Abstract
Context
Lost glucagon-like peptide 1 receptor (GLP-1R) function affects human physiology.
Objective
This work aimed to identify coding nonsynonymous GLP1R variants in Danish individuals to link their in vitro phenotypes and clinical phenotypic associations.
Methods
We sequenced GLP1R in 8642 Danish individuals with type 2 diabetes or normal glucose tolerance and examined the ability of nonsynonymous variants to bind GLP-1 and to signal in transfected cells via cyclic adenosine monophosphate (cAMP) formation and β-arrestin recruitment. We performed a cross-sectional study between the burden of loss-of-signaling (LoS) variants and cardiometabolic phenotypes in 2930 patients with type 2 diabetes and 5712 participants in a population-based cohort. Furthermore, we studied the association between cardiometabolic phenotypes and the burden of the LoS variants and 60 partly overlapping predicted loss-of-function (pLoF) GLP1R variants found in 330 566 unrelated White exome-sequenced participants in the UK Biobank cohort.
Results
We identified 36 nonsynonymous variants in GLP1R, of which 10 had a statistically significant loss in GLP-1–induced cAMP signaling compared to wild-type. However, no association was observed between the LoS variants and type 2 diabetes, although LoS variant carriers had a minor increased fasting plasma glucose level. Moreover, pLoF variants from the UK Biobank also did not reveal substantial cardiometabolic associations, despite a small effect on glycated hemoglobin A1c.
Conclusion
Since no homozygous LoS nor pLoF variants were identified and heterozygous carriers had similar cardiometabolic phenotype as noncarriers, we conclude that GLP-1R may be of particular importance in human physiology, due to a potential evolutionary intolerance of harmful homozygous GLP1R variants.
Keywords: functional study, genetic association, GLP-1R, glucagon-like peptide 1 receptor, type 2 diabetes
Agonists of the glucagon-like peptide 1 receptor (GLP-1R), a class B1 G protein–coupled receptor (GPCR), have been approved for treatment both of type 2 diabetes and obesity in the United States and Europe (1). GLP-1R is expressed in pancreatic β cells, the intestines, stomach, brain, kidneys, skeletal muscle, heart, and blood vessel endothelium (2). Binding of GLP-1 to its receptor is initiated with an interaction between the α-helical part of GLP-1 within the extracellular part of the receptor, enabling a docking of the N-terminus into the binding pocket of the receptor (3). Subsequent signaling occurs through an intracellular coupling to the Gαs-protein, which leads to a downstream cascade of signaling, increasing levels of cyclic adenosine monophosphate (cAMP) (4). A concomitant conformational change in the receptor allows for phosphorylation mediated by the GPCR kinases (GRKs) that subsequently leads to recruitment of β-arrestins as part of receptor desensitization (5). These β-arrestins—like cAMP—also may be important for insulin secretion through the GLP-1R (6-8).
Previous studies of the coding GLP1R variation found a low-frequency variant, A316T (minor allele frequency [MAF] 1.4%) was associated with lower fasting glucose levels, lower risk of type 2 diabetes, and decreased glucose-stimulated insulin secretion (9-11). Similarly, the R131Q variant was associated with lower fasting glucose levels and lower risk of type 2 diabetes (12). Furthermore, genome-wide association studies showed common intronic and missense variants in the GLP1R locus, including P7L, to be associated with type 2 diabetes (13, 14). However, the molecular phenotypes of the coding GLP1R variants are unclear and have not been studied in detail (15).
We aimed to elucidate the functional and physiological consequences of the coding GLP1R variants. We sequenced GLP1R in 8642 Danish individuals to map genetic GLP1R variations, and subsequently investigated them for receptor signaling and GLP-1 binding properties. Finally, we linked these functional consequences to human physiology with respect to type 2 diabetes and cardiometabolic traits.
Materials and Methods
Danish Study Cohorts and the UK Biobank Cohort
This cross-sectional study included targeted resequencing of 8642 individuals from 2 Danish cohorts: 1) individuals without known diabetes from the Danish population-based Inter99 study (aged 30-60 years), with detailed biochemical, anthropometric, and self-reported phenotypical characteristics (16, 17) (ClinicalTrials.gov identifier: NCT00289237). In total, 5712 individuals were available for analysis, 1378 with prediabetes and 4334 with normal glucose tolerance, defined in accordance with World Health Organization 1999 criteria; 2) a total of 2930 individuals with type 2 diabetes from the Danish Centre for Strategic Research in Type 2 Diabetes (DD2) study, a prospective, population-based cohort of newly diagnosed type 2 diabetes patients with anthropometric, biochemical, and disease history phenotypes (18).
The UK Biobank (RRID:SCR_012815) is a large population-based prospective study. We included 469 835 UK Biobank participants with available exome-sequencing data (aged 40-69 years) and detailed genotypic and phenotypic information, and longitudinal follow-up of health and disease progression (19). To find prevalent cases of type 2 diabetes in the UK Biobank, we implemented the Eastwood algorithm, which assigns presence of type 2 diabetes based on baseline self-reported data, and secondary care data (20).
Sequencing in the Danish Cohorts
GLP1R was sequenced using solution-based target region capture followed by next-generation sequencing (NGS) of the coding region, as part of a panel of 265 genes involved in the development of diabetes and obesity. The methods for DNA extraction, capture of the targeted region, and NGS have been described in detail previously (21). In short, polymerase chain reaction–amplified DNA libraries were sequenced using the Illumina HiSeq2000 Analyzer (RRID:SCR_020132). Reads were aligned to the GRCh37/hg19 human reference genome (UCSC Genome Browser) using the Burrows-Wheeler Aligner (RRID:SCR_010910), and variants were called using the Genome Analysis Toolkit (RRID:SCR_001876) (22). Individual genotypes with genotype quality less than 20, read depth less than 20, allelic depth ratio less than 0.2 or allelic depth ratio with a binomial test P value less than 5e-7 were set to missing. Variants with call rate less than 80%, mean depth less than 20, and Hardy Weinberg P value less than 5e-7 in the subset of Inter99 individuals were removed. Individuals with mean depth less than 20, call rate less than 80%, outlying heterozygosity, non-European ancestry, duplicates, and genotype sex differing from registered sex and not matching previous genotype data were excluded. GLP1R was covered with a minimum per-base mean depth of 30× and a median per-base mean coverage for the target region of 174×.
Anthropometric and Biochemical Analyses in Inter99
Body weight, height, waist circumference, hip circumference, body mass index (BMI), and waist/hip ratio were available in Inter99 data. All participants (except diabetes patients) underwent a 75-g oral glucose tolerance test after an overnight fast, with blood samples drawn (at fasting, and after 30 and 120 minutes) for measurement of serum insulin and plasma glucose levels. Concentrations of serum triglycerides, high-density lipoprotein (HDL) cholesterol, low-density lipoprotein (LDL) cholesterol, total cholesterol, and glycated hemoglobin A1c (HbA1c) were also measured. Blood pressure was measured twice, providing a mean value. For details, see (16, 17).
We calculated the following indices: insulinogenic index (s-insulin30min – s-insulin0min)/p-glucose30min; corrected insulin response ((100 × s-insulin30min)/(p-glucose30min × (p-glucose30min − 3.89))); the β-cell function insulin sensitivity glucose tolerance test (BIGTT)—acute insulin response (23); Homeostasis Model Assessment of Insulin Resistance (HOMA-IR) ((p-glucose0min × s-insulin0min)/22.5); Insulin Sensitivity Index Matsuda (24); and BIGTT-sensitivity index (23).
Quality Control of the UK Biobank Exome Sequencing Data
We analyzed the 469 835 individuals with available exome-sequencing data, accessed from the UK Biobank through application 32683. The pVCF file including GLP1R was loaded into Hail v.0.2.69 (25). In addition to initial quality control, variants with a genotype quality of less than 20, variants that did not meet the allele balance filter of 0.9 or greater (homozygous variant) and 0.2 or less (heterozygous variants) and variants with read depth less than 7 (single-nucleotide variations, formerly single-nucleotide polymorphisms [SNP]) or 10 (indel) were set to missing. Variants with a call rate of 95% or less and variants for which less than 90% of all genotypes for that variant had a read depth less than 10 (ukb23158_500k_OQFE.90pct10dp_qc_variants.txt) were removed. Based on variables provided by UK Biobank, the following sample filters were applied; non-White individuals and individuals with sex chromosome aneuploidy or high heterozygosity and missing rate were excluded. For related individuals, third degree or closer, we used Hail's maximal_independent_set to keep the highest number of unrelated individuals, opting to keep carriers of loss-of-signaling (LoS) variants. In total, 330 566 individuals were available in the final data set.
Variant Annotation, Selection, and Prediction
For both the Danish and UK Biobank sequencing data, the GLP1R (NM_002062.4) exon locations (from https://genome.ucsc.edu/cgi-bin/hgTables) with additional 50 base pair overhangs (Supplementary Table S1 (26)) were used to extract variants for annotation (27, 28). Before annotation, the Danish data set (build37) was lifted to build38 using liftOver (RRID:SCR_018160). Subsequent filtering of synonymous, splice region, intron, and 3′ untranslated region variants left variants eligible for further analysis. Variants were additionally annotated using dbNSFPv4.0a (RRID:SCR_005178) (28-30) to evaluate the potential damaging effect of the variants. Based on masks described by Flannick et al. (31), we defined predicted loss of function (pLoF) as variants that passed either LofTee = high confidence (HC) (M1) or VEST4 greater than 90% and CADD greater than 90% and DANN greater than 90% and Eigen-raw greater than 90% and Eigen-PC-raw greater than 90% (M2) or FATHMM pred = D and FATHMM-MKL pred = D and PROVEAN pred = D and MetaSVM pred = D and MetaLR pred = D and MCAP greater than 0.025 (M3) or PolyPhen HDIV pred = D and PolyPhen HVAR pred = D and SIFT pred = del and LRT pred = D and MutTaster pred (M4). Some of these algorithms were unable to evaluate certain mutations such as frameshift, stop codons, and in-frame deletions. Non-HC LofTee variants without any other information were excluded from the pLoF classification, whereas in general the pLoF algorithm was coded to prevent absence of pLoF classification in case of partly missing information.
Introduction of the 36 Variants Into the Glucagon-like Peptide 1 Receptor
The 36 variants were introduced into the GLP-1R (Ensembl canonical transcript [ENST00000373256.4]) through site-directed mutagenesis using primers designed by the program Geneious R66 (RRID:SCR_010519) and purchased from TAG. All mutations were verified and sequenced by Eurofins.
Transfection and Tissue Culture
COS-7 cells (NCBI_Iran catalog No. C143, RRID:CVCL_0224) were cultured at 10% CO2 and 37 °C in Dulbecco's modified Eagle’s medium 1885 supplemented with 10% fetal bovine serum, 2 mmol/L glutamine, 180 units/mL penicillin, and 45 g/mL streptomycin. HEK293 cells (CLS catalog No. 300192/p777_HEK293, RRID:CVCL_0045) were cultured at 10% CO2 and 37 °C in Dulbecco's modified Eagle’s medium GlutaMAX-I supplemented with 10% fetal bovine serum, 180 units/mL penicillin, and 45 g/mL streptomycin. Transient transfection was performed using the calcium phosphate precipitation method using the plasmid pcDNA3.1(+) (Thermo Fisher, catalog No. V79020, pcDNATM3.1(+) Mammalian Expression Vector) (32). HEK-293 cells and COS-7 cells were purchased from ATCC (CRL-1552 and CRL-1651, respectively). Cell line authentication was guaranteed by the sources from which the cells were purchased. Before and during tissue culture, all eukaryotic cell lines were tested for negativity to mycoplasma on a regular basis.
Cyclic Adenosine Monophosphate Measurements
Transiently transfected COS-7 cells were seeded in 96-well plates 1 day after transfection at a density of 35 000 cells/well in white plates. The following day, the cells were washed twice with HEPES-buffered saline buffer (HBS) and incubated with HBS and 1 mmol/L 3-isobutyl-1-methylxanthine for 30 minutes at 37 °C (33). GLP-1 (purchased from Caslo Aps) was added and incubated for 30 minutes at 37 °C. The HitHunter cAMP XS assay (DiscoveRx) was carried out according to the manufacturer's instructions. In vitro pharmacological analyses with determination of potency (EC50) and efficacy (Emax) were carried out with GraphPad Prism 9 (RRID:SCR_002798). The data were interpolated using a standard curve to determine absolute cAMP levels before being normalized to wild-type (WT). Sigmoid curves were fitted logistically with a Hillslope of 1.0.
Homologous Competition Binding
Transiently transfected COS-7 cells were seeded in clear 96-well plates 1 day after transfection at a density of 5000 to 45 000 cells/well, adjusted to obtain 5% to 10% specific binding of the radioligand. The following day, cells were washed twice with binding buffer (50 mmol/L HEPES-buffer [pH 7.2] supplemented with 0.5% bovine serum albumin) and assayed by competition binding for 3 hours at 4 °C using 15 to 40 pmol/L 125I-GLP-1(7-36)NH2 (Phoenix Pharmaceuticals), as well as unlabeled GLP-1(7-36)NH2 in binding buffer. After incubation, the cells were washed twice in ice-cold binding buffer and lysed using 200 mmol/L NaOH with 1% sodium dodecyl sulfate for 30 minutes. The samples were counted using Wallac Wizard 1470 Gamma Counter. In vitro pharmacological analysis with determination of affinity (IC50) was carried out using GraphPad Prism 9. Competitive binding data were normalized to the maximum specific response of WT GLP-1R for each individual experiment. Sigmoid curves were fitted logistically with a Hillslope of 1.0. Bmax was calculated using the competitive binding curves and the following equation: Bmax = (B0 × IC50)/[L], where B0 is the total specific binding, and [L] is the ligand concentration. Kd was calculated as IC50 – [L].
β-Arrestin Recruitment Study
HEK293 cells were transiently transfected with the GLP-1R, the donor Rluc3-Arrestin-2-Sp1, the acceptor mem-linker-citrine-SH3, and the GRK6 to facilitate β-arrestin 2 recruitment (34). One day before the transient transfection, the cells were seeded in tissue culture 6-well plates (0.9 × 106 to 1 × 106 cells/well). Two days following transfection, the cells were washed with phosphate-buffered saline (PBS) and resuspended in PBS with 5 mmol/L glucose. Next, 85 µL of the cell suspension solution was added to each well of a white 96-well plate, followed by the addition of PBS with 5 µmol/L coelenterazine-h. Following 10-minute incubation, increasing concentrations of GLP-1 were added and incubated for an additional 10 minutes. The BRET assay (DiscoveRx) was carried out according to the manufacturer's instructions. In vitro pharmacological analyses with determination of EC50 and Emax were carried out using GraphPad Prism 9. The data were normalized to the maximal response (Emax) detected with WT GLP-1R. Sigmoid curves were fitted logistically with a Hillslope of 1.0.
Gene-based Association Analysis
We used logistic and linear regression models, in which carriers represented individuals carrying any of the LoS/pLoF variants. Additionally, we used SKAT (v2.0.1, RRID:SCR_009396) burden tests (collapsing genetic variants into one pool) (35) with equal weights for LoS variants and SKAT-optimal for pLoF variants (36). All quantitative traits were rank-normalized before analyses. All tests were adjusted for sex, age, age squared, and 4 (Danish cohorts) or 20 (UK Biobank) principal components, and in addition BMI where mentioned. We used R statistical software (v4.0.2, RRID:SCR_001905) (37). In case of missing genotype data, imputation was applied. In each analysis, individuals with missing phenotype or missing covariates were excluded.
We calculated the least detectable effect (LDE) of a 2-sided t test for standardized quantitative traits with unequal group-sizes (LoScarriers = 379 vs LoSnoncarriers = 329,138, and pLoFcarriers = 261 vs pLoFnoncarriers = 329 256). An LDE of 0.14 and 0.18 was required for the LoS variant analyses and the pLoF variant analyses, respectively, to achieve 80% power. Finally, we calculated the LDE for the type 2 diabetes analyses and found that odds ratios (ORs) of 1.76 and 1.96 were required to achieve 80% power of the LoS variant analysis and the pLoF variant analysis, respectively (assuming the noncarrier prevalence to be 5%).
Mutational Constraint of the Glucagon-like Peptide 1 Receptor
For variants divided into 3 categories—synonymous, missense and pLoF—we obtained the ratio of the observed over expected number of single-nucleotide variants in GLP1R (Ensembl canonical transcript [ENST00000373256.4]) from the GnomAD database (v.2.1.1, RRID:SCR_014964) covering 125 748 exome and 15 708 whole-genome sequences (38).
Results
Targeted Sequencing Revealed 36 Nonsynonymous Variants in the Glucagon-like Peptide 1 Receptor
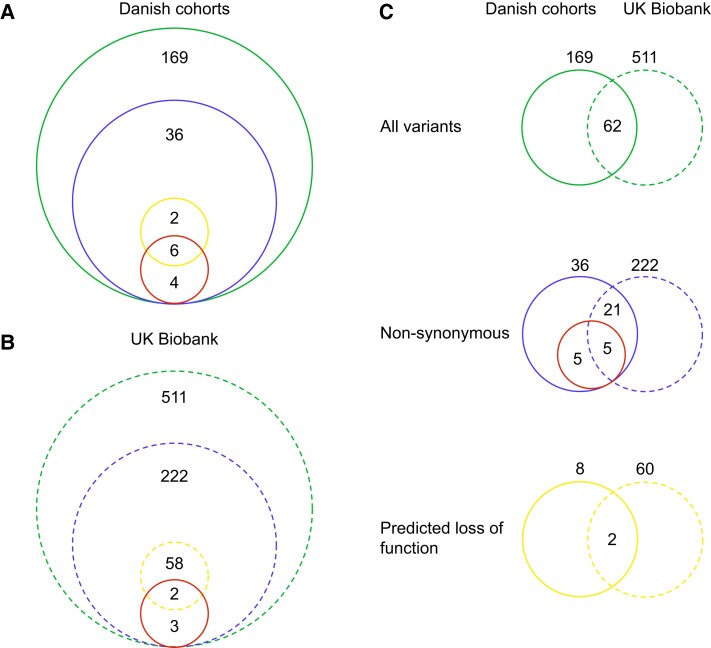
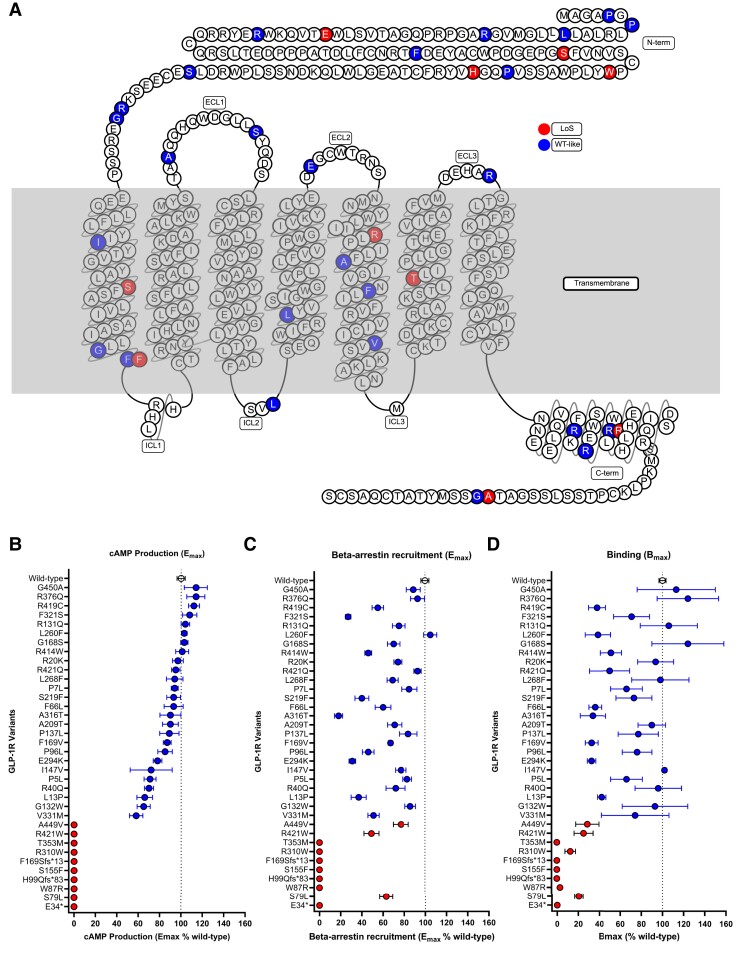
We identified 169 variants through targeted sequencing of 8642 Danish individuals, including 36 nonsynonymous variants (Fig. 1A; Supplementary Table S2 (26)), among which 1 introduced an early stop codon, 2 were frameshift mutations, and the remaining 33 mutations were classified as missense. Eighteen of the nonsynonymous variants were localized in the extracellular regions, 11 in the transmembrane regions, and 7 in the intracellular part of the receptor (Fig. 2A). Only 3 missense variants in the Danish study population (combining both Danish cohorts) were common (P7L, G168S, and L260F; MAF > 5%), one was a low-frequency variant (A316T, MAF 2%), and the rest were rare variants (MAF ≤ 0.1%) (Supplementary Table S2 (26)).
Figure 1.
Overview of GLP1R variants found in the Danish and UK Biobank cohorts. The green circle shows the total number of variants in exome regions with 50 bp overhangs, the blue circle shows the nonsynonymous variants, the yellow circle displays the number of predicted loss-of-function variants, and the red circle shows in vitro determined loss-of-signaling variants. A, Danish cohorts (solid line); B, UK Biobank (dashed line); C, Comparison of the variants found in the Danish cohorts and the UK biobank cohort. The numbers outside the circles represent the total number of variants within a given group.
Figure 2.
Localization of the 36 GLP1R variants from the Danish study cohorts and the consequences of the variants on receptor signaling. Blue, WT-like; red, loss-of-signaling (loss of cAMP signaling). A, Snake plot of the 36 variants; B, cAMP production compared to WT in percentage; C, β-Arrestin 2 recruitment compared to WT in percentage; D, Binding compared to WT in percentage. cAMP, cyclic adenosine monophosphate; ECL, extracellular loop; ICL, intracellular loop; WT, wild-type.
Molecular Pharmacological Profiling Identified 10 Loss-of-Signaling Variants
We identified 26 variants (∼ 72%) that displayed cAMP production similar to WT GLP-1R (EC50 of 97-pmol/L), defined as “WT-like” with less than a 5-fold change in potency (EC50, 13-370 pmol/L) and an Emax of at least 50% of the WT Emax (Supplementary Table S3 (26); see Fig. 2 and Supplementary Fig. S1 (26)). However, 10 variants showed no cAMP production and were denoted as LoS (Fig. 2B, Supplementary Table S3 (26)).
Seven Variants Showed Complete Loss of β-Arrestin Recruitment
Overall, the GLP-1R variants showed different abilities to recruit β-arrestin 2 (Fig. 2C, Supplementary Table S3, and Supplementary Fig. S1 (26)). The 26 WT-like cAMP-signaling variants also showed WT-like β-arrestin 2 recruitment (EC50, 1.2 nmol/L) with EC50 values varying from 0.4 nmol/L to 5.5 nmol/L (Supplementary Table S3 (26)). The efficacies varied from 58% to 114% of WT GLP-1R with an average Emax of 90.2% of WT ± 6.6% (SEM).
Three of the 10 LoS variants (S79L, R421W, and A449V) maintained β-arrestin 2 recruitment with EC50 values varying from 0.5 nmol/L to 2.2 nmol/L and therefore were affected selectively in one pathway over another. The remaining 7 variants (E34*, W87R, H99Qfs*83, S155F, F169Sfs*13, R310W, and T353M) completely lost β-arrestin 2 recruitment.
Five Variants Had Complete Loss of Receptor Binding
Compared to the WT GLP-1R with Kd of 8.1 nmol/L, native GLP-1 showed, as expected, a similar binding profile in the 26 variants with maintained cAMP production (Kd values varied from 1.8 to 18 nmol/L) (Supplementary Table S3 (26)). Bmax values varied from 33.5% to 124.4% for WT. Of the 10 LoS variants, 5 variants (E34*, H99Qfs*83, S155F, F169Sfs*13, and T353M) showed complete loss of binding (Fig. 2D). The remaining 5 LoS variants (S79L, W87R, R310W, R421W, and A449V) maintained GLP-1 binding, with Kd values varying from 3.5 to 15 nmol/L (Supplementary Fig. S1 (26)) and Bmax values from 3.0% to 70.8% of WT (Fig. 2D).
Loss-of-Signaling Glucagon-like Peptide 1 Receptor Variants Was Not Associated With Type 2 Diabetes or Phenotypes of Importance for Cardiometabolic Diseases
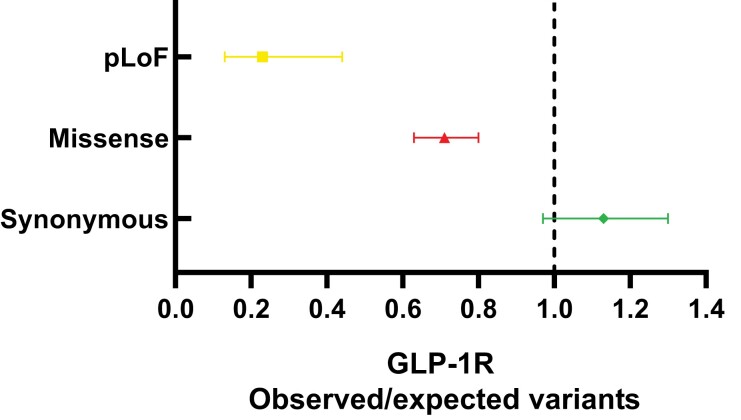
Next, we investigated the tolerance of the GLP-1R to certain classes of variations by the ratio of observed over expected (O/E) number of GLP1R variants from the GnomAD database (38). The O/E-ratio is a measure of how tolerant a gene is to the specific variation in the gene. For synonymous variants, we observed slighty more observed variants than expected (O/E-ratio of 1.13; 90% CI, 0.97-1.3). For missense and pLoF variants, we observed fewer variants than expected, yielding O/E-ratios of 0.71; 90% CI, 0.63 to 0.8; and 0.23; 90% CI, 0.13 to 0.44, respectively (Fig. 3). The low O/E-ratios for missense and pLoF variants in the GLP1R shows that the receptor is under strong selection against these classes of variations.
Figure 3.
Ratios of observed over expected variants for 3 categories of variants in the GLP1R. The observed (Danish cohorts) over expected (GnomAD) ratio of predicted loss-of-function (pLoF) variants, missense variants, and synonymous variants in GLP1R. Error bars represent 90% confidence intervals. The dashed line represents the null hypothesis; the observed number of variants = the expected number of variants.
In fact, the majority of the 10 LoS variants were extremely rare (MAF < 0.001), and all were exclusively presented as heterozygous in 27 individuals from the Danish cohorts (N = 7264, Supplementary Table S2 (26)).
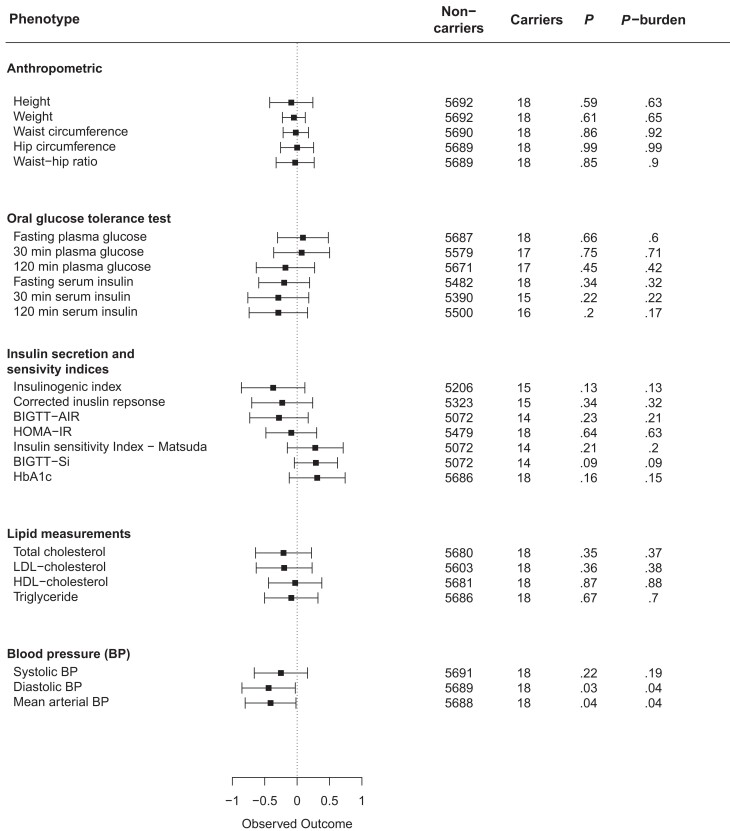
To evaluate the physiological effect of lost GLP-1R signaling, we evaluated whether the 10 LoS variants affected type 2 diabetes risk and cardiometabolic phenotypes. Among individuals with type 2 diabetes, 15 were carriers of LoS variants and 2915 were noncarriers compared to 12 carriers and 4322 noncarriers in the control group. Using a general linear model and a burden test, we observed no difference in type 2 diabetes risk for GLP1R LoS carriers vs noncarriers (OR = 2.23; CI, 0.75-6.54; Pglm = .14; Pburden = .24). Similarly, we observed no associations with changes in any of the cardiometabolic phenotypes (Fig. 4) in the Inter99 cohort (N = 5694; 7 of the 10 LoS variants were represented in 18 carriers). Adjustment for BMI did not change the associations (Supplementary Fig. S2 (26)). Next, we evaluated the LoS variants that were also predicted as pLoF variants (E34*, W87R, H99Qfs*83, F169Sfs*13, R310W, and T353M) because these variants may be most influential. Among individuals with type 2 diabetes, 2 were carriers of the LoS variants predicted as pLoF and 2928 were noncarriers, and among controls, 3 were carriers and 4330 were noncarriers. We found no association with type 2 diabetes (OR = 1.60; CI, 0.11-11.81; Pglm = .90; Pburden = .91; ORBMIadj = 1.21; CIBMIadj, 0.10-14.09; Pglm-BMIadj = .88; Pburden-BMIadj = .72), although we observed a minor increase in fasting plasma glucose (FPG) level (lost after BMI adjustment), and a decreased glucose-stimulated insulin response (insulinogenic index). Adjusting for BMI further revealed a marginal decreased LDL-cholesterol level and glucose-stimulated 30-min serum insulin level (Supplementary Fig. S3 (26)).
Figure 4.
Quantitative trait analyses of loss-of-signaling (LoS) GLP1R variants in the Inter99 cohort. Noncarriers = number of individuals with phenotype information who do not carry any of the variants; Carriers = number of individuals with the phenotype who carry an LoS variant. BIGTT-AIR, β-cell function insulin sensitivity glucose tolerance test—acute insulin response, β-cell function index; BIGTT-Si, β-cell function insulin sensitivity glucose tolerance test—insulin sensitivity index; BP, blood pressure; HbA1c, glycated hemoglobin A1c; HDL, high-density lipoprotein; HOMA-IR, Homeostatic Model Assessment of Insulin Resistance; LDL, low-density lipoprotein; P, P value of the linear regression model; P-burden, P value of burden test.
Comparing the Overlap Between Glucagon-like Peptide 1 Receptor Loss-of-Signaling and Predicted Loss-of-Function Variants
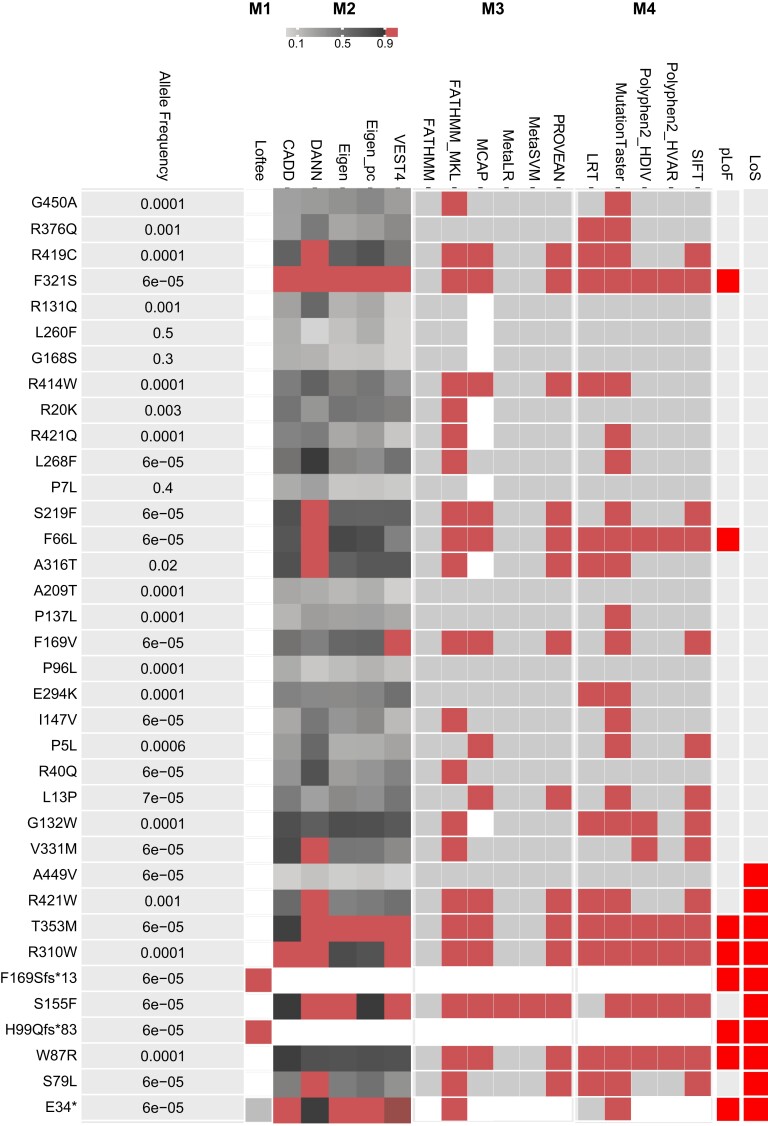
To evaluate how well the in vitro molecular implications of GLP1R variants correspond with pLoF, we classified variants based on aggregated scores (masks) of 17 variant effect predictor algorithms (Fig. 5). We defined the variant as a pLoF variant if all algorithms within any mask 1 to 4 (M1-M4) passed, and subsequently compared the variant with our in vitro results (see Fig. 2). Six of the 10 LoS variants were also pLoF variants (E34*, W87R, H99Qfs*83, F169Sfs*13, R310W, and T353M), whereas 4 were not recognized as pLoF. In contrast, 2 variants (F66L and F321S) were predicted to be pLoF but not classified as LoS in the in vitro studies (see Fig. 5).
Figure 5.
Binary heat map showing in silico prediction of severity of the 36 GLP1R variants in the Danish population. The left panel shows a list of each GLP1R variant with its minor allele frequency (MAF) in the Danish study cohorts (combined data set) sorted according to its cyclic adenosine monophosphate (cAMP) production (Fig. 2). Panel M1 shows whether the variant is a predicted loss-of-function (pLoF, red) variant by the Loss-of-Function Transcript Effect Estimator (Loftee) high-confidence (HC) mask. Panel M2-M4 shows the predicted deleteriousness of each variant for prediction algorithms grouped in masks. A red box indicates variants passing the specific algorithm as LoF. The masks consist of the following algorithms: M2: VEST4, CADD, DANN, Eigen-raw, and Eigen-PC-raw; M3: FATHMM, FATHMM-MKL, PROVEAN, MetaSVM, MetaLR, and MCAP; and M4: PolyPhen2 HDIV, PolyPhen HVAR, SIFT, LRT, and MutationTaster. Panel pLoF; predicted LoF variants (bright red). Panel LoS; loss-of-signaling variants from the in vitro studies (bright red). White color indicates no information (NA).
Analysis of Type 2 Diabetes Risk in the UK Biobank Confirmed That Loss of Glucagon-like Peptide 1 Receptor Signaling Is Not a Major Contributor to Type 2 Diabetes
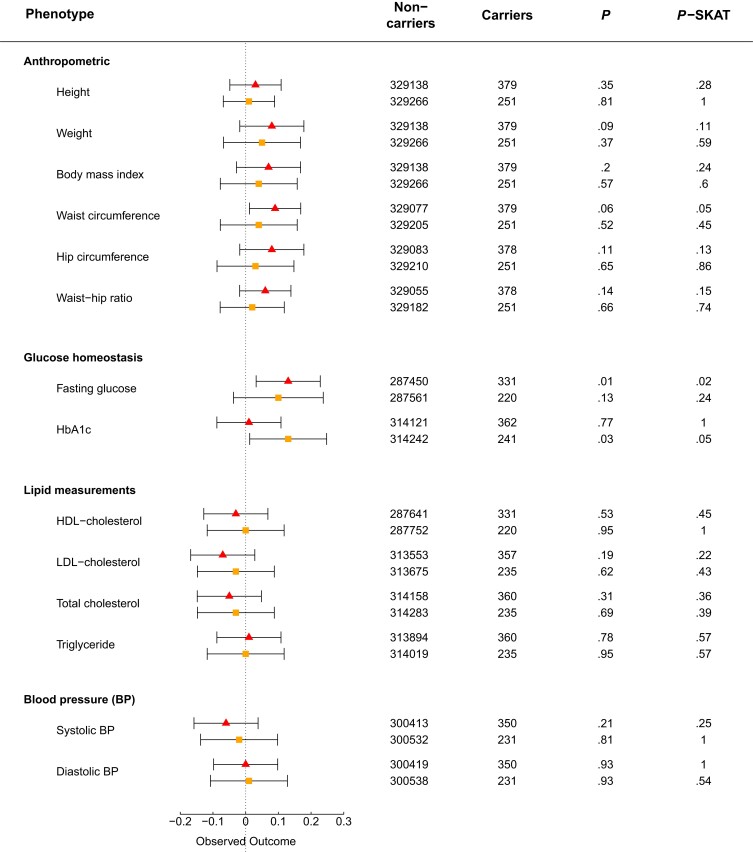
We examined data from 330 566 unrelated individuals of White origin with available exome-sequencing data from the UK Biobank cohort to improve statistical power for type 2 diabetes risk analysis compared to that in Danish cohorts. In total, 511 variants were identified in GLP1R, including 222 nonsynonymous variants (Fig. 1B and Supplementary Table S4 (26)). Of these, 26 variants overlapped with the 36 Danish nonsynonymous GLP1R variants in the present study, of which 5 were LoS variants (Fig. 1C and Supplementary Table S2 (26)). Among individuals with type 2 diabetes, 20 carried LoS variants and 15 761 were noncarriers. Among controls, there were 360 LoS variant carriers and 312 636 noncarriers. We observed no association between being a heterozygous carrier of an LoS variant and change in type 2 diabetes risk (OR = 1.12; CI, 0.70-1.78; P-logistic regression = .63, P-SKAT = .63; ORBMI-adj = 1.07; CIBMI-adj, 0.66-1.72; P-logistic regressionBMI-adj = .79, and P-SKATBMI-adj = 1.00). Among the cardiometabolic phenotypes, we observed a minor increase of the FPG level and waist circumference in only LoS variant carriers (Fig. 6).
Figure 6.
Quantitative trait analyses of loss-of-signaling (LoS) and predicted loss-of-function (pLoF) GLP1R variants in the UK Biobank. Noncarriers = the number of individuals with phenotype information who do not carry any of the variants; Carriers = the number of individuals with the phenotype who carry a loss-of-function variant. The triangle represents the LoS variant analyses. The square represents the pLoF variant analyses. BP, blood pressure; HbA1c, glycated hemoglobin A1c; HDL, high-density lipoprotein; LDL, low-density lipoprotein; LoS, loss of signaling; P, P value of the general linear regression model; pLoF, predicted loss-of-function; P-SKAT, P value of SKAT (LoS variants) or SKATO (pLoF variants) tests.
Since only 5 Danish LoS variants were present in the UK Biobank, of which only 2 were also pLoF variants, we evaluated the 222 nonsynonymous UK Biobank variants based on the aggregated scores of 17 variant effect predictor algorithms, classifying 60 variants as pLoF (Supplementary Fig. S4 (26)). We observed fewer pLoF carriers (∼ 0.08%) in the UK Biobank compared to LoS carriers in the UK Biobank (0.10%) and the Danish population (∼ 0.30%), possibly due to the higher MAF of the LoS variants compared to the pLoF variants. Among individuals with type 2 diabetes, 14 were carriers of the 60 pLoF variants, and 15 767 were noncarriers, compared to 235 carriers and 312 761 noncarriers in the control group. We observed no difference in type 2 diabetes prevalence between GLP1R pLoF variant carriers and noncarriers (OR = 1.25; CI, 0.72-2.18; P-logistic regression = .43, and P-SKATO = .56; ORBMI-adj = 1.07; CIBMI-adj, 0.59-1.92; P-logistic regressionBMI-adj = .82, and P-SKATOBMI-adj = .41). Similarly, we found no statistically significant associations between carrying pLoF variants and cardiometabolic traits, despite a marginal association with HbA1c (see Fig. 6).
Discussion
We mapped coding GLP1R variations in the Danish population to select and characterize their functional effect and possible effect on cardiometabolic phenotypes in humans. We identified 36 coding variants, among which 10 showed impaired cAMP signaling (LoS). We further observed an indication of selection against missense and pLoF variants of the GLP1R, as we identified fewer missense and pLoF variants than expected. These data suggest the existence of an evolutionary selection against damaging variants in the GLP1R, potentially explaining the superiority of heterozygous GLP1R variant carriers, as observed for the 10 LoS variants. The 10 GLP1R LoS variants include E34*, S79L, W87R, H99Qfs*83, S155F, F169Sfs*13, R310W, T353M, R421W, and A449V. Surprisingly, combined analyses of these 10 LoS variants revealed no apparent effect on type 2 diabetes or additional cardiometabolic phenotypes, in contrast to LoS variants of the glucose-dependent insulinotropic polypeptide (GIP) receptor and the glucagon receptor (39-41). We evaluated all LoS variants and pLoF variants as pooled groups of variants but both methods have limitations. Interestingly, evaluating the LoS variants predicted as pLoF variants revealed a minor increased FPG level, which might be driven by BMI rather than the variants, and a marginal decrease of LDL cholesterol, glucose-stimulated 30-minute insulin level, and insulin secretion in the Danish Inter99 cohort. However, none of the other indices for insulin secretion were affected. The study of the LoS variants and pLoF variants in the UK Biobank cohort validated the missing associations with cardiometabolic phenotypes. We did observe a minor increase of FPG levels and waist circumference, and HbA1c in LoS variant carriers and pLoF variant carriers, respectively, but if we adjusted for multiple testing, we would lose these associations.
Three LoS variants (S79L, R421W, and A449V) had altered cAMP signaling despite maintained binding and β-arrestin 2 recruitment, suggesting selective impairment of Gαs coupling. A naturally occurring GLP1R variant, R421C, previously showed no effect on cAMP signaling (42), and we observed altered cAMP signaling only when shifting to a tryptophan, but not to glutamine. Thus, a large tryptophan in helix 8 may sterically block the ability of the receptor to enter active states capable of Gαs binding. Variants at positions S79 and A449 were not previously identified. S79L positions in the receptor's N-terminus in the first β-sheet, potentially affecting the interaction with receptor activity modifying proteins, hence the G protein–coupling (43). The fact that A449V is a LoS variant could be due to an alteration of the latching-on with β-arrestins, hence an increased desensitization compared to WT. Moreover, 2 LoS variants (W87R and R310W) had altered cAMP signaling and β-arrestin 2 recruitment, but maintained binding, suggesting the presence of the variant GLP-1R at the cell surface with overall impaired activity. Previously, shifting to alanine at position 87 showed no altered GLP-1 binding or cAMP signaling (44), indicating that adding a positive charge by the shift to arginine, as in the present study, may explain the functional difference. R310W, in the top of transmembrane helix 5, was previously shown to reduce efficacy (45), supporting our finding of maintained GLP-1 affinity but decreased signaling. The last 5 LoS variants (E34*, H99Qfs*83, S155F, F169Sfs*13, and T353M) showed no GLP-1 binding, cAMP formation, or β-arrestin recruitment, as expected for the 3 variants with stop codon introduction. A shift to alanine at position 155 affected cAMP production and GLP-1 affinity (46), consistent with S155 being a conserved residue throughout the class B GPCRs. An alanine mutation at position 353 also resulted in altered cAMP production (46), supporting the importance of this position.
Interestingly, 4 of the 36 GLP1R variants (P7L, R131Q, G168S, and A316T) were associated with glycemic phenotypes in various populations (12, 14, 47, 48), but showed WT-like GLP-1R signaling in the present study. For instance, P7L and R131Q were associated with risk of type 2 diabetes (12, 14), and G168S was nominally associated with decreased insulin secretion in healthy individuals (12) and showed less HbA1c reduction after GLP-1R agonist therapy in type 2 diabetes patients (47). G168S may therefore be important for the action of GLP-1R agonists. In contrast, the low-frequency A316T variant showed decreased β-arrestin recruitment (18% of WT) but WT-like signaling and binding properties (49), but was associated with an improved glycemic profile, including lower risk of type 2 diabetes and coronary heart disease (47, 48, 50). It is particularly interesting that functioning coding GLP-1R variants influence metabolic traits, whereas the LoS variants had no or minor effects. However, this may be explained by our assessments of receptor activity in established laboratory cell lines (COS-7 and HEK-293 cells rather than β cells) and by altered receptor properties that were not determined in our study such as calcium signaling, agonist residence time, and receptor recycling properties (51).
In this study, none of the 10 LoS variants were statistically significantly associated with type 2 diabetes or cardiometabolic phenotypes in the Danish cohorts, despite rather considerable effect sizes. With these insignificant effect sizes and the obvious broad CIs for each evaluated phenotype, it implies that we were statistically underpowered, and we should therefore interpret the results with caution. We sought to circumvent this by including an analysis of the Danish LoS variants involving the large UK Biobank cohort. However, only 5 Danish LoS variants were present, and we therefore also used in silico prediction of variant severity. First, we compared the in vitro and in silico functionality in the Danish GLP1R variants. The in silico prediction did not classify S79L, S155W, R421W, or A449V as pLoF variants despite the lost cAMP signaling. Interestingly, for A449V all 17 prediction algorithms classified the variant as functional. However, A449V is positioned in the C-terminal tail of the protein, which may not be considered as crucial by the prediction tools we used. For S79L, S155W, and R421W, a subset of the 17 algorithms predicted LoF, suggesting that we were too stringent with the prediction masks, hence loose damaging variants. At the same time, F321S and F66L were predicted to be pLoF but had normal cAMP signaling, suggesting that relaxing the criteria may lead to identification of false pLoF variants. Thus, we proceeded with the prediction of pLoF in the UK Biobank cohort and confirmed the missing association with type 2 diabetes and cardiometabolic phenotypes. However, we did observe a nominally higher HbA1c level in LoS variant carriers in the Danish cohorts, with a weak signal for the pLoF variants in the UK Biobank. Thus, lost GLP-1R signaling may have a small effect on HbA1c level but further studies are needed to confirm that this is true. Furthermore, the LoS variants predicted as pLoF were marginally associated with a decreased glucose-stimulated insulin secretion in the Danish cohorts, but these observations could not be replicated in the UK Biobank. Given the inadequate number of carriers, hence limited statistical power, these results must be interpreted with care. Thus, more in-depth studies on the β-cell function and insulin sensitivity are required to pursue whether lost GLP-1R signaling affects these parts of the glucose regulation.
Since all the LoS and pLoF variants (in both the Danish cohorts and the UK Biobank) were presented only in heterozygous form, the lack of effect could be caused by allelic compensation in which the physiological phenotype is rescued by the WT, or by other compensatory mechanisms. This would conceal a dysfunctional GLP-1 system, such as the incretin effects of the GIP system. Hence, the GIP system may substitute for the disrupted GLP-1 system, as observed in mice (51). Furthermore, our results may support the recent hypothesis that the GLP-1 and GLP-1R system modifies central parts of human life history, such as energy maintenance and defense against pathogens, by which, that is, hyperglycemia and expansion of fat mass may lead to inappropriate activation of the immune system, conjointly increasing the risk of cardiovascular complications (52). Beyond the glucose-lowering and weight-loss effects, GLP-1R agonism has shown promising cardiovascular benefits in patients with metabolic dysfunction (53, 54). Thereby, GLP-1R agonist may rebalance human life history by acting on body weight and inflammation. Altogether, the beneficial effects on the cardiometabolic system by activation of the GLP-1R system may signify selection against fatal variants in GLP1R.
In conclusion, our results suggest that being a heterozygous carrier of LoS variants has no or minor implication for the here examined phenotypes related to cardiometabolism, although this may not be true for homozygous carriers. Only 4 Danish GLP1R variants were homozygous and all with WT-like GLP-1R function, suggesting an evolutionary selection against homozygous LoS variants, potentially due to fatal physiological implications. The GLP-1 system therefore may have an intolerance to damaging variants, supporting its importance in human physiology.
Acknowledgments
We deeply thank Maibritt Sigvardt Baggesen and Søren Petersen for their help with the in vitro studies.
Abbreviations
- BIGTT
β-cell function insulin sensitivity glucose tolerance test
- BMI
body mass index
- cAMP
cyclic adenosine monophosphate
- DD2
Danish Centre for Strategic Research in Type 2 Diabetes
- EC50
determination of potency
- Emax
determination of efficacy
- FPG
fasting plasma glucose
- GIP
glucose-dependent insulinotropic polypeptide
- GLP-1
glucagon-like peptide 1
- GLP-1R
glucagon-like peptide 1 receptor
- GPCR
G protein–coupled receptor
- GRK
G protein–coupled receptor kinase
- HbA1c
glycated hemoglobin A1c
- HBS
HEPES-buffered saline
- HC
high confidence
- HDL
high-density lipoprotein
- IC50
determination of affinity
- LDE
least detectable effect
- LDL
low-density lipoprotein
- LoF
loss-of-function
- LoS
loss-of-signaling
- MAF
minor allele frequency
- NGS
next-generation sequencing
- O/E
observed over expected ratio
- PBS
phosphate-buffered saline
- pLoF
predicted loss-of-function
- WT
wild-type
Contributor Information
Josefine U Melchiorsen, Department of Biomedical Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Kimmie V Sørensen, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Jette Bork-Jensen, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Hüsün S Kizilkaya, Department of Biomedical Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Lærke S Gasbjerg, Department of Biomedical Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Alexander S Hauser, Department of Drug Design and Pharmacology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2100, Denmark.
Jørgen Rungby, Department of Clinical Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Henrik T Sørensen, Department of Clinical Epidemiology, Aarhus University, Aarhus 8800, Denmark; Department of Epidemiology, Boston University, Boston, MA 02118, USA.
Allan Vaag, Steno Diabetes Center Copenhagen, Herlev Hospital, Herlev 2730, Denmark.
Jens S Nielsen, Steno Diabetes Center Odense, Odense University Hospital, Odense 5000, Denmark.
Oluf Pedersen, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark; Center for Clinical Metabolic Research, Gentofte Hospital, Hellerup 2900, Denmark.
Allan Linneberg, Department of Clinical Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark; Center for Clinical Research and Prevention, Copenhagen University Hospital—Bispebjerg and Frederiksberg, Frederiksberg 2000, Denmark.
Bolette Hartmann, Department of Biomedical Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Anette P Gjesing, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Jens J Holst, Department of Biomedical Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark; Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Torben Hansen, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Mette M Rosenkilde, Department of Biomedical Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Niels Grarup, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Funding
This work was supported by the Novo Nordisk Foundation (NNF 21OC0070347 to M.M.R.), the EFSD/Lilly European Diabetes Research Programme (to M.M.R.), and the Danish Diabetes Academy (NNF17SA0031406 to H.S.K.). The Novo Nordisk Foundation Center for Basic Metabolic Research (https://cbmr.ku.dk/) at the University of Copenhagen is an independent research facility that is partly funded by an unrestricted grant (NNF18CC0034900) donated by the Novo Nordisk Foundation. It was used to support K.V.S., J.B.J., O.P., B.H., A.P.G., J.J.H., T.H., and N.G.
Disclosures
The authors have nothing to declare. The authors have no conflicts of interest that may bias the current work or conclusion.
Data Availability
Some or all data sets generated during and/or analyzed during the present study are not publicly available but are available from the corresponding authors on reasonable request; the DD2 data set may be obtained by a third party after completing and submitting an application (https://dd2.dk/forskning/ansoeg-om-data) to Research Manager Kurt Højlund at Kurt.Hoejlund@rsyd.dk; and UK Biobank data sets are available on request by submitting the application form available at https://www.ukbiobank.ac.uk/enable-your-research.
References
- 1. Wilding JPH, Batterham RL, Calanna S, et al. . Once-weekly semaglutide in adults with overweight or obesity. N Engl J Med. 2021;384(11):989‐1002. [DOI] [PubMed] [Google Scholar]
- 2. Holst JJ, Deacon CF, Vilsbøll T, Krarup T, Madsbad S. Glucagon-like peptide-1, glucose homeostasis and diabetes. Trends Mol Med. 2008;14(4):161‐168. [DOI] [PubMed] [Google Scholar]
- 3. Schwartz TW, Frimurer TM. Full monty of family B GPCRs. Nat Chem Biol. 2017;13(8):819‐821. [DOI] [PubMed] [Google Scholar]
- 4. Doyle ME, Egan JM. Mechanisms of action of glucagon-like peptide 1 in the pancreas. Pharmacol Ther. 2007;113(3):546‐593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hager MV, Clydesdale L, Gellman SH, Sexton PM, Wootten D. Characterization of signal bias at the GLP-1 receptor induced by backbone modification of GLP-1. Biochem Pharmacol. 2017;136:99‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sonoda N, Imamura T, Yoshizaki T, Babendure JL, Lu JC, Olefsky JM. β-Arrestin-1 mediates glucagon-like peptide-1 signaling to insulin secretion in cultured pancreatic β cells. Proc Natl Acad Sci U S A. 2008;105(18):6614‐6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Girada SB, Kuna RS, Bele S, et al. . Gαs regulates glucagon-like peptide 1 receptor-mediated cyclic AMP generation at Rab5 endosomal compartment. Mol Metab. 2017;6(10):1173‐1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van der Velden WJC, Smit FX, Christiansen CB, et al. . GLP-1 Val8: a biased GLP-1R agonist with altered binding kinetics and impaired release of pancreatic hormones in rats. ACS Pharmacol Transl Sci. 2021;4(1):296‐313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mahajan A, Sim X, Ng HJ, et al. . Identification and functional characterization of G6PC2 coding variants influencing glycemic traits define an effector transcript at the G6PC2-ABCB11 locus. PLoS Genet. 2015;11(1):e1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wessel J, Chu AY, Willems SM, et al. . Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat Commun. 2015;6(1):5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scott RA, Freitag DF, Li L, et al. ; CVD50 Consortium; GERAD_EC Consortium; Neurology Working Group of the Cohorts for Heart; . et al. A genomic approach to therapeutic target validation identifies a glucose-lowering GLP1R variant protective for coronary heart disease. Sci Transl Med. 2016;8(341):341ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kwak SH, Chae J, Lee S, et al. . Nonsynonymous variants in PAX4 and GLP1R are associated with type 2 diabetes in an East Asian population. Diabetes. 2018;67(9):1892‐1902. [DOI] [PubMed] [Google Scholar]
- 13. Spracklen CN, Horikoshi M, Kim YJ, et al. . Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature. 2020;582(7811):240‐245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vujkovic M, Keaton JM, Lynch JA, et al. ; HPAP Consortium; Regeneron Genetics Center; VA Million Veteran Program . Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet. 2020;52(7):680‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Graaf C, Donnelly D, Wootten D, et al. . Glucagon-like peptide-1 and its class B G protein–coupled receptors: a long march to therapeutic successes. Pharmacol Rev. 2016;68(4):954‐1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glümer C, Jørgensen T, Borch-Johnsen K. Prevalences of diabetes and impaired glucose regulation in a Danish population. Diabetes Care. 2003;26(8):2335‐2340. [DOI] [PubMed] [Google Scholar]
- 17. Jørgensen T, Borch-Johnsen K, Thomsen TF, Ibsen H, Glümer C, Pisinger C. A randomized non-pharmacological intervention study for prevention of ischaemic heart disease: baseline results Inter99. Eur J Cardiovasc Prev Rehabil. 2003;10(5):377‐386. [DOI] [PubMed] [Google Scholar]
- 18. Nielsen JS, Thomsen RW, Steffensen C, Christiansen JS. The Danish Centre for Strategic Research in Type 2 Diabetes (DD2) study: implementation of a nationwide patient enrollment system. Clin Epidemiol. 2012;4(Suppl 1):27‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bycroft C, Freeman C, Petkova D, et al. . The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203‐209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eastwood SV, Mathur R, Atkinson M, et al. . Algorithms for the capture and adjudication of prevalent and incident diabetes in UK Biobank. PLoS One. 2016;11(9):e0162388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao R, Liu Y, Gjesing AP, et al. . Evaluation of a target region capture sequencing platform using monogenic diabetes as a study-model. BMC Genet. 2014;15(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McKenna A, Hanna M, Banks E, et al. . The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297‐1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hansen T, Drivsholm T, Urhammer SA, et al. . The BIGTT test: a novel test for simultaneous measurement of pancreatic beta-cell function, insulin sensitivity, and glucose tolerance. Diabetes Care. 2007;30(2):257‐262. [DOI] [PubMed] [Google Scholar]
- 24. Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22(9):1462‐1470. [DOI] [PubMed] [Google Scholar]
- 25. Hail Team . Hail 0.2.69. https://github.com/hail-is/hail
- 26. Melchiorsen JU, Sørensen KV, Bork-Jensen J, et al. . Supplementary material for “Rare heterozygous loss-of-function variants in the human GLP-1 receptor are not associated with cardiometabolic phenotypes.” Published 2023. Deposited May 30, 2023. https://github.com/kimmievsorensen/JCEM_Melchiorsen_et_al.git [DOI] [PMC free article] [PubMed]
- 27. McLaren W, Gil L, Hunt SE, et al. . The Ensembl Variant Effect Predictor. Genome Biol. 2016;17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32(8):894‐899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu X, Li C, Mou C, Dong Y, Tu Y. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020;12(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dong C, Wei P, Jian X, et al. . Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24(8):2125‐2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Flannick J, Mercader JM, Fuchsberger C, et al. ; DiscovEHR Collaboration; CHARGE; LuCamp ; et al. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature. 2019;570(7759):71‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Steen A, Thiele S, Guo D, Hansen LS, Frimurer TM, Rosenkilde MM. Biased and constitutive signaling in the CC-chemokine receptor CCR5 by manipulating the interface between transmembrane helices 6 and 7. J Biol Chem. 2013;288(18):12511‐12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hassing HA, Fares S, Larsen O, et al. . Biased signaling of lipids and allosteric actions of synthetic molecules for GPR119. Biochem Pharmacol. 2016;119:66‐75. [DOI] [PubMed] [Google Scholar]
- 34. Donthamsetti P, Quejada JR, Javitch JA, Gurevich VV, Lambert NA. Using bioluminescence resonance energy transfer (BRET) to characterize agonist-induced arrestin recruitment to modified and unmodified G protein-coupled receptors. Curr Protoc Pharmacol. 2015;70(1):2.14.1‐2.14.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seunggeun L, Zhangchen Z, with contributions from LM and MW. SKAT: SNP-set (sequence) Kernel association test. R package version 2.0.1. https://CRAN.R-project.org/package=SKAT
- 36. Lee S, Emond MJ, Bamshad MJ, et al. ; NHLBI GO Exome Sequencing Project—ESP Lung Project Team. . Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91(2):224‐237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2022. https://www.R-project.org [Google Scholar]
- 38. Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434‐443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kizilkaya HS, Sørensen KV, Kibsgaard CJ, et al. . Loss of function glucose-dependent insulinotropic polypeptide receptor variants are associated with alterations in BMI, bone strength and cardiovascular outcomes. Front Cell Dev Biol. 2021;9:749607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Akbari P, Gilani A, Sosina O, et al. ; Regeneron Genetics Center; DiscovEHR Collaboration . Sequencing of 640,000 exomes identifies GPR75 variants associated with protection from obesity. Science. 2021;373(6550):eabf8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van der Velden WJC, Lindquist P, Madsen JS, et al. . Molecular and in vivo phenotyping of missense variants of the human glucagon receptor. J Biol Chem. 2022;298(2):101413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fortin JP, Schroeder JC, Zhu Y, Beinborn M, Kopin AS. Pharmacological characterization of human incretin receptor missense variants. J Pharmacol Exp Ther. 2010;332(1):274‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Routledge SJ, Ladds G, Poyner DR. The effects of RAMPs upon cell signalling. Mol Cell Endocrinol. 2017;449:12‐20. [DOI] [PubMed] [Google Scholar]
- 44. Wilmen A, Van EB, Göke B, Göke R. Five out of six tryptophan residues in the N-terminal extracellular domain of the rat GLP-1 receptor are essential for its ability to bind GLP-1. Peptides (NY). 1997;18(2):301‐305. [DOI] [PubMed] [Google Scholar]
- 45. Dods RL, Donnelly D. The peptide agonist-binding site of the glucagon-like peptide-1 (GLP-1) receptor based on site-directed mutagenesis and knowledge-based modelling. Biosci Rep. 2015;36(1):e00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wootten D, Simms J, Miller LJ, Christopoulos A, Sexton PM. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc Natl Acad Sci U S A. 2013;110(13):5211‐5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dawed AY, Mari A, Brown A, et al. ; DIRECT Consortium . Pharmacogenomics of GLP-1 receptor agonists: a genome-wide analysis of observational data and large randomised controlled trials. Lancet Diabetes Endocrinol. 2023;11(1):33‐41. [DOI] [PubMed] [Google Scholar]
- 48. Sathananthan A, Man CD, Micheletto F, et al. . Common genetic variation in GLP1R and insulin secretion in response to exogenous GLP-1 in nondiabetic subjects. Diabetes Care. 2010;33(9):2074‐2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koole C, Wootten D, Simms J, et al. . Polymorphism and ligand dependent changes in human glucagon-like peptide-1 receptor (GLP-1R) function: allosteric rescue of loss of function mutation. Mol Pharmacol. 2011;80(3):486‐497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van der Velden WJC, Heitman LH, Rosenkilde MM. Perspective: implications of ligand–receptor binding kinetics for therapeutic targeting of G protein-coupled receptors. ACS Pharmacol Transl Sci. 2020;3(2):179‐189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pederson RA, Satkunarajah M, McIntosh CH, et al. . Enhanced glucose-dependent insulinotropic polypeptide secretion and insulinotropic action in glucagon-like peptide 1 receptor –/– mice. Diabetes. 1998;47(7):1046‐1052. [DOI] [PubMed] [Google Scholar]
- 52. Avogaro A, de Kreutzenberg SV, Morieri ML, Fadini GP, Del Prato S. Glucose-lowering drugs with cardiovascular benefits as modifiers of critical elements of the human life history. Lancet Diabetes Endocrinol. 2022;10(12):882‐889. [DOI] [PubMed] [Google Scholar]
- 53. Palmer SC, Tendal B, Mustafa RA, et al. . Sodium-glucose cotransporter protein-2 (SGLT-2) inhibitors and glucagon-like peptide-1 (GLP-1) receptor agonists for type 2 diabetes: systematic review and network meta-analysis of randomised controlled trials. BMJ. 2021;372:m4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sattar N, Lee MMY, Kristensen SL, et al. . Cardiovascular, mortality, and kidney outcomes with GLP-1 receptor agonists in patients with type 2 diabetes: a systematic review and meta-analysis of randomised trials. Lancet Diabetes Endocrinol. 2021;9(10):653‐662. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data sets generated during and/or analyzed during the present study are not publicly available but are available from the corresponding authors on reasonable request; the DD2 data set may be obtained by a third party after completing and submitting an application (https://dd2.dk/forskning/ansoeg-om-data) to Research Manager Kurt Højlund at Kurt.Hoejlund@rsyd.dk; and UK Biobank data sets are available on request by submitting the application form available at https://www.ukbiobank.ac.uk/enable-your-research.