36I for pSPIN and pSPIN-R) in the 17-bp space between the 5′ end of the mini-Tn and the MmeI digestion site. By digesting the entirety of the adaptor-ligated gDNA elution with 20 U of HindIII or Bsu36I in a 34.4-μl reaction for 2 h at 37 °C, before a heat inactivation step at 65 °C for 20 min, we were able to reduce contamination of donor sequences within the NGS libraries. DNA clean-up using magnetic beads was then performed.
Eluted DNA was then amplified in a PCR-1 step, where adaptor-ligated transposons were enriched using a universal i5-adaptor primer and a transposon-specific primer with a 5′ overhang containing a universal i7 adaptor. In a 25-μl PCR-1 reaction, 16.7 μl of HindIII/Bsu36I-digested gDNA was mixed with 200 μM dNTPs, 0.5 μM primers, 1× Q5 reaction buffer and 0.5 U of Q5 DNA Polymerase (NEB). Amplification proceeded for 25 cycles at an annealing temperature of 66 °C. Reaction products were used as template and diluted 20-fold into a second 20-μl PCR reaction (PCR-2) with indexed p5/p7 Illumina primers. The PCR-2 reaction was subjected to ten amplification cycles with an annealing temperature of 65 °C, after which analytical gel electrophoresis was performed to verify amplification for each library. Barcoded reactions were pooled and resolved by 2.5% agarose gel electrophoresis, followed by isolation of DNA using the Gel Extraction Kit (Qiagen), and NGS libraries were quantified by qPCR using the NEBNext Library Quant Kit (NEB). Illumina sequencing was performed with a NextSeq mid-output kit with 150-cycle single-end reads and automated adaptor trimming and demultiplexing (Illumina).
For pSPIN libraries involving a spike-in, 10 μl of a 0.02 ng μl−1 spike-in plasmid was added to each 1 μg of DNA sample before MmeI digestion, and library preparation proceeded as described above. The plasmid contains a full-size MmeI-mini-Tn but without a Bsu36I restriction site in the 17-bp fingerprint region; this fingerprint, therefore, survives the Bsu36I donor digestion step for pSPIN libraries and provides a constant ‘contamination’ in the final library to control for sequencing depth.
Random fragmentation library prep and sequencing
BL21(DE3) cells were transformed with Vch-pSPIN or Sho-pSPIN or were co-transformed with pHelper and pDonor for ShCAST. Transformation, incubation and gDNA extraction with the Wizard Genomic DNA Purification Kit (Promega) were performed as described previously.
Roughly 2.5 μg of gDNA was fragmented for 14 min following the NEBNext dsDNA fragmentase protocol. The fragmentation reactions were purified using 1.4× Mag-Bind TotalPure NGS (Omega) beads with an elution step in 30 μl of 1× TE. Approximately 1 μg of the fragmented DNA was used for end preparation, adapter ligation and USER cleavage, according to the NEBNext Ultra II DNA Library Prep Kit for Illumina protocol. Reactions were purified using 1.2× magnetic beads with an elution step in 30 μl of DI water.
To reduce the number of fragments deriving from the mini-Tn on the donor plasmid, the samples were digested with restriction enzymes (VchINT: KpnI and Bsu36I; ShoINT: PstI and HindIII; ShCAST: NcoI and AvrII) overnight at 37 °C. The reactions were then purified using 1.2× magnetic beads with 30 μl of DI water elution.
PCR-1 reactions were performed using Q5 Polymerase (NEB) in a 20-μl reaction containing 200 μM dNTPs, 0.5 μM of each primer and 30 ng of input DNA. Transposon-containing fragments were amplified over 20 PCR cycles using a transposon-specific primer carrying an i5 adapter and an i7-specific primer. A second PCR reaction (PCR-2) was used to add specific Illumina index sequences to the i5 and i7 adapters over ten PCR cycles in a 25-μl reaction, with 1.25 μl from PCR-1 as the template DNA.
Samples were purified using the Qiagen PCR Clean-up Kit, and DNA concentrations were measured using a DeNovix spectrometer. The amount of DNA was normalized, and samples were combined. The pooled libraries were then quantified using the NEBNext Library Quant Kit, and Illumina sequencing was performed as described above.
Analysis of NGS data
Analysis of all Tn-seq and random fragmentation sequencing data was performed using a custom Python pipeline. Demultiplexed raw reads were filtered to remove reads where less than half of the bases passed a Phred quality score of 20 (Q20, corresponding to >1% base miscalling). Reads that contained the 15-bp 5′-terminal sequence of the mini-Tn R end (allowing up to one mismatch) were then selected, and the 17-bp sequence directly upstream of this R-end sequence was extracted. This 17-bp ‘fingerprint’ sequence corresponds to the distance from the R end to the MmeI digestion site and contains the genomic sequence context in which the mini-Tn is found (Supplementary Fig. 3a). Reads without sufficient length to extract a 17-bp fingerprint were removed from analysis. For each random fragmentation sample, because the two transposon ends were amplified and sequenced as two separate libraries, extraction of fingerprints from reads was performed separately for the R and L transposon ends.
Fingerprint sequences were aligned to reference genomes of the corresponding species and strain, depending on each specific library. The full list of strains, species and corresponding reference genome accession identifiers is provided in Supplementary Table 6. Reference genomes for E. coli and P. putida were obtained from published National Center for Biotechnology Information (NCBI) genomes, whereas our K. oxytoca parent strain was sequenced and assembled de novo using whole-genome SMRT-seq to obtain the reference genome (see below for the SMRT-seq method). Alignment to the reference genome was performed using the Bowtie2 alignment library65. Perfect mapping was used for alignment, and only reads that aligned exactly once to the reference genome were used for downstream analyses. Fingerprints that did not map to the reference genome were screened for sequences corresponding to undigested donor contamination or for fingerprints mapping downstream of the CRISPR array on the donor plasmid, which correspond to self-targeting events (Fig. 5d,e). For cases where a spike-in plasmid was used, the number of fingerprints containing the spike-in sequence was also determined.
Bowtie2 alignment outputs were used to generate genome-wide integration distributions, and the number of reads corresponding to integration events at each position across the reference genome was plotted. For visualization purposes, these positions were grouped into 456 separate 10-kb bins, and peaks were plotted as a percentage of total mapping reads. This analysis was performed similarly for each random fragmentation library by combining R-end and L-end fingerprints before alignment and plotting. In cases where a spike-in was used, peaks were further normalized by the number of spike-in fingerprints detected, and the plot for each non-targeting control was scaled similarly to the corresponding targeting sample.
Integration site distance distribution plots were generated from Bowtie2 alignments by plotting the number of reads versus the distance between the 3′ end of the target site and the site of insertion deduced from the reads, at single-bp resolution. The on-target percentage was calculated as the percentage of reads corresponding to integration events within a 100-bp window centered at the integration site with the largest number of reads. The integration orientation bias is defined as the ratio of number of reads corresponding to T-RL insertions to those corresponding to T-LR insertions. For random fragmentation libraries, alignments for this analysis were performed separately for R-end and L-end fingerprints, and the results were combined to generate the plot.
We note that our Tn-seq sequencing is susceptible to potential biases arising from differences in MmeI digestion efficiency at each site and in ligation efficiencies of 3′-terminal NN overhang adapters, which were not taken into account.
Pacific Biosciences SMRT-seq and analysis
gDNA samples for library preparation were extracted from overnight LB cultures using the Wizard Genomic DNA Purification Kit (Promega), as described above. Multiplexed microbial whole-genome SMRTbell libraries were prepared, as recommended by the manufacturer (Pacific Biosciences). Briefly, 2 μg of high-molecular-weight gDNA from each sample (n = 12 per pool) was sheared using a g-TUBE to ~10 kb (Covaris). These sheared gDNA samples were then used as input for SMRTbell preparation using the Template Preparation Kit 1.0, where each sample was treated with a DNA Damage Repair and End Repair mix to repair nicked DNA and repair blunt ends. Barcoded SMRTbell adapters were ligated onto each sample to complete SMRTbell library construction, and these libraries were then pooled equimolarly, with a final multiplex of 12 samples per pool. The pooled libraries were then treated with exonuclease III and VII to remove any unligated gDNA and cleaned up with 0.45× AMPure PB beads to remove small fragments and excess reagents (Pacific Biosciences). The completed 12-plex pool was annealed to sequencing primer V3 and bound to sequencing polymerase 2.0, before being sequenced using one SMRT Cell 8M on the Sequel II system with a 20-h movie.
After data collection, the raw sequencing reads were demultiplexed according to their corresponding barcodes using the Demultiplex Barcodes tool found within the SMRTLink analysis suite, version 8.0. Demultiplexed subreads were downsampled ten-fold by random downsampling and assembled de novo using the Hierarchical Genome-Assembly Process tool, version 4.0, using the following parameters: Aggressive mode = off, Downsampling factor = 0, Minimum mapped length = 50 bp, Seed coverage = 30, Consensus algorithm = best, Seed length cutoff = −1, Minimum mapped concordance = 70%.
Subread mapping and structural variant analysis were performed using the PB-SV tool within SMRTLink 8.0, using the BL21(DE3) genome (accession CP001509.3) as reference, with the following parameters: Minimum SV length = 20 bp, Minimum reads supporting variant for any one sample = 2, Minimum mapped length = 50 bp, Minimum length of copy number variant = 1,000 bp, Minimum reads supporting variant (total over all samples) = 2, Minimum % of reads supporting variant for any one sample = 20%, Minimum mapped concordance = 70%. VCF outputs were used to generate the SV analysis results shown in Supplementary Table 2, and BAM alignments were visualized with IGV to generate genome-deletion coverage plots (Supplementary Fig. 9). We found no evidence of cointegrate products for Vch INTEGRATE in this study, consistent with transposition proceeding through a cut-and-paste pathway dependent on both TnsA and TnsB35,66,67.
For the coverage plot of the 10-kb insertion (Supplementary Fig. 6c), circular consensus sequence reads were generated with SMRTLink 8.0 and filtered using a custom Python script to obtain only reads containing 20 bp of the R end and/or L end of the mini-Tn. These filtered reads were then aligned to an artificial reference genome, in which the entire 10-kb mini-Tn was computationally inserted 49 bp downstream of the crRNA-4 target sequence of the CP001509.3 reference genome. Alignments were performed using Geneious Prime at medium sensitivity with no fine-tuning.
Animal ethics statement
All animal experiments were performed in compliance with Columbia University Medical Center Institutional Animal Care and Use Committee protocols AC-AAAU6464 and AC-AAAU1460.
Isolation of live mouse gut bacteria
Conventionally raised 7-week-old B6-albino and BALB/C female mice (Taconic Biosciences) were the source of the two different types of mammalian gut complex communities used in this study. Mice were housed with 12-hour light/12-hour dark cycles, with a temperature of 65–75 °F (~18–23 °C) and 40–60% humidity. Fresh fecal pellets were collected from mice, and live gut bacteria were isolated by mechanical homogenization. Briefly, 250 μl of PBS was added to previously weighed pellets in a microcentrifuge tube. Pellets were thoroughly mechanically disrupted with a motorized pellet pestle, and then 750 μl of PBS was added. The disrupted pellets in PBS were then subjected to four iterations of vortex mixing for 15 s at medium speed, centrifugation at 1,000 r.p.m. for 30 s at room temperature, recovery of 750 μl of supernatant in a new tube and replacement of that volume of PBS before the next iteration. The resulting 3 ml of isolated cells was pelleted by centrifugation at 4,000g for 5 min at room temperature; the supernatant was discarded; and cells were resuspended in 0.5–1.0 ml of PBS. All gut bacteria isolations were performed in an anaerobic chamber (Coy Laboratory Products).
Ex vivo conjugation using INTEGRATE to target specific strains in natural complex communities
Before conjugation, donor strains harboring conjugative pSPIN vectors were grown from a single colony in 5 ml of LB-Lennox media (BD) supplemented with 50 μg ml−1 of kanamycin and 50 μM DAP at 37 °C overnight (~10 h). The recipient community was isolated anaerobically from fresh mouse feces as described above, immediately before conjugation. Donor cells were washed three times in PBS and quantified by OD600, whereas fecal bacteria were quantified by flow cytometry using SYTO 9 staining. Then, either 108 or 107 donor cells (E. coli strain EcGT2 containing pSPIN) and 108 target cells (K. oxytoca strain M5a1) were mixed with 109 fecal bacteria cells, pelleted by centrifugation at 4,000g and resuspended in 10–20 μl of PBS. The mixtures were spotted on MGAM + 2% agar plates supplemented with 50 μM DAP and incubated at 37 °C anaerobically for 24 h. After conjugation, cells were scraped from the plate into 1 ml of PBS and plated on LB-Lennox agar and LB-Lennox 2% agar supplemented with 50 μg ml−1 of kanamycin at different dilutions.
Metagenomic 16S sequencing
gDNA from fecal bacterial extraction was isolated using mechanical lysis with 0.1 mm Zirconia beads (BioSpec) and subsequently purified with SPRI beads (AMPure). PCR amplification of the 16S ribosomal RNA (rRNA) V4 region and multiplexed barcoding of samples were performed in accordance with previous protocols. The V4 region of the 16S rRNA gene was amplified with customized primers according to the method described by Kozich et al.68, with the following modifications: 1) alteration of 16S primers to match updated EMP 505f and 806rB primers and 2) use of Nextera XT indices such that each index pair was separated by a Hamming distance of >2, so that Illumina low-plex pooling guidelines could be used. Sequencing was done with the Illumina MiSeq system (300V2 kit) immediately before the experiment (T0) and after 24 h (T24).
Analysis of 16S NGS data
The composition of the communities for each sample was determined from 16S sequencing data via DADA2 pipeline69 to generate the amplicon sequence variance (ASV) tables and calculate relative abundances. Phyloseq70 and the Silva database (https://www.arb-silva.de/) were used to assign the taxonomy. In the MiSeq run, two blank controls with sterile water as input material were included to check for contaminants in the reagents and to filter out contaminant ASVs, if present. Reads mapping to non-bacterial DNA (for example, mitochondria, plastids or other eukaryotic DNA) were also excluded from the analysis. Only ASVs with more than 15,000 reads and present in more than 1% of the samples were considered in the downstream analysis.
Quantification of site-specific transposition efficiency in bacterial communities
Different dilutions from the community conjugations were plated on LB agar with 50 μg ml−1 of kanamycin selection for pSPIN. Between 40 and 66 colonies were picked for each single experiment (~15–20 colonies per replicate, to capture at least 5% efficiency), and transposon–genome junction PCRs and 16S PCRs were run for each single colony. Junction PCRs were analyzed by 1% agarose gel electrophoresis to confirm integration events, and 16S Sanger sequencing confirmed that each colony was K. oxytoca.
Statistics and reproducibility
Analytical PCRs resolved by agarose gel electrophoresis produced similar results in three independent replicates (Figs. 3b-d, 4h and 5b and Supplementary Figs. 5c,e, 7a and 14c) or were analyzed by gel electrophoresis once (Supplementary Fig. 7c) and verified using qPCR for three independent replicates (Supplementary Fig. 7e).
Supplementary Material
Acknowledgements
We thank N. Jaber for laboratory support, J. Bondy-Denomy for discussions, L.F. Landweber for qPCR instrument access, J. Mohabir for assistance with NGS read alignment, the JP Sulzberger Columbia Genome Center for NGS support and M.L. Smith, I. Oussenko and the Genomics Technology Laboratory at the Icahn School of Medicine at Mount Sinai for SMRT sequencing. H.H.W. acknowledges funding support for this work from the National Science Foundation (MCB-1453219), the National Institutes of Health (1U01GM110714 and 1R01AI132403), the Office of Naval Research (N00014-17-1-2353) and the Burroughs Wellcome Fund (PATH1016691). C.R. is supported by a Junior Fellows Scholarship from the Simons Society of Fellows. S.H.S. acknowledges a generous startup package from the Columbia University Irving Medical Center Dean’s Office and the Vagelos Precision Medicine Fund.
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41587-020-00745-y.
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Competing interests
P.L.H.V., S.E.K. and S.H.S. are inventors on patents and patent applications related to CRISPR–Cas systems and uses thereof. H.H.W. is a scientific advisor to SNIPR Biome. S.H.S. is a co-founder and scientific advisor to Dahlia Biosciences and an equity holder in Dahlia Biosciences and Caribou Biosciences.
Supplementary information is available for this paper at https://doi.org/10.1038/s41587-020-00745-y.
Data availability
NGS data are available in the NCBI Sequence Read Archive (BioProject accession code PRJNA668381). Published genomes used for analyses were obtained from the NCBI (accessions codes CP001509.3, U00096.3, CP009273.1 and AE015451.2). Datasets generated and analyzed in the current study, as well as custom scripts used for the described data analyses, are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
Custom Python scripts used for the described NGS data analyses are available online via GitHub (https://github.com/sternberglab/Vo_etal_2020). The INTEGRATE guide RNA design tool and associated documentation are available online via GitHub (https://github.com/sternberglab/INTEGRATE-guide-RNA-tool).
References
- 1.Dunbar CE et al. Gene therapy comes of age. Science 359, eaan4672 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Gelvin SB Integration of Agrobacterium T-DNA into the plant genome. Annu. Rev. Genet 51, 195–217 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Davy AM, Kildegaard HF & Andersen MR Cell factory engineering. Cell Syst. 4, 262–275 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Brophy JAN et al. Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nat. Microbiol 3, 1043–1053 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Miyazaki R & van der Meer JR A new large-DNA-fragment delivery system based on integrase activity from an integrative and conjugative element. Appl. Environ. Microbiol 79, 4440–4447 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martínez-García E & de Lorenzo V Transposon-based and plasmid-based genetic tools for editing genomes of gram-negative bacteria. Methods Mol. Biol 813, 267–283 (2012). [DOI] [PubMed] [Google Scholar]
- 7.van Opijnen T, Bodi KL & Camilli A Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6, 767–772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang HH et al. Genome-scale promoter engineering by coselection MAGE. Nat. Methods 9, 591–593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharan SK, Thomason LC, Kuznetsov SG & Court DL Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc 4, 206–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Buchholz F, Muyrers JP & Stewart AF A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet 20, 123–128 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Baba T et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotta-de-Almeida V, Schonhoff S, Shibata T, Leiter A & Snapper SB A new method for rapidly generating gene-targeting vectors by engineering BACs through homologous recombination in bacteria. Genome Res. 13, 2190–2194 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datsenko KA & Wanner BL One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang W, Bikard D, Cox D, Zhang F & Marraffini LA RNA-guided editing of bacterial genomes using CRISPR–Cas systems. Nat. Biotechnol 31, 233–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K et al. Defining synonymous codon compression schemes by genome recoding. Nature 539, 59–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sukhija K et al. Developing an extended genomic engineering approach based on recombineering to knock-in heterologous genes to Escherichia coli genome. Mol. Biotechnol 51, 109–118 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Wang HH et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vento JM, Crook N & Beisel CL Barriers to genome editing with CRISPR in bacteria. J. Ind. Microbiol. Biotechnol 46, 1327–1341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun 8, 15179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosicki M, Tomberg K & Bradley A Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol 36, 765–771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wannier TM et al. Improved bacterial recombineering by parallelized protein discovery. Proc. Natl Acad. Sci. USA 117, 13689–13698 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corts AD, Thomason LC, Gill RT & Gralnick JA A new recombineering system for precise genome-editing in Shewanella oneidensis strain MR-1 using single-stranded oligonucleotides. Sci. Rep 9, 39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters JM et al. Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nat. Microbiol 4, 244–250 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.St-Pierre F et al. One-step cloning and chromosomal integration of DNA. ACS Synth. Biol 2, 537–541 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Tellier M, Bouuaert CC & Chalmers R Mariner and the ITm superfamily of transposons. Microbiol. Spectr 3, MDNA3–0033–2014 (2015). [DOI] [PubMed] [Google Scholar]
- 26.van Opijnen T & Camilli A Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol 11, 435–442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haniford DB & Ellis MJ Transposons Tn10 and Tn5. Microbiol. Spectr 3, MDNA3–0002–2014 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Goodall ECA et al. The essential genome of Escherichia coli K-12. Mbio 9, e02096–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen SP & Wang HH An engineered Cas-transposon system for programmable and site-directed DNA transpositions. CRISPR J. 2, 376–394 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhatt S & Chalmers R Targeted DNA transposition in vitro using a dCas9-transposase fusion protein. Nucleic Acids Res. 6, 7–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enyeart PJ, Mohr G, Ellington AD & Lambowitz AM Biotechnological applications of mobile group II introns and their reverse transcriptases: gene targeting, RNA-seq, and non-coding RNA analysis. Mob. DNA 5, 2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esvelt KM & Wang HH Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol 9, 641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perutka J, Wang W, Goerlitz D & Lambowitz AM Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J. Mol. Biol 336, 421–439 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Karberg M et al. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat. Biotechnol 19, 1162–1167 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Klompe SE, Vo PLH, Halpin-Healy TS & Sternberg SH Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Peters JE, Makarova KS, Shmakov S & Koonin EV Recruitment of CRISPR–Cas systems by Tn7-like transposons. Proc. Natl Acad. Sci. USA 114, E7358–E7366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halpin-Healy TS, Klompe SE, Sternberg SH & Fernández IS Structural basis of DNA targeting by a transposon-encoded CRISPR–Cas system. Nature 577, 271–274 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Faure G et al. CRISPR–Cas in mobile genetic elements: counter-defence and beyond. Nat. Rev. Microbio 17, 513–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters JE Targeted transposition with Tn7 elements: safe sites, mobile plasmids, CRISPR/Cas and beyond. Mol. Microbiol 112, 1635–1644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strecker J et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chavez M & Qi LS Site-programmable transposition: shifting the paradigm for CRISPR–Cas systems. Mol. Cell 75, 206–208 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Hou Z & Zhang Y Inserting DNA with CRISPR. Science 365, 25–26 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Ronda C, Chen SP, Cabral V, Yaung SJ & Wang HH Metagenomic engineering of the mammalian gut microbiome in situ. Nat Meth 16, 167–170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stellwagen AE & Craig NL Avoiding self: two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J. 16, 6823–6834 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greene EC & Mizuuchi K Target immunity during Mu DNA transposition. Transpososome assembly and DNA looping enhance MuA-mediated disassembly of the MuB target complex. Mol. Cell 10, 1367–1378 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Hagemann AT & Craig NL Tn7 transposition creates a hotspot for homologous recombination at the transposon donor site. Genetics 133, 9–16 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin MT et al. In Methods in Enzymology Isotope Labeling of Biomolecules—Labeling Methods Vol. 565 (Ed. Kelman Z) 45–66 (Academic Press, 2015). [Google Scholar]
- 48.Hickman AB & Dyda F DNA transposition at work. Chem. Rev 116, 12758–12784 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abbas AF, Al-Saadi AGM & Alkhudhairy MK Biofilm formation and virulence determinants of Klebsiella oxytoca clinical isolates from patients with colorectal cancer. J. Gastrointest. Cancer 51, 855–860 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Kim D-K et al. Metabolic engineering of a novel Klebsiella oxytoca strain for enhanced 2,3-butanediol production. J. Biosci. Bioeng 116, 186–192 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Loeschcke A & Thies S Pseudomonas putida—a versatile host for the production of natural products. Appl. Microbiol. Biotechnol 99, 6197–6214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikel PI & de Lorenzo V Pseudomonas putida as a functional chassis for industrial biocatalysis: from native biochemistry to trans-metabolism. Metab. Eng 50, 142–155 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Sun J et al. Genome editing and transcriptional repression in Pseudomonas putida KT2440 via the type II CRISPR system. Microb. Cell Fact 17, 41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wirth NT, Kozaeva E & Nikel PI Accelerated genome engineering of Pseudomonas putida by I-SceI-mediated recombination and CRISPR–Cas9 counterselection. Microb. Biotechnol 13, 233–249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai SQ & Joung JK Defining and improving the genome-wide specificities of CRISPR–Cas9 nucleases. Nat. Rev. Genet 17, 300–312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y et al. Multicopy chromosomal integration using CRISPR-associated transposases. ACS Synth. Biol 9, 1998–2008 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Yu BJ & Kim C Minimization of the Escherichia coli genome using the Tn5-targeted Cre/loxP excision system. Nat. Biotechnol 20, 1018–1023 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Adiego-Pérez B et al. Multiplex genome editing of microorganisms using CRISPR–Cas. FEMS Microbiol. Lett 366, fnz086 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horlbeck MA et al. Mapping the genetic landscape of human cells. Cell 174, 953–967(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bassalo MC et al. Rapid and efficient one-step metabolic pathway integration in E. coli. ACS Synth. Biol 5, 561–568 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Rubin BE et al. Targeted genome editing of bacteria within microbial communities. Preprint at bioRxiv 10.1101/2020.07.17.209189 (2020). [DOI] [Google Scholar]
- 62.Valderrama JA, Kulkarni SS, Nizet V & Bier E A bacterial gene-drive system efficiently edits and inactivates a high copy number antibiotic resistance locus. Nat. Commun 10, 5726–5728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duque E et al. Identification and elucidation of in vivo function of two alanine racemases from Pseudomonas putida KT2440. Environ. Microbiol. Rep 9, 581–588 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Aparicio T, de Lorenzo V & Martínez-García E CRISPR/Cas9-enhanced ssDNA recombineering for Pseudomonas putida. Microb. Biotechnol 12, 1076–1089 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rice PA, Craig NL & Dyda F Comment on ‘RNA-guided DNA insertion with CRISPR-associated transposases’. Science 368, eabb2022 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Strecker J, Ladha A, Makarova KS, Koonin EV & Zhang F Response to comment on ‘RNA-guided DNA insertion with CRISPR-associated transposases’ Science 368, eabb2920 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Kozich JJ, Westcott SL, Baxter NT, Highlander SK & Schloss PD Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Callahan BJ et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McMurdie PJ & Holmes S phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS data are available in the NCBI Sequence Read Archive (BioProject accession code PRJNA668381). Published genomes used for analyses were obtained from the NCBI (accessions codes CP001509.3, U00096.3, CP009273.1 and AE015451.2). Datasets generated and analyzed in the current study, as well as custom scripts used for the described data analyses, are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Custom Python scripts used for the described NGS data analyses are available online via GitHub (https://github.com/sternberglab/Vo_etal_2020). The INTEGRATE guide RNA design tool and associated documentation are available online via GitHub (https://github.com/sternberglab/INTEGRATE-guide-RNA-tool).