Abstract
Among the various bacterial infections, tuberculosis (TB) remains a life-threatening infectious disease responsible as the most significant cause of mortality and morbidity worldwide. The co-infection of human immunodeficiency virus (HIV) in association with TB burdens the healthcare system substantially. Notably, M.tb possesses defence against most antitubercular antibiotic drugs, and the efficacy of existing frontline anti-TB drugs is waning. Also, new and recurring cases of TB from resistant bacteria such as multidrug-resistant TB (MDR), extensively drug-resistant TB (XDR), and totally drug-resistant TB (TDR) strains are increasing. Hence, TB begs the scientific community to explore the new therapeutic class of compounds with their novel mechanism. M.tb requires iron from host cells to sustain, grow, and carry out several biological processes. M.tb has developed strategic methods of acquiring iron from the surrounding environment. In this communication, we discuss an overview of M.tb iron-scavenging tools. Also, we have summarized recently identified MbtA and MbtI inhibitors, which prevent M.tb from scavenging iron. These iron-scavenging tool inhibitors have the potential to be developed as anti-TB agents/drugs.
Introduction
TB is caused by Mycobacterium tuberculosis (M.tb) and remains the world's leading infectious killer disease responsible for significant mortality. TB usually infects the lungs and other body parts, including the kidney, spine, and brain.1 During TB infection, M.tb has periods of active growth and heightened metabolic inactivity.1,2 According to WHO Report 2022, approximately 10.6 million people fall ill due to active TB and 150 359 due to drug-resistant TB. Among HIV-negative people, TB illness claimed over 1.6 million lives. The co-infection of HIV with TB significantly burdens the healthcare system and claimed 187 000 deaths.3,4M.tb bacilli are phagocytozed by alveolar macrophages, eliminating most of them. At the same time, some bacilli may survive in the early phagosomal compartment and avoid immune response. Hence, the bacilli can live in the host without exhibiting symptoms, a latent TB infection. The regrowth of the bacilli or poor management of the bacilli by the immune system can result in a further disease outbreak. When an infected individual shows TB symptoms, it is called active infection.5–7 Persisters are the subpopulation of M.tb bacilli, which can survive antibiotic treatment. Nonetheless, persisters are genetically identical to drug-susceptible bacteria and continue to be non-replicating with low or no nullification in their metabolic activity. They may be responsible for latent TB infections.8 Here, the challenges lie in elucidating the mycobacterial persistence and latency biological mechanisms.
The current standard antitubercular regimen for drug-susceptible TB disease consists of a two-month induction phase, which includes isoniazid (INH), rifampin (RIF), pyrazinamide (PZA), and ethambutol (EMB), followed by a four-month consolidation phase with at least INH and RIF to eradicate the dormant slow-growing bacteria.9 Recent decades have seen a fast spread in multidrug-resistant bacteria such as MDR-TB, XDR-TB, and TDR-TB due to the misuse and abuse of anti-TB medications. At the same time, an increase in the resistance has augmented the virulence of the bacilli.7,9
The current first-line anti-TB regimen has drawbacks, including poor adherence to the treatment regimen, long duration, complexity, side effects, and drug resistance. Also, the standard MDR-TB treatment course requires expensive combination therapy with second-line medications for 18–24 months.1 Further, antiretroviral and anti-TB medication interactions make TB therapy more difficult for HIV-positive patients. It is incredibly challenging to modify drug dosages to safely and efficiently treat people with TB and HIV infections, especially in settings with limited resources. Another challenge is that the detection and diagnosis of TB are not rapid and have to rely on chest radiography and sputum smear microscopy. The identification of the strains is another daunting task. Moreover, identifying HIV-related drug-resistant TB is tricky, costly, time-consuming, and technically demanding.10,11 The next challenge is identifying new chemical scaffolds from cell-based or phenotypic-based screening.12
The tuberculosis infections associated with these drug-resistant M.tb strains pose serious threats worldwide. In 2019, the first 6-month regimen comprising bedaquiline (BDQ), pretomanid, and linezolid were approved for MDR and XDR TB treatment.13
According to the Centers for Disease Control and Prevention (CDC), MDR-TB strains resist at least INH and RIF, the two most potent first-line anti-TB drugs.14 XDR-TB strains are a rare type of MDR TB that are resistant to INH and RIF, plus any fluoroquinolone (FQ) and at least one of three injectable second-line drugs (i.e., amikacin (AMI), kanamycin (KAN), or capreomycin (CAP)). TDR-TB refers to M.tb strains that show in vitro resistance to all the first and second-line anti-TB drugs.15 WHO recommended daily treatment of drug-susceptible TB with an initial two months intensive phase of drugs (INH, RIF, PZA, and ethambutol), continued by dual therapy of INH and RIF for the last four months.16 WHO recommended treating RIF-resistant and multidrug-resistant TB with core and non-core drugs. The core drugs comprise group A: fluoroquinolones (levofloxacin, moxifloxacin, and gatifloxacin), group B: second-line injectables drugs (amikacin, capreomycin, kanamycin, and streptomycin), Group C: ethionamide or prothionamide, cycloserine, or terizidone, linezolid, clofazimine, and group D: add-on drugs (not part of the core multidrug-resistant regimen), which is further categorized in D1 (PZA, EMB, high-dose INH), D2 (bedaquiline (BDQ), delamanid), and D3 (para-aminosalicylic acid, imipenem plus cilastatin (requires clavulanate), meropenem (requires clavulanate), amoxicillin plus clavulanate, and thioacetazone).15,17 INH-monoresistant M.tb is treated by substituting INH with a later-generation FQ (levofloxacin or moxifloxacin) as part of a standard treatment regimen (RIF, PZA, and EMB).18 To prevent resistance in bacilli and the complete eradication of TB, a combination of anti-TB therapy is needed, which should act against active and latent M.tb.19 There is an urgent need for medications with novel modes of action, which could be effective against susceptible and resistant strains of mycobacteria.20,21 In recent years, substantial progress has been made in identifying novel targets and inhibitors for treating TB in humans. The anti-TB agents/drugs in clinical development are discussed below.
New approaches for treating TB
The current anti-TB regimen requires prolonged treatment, contributing to noncompliance among patients. Also, first-line and second-line anti-TB drugs are associated with several side effects.22 TB therapy with direct and prolonged release would benefit TB patients. Notably, inhalation therapy provides several benefits, such as the drug will reach the target site directly, evading first-pass metabolism, and thus it will reduce the systemic side effects. In addition, the inhalation of antibiotics will also have local impacts. However, it would likely give no benefit, nor would it achieve adequate therapeutic serum concentrations in the case of extrathoracic disease.23 Nanoparticle-based delivery systems have shown convincing results in treating chronic TB infections. The controlled and sustained release of drugs will have the advantages of anti-TB nanoparticle-based medicines over free drugs as it will reduce the dosage frequency and resolve poor patient compliance.24,25 In addition, inhalational liposome anti-TB formulation is one of the future alternatives for sustained and onsite drug release.26
M.tb can live in the host as dormant bacilli within macrophages. Bacilli enter a dormant condition during latent TB infection and exhibit antibiotic tolerance.27 Notably, most conventional drugs are effective against active bacilli but fail to show action against persistent bacilli.27M.tb gene expression promotes its adaptation and survival in vitro and in the host. Importantly, two-component regulatory system (TCS) is a mechanism maintained by M.tb to detect changes in the environment cue and the modulation of gene expression.28 These systems integrate with multiple stimuli, coordinating global gene expression changes. Usually, M.tb possess many 2CRSs, which respond to specific signals and allow adaptive responses.29 TCS comprises a sensor histidine kinase (HK) and a response regulator (RR). The change in the external environment resulted in HK activation, which further autophosphorylates to a conserved histidine residue. Furthermore, HK mediates phosphotransfer to a conserved aspartate residue on the RR. Moreover, the phosphorylation of RR leads to DNA binding and promotion of the transcription factor.29M.tb has 11 paired TCS and several orphaned sensor kinases and response regulators. Also, several M.tb TCS, including DosRS, MprAB, PhoPR, PrrAB, and SenX3-RegX3, are required for virulence in macrophages or animal infection models of TB.30,31 Anti-TB therapy, which targets the TCS, will disrupt environmental sensing and may sensitize pathogens to clearance by the immune system. Thus, this TCS targeting therapy, in combination with primary antibiotics, can eradicate TB.30
The resuscitation-promoting factor (RPF) stimulates the resuscitation of M.tb from dormancy. A study demonstrated that in M. luteus, a picomolar concentration of RPF is needed to resurrect the growth of dormant bacilli. In this regard, the previous strategy suggests that potent inhibitors of Rpfs will not allow latent TB and avoid the reactivation of the sleeping pathogen. But this strategy has a drawback: it hides latent TB rather than treating or eradicating it. In contrast, Seidi et al. proposed that administering RPF (to awaken the latent M.tb bacilli) with high serum levels of antibiotics can cure and eradicate TB.32,33
An effective immune system is essential to eradicate latent M.tb infections. In a host-directed therapy, the stimulation of the immune response will improve the clinical outcomes in TB patients. Tumor necrosis factor (TNF) is essential for resistance against intracellular bacterial infections but is also associated with human immunopathologies. TNF blockers ameliorate the inflammatory conditions but compromise the host's immunity to TB. At the same time, the concurrent administration of TNF blockers with standard anti-TB drugs has shown improvement in treating TB infections in patients.34,35 Also, the use of a patient's own bone marrow-derived mesenchymal stromal cells (MSCs) as an adjunct cellular therapy seems to be a viable treatment option against MDR and XDR-TB as MSCs modulate the immune responses and provide anti-inflammatory and tissue repairing effects on damaged tissues.36
Anti-tubercular drug targets
Electron transport chain (ETC) and ATP synthase
In M.tb, two NADH dehydrogenases are present in the mycobacteria: Ndh and NdhA. Type I NADH dehydrogenase is homologous to the complex I in mitochondria and is unnecessary for bacterial growth.37,38 Electrons derived from NADH are released into the electron transport chain by NADH dehydrogenase, which reduces the menaquinone pool (MK/MKH2). Also, the menaquinone pool can be reduced by electron donors via succinate dehydrogenase (SDH). At the same time, two succinate dehydrogenase enzymes (Sdh-1 and Sdh-2) and one fumarate reductase carried out the reverse reaction. The electrons from the menaquinone pool can be transferred to the cytochrome bc1 complex. In mycobacteria, the cytochrome bc1 complex forms a supercomplex (consisting of subunits QcrA-C), which moves the electrons onto oxygen by the terminal cytochrome aa3-type oxidase (CtaC-F).39 QcrB is the cytochrome b subunit of the cytochrome bc1, an essential part of a super complex terminal oxidase in the ETC.40 Also, oxygen alternatively can be reduced by a cytochrome bd-type terminal oxidase, which directly accepts electrons from the menaquinone pool.40 Overall, in the electron transport chain of mycobacteria, NADH or succinate donates electrons to membrane protein complexes that subsequently pass these electrons to terminal oxidases or reductases through the electron-carrier menaquinone.39 This activity leads to the pumping of protons from the cytoplasm to the periplasm, which generates a transmembrane pH gradient and contributes to the membrane potential. Furthermore, this force drives the rotation of the F-ATP synthase c-ring, which consequently drives ATP (adenosine triphosphate) synthesis. On an average, M.tb exhibits a proton motive force (PMF) of −110 mV.41 The F1Fo-ATP synthase is one of the essential enzymes critical for energy production in both the mycobacteria's proliferating aerobic and hypoxic dormant stages. ATP synthase enzyme utilizes the energy stored in the form of PMF, i.e., electrochemical potential difference of the transmembrane for the production of ATP.42 At low PMF, in an environment with limited oxygen concentration, this reaction is reversed; ATP synthase can catalyze the hydrolysis of ATP to give ADP (adenosine diphosphate) and the inorganic phosphate to maintain PMF across the membrane.43 ATP synthase is a multisubunit complex consisting of membrane-embedded Fo (subunits ab2c10–15) and a cytoplasmic domain F1 (subunits α3β3γδε), joined with a central (γ-subunit and ε-subunit) and a peripheral stalk (β-subunits). ATP synthase is one of the smallest nanomotors, which rotates its central rotor clockwise to synthesize ATP from ADP and inorganic phosphate and counter-clockwise to hydrolyze ATP.44 Notably, the three α- and three β-subunits are arranged alternately to form a spherical structure and in the β-subunits where the catalytic ATP synthesis or hydrolysis activity occurs.41 The respiratory ATP synthase is required for the optimal growth of mycobacteria. Thus, targeting ETC and ATP synthase is a validated target in M.tb.37,45
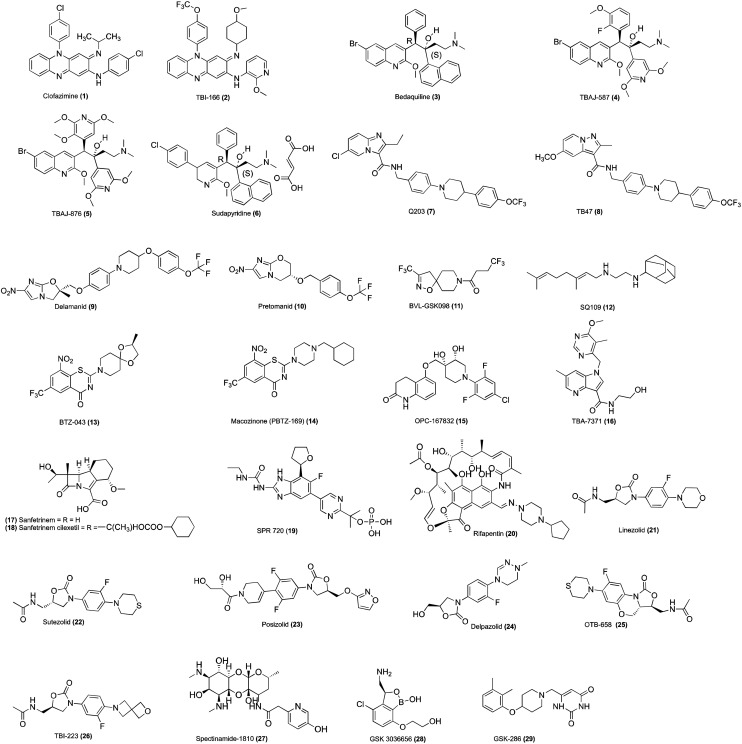
Clofazimine (1, Fig. 1) belongs to the riminophenazine class. It is a prodrug reduced by the Mycobacterial enzyme NADH-quinone oxidoreductase type II (NDH-2). Clofazimine is a competitive inhibitor of menaquinone (MK-4) and checks the point of entry of electrons into the respiratory chain.46–49 It also leads to the spontaneous re-oxidation of the reduced clofazimine by oxygen, producing reactive oxygen species (ROS), most probably O2−, contributing to antimycobacterial activity.47,50 It is a well-known antileprotic agent demonstrating antimycobacterial activity against slowly and rapidly growing mycobacterium species.51–54 Clofazimine is registered in the Chinese Clinical Trial Registry in background treatment regimens for treating XDR-TB in humans.55 TBI-166 (2, Fig. 1) is a potent congener of clofazimine and displayed activity against drug-sensitive, MDR-TB, and intracellular M.tb.48,56
Fig. 1. Chemical structures of anti-TB agents/drugs under clinical evaluation.
Bedaquiline (3, Fig. 1) congeners such as TBAJ-587 (4, Fig. 1), TBAJ-876 (5, Fig. 1), and sudapyridine (WX-081, 6, Fig. 1) showed broad-spectrum antimycobacterial activity by inhibiting mycobacterial F-ATP synthase.57,58 TBAJ-587, TBAJ-876, and sudapyridine are currently in preclinical development for treating TB.57–60 Notably, TBAJ-587 and TBAJ-876 showed antimycobacterial activity against replicating and non-replicating M.tb H37Rv.57 Sudapyridin displayed antimycobacterial activity against MDR-TB strains and demonstrated excellent pharmacokinetic parameters in animals, better lung exposure, and lower QTc prolongation than bedaquiline.59,60
Q203 (7, Fig. 1) chemically belongs to the imidazopyridine amide class of compounds, which showed antimycobacterial activity by targeting the respiratory cytochrome bc1 complex.61 The cytochrome bc1 complex is an essential component of the respiratory ETC, and the inhibition leads to the depletion of ATP, resulting in cell death.62 TB47 (8, Fig. 1) is a congener of Q203, which targets QcrB in the electron transport chain. It showed synergistic and sterilizing activity in combination with clofazimine. The cryo-EM structures revealed Q203 and TB47 bound to the quinol-binding site and thus blocked the binding of quinone at the Qo site; therefore, it inhibits electron transfer in M.tb.63 Q203 inhibits the growth of MDR-TB and XDR-TB clinical isolates in the culture broth medium and a mouse model of TB.61 It also showed inhibitory activity against M. ulcerans.62,64
Fatty acid synthase (I) & (II) and polyketide synthase 13 (Pks13)
Mycolic acids are biosynthesized by mixed fatty acid synthase (FAS) and polyketide synthase (PKS) enzymes. Mycolic acids are essential in maintaining virulence and for the survival of M.tb within the macrophages.65 Mycolate compounds, including trehalose monomycolate (TMM), trehalose dimycolate (TDM), and arabinogalactan-mycolate, constitute significant components of the mycobacterial cell wall.66 FAS I perform the de novo synthesis of C16–C18 and C24–C26 fatty acids in a bimodal fashion, and type II multienzyme complex FAS II catalyzes the processive addition of multiple malonate units and carries out the extension in the length of the fatty acids onto the products of FAS-I C16–C18 to generate the long-chain meromycolic acid (C48–C64). FAS-II multienzyme complex includes enzymes KasA and KasB, MabA and (3R)-hydroxyacyl-ACP dehydratases, HadAB, HadBC, and the enoyl-reductase InhA.67 FadD32 is essential for activating meromycolic acids to facilitate condensation with a shorter fatty acid, resulting in the final β-keto-alpha alkyl mycolic acid.21 FadD32 carried out the two enzymatic functions, i.e., fatty acyl-AMP ligase (FAAL) activity, wherein it adenylates meromycolic acids produced by the fatty acid synthase II biosynthetic pathway and fatty acyl ACP synthetase (FAAS) activity, which transfer the activated intermediate to the ACP domain of the condensing enzyme PKS13. HadD is a fatty acid synthase type II protein, which has a role in the synthesis of full-size alpha- and epoxy-MAs during the late FAS-II elongation cycles.68 Also, HadD performs the catalysis of 3-hydroxyacyl dehydration at the late stage of FAS-II elongation cycles during keto-MA biosynthesis, and its deletion produces a marked change in keto-MA content, size distribution, and production of full-size molecules observed in the M.tb.69Mycobacterium smegmatis having hadD knockout produces only the medium-size alpha-MAs.68 Pks13 performs the condensation reaction between two long-chain fatty acyls (meromycolyl-AMP and carboxyl-acyl-CoA) to produce α-alkyl-β-ketoacyl derivatives, the precursors of mycolic acids.70 Further, Pks13 performs an acyltransferase activity and transfers the mycolate precursors to trehalose, forming TMM, a common precursor of the mycolate-containing compounds.70–72 Isoxyl and thioacetazone showed antimycobacterial activity by inhibiting the dehydratase HadAB of the type II FAS.73
Nitroimidazoles are used for treating anaerobic bacterial and parasitic infections. The synthetic modifications of metronidazole led to OPC-6783 (delamanid, 9, Fig. 1) and PA-824 (pretomanid, 10, Fig. 1). Delamanid and pretomanid are prodrugs and require intracellular activation for their biological function. M.tb reduces the nitro group to generate active species with antimycobacterial activity. The activation of delamanid and pretomanid depends on the actions of the reduced deazaflavin cofactor F420 and activating enzyme F420-dependent glucose-6-phosphate dehydrogenase and nitroreductase gene product of Rv3547.74 Delamanid showed antitubercular activity by inhibiting methoxy-mycolic acid and ketomycolic acid. In contrast, pretomanid inhibits the synthesis of cell wall lipids and proteins. Delamanid is active against MDR-TB and exhibits significant bactericidal activity against replicating and dormant bacilli in the in vivo guinea pig model of chronic TB infection.75 Delamanid is recommended by the European Union (EU) for its use in combination with an optimized background anti-TB regimen (OBR) in patients with MDR-TB.76 In 2019, the US FDA approved pretomanid as part of the BPaL (BDQ, pretomanid, and linezolid) regimen for treating adult patients suffering from pulmonary XDR or treatment-intolerant or non-responsive MDR-TB.77
InhA (enoyl-acyl carrier protein reductase)
InhA is an enoyl-acyl carrier protein (ACP) reductase, which is a critical component of the FAS-II system and performs the reduction of the double bond at position two of growing fatty acid chains that are linked to ACP.78 INH active metabolite isonicotinyl acyl radical reacts covalently with the NAD cofactor to form INH-NADH adduct, which inhibits the ACP reductase of FAS type II, which leads to the accumulation of long-chain fatty acids.22,79–81 Ethionamide is a structural analogue of INH and a prodrug, which requires activation by ethA-encoded mono-oxygenase.82 The activated ETH block enoyl reductase InhA and is followed by mycolic acid synthesis inhibition. Mycobacteria having mutations in the ethR gene reduced the synthesis of EthA and thus decreased ETH activation. SMARt-420 (small molecule aborting resistance-420) belongs to the spiroisoxazoline class and is devoid of anti-TB activity. It is given in conjunction with ethionamide to increase the sensitivity of M.tb toward ETH and completely reverse its resistance.83,84 On a similar line, BVL-GSK908 (11, Fig. 1) is used in association with ETH/prothionamide to increase the sensitivity of ETH/prothionamide. The combination of BVL-GSK908 with ETH/prothionamide has been developed to treat pulmonary TB.85
MmpL3
The mycobacterial membrane protein large (MmpL) proteins belong to the resistance nodulation and cell division (RND) superfamily transporters.37M.tb genome encodes 13 MmpL proteins (MmpL1–13), among which MmpL3 translocates TMM across the inner membrane.67,86 The transported glycolipids TMM act as a mycolic acid donor. The Ag85 complex enzyme, by its acyltransferase action, performs the synthesis of TDM and arabinogalactan-mycolates. These glycolipids form the significant components of the mycomembrane.87 MmpL3 depletion in the Mycobacterium smegmatis demonstrated the loss of cell wall mycolylation and TDM levels with a concomitant increase in TMM.88 The silencing of the MmpL3 in M.tb led to hypersusceptibility to MmpL3 inhibitors and rapid bactericidal effect on actively replicating cells in vitro and also reduced bacterial loads in the mouse lungs.89 Moreover, the downregulation of MmpL3 leads to the ceasing of cell division and rapid cell death.90 SQ109 (12, Fig. 1) is a diamine analogue of ethambutol, which showed antimycobacterial activity by multiple mechanisms. SQ109 block the MmpL3 transport, leading to the failure in the mycolate attachment to the arabinogalactan. It also disrupts the cell wall assembly and interferes with the mycolic acids within the cell wall of M.tb. In addition, it disrupts the PMF in the mycobacteria.91,92
DprE1
The gene dprE1 (rv3790) encodes DprE1, an essential flavoenzyme in the M.tb and involved in cell wall synthesis.93 The decaprenylphosphoryl-β-d-arabinofuranose (DPA) is needed for the biosynthesis of arabinogalactan. The decaprenylphosphoryl-β-d-ribofuranose 2′-oxidase (DprE1) works in concert with decaprenylphosphoryl-2-keto-d-ery-thropentose reductase (DprE2) in two steps to give DPA. Initially, decaprenylphosphoryl-β-d-ribose (DPR) is converted to decaprenylphosphoryl-2-keto-β-d-erythro-pentofuranose (DPX) by DprE1 and then subsequently DPX is converted into DPA by DprE2. DPA acts as a substrate for arabinosyltransferase, and the synthesis of d-arabinofuranose (Araf) residues is a building block of arabinogalactan polysaccharides.37,94 DprE1 is essential for the growth and survival of M.tb, which is confirmed by the DprE1 knockout strain.95 BTZ043 (13, Fig. 1) and PBTZ169 (14, Fig. 1) chemically belong to the benzothiazone (BTZ) class. BTZ043 and PBTZ169 are prodrugs and require an aromatic nitro group for the activation to bind to the DprE1.96,97 Mycobacteria reduces the nitroaromatic group into nitrosoarene. The cysteine residue of DprE1 reacts with the nitroso group to form a semimercaptal adduct. Thus, on attachment to DprE1, it inactivates DprE1.98 OPC-167832 (15, Fig. 1) is chemically 3,4-dihydrocarbostyril, which showed antimycobacterial activity in in vitro and in vivo mouse models of chronic TB infections.99 Moreover, OPC-167832, in combination with other drugs such as BDQ, FQs, and PZA, showed a significant reduction of the bacterial colony-forming units (CFUs) in the lung and spleen.99 TBA-7371 (16, Fig. 1) belong to the azaindoles class, and unlike benzothiazone, it does not form a covalent bond with DprE1. TBA-7371 shows selective bactericidal activity against M.tb and Mycobacterium smegmatis. It also demonstrated efficacy in treating TB in BALB/c mice of acute and chronic infection models.100
Penicillin-binding proteins (PBPs)
Classical β-lactam, such as penicillins, are potent inhibitors of PBPs, which synthesize the peptidoglycan layer of the bacterial cell wall.101Mycobacterium shows resistance to antibiotics by modifying or degrading the antibiotics. The modification of the antibiotic by Mycobacterium modifying enzymes prevents the binding of the antibiotic to the target protein.102 For example, Mycobacterium produces an ambler class-A β-lactamase encoded by the blaC gene, which is responsible for the poor activity of the β-lactam antibiotics. The ambler class-A β-lactamase is present in M. smegmatis and M. fortuitum.103 Sanfetrinem (17, Fig. 1) is the first tricyclic β-lactam compound. Sanfetrinem cilexetil (GV-104326, 18, Fig. 1) is the oral prodrug of sanfetrinem and exhibits bactericidal activity against Gram-negative and Gram-positive bacteria.104 Notably, unlike other β-lactam, sanfetrinem is stable to β-lactamases and human renal dehydropeptidase (DHP).105 It has been repurposed for treating TB in humans. It does not show any significant change in MIC with or without clavulanate.106
DNA gyrase
DNA inside the cell is in supercoiled form, which has to be relaxed before initiating new DNA strand synthesis.38,107 In eukaryotic and prokaryotic organisms, type II DNA topoisomerases play a critical role in maintaining DNA function. M.tb DNA gyrase is a heterotetramer (A2B2), which consists of two subunits domains, i.e., gyrase A (encoded by gyrA gene) and gyrase B (encoded by gyrB gene), and are involved in the maintenance of the DNA topology.108 During the catalytic cycle, GyrA carries out bound DNA's catalysis breakage and religation (G-segment). In contrast, GyrB captures a second DNA segment (T-segment) and drives a series of conformational changes via ATP hydrolysis, which induces a G-segment break. DNA gyrase carries out several cellular reactions, including the restoration of the removal of negative supercoiling of closed circular double-stranded DNA, fork movement, and resolution of catenated DNA.38,109
Fluoroquinolones (FQs) showed antimycobacterial activity by interacting with the GyrA subunit of DNA gyrase and topoisomerase IV. Further, FQs block DNA replication, which leads to the accumulation of double-stranded DNA fragments. FQs such as levofloxacin and gatifloxacin are essential second-line drugs for treating MDR-TB. SPR719 is a novel aminobenzimidazole, an inhibitor of gyrase B that targets ATPase subunits, resulting in the growth inhibition of drug-susceptible and MDR-TB. In addition, it can reduce mycobacterial burdens in the lungs of infected mice.110 SPR719 showed potent activity against multiple clinical strains of nontuberculous mycobacteria (NTM), including Mycobacterium avium complex and M. abscessus.111 SPR720 (19, Fig. 1) is a phosphate prodrug of SPR719 and is currently being developed for treating infections caused by nontuberculous mycobacteria.112
RNA polymerase
In protein synthesis, transcription is a critical step in which mRNA is synthesized from the DNA template through initiation, chain elongation, and chain termination processes.
The transcription process is conducted via DNA-directed RNA polymerase (RNAP) enzyme.113 RNAP is of two forms, i.e., RNAP core (subunit composition: 2α, β, β′, and ω) and RNAP holo (RNAP core + σ). The RNAP core is responsible for RNA synthesis but cannot initiate transcription from the promoter.114 The inhibition of the RNAP activity is a validated drug target for killing the bacteria.113 Rifamycins, including rifampin, rifapentine, rifabutin, and rifamixin, showed antibacterial function by binding to and inhibiting bacterial RNAP. Notably, these rifamycins bind to an adjacent site on the RNAP active center and prevent the extension of RNA chains beyond a length of 2–3 nucleotides.115 Rifapentine is a cyclopentyl-substituted rifamycin; currently, it entered phase-3 clinical trials for treating TB.116 Rifabutin (20, Fig. 1) is another congener of rifamycin, which is used to treat TB in patients co-infected with HIV and receiving a combination of antiretroviral therapy.117,118 It exhibits antibacterial activity against NTM species, including M. abscessus subsp. abscessus ATCC 19977, M. abscessus subsp. bolletii CCUG 50184-T, and M. abscessus subsp. massiliense CCUG 48898-T.119
Protein synthesis
The prokaryotic ribosome consists of a smaller subunit (30S) and a larger subunit (50S). The 30S subunit comprises 16S rRNA chains and 20 proteins, and the 50S subunit of 5S rRNA chains and 34 proteins. These subunits and several factors contribute to mRNA translation onto a polypeptide chain in three major stages: initiation, elongation, and termination.120 Protein synthesis inhibitors interact with the ribosomal RNA, which induces conformational changes, thus disrupting the ribosome components' optimal arrangements and the ribosome's inability to synthesise proteins.121 Oxazolidinone showed antibacterial activity by binding to the 23S rRNA in the catalytic site of the 50S ribosome, which prevents the formation of a functional 70S initiation complex.122 Linezolid (21, Fig. 1) belongs to the oxazolidinone class. It is used to treat Gram-positive bacterial infection, the only marketed oxazolidinone, which has been used off-label in combination regimens to treat MDR-TB.122 PNU-100480 (sutezolid, 22, Fig. 1) and AZD-5847 (posizolid, 23, Fig. 1) are linezolid analogues with better activity against M.tb. Sutezolid is highly effective against both drug-susceptible and drug-resistant isolates of M.tb.123,124 Posizolid exhibits improved in vitro bactericidal activity against extracellular and intracellular M.tb compared to that of linezolid. Moreover, posizolid showed superior killing kinetics in broth media and macrophages than linezolid.125 Delpazolid (LCB01-0371, 24, Fig. 1) is a new oxazolidinone effective against several M. abscessus strains in vitro and in a macrophage infection model. Moreover, in the murine model, delpazolid showed similar efficacy to linezolid in the lungs.126 Delpazolid is in phase II clinical trials for pulmonary tuberculosis.102,127 OTB-658 (25, Fig. 1) exhibits better anti-TB activity in vitro and in vivo and safety profile than linezolid, and it is a promising compound, which can replace linezolid in TB treatment.128 TBI-223 (26, Fig. 1) is a constrained oxazolidinone (26, Fig. 1), which showed a safer pharmacokinetic profile than linezolid and is in phase I clinical stage of the development for treating TB.128
Spectinomycin (27, Fig. 1) is an aminoglycoside antibiotic with potent bacterial protein synthesis inhibition with poor antitubercular effects. M.tb with the overexpressed efflux pump Rv1258c results in decreased activity, thus limiting its application in treating TB. The synthetic modified spectinamides with a pyridyl side chain blocked the efflux of spectinamides and increased its concentration in the cells. It also exhibits increased interactions with the ribosome. Spectinamides are active against MDR-TB and XDR-TB strains.129 Among them, spectinamide-1810 selectively inhibits ribosomes, has a good safety profile, and is effective in several MDR-TB and XDR-TB infections mouse models.130–132 Also, it has an excellent pharmacokinetic and safety profile.133 It is currently in the early phase of TB drug development.129,134
GSK 3036656 (GSK 656, GSK 070, 28, Fig. 1) belongs to the 3-aminomethyl benzoxaborole scaffold, which showed anti-mycobacterial activity by inhibiting amino acyl-tRNA synthetases (AARSs).135 It interferes with M.tb LeuRS with an IC50 of 0.20 μM and against M.tb H37Rv, and it shows a MIC of 0.08 μM. Importantly, it exhibits remarkable pharmacokinetic (PK) profiles and efficacy against M.tb in the mouse infections model of TB. It is well tolerated after single and multiple doses, with no reports of serious adverse events in humans.136
Cholesterol catabolism
Cholesterol is an essential carbon source during latent TB infections. M.tb uses cholesterol as a unique source of carbon and energy. The cholesterol catabolism contributes to propionyl-CoA, a precursor that is incorporated into lipid virulence factors.137 The cholesterol catabolism is essential for M.tb persistence in the lungs of chronically-infected animals.138M.tb uses cholesterol genes, which can utilize host sterol for infection and persistence.139 The gene upregulation was seen in Mycobacterium smegmatis fed with cholesterol.138 GSK-286 (GSK 2556286, 29, Fig. 1) acts on mycobacterial cholesterol catabolism and inhibits intracellular M.tb H37Rv. Currently, GSK-286 is in preclinical evaluation for treating TB.140
Caseinolytic protease C1 (ClpP1)
M.tb harbors proteolytic machinery consisting of caseinolytic protein C1 (ClpP1) and caseinolytic protein C2 (ClpP2) subunits and hexameric ring-like ATP-dependent unfoldases composed of ClpX or ClpC1.141 ClpC1 is responsible for maintaining protein homeostasis in the mycobacteria and is identified as essential for cell growth. Notably, the mycobacteria without functional ClpC1 lead to the reduction of the procellular protein degradation.142,143 The disruption of the mycobacterial proteasome's proteolysis mechanism affects the mycobacteria's growth. Thus, ClpP1 is one of the essential targets of M.tb. Compounds such as rufomycins, Ilamycins, and ecumicin showed antitubercular activity by binding to ClpC1.144–147
LipY
M.tb harbors several genes encoding PE/PPE proteins, which have a potential role in pathogen virulence.37 The members of the PE-PPE family are designated according to the presence of either a conserved proline–glutamic acid (PE) or proline–proline–glutamic acid (PPE) motif within the highly conserved N-terminal domain of the protein, which is approximately 100 or 180 residues long, respectively. The PE-PPE family protein is involved in the pathogenesis of the M.tb.148 The PE/PPE proteins are secreted via type VII secretion systems known as the ESX secretion systems.149 LipY is also a PE protein with a C-terminal triglyceride lipase domain.150 LipY breaks down the host-derived triglycerides in lipid droplets and stored triglycerides that act as a source of energy during reactivation from the dormancy. It was observed that under nutrient starvation and oxygen-depleted conditions, extensive LipY-induced breakdown of triglycerides was observed in the M.tb.151 At the same time, the LipY-disrupted mutant showed a decrease in the hydrolysis of triglycerides. Also, the overexpression of LipY was found to increase M.tb virulence and showed increased mortality, weight loss, and bacterial loads in mice infected with TB. It also downregulates host immunity.150,152 Currently, it is a prospective drug/vaccine candidate for treating tuberculosis.
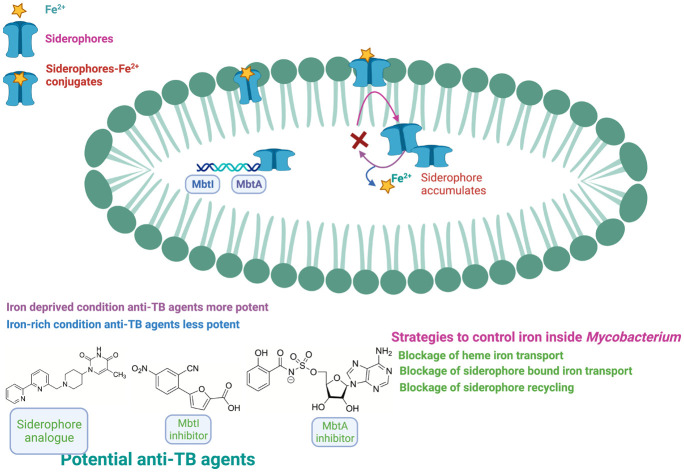
Importance of iron in Mycobacterium
The human body enzymes require metal as cofactors for their activity and are referred to as physiologically active metals (biometals). Micronutrients such as iron, zinc, copper, and magnesium are necessary for M.tb to carry out their physiological activities.153–155 The iron-binding proteins in the vertebrate hosts keep up low levels of free ferric iron in the serum and body fluids of 10−24 M. The host uses iron to maintain a critical role in numerous cellular functions, including amino acid biosynthesis, DNA biosynthesis, and glycolysis.154,155 Iron is essential in maintaining vital biological processes in M.tb. Moreover, M.tb requires iron as an essential micronutrient to sustain its growth in animal and hosts. Iron participates in oxidation–reduction reactions in electron transport by transitioning between the oxidized form ferric (Fe3+) and reduced form ferrous (Fe2+).156,157 The low iron concentration in M.tb can lead to environmental stress. Thus, it suggests that iron is essential for M.tb. The two types of iron storage proteins are present in the mycobacteria, i.e., bacterioferritin and heme-free ferritins, and the absence of these iron storage proteins leads to iron in excess, causing toxicity to mycobacteria and increases the susceptibility to antibiotics.157
Biosynthesis of Mycobacterium siderophores
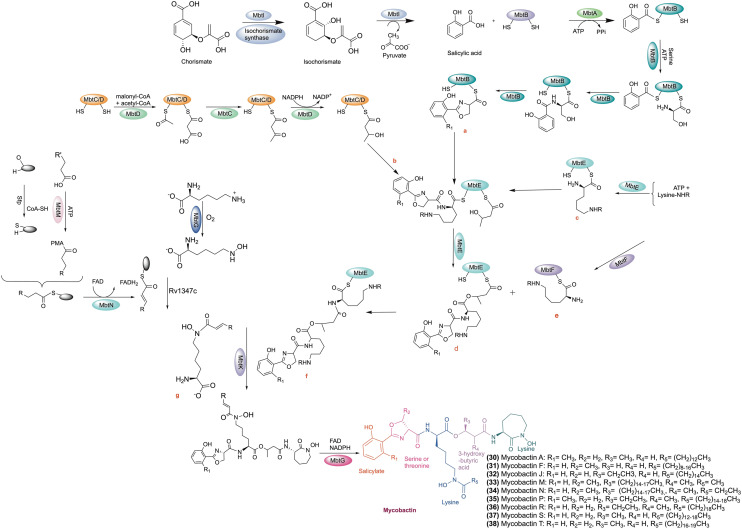
Iron is available as insoluble ferric oxide/hydroxide complexes in the environment. As a result, pathogenic microorganisms have evolved to sequester iron from the environment. Pathogens have developed strategies to obtain sufficient iron to support its growth by the acidification of the surrounding medium, using ferric oxidoreductases/permeases, secretion of hemolysins, transferrin and heme uptake, and siderophore secretion.154,158M.tb biosynthesize siderophores (‘sidirou foreas’ in Greek: iron carrier) to chelate iron for survival in extreme iron-deprived conditions. M.tb balances the excess iron and limits inside the host cell and thus can maintain its growth and TB infection in the host.159 Siderophores are low molecular weight organic compounds produced by numerous bacteria and fungi, which chelate the ferric iron. In M.tb, siderophore is one of the critical iron-scavenging machinery to fulfil the iron requirement from the host.159 There are two kinds of enzymatic machinery in the Mycobacterium. The biosynthesis of siderophores in Mycobacterium is either nonribosomal peptide synthetase-dependent (NRPS) or NRPS-independent.160 NRPS involves modular multienzymes for adenylation, thiolation, and condensation. Two distinct regions of the M.tb genome, i.e., the mbt-1 (mbtA–J) and mbt-2 (mbtK–N) loci, are involved in the siderophore biosynthesis.157,161 A collection of proteins encoded by the mbt-1 group (mbtA–mbtJ) of genes work together to form the scaffold of mycobactin. Four genes (mbtK–mbtN) of the mbt-2 group work together to activate the lipid residue and attach it to the growing platform of mycobactin. Enzymes MbtA–F consists of a mixed nonribosomal peptide synthetase polyketide assembly line synthesizing the mycobactin scaffold.161 MbtI and the isochorismate synthase together synthesize isochorismate from chorismite. Then, the isochorismate is converted into salicylic acid by the MbtI.162 MbtA carries out the integration of salicylic acid into the mycobactin core scaffold in two steps. MbtA, the salicyl-AMP ligase/salicyl-S-ArCP synthetase, activates and transfers the salicylate to the ACP (acyl carrier protein) site of MbtB to form salicyl-S-ACP. The aryl acid adenylation enzyme (AAAE) starts the first step of mycobactin biosynthesis using ATP to activate salicylic acid. This results in an adenylated intermediate, which is then passed to the thiolation domain.163 The adenylate acylation occurs in the second stage when AAAE transfers the acyl moiety to a nucleophilic sulphur atom on an aryl carrier domain. This action produces adenosine monophosphate and an aryl-capped siderophore (ACP).163 Then, salicyl-S-ACP linked to Ser/Thr are cyclized into a phenyloxazoline ring.162 Subsequently, a lysine activated by peptide synthetase MbtE or MbtF is attached to the peptide chain. Then, malonyl-CoA and acetyl-CoA are incorporated into the lysine residue by polyketide synthase MbtC/D. The developing molecule is then concentrated by MbtE using a modified l-lysine, and MbtD follows by adding a β-keto group to produce β-hydroxybutyrate. The first lysine is esterified at the α-carboxy with a β-hydroxy acid, and then the second lysine moiety cyclized to give a seven-membered lactam ring. Finally, MbtF joins the intermediate molecule with a different modified lysine, and the mycobactin is released upon the cyclization of this terminal moiety.164 Further supplementary changes, such as lipidation, are catalyzed by MbtK and N-hydroxylation of the lysine residue catalyzed by MbtG, which leads to the completion of mycobactins biosynthesis.165
A study suggests that the siderophore biosynthesis failed when the Mbt-B gene was replaced with a hygromycin-resistance cassette in M.tb H37Rv, and thus M.tb failed to survive in low iron conditions.163 Notably, MbtB gene deletion attenuates M.tb growth in THP-1 cells and changes the colony morphology. Also, M.tb with the MbtB gene deleted showed growth defects in liquid fermentation and macrophages. The above finding suggests that mycobactins play an essential role in the survival and virulence of M.tb.166,167 In another study, the disruption of mycobactin biosynthesis in M.tb displayed an altered colony morphology and attenuated growth in broth culture and in macrophages. Moreover, M.tb ΔmbtE (unable to synthesize mycobactins) demonstrated reduced virulence in guinea pigs.168
A study by Zhang et al. demonstrated that the rv0455c gene is necessary for M.tb growth in a low-iron medium. Also, the M.tb mutant with the deleted rv0455c gene showed reduced secretion of carboxymycobactins and mycobactins. In addition, the lack rv0455c gene is responsible for siderophore toxicity.169 The accumulation of intracellular siderophores is toxic to the M.tb.170 Moreover, M.tb with the deleted rv0455c gene failed to replicate inside the mice.169 The biosynthesis of mycobactin is shown in Scheme 1.171
Scheme 1. Biosynthesis of mycobactin. Chemical structures of mycobactin A, F, J, M, N, P, R, S, and T.
Mycobacterium iron-scavenging tools
Siderophores competitively bind to the host iron-transport proteins transferrin with Kd of ∼10−20 M.172 The high affinity of siderophores for ferric iron allows them to scavenge iron from several sources, including minerals in the soil, marine and freshwater, plants, or other organisms.160 Mycobacterial siderophores are divided according to the source derived, i.e., from non-pathogenic or pathogenic mycobacteria. M.tb and M. smegmatis use three types of siderophores tools to chelate iron, i.e., carboxymycobactin, mycobactin, and exochelin. Mycobactin is associated with the cell envelope, facilitating the entry and transport of iron via mycomembrane into the cytoplasm, whereas carboxymycobactin and exochelin are secreted outside the cells as iron chelators, which acquire iron from the extracellular environment and transport it to the cytoplasm.173Mycobacterium has diversified types of siderophores, which contain several functional groups such as (–N(OH)–CO–), dihydroxybenzoic acid, catecholate, and polydentate phenolate/nitrogen heterocycle/carboxylate combinations. Based on siderophore's ligand donation ability to coordinate Fe(iii), it is classified as hydroxamic acids, catechols, α-hydroxy acids, and aryl oxazolines.174–176 Based on the presence or absence of a 2-hydroxyphenyloxazoline ring, further mycobacterial siderophores are sub-classified into two structural classes, namely, salicylate-based hydrophobic mycobactins and water-soluble carboxymycobactins.20,157 Salicylate-based mycobactins have a primary nucleus of a 2-hydroxyphenyloxazoline moiety (derived from salicylic acid) linked to an amide bond by an acylated ε-N-hydroxylysine residue. The hydroxylated lysine residue has an alkyl chain of 10–21 carbons, and the length of the alkyl chains varies among the different species of mycobacteria.173 Due to this long alkyl chain, mycobactins are lipophilic and associated with the hydrophobic cell wall. Some functional groups, such as hydroxamic acid (N–OH) of the ε-N-hydroxylysine, the phenolate oxygen atom, and the nitrogen atom of the oxazoline moiety, are responsible for chelating Fe3+. Mycobactin acts as a growth factor for mycobacteria and shows a high affinity of (∼1030 KD) for the metal ion Fe3+ (oxidized form) and poor binding for Fe2+ (reduced form).177 The chemical structures of mycobactins are given in Scheme 1.27
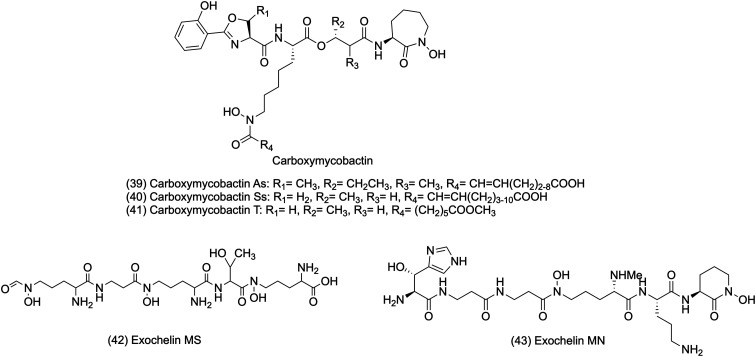
Carboxymycobactin is an extracellular siderophore containing an alkyl chain of 2–9 carbons and carrying a shorter acyl chain that terminates with a carboxylic acid or methyl chain. Iron chelation requires polarity and solubility, which are enhanced by the terminal methyl ester motif of carboxymycobactin.157 Carboxymycobactin has a high affinity for Fe3+ and can remove insoluble and protein-bound iron, but it also competes with iron-binding molecules in the aqueous growth media.161,178 The chemical structures of carboxymycobactins are shown in Fig. 3.
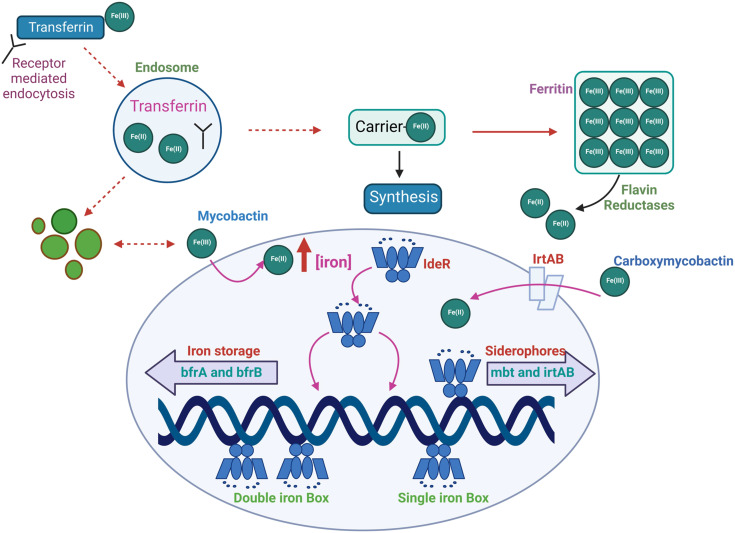
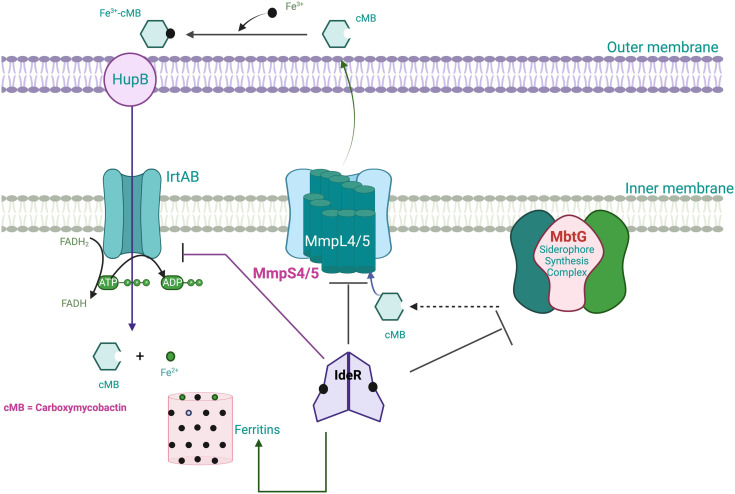
Fig. 3. IdeR plays a central role in regulating the iron homeostasis process. M.tb enhances the expression of the iron-dependent regulator (IdeR) under low iron environments, which leads to a rise in intracellular iron. The transfer of Fe(iii) inside the macrophage is performed via receptor-mediated endocytosis. The IrtAB transporter transports ferri-carboxymycobactin complexes across the cell membrane. Further, the receptor transferrin complex is internalized into the endosome, where the pH is maintained at 5.5 by an ATP-dependent pump. Lowering the pH promotes Fe(iii) reduction and the release of iron from transferrin. Then, iron(ii) is transferred to ferritin, oxidized, stored as iron(iii), or utilized to make proteins that need iron. Mycobacteria may incorporate Fe(ii) into their proteins by reducing Fe(iii) in ferritin through extracellular reductases. The ferric mycobactin reductase converts the Fe(iii) that mycobactin transports to Fe(ii). With the increase in Fe(ii), IdeR attaches to a single iron box repressing the synthesis of many genes, including siderophores and IrtAB transporters. Also, in surplus iron inside M.tb, IdeR promotes the synthesis of iron storage protein bacterioferritin.184,188.
Exochelins are high-affinity iron-binding molecules present in the cell wall of mycobacteria. Early studies created confusion by referring to the term exochelin for all siderophores isolated from saprophytic and pathogenic mycobacteria. But later, different core structures of exochelin for pathogenic and saprophytic mycobacteria were confirmed. The extracellular siderophores from saprophytic mycobacteria are called exochelins.173 It scavenges iron from the host-iron binding molecules and transfers it to the mycobactins associated with the cell wall. Exochelins are similar to the mycobactin core structure with shorter alkyl side chains that terminate with either a carboxyl group or a methyl ester containing three amino acid residues, two N-hydroxylysines, and one serine or threonine. Also, exochelins are smaller than mycobactins due to their shorter alkyl side chain. However, the exochelins differ from mycobactins in polarity and primarily depend upon the length and modification of an alkyl side chain.179M. smegmatis and Mycobacterium neoaurum synthesize exochelins of linear penta- and hexapeptides, in which hydroxamate groups are responsible for the significant iron-binding centers.157M. smegmatis synthesize exochelin MS, a water-soluble pentapeptide. Exochelin MS is a formylated pentapeptide (N-(δ-N-formyl, δN-hydroxy-R-ornithyl)-β-alaninyl-δN-hydroxy-R-ornithinyl-R-allo-threoninyl-δN-hydroxy-S-ornithine) having three hydroxamic acid groups for iron chelation. Exochelin MS comprises one residue of N-hydroxyformylornithine, two residues of N-hydroxyornithine, one residue of β-alanine, and one of d-threonine.180 Exochelin MN is also an extracellular siderophore produced by non-pathogenic Mycobacterium neoaurum, which efficiently transports iron in its cells. Also, exochelin MN is used by pathogenic M. leprae.181,182 Exochelins of M. neoaurum contain water-soluble moiety and a formylated hexapeptide with an unusual β-hydroxyhistidine and an N-methyl group residue that forms one of the three iron binding pairs of ligands. The exochelin MN chelates iron using its two cis-hydroxamate motifs, the hydroxyl, and imidazole nitrogen of the β-OH-histidine of the molecule.179 The chemical structures of exochelins are shown in Fig. 2.157,170
Fig. 2. Chemical structures of carboxymycobactins and exochelins.
Iron homeostasis in Mycobacterium
Iron-dependent regulator (IdeR)
In mycobacteria, the cellular Fe is maintained by the Fe-dependent transcriptional regulator IdeR. Notably, Mtb lacking IdeR demonstrated unrestricted iron uptake and deficient iron storage, which results in toxic iron overloading inside the cell.183 The IdeR plays a central role in regulating iron homeostasis and controlling the central siderophore synthesis in M.tb. Iron is maintained in M.tb by controlling gene expression, which turns the iron-dependent transcription machinery on and off according to the requirement.184 In low iron conditions, M.tb increases the gene expression of the IdeR. The diphtheria toxin repressor of the DxtR family is a regulatory protein that regulates iron homeostasis in M.tb.20 In low iron conditions, IdeR will dissociate from its target DNA, which results in the siderophore synthesis to acquire iron and repress the synthesis of iron storage proteins.185 When there is an excess of iron, IdeR attaches to a single iron box in M.tb, which suppresses the iron acquisition machinery of the bacilli by deactivating iron acquisition (MbtB gene) and irtA (iron transport). Also, in high iron conditions, IdeR binds to a double iron box and activates the synthesis of iron storage proteins by upregulating the iron storage protein bacterioferritin.186 In M.tb, the genes (bfrA and bfrB) are responsible for Fe storage. In contrast, in the case of iron deficiency, IdeR loses its ability to bind to the target promoter region, which promotes the synthesis of mycobactins and inhibits the repression of the synthesis of iron storage proteins.186,187 A recent study suggests the IdeR induces bfrB by antagonizing the repressor activity of Lsr2.183 IdeR-dependent iron regulation in M.tb is shown in Fig. 3.
Carboxymycobactin transport
To get iron from the surrounding environment, mycobacterium secretes hydrophilic deferri-carboxymycobactin via Msp porins. Ferri-carboxymycobactin is also transported by Msp porins back through the mycobacterial outer membrane.173,189,190
Desferri-carboxymycobactin, on chelation with Fe3+, produces ferri-carboxymycobactin, an insoluble or protein-bound iron. Iron from ferri-carboxymycobactin is believed to be transferred to cell wall-associated mycobactin with HupB, a 28 kDa iron-regulated cell wall-associated protein acting as the iron transporter or is delivered to the plasma membrane-bound protein complex, IrtAB (iron-regulated transporter A and B).172,191 According to a study HupB is elevated in response to iron deficiency and acts as a transcriptional activator for mycobactin production, which is necessary for the pathogen to thrive inside macrophages. The experimental work demonstrates that M.tb (WT M.tb H37Rv) and not the hupB KO mutant (M.tb DhupB) carried out Fe-carboxymycobactin uptake, indicating that HupB is required for ferri-carboxymycobactin uptake.55,192 At the periplasmic space, ferri-carboxymycobactin delivers its iron to the mycobactin-associated cell wall or the protein complex IrtAB bound to the plasma membrane. IrtAB comprises the membrane proteins IrtA and IrtB, which belongs to the ATP-binding cassette (ABC) transporter encoded by the genes irtA and irtB (Rv1348 and Rv1349), respectively.193 IrtAB's role as the iron importer is hampered by the inactivation of this gene pair, which prevents ferri-carboxymycobactin from being used by the mycobacteria. It has been demonstrated that the IrtA-N-terminal NTD's domain binds FAD-reduced Fe3+ to Fe2+, facilitating iron uptake.194 Fe2+ is transported into the cytoplasm by an energy-dependent mechanism, where it is used for various metabolic activities, and excess irons are stored in the form of bacterioferritin A (BfrA) and bacterioferritin B (BfrB).192 The export system of carboxymycobactin is carried out by the membrane complexes of mycobacterial membrane proteins Large5/S5 (MmpL5/MmpS5) and MmpL4/MmpS4 proteins, where the MmpLs are, and the MmpSs are tethered to the membrane and are thought to be transported accessory proteins.195 Sandhu et al. demonstrated from a dynamic simulation study, that MmpL5's role is to carry out the uptake of mycobactin from the cytoplasm and its release into the periplasmic space, and MmpS5 to facilitate the periplasmic release of mycobactin and enhancement in the transport function of MmpL5.196 A recent study showed that when mmpL4/S4/S5 or mmpL5/S4/S5 is disrupted in M.tb, then the MBT/cMBT production and secretion are impaired, which leads to further growth defects in low iron environments.197,198 A schematic representation depicting carboxymycobactin transport in M.tb under iron-limited conditions and their control by ItrAB importer and MmpL transporters is shown in Fig. 4.
Fig. 4. A schematic figure depicting carboxymycobactin (cMB) is secreted by M.tb under iron-limited conditions. The MmpL4/5 transporter, in association with the MmpS4 and MmpS5 membrane-associated proteins, is involved in the secretion of the carboxymycobactins. Once carboxymycobactins are secreted, they chelate Fe3+ and travel back across the outer membrane and periplasm via the inner membrane importer IrtAB to reach the cytoplasm. Using the FAD-binding domain of IrtA, ferric iron can reduce to ferrous iron in the cytosol and thus dissociate the iron–siderophore complex. The released ferrous irons are used, and excess ferrous irons are stored as ferritins. The binding of irons to the regulator IdeR activates its DNA binding activity, and the expression of several genes is suppressed, which are responsible for siderophores synthesis, secretion, and transport. Simultaneously, iron storage is activated by IdeR-Fe2+ binding to the promoters of ferritins (ferritin and bacterioferritin), which prevents iron-mediated toxicity; thus, iron homeostasis is maintained.200.
Once iron is trafficked, carboxymycobactins extract iron from the lactoferrin/transferrin, and mycobactins on the bacterial membrane transfer the iron into the cytoplasm.178 If mycobactin fails to transport the iron, carboxymycobactin transports iron in the cytoplasm through the IrtAB transporter, and iron is released from the endosome through a reduction and acidification process. The secreted siderophores carboxymycobactin and exochelin are imported and exported via unique mechanisms and machinery. Pathogenic mycobacteria dominantly follow carboxymycobactin transport, while saprophytic species follow exochelin transportation.173,189,199
Exochelins transport and secretion system
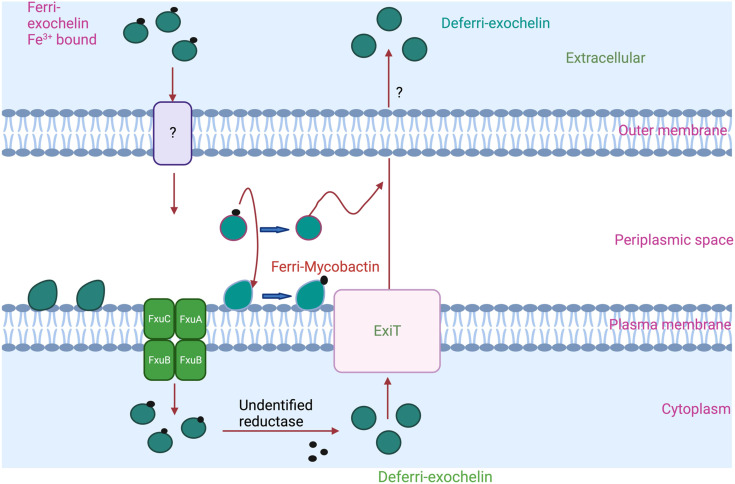
Exochelins are expected to transfer iron to other high-affinity iron-binding molecules in the bacterial cell wall and mycobactins for further utilization by the bacilli. Exochelins are smaller than mycobactins due to their shorter alkyl side chain, and the side chain terminates in a methyl ester.201 Exochelins are more polar than mycobactins; hence, they are soluble in the extracellular milieu of the mycobacteria in which they bind to the host. Exochelins are not biosynthesized in the M.tb. Non-pathogenic bacteria, such as Mycobacterium neoaurum and Mycobacterium smegmatis, mainly biosynthesize exochelins.200 In M. smegmatis, exochelin MS biosynthesis genes fxbA, fxbB, and fxbC and the transport gene exiT are involved in the synthesis, assembly, and transport of exochelin MS. Among these, the fxbA gene is responsible for the formylation of the pentapeptide, a step necessary for exochelin MS biosynthesis, and fxbB and fxbC genes produce large proteins of 257 and 497 kDa, respectively, which catalyze the non-ribosomal peptide synthesis of exochelin MS. The exiT gene encoding a protein is involved in exporting exochelin MS.180 ExiT, an ABC transporter protein, may export the deferri-exochelin. ExiT transporter shares the same operon as exochelin synthetase genes fxbB and fxbC.37 When ferri-exochelin binds to extracellular iron, it can be taken up by unidentified receptors on M.tb outer membrane and transported into the periplasmic space.202 There, ferri-exochelin can combine with FxuD, the ferri-exochelin carrier, and transfer iron to mycobactin or the FxuA, FxuB, and FxuC ferric-exochelin importer membrane complex. A schematic representation depicting exochelin import and export by M. smegmatis is shown in Fig. 5.
Fig. 5. A schematic representation depicting exochelin import and export by M. smegmatis. An ExiT ABC transporter protein may export the deferri-exochelin. The ferri-exochelin is delivered to the plasma membrane's FxuA–B–C complex after being recognized by an unidentified protein (on the outer membrane, indicated in the image by a “?”; yet to be identified). Inside the cell envelope, ferri-exochelin may transfer iron to mycobactin or another type of high-affinity iron-binding molecule in the bacterial cell wall. An unidentified reductase reduced the ferric ions and released them for utilization. Further deferri-exochelin exported outside the cell.173.
M.tb encodes five types VII secretion systems, i.e., ESX-1–ESX-5, responsible for growth and pathogenesis.203 ESX-1 is responsible for host-cell interactions, whereas the iron transfer has been connected to the ESX-3 secretion pathway. In other bacteria, the secretion system is responsible for the antibiotic resistance.204 The experimental data suggest that the ESX-3 secretion pathway is necessary for M.tb survival inside macrophages,205 whereas M. smegmatis require ESX-3 for siderophore-mediated iron acquisition.205 Also, a study revealed that ESX-3 mutants could not uptake siderophore-bound iron, which finally led to an accumulation of cell-associated mycobactin siderophores. In addition, ESX-3 T7SS secretes specific effectors, which have a potential role in virulence modulation in an iron-dependent fashion. It is stated that ESX-3 functions in iron acquisition via the mycobactin pathway.205 Thus, ESX-3 is vital for utilizing iron-bound mycobactins in the Mycobacterium species.203
M.tb scavenging iron inside the host cells
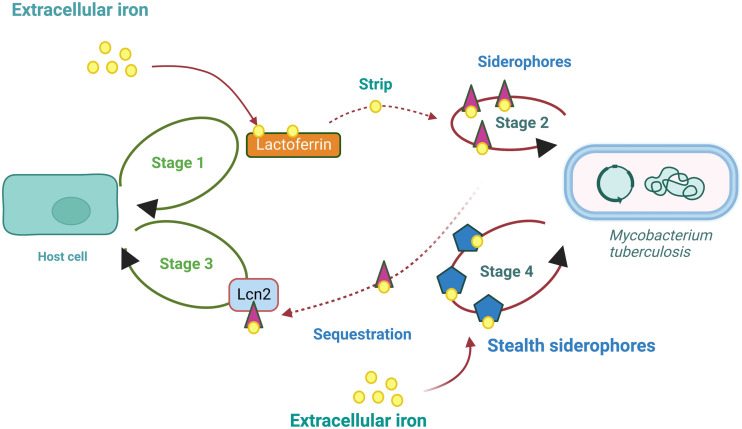
Host secretes lactoferrin to sequester readily available iron and store them, thus preventing the pathogen from acquiring iron.206,207 Lactoferrin is the host-secreting iron-binding protein, which sequesters iron. Iron linked to transferrin or lactoferrin enters macrophages by attaching to their respective membrane receptors. M.tb produces iron chelators (siderophores), which showed a higher affinity for iron and striped iron from host cells' iron carriers proteins such as lactoferrin (Lf) and transferrin (Tf).163,208 According to a recent study, M.tb, seized and internalized the host iron carrier protein, and it pushes numerous housekeeping proteins to the cell surface.209 In response, host cells secrete lipocalin 2 (Lcn2) to sequester the irons from the pathogens. Lcn2 is stored in the neutrophils and secreted rapidly as a part of first-response to infections. Some pathogens have evolved to have modified, Lcn2-resistant siderophores called ‘stealth siderophores’. The stealth siderophores are chemically modified that cannot bind to Lcn2. As the Lcn2 cannot attach to the stealth siderophores, irons are not sequestered by the host.189
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a glycolytic enzyme, is a conserved multifunctional protein identified as a virulence factor.210 GAPDH works as a dual receptor for transferrin and lactoferrin. GAPDH sequesters human lactoferrin, which is abundantly available in lung fluid, unlike transferrin, which is prevalent in plasma. Lf-GAPDH has a greater affinity of 31.7 ± 1.68 nM, whereas Tf-GAPDH has 160 ± 24 nM.210,211 In response to siderophores, siderophore-binding protein lipocalin 2 (important in innate immunity) is secreted by host cells to neutralize the siderophore and prevent pathogen reuptake. Siderocalins are atypical lipocalins able to capture siderophores with high affinity, and they stop iron absorption in the bacteria.212,213 Cyclic diguanylate monophosphate (c-di-GMP) is a secondary messenger biosynthesized in bacteria. Notably, the c-di-GMP directly binds to siderocalin and inhibits its antibacterial activity. Also, c-di-GMP makes siderocalin non-competitive for siderophores. These results suggest that siderocalin can act as a receptor for c-di-GMP. In Escherichia coli, in vitro growth is substantially less inhibited by siderocalin when a specific c-di-GMP binds to it. As c-di-GMP competes for the receptor siderocalin, the same strategy was used to study M. tuberculosis H37Ra growth in iron-limiting conditions. Interestingly, c-di-GMP reduces siderocalin-mediated inhibition of M.tb. Thus, c-di-GMP signalling may be a valid alternative target to stop bacteria from acquiring iron.213 A figure depicting M.tb scavenging iron inside the host cells is shown in Fig. 6.
Fig. 6. A schematic representation depicting M.tb scavenging iron in the host cells.189.
Abreu et al. demonstrated that IFN-mediated nitric oxide production enhanced ferroportin expression dramatically in murine macrophages, significantly reducing intracellular bacterial proliferation. IFN significantly increases ferroportin expression and decreases hepcidin secretion in human macrophages, inhibiting the growth of intracellular bacterial pathogens by reducing iron availability.214 Hepatocytes' ability to express hepcidin is hampered by the highly sulfated glycosaminoglycan heparin, reducing intracellular iron availability. Moreover, heparin dramatically lowers hepcidin expression in macrophages harboring M.tb bacilli. In addition, macrophages treated with heparin express more ferroportin than macrophages not treated with heparin, encouraging iron export and reducing the amount of iron available to intracellular bacilli.215
MbtI inhibitors
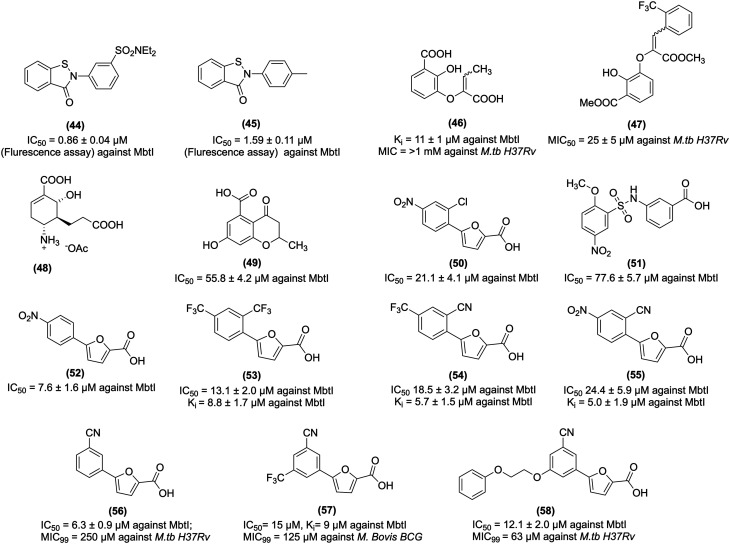
Vasan et al. screened a total of 104 802 compounds for MbtI inhibition activity. Among the screened compounds, benzisothiazolone analogues (44 and 45, Fig. 7) showed potent activity. 44 inhibits MbtI with IC50 values of 0.86 μM (fluorescence assay) and 0.96 μM (HPLC assay), and 45 inhibits the same enzyme MbtI with an IC50 value of 1.59 μM (fluorescence assay), respectively. These results suggest that 44 and 45 are tightly bound with MbtI by irreversible inhibition, as further evidenced by the high Hill slopes results. Further investigation revealed that these compounds are not suitable for the drug candidate, the reason being their non-specific labeling of the cysteine residue. Thus these compounds are categorized in substructure as a pan-assay-interference compound (PAINS).216
Fig. 7. MbtI inhibitors as anti-tubercular agents.
Chi et al. studied MbtI inhibitors based on the enzyme isochorismate having hydrophobic substitution appended to the terminal alkene of the enolpyruvyl group. Notably, their study demonstrated a switch in the binding mode at the MbtI active site for the inhibitors possessing a substituted enolpyruvyl group relative to the parent molecule. Further in silico analysis suggests that the change in the binding mode and potency, is due to the substituents on the conformational landscape of the core inhibitor structure.217 Manos-Turvey et al. synthesized gallate scaffold as the chorismatase transition state (TS) analogues. Among the designed analogues, the representative compound (46, Fig. 7) is a 2,3-dihydroxybenzoate scaffold with the most promising inhibitory activity, with a Ki value of 11 ± 1 μM against MbtI. However, 46 exhibited poor anti-tubercular activity in whole-cell screens against M.tb, with MIC50 value >1 mM.217 In other work, the plasticity of the MbtI active site was probed with a library of synthesized compounds based on a 2,3-dihydroxybenzoate scaffold with a range of substituted phenylacrylate side chains appended to the C3 position. The representative compound (47, Fig. 7) showed significant in vitro antitubercular activity with an MIC50 value of 25 ± 5 μM against M.tb H37Ra. At the same time, compound 47 hydrolyzed ester showed MbtI enzyme inhibition activity with a Ki value of 125 ± 13 μM.218
Liu et al. designed an isochorismatase transition state (TS) analogue (48, Fig. 7) for the partial reaction of MbtI. 48 exhibited suboptimal MbtI inhibition activity (i.e., <10%) at a concentration of 100 μM. A docking study of 48 with MbtI revealed the introduction of a methylene moiety as a bio-isosteric replacement for the C-5 oxygen atom of chorismite, which resulted in the loss of a critical hydrogen bond with the Arg405 and protonated C-4 amino group, exhibiting repulsive electrostatic interaction with Arg405 and thus yielding an inactive analogue.219
Pini et al. synthesized chromane and 4-oxochromane motifs bearing a carboxylic moiety at the C5 position. The author incorporated the chromanone nucleus as a vital scaffold, expecting that these compounds would mimic the first transition state of the isochorismate partial nucleus. The synthesized analogues (49, Fig. 7) showed promising activity against MbtI with an IC50 value of 55.8 μM. Molecular modelling studies revealed that 49 at the MbtI active site exhibits H-bond interaction with the nitrogen backbone of G241, and the aromatic ring shows a cation–π interaction with K438. In addition, the hydroxyl group forms a stable H-bond with the oxygen backbone of G270. The carbonyl oxygen forms a H-bond with the charged nitrogen of K205, and methyl substitution shows lipophilic interaction with the side chains of L402 and L404, respectively.220
Chiarelli et al. carried out receptor-based virtual screening of the enamine database, which led to the identification of compounds 5-(2-chloro-4-nitrophenyl)furan-2-carboxylic acid (50 and 51, Fig. 7). 50 and 51 showed MbtI enzyme inhibitory activity with IC50 values of 21.1 and 77.6 μM, respectively. Further, they explored 50 and synthesized its analogues to study SAR. Notably, the modified representative compound (52, Fig. 7) showed activity against the MbtI enzyme with an IC50 value and Ki value of 7.6 ± 1.6 μM and 5.3 ± 0.6 μM, respectively. Moreover, 52 showed activity against M.tb H37Rv with an MIC99 value of 156 μM. The SAR study revealed that 50 contains a nitrophenyl moiety, which acts as a bioisosteric of the salicylic moiety of methyl-AMT, and thus it confirms siderophore biosynthesis inhibition and antimycobacterial activity.221
Chiarelli et al. explored the MbtI hit compound 50 for better activity and performed the modification by replacing the nitro group. They also evaluated the modified compounds against MbtI enzyme activity. 50 showed activity against MbtI with an IC50 value of 21.1 μM, whereas the synthesized 5-phenylfuran-2-carboxylic derivatives (53, 54, and 55, Fig. 7) displayed Ki against MbtI with values of 8.8, 5.7, and 5.0 μM, respectively. Moreover, 53, 54, and 55 inhibited M.tb H37Rv with IC50 values of 13.1 ± 2.0, 18.5 ± 3.2, and 24.4 ± 5.9 μM, respectively. 54, possessing additional p-CF3 substitution, showed better antitubercular activity than 55, having p-NO2 substitution. Notably, 53, 54, and 55 showed no toxicity to human MRC-5 fibroblast cells. Thus, these compounds are safe and can be evaluated further in an in vivo model of TB infections.222
Mori et al. reported structural insight into MbtI. Their study revealed the Mg2+ independent binding mode and the crucial role of the Mg2+ cofactor in the catalysis and stability of the MbtI-Mg2+-salicylate ternary complex. The previous study revealed that 5-phenylfuroic acid having electron-withdrawing substituents such as –CF3 and NO2 are potent compounds and showed activity against MbtI. In contrast, electron-donating functions such as hydroxyl, methyl, and amino group abolished or significantly weakened the MbtI inhibition activity. The representative active compound (56, Fig. 7), with a m-CN group, inhibits MbtI with a Ki value of 3.1 ± 1.0 μM and an IC50 value of 6.3 ± 0.9 μM in the MbtI inhibition assay. The co-crystal structure of ligand 56 with MbtI highlighted the importance of essential residues for ligand binding, such as Thr361, Tyr385, Arg405, and Lys438. In addition, interaction with Lys205, a vital amino acid, is involved in the first step of the catalytic reaction. It also supported the role of Mg2+ ion as a cofactor and stabilized the enzymes in the close conformation. The crystal structure of the ternary complex MbtI with Mg2+ and salicylate showed that the product of the catalytic reaction chelates the metal ion with carboxylic acid and forms additional hydrogen bonds with Gly270, Gly241, and Thr271. Thus, the above result paved the way for the rational design of improved MbtI inhibitors.223
From earlier work on MbtI inhibitors, the SAR study revealed that 5-phenylfuroic acid having electron-withdrawing substituents are essential for MbtI enzyme inhibition and antitubercular activity. Also, removing the substituents from the phenyl ring led to a loss in activity. Mori et al. studied the importance of furan ring and replaced it with other heterocyclic cores such as thiophene, thiazole, oxazole, imidazole, 1,3,4-oxadiazole, and 1,2,3-triazole. However, these heterocyclic modifications were less active than 50. Revisiting the chemical structure 50 led to identifying a compound (57, Fig. 7) that showed potent activity against MbtI with IC50 and Ki values of 15 and 9 μM, respectively. 57 inhibits M. bovis BCG strain with an MIC99 value of 125 μM in the whole-cell assay. In silico study revealed that 57 showed H-bonds interaction with MbtI enzymes through its carboxylic group with Tyr385, Arg405, and an ordered water molecule. The oxygen of the furan interacts with Arg405, while the phenyl ring forms a cation-π interaction with Lys438 and a van der Waals contact with Thr361.224
Mori et al. explored the MbtI co-crystal ligand 56 and highlighted the development of improved agents acting against MbtI. The 5th position of the phenyl ring of 5-(3-cyanophenyl)furan-2-carboxylic acid was modified to increase the cell wall permeability lipophilic character. The representative synthesized compound (58, Fig. 7) showed potent antitubercular activity against M. bovis BCG with a MIC99 value of 63 μM and inhibited MbtI with an IC50 value of 12.1 ± 2.0 μM. SAR study revealed that the side chain linked to the phenyl moiety improves the in vitro antimycobacterial activity. Further, 58 showed no cell cytotoxicity against MPI-2_EeGFP mAMs cells at a concentration of 250 mM; thus, it can be considered safe for further studies.225
MbtA inhibitors
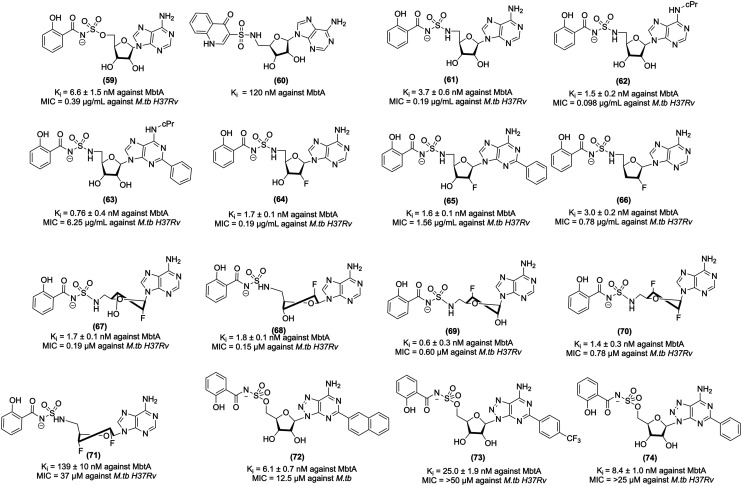
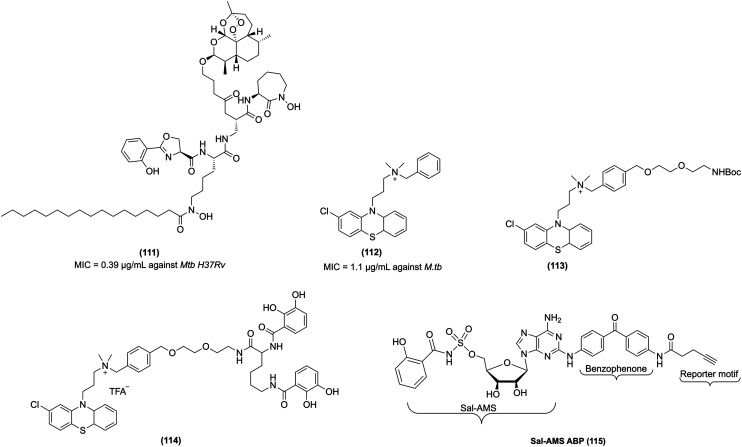
5′-O-[N-(Salicyl)sulfamoyl]adenosine (Sal-AMS, 59, Fig. 8) is a bisubstrate nucleoside antibiotic, which targets iron acquisition through the inhibition of aryl acid adenylating enzymes (AAAEs). Sal-AMS inhibited several pathogenic bacteria, including MbtA from M.tb, EntE from Escherichia coli and Klebsiella pneumoniae, BasE from Acinetobacter baumannii, YbtE from Yersinia pestis, VibE from Vibrio cholerae, and PchA from Pseudomonas aeruginosa.226 Lun et al. carried out a pharmacokinetic study of 59 that specifically targets MbtA by mimicking the salicyl-AMP intermediate. In vitro studies confirmed that salicyl-AMS intermediate inhibits MbtA and M. tuberculosis H37Rv growth with an MIC value of 0.5 μg mL−1. It demonstrated activity against MbtA with a Ki value of 6.6 nM. This study further showed an intraperitoneal injection of 59 in a TB-infected acute mice model over four weeks (intraperitoneal administration, 5.6 or 16.7 mg kg−1 dose) yielded much better pharmacokinetic parameter values than oral administration and significantly inhibited M.tb growth in the mouse lung. In addition, salicyl-AMS is nontoxic to the murine leukaemia cell line P388 at concentrations of >200 μM and do not show any toxicity in Chinese hamster ovary cells at concentrations of up to 500 μM.227 Despite the excellent antitubercular activity of 59, it suffers from sub-optimal drug disposition properties, resulting in a short half-life (t1/2), low exposure (AUC), and low bioavailability. To improve the bioavailability of Sal-AMS, Engelhart et al. carried out the computational analysis and structural studies and, based on the pharmacophoric features, designed and synthesized conformationally constrained analogues of Sal-AMS (by the removal of two rotatable bonds and ionized sulfamate group). Based on the analysis, chromone-, quinolone-, and benzoxazinone-3-sulfonamide derivatives of Sal-AMS were synthesized. The biochemical studies with MbtA revealed that the negative charge of Sal-AMS is essential for the activity as chromone and benzoxazine analogues, which lack an ionizable function in the heterocycle and displayed substantially reduced potency, whereas quinolone (60, Fig. 8), which contains an ionizable NH moiety at N-1, is only 18-fold less active than Sal-AMS toward MbtA. Also, quinolone modification improved ClogP and tPSA values relative to Sal-AMS. However, the quinolone derivative is inactive against M.tb in a whole-cell assay and exhibits an MIC value greater than 50 μM. Notably, 60 showed >128-fold loss in activity relative to Sal-AMS with a Ki value of 120 nM against an MbtA enzyme. 60 is inactive against M.tb probably due to its reduced cellular accumulation.228
Fig. 8. MbtA inhibitors as anti-tubercular agents.
Nelson et al. performed strategic modifications of Sal-AMS, which included conversion into prodrugs, increasing the pKa of the acyl–sulfonyl moiety, modulation of the lipophilicity, and introduction of fluorine into the molecule. The modified prodrugs were unsuccessful as these compounds underwent rapid cleavage to give 59. In the next attempt, increasing the pKa of the acyl–sulfonyl nitrogen linker was performed. The representative compound (61, Fig. 8) with the increased pKa showed 3-fold improved bioavailability relative to 59 and 50% increase in oral exposure and a more than 3-fold higher Cmax/MIC ratio. The third approach was to increase the lipophilicity by incorporating a fluorine atom at the C-2′ of the nucleoside in analogues, which led to compounds 62–66 (Fig. 8). 62–66 were the most pronounced improvements in oral exposure, primarily through an increase in the terminal half-life.229 In another study, Dawadi et al. further explored the contribution of the fluorinated ribose unit of the MbtA nucleoside inhibitors for anti-tubercular activity, enzyme inhibition efficacy, and drug disposition profile. In addition, the introduction of fluorine atoms at both 2′ and 3′ positions of the glycosyl unit of the nucleoside. Incorporating one fluorine unit resulted in compounds (67, 68, and 69, Fig. 8), which showed activity with an MIC value of 0.19, 0.15, and 0.60 μM, respectively, against the M.tb H37Rv strain, while the introduction of two fluorine units changes the stereoelectronic properties of the nucleoside's ribose moiety, orienting the sugar into a 2′-endo, 3′-exo(south), or a 3′-endo, 2′-exo(north) conformation. Importantly, SAR revealed the compound (70, Fig. 8), which had the ribose unit in the north (C3′-endo), maintained potent biochemical and whole-cell activity. In contrast, improved pharmacokinetic properties were associated with the C2′- endo conformation compound (71, Fig. 8), which had south (C2′-endo) and showed enhanced pharmacokinetic properties. 70 and 71 showed activity against M.tb H37Rv strain with MIC values of 0.78 and 37 μM, respectively. Notably, the overall fluorination of SAL-AMS resulted in a dramatic increase in the pharmacokinetic properties, increasing the half-life by 25-fold, oral exposure by 75-fold, and oral bioavailability by 10-fold.230
The biosynthetic pathway of mycobactins for constructing the conserved peptidic core uses a mixed nonribosomal peptide synthetase-polyketide synthase (NRPS-PKS) assembly line of six proteins designated MbtA through MbtF. Considering the importance of N-(salicyl)sulfamoyl moiety, which is essential for anti-tubercular and MbtA inhibition activity, Krajczyk et al. designed analogues of Sal-AMS and performed conservative substitutions to the salicyl ring. Krajczyk et al. designed analogues of Sal-AMS. The introduction of the 2-naphthyl substituent in Sal-AMS led to the potent compound 8-aza-3-deazaadenine (72, Fig. 8), which exhibits a Ki value of 6.1 ± 0.7 nM against MbtA and inhibits M.tb H37Rv with an MIC value of 12.5 μM. In contrast, the p-trifluoromethyl substituted compound (73, Fig. 8) exhibits a Ki value of 25.0 ± 1.9 against MbtA and inhibits M.tb H37Rv with an MIC value of 50 μM. The 2-phenyl-Sal-AMS derivative (74, Fig. 8) exhibited activity under the iron-deprived condition against MbtA with a Ki value of 8.4 ± 1.0 nM and inhibits M.tb H37Rv MIC value of 25 μM.231
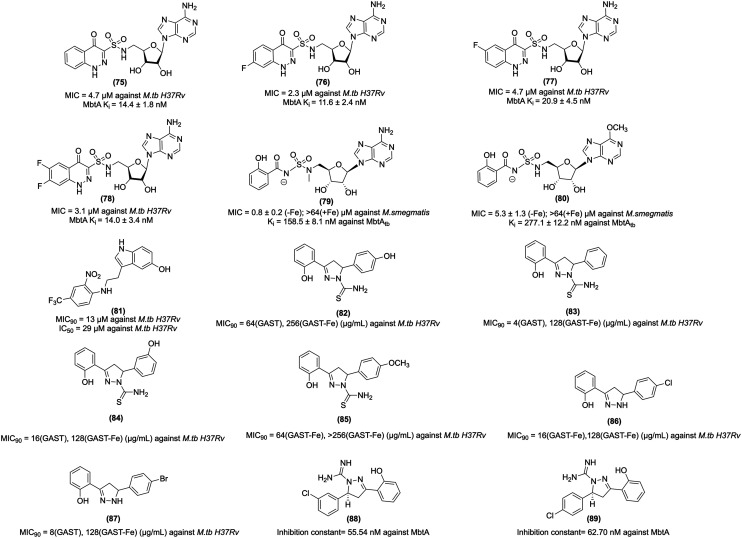
Earlier work on MbtA suggests that the formal negative charge of the acyl-sulfamate linker in Sal-AMS is critical for binding to MbtA, confirmed by the computational and crystallographic analysis. The presence of a formal negative charge is a detrimental effect on the passive diffusion of Sal-AMS across mammalian membranes. Thus, optimum pKa is required to maintain potent antimycobacterial activity and acceptable membrane permeability. Keeping these parameters in view, Dawadi et al. carried out the bioisosteric replacement of the salicyl-sulfamoyl of Sal-AMS and synthesized conformationally constrained analogues using the cinnolone scaffold. Cinnolone-containing compound (75, Fig. 9) showed antitubercular activity against M.tb H37Rv with an MIC value of 4.7 μM. The halogenated 7-fluorocinnolone analogue (76, 77, and 78, Fig. 9) showed improved anti-tubercular activity with MIC values of 2.3, 4.7, and 3.1 μM against M.tb H37Rv under iron-deprived conditions. Moreover, 76 inhibited the biosynthesis of the lipophilic mycobactin-T, and the water-soluble carboxymycobactins. 76 demonstrated a better pharmacokinetic profile with 0.57 L kg−1 than Sal-AMS with 0.079 L kg−1. Furthermore, the cinnolone compounds exhibited less toxicity against Vero and HepG2 cell lines. The above results suggest that these compounds have potential antitubercular activity and can be studied further in in vivo models of tuberculosis.232 Bythrow et al. synthesized salicyl-AMS analogues, i.e., Sal-AMSNMe (79, Fig. 9) and Sal-6-MeO-AMSN (80, Fig. 9). Sal-AMS inhibit M. smegmatis ΔEM-pMbtAtb with a MIC value of 0.8 μg mL−1, whereas 79 and 80 inhibit M. smegmatis ΔEM-pMbtAtb with MIC values of 0.8 and 5.3 μg mL−1, respectively. 79 showed improved MtbAtb inhibition due to its cisoid conformation. Notably, 79 and 80 exhibit substantial post-antibiotic effects.233
Fig. 9. MbtA inhibitors as anti-tubercular agents.
Ferguson et al. screened a 3200-member library containing lead-like structurally diverse compounds against M. tuberculosis for whole-cell inhibitory activity. With fluorescence-based thermal shift assay and NMR-based Water-LOGSY and saturation transfer difference (STD) experiments, 846 compounds were identified, which inhibited the tubercle bacilli. The identified lead molecule 5-hydroxy-indol-3-ethylamino-(2-nitro-4-trifluoromethyl)benzene (81, Fig. 9) inhibits M.tb H37Rv with a MIC90 value of 13 μM and the mode of action to be MbtA inhibition. Moreover, 81 inhibited intracellular M.tb with an IC90 value of 9.3 μM. 81 is moderately toxic to raw 264.7 cells with an IC50 value of 29 μM, which suggests the limitation of the compound. Further in silico docking studies revealed that Sal-AMS and 81 occupy different binding orientations in the MbtA protein of Mycobacterium smegmatis. Sal-AMS is positioned with the salicyl group occupying a deep pocket in the protein and stacking with Phe237. The purine ring lies adjacent to Arg426 near the protein surface, and sulphonamide forms hydrogen bond interactions with His235 and Gly325, whereas 81 is positioned with its indole ring over Phe324, the phenyl nitro substituent interacts with Gly192 and the amine NH of the ligand is predicted to form a hydrogen bond with Gly325. Notably, 81 and SAL-AMS demonstrated a binding energy of −9.3 and −8.2 kcal mol−1, respectively, at the MbtA active site.234
Shyam et al. designed and synthesized a small library of mycobactin analogues retaining diaryl-substituted pyrazoline as the basic scaffold of mycobactin. Compounds (82, 83, 84, and 85, Fig. 9) with thiocarbamoyl substitution at the pyrazoline N1 were active against M.tb under iron-deprived conditions and simultaneously inactive against M. smegmatis. Diaryl-substituted pyrazoline with acetyl substitutions were inactive against both M.tb and M. smegmatis. Moreover, the pyrazoline compounds (86 and 87, Fig. 9) without any substitution at the N1 position were found to be active against both M. smegmatis and M.tb86 showed activity in glycerol-alanine-salts (GAST) and GAST-Fe media against M.tb H37Rv with MIC90 values of 16 and 128 μg mL−1, and in M. smegmatis with MIC90 values of 4 and 64 μg mL−1. 87 showed activity in GAST and GAST-Fe media against M.tb H37Rv with MIC90 values of 8 and 128 μg mL−1 and M. smegmatis with MIC90 values of 4 and 64 μg mL−1, respectively. Thermofluorimetric analysis and molecular dynamics simulations revealed the plausible modes of action of these compounds by inhibiting MbtA and siderophore production in mycobacteria. Interestingly, 86 and 87 demonstrated efflux pump inhibition in M. smegmatis, highlighting their potential in reversing drug resistance.235
Rakshit et al. designed a 12-member small library of mycobactin analogues retaining the diaryl-substituted pyrazoline as the primary scaffold. The docking of the designed molecules was performed in the active site of the MbtA receptor (by analogy with the related structure, PDB: 1MDB) to evaluate the binding modes and inhibitory profiles. The docking results showed compounds (88 and 89, Fig. 9) as potent MbtA inhibitors, which could serve as good leads. The docking score in the binding pocket of the target MbtA protein of compounds showed −9.90 and −9.83 Kcal mol−1, respectively. Moreover, 88 and 89 showed MbtA inhibition constants values of 55.54 and 62.70 nM, respectively. Further, the in silico study revealed that both the compounds form H-bond interactions Gly460, Thr462, and Ala356 of MbtA. In toxicity studies, the maximum tolerated dosage (human) range was between −0.127 and −0.199 log mg kg−1 per day. 88 and 89 showed acceptable pharmacokinetic profiles and low toxicity profiles.236
TMPK inhibitors
M. tuberculosis thymidine monophosphate kinase (MtbTMPK) is the last specific enzyme in the biosynthesis of thymidine triphosphate, which is essential for mycobacterial growth. The earlier discovered potent MtbTMPK inhibitors showed poor antimycobacterial activity due to insignificant bacterial uptake; to address this issue, Jian et al. introduced a simplified Fe-chelating siderophore motif analogue (90, Fig. 10) that showed MtbTMPK enzyme activity with an IC50 value of 34 ± 5 μM and inhibited M.tb with an MIC value of 12.5 μM. The further modification of 90 with an imidazo[1,2-a]pyridine or 3,5-dinitrobenzamide scaffold afforded analogues (91, 92 and 93, Fig. 10) with moderate MtbTMPK enzyme inhibitory potency. 91 inhibits M.tb H37Rv with MIC values of 4.7 μM in 7H9/glucose, and 2.3 μM in GAST (Fe (+)), and 92 inhibits M.tb H37Rv with MIC values of 9.4 μM in 7H9/glucose, and 0.78 μM in GAST (Fe (+)), and 1.2 μM in GAST (Fe (−)), and 93 inhibits M.tb H37Rv with MIC values of 9.4 μM in 7H9/glucose, and 9.4 μM in GAST (Fe (+)), and 6.25 μM in GAST (Fe (−)), respectively.237
Fig. 10. TMPK, IdeR, ESX-3 inhibitors, and siderophore structural intermediate as anti-tubercular agents.
IdeR inhibitors
Rohilla et al. performed a virtual screening of the NCI database against the IdeR DNA binding domain, followed by inhibition studies using electrophoretic mobility shift assay (EMSA). Among the identified compounds NSC 281033 (94, Fig. 10) and NSC 12453 (95, Fig. 10) showed IdeR DNA binding activity with IC50 values of 2.4 and 1 μg mL−1, respectively. Compounds were synthesized based on the essential features for IdeR inhibition and a five-point pharmacophore model. The representative compound 96 showed antitubercular activity against M.tb with the MIC90 value of 17.5 μg mL−1. Also, 96 showed negligible cytotoxicity in THPI, hek, mdck, and HepG2 cell lines. Further, this compound should be evaluated for antitubercular activity in an in vivo model of tuberculosis infections.187
SAL-AMS prodrug
59 is a nucleoside antibiotic that exhibits potent activity against M.tb by inhibiting siderophore biosynthesis under iron-deficient conditions in the host. However, it has poor drug disposition properties, resulting in poor bioavailability and rapid clearance. To improve its bioavailability, Dawadi et al. synthesized lipophilic ester prodrugs containing linear and α-branched alkanoyl groups from two to twelve carbons at the 3′-position of a 2′-fluorinated analogue of (59). Derivatives with ester linkages include (97, 98, 99, 101, 102, 103, and 104Fig. 10). The synthesized prodrugs showed remarkable stability in mouse, rat, and human serum. However, these ester prodrugs showed reduced oral bioavailability due to reduced permeation in the apical-to-basolateral direction and enhanced permeation in the basolateral-to-apical order relative to the parent compound, resulting in a 5–28 times higher efflux ratio.238
ESX-3 function modulators
The ESX-3 gene cluster present in mycobacteria contains a group of 11 genes. These genes encode the ESX-3 type VII secretion system. Notably, ESX-3 is involved in iron transfer. The exochelin is biosynthesized in M. smegmatis in addition to the mycobactins, and it requires ESX-3 for siderophore-mediated iron acquisition.205 Dragset et al. evaluated pyrazolopyrimidinone (3-(4-chlorophenyl)-5-(cyclohexylmethyl)pyrazolo[1,5-α]pyrimidin-7(4H)-one) (105, Fig. 10) against ESX-3 function. (3-(4-Chlorophenyl)-5-(cyclohexylmethyl)pyrazolo[1,5-α]pyrimidin-7(4H)-one)chelate (106, Fig. 10) with Fe2+ is shown in Fig. 10. The study revealed that 105 restricts mycobacterial growth by targeting ESX-3 and thus prevents iron uptake in Mycobacterium. This finding demonstrates that 105 restricts mycobacterial growth by chelating intrabacterial iron and depriving the cells of available iron storage. Further, in vivo studies are needed to evaluate the therapeutic potential and safety of compound 105.239
Siderophores structural intermediates inhibitors
M.tb and Yersinia pestis produce siderophores with scaffolds of nonribosomal peptide polyketide origin. Keeping this in view, Ferreras et al. synthesized compounds resembling the structural features of M.tb and Yersinia pestis siderophores. The synthesized compounds, including 3,5-diaryl pyrazoline (DAP) and 2(E)-2-benzylidene-N-hydroxyhydrazine carbo(ox/thio/oximid)amide (BHHC) derivatives, were evaluated for growth inhibitory activity against M.tb and Yersinia pestis in iron-limiting media, which mimic the iron-scarcity condition that the pathogens encounter in the host and standard iron-rich media. The representative compounds (107, 108, 109, and 110, Fig. 10) showed activity against M.tb with MIC90 values in GASTD + Fe 16, 16, 6, and 9 μM and in GASTD with MIC99 values of 16, 16, 7, and 10 μM, respectively. Encouragingly, inhibitors from the DAP and BHHC derivatives were bactericidal against M.tb (>99% killing relative to inoculum) at 1–2 X MICGASTD concentrations. Notably, the examination of IC50GASTD + Fe/IC50GASTD and MICGASTD+Fe/MICGASTD ratios revealed that these inhibitors had no increased potency in the iron-limiting medium. Some of the compounds from the series DAP and BHHC showed toxicity, which is the limitation of the compound for further development as a drug candidate.240
M.tb siderophore applications
Siderophores as antitubercular agents
Artemisinin is a naturally derived drug used for the treatment of malaria. Artemisinin demonstrates antimalarial activity by generating oxyradicals that can kill malarial parasites. It was hypothesized that artemisinin, on internalization, can release free radicals and kill mycobacteria. Interestingly, the synthetic modification of mycobactin with artemisinin conjugate (111, Fig. 11) showed potent antitubercular activity with an MIC value of 0.39 μg mL−1 against M.tb H37Rv. Moreover, it showed activity against MDR-TB strains with MIC values in the range of 1.25–0.16 μg mL−1 and XDR-TB strains with MIC values in the range of 0.625–0.078 μg mL−1.241
Fig. 11. Chemical structures of siderophores conjugated drugs and SAL-AMS ABP.
Thioridazine scaffold (112, Fig. 11) is reported to possess activity against M.tb, multidrug-resistant and latent TB.242 Further, it has been demonstrated that phenothiazine scaffold enhanced its anti-TB activity because of its localization and increased its effective concentration at its target site, the membrane-bound oxidoreductase NADH-2.243,244 Tarapdar et al. synthesized phenothiazine–siderophore conjugates (113 and 114Fig. 11). In M. smegmatis, the siderophore conjugates showed an equipotent MIC value compared to the parent phenothiazine derived from it. Currently, the synthesis of a library of mono-, bis-, and tris-catechol phenothiazine-siderophore conjugates are under biological evaluation and development process.245
Gokran et al. evaluated the antitubercular activity of exogenous iron chelators, exochelin-MS and deferoxamine-B against MDR-TB and pyrazinamide-resistant M.tb isolates. Interestingly, exochelin-MS alone inhibited some MDR-TB isolates, whereas deferoxamine-B alone inhibited most of the MDR-TB isolates. Against seven pyrazinamide-resistant M.tb, deferoxamine-B and exochelin-MS both alone inhibited five strains. Notably, deferoxamine-B, combined with isoniazid, rifampin, and pyrazinamide, decreased its MIC value; in contrast, exochelin-MS did not change the MIC. Thus, this finding suggests that exochelin-MS and deferoxamine-B can be used as adjuvants with the primary antibiotic to treat MDR-TB in humans. Also, these siderophores should be evaluated against other MDR-TB strains, and their association with the first-line anti-TB should be assessed.246
Activity-based chemical prob
SAL-AMP is the intermediate in the biosynthesis of mycobactins. Based on the SAL-AMP structure, the Aldrich group and others developed SAL-AMS as a potent nanomolar bisubstrate inhibitor of MbtA, which also shows excellent antitubercular activity. Interestingly, this inhibitor showed potent MIC under both iron-deficient and iron-replete conditions, suggesting possible off-targets of this compound because mycobactin biosynthesis is not required in the presence of iron. The same group designed an activity-based chemical probe to understand the mechanism of SAL-AMS in M.tb comprehensively. Sal-AMS ABP (115, Fig. 11) is an activity-based probe for MbtA, and this developed probe has three critical motifs: binding, reactive, and reporter motif. The binding motif of Sal-AMS strongly binds to MbtA. Based on the favorable SAR, benzophenone was attached as a reactive group at the C-2 position of the adenosine ring, followed by linking alkyne as the reporter motif. Significantly, Sal-AMS ABP inhibits MbtA with an apparent binding affinity of 0.94 nM. In iron-deficient conditions, Sal-AMS ABP and Sal-AMS showed activity against M.tb H37Rv with MIC values of 3–6 and 0.39 μM for, whereas in iron-rich conditions, Sal-AMS ABP and Sal-AMS showed activity against M.tb H37Rv with MIC values of 50 and 1.56 μM, respectively. Sal-AMS ABP was demonstrated to label E. coli lysates harboring MbtA overexpressing plasmid. Moreover, Sal-AMS ABP showed labeling in the purified MbtA and within mycobacterial lysate. The labeling was entirely inhibited by preincubation with Sal-AMS, which confirms the specific nature of the labeling. The discovery of Sal-AMS ABP provides a prototypical core scaffold for creating AfBPs to profile other adenylating enzymes in M.tb or adenylating enzymes present in other pathogenic bacteria.247
Conclusion
The current anti-TB therapy includes multiple antibiotics and it requires long-duration treatment. Notably, the ineffectiveness of most first-line anti-TB drugs has given rise to MDR-TB strains. Moreover, prolonged treatment, adverse effects associated with antibiotics, drug interaction, and socioeconomic factors contribute to noncompliance among patients. M.tb requires iron from host cells to sustain and support its growth and to carry out several biological processes, and at the same time low iron concentration inside M.tb leads to environmental stress. In this review, we shed light on iron homeostasis in mycobacteria, and its iron-scavenging tools, including mycobactin, carboxymycobactin, and exochelin. The most exciting aspect covered in this review is the complexity of iron homeostasis in the mycobacteria, which play a critical role in M.tb growth and survival. Then, we have summarized the MbtA and MbtI inhibitors, which prevent M.tb from acquiring iron from host cells and the recent application of siderophore development. We strongly believe that these iron-scavenging tools inhibitors have the potential to be developed as anti-TB agents. However, no anti-TB drugs have been developed to target iron-scavenging tools. Furthermore, in-depth studies are needed to develop an anti-TB medicine targeting Mycobacterium iron-scavenging tools.
Abbreviations
- ABC
ATP-binding cassette
- ABP
Activity-based probe
- ACP
Enoyl-acyl carrier protein
- AAAE
Aryl acid adenylation enzyme
- ABPP
Activity-based protein profiling
- AfBPP
Affinity-based protein profiling
- AMI
Amikacin
- AMP
Adenosine monophosphate
- Araf
d-Arabinofuranose
- ATP
Adenosine triphosphate
- BCG
Bacillus calmette guerin
- BDQ
Bedaquiline
- BPaL
BDQ, pretomanid and linezolid
- CAP
Capreomycin
- c-di-GMP
Cyclic diguanylate monophosphate
- CFUs
Colony forming units
- DHP
Dehydropeptidase
- DPA
Decaprenylphosphoryl-β-d-arabinofuranose
- DprE1
Decaprenylphosphoryl-β-d-ribofuranose 2′-oxidase
- DprE2
Decaprenylphosphoryl-2-keto-d-ery-thropentose reductase
- DNA
Deoxyribonucleic acid
- DPX
Decaprenylphosphoryl-2-keto-β-d-erythro-pentofuranose
- DxtR
Diphtheria toxin repressor
- EMB
Ethambutol
- EMSA
Electrophoretic mobility shift assay
- ETC
Electron transport chain
- FAAL
Fatty acyl-AMP ligase
- FAAS
Fatty acyl ACP synthetase
- FAS
Fatty acid synthase
- FQs
Fluoroquinolones
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GAST
Glycerol-alanine-salts
- HIV
Human immunodeficiency virus
- HK
Histidine kinase
- IdeR
Iron-dependent regulator
- INH
Isoniazid
- KAN
Kanamycin
- MABA
Microplate alamar blue assay
- MDR
Multidrug-resistant
- MK/MKH2
Menaquinone pool
- MK4
Menaquinone
- MmpL
Mycobacterial membrane protein large
- MSCs
Mesenchymal stromal cells
- Msm
Mycobacterium smegmatis
- MTBC
Mycobacterium tuberculosis complex
- M.tb
Mycobacterium tuberculosis
- NRPS
Nonribosomal peptide synthetase-dependent
- OBR
Optimized background anti-TB regimen
- PBPs
Penicillin-binding proteins
- PKS
Polyketide synthase
- PMF
Proton motive force
- PZA
Pyrazinamide
- RIF
Rifampin
- ROS
Reactive oxygen species
- RPF
Resuscitation-promoting factor
- RR
Response regulator
- SMARt-420
Small molecule aborting resistance-420
- SDH
Succinate dehydrogenase
- TDM
Trehalose dimycolate
- TMM
Trehalose monomycolate
- TMPK
Thymidine monophosphate kinase
- TNF
Tumor necrosis factor
- TB
Tuberculosis
- TCS
Two-component regulatory systems
- TDR
Totally drug-resistant
- XDR
Extensively drug-resistant
Conflicts of interest
There is no conflict of interest to declare.
Supplementary Material
Acknowledgments
We thank the Director, NIPER-Hyderabad, for providing the necessary research facilities. Amruta Patil is thankful to NIPER Hyderabad for providing a fellowship. All figures were created in the BioRender software.
Biographies
Biography
Gautam Kumar.

Gautam Kumar is an Assistant Professor at the Department of Natural Products, Chemical Sciences, NIPER-Hyderabad. He studied B. Pharm. and M. Pharm. from Jamia Hamdard, Hamdard University, New Delhi, India. He did his PhD dissertation in Natural Products at the Department of Natural Products (NIPER-Mohali), Punjab, India. During his PhD, he was involved in the synthesis of heterocyclic compounds as potential antitubercular agents. He conducted his post-doctoral research at IIT Bombay, India. During his post-doctoral research, he developed “clickable” chemical probes based on Mycobacterium tuberculosis virulence-associated glycolipid for real-time proteomics and imaging. Currently, he leads a lab in the area of Natural Products at NIPER-Hyderabad with a focus on bioassay-guided fractionation of extracts, isolation of natural products, phytopharmaceutical formulation development, and synthesis of biologically active molecules.
Biography
Patil Amruta Adhikrao.

Patil Amruta Adhikrao pursued a B. Pharm. degree from the Government College of Pharmacy, Karad, Maharashtra. She did a MS (Pharm) in Natural Products from the National Institute of Pharmaceutical Education and Research (NIPER), Hyderabad. During her B. Pharma, she did a project on the “Ethnobotanical assessment of indigenous plants used as sunscreen”, which involved formulation, characterization, and evaluation of its cosmetic value. During her MS, she did her work on the synthesis of benzimidazole derivatives as antitubercular agents. This work includes the synthesis, characterization, and evaluation of benzimidazole derivatives against Mycobacterium tuberculosis.
References
- Momin M. A. M. Tucker I. G. Das S. C. Int. J. Pharm. 2018;550:398–417. doi: 10.1016/j.ijpharm.2018.08.061. [DOI] [PubMed] [Google Scholar]
- Reuter A. Hughes J. Furin J. Lancet. 2019;394:967–978. doi: 10.1016/S0140-6736(19)32045-8. [DOI] [PubMed] [Google Scholar]
- Hunter R. L. Tuberculosis. 2016;97:8–17. doi: 10.1016/j.tube.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski A. Jansson M. Sköld M. Rottenberg M. E. Källenius G. PLoS Pathog. 2012;8:1–7. doi: 10.1371/journal.ppat.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty S. Ramesh M. Singh-Pillay A. Soliman M. E. S. Bioorg. Med. Chem. Lett. 2017;27:370–386. doi: 10.1016/j.bmcl.2016.11.084. [DOI] [PubMed] [Google Scholar]
- Kiazyk S. Ball T. Can. Commun. Dis. Rep. 2017;43:62–66. doi: 10.14745/ccdr.v43i34a01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews J. R. Noubary F. Walensky R. P. Cerda R. Losina E. Horsburgh C. R. Clin. Infect. Dis. 2012;54:784–791. doi: 10.1093/cid/cir951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S. Njikan S. Kumar A. Early J. V. Parish T. Microbiology. 2019;165:492–499. doi: 10.1099/mic.0.000760. [DOI] [PubMed] [Google Scholar]
- Nguta J. M. Appiah-Opong R. Nyarko A. K. Yeboah-Manu D. Addo P. G. A. Int. J. Mycobact. 2015;4:165–183. doi: 10.1016/j.ijmyco.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Weyer K. Mirzayev F. Migliori G. B. Van Gemert W. D'Ambrosio L. Zignol M. Floyd K. Centis R. Cirillo D. M. Tortoli E. Gilpin C. De Dieu Iragena J. Falzon D. Raviglione M. Eur. Respir. J. 2013;42:252–271. doi: 10.1183/09031936.00157212. [DOI] [PubMed] [Google Scholar]
- Cheon S. A. Cho H. H. Kim J. Lee J. Kim H. J. Park T. J. J. Microbiol. Methods. 2016;123:51–61. doi: 10.1016/j.mimet.2016.02.007. [DOI] [PubMed] [Google Scholar]
- Manjunatha U. H. Smith P. W. Bioorg. Med. Chem. 2015;23:5087–5097. doi: 10.1016/j.bmc.2014.12.031. [DOI] [PubMed] [Google Scholar]
- Conradie F. Diacon A. H. Ngubane N. Howell P. Everitt D. Crook A. M. Mendel C. M. Egizi E. Moreira J. Timm J. McHugh T. D. Wills G. H. Bateson A. Hunt R. Van Niekerk C. Li M. Olugbosi M. Spigelman M. N. Engl. J. Med. 2020;382:893–902. doi: 10.1056/NEJMoa1901814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins H. E. Tolman A. W. Yuen C. M. Parr J. B. Keshavjee S. Pérez-Vélez C. M. Pagano M. Becerra M. C. Cohen T. Lancet. 2014;383:1572–1579. doi: 10.1016/S0140-6736(14)60195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennassiri W. Jaouhari S. Cherki W. Charof R. Filali-Maltouf A. Lahlou O. J. Global Antimicrob. Resist. 2017;11:75–80. doi: 10.1016/j.jgar.2017.07.002. [DOI] [PubMed] [Google Scholar]
- Tiberi S. du Plessis N. Walzl G. Vjecha M. J. Rao M. Ntoumi F. Mfinanga S. Kapata N. Mwaba P. McHugh T. D. Ippolito G. Migliori G. B. Maeurer M. J. Zumla A. Lancet Infect. Dis. 2018;18:1–16. doi: 10.1016/S1473-3099(18)30110-5. [DOI] [PubMed] [Google Scholar]
- W. H. Organization, Consolidated Guidelines on Tuberculosis Treatment, 2020
- Lange C. Dheda K. Chesov D. Mandalakas A. M. Udwadia Z. Horsburgh C. R. Lancet. 2019;394:953–966. doi: 10.1016/S0140-6736(19)31882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienhardt C. Raviglione M. Spigelman M. Hafner R. Jaramillo E. Hoelscher M. Zumla A. Gheuens J. J. Infect. Dis. 2012;205:241–249. doi: 10.1093/infdis/jis034. [DOI] [PubMed] [Google Scholar]
- Hameed S. Pal R. Fatima Z. Open Microbiol. J. 2015;9:91–97. doi: 10.2174/1874285801509010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington S. Hung D. T. ACS Infect. Dis. 2018;4:696–714. doi: 10.1021/acsinfecdis.7b00255. [DOI] [PubMed] [Google Scholar]
- Riccardi G. Pasca M. R. J. Antibiot. 2014;67:655–659. doi: 10.1038/ja.2014.109. [DOI] [PubMed] [Google Scholar]
- Gill C. M. Dolan L. Piggott L. M. McLaughlin A. M. Breathe. 2022;18:210149. doi: 10.1183/20734735.0149-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasiruddin M. Neyaz M. K. Das S. Tuberc. Res. Treat. 2017;2017:1–12. doi: 10.1155/2017/4920209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil K. Bagade S. Bonde S. Sharma S. Saraogi G. Biomed. Pharmacother. 2018;99:735–745. doi: 10.1016/j.biopha.2018.01.115. [DOI] [PubMed] [Google Scholar]
- Larsson L.-O. Int. J. Mycobact. 2016;5:S29–S30. doi: 10.1016/j.ijmyco.2016.09.032. [DOI] [PubMed] [Google Scholar]
- Glaus F. and Altmann K., 2015, 1937–1940 [DOI] [PubMed]
- Bretl D. J. Demetriadou C. Zahrt T. C. Microbiol. Mol. Biol. Rev. 2011;75:566–582. doi: 10.1128/MMBR.05004-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish T., in Molecular Genetics of Mycobacteria, ed. G. F. Hatfull and W. R. Jacobs Jr., 2014, vol. 2, pp. 209–233 [Google Scholar]
- Zheng H. Abramovitch R. B. Future Med. Chem. 2020;12:457–467. doi: 10.4155/fmc-2019-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse P. J. Karakousis P. C. Klinkenberg L. G. Kesavan A. K. Ly L. H. Allen S. S. Grosset J. H. Jain S. K. Lamichhane G. Manabe Y. C. McMurray D. N. Nuermberger E. L. Bishai W. R. Infect. Immun. 2009;77:1230–1237. doi: 10.1128/IAI.01117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidi K. Jahanban-Esfahlan R. J. Med. Hypotheses Ideas. 2013;7:69–74. doi: 10.1016/j.jmhi.2013.04.002. [DOI] [Google Scholar]
- Gan Y. Yao Y. Guo S. Med. Hypotheses. 2015;84:477–480. doi: 10.1016/j.mehy.2015.01.046. [DOI] [PubMed] [Google Scholar]
- Wallis R. S. van Vuuren C. Potgieter S. Clin. Infect. Dis. 2009;48:1429–1432. doi: 10.1086/598504. [DOI] [PubMed] [Google Scholar]
- Mayanja-Kizza H. Jones-Lopez E. Okwera A. Wallis R. S. Ellner J. J. Mugerwa R. D. Whalen C. C. J. Infect. Dis. 2005;191:856–865. doi: 10.1086/427995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A. Chakaya J. Centis R. D'Ambrosio L. Mwaba P. Bates M. Kapata N. Nyirenda T. Chanda D. Mfinanga S. Hoelscher M. Maeurer M. Migliori G. B. Lancet Respir. Med. 2015;3:220–234. doi: 10.1016/S2213-2600(15)00063-6. [DOI] [PubMed] [Google Scholar]
- Saxena A. K. Singh A. Curr. Top. Med. Chem. 2019;19:337–355. doi: 10.2174/1568026619666190219105722. [DOI] [PubMed] [Google Scholar]
- Nandi S., Saxena M. and Saxena A. K., in Integrated Science, ed. N. Rezaei, Springer International Publishing, Cham, 2023, pp. 429–457 [Google Scholar]
- Urban M. Šlachtová V. Brulíková L. Eur. J. Med. Chem. 2021;212:113139. doi: 10.1016/j.ejmech.2020.113139. [DOI] [PubMed] [Google Scholar]
- Iqbal I. K. Bajeli S. Akela A. K. Kumar A. Pathogens. 2018;7:1–30. doi: 10.3390/pathogens7010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard M. Bald D. Ingmer H. J. Global Antimicrob. Resist. 2022;29:29–41. doi: 10.1016/j.jgar.2022.01.026. [DOI] [PubMed] [Google Scholar]
- Lu P. Lill H. Bald D. Biochim. Biophys. Acta, Bioenerg. 2014;1837:1208–1218. doi: 10.1016/j.bbabio.2014.01.022. [DOI] [PubMed] [Google Scholar]
- Haagsma A. C. Driessen N. N. Hahn M.-M. Lill H. Bald D. FEMS Microbiol. Lett. 2010;313:68–74. doi: 10.1111/j.1574-6968.2010.02123.x. [DOI] [PubMed] [Google Scholar]
- Narang R. Kumar R. Kalra S. Nayak S. K. Khatik G. L. Kumar G. N. Sudhakar K. Singh S. K. Eur. J. Med. Chem. 2019;182:111644. doi: 10.1016/j.ejmech.2019.111644. [DOI] [PubMed] [Google Scholar]
- Bald D. Koul A. FEMS Microbiol. Lett. 2010;308:1–7. doi: 10.1111/j.1574-6968.2010.01959.x. [DOI] [PubMed] [Google Scholar]
- Yano T. Kassovska-Bratinova S. Shin Teh J. Winkler J. Sullivan K. Isaacs A. Schechter N. M. Rubin H. J. Biol. Chem. 2011;286:10276–10287. doi: 10.1074/jbc.M110.200501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechartier B. Cole S. T. Antimicrob. Agents Chemother. 2015;59:4457–4463. doi: 10.1128/AAC.00395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. Zheng M. Wang B. Fu L. Zhao W. Li P. Xu J. Zhu H. Jin H. Yin D. Huang H. Upton A. M. Ma Z. Antimicrob. Agents Chemother. 2011;55:5185–5193. doi: 10.1128/AAC.00699-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosset J. H. Tyagi S. Almeida D. V. Converse P. J. Li S. Y. Ammerman N. C. Bishai W. R. Enarson D. Trébucq A. Am. J. Respir. Crit. Care Med. 2013;188:608–612. doi: 10.1164/rccm.201304-0753OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholo M. C. Mothiba M. T. Fourie B. Anderson R. J. Antimicrob. Chemother. 2017;72:338–353. doi: 10.1093/jac/dkw426. [DOI] [PubMed] [Google Scholar]
- Luo J. Yu X. Jiang G. Fu Y. Huo F. Ma Y. Wang F. Shang Y. Liang Q. Xue Y. Huang H. Antimicrob. Agents Chemother. 2018;62:1–9. doi: 10.1128/AAC.00072-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaeffle H. O. I. Alameer R. M. Marshall M. H. Houpt E. R. Albon D. P. Heysell S. K. Pulm. Pharmacol. Ther. 2021;70:102058. doi: 10.1016/j.pupt.2021.102058. [DOI] [PubMed] [Google Scholar]
- van Ingen J. Totten S. E. Helstrom N. K. Heifets L. B. Boeree M. J. Daley C. L. Antimicrob. Agents Chemother. 2012;56:6324–6327. doi: 10.1128/AAC.01505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaschewski B. Verma D. Pennings L. J. Zimmerman M. Ye Q. Gadawa J. Dartois V. Ordway D. van Ingen J. Ufer S. Stapleton K. Hofmann T. J. Cystic Fibrosis. 2019;18:714–720. doi: 10.1016/j.jcf.2019.05.013. [DOI] [PubMed] [Google Scholar]
- Wang Q. Pang Y. Jing W. Liu Y. Wang N. Yin H. Zhang Q. Ye Z. Zhu M. Li F. Liu P. Wu T. Chen W. Wu W. Qin Z. Qiu C. Deng Q. Xu T. Wang J. Guo R. Du Y. Wang J. Huang H. Chen X. Chu N. Antimicrob. Agents Chemother. 2018;62:1–9. doi: 10.1128/AAC.02149-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. L. J. Wang B. Fu L. Zhu H. Guo S. Huang H. Yin D. Zhang Y. Antimicrob. Agents Chemother. 2019;63:1–12. doi: 10.1128/AAC.02155-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H. S. Tong A. S. T. Choi P. J. Blaser A. Conole D. Franzblau S. G. Lotlikar M. U. Cooper C. B. Upton A. M. Denny W. A. Palmer B. D. Bioorg. Med. Chem. 2019;27:1292–1307. doi: 10.1016/j.bmc.2019.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarathy J. P. Ragunathan P. Shin J. Cooper C. B. Upton A. M. Grüber G. Dick T. Antimicrob. Agents Chemother. 2019;63:1–11. doi: 10.1128/AAC.01191-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. Luo W. Xu D. Guo F. Yang M. Zhu Y. Shen L. Chen S. Tang D. Li L. Li Y. Wang B. Franzblau S. G. Ding C. Z. Bioorg. Med. Chem. Lett. 2022;71:128824. doi: 10.1016/j.bmcl.2022.128824. [DOI] [PubMed] [Google Scholar]
- Yao R. Wang B. Fu L. Li L. You K. Li Y.-G. Lu Y. Microbiol. Spectrum. 2022;10:1–11. doi: 10.1128/spectrum.02477-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethe K. Bifani P. Jang J. Kang S. Park S. Ahn S. Jiricek J. Jung J. Jeon H. K. Cechetto J. Christophe T. Lee H. Kempf M. Jackson M. Lenaerts A. J. Pham H. Jones V. Seo M. J. Kim Y. M. Seo M. Seo J. J. Park D. Ko Y. Choi I. Kim R. Kim S. Y. Lim S. Yim S. A. Nam J. Kang H. Kwon H. Oh C. T. Cho Y. Jang Y. Kim J. Chua A. Tan B. H. Nanjundappa M. B. Rao S. P. S. Barnes W. S. Wintjens R. Walker J. R. Alonso S. Lee S. Kim J. Oh S. Oh T. Nehrbass U. Han S. J. No Z. Lee J. Brodin P. Cho S. N. Nam K. Kim J. Nat. Med. 2013;19:1157–1160. doi: 10.1038/nm.3262. [DOI] [PubMed] [Google Scholar]
- Scherr N. Bieri R. Thomas S. S. Chauffour A. Kalia N. P. Schneide P. Ruf M. T. Lamelas A. Manimekalai M. S. S. Grüber G. Ishii N. Suzuki K. Tanner M. Moraski G. C. Miller M. J. Witschel M. Jarlier V. Pluschke G. Pethe K. Nat. Commun. 2018;9:1–9. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S. Wang W. Zhou X. Zhang Y. Lai Y. Tang Y. Xu J. Li D. Lin J. Yang X. Ran T. Chen H. Guddat L. W. Wang Q. Gao Y. Rao Z. Gong H. eLife. 2021;10:1–24. doi: 10.7554/eLife.69418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida D. V. Converse P. J. Omansen T. F. Tyagi S. Tasneen R. Kim J. Nuermberger E. L. Antimicrob. Agents Chemother. 2020;64(6):1–10. doi: 10.1128/AAC.00259-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quémard A. Trends Microbiol. 2016;24:725–738. doi: 10.1016/j.tim.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Lovewell R. R. Sassetti C. M. VanderVen B. C. Curr. Opin. Microbiol. 2016;29:30–36. doi: 10.1016/j.mib.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Rayasam G. V. Expert Opin. Ther. Targets. 2014;18:247–256. doi: 10.1517/14728222.2014.859677. [DOI] [PubMed] [Google Scholar]
- Lefebvre C. Boulon R. Ducoux M. Gavalda S. Laval F. Jamet S. Eynard N. Lemassu A. Cam K. Bousquet M. P. Bardou F. Burlet-Schiltz O. Daffé M. Quémard A. Sci. Rep. 2018;8:1–15. doi: 10.1038/s41598-018-24380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre C. Frigui W. Slama N. Lauzeral-Vizcaino F. Constant P. Lemassu A. Parish T. Eynard N. Daffé M. Brosch R. Quémard A. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavalda S. Bardou F. Laval F. Bon C. Malaga W. Chalut C. Guilhot C. Mourey L. Daffé M. Quémard A. Chem. Biol. 2014;21:1660–1669. doi: 10.1016/j.chembiol.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Irfan A. Faisal S. Zahoor A. F. Noreen R. Al-Hussain S. A. Tuzun B. Javaid R. Elhenawy A. A. Zaki M. E. A. Ahmad S. Abdellattif M. H. Pharmaceuticals. 2023;16:1–19. doi: 10.3390/ph16060829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altharawi A. Alossaimi M. A. Alanazi M. M. Alqahatani S. M. Tahir ul Qamar M. Sci. Rep. 2023;13:7014. doi: 10.1038/s41598-023-34222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzegorzewicz A. E. Eynard N. Quémard A. North E. J. Margolis A. Lindenberger J. J. Jones V. Korduláková J. Brennan P. J. Lee R. E. Ronning D. R. McNeil M. R. Jackson M. ACS Infect. Dis. 2016;1:91–97. doi: 10.1021/id500032q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair H. A. Scott L. J. Drugs. 2015;75:91–100. doi: 10.1007/s40265-014-0331-4. [DOI] [PubMed] [Google Scholar]
- Chen X. Hashizume H. Tomishige T. Nakamura I. Matsuba M. Fujiwara M. Kitamoto R. Hanaki E. Ohba Y. Matsumoto M. Antimicrob. Agents Chemother. 2017;61:1–11. doi: 10.1128/AAC.02402-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan N. J. Lo J. H. Drugs. 2014;74:1041–1045. doi: 10.1007/s40265-014-0241-5. [DOI] [PubMed] [Google Scholar]
- Keam S. J. Drugs. 2019;79:1797–1803. doi: 10.1007/s40265-019-01207-9. [DOI] [PubMed] [Google Scholar]
- Rožman K. Sosič I. Fernandez R. Young R. J. Mendoza A. Gobec S. Encinas L. Drug Discovery Today. 2017;22:492–502. doi: 10.1016/j.drudis.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Unissa A. N. Subbian S. Hanna L. E. Selvakumar N. Infect., Genet. Evol. 2016;45:474–492. doi: 10.1016/j.meegid.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Laborde J. Deraeve C. Lecoq L. Sournia-Saquet A. Stigliani J. L. Orena B. S. Mori G. Pratviel G. Bernardes-Génisson V. ChemistrySelect. 2016;1:172–179. doi: 10.1002/slct.201600040. [DOI] [Google Scholar]
- Kumar G. Krishna V. S. Sriram D. Jachak S. M. Eur. J. Med. Chem. 2018;156:871–884. doi: 10.1016/j.ejmech.2018.07.047. [DOI] [PubMed] [Google Scholar]
- Thee S. Garcia-Prats A. J. Donald P. R. Hesseling A. C. Schaaf H. S. Tuberculosis. 2016;97:126–136. doi: 10.1016/j.tube.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Blondiaux N. Moune M. Desroses M. Frita R. Flipo M. Mathys V. Soetaert K. Kiass M. Delorme V. Djaout K. Trebosc V. Kemmer C. Wintjens R. Wohlkönig A. Antoine R. Huot L. Hot D. Coscolla M. Feldmann J. Gagneux S. Locht C. Brodin P. Science. 2017;355:1206–1211. doi: 10.1126/science.aag1006. [DOI] [PubMed] [Google Scholar]
- Wohlkönig A. Remaut H. Moune M. Tanina A. Meyer F. Desroses M. Steyaert J. Willand N. Baulard A. R. Wintjens R. Biochem. Biophys. Res. Commun. 2017;487:403–408. doi: 10.1016/j.bbrc.2017.04.074. [DOI] [PubMed] [Google Scholar]
- Lociuro S., Press RELEASE BioVersys, 2020, pp. 1–3
- Umare M. D. Khedekar P. B. Chikhale R. V. ChemMedChem. 2021;16:3136–3148. doi: 10.1002/cmdc.202100359. [DOI] [PubMed] [Google Scholar]
- Bolla J. R. Biochem. Soc. Trans. 2020;48:1463–1472. doi: 10.1042/BST20190950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela C. Rittmann D. Singh A. Krumbach K. Bhatt K. Eggeling L. Besra G. S. Bhatt A. Chem. Biol. 2012;19:498–506. doi: 10.1016/j.chembiol.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Obregón-Henao A. Wallach J. B. North E. J. Lee R. E. Gonzalez-Juarrero M. Schnappinger D. Jackson M. Antimicrob. Agents Chemother. 2016;60:5198–5207. doi: 10.1128/AAC.00826-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degiacomi G. Benjak A. Madacki J. Boldrin F. Provvedi R. Palù G. Kordulakova J. Cole S. T. Manganelli R. Sci. Rep. 2017;7:1–8. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A. Arnoult E. Lounis N. Guillemont J. Andries K. Nature. 2011;469:483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- Foss M. H. Pou S. Davidson P. M. Dunaj J. L. Winter R. W. Pou S. Licon M. H. Doh J. K. Li Y. Kelly J. X. Dodean R. A. Koop D. R. Riscoe M. K. Purdy G. E. ACS Infect. Dis. 2016;2:500–508. doi: 10.1021/acsinfecdis.6b00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton J. Foo C. S. Y. Cole S. T. Drug Discovery Today. 2017;22:526–533. doi: 10.1016/j.drudis.2016.09.014. [DOI] [PubMed] [Google Scholar]
- Imran M. Khan S. A. Asdaq S. M. B. Almehmadi M. Abdulaziz O. Kamal M. Alshammari M. K. Alsubaihi L. I. Hussain K. H. Alharbi A. S. Alzahrani A. K. J. Infect. Public Health. 2022;15:1097–1107. doi: 10.1016/j.jiph.2022.08.016. [DOI] [PubMed] [Google Scholar]
- Chhabra S. Kumar S. Parkesh R. ACS Omega. 2021;6:14430–14441. doi: 10.1021/acsomega.1c01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi G. Pasca M. R. Chiarelli L. R. Manina G. Mattevi A. Binda C. Appl. Microbiol. Biotechnol. 2013;97:8841–8848. doi: 10.1007/s00253-013-5218-x. [DOI] [PubMed] [Google Scholar]
- Hoagland D. T. Liu J. Lee R. B. Lee R. E. Adv. Drug Delivery Rev. 2016;102:55–72. doi: 10.1016/j.addr.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhale R. V. Barmade M. A. Murumkar P. R. Yadav M. R. J. Med. Chem. 2018;61:8563–8593. doi: 10.1021/acs.jmedchem.8b00281. [DOI] [PubMed] [Google Scholar]
- Activity A. Inhibitor D. Hariguchi N. Chen X. Hayashi Y. Kawano Y. Fujiwara M. Matsuba M. Antimicrob. Agents Chemother. 2020;64:1–13. doi: 10.1128/AAC.02020-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirude P. S. Shandil R. Sadler C. Naik M. Hosagrahara V. Hameed S. Shinde V. Bathula C. Humnabadkar V. Kumar N. Reddy J. Panduga V. Sharma S. Ambady A. Hegde N. Whiteaker J. McLaughlin R. E. Gardner H. Madhavapeddi P. Ramachandran V. Kaur P. Narayan A. Guptha S. Awasthy D. Narayan C. Mahadevaswamy J. Vishwas K. Ahuja V. Srivastava A. Prabhakar K. Bharath S. Kale R. Ramaiah M. Choudhury N. R. Sambandamurthy V. K. Solapure S. Iyer P. S. Narayanan S. Chatterji M. J. Med. Chem. 2013;56:9701–9708. doi: 10.1021/jm401382v. [DOI] [PubMed] [Google Scholar]
- Jaganath D. Lamichhane G. Shah M. Int. J. Tuberc. Lung Dis. 2016;20:1436–1447. doi: 10.5588/ijtld.16.0498. [DOI] [PubMed] [Google Scholar]
- Luthra S. Rominski A. Sander P. Front. Microbiol. 2018;9:1–13. doi: 10.3389/fmicb.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J. F. Mobashery S. Cold Spring Harbor Perspect. Med. 2016;6:1–20. doi: 10.1101/cshperspect.a025221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gamal M. I. Brahim I. Hisham N. Aladdin R. Mohammed H. Bahaaeldin A. Eur. J. Med. Chem. 2017;131:185–195. doi: 10.1016/j.ejmech.2017.03.022. [DOI] [PubMed] [Google Scholar]
- Vilchèze C. Appl. Sci. 2020;10:1–35. [Google Scholar]
- Lange C. Alghamdi W. A. Al-Shaer M. H. Brighenti S. Diacon A. H. DiNardo A. R. Grobbel H. P. Gröschel M. I. von Groote-Bidlingmaier F. Hauptmann M. Heyckendorf J. Köhler N. Kohl T. A. Merker M. Niemann S. Peloquin C. A. Reimann M. Schaible U. E. Schaub D. Schleusener V. Thye T. Schön T. J. Intern. Med. 2018;284:163–188. doi: 10.1111/joim.12780. [DOI] [PubMed] [Google Scholar]
- Stokes S. S. Vemula R. Pucci M. J. ACS Infect. Dis. 2020;6:1323–1331. doi: 10.1021/acsinfecdis.0c00025. [DOI] [PubMed] [Google Scholar]
- Kashyap A. Singh P. K. Silakari O. Tuberculosis. 2018;113:43–54. doi: 10.1016/j.tube.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Piton J. Petrella S. Delarue M. André-Leroux G. Jarlier V. Aubry A. Mayer C. PLoS One. 2010;5:e12245. doi: 10.1371/journal.pone.0012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locher C. P. Jones S. M. Hanzelka B. L. Perola E. Shoen C. M. Cynamon M. H. Ngwane A. H. Wiid I. J. Van Helden P. D. Betoudji F. Nuermberger E. L. Thomson J. A. Antimicrob. Agents Chemother. 2015;59:1455–1465. doi: 10.1128/AAC.04347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown-Elliott B. A. Rubio A. Wallace R. J. Antimicrob. Agents Chemother. 2018;62:1–6. doi: 10.1128/AAC.01503-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcik M. Tomašič T. Zidar N. Zega A. Kikelj D. Mašič L. P. Ilaš J. Expert Opin. Ther. Pat. 2019;29:171–180. doi: 10.1080/13543776.2019.1575362. [DOI] [PubMed] [Google Scholar]
- Stephanie F. Tambunan U. S. F. Siahaan T. J. Life. 2022;12:1774. doi: 10.3390/life12111774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R. Rudra P. Prajapati R. K. Sengupta S. Mukhopadhyay J. Tuberculosis. 2014;94:397–404. doi: 10.1016/j.tube.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Srivastava A. Talaue M. Liu S. Degen D. Ebright R. Y. Sineva E. Chakraborty A. Druzhinin S. Y. Chatterjee S. Mukhopadhyay J. Ebright Y. W. Zozula A. Shen J. Sengupta S. Niedfeldt R. R. Xin C. Kaneko T. Irschik H. Jansen R. Donadio S. Connell N. Ebright R. H. Curr. Opin. Microbiol. 2011;14:532–543. doi: 10.1016/j.mib.2011.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino C., Andrade J. M., Ferreira I. M., Almeida J. F. and Rijo P., Lead molecules from natural products: Insight into tubercular targets; Studies in Natural Products Chemistry, 2020, vol. 65
- Alfarisi O. Alghamdi W. A. Al-Shaer M. H. Dooley K. E. Peloquin C. A. Expert Rev. Clin. Pharmacol. 2017;10:1027–1036. doi: 10.1080/17512433.2017.1366311. [DOI] [PubMed] [Google Scholar]
- Sirgel F. A. Warren R. M. Böttger E. C. Klopper M. Victor T. C. van Helden P. D. PLoS One. 2013;8:e59414. doi: 10.1371/journal.pone.0059414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz D. B. Low J. L. Wu M.-L. Gengenbacher M. Teo J. W. P. Dartois V. Dick T. Antimicrob. Agents Chemother. 2017;61:1–10. doi: 10.1128/AAC.00155-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy L. S. Xie Y. Tor Y. Wiley Interdiscip. Rev.: RNA. 2011;2:209–232. doi: 10.1002/wrna.60. [DOI] [PubMed] [Google Scholar]
- Kumar N. Sharma S. Kaushal P. S. Mol. Aspects Med. 2021;81:101002. doi: 10.1016/j.mam.2021.101002. [DOI] [PubMed] [Google Scholar]
- Leach K. L. Brickner S. J. Noe M. C. Miller P. F. Ann. N. Y. Acad. Sci. 2011;1222:49–54. doi: 10.1111/j.1749-6632.2011.05962.x. [DOI] [PubMed] [Google Scholar]
- Alffenaar J. W. C. Van Der Laan T. Simons S. Van Der Werf T. S. Van De Kasteele P. J. De Neeling H. Van Soolingen D. Antimicrob. Agents Chemother. 2011;55:1287–1289. doi: 10.1128/AAC.01297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip P. C. W. Kam K. M. Lam E. T. K. Chan R. C. Y. Yew W. W. Int. J. Antimicrob. Agents. 2013;42:96–97. doi: 10.1016/j.ijantimicag.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Balasubramanian V. Solapure S. Iyer H. Ghosh A. Sharma S. Kaur P. Deepthi R. Subbulakshmi V. Ramya V. Ramachandran V. Balganesh M. Wright L. Melnick D. Butler S. L. Sambandamurthy V. K. Antimicrob. Agents Chemother. 2014;58:495–502. doi: 10.1128/AAC.01903-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. S. Choe J. H. Kim Y. J. Yang C. Kwon H. Jeong J. Kim G. Jo E. Cho Y. Jang J. Antimicrob. Agents Chemother. 2017;61:1–12. doi: 10.1128/AAC.02752-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen M. D. Herrmann J.-L. Kremer L. Nat. Rev. Microbiol. 2020;18:392–407. doi: 10.1038/s41579-020-0331-1. [DOI] [PubMed] [Google Scholar]
- Liu H. Zhu H. Fu L. Zhang W. Chen X. Wang B. Guo S. Ding Y. Wang N. Li D. Lu Y. Antimicrob. Agents Chemother. 2023;67:1–9. doi: 10.1128/aac.01399-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. E. Hurdle J. G. Liu J. Bruhn D. F. Matt T. Scherman M. S. Vaddady P. K. Zheng Z. Qi J. Akbergenov R. Das S. Madhura D. B. Rathi C. Trivedi A. Villellas C. Lee R. B. Rakesh Waidyarachchi S. L. Sun D. McNeil M. R. Ainsa J. A. Boshoff H. I. Gonzalez-Juarrero M. Meibohm B. Böttger E. C. Lenaerts A. J. Nat. Med. 2014;20:152–158. doi: 10.1038/nm.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Bruhn D. F. Lee R. B. Zheng Z. Janusic T. Scherbakov D. Scherman M. S. Boshoff H. I. Das S. Rakesh Waidyarachchi S. L. Brewer T. A. Gracia B. Yang L. Bollinger J. Robertson G. T. Meibohm B. Lenaerts A. J. Ainsa J. Böttger E. C. Lee R. E. ACS Infect. Dis. 2017;3:72–88. doi: 10.1021/acsinfecdis.6b00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. F. K. Laughon B. E. McHugh T. D. Lipman M. Curr. Opin. Pulm. Med. 2019;25:271–280. doi: 10.1097/MCP.0000000000000570. [DOI] [PubMed] [Google Scholar]
- Kumar G. Kapoor S. Bioorg. Med. Chem. 2023;81:117212. doi: 10.1016/j.bmc.2023.117212. [DOI] [PubMed] [Google Scholar]
- Wagh S. Rathi C. Lukka P. B. Parmar K. Temrikar Z. Liu J. Scherman M. S. Lee R. E. Robertson G. T. Lenaerts A. J. Meibohm B. Antimicrob. Agents Chemother. 2021;65:1–15. doi: 10.1128/AAC.01744-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhura D. B. Liu J. Meibohm B. Lee R. E. MedChemComm. 2016;7:114–117. doi: 10.1039/C5MD00349K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palencia A. Li X. Bu W. Choi W. Ding C. Z. Easom E. E. Feng L. Hernandez V. Houston P. Liu L. Meewan M. Mohan M. Rock F. L. Sexton H. Zhang S. Zhou Y. Wan B. Wang Y. Franzblau S. G. Woolhiser L. Gruppo V. Lenaerts A. J. O'Malley T. Parish T. Cooper C. B. Waters M. G. Ma Z. Ioerger T. R. Sacchettini J. C. Rullas J. Angulo-Barturen I. Pérez-Herrán E. Mendoza A. Barros D. Cusack S. Plattner J. J. Alley M. R. K. Antimicrob. Agents Chemother. 2016;60:6271–6280. doi: 10.1128/AAC.01339-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vjecha M. J. Tiberi S. Zumla A. Nat. Rev. Drug Discovery. 2018;17:607–608. doi: 10.1038/nrd.2018.28. [DOI] [PubMed] [Google Scholar]
- Abuhammad A. Br. J. Pharmacol. 2017;174:2194–2208. doi: 10.1111/bph.13694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhía I. Galán B. Kendall S. L. Stoker N. G. García J. L. Environ. Microbiol. Rep. 2012;4:168–182. doi: 10.1111/j.1758-2229.2011.00314.x. [DOI] [PubMed] [Google Scholar]
- Ouellet H. Johnston J. B. de Montellano P. R. O. Trends Microbiol. 2011;19:530–539. doi: 10.1016/j.tim.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkaran A. C., Biswas R. and Mohan C. G., in Structural Bioinformatics: Applications in Preclinical Drug Discovery Process, 2019, pp. 307–346 [Google Scholar]
- Kumar G. C A. Drug Dev. Res. 2023:1–26. doi: 10.1002/ddr.22063. [DOI] [PubMed] [Google Scholar]
- Vasudevan D. Rao S. P. S. Noble C. G. J. Biol. Chem. 2013;288:30883–30891. doi: 10.1074/jbc.M113.493767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmaier U. Junk L. Mar. Drugs. 2021;19:1–27. doi: 10.3390/md19080446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. Suh J. W. J. Ind. Microbiol. Biotechnol. 2016;43:205–212. doi: 10.1007/s10295-015-1709-3. [DOI] [PubMed] [Google Scholar]
- Zhou B. Shetye G. Yu Y. Santarsiero B. D. Klein L. L. Abad-Zapatero C. Wolf N. M. Cheng J. Jin Y. Lee H. Suh J. W. Lee H. Bisson J. Mcalpine J. B. Chen S. N. Cho S. H. Franzblau S. G. Pauli G. F. J. Nat. Prod. 2020;83:657–667. doi: 10.1021/acs.jnatprod.9b01095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C. Liu Z. Zhu X. Fan Z. Huang X. Wu Q. Zheng X. Qin X. Zhang T. Zhang H. Ju J. Ma J. J. Nat. Prod. 2020;83:1646–1657. doi: 10.1021/acs.jnatprod.0c00151. [DOI] [PubMed] [Google Scholar]
- Gao W. Kim J. Y. Anderson J. R. Akopian T. Hong S. Jin Y. Y. Kandror O. Kim J. W. Lee I. A. Lee S. Y. McAlpine J. B. Mulugeta S. Sunoqrot S. Wang Y. Yang S. H. Yoon T. M. Goldberg A. L. Pauli G. F. Suh J. W. Franzblau S. G. Cho S. Antimicrob. Agents Chemother. 2015;59:880–889. doi: 10.1128/AAC.04054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza C. Kishore U. Tsolaki A. G. Immunobiology. 2023;228:1–18. doi: 10.1016/j.imbio.2022.152321. [DOI] [PubMed] [Google Scholar]
- Daleke M. H. Cascioferro A. de Punder K. Ummels R. Abdallah A. M. van der Wel N. Peters P. J. Luirink J. Manganelli R. Bitter W. J. Biol. Chem. 2011;286:19024–19034. doi: 10.1074/jbc.M110.204966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett C. K. Broadwell L. J. Hayne C. K. Neher S. B. PLoS One. 2015;10:1–14. doi: 10.1371/journal.pone.0135447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A. K. Roy K. K. Singh S. Vishnoi S. P. Kumar A. Kashyap V. K. Kremer L. Srivastava R. Srivastava B. S. Int. J. Antimicrob. Agents. 2013;42:27–35. doi: 10.1016/j.ijantimicag.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Singh V. K. Srivastava M. Dasgupta A. Singh M. P. Srivastava R. Srivastava B. S. Tuberculosis. 2014;94:252–261. doi: 10.1016/j.tube.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Ward S. K. Abomoelak B. Hoye E. A. Steinberg H. Talaat A. M. Mol. Microbiol. 2010;77:1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow A. Sule P. Odonnell T. J. Burger A. Mattila J. T. Antonio B. Vergara K. Marcantonio E. Garry Adams L. James N. Williams P. G. Cirillo J. D. Prisic S. PLoS Pathog. 2021;17:1–24. doi: 10.1371/journal.ppat.1009570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. Ahn Y.-M. Jonnala S. Oh S. Fisher J. M. Goodwin M. B. Ioerger T. R. Via L. E. Bayliss T. Green S. R. Ray P. C. Wyatt P. G. Barry C. E. Boshoff H. I. Antimicrob. Agents Chemother. 2019;63:1–14. doi: 10.1128/AAC.01006-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai K. Meydani S. N. Urassa W. Wu D. Mugusi F. M. Saathoff E. Bosch R. J. Villamor E. Spiegelman D. Fawzi W. W. Epidemiol. Infect. 2014;142:1505. doi: 10.1017/S0950268813002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sritharan M. J. Bacteriol. 2016;198:2399–2409. doi: 10.1128/JB.00359-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J. W. Caza M. Front. Cell. Infect. Microbiol. 2013;3:80. doi: 10.3389/fcimb.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayambhu G. Bruno M. Gulick A. M. Pfeifer B. A. Curr. Opin. Biotechnol. 2021;69:242–251. doi: 10.1016/j.copbio.2021.01.021. [DOI] [PubMed] [Google Scholar]
- Carroll C. S. Moore M. M. Crit. Rev. Biochem. Mol. Biol. 2018;53:356–381. doi: 10.1080/10409238.2018.1476449. [DOI] [PubMed] [Google Scholar]
- Wells R. M. Jones C. M. Xi Z. Speer A. Danilchanka O. Doornbos K. S. Sun P. Wu F. Tian C. Niederweis M. PLoS Pathog. 2013;9:e1003120. doi: 10.1371/journal.ppat.1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. He J. Xie L. Chen T. Xie J. Cell. Physiol. Biochem. 2013;31:1–13. doi: 10.1159/000343343. [DOI] [PubMed] [Google Scholar]
- Patel K. Butala S. Khan T. Suvarna V. Sherje A. Dravyakar B. Eur. J. Med. Chem. 2018;157:783–790. doi: 10.1016/j.ejmech.2018.08.030. [DOI] [PubMed] [Google Scholar]
- Kem M. P. Butler A. BioMetals. 2015;28:445–459. doi: 10.1007/s10534-015-9827-y. [DOI] [PubMed] [Google Scholar]
- Kurth C. Kage H. Nett M. Org. Biomol. Chem. 2016;14:8212–8227. doi: 10.1039/C6OB01400C. [DOI] [PubMed] [Google Scholar]
- Hu Z. Zhang W. ACS Infect. Dis. 2020;6:25–33. doi: 10.1021/acsinfecdis.9b00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena L. S. Rajni T. FEBS J. 2010;277:2416–2427. doi: 10.1111/j.1742-4658.2010.07666.x. [DOI] [PubMed] [Google Scholar]
- Reddy P. V. Puri R. V. Chauhan P. Kar R. Rohilla A. Khera A. Tyagi A. K. J. Infect. Dis. 2013;208:1255–1265. doi: 10.1093/infdis/jit250. [DOI] [PubMed] [Google Scholar]
- Zhang L. Kent J. E. Whitaker M. Young D. C. Herrmann D. Aleshin A. E. Ko Y.-H. Cingolani G. Saad J. S. Moody D. B. Marassi F. M. Ehrt S. Niederweis M. Nat. Commun. 2022;13:2255. doi: 10.1038/s41467-022-29873-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. M. Wells R. M. Madduri A. V. R. Renfrow M. B. Ratledge C. Moody D. B. Niederweis M. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1945–1950. doi: 10.1073/pnas.1311402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M. D. Rush J. S. Thomas M. G. J. Bacteriol. 2012;194:2809–2818. doi: 10.1128/JB.00088-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro M. Simões M. Environ. Chem. Lett. 2019;17:1485–1494. doi: 10.1007/s10311-019-00887-9. [DOI] [Google Scholar]
- Fang Z. Sampson S. L. Warren R. M. Gey Van Pittius N. C. Newton-Foot M. Tuberculosis. 2015;95:123–130. doi: 10.1016/j.tube.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Juárez-Hernández R. E., Zhu H. and Miller M. J., SpringerBriefs in Molecular Science, Siderophore-Mediated Iron Acquisition: Target for the Development of Selective Antibiotics Towards Mycobacterium tuberculosis, 2013, pp. 65–88
- Li K. Steven W. C. BioMetals. 2016;29:377–388. doi: 10.1007/s10534-016-9935-3. [DOI] [PubMed] [Google Scholar]
- Sah S. Singh R. Agriculture. 2015;61:97–114. [Google Scholar]
- Choo S. W. Dutta A. Wong G. J. Wee W. Y. Ang M. Y. Siow C. C. PLoS One. 2016;11:1–19. doi: 10.1371/journal.pone.0150413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyrolles O. Wolschendorf F. Mitra A. Niederweis M. Immunol. Rev. 2015;264:249–263. doi: 10.1111/imr.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J. Xie J. Acta Pharm. Sin. B. 2011;1:8–13. doi: 10.1016/j.apsb.2011.04.008. [DOI] [Google Scholar]
- Gokarn K. Pal R. B. Sarangdhar V. Mol. Biol. 2016;05:1–5. doi: 10.4172/2168-9547.1000176. [DOI] [Google Scholar]
- Kumar N. Sritharan M. J. Appl. Microbiol. 2020;129:1733–1743. doi: 10.1111/jam.14728. [DOI] [PubMed] [Google Scholar]
- Bento C. M. Gomes M. S. Silva T. Antibiotics. 2020;9:1–25. doi: 10.3390/antibiotics9010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurthkoti K. Tare P. Paitchowdhury R. Gowthami V. N. Garcia M. J. Colangeli R. Chatterji D. Nagaraja V. Rodriguez G. M. Mol. Microbiol. 2015;98:864–877. doi: 10.1111/mmi.13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold F. M. Weber M. S. Gonda I. Gallenito M. J. Adenau S. Egloff P. Zimmermann I. Hutter C. A. J. Hürlimann L. M. Peters E. E. Piel J. Meloni G. Medalia O. Seeger M. A. Nature. 2020;580:413–417. doi: 10.1038/s41586-020-2136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. Hu X. Chai X. Ye Q. Pang J. Li D. Hou T. Drug Discovery Today. 2022;27:326–336. doi: 10.1016/j.drudis.2021.09.003. [DOI] [PubMed] [Google Scholar]
- Kwofie S. K. Enninful K. S. Yussif J. A. Asante L. A. Adjei M. Kan-Dapaah K. Tiburu E. K. Mensah W. A. Miller W. A. Mosi L. Wilson M. D. Molecules. 2019;24:1–21. doi: 10.3390/molecules24122299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohilla A. Khare G. Tyagi A. K. Sci. Rep. 2017;7:1–14. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfeli R. Beeson C. Infect. Disord.: Drug Targets. 2007;7:213–220. doi: 10.2174/187152607782110031. [DOI] [PubMed] [Google Scholar]
- Wilson B. R. Bogdan A. R. Miyazawa M. Hashimoto K. Tsuji Y. Trends Mol. Med. 2016;22:1077–1090. doi: 10.1016/j.molmed.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. M. Niederweis M. J. Bacteriol. 2010;192:6411–6417. doi: 10.1128/JB.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzón-Posse F. Quevedo-Acosta Y. Mahecha-Mahecha C. Acosta-Guzmán P. Eur. J. Org. Chem. 2019;2019:7747–7769. doi: 10.1002/ejoc.201901257. [DOI] [Google Scholar]
- Choudhury M. Koduru T. N. Kumar N. Salimi S. Desai K. Prabhu N. P. Sritharan M. BioMetals. 2021;34:511–528. doi: 10.1007/s10534-021-00292-2. [DOI] [PubMed] [Google Scholar]
- Ferreira D. Seca A. M. L. Pinto D. C. G. A. Silva A. M. S. J. Proteomics. 2016;145:153–166. doi: 10.1016/j.jprot.2016.04.006. [DOI] [PubMed] [Google Scholar]
- Chao A. Sieminski P. J. Owens C. P. Goulding C. W. Chem. Rev. 2019;119:1193–1220. doi: 10.1021/acs.chemrev.8b00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalut C. Tuberculosis. 2016;100:32–45. doi: 10.1016/j.tube.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Sandhu P. Akhter Y. J. Inorg. Biochem. 2017;170:75–84. doi: 10.1016/j.jinorgbio.2017.02.013. [DOI] [PubMed] [Google Scholar]
- McLean K. J. Munro A. W. Drug Discovery Today. 2017;22:566–575. doi: 10.1016/j.drudis.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Melly G. Purdy G. E. Microorganisms. 2019;7:1–16. doi: 10.3390/microorganisms7030070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyam M. Shilkar D. Verma H. Dev A. Sinha B. N. Brucoli F. Bhakta S. Jayaprakash V. J. Med. Chem. 2021;64:71–100. doi: 10.1021/acs.jmedchem.0c01176. [DOI] [PubMed] [Google Scholar]
- Rodriguez G. M. and Neyrolles O., in Molecular Genetics of Mycobacteria, ASM Press, Washington, DC, USA, 2015, pp. 377–387 [Google Scholar]
- Hider R. C. Kong X. Nat. Prod. Rep. 2010;27:637–657. doi: 10.1039/B906679A. [DOI] [PubMed] [Google Scholar]
- Leon-Sicairos N. Reyes-Cortes R. Guadrón-Llanos A. M. Madueña-Molina J. Leon-Sicairos C. Canizalez-Román A. BioMed Res. Int. 2015;2015:1–17. doi: 10.1155/2015/476534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufariello J. A. M. Chapman J. R. Kerantzas C. A. Wong K. W. Vilchèze C. Jones C. M. Cole L. E. Tinaztepe E. Thompson V. Fenyö D. Niederweis M. Ueberheide B. Philips J. A. Jacobs W. R. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E348–E357. doi: 10.1073/pnas.1523321113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudaher E. Shaffer C. L. MedChemComm. 2019;10:682–692. doi: 10.1039/C9MD00076C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan Siegrist M. Steigedal M. Ahmad R. Mehra A. Dragset M. S. Schuster B. M. Philips J. A. Carr S. A. Rubin E. J. MBio. 2014;5:1–10. doi: 10.1128/mBio.01073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Actor J. K. Mediators Inflammation. 2015;2015:15–20. doi: 10.1155/2015/409596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl S. L. Woodworth J. S. Lerche C. J. Cramer E. P. Nielsen P. R. Moser C. Thomsen A. R. Borregaard N. Cowland J. B. Front. Immunol. 2018;9:1–16. doi: 10.3389/fimmu.2018.02717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boradia V. M. Malhotra H. Thakkar J. S. Tillu V. A. Vuppala B. Patil P. Sheokand N. Sharma P. Chauhan A. S. Raje M. Raje C. I. Nat. Commun. 2014;5:4730. doi: 10.1038/ncomms5730. [DOI] [PubMed] [Google Scholar]
- Banerjee S. Farhana A. Ehtesham N. Z. Hasnain S. E. Infect., Genet. Evol. 2011;11:825–838. doi: 10.1016/j.meegid.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Chauhan A. S. Rawat P. Malhotra H. Sheokand N. Kumar M. Patidar A. Chaudhary S. Jakhar P. Raje C. I. Raje M. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep18465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra H. Patidar A. Boradia V. M. Kumar R. Nimbalkar R. D. Kumar A. Gani Z. Kaur R. Garg P. Raje M. Raje C. I. Front. Cell. Infect. Microbiol. 2017;7:1–15. doi: 10.3389/fcimb.2017.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudevylle N. Geist L. Hötzinger M. Hartl M. Kontaxis G. Bister K. Konrat R. J. Biol. Chem. 2010;285:41646–41652. doi: 10.1074/jbc.M110.123331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Cui T. Hu L. Wang Z. Li Z. He Z. G. Nat. Commun. 2015;6:1–9. doi: 10.1038/ncomms9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu R. Essler L. Giri P. Quinn F. PLoS One. 2020;15:1–19. doi: 10.1371/journal.pone.0240949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu R. Essler L. Loy A. Quinn F. Giri P. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-25480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan M. Neres J. Williams J. Wilson D. J. Teitelbaum A. M. Remmel R. P. Aldrich C. C. ChemMedChem. 2010;5:2079–2087. doi: 10.1002/cmdc.201000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi G. Manos-Turvey A. O'Connor P. D. Johnston J. M. Evans G. L. Baker E. N. Payne R. J. Lott J. S. Bulloch E. M. M. Biochemistry. 2012;51:4868–4879. doi: 10.1021/bi3002067. [DOI] [PubMed] [Google Scholar]
- Manos-Turvey A. Cergol K. M. Salam N. K. Bulloch E. M. M. Chi G. Pang A. Britton W. J. West N. P. Baker E. N. Lott J. S. Payne R. J. Org. Biomol. Chem. 2012;10:9223–9236. doi: 10.1039/C2OB26736E. [DOI] [PubMed] [Google Scholar]
- Liu Z. Liu F. Aldrich C. C. J. Org. Chem. 2015;80:6545–6552. doi: 10.1021/acs.joc.5b00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pini E. Poli G. Tuccinardi T. Chiarelli L. Mori M. Gelain A. Costantino L. Villa S. Meneghetti F. Barlocco D. Molecules. 2018;23:1506. doi: 10.3390/molecules23071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarelli L. R. Mori M. Barlocco D. Beretta G. Gelain A. Pini E. Porcino M. Mori G. Stelitano G. Costantino L. Lapillo M. Bonanni D. Poli G. Tuccinardi T. Villa S. Meneghetti F. Eur. J. Med. Chem. 2018;155:754–763. doi: 10.1016/j.ejmech.2018.06.033. [DOI] [PubMed] [Google Scholar]
- Chiarelli L. R. Mori M. Beretta G. Gelain A. Pini E. Sammartino J. C. Stelitano G. Barlocco D. Costantino L. Lapillo M. Poli G. Caligiuri I. Rizzolio F. Bellinzoni M. Tuccinardi T. Villa S. Meneghetti F. J. Enzyme Inhib. Med. Chem. 2019;34:823–828. doi: 10.1080/14756366.2019.1589462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M. Stelitano G. Gelain A. Pini E. Chiarelli L. R. Sammartino J. C. Poli G. Tuccinardi T. Beretta G. Porta A. Bellinzoni M. Villa S. Meneghetti F. J. Med. Chem. 2020;63:7066–7080. doi: 10.1021/acs.jmedchem.0c00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M. Stelitano G. Chiarelli L. R. Cazzaniga G. Gelain A. Barlocco D. Pini E. Meneghetti F. Villa S. Pharmaceuticals. 2021;14:155. doi: 10.3390/ph14020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M. Stelitano G. Griego A. Chiarelli L. R. Cazzaniga G. Gelain A. Pini E. Camera M. Canzano P. Fumagalli A. Scarpa E. Cordiglieri C. Rizzello L. Villa S. Meneghetti F. Pharmaceuticals. 2022;15:992. doi: 10.3390/ph15080992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreras J. A. Ryu J.-S. Di Lello F. Tan D. S. Quadri L. E. N. Nat. Chem. Biol. 2005;1:29–32. doi: 10.1038/nchembio706. [DOI] [PubMed] [Google Scholar]
- Lun S. Guo H. Adamson J. Cisar J. S. Davis T. D. Chavadi S. S. Warren J. D. Quadri L. E. N. Tan D. S. Bishai W. R. Antimicrob. Agents Chemother. 2013;57:5138–5140. doi: 10.1128/AAC.00918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhart C. A. Aldrich C. C. J. Org. Chem. 2013;78:7470–7481. doi: 10.1021/jo400976f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. M. Viswanathan K. Dawadi S. Duckworth B. P. Boshoff H. I. Barry C. E. Aldrich C. C. J. Med. Chem. 2015;58:5459–5475. doi: 10.1021/acs.jmedchem.5b00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawadi S. Viswanathan K. Boshoff H. I. Barry C. E. Aldrich C. C. J. Org. Chem. 2015;80:4835–4850. doi: 10.1021/acs.joc.5b00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajczyk A. Zeidler J. Januszczyk P. Dawadi S. Boshoff H. I. Barry C. E. Ostrowski T. Aldrich C. C. Bioorg. Med. Chem. 2016;24:3133–3143. doi: 10.1016/j.bmc.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawadi S. Boshoff H. I. M. Park S. W. Schnappinger D. Aldrich C. C. ACS Med. Chem. Lett. 2018;9:386–391. doi: 10.1021/acsmedchemlett.8b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bythrow G. V. Mohandas P. Guney T. Standke L. C. Germain G. A. Lu X. Ji C. Levendosky K. Chavadi S. S. Tan D. S. Quadri L. E. N. Biochemistry. 2019;58:833–847. doi: 10.1021/acs.biochem.8b01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L. Wells G. Bhakta S. Johnson J. Guzman J. Parish T. Prentice R. A. Brucoli F. ChemMedChem. 2019;14:1735–1741. doi: 10.1002/cmdc.201900217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyam M. Verma H. Bhattacharje G. Mukherjee P. Singh S. Kamilya S. Jalani P. Das S. Dasgupta A. Mondal A. Das A. K. Singh A. Brucoli F. Bagnéris C. Dickman R. Basavanakatti V. N. Naresh Babu P. Sankaran V. Dev A. Sinha B. N. Bhakta S. Jayaprakash V. J. Med. Chem. 2022;65:234–256. doi: 10.1021/acs.jmedchem.1c01349. [DOI] [PubMed] [Google Scholar]
- Rakshit G., Murtuja S. and Jayaprakash V., in ECSOC-25, MDPI, Basel Switzerland, 2021, vol. 8, p. 62 [Google Scholar]
- Jian Y. Merceron R. De Munck S. Forbes H. E. Hulpia F. Risseeuw M. D. P. Van Hecke K. Savvides S. N. Munier-Lehmann H. Boshoff H. I. M. Van Calenbergh S. Eur. J. Med. Chem. 2020;15:1–14. doi: 10.1016/j.ejmech.2020.112659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawadi S. Kawamura S. Rubenstein A. Remmel R. Aldrich C. C. Bioorg. Med. Chem. 2016;24:1314–1321. doi: 10.1016/j.bmc.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragset M. S. Poce G. Alfonso S. Padilla-Benavides T. Ioerger T. R. Kaneko T. Sacchettini J. C. Biava M. Parish T. Argüello J. M. Steigedal M. Rubin E. J. Antimicrob. Agents Chemother. 2015;59:2256–2264. doi: 10.1128/AAC.05114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreras J. A. Gupta A. Amin N. D. Basu A. Sinha B. N. Worgall S. Jayaprakash V. Quadri L. E. N. Bioorg. Med. Chem. Lett. 2011;21:6533–6537. doi: 10.1016/j.bmcl.2011.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. J. Walz A. J. Zhu H. Wu C. Moraski G. Möllmann U. Tristani E. M. Crumbliss A. L. Ferdig M. T. Checkley L. Edwards R. L. Boshoff H. I. J. Am. Chem. Soc. 2011;133:2076–2079. doi: 10.1021/ja109665t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison A. T. Abouelhassan Y. Kallifidas D. Bai F. Ukhanova M. Mai V. Jin S. Luesch H. Huigens R. W. Angew. Chem. 2015;127:15032–15036. doi: 10.1002/ange.201508155. [DOI] [PubMed] [Google Scholar]
- Madak J. Neamati N. Curr. Top. Med. Chem. 2015;15:745–766. doi: 10.2174/1568026615666150302105622. [DOI] [PubMed] [Google Scholar]
- Dunn E. A. Roxburgh M. Larsen L. Smith R. A. J. McLellan A. D. Heikal A. Murphy M. P. Cook G. M. Bioorg. Med. Chem. 2014;22:5320–5328. doi: 10.1016/j.bmc.2014.07.050. [DOI] [PubMed] [Google Scholar]
- Tarapdar A. Norris J. K. S. Sampson O. Mukamolova G. Hodgkinson J. T. Beilstein J. Org. Chem. 2018;14:2646–2650. doi: 10.3762/bjoc.14.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokarn K. Pal R. B. BMC Complementary Altern. Med. 2017;17:1–8. doi: 10.1186/s12906-016-1505-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth B. P. Wilson D. J. Nelson K. M. Boshoff H. I. Barry C. E. Aldrich C. C. ACS Chem. Biol. 2012;7:1653–1658. doi: 10.1021/cb300112x. [DOI] [PMC free article] [PubMed] [Google Scholar]