Abstract
Background:
Lack of anatomy recognition represents a clinically relevant risk in abdominal surgery. Machine learning (ML) methods can help identify visible patterns and risk structures; however, their practical value remains largely unclear.
Materials and methods:
Based on a novel dataset of 13 195 laparoscopic images with pixel-wise segmentations of 11 anatomical structures, we developed specialized segmentation models for each structure and combined models for all anatomical structures using two state-of-the-art model architectures (DeepLabv3 and SegFormer) and compared segmentation performance of algorithms to a cohort of 28 physicians, medical students, and medical laypersons using the example of pancreas segmentation.
Results:
Mean Intersection-over-Union for semantic segmentation of intra-abdominal structures ranged from 0.28 to 0.83 and from 0.23 to 0.77 for the DeepLabv3-based structure-specific and combined models, and from 0.31 to 0.85 and from 0.26 to 0.67 for the SegFormer-based structure-specific and combined models, respectively. Both the structure-specific and the combined DeepLabv3-based models are capable of near-real-time operation, while the SegFormer-based models are not. All four models outperformed at least 26 out of 28 human participants in pancreas segmentation.
Conclusions:
These results demonstrate that ML methods have the potential to provide relevant assistance in anatomy recognition in minimally invasive surgery in near-real-time. Future research should investigate the educational value and subsequent clinical impact of the respective assistance systems.
Keywords: artificial intelligence, laparoscopy, minimally invasive surgery, surgical anatomy, surgical data science, surgical innovation
Introduction
Highlights
Machine learning models to reduce surgical risks that precisely identify 11 anatomical structures: abdominal wall, colon, intestinal vessels (inferior mesenteric artery and inferior mesenteric vein with their subsidiary vessels), liver, pancreas, small intestine, spleen, stomach, ureter, and vesicular glands.
Large training dataset of 13 195 real-world laparoscopic images with high-quality anatomy annotations.
Similar performance of individual segmentation models for each structure and combined segmentation models in identifying intra-abdominal structures, and similar segmentation performance of DeepLabv3-based and SegFormer-based models.
DeepLabv3-based models are capable of near-real-time operation while SegFormer-based models are not, but SegFormer-based models outperform DeepLabv3-based models in terms of accuracy and generalizability.
All models outperformed at least 26 out of 28 human participants in pancreas segmentation, demonstrating their potential for real-time assistance in recognizing anatomical landmarks during minimally invasive surgery.
Computer vision describes the computerized analysis of digital images aiming at the automation of human visual capabilities, most commonly using machine learning (ML) methods, in particular deep learning. This approach has transformed medicine in recent years, with successful applications including computer-aided diagnosis of colonic polyp dignity in endoscopy1,2, detection of clinically actionable genetic alterations in histopathology3, and melanoma detection in dermatology4. The availability of large amounts of training data is the defining prerequisite for the successful application of deep learning methods. With the establishment of laparoscopy as the gold standard for a variety of surgical procedures5–8 and the increasing availability of computing resources, these concepts have gradually been applied to abdominal surgery. The overwhelming majority of research efforts in the field of Artificial Intelligence (AI)-based analysis of intraoperative surgical imaging data (i.e. video data from laparoscopic or open surgeries) has focused on classifying images with respect to the presence and/or location of previously annotated surgical instruments or anatomical structures9–13 or on analysis of surgical proficiency14–16 based on recorded procedures. However, almost all research endeavors in the field of computer vision in laparoscopic surgery have concentrated on preclinical stages, and to date, no AI model based on intraoperative surgical imaging data could demonstrate a palpable clinical benefit17,18. Among the studies closest to clinical application are recent works on the identification of instruments and hepatobiliary anatomy during cholecystectomy for automated assessment of the critical view of safety13 and on the automated segmentation of safe and unsafe preparation zones during cholecystectomy19.
In surgery, patient outcome heavily depends on the experience and performance of the surgical team20,21. In a recent analysis of Human Performance Deficiencies in major cardiothoracic, vascular, abdominal transplant, surgical oncology, acute care, and general surgical operations, more than half of the cases with postoperative complications were associated with identifiable human error. Among these errors, lack of recognition (including misidentified anatomy) accounted for 18.8%, making it the most common Human Performance Deficiency overall22. Examples of complications directly related to anatomical misperception are iatrogenic lesions to the ureter in gynecologic procedures23 and pancreatic injuries during splenic flexure mobilization in colorectal surgery24. While AI-based systems identifying anatomical risk and target structures would theoretically have the potential to alleviate this risk, limited availability and diversity of (annotated) laparoscopic image data drastically restrict the clinical potential of such applications in practice.
To advance and diversify the applications of computer vision in laparoscopic surgery, we have recently published the Dresden Surgical Anatomy Dataset25, providing 13 195 laparoscopic images with high-quality26, expert-reviewed annotations of the presence and exact location of 11 intra-abdominal anatomical structures: abdominal wall, colon, intestinal vessels (inferior mesenteric artery and inferior mesenteric vein with their subsidiary vessels), liver, pancreas, small intestine, spleen, stomach, ureter and vesicular glands. Here, we present the first study based on this dataset and present ML models to assist in precisely delineating anatomical structures, aiming to reduce surgical risks. Specifically, we evaluate the automated detection and localization of organs and anatomical structures in laparoscopic view using two state-of-the-art model architectures: DeepLabv3 and SegFormer. To assess the clinical value of the presented ML models, we compare algorithm segmentation performance to that of humans using the example of delineation of the pancreas.
Methods
Patient cohort
Video data from 32 robot-assisted anterior rectal resections or rectal extirpations were gathered at the trial center between February 2019 and February 2021. All included patients had a clinical indication for the surgical procedure recommended by an interdisciplinary tumor board. Patients were not specifically selected with respect to demographic or physical parameters (i.e. age, sex, body mass index, comorbidities, previous surgical procedures) or disease-specific criteria (i.e. indication, disease stage). Respective details of the underlying patient cohort have been published previously25. The procedures were performed using the da Vinci Xi system (Intuitive Surgical, Sunnyvale, CA, USA) with a standard Da Vinci Xi/X Endoscope with Camera (8 mm diameter, 30 degree angle, Intuitive Surgical, Sunnyvale, California, USA, Item code 470057). Surgeries were recorded using the CAST system (Orpheus Medical GmBH, Frankfurt a.M., Germany). Each record was saved at a resolution of 1920×1080 pixels in MPEG-4 format.
Dataset
Based on the full-length surgery recordings and respective temporal annotations of organ visibility, individual image frames were extracted and annotated as described previously25. In brief, three independent annotators with substantial experience in robot-assisted rectal surgery created pixel-wise annotations, which were subsequently reviewed by a surgeon with 4 years of experience in robot-assisted rectal surgery. A detailed description of the annotation process, including underlying annotation protocols as well as analyses of annotator agreement and technical parameters, has been published previously25. To guarantee the real-world applicability of ML models trained on the dataset, images with perturbations such as blurring due to camera movements, soiling of the lens, and presence of blood or smoke were not specifically excluded. However, the annotation protocols advised annotators to only annotate structures in soiled and blurry images if the respective structures were clearly delineable. The resulting Dresden Surgical Anatomy Dataset comprises 13 195 distinct images with pixel-wise segmentations of 11 anatomical structures: abdominal wall, colon, intestinal vessels (inferior mesenteric artery and inferior mesenteric vein with their subsidiary vessels), liver, pancreas, small intestine, spleen, stomach, ureter, and vesicular glands. Moreover, the dataset comprises binary annotations of the presence of each of these organs for each image. The dataset is publicly available via the following link: https://doi.org/10.6084/m9.figshare.21702600.
For ML purposes, the Dresden Surgical Anatomy Dataset was split into training, validation, and test data as follows (Fig. 1):
Training set (at least 12 surgeries per anatomical structure): surgeries 1, 4, 5, 6, 8, 9, 10, 12, 15, 16, 17, 19, 22, 23, 24, 25, 27, 28, 29, 30, 31.
Validation set (3 surgeries per anatomical structure): surgeries 3, 21, 26.
Test set (5 surgeries per anatomical structure): surgeries 2, 7, 11, 13, 14, 18, 20, 32.
Figure 1.
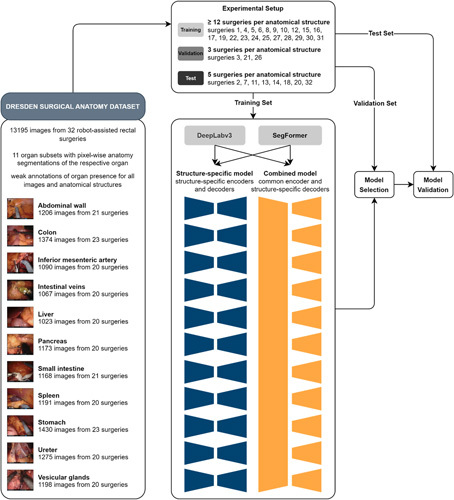
Schematic illustration of the structure-specific and combined machine learning (ML) models used for semantic segmentation. The Dresden Surgical Anatomy Dataset was split into a training, a validation, and a test set. For spatial segmentation, two sets of ML models – a structure-specific model with individual encoders and decoders, and a combined model with a common encoder and structure-specific decoders – were trained for DeepLabv3-based and SegFormer-based model architectures.
This split is proposed for future works using the Dresden Surgical Anatomy Dataset to reproduce the variance of the entire dataset within each subset, and to ensure comparability regarding clinical variables between the training, the validation, and the test set. Surgeries for the test set were selected to minimize variance regarding the number of frames over the segmented classes. Out of the remaining surgeries, the validation set was separated from the training set using the same criterion.
Structure-specific semantic segmentation models
To segment each anatomical structure, a separate convolutional neural network for the segmentation of individual structures was trained. Specifically, we trained and compared two different architectures: a Deeplabv327 model with a ResNet50 backbone with default PyTorch pretraining on the COCO dataset28 and a SegFormer29 model pretrained on the Cityscapes dataset30. The networks were trained using cross-entropy loss and the AdamW optimizer31 for 100 epochs with a starting learning rate of 10−4 and a linear learning rate scheduler, decreasing the learning rate by 0.9 every 10 epochs. For data augmentation, we applied random scaling and rotation, as well as brightness and contrast adjustments. The final model for each organ was selected via the Intersection-over-Union (IoU, Supplementary Fig. 1, Supplemental Digital Content 1, http://links.lww.com/JS9/A792) on the validation dataset and evaluated using the Dresden Surgical Anatomy Dataset with the abovementioned training-validation-test split (Fig. 1).
Segmentation performance was assessed using F1 score, IoU, precision, recall, and specificity on the test folds. These parameters are commonly used technical measures of prediction exactness, ranging from 0 (least exact prediction) to 1 (entirely correct prediction without any misprediction, Supplementary Fig. 1, Supplemental Digital Content 1, http://links.lww.com/JS9/A792).
Combined semantic segmentation models
A convolutional neural network with a common encoder and 11 decoders for combined segmentation of the 11 anatomical structures was trained. As for the structure-specific models, DeepLabv3-based27 and SegFormer-based29 models were used. For DeepLabv3, a shared ResNet50 backbone with default PyTorch pretraining on the COCO dataset28 was used. For each class, a DeepLabv3 decoder was then run on the features extracted from a given image by the backbone. Similarly, for SegFormer, an encoder pretrained on the Cityscapes dataset30 was combined with 11 decoders.
As the images are only annotated for binary classes, the loss is only calculated for every pixel in images in which the structure associated with the current decoder is annotated. For images in which the associated class is not annotated, only the pixels that are annotated as belonging to another class are included in the loss, for example pixels that were annotated as belonging to the class ‘liver’ can be used as negative examples for the class ‘pancreas’. The remaining training procedure was identical to the structure-specific model. The models were trained and evaluated using the Dresden Surgical Anatomy Dataset with the abovementioned training-validation-test split (Fig. 1).
Segmentation performance was assessed using the F1 score, IoU, precision, recall, and specificity on the test folds.
Evaluation of the semantic segmentation models on an external dataset
To explore generalizability, structure-specific and combined models based on both architectures (DeepLabv3 and SegFormer) were deployed to laparoscopic image data from the publicly available LapGyn4 dataset32. Models were separately deployed for full-scene segmentations, and their performance was visually compared.
Comparative evaluation of algorithmic and human performance
To determine the clinical potential of automated segmentation of anatomical risk structures, the segmentation performance of 28 humans was compared to that of the structure-specific and the combined semantic segmentation models using the example of the pancreas. The local Institutional Review Board reviewed and approved this study (approval number: BO-EK-566122021). All participants provided written informed consent to anonymous study participation, data acquisition and analysis, and publication. In total, 28 participants (physician and non-physician medical staff, medical students, and medical laypersons) marked the pancreas in 35 images from the Dresden Surgical Anatomy Dataset25 with bounding boxes. These images originated from 26 different surgeries, and the pancreas was visible in 16 of the 35 images. Each of the previously selected 35 images was shown once, the order being arbitrarily chosen but identical for all participants. The open-source annotation software Computer Vision Annotation Tool (CVAT) was used for annotations. In cases where the pancreas was seen in multiple, non-connected locations in the image, participants were asked to create separate bounding boxes for each area.
Based on the structure-specific and the combined semantic segmentation models, axis-aligned bounding boxes marking the pancreas were generated in the 35 images from the pixel-wise segmentation. To guarantee that the respective images were not part of the training data, four-fold cross-validation was used, that is the origin surgeries were split into four equal-sized batches, and algorithms were trained on three batches that did not contain the respective origin image before being applied to segmentation.
To compare human and algorithm performance, the bounding boxes created by each participant and the structure-specific, as well as the combined semantic segmentation models, were compared to bounding boxes derived from the Dresden Surgical Anatomy Dataset, which were defined as ground truth. IoU between the manual or automatic bounding box and the ground truth was used to compare segmentation accuracy.
Results
ML-based anatomical structure segmentation in structure-specific models
Structure-specific multilayer convolutional neural networks (Fig. 1) based on two different semantic segmentation architectures termed DeepLabv3 and SegFormer, were trained to segment the abdominal wall, the colon, intestinal vessels (inferior mesenteric artery and inferior mesenteric vein with their subsidiary vessels), the liver, the pancreas, the small intestine, the spleen, the stomach, the ureter, and vesicular glands (Supplementary Table 1, Supplemental Digital Content 1, http://links.lww.com/JS9/A792). Table 1 displays technical metrics of overlap between the annotated ground truth and the model predictions (mean F1 score, IoU, precision, recall, and specificity) for individual anatomical structures as predicted by the structure-specific algorithms on the test data.
Table 1.
Summary of performance metrics for anatomical structure segmentation using DeepLabv3-based (A) and SegFormer-based (B) structure-specific models on the test dataset (for each metric, mean and standard deviation are displayed).
| Anatomical structure | F1 score | IoU | Precision | Recall | Specificity | |
|---|---|---|---|---|---|---|
| A. DeepLabv3 | Abdominal wall | 0.90±0.10 | 0.83±0.14 | 0.89±0.14 | 0.93±0.07 | 0.97±0.04 |
| Colon | 0.79±0.20 | 0.69±0.22 | 0.80±0.21 | 0.82±0.21 | 0.97±0.05 | |
| Inferior mesenteric artery | 0.54±0.26 | 0.41±0.22 | 0.55±0.25 | 0.67±0.33 | 0.99±0.01 | |
| Intestinal veins | 0.54±0.33 | 0.44±0.29 | 0.70±0.26 | 0.56±0.36 | 1.00±0.00 | |
| Liver | 0.80±0.23 | 0.71±0.25 | 0.85±0.21 | 0.81±0.24 | 0.98±0.03 | |
| Pancreas | 0.37±0.32 | 0.28±0.27 | 0.59±0.37 | 0.37±0.36 | 1.00±0.01 | |
| Small intestine | 0.87±0.14 | 0.80±0.18 | 0.87±0.16 | 0.91±0.15 | 0.97±0.04 | |
| Spleen | 0.79±0.23 | 0.69±0.24 | 0.74±0.22 | 0.90±0.24 | 0.99±0.01 | |
| Stomach | 0.71±0.24 | 0.60±0.25 | 0.65±0.25 | 0.89±0.21 | 0.98±0.02 | |
| Ureter | 0.47±0.30 | 0.36±0.25 | 0.53±0.28 | 0.57±0.39 | 1.00±0.00 | |
| Vesicular glands | 0.40±0.25 | 0.28±0.21 | 0.37±0.28 | 0.62±0.35 | 0.97±0.03 | |
| B. SegFormer | Abdominal wall | 0.91±0.11 | 0.85±0.15 | 0.90±0.14 | 0.94±0.09 | 0.98±0.03 |
| Colon | 0.77±0.21 | 0.66±0.22 | 0.73±0.22 | 0.87±0.22 | 0.95±0.07 | |
| Inferior mesenteric artery | 0.60±0.23 | 0.46±0.21 | 0.58±0.25 | 0.73±0.29 | 0.99±0.01 | |
| Intestinal veins | 0.65±0.25 | 0.52±0.24 | 0.62±0.27 | 0.76±0.27 | 1.00±0.00 | |
| Liver | 0.83±0.21 | 0.75±0.24 | 0.82±0.23 | 0.88±0.18 | 0.98±0.03 | |
| Pancreas | 0.47±0.32 | 0.37±0.28 | 0.61±0.36 | 0.48±0.36 | 0.99±0.01 | |
| Small intestine | 0.89±0.13 | 0.83±0.17 | 0.87±0.16 | 0.95±0.10 | 0.97±0.04 | |
| Spleen | 0.85±0.19 | 0.78±0.21 | 0.80±0.19 | 0.95±0.16 | 1.00±0.01 | |
| Stomach | 0.75±0.27 | 0.66±0.28 | 0.76±0.25 | 0.82±0.29 | 0.99±0.01 | |
| Ureter | 0.58±0.27 | 0.46±0.24 | 0.53±0.26 | 0.74±0.32 | 0.99±0.01 | |
| Vesicular glands | 0.43±0.26 | 0.31±0.22 | 0.40±0.28 | 0.63±0.35 | 0.97±0.03 |
Out of the analyzed segmentation models based on DeepLabv3, performance was lowest for vesicular glands (mean IoU: 0.28±0.21), the pancreas (mean IoU: 0.28±0.27), and the ureter (mean IoU: 0.36±0.25), while excellent predictions were achieved for the abdominal wall (mean IoU: 0.83±0.14) and the small intestine (mean IoU: 0.80±0.18) (Supplementary Fig. 1, Supplemental Digital Content 1, http://links.lww.com/JS9/A792). In segmentation of the pancreas, the ureter, vesicular glands, and intestinal vessel structures, there was a relevant proportion of images with no detection or no overlap between prediction and ground truth, while for all remaining anatomical structures, this proportion was minimal (Fig. 2A). While the images, in which the highest IoUs were observed, mostly displayed large organ segments that were clearly visible (Fig. 2B), the images with the lowest IoU were of variable quality with confounding factors such as blood, smoke, soiling of the endoscope lens, or pictures blurred by camera shake (Fig. 2C). While overall segmentation performance of both architectures was similar for structure-specific models, SegFormer-based models showed a trend toward better performance than DeepLabv3-based models in segmentation of the pancreas, the spleen, and the ureter (Table 1, Fig. 2, Supplementary Fig. 2, Supplemental Digital Content 1, http://links.lww.com/JS9/A792).
Figure 2.
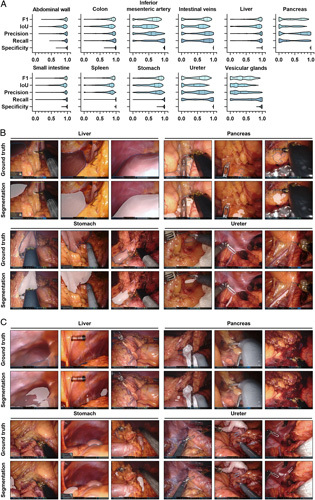
Pixel-wise organ segmentation with DeepLabv3-based structure-specific models trained on the respective organ subsets of the Dresden Surgical Anatomy Dataset. (A) Violin plot illustrations of performance metrics for DeepLabv3-based structure-specific segmentation models on the test dataset. The median and quartiles are illustrated as solid and dashed lines, respectively. (B) Example images from the test dataset with the highest IoUs for liver, pancreas, stomach, and ureter segmentation with DeepLabv3-based structure-specific segmentation models. Ground truth is displayed as blue line (upper panel), model segmentations are displayed as white overlay (lower panel). (C) Example images from the test dataset with the lowest IoUs for liver, pancreas, stomach, and ureter segmentation with DeepLabv3-based structure-specific segmentation models. Ground truth is displayed as blue line (upper panel), model segmentations are displayed as white overlay (lower panel). IoU, Intersection-over-Union.
To determine the models’ capabilities to operate in real-time (frame rates of >20 frames per second), we determined their inference times per image. For the DeepLabv3-based structure-specific models, inference on a single image with a resolution of 640×512 pixels required, on average, 28 ms on an Nvidia A5000, resulting in a frame rate of almost 36 frames per second. In contrast, the SegFormer-based structure-specific semantic segmentation models operated considerably slower at an inference time of 53 ms per image, resulting in a frame rate of 18 frames per second. This runtime includes one decoder, meaning that only the segmentation for one anatomical class (organ or structure) is included.
ML-based anatomical structure segmentation in combined models
In contrast to structure-specific models, models with a mutual encoder and organ-specific decoders could facilitate the identification of multiple organs at once, with the potential benefit of faster operation for multiple classes instead of sequential operation of several class-specific models. Therefore, combined models for both semantic segmentation architectures – DeepLabv3 and SegFormer – were trained using annotated images from the Dresden Surgical Anatomy Dataset across anatomical structure classes (Fig. 1, Supplementary Table 2, Supplemental Digital Content 1, http://links.lww.com/JS9/A792). Table 2 displays the mean F1 score, IoU, precision, recall, and specificity for anatomical structure segmentation in the combined model.
Table 2.
Summary of performance metrics for anatomical structure segmentation using the DeepLabv3-based (A) and SegFormer-based (B) combined models (common encoder with structure-specific decoders) on the test dataset (for each metric, mean and standard deviation are displayed).
| Anatomical structure | F1 score | IoU | Precision | Recall | Specificity | |
|---|---|---|---|---|---|---|
| A. DeepLabv3 | Abdominal wall | 0.86±0.11 | 0.77±0.15 | 0.81±0.15 | 0.95±0.09 | 0.95±0.04 |
| Colon | 0.75±0.19 | 0.63±0.21 | 0.71±0.18 | 0.84±0.23 | 0.95±0.04 | |
| Inferior mesenteric artery | 0.53±0.25 | 0.40±0.21 | 0.52±0.22 | 0.68±0.32 | 0.99±0.01 | |
| Intestinal veins | 0.46±0.32 | 0.35±0.27 | 0.70±0.23 | 0.48±0.36 | 1.00±0.00 | |
| Liver | 0.65±0.34 | 0.57±0.33 | 0.76±0.23 | 0.69±0.38 | 0.98±0.03 | |
| Pancreas | 0.32±0.30 | 0.23±0.24 | 0.61±0.33 | 0.32±0.35 | 0.99±0.01 | |
| Small intestine | 0.81±0.19 | 0.72±0.21 | 0.81±0.17 | 0.87±0.23 | 0.96±0.03 | |
| Spleen | 0.78±0.24 | 0.69±0.24 | 0.76±0.18 | 0.89±0.26 | 0.99±0.01 | |
| Stomach | 0.63±0.32 | 0.53±0.29 | 0.68±0.23 | 0.74±0.37 | 0.98±0.02 | |
| Ureter | 0.40±0.28 | 0.29±0.22 | 0.44±0.27 | 0.56±0.40 | 0.99±0.01 | |
| Vesicular glands | 0.42±0.27 | 0.30±0.23 | 0.41±0.30 | 0.56±0.36 | 0.98±0.02 | |
| B. SegFormer | Abdominal wall | 0.76±0.24 | 0.66±0.24 | 0.78±0.15 | 0.86±0.28 | 0.92±0.07 |
| Colon | 0.64±0.27 | 0.52±0.24 | 0.66±0.19 | 0.78±0.34 | 0.93±0.06 | |
| Inferior mesenteric artery | 0.40±0.24 | 0.28±0.18 | 0.37±0.21 | 0.68±0.38 | 0.97±0.02 | |
| Intestinal veins | 0.43±0.33 | 0.33±0.27 | 0.63±0.24 | 0.52±0.41 | 0.99±0.01 | |
| Liver | 0.62±0.35 | 0.53±0.32 | 0.76±0.19 | 0.71±0.40 | 0.95±0.07 | |
| Pancreas | 0.35±0.32 | 0.26±0.25 | 0.56±0.28 | 0.43±0.41 | 0.99±0.01 | |
| Small intestine | 0.78±0.19 | 0.67±0.20 | 0.74±0.14 | 0.90±0.23 | 0.94±0.06 | |
| Spleen | 0.71±0.24 | 0.59±0.23 | 0.65±0.21 | 0.89±0.25 | 0.99±0.01 | |
| Stomach | 0.65±0.32 | 0.55±0.29 | 0.71±0.22 | 0.75±0.36 | 0.98±0.02 | |
| Ureter | 0.38±0.29 | 0.27±0.23 | 0.43±0.27 | 0.55±0.40 | 0.99±0.01 | |
| Vesicular glands | 0.38±0.25 | 0.26±0.20 | 0.32±0.25 | 0.66±0.35 | 0.96±0.03 |
AI, artificial intelligence; IoU, Intersection-over-Union; SD, standard deviation.
The performance of the combined model based on DeepLabv3 was overall similar to that of structure-specific models (Table 1), with highest segmentation performance for the abdominal wall (mean IoU: 0.77±0.15) and the small intestine (mean IoU: 0.72±0.21), and the lowest performance for the pancreas (mean IoU: 0.23±0.29), the ureter (IoU: 0.29±0.22) and vesicular glands (IoU: 0.30±0.23) (Supplementary Fig. 1, Supplemental Digital Content 1, http://links.lww.com/JS9/A792). In comparison to the respective structure-specific models, the combined DeepLabv3-based model performed notably weaker in liver segmentation, while performance for the other anatomical structures was similar. The proportion of images for which the combined DeepLabv3-based model could not create a prediction or for which predictions showed no overlap with the ground truth at all was largest in the ureter, the pancreas, the stomach, the abdominal vessel structures, and the vesicular glands (Fig. 3A). Similar to the DeepLabv3-based structure-specific models, trends toward an impact of segment size, uncommon angles of vision, endoscope lens soiling, blurry images, and presence of blood or smoke were seen when comparing image quality of well-predicted images (Fig. 3B) to images with poor or no prediction (Fig. 3C). Similar to the structure-specific models, segmentation performance of the SegFormer-based combined segmentation model was, overall, similar to that of DeepLabv3-based models. For segmentation of the spleen, there was a trend toward weaker performance of SegFormer-based models than DeepLabv3-based combined models (Table 2, Fig. 3, Supplementary Fig. 3, Supplemental Digital Content 1, http://links.lww.com/JS9/A792).
Figure 3.
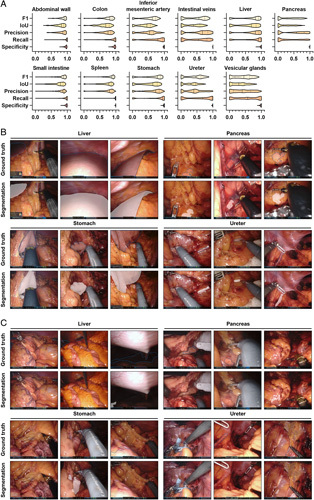
Pixel-wise organ segmentation with the DeepLabv3-based combined model trained on the Dresden Surgical Anatomy Dataset across anatomical structure classes with a common encoder and structure-specific decoders. (A) Violin plot illustrations of performance metrics for the DeepLabv3-based combined segmentation model on the test dataset. The median and quartiles are illustrated as solid and dashed lines, respectively. (B) Example images from the test dataset with the highest IoUs for liver, pancreas, stomach, and ureter segmentation with the DeepLabv3-based combined segmentation model. Ground truth is displayed as blue line (upper panel), model segmentations are displayed as white overlay (lower panel). (C) Example images from the test dataset with the lowest IoUs for liver, pancreas, stomach, and ureter segmentation with the DeepLabv3-based combined segmentation model. Ground truth is displayed as blue line (upper panel), model segmentations are displayed as white overlay (lower panel). IoU, Intersection-over-Union.
For the DeepLabv3-based combined models, inference on a single image with a resolution of 640×512 pixels required, on average, 71 ms on an Nvidia A5000, resulting in a frame rate of about 14 frames per second. As for structure-specific models of both architectures, SegFormer-based combined semantic segmentation models operated considerably slower at an inference time of 102 ms per image, resulting in a frame rate of about 10 frames per second. This runtime includes all 11 decoders, meaning that segmentations for all anatomical classes (organs or structures) are included.
Performance of ML models on an external laparoscopic image dataset
To evaluate model robustness on an external dataset, we deployed the different organ segmentation models onto the publicly available LapGyn4 dataset32 and qualitatively compared their performance. Overall, the combined models better reflected true anatomical constellations than the structure-specific models that generally lacked specificity. With respect to model architecture, the SegFormer-based segmentations were considerably more robust than the DeepLabv3-based models. Common mispredictions included confusion about liver and spleen, misinterpretation of organs that were not part of the training dataset (i.e. the gallbladder), and poor segmentation performance on less common images (i.e. extreme close-ups) (Fig. 4).
Figure 4.
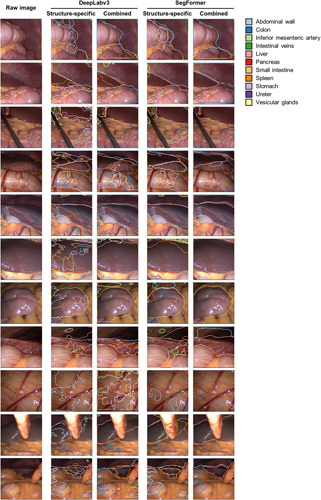
Comparison of DeepLabv3-based and SegFormer-based structure-specific and combined segmentation model performance on an external laparoscopic image dataset (LapGyn4). Models were deployed to the publicly available LapGyn4 dataset of non-semantically segmented images from gynecological procedures in conventional laparoscopic technique. Model segmentations for each organ are displayed. For the structure-specific models, segmentations of the 11 individual segmentation models are overlayed in one image. The figure shows representative images from the dataset.
In summary, the SegFormer-based combined semantic segmentation model resulted in robust segmentations reproducing the true underlying anatomy. The remaining segmentation models provided substantially less specific and less robust segmentation outputs on the external dataset.
Performance of ML models in relation to human performance
To approximate the clinical value of the previously described algorithms for anatomical structure segmentation, the performances of the DeepLabv3-based and SegFormer-based structure-specific and the combined models were compared to that of a cohort of 28 physicians, medical students and persons with no medical background (Fig. 5A), and different degrees of experience in laparoscopic surgery (Fig. 5B). A vulnerable anatomical structure24 with – measured by classical metrics of overlap (Tables 1 and 2) – comparably weak segmentation performance of the trained algorithms, the pancreas was selected as an example.
Figure 5.
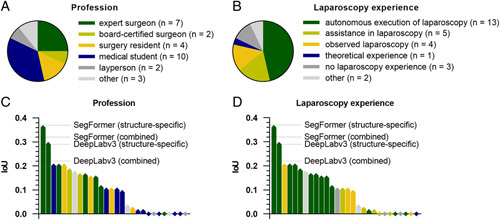
Comparison of pancreas segmentation performance of the structure-specific and the combined semantic segmentation models with a cohort of 28 human participants. (A) Distribution of medical and non-medical professions among human participants. (B) Distribution of laparoscopy experience among human participants. (C) Waterfall chart displaying the average pancreas segmentation IoUs of participants with different professions as compared to the IoU generated by the structure-specific and the combined semantic segmentation models. (D) Waterfall chart displaying the average pancreas segmentation IoUs of participants with varying laparoscopy experience as compared to the IoU generated by the structure-specific and the combined semantic segmentation models. IoU, Intersection-over-Union.
Comparing bounding box segmentations of the pancreas of human annotators, the medical and laparoscopy-specific experience of participants was mirrored by the respective IoUs describing the overlap between the pancreas annotation and the ground truth. The pancreas-specific segmentation models based on DeepLabv3 (IoU: 0.29) and SegFormer (IoU: 0.37), as well as the combined segmentation models based on DeepLabv3 (IoU: 0.21) and SegFormer (IoU: 0.32) outperformed at least 26 out of the 28 human participants (Fig. 5C, D). Overall, these results demonstrate that the developed models have clinical potential to improve the recognition of vulnerable anatomical structures in laparoscopy.
Discussion
In surgery, misinterpretation of visual cues can result in objectifiable errors with serious consequences22. ML models could augment the identification of anatomical structures during minimally invasive surgery and thereby contribute to a reduction of surgical risks. However, data scarcity and suboptimal dataset quality, among other factors, drastically restrict the clinical impact of applications in the field of surgical data science17,33–37. Based on a robust public dataset providing 13 195 laparoscopic images with segmentations of 11 intra-abdominal anatomical structures, this study explores the potential of ML for automated segmentation of these organs and compares algorithmic segmentation quality to that of humans with varying experience in minimally invasive abdominal surgery.
In summary, the presented findings suggest that ML-based segmentation of intra-abdominal organs and anatomical structures is possible and has the potential to provide clinically valuable information. At an average runtime of 71 ms per image, corresponding to a frame rate of 14 frames per second, the combined DeepLabv3-based model would facilitate near-real-time identification of 11 anatomical structures. In contrast, the SegFormer-based model is further from real-time performance at a runtime of 102 ms per image, resulting in a frame rate of less than 10 frames per second. These runtimes mirror the performances of non-optimized versions of the models, which can be significantly improved using methods such as TensorRT from Nvidia. However, with respect to generalizability and robustness, we observed substantially more accurate segmentation performance of the SegFormer-based models as compared to the DeepLabv3-based models when deployed to an external conventional laparoscopic dataset. Moreover, the structure-specific models exhibited a lack of accuracy and anatomical coherence, which can be explained by their organ-specific training process.
Measured by classical metrics of overlap between segmentation and ground truth, predictions were, overall, better for large and similar-appearing organs such as the abdominal wall, the liver, the stomach, and the spleen as compared to smaller and more diverse-appearing organs such as the pancreas, the ureter, or vesicular glands. Furthermore, poor image quality (i.e. images blurred by camera movements, presence of blood or smoke in images) was linked to lower accuracy of ML-based segmentations. Consequently, it is likely that a better nominal performance of the ML models could be achieved through a selection of images from the early phases of the surgery, in which such perturbations are not present. However, we purposely did not exclude images with suboptimal image quality, as the selection on image level would introduce bias and thereby limit applicability. In this context, selection on the patient level and on the image level is a common challenge in computer vision38, which can lead to skewed reporting of outcomes and poor performance on real-world data34. Overall, our findings on the influence of image quality on segmentation performance imply that computer vision studies in laparoscopy should be carefully interpreted, taking representativity and potential selection of underlying training and validation data into consideration.
Measured by classical metrics of overlap (e.g. IoU, F1 score, precision, recall, specificity) that are commonly used to evaluate segmentation performance, the structure-specific models and the combined models provided comparable segmentation performances on the internal test dataset. Interpretation of such metrics of overlap, however, represents a major challenge in computer vision applications in medical domains such as dermatology and endoscopy39–41 as well as non-medical domains such as autonomous driving42. In the specific use case of laparoscopic surgery, evidence suggests that such technical metrics alone are not sufficient to characterize the clinical potential and utility of segmentation algorithms37,43. In this context, the subjective clinical utility of a bounding box-based detection system recognizing the common bile duct and the cystic duct at average precisions of 0.32 and 0.07, respectively, demonstrated by Tokuyasu et al., supports this hypothesis12. In colorectal surgery, anatomical misinterpretation during splenic flexure mobilization can result in iatrogenic lesions in the pancreas24. In the presented analysis, the trained structure-specific and combined ML algorithms outperformed all human participants in the specific task of bounding box segmentation of the pancreas, except for two surgical specialists with over 10 years of experience. This suggests that even structures such as the pancreas with seemingly poor segmentation quality (segmentation IoU of the best-performing model: 0.37±0.28 in the test set) have the potential to provide clinically valuable help in anatomy recognition. In this context, analysis of additional anatomical risk structures (i.e. ureters and blood vessels) and inclusion of more advanced personnel in future comparison studies will help better define the models’ capabilities in comparison with (expert) surgeons. Notably, the best average IoUs for pancreas segmentation achieved in this comparative study were 0.37 (for the SegFormer-based structure-specific model) and 0.36 (for the best human participant), which would both be considered less reliable segmentation quality measures on paper. This encourages further discussion about metrics for segmentation quality assessment in clinical AI. In the future, the potential of the described dataset25 and organ segmentation algorithms could be exploited for educational purposes44,45, for guidance systems facilitating real-time detection of risk and target structures19,43,46,47, or as an auxiliary function integrated with more complex surgical assistance systems, such as guidance systems relying on automated liver registration48.
The limitations of this work are mostly related to the dataset and general limitations of ML-based segmentation: First, the Dresden Surgical Anatomy Dataset is a monocentric dataset based on 32 robot-assisted rectal surgeries. Therefore, the images used for algorithm training and validation originate from one set of hardware and display organs from specific angles. As a consequence, given the lack of a laparoscopic image dataset with similarly rigorous organ annotations, the generalizability and transferability of the presented findings to other centers and other minimally invasive abdominal surgeries, particularly non-robotic procedures, could only be qualitatively investigated. Second, annotations were required for training of ML algorithms, potentially inducing some bias toward the way that organs were annotated in the resulting models. With respect to annotation quality, three individual annotations of each anatomical structure were reviewed by a single surgical expert. This represents a major limitation of the underlying dataset, which is reasoned in the time-consuming and effortful annotation process, making the inclusion of more expert surgeons unfeasible. Given that annotations were based on specific annotation protocols, including images49 and all annotators had a medical background with several years of experience in the field of human anatomy25, the quality of annotations can be considered high, despite the limited experience of the reviewing surgeon (4 years of experience in robot-assisted rectal surgery). This is particularly true when comparing the underlying dataset with other datasets commonly used in surgical data science that are often based on single annotations carried out by individuals without domain knowledge17,26,50. Still, the way that organs are annotated may differ from individual healthcare professionals’ way of recognizing an organ. This is particularly relevant for organs such as the ureters or the pancreas, which often appear covered by layers of tissue. Here, computer vision-based algorithms that solely consider the laparoscopic images provided by the Dresden Surgical Anatomy Dataset for the identification of risk structures will only be able to identify an organ once it is visible. For an earlier recognition of such hidden risk structures, more training data with meaningful annotations would be necessary. Importantly, the presented comparison to human performance focused on the segmentation of visible anatomy as well, neglecting that humans (and possibly computers, too) could already identify a risk structure hidden underneath layers of tissue. Third, the dataset only includes individual annotated images. In some structures, such as the ureter, video data offers considerably more information than still image data. In this context, it is conceivable that incorporation of temporal aspects could result in major improvements in both human and algorithm recognition performance.
While the presented ML models show promise in improving the identification of anatomical structures in laparoscopy, their clinical utility still needs to be explored. Successful adoption of new technologies in surgery depends on factors beyond segmentation performance, runtime and generalizability, such as visualization of intraoperative decision support51, human–machine interaction52, and interface design. Therefore, interdisciplinary collaboration is critical to better understand respective surgeon needs. Moreover, prospective trials are needed to determine the impact of these factors on clinical outcomes. The existing limitations notwithstanding, the presented study represents an important addition to the growing body of research on medical image analysis in laparoscopic surgery, particularly by linking technical metrics to human performance.
In conclusion, this study demonstrates that ML methods have the potential to provide clinically relevant near-real-time assistance in anatomy recognition in minimally invasive surgery. This study is the first to use the recently published Dresden Surgical Anatomy Dataset, providing baseline algorithms for organ segmentation and evaluating the clinical relevance of such algorithms by introducing more clinically meaningful comparators beyond classical computer vision metrics. Future research should investigate other segmentation methods, the potential to integrate high-level anatomical knowledge into segmentation models38, the transferability of these results to other surgical procedures, and the clinical impact of real-time surgical assistance systems and didactic applications based on automated segmentation algorithms. Furthermore, seeing that the DeepLabv3-based models outperform the SegFormer-based models in terms of runtime but are lacking in accuracy and generalizability, future research could focus on combining the two in order to harness the best of both worlds.
Ethical approval
All experiments were performed in accordance with the ethical standards of the Declaration of Helsinki and its later amendments. The local Institutional Review Board (ethics committee at the Technical University Dresden) reviewed and approved this study (approval numbers: BO-EK-137042018 and BO-EK-566122021). The trial was registered on clinicaltrials.gov (trial registration ID: NCT05268432). Written informed consent to laparoscopic image data acquisition, data annotation, data analysis, and anonymized data publication was obtained from all participants. Before publication, all data was anonymized according to the general data protection regulation of the European Union.
Sources of funding
This work has been funded by the Else Kröner Fresenius Center for Digital Health (EKFZ), Dresden, Germany (project ‘CoBot’), by the German Research Foundation DFG within the Cluster of Excellence EXC 2050: ‘Center for Tactile Internet with Human-in-the-Loop (CeTI)’ (project number 390696704) and by the German Federal Ministry of Health (BMG) within the ‘Surgomics’ project (grant number BMG 2520DAT82). Furthermore, FRK received funding from the Medical Faculty of the Technical University Dresden within the MedDrive Start program (grant number 60487) and from the Joachim Herz Foundation (Add-On Fellowship for Interdisciplinary Life Science). FMR received a doctoral student scholarship from the Carus Promotionskolleg Dresden.
Author contribution
F.R.K., J.W., M.D., S.S., and S.B.: conceptualized the study; F.R.K., F.M.R., and M.C.: collected and annotated clinical and video data and contributed to data analysis; A.C.J., S.L., and S.B.: implemented and trained the neural networks and contributed to data analysis; J.W., M.D., and S.S.: supervised the project, provided infrastructure and gave important scientific input; F.R.K.: drafted the initial manuscript text. All authors reviewed, edited, and approved the final manuscript.
Conflicts of interest disclosure
The authors declare that they have no conflicts of interest.
Guarantor
Fiona R. Kolbinger and Sebastian Bodenstedt.
Data availability
The Dresden Surgical Anatomy Dataset is publicly available via the following link: https://doi.org/10.6084/m9.figshare.21702600. All other data generated and analyzed during the current study are available from the corresponding authors on reasonable request. To gain access, data requestors will need to sign a data access agreement.
Code availability
The most relevant scripts used for dataset compilation are publicly available via the following link: https://zenodo.org/record/6958337#.YzsBdnZBzOg. The code used for segmentation algorithms is available at https://gitlab.com/nct_tso_public/anatomy-recognition-dsad.
Supplementary Material
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal's website, www.lww.com/international-journal-of-surgery.
Published online 31 July 2023
Contributor Information
Fiona R. Kolbinger, Email: fiona.kolbinger@uniklinikum-dresden.de.
Franziska M. Rinner, Email: franziska.rinner@uniklinikum-dresden.de.
Alexander C. Jenke, Email: alexander.jenke@nct-dresden.de.
Matthias Carstens, Email: matthias.carstens@uniklinikum-dresden.de.
Stefanie Krell, Email: stefanie.krell@nct-dresden.de.
Stefan Leger, Email: stefan.leger@zeiss.de.
Marius Distler, Email: marius.distler@uniklinikum-dresden.de.
Jürgen Weitz, Email: juergen.weitz@uniklinikum-dresden.de.
Stefanie Speidel, Email: stefanie.speidel@nct-dresden.de.
Sebastian Bodenstedt, Email: sebastian.bodenstedt@nct-dresden.de.
References
- 1.Wang P, Liu X, Berzin TM, et al. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol Hepatol 2020;5:343–351. [DOI] [PubMed] [Google Scholar]
- 2.Wang P, Berzin TM, Glissen Brown JR, et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut 2019;68:1813–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kather JN, Heij LR, Grabsch HI, et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer 2020;1:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simillis C, Lal N, Thoukididou SN, et al. Open versus laparoscopic versus robotic versus transanal mesorectal excision for rectal cancer: a systematic review and network meta-analysis. Ann Surg 2019;270:59–68. [DOI] [PubMed] [Google Scholar]
- 6.Zhao JJ, Syn NL, Chong C, et al. Comparative outcomes of needlescopic, single-incision laparoscopic, standard laparoscopic, mini-laparotomy, and open cholecystectomy: a systematic review and network meta-analysis of 96 randomized controlled trials with 11,083 patients. Surgery 2021;170:994–1003. [DOI] [PubMed] [Google Scholar]
- 7.Luketich JD, Pennathur A, Awais O, et al. Outcomes after minimally invasive esophagectomy: review of over 1000 patients. Ann Surg 2012;256:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomson JE, Kruger D, Jann-Kruger C, et al. Laparoscopic versus open surgery for complicated appendicitis: a randomized controlled trial to prove safety. Surg Endosc 2015;29:2027–2032. [DOI] [PubMed] [Google Scholar]
- 9.Islam M, Atputharuban DA, Ramesh R, et al. Real-time instrument segmentation in robotic surgery using auxiliary supervised deep adversarial learning. IEEE Robot Autom Lett 2019;4:2188–2195. [Google Scholar]
- 10.Roß T, Reinke A, Full PM, et al. Comparative validation of multi-instance instrument segmentation in endoscopy: results of the ROBUST-MIS 2019 challenge. Med Image Anal 2020;70:101920. [DOI] [PubMed] [Google Scholar]
- 11.Shvets AA, Rakhlin A, Kalinin AA, et al. Automatic instrument segmentation in robot-assisted surgery using deep learning. In: Proceedings – 17th IEEE International Conference on Machine Learning and Applications, ICMLA 2018. Institute of Electrical and Electronics Engineers Inc.; 2019.624–8.
- 12.Tokuyasu T, Iwashita Y, Matsunobu Y, et al. Development of an artificial intelligence system using deep learning to indicate anatomical landmarks during laparoscopic cholecystectomy. Surg Endosc 2020;35:1651–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mascagni P, Vardazaryan A, Alapatt D, et al. Artificial intelligence for surgical safety: automatic assessment of the critical view of safety in laparoscopic cholecystectomy using deep learning. Ann Surg 2022;275:955–961. [DOI] [PubMed] [Google Scholar]
- 14.Jin A, Yeung S, Jopling J, et al. Tool detection and operative skill assessment in surgical videos using region-based convolutional neural networks. Proc - 2018 IEEE Winter Conf Appl Comput Vision, (WACV). Lake Tahoe, NV, USA; 2018:691–9. [Google Scholar]
- 15.Funke I, Bodenstedt S, Oehme F, et al. Using 3D convolutional neural networks to learn spatiotemporal features for automatic surgical gesture recognition in video. Med Image Comput Comput Assist Interv – MICCAI 2019 Lect Notes Comput Sci 2019;11768:467–475. [Google Scholar]
- 16.Lavanchy JL, Zindel J, Kirtac K, et al. Automation of surgical skill assessment using a three-stage machine learning algorithm. Sci Rep 2021;11:5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maier-Hein L, Eisenmann M, Sarikaya D, et al. Surgical data science – from concepts toward clinical translation. Med Image Anal 2022;76:102306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolbinger FR, Bodenstedt S, Carstens M, et al. Artificial Intelligence for context-aware surgical guidance in complex robot-assisted oncological procedures: An exploratory feasibility study. Eur J Surg Oncol 2023. 10.1016/j.ejso.2023.106996. [DOI] [PubMed] [Google Scholar]
- 19.Madani A, Namazi B, Altieri MS, et al. Artificial intelligence for intraoperative guidance: using semantic segmentation to identify surgical anatomy during laparoscopic cholecystectomy. Ann Surg 2022;276:363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fecso AB, Szasz P, Kerezov G, et al. The effect of technical performance on patient outcomes in surgery. Ann Surg 2017;265:492–501. [DOI] [PubMed] [Google Scholar]
- 21.Mazzocco K, Petitti DB, Fong KT, et al. Surgical team behaviors and patient outcomes. Am J Surg 2009;197:678–685. [DOI] [PubMed] [Google Scholar]
- 22.Suliburk JW, Buck QM, Pirko CJ, et al. Analysis of human performance deficiencies associated with surgical adverse events. JAMA Netw Open 2019;2:e198067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adelman MR, Bardsley TR, Sharp HT. Urinary tract injuries in laparoscopic hysterectomy: a systematic review. J Minim Invasive Gynecol 2014;21:558–566. [DOI] [PubMed] [Google Scholar]
- 24.Freund MR, Kent I, Horesh N, et al. Pancreatic injuries following laparoscopic splenic flexure mobilization. Int J Colorectal Dis 2022;37:967–971. [DOI] [PubMed] [Google Scholar]
- 25.Carstens M, Rinner FM, Bodenstedt S, et al. The Dresden Surgical Anatomy Dataset for abdominal organ segmentation in surgical data science. Sci Data 2023;10:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joskowicz L, Cohen D, Caplan N, et al. Inter-observer variability of manual contour delineation of structures in CT. Eur Radiol 2019;29:1391–1399. [DOI] [PubMed] [Google Scholar]
- 27.Chen L-C, Papandreou G, Schroff F, et al. Rethinking Atrous Convolution for Semantic Image Segmentation. 2017. Accessed 10 October 2022.https://arxiv.org/abs/1706.05587v3 [Google Scholar]
- 28.Lin TY, Maire M, Belongie S, et al. Microsoft COCO: Common Objects in Context. Lecture Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 2014;8693:740–755. Accessed 11 November 2022.https://arxiv.org/abs/1405.0312v3 [Google Scholar]
- 29.Xie E, Wang W, Yu Z, et al. SegFormer: simple and efficient design for semantic segmentation with transformers. Adv Neural Inf Process Syst 2021;34:12077–12090. [Google Scholar]
- 30.Cordts M, Omran M, Ramos S, et al. The Cityscapes Dataset for Semantic Urban Scene Understanding. Proc IEEE Conf Comput Vis Pattern Recognit 2016. [Google Scholar]
- 31.Loshchilov I, Hutter F. Decoupled Weight Decay Regularization. 7th International Conference Learn Represent ICLR 2019 . 2017. Accessed 9 November 2022.https://arxiv.org/abs/1711.05101v3 [Google Scholar]
- 32.Leibetseder A, Petscharnig S, Primus MJ, et al. LapGyn4: a dataset for 4 automatic content analysis problems in the domain of laparoscopic gynecology. MMSys’18: Proceedings of the 9th ACM Multimedia Systems Conference, 2018. Accessed 19 July 2021. 10.1145/3204949.3208127 [DOI]
- 33.Reddy CL, Mitra S, Meara JG, et al. Artificial Intelligence and its role in surgical care in low-income and middle-income countries. Lancet Digit Heal 2019;1:e384–e386. [DOI] [PubMed] [Google Scholar]
- 34.Moglia A, Georgiou K, Georgiou E, et al. A systematic review on artificial intelligence in robot-assisted surgery. Int J Surg 2021;95:106151. [DOI] [PubMed] [Google Scholar]
- 35.Anteby R, Horesh N, Soffer S, et al. Deep learning visual analysis in laparoscopic surgery: a systematic review and diagnostic test accuracy meta-analysis. Surg Endosc 2021;35:1521–1533. [DOI] [PubMed] [Google Scholar]
- 36.Kuo RYL, Harrison CJ, Jones BE, et al. Perspectives: a surgeon’s guide to machine learning. Int J Surg 2021;94:106133. [DOI] [PubMed] [Google Scholar]
- 37.Reinke A, Tizabi MD, Sudre CH, et al. Common Limitations of Image Processing Metrics: A Picture Story. 2021. Accessed 13 May 2022.https://arxiv.org/abs/2104.05642v4
- 38.Jin C, Udupa JK, Zhao L, et al. Object recognition in medical images via anatomy-guided deep learning. Med Image Anal 2022;81:102527. [DOI] [PubMed] [Google Scholar]
- 39.Renard F, Guedria S, Palma N, et al. Variability and reproducibility in deep learning for medical image segmentation. Sci Rep 2020;10:13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powers DMW, Ailab. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. 2020. Accessed 16 October 2022.https://arxiv.org/abs/2010.16061v1
- 41.Parikh RB, Teeple S, Navathe AS. Addressing bias in artificial intelligence in health care. JAMA 2019;322:2377–2378. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Mehta S, Caspi A. Rethinking Semantic Segmentation Evaluation for Explainability and Model Selection. 2021. Accessed 27 July 2021.https://arxiv.org/abs/2101.08418v1
- 43.Hashimoto DA, Rosman G, Witkowski ER, et al. Computer vision analysis of intraoperative video: automated recognition of operative steps in laparoscopic sleeve gastrectomy. Ann Surg 2019;270:414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu YY, Mazer LM, Yule SJ, et al. Complementing operating room teaching with video-based coaching. JAMA Surg 2017;152:318–325. [DOI] [PubMed] [Google Scholar]
- 45.Mizota T, Anton NE, Stefanidis D. Surgeons see anatomical structures faster and more accurately compared to novices: development of a pattern recognition skill assessment platform. Am J Surg 2019;217:222–227. [DOI] [PubMed] [Google Scholar]
- 46.Ward TM, Mascagni P, Ban Y, et al. Computer vision in surgery. Surgery 2022;169:1253–1256. [DOI] [PubMed] [Google Scholar]
- 47.Chopra H, Baig AA, Arora S, et al. Artificial intelligence in surgery: modern trends. Int J Surg 2022;106:106883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Docea R, Pfeiffer M, Bodenstedt S, et al. Linte CA, Siewerdsen JH. Simultaneous localisation and mapping for laparoscopic liver navigation : a comparative evaluation study. Medical Imaging 2021: Image-Guided Procedures, Robotic Interventions, and Modeling. SPIE; 2021:8. [Google Scholar]
- 49.Rädsch T, Reinke A, Weru V, et al. Labelling instructions matter in biomedical image analysis. Nat Mach Intell 2023;5:273–283. [Google Scholar]
- 50.Freeman B, Hammel N, Phene S, et al. Iterative Quality Control Strategies for Expert Medical Image Labeling. Proc AAAI Conf Hum Comput Crowdsourcing 2021;9:60–71. [Google Scholar]
- 51.Shaalan D, Jusoh S. Visualization in Medical System Interfaces: UX Guidelines. Proceedings of the 12th International Conference on Electronics, Computers and Artificial Intelligence (ECAI) 2020, 1 June 2020.
- 52.Henry KE, Kornfield R, Sridharan A, et al. Human–machine teaming is key to AI adoption: clinicians’ experiences with a deployed machine learning system. npj Digit Med 2022;5:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Dresden Surgical Anatomy Dataset is publicly available via the following link: https://doi.org/10.6084/m9.figshare.21702600. All other data generated and analyzed during the current study are available from the corresponding authors on reasonable request. To gain access, data requestors will need to sign a data access agreement.


