Abstract
Imaging is often a first-line method for diagnostics and treatment. Radiological workflows increasingly mine medical images for quantifiable features. Variability in device/vendor, acquisition protocol, data processing, etc., can dramatically affect quantitative measures, including radiomics. We recently developed a method (PixelPrint) for 3D-printing lifelike computed tomography (CT) lung phantoms, paving the way for future diagnostic imaging standardization. PixelPrint generates phantoms with accurate attenuation profiles and textures by directly translating clinical images into printer instructions that control density on a voxel-by-voxel basis. The present study introduces a library of 3D printed lung phantoms covering a wide range of lung diseases, including usual interstitial pneumonia with advanced fibrosis, chronic hypersensitivity pneumonitis, secondary tuberculosis, cystic fibrosis, Kaposi sarcoma, and pulmonary edema. CT images of the patient-based phantom are qualitatively comparable to original CT images, both in texture, resolution and contrast levels allowing for clear visualization of even subtle imaging abnormalities. The variety of cases chosen for printing include both benign and malignant pathology causing a variety of alveolar and advanced interstitial abnormalities, both clearly visualized on the phantoms. A comparison of regions of interest revealed differences in attenuation below 6 HU. Identical features on the patient and the phantom have a high degree of geometrical correlation, with differences smaller than the intrinsic spatial resolution of the scans. Using PixelPrint, it is possible to generate CT phantoms that accurately represent different pulmonary diseases and their characteristic imaging features.
Keywords: Computed tomography, 3D printing, image quality phantoms, quality assurance, lung
1. INTRODUCTION
In research and clinical practice, anthropomorphic phantoms are utilized to develop, optimize, and evaluate medical imaging hardware and software. Typically, these phantoms are made using homogeneous materials that replicate tissue properties relevant to the imaging modality, such as x-ray attenuation coefficients for computed tomography. Realistic patient-based phantoms have many advantages for clinical and research purposes, such as optimizing imaging protocols and providing target data for artificial intelligence algorithms used in denoising or artifact correction. Although a wide variety of commercial phantoms are available, there is a shortage of patient-based phantoms capable of accurately representing the quantitative imaging characteristics and textures found in clinical images.
Over the past decade, 3D-printing of phantoms that represent the x-ray attenuation and texture of various tissues, anatomies, and diseases has been extensively explored. Despite being close to clinical imaging characteristics, phantoms produced with previous techniques still have limitations. As a result of segmenting regions and converting them to surface models, abrupt and unrealistic transitions between homogeneous regions are produced within printed phantoms, compromising spatial resolution and textural details. To overcome the limitations described above, we recently introduced PixelPrint [1][2]. PixelPrint directly translates DICOM image data into printer instructions that continuously control the printed material density by varying the printer speed on a voxel-by-voxel basis, while maintaining a constant filament extrusion rate. We previously presented quantitative comparisons between clinical CT slices and 3D-printed phantoms [3].
We present here an enhancement of PixelPrint and, simultaneously, a library of 3D printed lung phantoms covering a wide range of pulmonary disorders. PixelPrint was used to create 3D phantoms based on six CT scans from patients with different types of lung diseases. A clinical CT scanner was used to scan the phantoms. Our findings demonstrate that PixelPrint can reproduce accurate pathological changes in lung tissue in CT phantoms. In addition to providing ground-truth for validation of novel algorithms, this library of phantoms may also be used to optimize imaging protocols between health care facilities.
2. MATERIALS AND METHODS
2.1. Data collection.
This retrospective study was approved by the Institutional Review Board (IRB). We retrospectively gathered CT data from the PACS system at the Hospital of the University of Pennsylvania. A total of six patient data sets were selected, representing usual interstitial pneumonia (UIP), chronic hypersensitivity pneumonitis (CHP), reactivation tuberculosis (TB), cystic fibrosis (CF), Kaposi sarcoma (KS) and moderately severe pulmonary edema (PE). Three of the patient CT scans were performed using Siemens SOMATOM Force, two with Siemens SOMATOM Definition, and one with Siemens SOMATOM Definition AS+ (all from Siemens Healthineers, Erlangen, Germany). For each indication, CT data was acquired according to standard departmental protocols.
2.2. Phantom Production.
Each patient’s clinical DICOM images were converted into instructions for 3D-printing using PixelPrint software. For each phantom, the lung was segmented and prepared for printing as a 15 or 20 cm diameter phantom. The phantoms presented in this study were printed using 1.75 mm diameter Polylactic Acid (PLA) filament (MakeShaper, Keene Village Plastics, Cleveland, OH, USA) on a Lulzbot TAZ 6 fused-filament printer (Fargo Additive Manufacturing Equipment 3D, LLC Fargo, ND, USA) with a brass nozzle with a diameter of 0.25 mm. A constant extrusion rate and a fixed layer height were used to print the phantoms. The printing speed ranged from 3 to 30 mm/s, producing filament line widths of 0.1 to 1.0 mm.
2.3. Data collection and acquisition.
Imaging was performed with a dual-layer spectral CT (IQon spectral CT, Philips Healthcare, Cleveland, Ohio, USA). In order to generate side-by-side comparisons, the resulting data were reviewed by an experienced thoracic radiologist. For comparison of the DICOM images between patient and phantom, mean HU values and standard deviations were calculated. Histograms were calculated from the patient and phantom data to compare the HU distributions.
3. RESULT
In Figures 1 to 5, CT images of the patient and the phantom scan were displayed and compared side-by-side. Coronal and sagittal views of the data were also included in sub-figure panels. As one can observe, PixelPrint is well suited to recreating lung parenchymal tissue structures which closely resemble pathological changes. Pathological processes involving primarily alveolar disease such as consolidations, cavitary changes or lung masses as well as primarily interstitial disease such as lung fibrosis or interstitial edema can be clearly identified and characterized on the phantoms. Figure 6 shows the histograms for the phantom of a patient and the original patient image. Over the typical lung tissue range, PixelPrint phantom images resembled the contrast of the original patient image and maintained the same levels. Due to the 10% and 100% cut-off, the signal accumulates at 0 and −800 HU in the CT phantoms. Figure 7 shows the Hounsfield unit measurement in five lung regions, with measuring difference less than 6 HU between patient and phantom.
Figure 1.
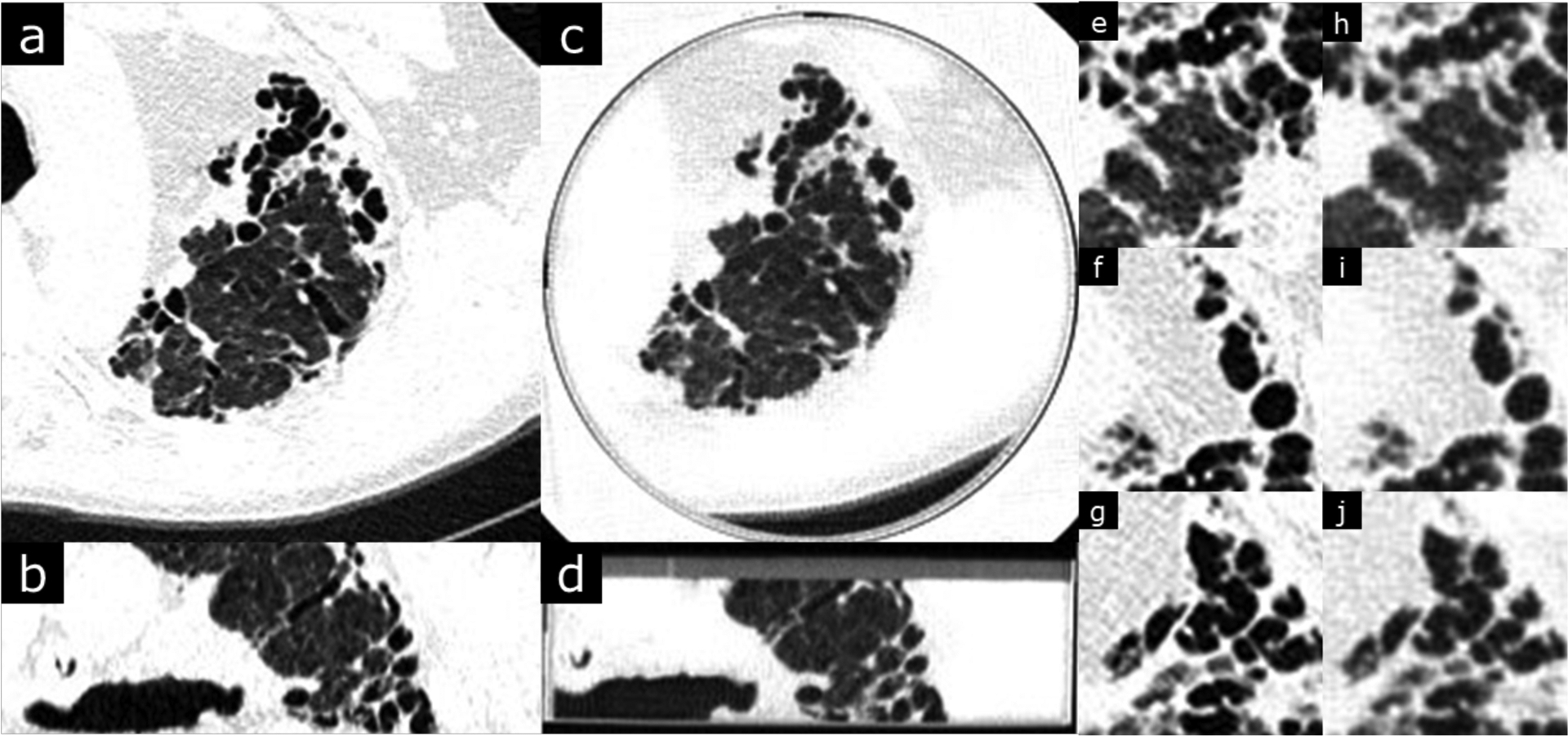
Usual interstitial pneumonia with advanced fibrosis. Original DICOM image resolution is 0.762 mm (field of view was 390 mm over 512 pixels). Window level is −500 and width is 1000 HU. b and d are coronal views of the patient and phantom images. Phantom images (c,d,h,i,j) clearly depicts the interstitial changes of UIP including traction bronchiectasis, honeycombing and coarse fibrotic bands with architectural distortion.
Figure 5.
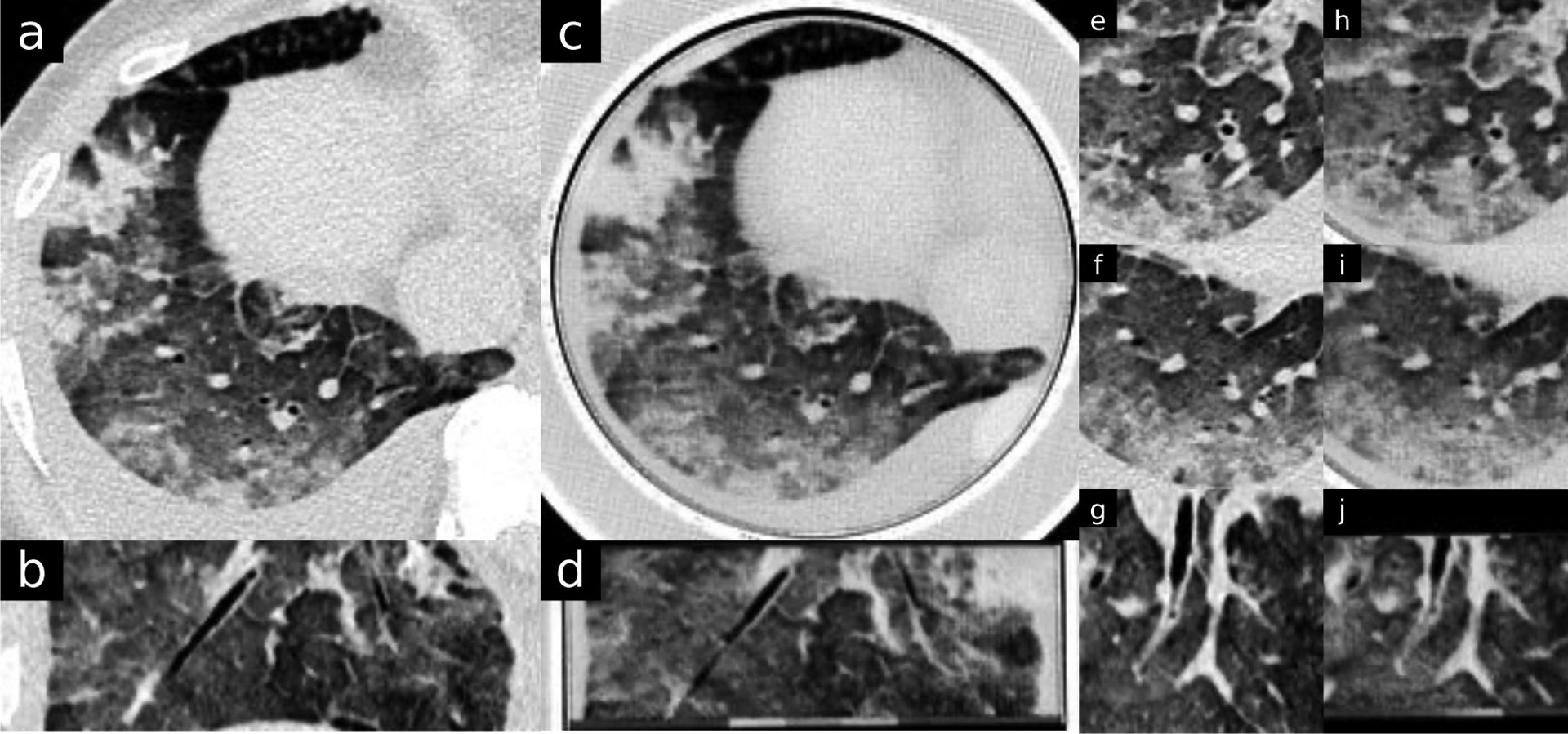
Moderate pulmonary edema. Original DICOM image resolution is 0.773 mm (field of view was 396 mm over 512 pixels). Window level is −330 and width is 1000 HU. Patient and phantom images are approximated to be the same location. b, d, g and j are sagittal views. Phantom images (c,d,h,i,j) clearly depicts typical findings of pulmonary edema with patchy mixed density opacities, interlobular septal thickening and pleural effusion.
Figure 6.
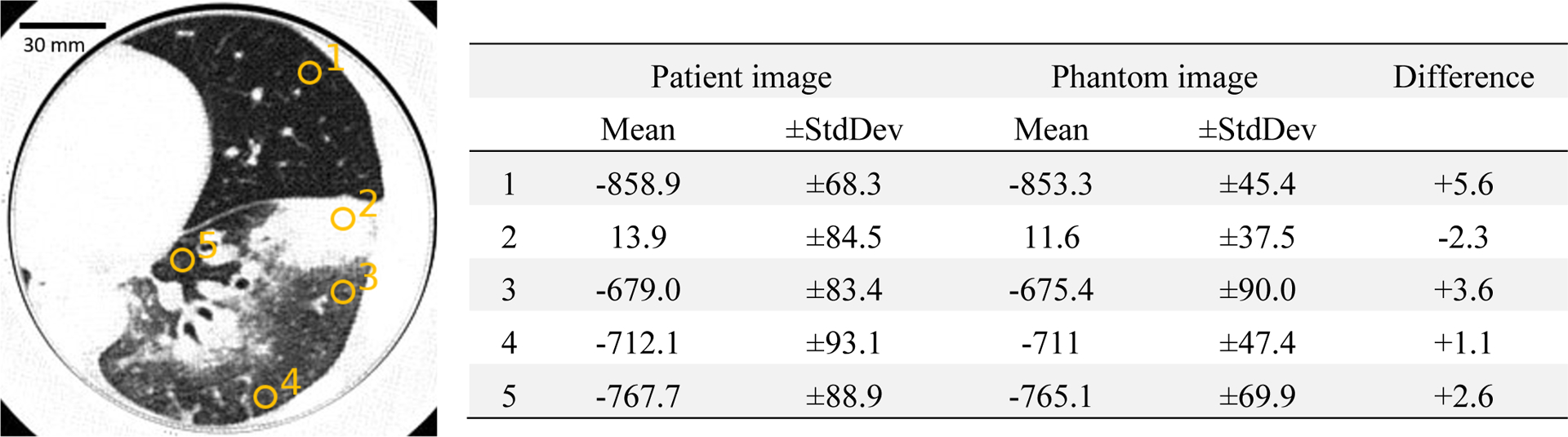
Measured mean and standard deviation of the patient and phantom images. Yellow circles indicate regions of interest measured in ImageJ. Window level is −500 and width is 1000 HU.
Figure 7.
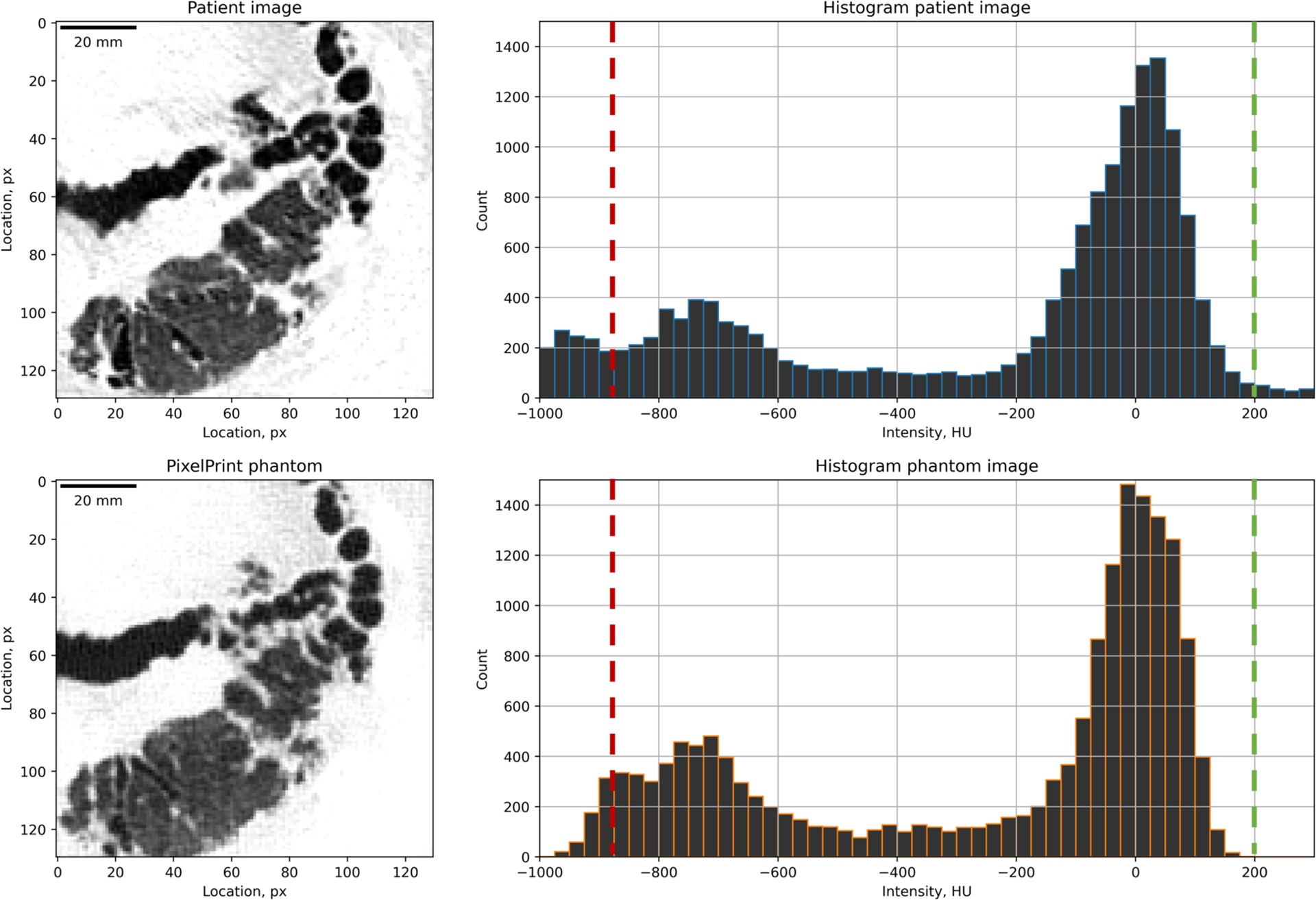
Histograms of the patient and phantom images. Histograms were computed from the lung areas shown as the left images (130px by 130px). Window level is −500 and width is 1000 HU. Two images were registered 2D-wise and assumed to be the same location. Histogram bin size is 25 HU. The green dash line at 200 HU was placed at the density of 100% PLA. The red dash line indicates the point where PLA lines are too thin.
4. DISCUSSION
Recently, we introduced a novel 3D-printing method, PixelPrint, which allows generation of patient-based phantoms with accurate anatomical geometry, image texture, and contrast values. This method allows direct conversion of CT DICOM images into 3D-printer instructions without requiring segmentation. In this work, we presented a further optimized version of PixelPrint as well as a library of phantoms covering a wide range of respiratory diseases. CT images of the patient-based phantoms in our collection are quantitatively and qualitatively comparable to original CT images, both in texture and contrast levels, and clinical features are clearly visible.
In the future, we plan to extend our library to different stages of various lung diseases as well as other conditions outside the lung. By using these phantoms in increasing numbers, they will become more valuable to the entire medical community and enable standardization, comparison, and assessment of advanced medical algorithms. The larger medical, academic, and industrial CT communities can obtain copies of all phantoms presented in this study, as well as phantoms based on specific CT images.
5. CONCLUSION
The present study illustrates the possibility of creating a library of 3D-printed patient-based lung phantoms with accurate organ geometry, image texture, and attenuation profiles. On the long run such a library of phantoms may aid the translation of quantifiable features, including radiomics. This may lead to a paradigm change for the development of novel CT hardware and software by enabling accelerated evaluation and validation with realistic patient-based data. Ultimately this will shape the clinical day-to-day routine and benefit patients with novel and standardized CT imaging.
Figure 2.
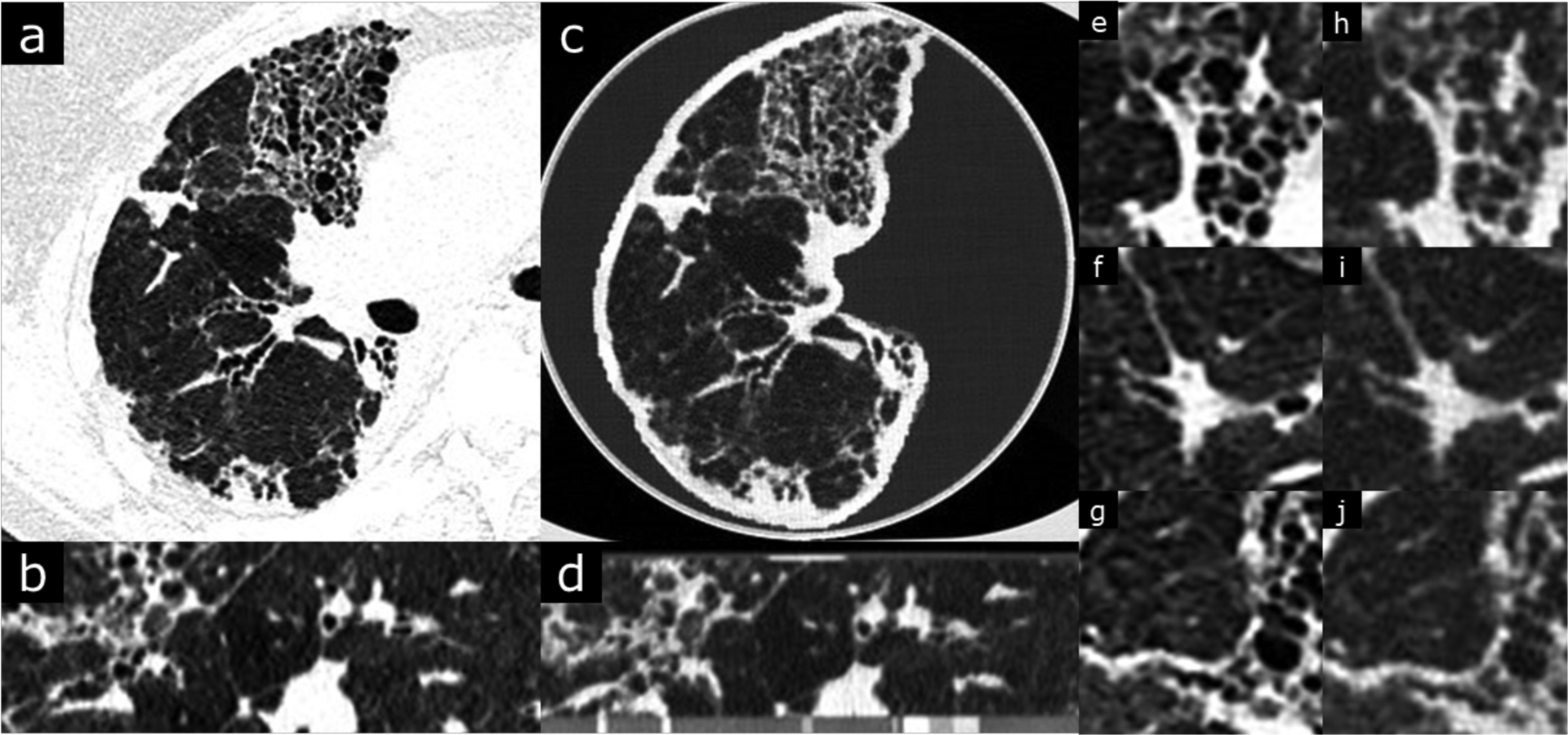
Chronic hypersensitivity pneumonitis with fibrotic change. Original DICOM image resolution is 0.684 mm (field of view was 350 mm over 512 pixels). Window level is −500 and width is 1000 HU. b and d are coronal views of the patient and phantom images. Phantom images (c,d,h,i,j) clearly depicts the interstitial changes including traction bronchiectasis, cystic change and volume loss. Typical areas of alveolar air trapping resulting in “mosaic” attenuation differences are also well visualized.
Figure 3.
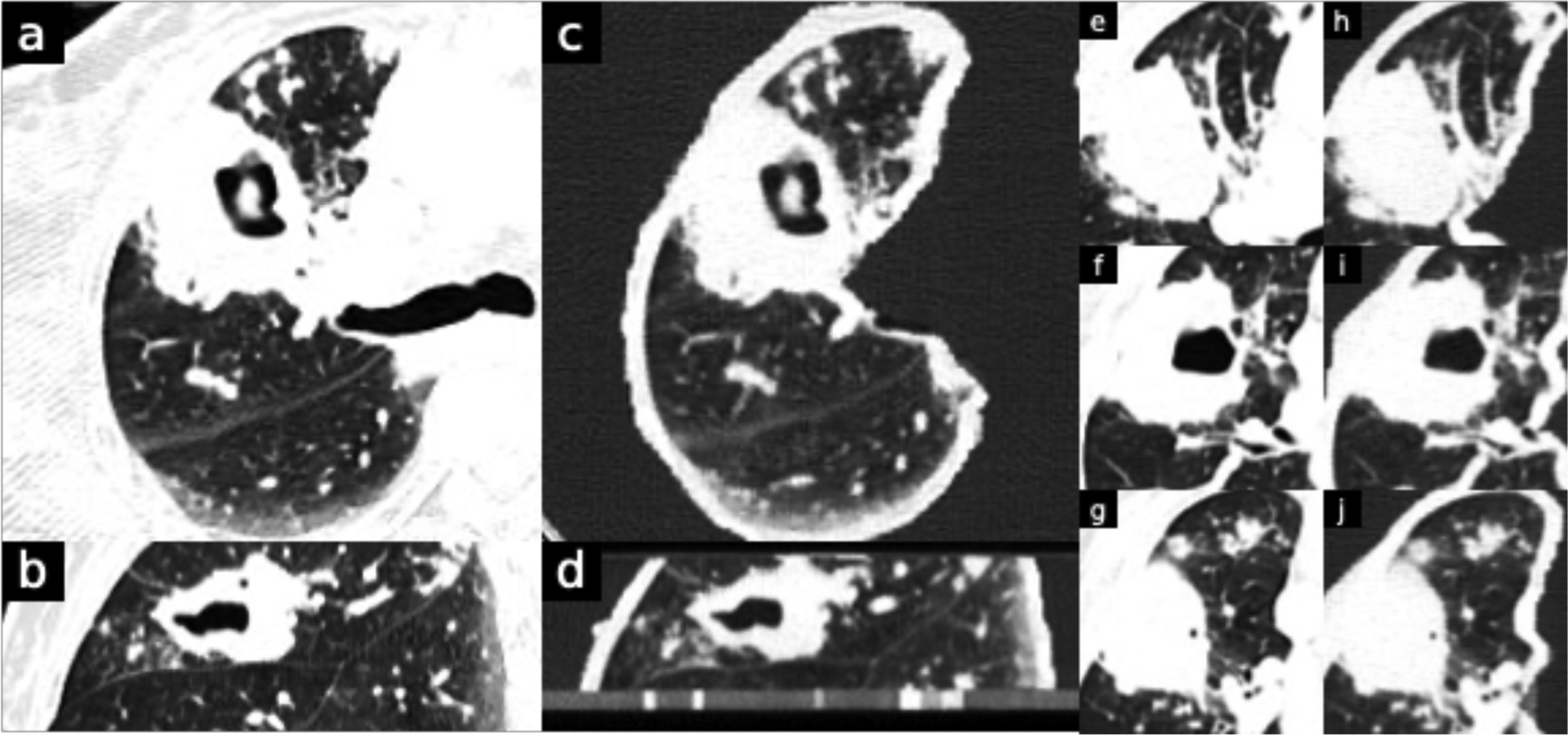
Reactivation Tuberculosis. Original DICOM image resolution is 0.813 mm (field of view was 416 mm over 512 pixels). Window level is −500 and width is 1000 HU. B and d are sagittal views. Phantom images (c,d,h,i,j) clearly depicts the lung cavitation with surrounding consolidation and clustered areas of centrilobular nodularity compatible with infectious bronchiolitis, all typical for reactivation tuberculosis.
Figure 4.
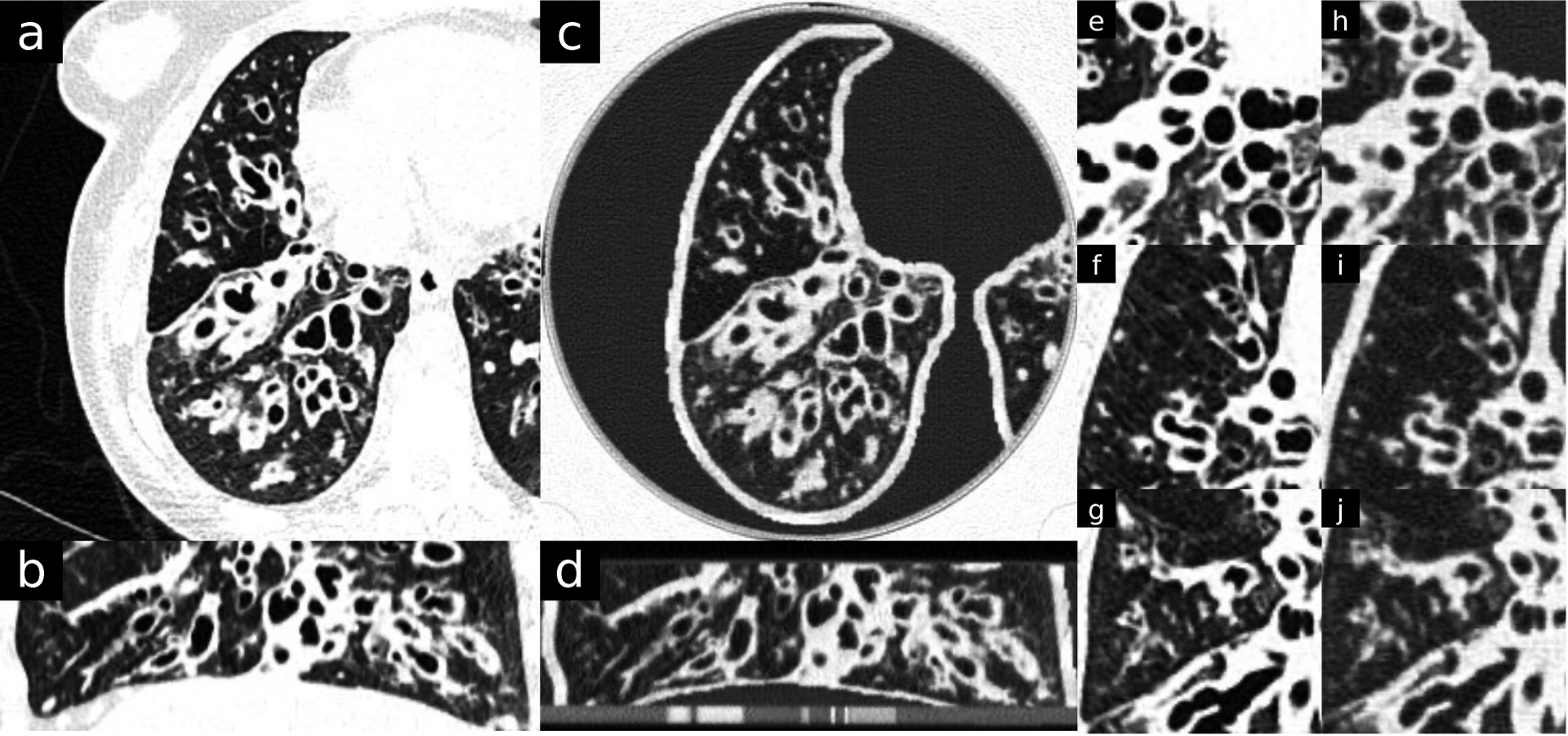
Cystic Fibrosis. Original DICOM image resolution is 0.705 mm (field of view was 361 mm over 512 pixels). Window level is −500 and width is 1000 HU. b and d are sagittal views. Phantom images (c,d,h,i,j) clearly depicts classic central bronchiectatic changes, with associated mucus plugging and small clusters of bronchiolitis.
ACKNOWLEDGMENT
We acknowledge support through the National Institutes of Health (R01EB030494, R01EB031592, & R01CA249538).
REFERENCES
- [1].Mei K, Geagan M, Roshkovan L, Litt HI, Gang GJ, Shapira N, Stayman JW and Noël PB, 2022. Three-dimensional printing of patient-specific lung phantoms for CT imaging: emulating lung tissue with accurate attenuation profiles and textures. Medical physics, 49(2), pp.825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mei K, Geagan M, Shapira N, Liu LP, Pasyar P, Gang GJ, … & Noël PB (2022, October). PixelPrint: three-dimensional printing of patient-specific soft tissue and bone phantoms for CT. In 7th International Conference on Image Formation in X-Ray Computed Tomography (Vol. 12304, pp. 545–550). SPIE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shapira N, Donovan K, Mei K, Geagan M, Roshkovan L, Gang G, Abed M, Linna NB, Cranston CP, Leary CN and Dhanaliwala AH, 2022. PixelPrint: Three-dimensional printing of realistic patient-specific lung phantoms for validation of computed tomography post-processing and inference algorithms. medRxiv, pp.2022–05. [Google Scholar]


