Abstract
Of a total of 147 erythromycin-resistant Streptococcus pneumoniae isolates, 64 (43.5%) were resistant to erythromycin, clindamycin, and streptogramin B (MLSB phenotype), 57 of which possessed the ermB gene. Eighty-two (55.8%) were resistant to erythromycin alone (M phenotype), 81 of which possessed the mefE gene. One was erythromycin and streptogramin B resistant but susceptible to clindamycin (MS phenotype) and possessed neither the erm gene nor the mefE gene.
The macrolide, lincosamide, and streptogramin B (MLSB) antimicrobials are three chemically distinct, functionally related drug classes which inhibit protein synthesis through alteration of the 50S subunit (2). Resistance may occur by target site modification, active efflux, or enzymatic inactivation.
Target modification arises through the presence of an erm methylase, which induces a conformational change in the ribosome. This results in inducible or constitutive cross-resistance to MLSB antimicrobials (MLSB phenotype) (2). Previously, target modification, due to ermAM(B), was the only reported mechanism of MLSB resistance in S. pneumoniae (10). More recently, however, a macrolide-specific efflux mechanism, encoded by mef, has been described (M phenotype) (9). In this study, we looked at the prevalence of these phenotypes in a large collection of strains and correlated it with the presence of the erm and mef genes.
A total of 5,029 clinical isolates of S. pneumoniae were obtained from 1993 to 1996 from a cross-Canada surveillance study involving 113 hospital and private laboratories in all 10 provinces. These strains were characterized by in vitro susceptibility testing and molecular techniques in order to determine the prevalence and mechanisms of macrolide resistance.
Susceptibility testing was carried out by broth microdilution with all isolates and disk diffusion with those isolates subsequently found to be erythromycin resistant, according to the National Committee for Clinical Laboratory Standards guidelines (4). An MLSB phenotype isolate was defined as an isolate that was found by disk diffusion to be resistant to erythromycin, clindamycin, and quinupristin (streptogramin B). An M phenotype isolate was an isolate found to be resistant to erythromycin alone. An MS phenotype isolate was an isolate found to be resistant to erythromycin and quinupristin. The antimicrobial concentration of the disks was 15 μg of erythromycin, 2 μg of clindamycin (Oxoid, Nepean, Ontario, Canada), and 7.5 μg of quinupristin (Rhone-Poulenc-Rorer, Collegeville, Pa.).
Genomic DNA was isolated as described by Smith et al. (7). Plasmid isolation was performed with the Quantum Prep plasmid miniprep kit (Bio-Rad, Mississauga, Ontario, Canada). Multiplex PCR was performed with primers specific for ermA, ermB (ermAM), ermC, and mefE as described by Sutcliffe et al. (8) for all erythromycin-resistant strains and eight random erythromycin-susceptible strains. Pulsed-field gel electrophoresis (PFGE) (3) with the CHEF DRII apparatus (Bio-Rad) and SmaI digestion were performed with 8 to 12 representative isolates of each of the following groups: MLSB phenotypes that were erm positive, M phenotypes that were mefE positive, susceptible strains that were erm negative and mefE negative, and resistant strains that were erm negative and mefE negative. Modifications included a lysis time of 2 h and the following electrophoretic parameters: pulse times of 0.2 to 35 s, a temperature of 14°C, and 200 V for 21 h. S. pneumoniae strains 3585 (ermAM), 02J1175 (mefE), and ATCC 49619 were used as controls. Southern blotting of the PFGE gels was performed as described previously, and blots were probed with ermAM and mefE amplicons (1). Dot blot hybridization was performed by standard methodology on the representative isolates with the addition of plasmid DNA from E. coli RN7951 (ermA) and S. aureus RN4220 (ermC), and blots were probed with ermA and ermC amplicons. To ensure genomic DNA integrity, 16S rRNA was amplified by PCR from S. pneumoniae 3585, under the conditions used for mefE and erm, and was used to confirm the presence of DNA. Probes were purified with the QIAquick PCR purification kit (Qiagen, Mississauga, Ontario, Canada). Hybridization and detection were performed by enhanced chemiluminescence with the ECL direct nucleic acid labelling system (Amersham Life Science, Oakville, Ontario, Canada).
Of 5,029 isolates tested, 147 (2.9%) were found to be erythromycin resistant by broth microdilution. Disk diffusion results (Table 1) demonstrated that 64 isolates were of the MLSB phenotype, 82 were of the M phenotype, and 1 was of the MS phenotype. PFGE demonstrated that of 42 strains tested, 41 were clonally distinct (data not shown).
TABLE 1.
Characterization of 147 erythromycin-resistant S. pneumoniae isolates used in this study
| Resistance phenotype | No. of strains | No. of isolates with resistance gene(s) detected
|
||
|---|---|---|---|---|
| ermA, -B, or -C | mefE | None | ||
| MLSB | 64 | 57 | 1a | 7 |
| M | 82 | 0 | 81 | 1 |
| MS | 1 | 0 | 0 | 1 |
One constitutive MLSB isolate possessed both erm and mefE genes.
Multiplex PCR with primers specific for ermA, ermB (ermAM), ermC, and mefE yielded results predictive of the observed MLSB and M phenotypes (Table 1). Of 64 strains possessing an MLSB phenotype, 57 yielded an erm amplicon. One isolate possessed both mefE and erm genes. In contrast, of 82 strains bearing the M phenotype, 81 possessed a mefE gene. Of nine strains lacking mef and erm, seven were of the MLSB phenotype, one was of the M phenotype, and one was of the MS phenotype.
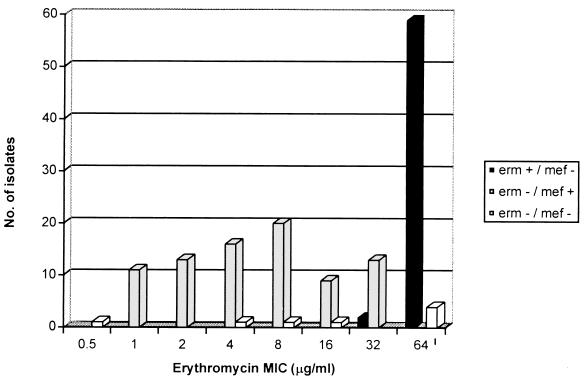
When tested by disk diffusion, broth microdilution results were 100% concordant for erythromycin and 97.3% concordant for clindamycin. For the latter, four (2.7%) of the isolates appeared clindamycin susceptible by broth microdilution but were fully resistant by disk diffusion and possessed an erm gene. Of the 64 isolates with an MLSB phenotype, 57 were found to have an erm gene, whereas, of the 82 isolates with an M phenotype, 81 were found to possess the mefE gene. Erythromycin resistance was generally found to be greater in the isolates with an erm gene (MIC at which 50% of the isolates are inhibited [MIC50], ≥64 μg/ml; MIC90, ≥64 μg/ml) than in those with a mefE gene (MIC50, 4.0 μg/ml; MIC90, 32 μg/ml) (Fig. 1).
FIG. 1.
Distribution of mefE and erm genes according to the MIC of erythromycin.
Hybridization studies of selected isolates supported the PCR findings in that only those positive for an erm gene by PCR hybridized with the ermB probe, but not with the ermA, ermC, and mefE probes, whereas, only those positive for mefE by PCR hybridized with a mefE probe. Erythromycin-susceptible strains and erythromycin-resistant strains, which were ermB mefE negative, did not hybridize with any of the probes. Erythromycin-resistant ermB-mefE-negative isolates were negative by probing for the ermA, -B, and -C and mefE genes. Thus, PCR was reliable in characterizing the two major mechanisms of resistance. The presence of nine isolates which did not yield amplicons by PCR and did not hybridize with erm and mefE suggests the presence of novel genes or mechanisms of resistance.
In this population-based study of clinical S. pneumoniae isolates from across Canada, we found that 55.8% of all macrolide-resistant isolates possessed the M phenotype, compared to 85 and 42% found by Sutcliffe et al. (9) and Shortridge et al. (6), respectively. Knowledge of the prevalence and type of macrolide resistance may have therapeutic implications in view of the different levels of resistance, depending on the resistance mechanism and the ability of the macrolides to concentrate at the site of some infections (Fig. 1) (5).
Acknowledgments
We gratefully acknowledge the following for providing control strains: Barry Kreiswirth (Public Health Research Institute, New York, N.Y.) for E. coli RN7951 and S. aureus RN4220, Joyce Sutcliffe (Groton, Connecticut) for S. pneumoniae 02J1175, and the Centers for Disease Control and Prevention (Atlanta, Ga.) for S. pneumoniae R6.
This work was supported in part by a grant from Pfizer Pharmaceutical and the Canadian Bacterial Diseases Network.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology, chapter 2, section 2.9.2. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 2.Fernandez-Munoz R, Monro R E, Torres-Pinedo R, Vasquez D. Substrate- and antibiotic-binding sites at the peptidyl-transferase centre of Escherichia coli ribosomes. Studies on the chloramphenicol, lincomycin and erythromycin sites. Eur J Biochem. 1971;23:185–193. doi: 10.1111/j.1432-1033.1971.tb01607.x. [DOI] [PubMed] [Google Scholar]
- 3.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Committee for Clinical Laboratory Standards. Approved standards M2-A6 and M7-A4. National Committee for Clinical Laboratory Standards, Villanova, Pa. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. [Google Scholar]
- 5.Rodvold K A, Gotfried M H, Danziger L H, Servi R J. Intrapulmonary steady-state concentrations of clarithromycin and azithromycin in healthy adult volunteers. Antimicrob Agents Chemother. 1997;41:1399–1402. doi: 10.1128/aac.41.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shortridge V D, Flamm R K, Ramer N, Beyer J, Tanaka S K. Novel mechanism of macrolide resistance in Streptococcus pneumoniae. Diagn Microbiol Infect Dis. 1996;26:73–78. doi: 10.1016/s0732-8893(96)00183-6. [DOI] [PubMed] [Google Scholar]
- 7.Smith A M, Klugman K P, Coffey T J, Spratt B G. Genetic diversity of penicillin-binding protein 2B and 2X genes from Streptococcus pneumoniae in South Africa. Antimicrob Agents Chemother. 1993;37:1938–1944. doi: 10.1128/aac.37.9.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]