Abstract
Purpose:
This study aimed to assess the amount and types of clinical genetic testing denied by insurance and the rate of diagnostic and candidate genetic findings identified through research in patients who faced insurance denials.
Methods:
Analysis consisted of review of insurance denials in 801 patients enrolled in a pediatric genomic research repository with either no previous genetic testing or previous negative genetic testing result identified through cross-referencing with insurance prior-authorizations in patient medical records. Patients and denials were also categorized by type of insurance coverage. Diagnostic findings and candidate genetic findings in these groups were determined through review of our internal variant database and patient charts.
Results:
Of the 801 patients analyzed, 147 had insurance prior-authorization denials on record (18.3%). Exome sequencing and microarray were the most frequently denied genetic tests. Private insurance was significantly more likely to deny testing than public insurance (odds ratio = 2.03 [95% CI = 1.38–2.99] P = .0003). Of the 147 patients with insurance denials, 53.7% had at least 1 diagnostic or candidate finding and 10.9% specifically had a clinically diagnostic finding. Fifty percent of patients with clinically diagnostic results had immediate medical management changes (5.4% of all patients experiencing denials).
Conclusion:
Many patients face a major barrier to genetic testing in the form of lack of insurance coverage. A number of these patients have clinically diagnostic findings with medical management implications that would not have been identified without access to research testing. These findings support re-evaluation of insurance carriers’ coverage policies.
Keywords: Access to testing, Denials, Genomic medicine, Insurance, Research testing
Introduction
Comprehensive genetic testing in the form of microarray, multigene panel testing, exome sequencing (ES), and, in some cases, genome sequencing (GS) has been available clinically for a decade or more, and the diagnostic utility of this testing, particularly in pediatric patient populations, has been well established.1–8 In pediatric cases with a suspected genetic condition and no prior molecular testing, microarray has a diagnostic rate of approximately 10%, whereas ES and GS are reported to have higher rates of up to 36% and 41%, respectively.9
Despite well-established diagnostic rates for many molecular findings in children with intellectual disability (ID), developmental delays, and/or congenital anomalies (CA), insurance denials continue to be a significant barrier to accessing genetic testing. For the majority of genetic conditions, there is no standard criteria used to determine the coverage eligibility by the insurance payers and it is instead left up to the individual discretion of each payer.10 Many insurance carriers, both private and public, follow National Comprehensive Cancer Center guidelines for hereditary cancer testing (particularly hereditary breast and ovarian cancer and Lynch syndrome)11–13; yet, despite recommendations from the American College of Medical Genetics and Genomics (ACMG) regarding ES in pediatric populations with ID, developmental delay, and CAs,14 there is not a similarly universal approach to insurance coverage in this population, with some insurance companies using this criteria and others not referencing it in their coverage policies.15–17 This lack of firm, universal criteria for testing can result in inconsistent insurance determinations across identical clinical presentations, contributing to inequitable access to genetic testing.18 Other policies dictate that a microarray must be completed and resulted out as non-diagnostic before ES will even be reviewed for approval. The testing strategy for patients with suspected genetic disorders does often entail microarray with reflex to ES; however, microarray is not always the most clinically appropriate first-line testing for patients in whom a genetic disorder is suspected.14,19
Direct cost of genetic testing is a major issue for patients after facing insurance denials. On average, the cost of clinical genetic testing ranges from a few hundred dollars for single variant Sanger sequencing to several thousand dollars for ES and GS.20,21 This is a prohibitively high out-of-pocket cost but similar in price to the cost of other services routinely covered by health insurance, such as blood panel tests for annual physicals, computed tomography scans, and magnetic resonance imaging.22–24
It is difficult to know how many patients who receive insurance denials for genetic testing have clinically actionable findings because they often will not receive the testing they need. As a result, there is little published literature about the diagnostic rate among insurance denials and subsequent effect on patient health care.25,26 In 2019, Reuter et al26 published a comprehensive review of genetic diagnostic rates among patients with insurance denials in the Undiagnosed Diseases Network (UDN). This study found a 35% diagnostic rate in their cohort. Although vitally important to the current genetic testing landscape, the UDN has a strict set of enrollment criteria, which includes requiring that all available clinical diagnostic workup be exhausted, a letter of recommendation to the study be provided from a health care provider, and a thorough review of the application be performed by an interdisciplinary team overseeing 7 UDN sites.26 This excludes a large subset of patients who may still need or qualify for genetic testing. In contrast, the Genomic Answers for Kids (GA4K) research study has much broader eligibility criteria in which any child with a known or suspected genetic condition can enroll with no requirement for previous genetic testing history or specific letters of recommendation.
Our objective with this research was to assess the rate of insurance denials in patients enrolled in GA4K and subsequently determine the rate of research-identified diagnostic and candidate genetic findings in the patients who experienced clinical insurance denials. We also sought to determine whether any significant difference in denial rate existed between the types of insurance payers: private, public (Kansas and Missouri Medicaid), and military. Our work adds to the existing literature on diagnostic rates among insurance denials by assessing this information in a cohort with broader selection criteria than existing studies and reporting on candidate findings in addition to diagnostic findings.
Materials and Methods
Cohort
Patients qualified for the GA4K study if they had a known or suspected genetic condition based on clinical presentation. Given that most such conditions manifest early in life, the majority of the GA4K cohort was pediatric. Based on these criteria, any patient who had undergone or was undergoing clinical genetic testing for any reason as well as those with confirmed diagnostic genetic testing was included in the study. In addition, a subset of patients not undergoing clinical testing but with a condition known to have possible genetic causes, such as autism, also qualified for GA4K. When available, parents and other relatives (both affected and unaffected) were also enrolled in GA4K to inform on variant segregation (de novo variants are known to represent a significant portion of diagnoses27). Patients are primarily recruited from Children’s Mercy Hospital outpatient clinics and inpatient floors, with some patients being recruited from outside Children’s hospitals. Enrolled patients underwent ES followed by GS, whole-genome genotyping, single-cell RNA analysis, and/or methylome analysis when indicated based on clinical presentation and previous results.28 Specifically, if a clinical diagnosis was found in the first-stage testing (ES) and no additional diagnosis was suspected, no additional testing was performed. All initial studies were conducted on germline blood and/or buccal DNA. The research testing described above was provided at no cost to the enrolled families. Families may enroll at any time during the clinical testing process, because research testing is separate from clinical testing.
We began our analysis by selecting 1333 successive patients from 1203 unrelated families enrolled in GA4K between 2011 and 2021. This included the 960 families reported on by Cohen et al28 and an additional 243 families analyzed after publication of the initial data. The cohort was then pared down to include only those patients with no clinical genetic testing (n = 118) or negative clinical genetic testing results (n = 683) before the study entry, thus excluding the patients who entered the GA4K study with a genetic diagnosis as well as patients who were undergoing clinical testing concurrent with study enrollment, resulting in a final subcohort of 801 patients for analysis.
Analysis
ES and GS analyses were completed through GA4K protocols as previously described.15 Human Phenotype Ontology terms annotated via PhenoTips pedigree software (Gene42 Inc) were used to assist in the initial prioritization of variants.29 After analysis, patients were divided into groups based on previous genetic testing history (no testing or negative testing result). Patient insurance prior-authorization information was available through patient chart review, whereas final determinations of insurance coverage after completion of testing were not available, because these are not documented in the patient chart and are in a separate financial clearance portal that our group did not have access to. Charts were also reviewed for any previous insurance denial at the time of prior-authorization. Prior-authorizations that were initially denied and then approved after appeal were included in this cohort and considered approvals for analysis. Patient insurance carriers were categorized as private, public, or military. Cases in which authorization was not required by insurance were considered approvals. Cases in which neither a denial nor an approval was clearly documented in prior-authorization, but testing was completed, were also considered approvals given that most families decline self-pay testing because of high out-of-pocket costs.
Patients found to have diagnostic findings via GA4K had their results clinically confirmed in the Children’s Mercy Clinical Laboratory Improvement Amendments–certified laboratory through the best applicable validated methods and clinically reported in real time for incorporation into clinical management. Results were determined to be clinically actionable if any medical management changes, including, but not limited to, preventative screening procedures, surgical intervention, avoidance of high-risk activities, and referral to other subspecialties to rule out other affected systems, had been or could be made based on the result.
An odds ratio was calculated to examine the significance of the relationship between insurance carrier type (private, public, military) and approval/denial identified during insurance prior-authorization.
Results
Demographics
Among the 801 patients, ages at the time of enrollment ranged from prenatal to 38 years (some patients were enrolled as adults but qualified if they had exhibited symptoms of a genetic condition since childhood). Breakdown of patients by sex revealed that 368 out of 801 patients (45.9%) were female and 433 (54.1%) were male. Our cohort consisted predominantly of individuals of White (non-Latino) background (n = 669, 83.5%), with a number of other racial backgrounds represented: native American/American-Indian (n = 2, 0.3%), Asian (n = 8, 0.9%), Black (n = 36, 4.5%), Hispanic/Latino (n = 31, 3.9%), multiracial (n = 45, 5.6%), native Hawaiian (n = 3, 0.4%), other (n = 3, 0.4%), and race unknown (n = 4, 0.5%).
Insurance denials
A total of 147 cases out of 801 (18.3%) had at least 1 previous insurance prior-authorization denial on file. Testing was separated into approvals, denials, and no testing ordered for private, public, and military insurance. In our cohort of 801 patients, 422 (52.7%) had private insurance coverage, 347 (43.3%) had public insurance coverage, 28 (3.5%) had military insurance coverage, and 4 (0.5%) had no coverage/self-paid for testing. In total, 81 patients had no genetic testing ordered before study entry, whereas the remaining 720 patients (374 private insurance, 323 public, 19 military, and 4 no coverage) had some clinical testing ordered by the clinical provider at some point before the study entry. A breakdown of denial rates in this group is shown in Figure 1. In the 720 patients with previous clinical genetic testing ordered, 147 (20.4%) had one or more insurance denials. Of the 147, 82 (55.8%) patients had at least 1 test denied but may have had other tests approved, whereas 65 out of 147 (44.2%) had all testing requests denied.
Figure 1. Percent of patients with insurance denials by payer type.
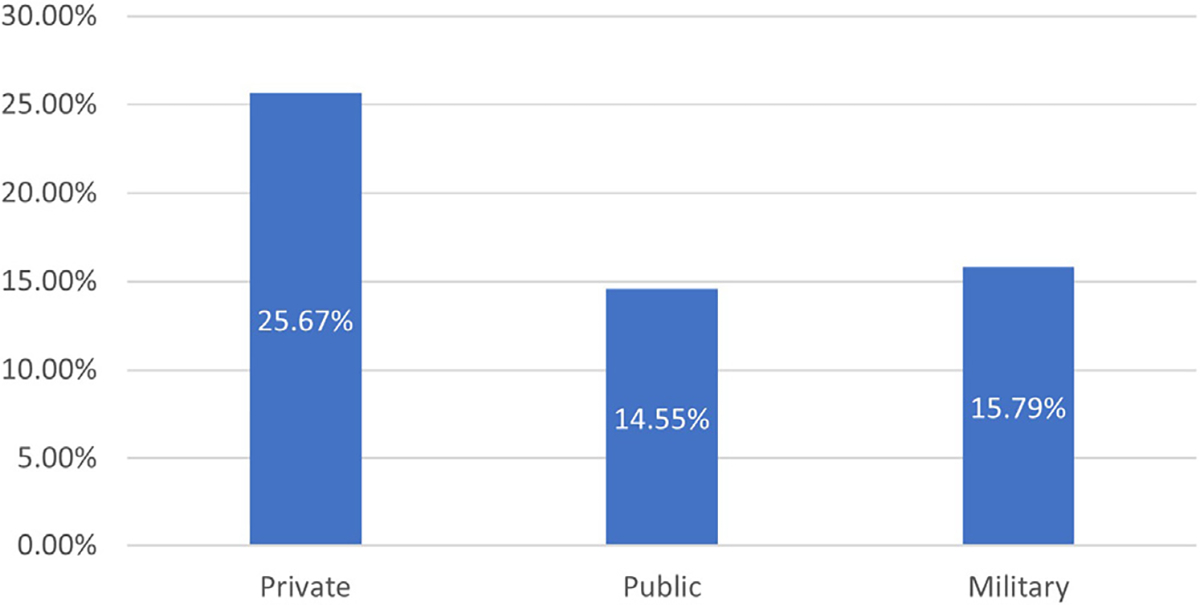
Breakdown by payer type showed that of 374 patients in our cohort who had any testing ordered, 25.7% (N = 96) of patients with private insurance, 14.6% (N = 47) of patients with public insurance, and 15.8% (N = 3) of patients with military insurance experienced denials.
The proportion of denials by type of testing is shown in Figure 2A, and the proportion of denials vs approvals for each test is shown in Figure 2B. Rates of denial were highest for ES (16.6%) and microarray (10.4%).
Figure 2. Breakdown of denials across testing types.
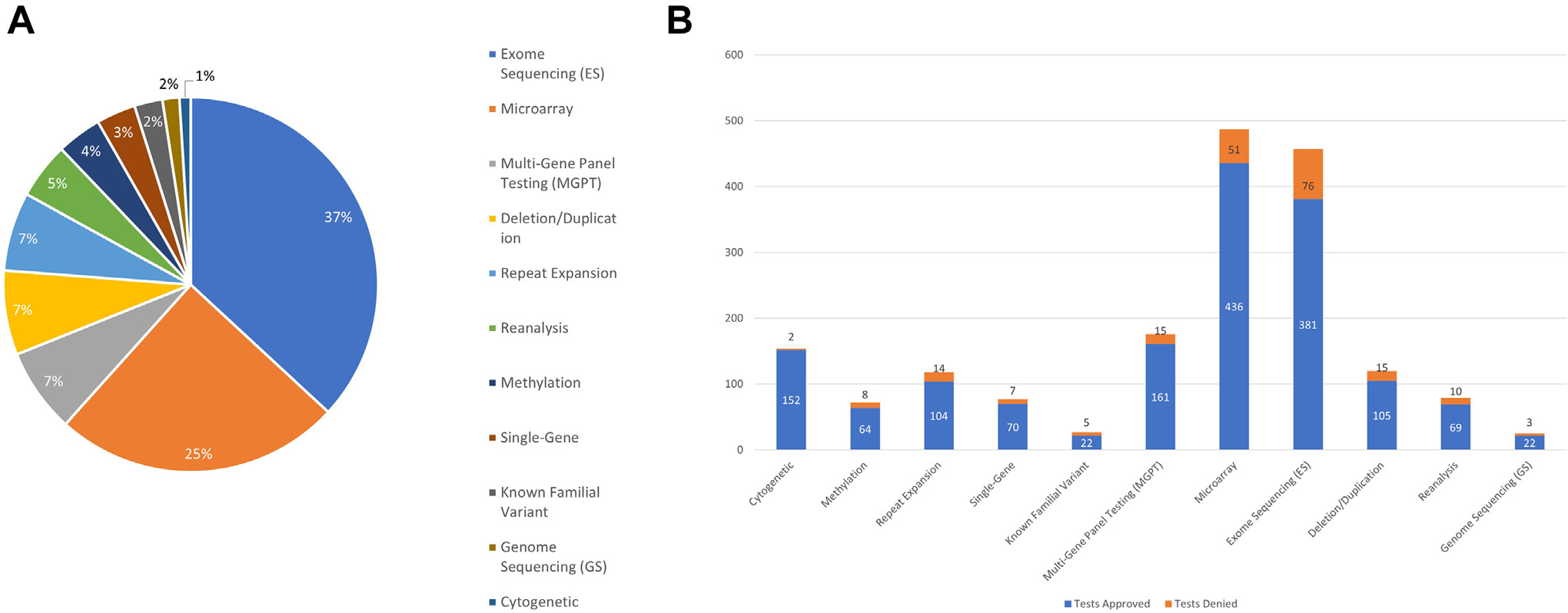
A. Denials by testing type. Exome sequencing (ES) and microarray together made up the majority of denied tests. B. Proportion of denials by testing type. The test with the highest denial rate to amount of tests ordered was known familial variant analysis with 5 tests denied of 27 cases ordered (18.5%). ES was the next highest with 76 tests denied out of 457 ordered (16.6%)
An odds ratio was calculated to determine if any significant difference existed between private and public insurance denials (patients with military insurance were too few to draw meaningful statistical differences from). This was done by removing cases in which no testing had ever been ordered, because these patients had neither approvals nor denials on record because of lack of testing orders. This resulted in a total of 96 patients with private insurance denials out of a total of 374 private insurance patients who had testing ordered and 47 patients with public insurance denials out of a totally of 323 public insurance patients who had testing ordered, with an odds ratio of 2.03 (95% CI = 1.38–2.99) P = .0003. A diagram of this patient breakdown can be viewed in Supplemental Figure 1.
Genetic findings in patients with insurance denials
Genetic findings identified after enrollment in the GA4K study were assessed among the 147 patients who experienced insurance denials. Findings were placed into 1 of the 6 categories: (1) pathogenic or likely pathogenic variant (according to ACMG/Association for Molecular Pathology criteria) in a gene with a well-described gene–disease relationship and fully explaining patient phenotype, (2) partial diagnosis: same criteria as diagnostic but not fully explaining the patient’s phenotype, indicating that a second diagnosis is suspected but was not identified, (3) gene of uncertain significance: strong candidate finding in a gene currently meeting limited criteria for a gene–disease relationship, (4) variant of uncertain significance: strong candidate finding not meeting pathogenic/likely pathogenic criteria in a gene with a well-described relationship to disease, (5) partial genotype: 1 pathogenic or likely pathogenic finding in a recessive gene strongly matching patient phenotype, or (6) negative: no candidate or diagnostic findings identified. The breakdown of these findings in patients receiving insurance denials is shown in Figure 3.
Figure 3.
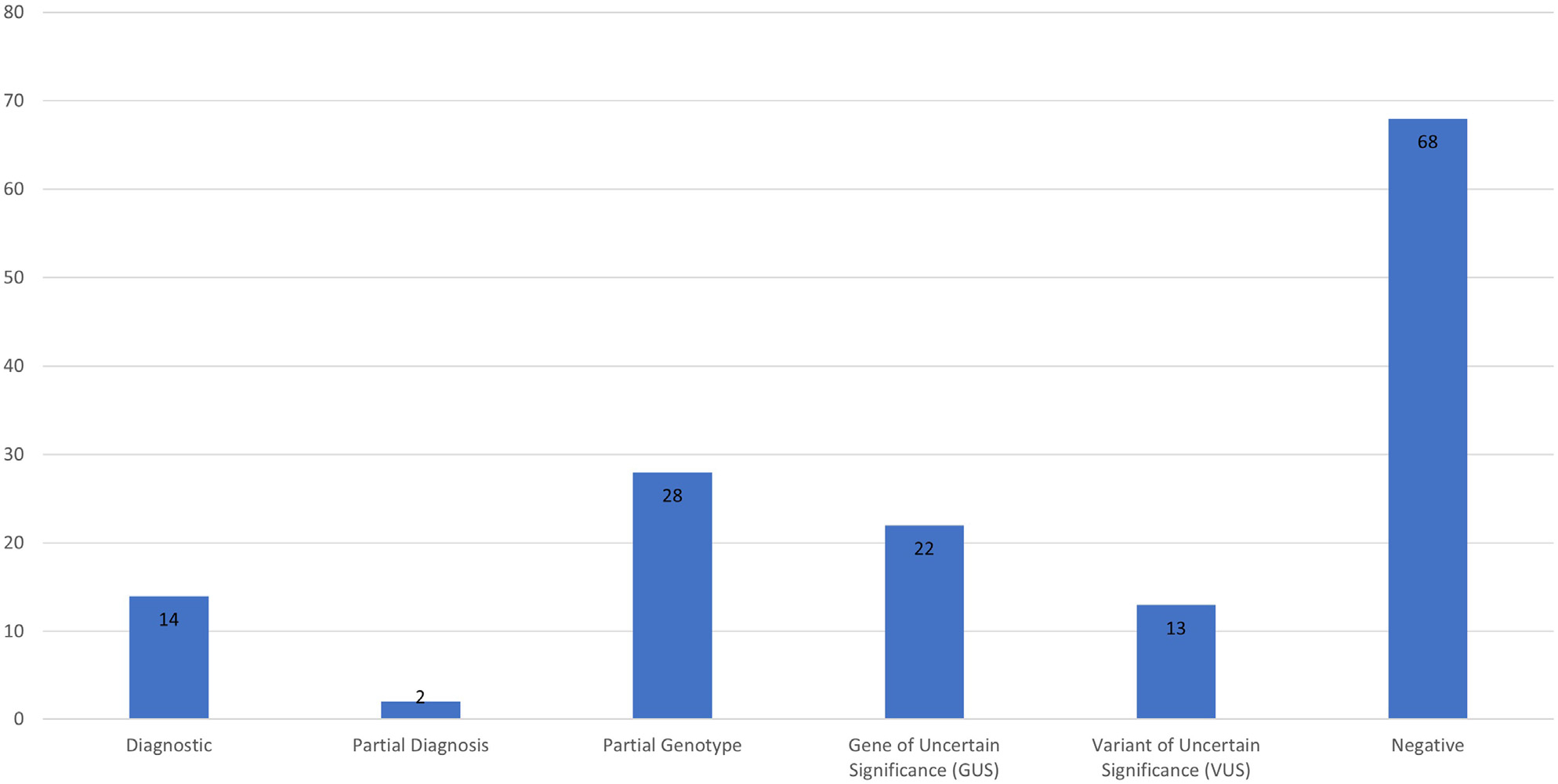
Genetic findings in cases with previous insurance denials.
Of these findings, some were identified through clinical testing ordered after enrollment in the study, whereas others were identified in the GA4K study itself. This is because some patients go on to have additional clinical testing after enrollment in GA4K, mostly through hospital financial assistance or philanthropy funding. These patients remained categorized as denials because of the fact that insurance did not cover the testing that was ultimately run.
Cross-referencing of patients with insurance denials to genetic findings in our analysis database identified 79 out of 147 (53.7%) patients with non-negative genetic testing results after enrollment in GA4K, 16 of which were pathogenic or likely pathogenic according to ACMG criteria, resulting in a diagnostic rate of 10.9% in this population. In 78 of the 79 cases, genetic findings were identified via ES, and in 45 of these 79 cases, (56.9%) ES was specifically one of the tests denied by insurance. A breakdown of the genetic variants and medical management changes among the 16 patients with diagnostic findings is shown in Table 1. Of these 16 cases, 8 (50%) had immediate medical management changes as a result of their diagnostic finding. This represents a management change in 5.4% (8/147) of the overall insurance denial subcohort.
Table 1.
Diagnostic findings in patients with previous insurance denials
| Patient | Gene | Isoform (GRCH38) | Variant | Genomic Reference Sequence (GRCH38) | Genomic Variant | Management Changes? | Describe | Tests Denied by Insurance | Method of Dx |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| 1 | ANKRD11 | NM_013275.5 | c.3931C>T p.(Arg1311Ter) | NC_000016.10 | g.89282611G>A | Y | Referral to additional subspecialties | ES | ES |
| 2 | ANKRD11 | NM_013275.5 | c.4384dup p.(Arg1462LysfsTer92) | NC_000016.10 | g.89282158dup | Y | Referral to additional subspecialties | Microarray, MGPT | ES |
| 3 | DDX3X | NM_001193416.3 | c.610A>C p.(Thr204Pro) | NC_000023.11 | g.41343282A>C | N | ES | ES | |
| 4 | FOXP1 | NM_032682.5 | c.1490G>C p.(Arg497Pro) | NC_000003.12 | g.70976981C>G | N | Microarray, ES | ES | |
| 5 | HCN1 | NM_021072.3 | c.1072T>C p.(Cys358Arg) | NC_000005.10 | g.45396650A>G | N | ES | ES | |
| 6 | KCNQ2 | NM_172107.2 | c.619C>T p.(Arg207Trp) | NC_000020.11 | g.63444730G>A | N | ES | ES | |
| 7 | NALCN | NM_052867.4 | c.4333A>C p.(Ile1445Leu) | NC_000013.11 | g.101068031T>G | N | Microarray, Repeat-expansion | ES | |
| 8 | NF1 | NM_000267.3 | c.1466A>G p.(Tyr489Cys) | NC_000017.11 | g.31214524A>G | Y | Additional screening | Single-gene | ES |
| 9 | NPC1 | NM_000271.4 | c.3570_3573dup p.(Ala1192Thrfs*67) c.1947+5G>C |
NC_000018.10 | g.23534464_23534467dup g.23544955C>G |
N | GS | ES | |
| 10 | PDE2A | NM_002599.4 | c.2434C>T p.(Gln812Ter) c.1962del p. (Tyr655ThrfsTer106) |
NC_000011.9 | g.72578932G>A g.72581445del |
Y | Access to additional rehabilitative services | Microarray | Karyotype, MGPT, ES |
| 11 | SCN4A | NM_000334.4 | c.3404G>A p.(Arg1135His) | NC_000017.11 | g.63947082C>T | Y | Medication changes, vitamin supplementation, access to additional rehabilitative services | ES | ES |
| 12 | SLC12A3 | NM_001126108.2 | c.482G>A p.(Trp161Ter) | NC_000016.10 | g.56868349G>A | Y | Access to mutation-positive contingent trials/therapies | Single-gene | ES |
| 13 | TRPV4 | NM_021625.5 | c.2389G>A p.(Glu797Lys) | NC_000012.12 | g.109784385C>T | Y | Changes to physical activity | MGPT, ES | ES |
| 14 | TSC2 | NM_000548.3 | c.2494C>T p.(His832Tyr) | NC_000016.10 | g.2071799del | Y | Additional screening | Del/dup (Exon array) | ES |
| 15 | TUBGCP4 | NM_014444.5 | c.1746G>T p.(Leu582=) | NC_000015.10 | g.43403697G>T | N | ES | ES | |
| 16 | N/A | N/A | arr[GRCh37] 7q34 (140433289_140486290)x1 | NC_000007.13 | g.140433289_140486290del | N | Repeat-expansion | Microarray | |
Del, deletion; dup, duplication; Dx, diagnosis; ES, exome sequencing; GS, genome sequencing; MGPT, multigene panel testing; N, no; N/A, not applicable; Y, yes.
Discussion
Our analysis supports the existing data that a significant number of patients are affected by insurance denials for genetic testing.26 This may have lasting clinical impact for these patients as well as financial, emotional, and reproductive impact for their families. Cases in which diagnostic genetic results do not lead to management changes, the families still benefit from the psychosocial impact of having an answer after a long diagnostic odyssey (average 5–6 years) and the ability to address concerns about recurrence risks.30,31 A genetic diagnosis can help alleviate long-standing concerns of guilt and blame associated with a child’s genetic condition and assist in connecting families with support groups; in fact, this is quoted by families as a major benefit of having a genetic diagnosis.30,31
There are a number of articles reporting on how insurance payers approach genetic testing and rates of approval in different genetic testing cohorts.25,32–35 Perhaps the most pertinent to our own work is a 2017 study by Smith et al25 assessing prior-authorization request outcomes in an outpatient setting in 2 children’s hospitals in Texas. This article provided a detailed look at the rates of approvals and denials by test category (cytogenetic, ES, mitochondrial, epigenetic, single/multigene panels) and documented the reasons for denials. Similar to our own results, Smith et al25 found that public payers were more likely to approve testing than private payers (85.5% vs 70.3%, P < .001). They suggested that one potential explanation for this is that public payers have more readily accessible and understandable policies for clinical providers to access. It remains uncertain whether this explanation applies to a significant portion of patients in our study and/or whether other factors may play a role. Private companies are for-profit, often publicly traded, and not government funded and therefore may have a higher incentive to be stricter in their coverage policies than public insurance payers. Our results also come with the caveat that authorization not required was considered an approval; therefore, it is possible that this skewed the approval rate for 1 or more payers.
Although Smith et al25 and others have reported on insurance payers and their relationship with clinical genetic testing,32–35 to our knowledge, the UDN is one of the only groups to publish similar data to ours on diagnostic rates, specifically, in patients who received insurance denials for clinical genetic testing.26 Their study accepted 686 patients from 2015 to 2017 of which 218 underwent ES. Of these 218 patients, 66 had documented clinical insurance denials. The UDN found a diagnosis in 23 out of 66 patients (35%), with medical management changes in 14 of the 23 diagnoses (61%).26 This rate differs from our own diagnostic rate, in part, because the eligibility criteria between the 2 programs differ considerably. The UDN has strict acceptance criteria, requiring letters of recommendation, exhaustive past genetic testing, and review of applications from a UDN interdisciplinary team.26 Conversely, our study accepts anyone with a known or suspected genetic condition regardless of past testing history. Patients are also able to self-refer to our study. Self-referrals are reviewed for medical eligibility, and all referrals are reviewed for research eligibility (child not in foster care, no legal issues preventing enrollment). This allows for a larger number of patients to be eligible but also results in a lower diagnostic rate because patients less likely to have a genetic diagnosis would be included. Therefore, our lower diagnostic rate and subsequent lower rate of medical management changes based on diagnoses were not unexpected.
Insurance companies have been slow to accept genetic testing as a standard of care, and genetic testing is still treated as experimental or investigational by some insurance companies. Although a handful of insurance companies have taken to hiring genetic counselors to assist in review of genetic testing requests, many do not have a specialized genetics professional reviewing these requests and whether this is by design or because of a lack of available genetics professionals is unclear. Microarray and ES were the most frequently denied tests in our cohort despite established guidelines from the ACMG recommending both tests as first-tier testing in patients with CAs and/or ID.14 This recommendation by the ACMG is backed by significant scientific evidence confirming the diagnostic benefit of both microarray and ES as well as the cost-savings benefit of more comprehensive tests.14 Considering that these ACMG recommendations were recently published (2021), it will, of course, take time for insurance companies to update their policies to reflect this; however, these recommendations should be considered when evaluating future genetic testing requests. Further work should focus on the individual context, such as whether the specialty of the referring provider plays a role in denial rate, within which the tests in our cohort were denied against the recommendation of medical professionals.
Our findings should be interpreted within the limitations of the study. As mentioned in our methods, because our focus was specifically on diagnostic rates among insurance denials, all cases not specifically documenting a denial during prior-authorization were considered approvals. This does not reflect possible denials after testing was completed or possible inpatient testing ordered in which prior-authorization was not required; therefore, it is possible the approval rate is skewed favorably for both private and public payers who have “authorization not required” policies. In addition, our study only addresses insurance as a barrier to testing coverage, however there are many other barriers, including socioeconomic, educational, and emotional barriers, not discussed here. Most of our cohort represents 1 children’s hospital in the Midwest in a relatively selected tertiary care setting and therefore may represent a conservative estimate of both denial rates and diagnostic rates in pediatric genetic testing populations. Our cohort was also predominantly White and therefore may not accurately represent insurance coverage and access to testing in other racial backgrounds.
This review of diagnostic rates among patients experiencing insurance denials in our selected pediatric genetic testing cohort provides an estimate of how many patients may be missing out on important genetic information, both diagnostic findings and candidate findings that may go on to become diagnostic. As we show, these results affect immediate medical management for a proportion of patients. Although other barriers to testing exist, it is clear that insurance coverage and discrepancies between payer policies remain a major barrier to diagnosis for many families. A constant re-evaluation of coverage policies for genetic testing and further involvement of genetics professionals in all steps of the process is necessary to ensure optimal clinical care for every patient.
Supplementary Material
Acknowledgments
We would like to thank the families for participating in our study. This work was made possible by the generous gifts to Children’s Mercy Research Institute and the Genomic Answers for Kids program at Children’s Mercy Kansas City.
Funding
This work was supported by a Clinical and Translational Science Awards Program grant from National Center for Advancing Translational Sciences awarded to the Frontiers Clinical and Translational Science Institute at the University of Kansas (# UL1TR002366). The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or National Center for Advancing Translational Sciences.
Footnotes
Conflict of Interest
The authors declare no conflicts of interest.
Ethics Declaration
All studies were approved by the Children’s Mercy Institutional Review Board (study # 11120514). Informed written consent was obtained from all participants before study inclusion.
Additional Information
The online version of this article (https://doi.org/10.1016/j.gim.2023.100020) contains supplementary material, which is available to authorized users.
Data Availability
Processed data for rare variants, de-identified pedigrees, and coded phenotypes are available to registered users through a cloud-hosted PhenoTips web user interface (https://phenotips-ga4k.cmh.edu). Access inquiries for investigators should be directed to GA4K@cmh.edu (including key to correlate study numbers used in this manuscript). Recurrent variants and their frequencies derived from >1000 alleles of HiFi-GS data are available at https://github.com/ChildrensMercyResearchInstitute/GA4K
References
- 1.Retterer K, Juusola J, Cho MT, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18(7):696–704. 10.1038/gim.2015.148 [DOI] [PubMed] [Google Scholar]
- 2.Ellison JW, Ravnan JB, Rosenfeld JA, et al. Clinical utility of chromosomal microarray analysis. Pediatrics. 2012;130(5):e1085–e1095. 10.1542/peds.2012-0568 [DOI] [PubMed] [Google Scholar]
- 3.Roberts JL, Hovanes K, Dasouki M, Manzardo AM, Butler MG. Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders or learning disability presenting for genetic services. Gene. 2014;535(1):70–78. 10.1016/j.gene.2013.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan CA, Topper S, Del Gaudio D, et al. Characterization of patients referred for non-specific intellectual disability testing: the importance of autosomal genes for diagnosis. Clin Genet. 2016;89(4):478–483. 10.1111/cge.12575 [DOI] [PubMed] [Google Scholar]
- 5.Scocchia A, Kangas-Kontio T, Irving M, et al. Diagnostic utility of next-generation sequencing-based panel testing in 543 patients with suspected skeletal dysplasia. Orphanet J Rare Dis. 2021;16(1):412. Published correction appears in Orphanet J Rare Dis. 2022;17(1):59. 10.1186/s13023-021-02025-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmquist R, Jenkins SM, Bentley D, et al. Evaluating use of changing technologies for rapid next-generation sequencing in pediatrics. Pediatr Res. 2022;92(5):1364–1369. 10.1038/s41390-022-01965-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malinowski J, Miller DT, Demmer L, et al. Systematic evidence-based review: outcomes from exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability. Genet Med. 2020;22(6):986–1004. 10.1038/s41436-020-0771-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y, Peng J, Liang D, et al. Genome sequencing demonstrates high diagnostic yield in children with undiagnosed global developmental delay/intellectual disability: a prospective study. Hum Mutat. 2022;43(5):568–581. 10.1002/humu.24347 [DOI] [PubMed] [Google Scholar]
- 9.Clark MM, Stark Z, Farnaes L, et al. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom Med. 2018;3(1):16. 10.1038/s41525-018-0053-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amendola LM, Hart MR, Bennett RL, et al. Insurance coverage does not predict outcomes of genetic testing: the search for meaning in payer decisions for germline cancer tests. J Genet Couns. 2019;28(6):1208–1213. 10.1002/jgc4.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cigna. Medical coverage policy. Genetic testing for hereditary cancer susceptibility syndromes. Cigna. 2022. Accessed October 22, 2022. https://static.cigna.com/assets/chcp/pdf/coveragePolicies/medical/mm_0518_coveragepositioncriteria_genetic_cancer_syndromes.pdf [Google Scholar]
- 12.Anthem BlueCross. Clinical UM guideline. BRCA genetic testing. Anthem. Published April 13, 2022. Accessed November 11, 2022 https://www.anthem.com/dam/medpolicies/abc/active/guidelines/gl_pw_e000235.html
- 13.Humana. Genetic testing medical coverage policy. OSF healthcare. 2017. Accessed November 22, 2022. https://www.osfhealthcare.org/media/filer_public/49/05/490514d9-6c92-4190-bd5d-45bba569653a/humana_2017_genetic_testing_medical_coverage_guidelines.pdf
- 14.Manickam K, McClain MR, Demmer LA, et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021;23(11):2029–2037. 10.1038/s41436-021-01242-6 [DOI] [PubMed] [Google Scholar]
- 15.Cigna. Medical coverage policy. Whole exome and whole genome sequencing for non-cancer indications. Cigna. 2023. Accessed December 13, 2022. https://static.cigna.com/assets/chcp/pdf/coveragePolicies/medical/mm_0519_coveragepositioncriteria_exome_genome_sequence.pdf [Google Scholar]
- 16.Anthem BlueCross. Medical policy. Whole genome sequencing, whole exome sequencing, gene panels, and molecular profiling. Anthem. Published December 28, 2022. Accessed January 6, 2023. https://www.anthem.com/dam/medpolicies/abc/active/policies/mp_pw_e000224.html
- 17.Humana. Whole genome/exome sequencing and genome-wide association studies. 2022. Accessed December 13, 2022. https://apps.humana.com/tad/tad_new/Search.aspx?criteria=exome&searchtype=freetext&policyType=both
- 18.Deverka PA, Dreyfus JC. Clinical integration of next generation sequencing: coverage and reimbursement challenges. J Law Med Ethics. 2014;42(Suppl 1):22–41. 10.1111/jlme.12160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vissers LELM, van Nimwegen KJM, Schieving JH, et al. A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet Med. 2017;19(9):1055–1063. 10.1038/gim.2017.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarze K, Buchanan J, Taylor JC, Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018;20(10):1122–1130. 10.1038/gim.2017.247 [DOI] [PubMed] [Google Scholar]
- 21.Blueprint Genetics. Pricing. Blueprint Genetics. 2022. Accessed September 8, 2022. https://blueprintgenetics.com/pricing/ [Google Scholar]
- 22.Schoen C, Osborn R, Squires D, Doty MM, Pierson R, Applebaum S. How health insurance design affects access to care and costs, by income, in eleven countries. Health Aff (Millwood). 2010;29(12):2323–2334. 10.1377/hlthaff.2010.0862 [DOI] [PubMed] [Google Scholar]
- 23.Duffy EL, Adler L, Ginsburg PB, Trish E. Prevalence and characteristics of surprise out-of-network bills from professionals in ambulatory surgery centers. Health Aff (Millwood). 2020;39(5):783–790. 10.1377/hlthaff.2019.01138 [DOI] [PubMed] [Google Scholar]
- 24.Papanicolas I, Woskie LR, Jha AK. Health care spending in the United States and other high-income countries. JAMA. 2018;319(10):1024–1039. Published correction appears in JAMA. 2018;319(17):1824. 10.1001/jama.2018.1150 [DOI] [PubMed] [Google Scholar]
- 25.Smith HS, Franciskovich R, Lewis AM, et al. Outcomes of prior authorization requests for genetic testing in outpatient pediatric genetics clinics. Genet Med. 2021;23(5):950–955. 10.1038/s41436-020-01081-x [DOI] [PubMed] [Google Scholar]
- 26.Reuter CM, Kohler JN, Bonner D, et al. Yield of whole exome sequencing in undiagnosed patients facing insurance coverage barriers to genetic testing. J Genet Couns. 2019;28(6):1107–1118. 10.1002/jgc4.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312(18):1880–1887. 10.1001/jama.2014.14604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen ASA, Farrow EG, Abdelmoity AT, et al. Genomic answers for children: dynamic analyses of >1000 pediatric rare disease genomes. Genet Med. 2022;24(6):1336–1348. 10.1016/j.gim.2022.02.007 [DOI] [PubMed] [Google Scholar]
- 29.Girdea M, Dumitriu S, Fiume M, et al. PhenoTips: patient phenotyping software for clinical and research use. Hum Mutat. 2013;34(8):1057–1065. 10.1002/humu.22347 [DOI] [PubMed] [Google Scholar]
- 30.Carmichael N, Tsipis J, Windmueller G, Mandel L, Estrella E. “Is it going to hurt?”: the impact of the diagnostic odyssey on children and their families. J Genet Couns. 2015;24(2):325–335. 10.1007/s10897-014-9773-9 [DOI] [PubMed] [Google Scholar]
- 31.Donohue KE, Dolan SM, Watnick D, et al. Hope versus reality: parent expectations of genomic testing. Patient Educ Couns. 2021;104(8):2073–2079. 10.1016/j.pec.2021.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trosman JR, Weldon CB, Slavotinek A, Norton ME, Douglas MP, Phillips KA. Perspectives of US private payers on insurance coverage for pediatric and prenatal exome sequencing: results of a study from the Program in Prenatal and Pediatric Genomic Sequencing (P3EGS). Genet Med. 2020;22(2):283–291. 10.1038/s41436-019-0650-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips KA, Trosman JR, Deverka PA, et al. Insurance coverage for genomic tests. Science. 2018;360(6386):278–279. 10.1126/science.aas9268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips KA, Deverka PA, Hooker GW, Douglas MP. Genetic test availability and spending: where are we now? Where are we going? Health Aff (Millwood). 2018;37(5):710–716. 10.1377/hlthaff.2017.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin GA, Trosman JR, Douglas MP, et al. Influence of payer coverage and out-of-pocket costs on ordering of NGS panel tests for hereditary cancer in diverse settings. J Genet Couns. 2022;31(1):130–139. 10.1002/jgc4.1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed data for rare variants, de-identified pedigrees, and coded phenotypes are available to registered users through a cloud-hosted PhenoTips web user interface (https://phenotips-ga4k.cmh.edu). Access inquiries for investigators should be directed to GA4K@cmh.edu (including key to correlate study numbers used in this manuscript). Recurrent variants and their frequencies derived from >1000 alleles of HiFi-GS data are available at https://github.com/ChildrensMercyResearchInstitute/GA4K


