Abstract
Large numbers of radiographic images are available in musculoskeletal radiology practices which could be used for training of deep learning models for diagnosis of knee abnormalities. However, those images do not typically contain readily available labels due to limitations of human annotations. The purpose of our study was to develop an automated labeling approach that improves the image classification model to distinguish normal knee images from those with abnormalities or prior arthroplasty. The automated labeler was trained on a small set of labeled data to automatically label a much larger set of unlabeled data, further improving the image classification performance for knee radiographic diagnosis. We used BioBERT and EfficientNet as the feature extraction backbone of the labeler and imaging model, respectively. We developed our approach using 7382 patients and validated it on a separate set of 637 patients. The final image classification model, trained using both manually labeled and pseudo-labeled data, had the higher weighted average AUC (WA-AUC 0.903) value and higher AUC values among all classes (normal AUC 0.894; abnormal AUC 0.896, arthroplasty AUC 0.990) compared to the baseline model (WA-AUC = 0.857; normal AUC 0.842; abnormal AUC 0.848, arthroplasty AUC 0.987), trained using only manually labeled data. Statistical tests show that the improvement is significant on normal (p value < 0.002), abnormal (p value < 0.001), and WA-AUC (p value = 0.001). Our findings demonstrated that the proposed automated labeling approach significantly improves the performance of image classification for radiographic knee diagnosis, allowing for facilitating patient care and curation of large knee datasets.
Keywords: EMR, Image classification, NLP, Knee, Osteoarthritis
Introduction
In modern radiology practices, large numbers of radiographic images are readily available for data-driven research in radiology [1–3]. Particularly, musculoskeletal (MSK) images are annotated for diagnosis of a wide range of pathologies and utilized by radiologists, orthopedics, and other advanced practitioners toward guiding patient management and improving patient care for knee abnormality diagnosis. However, structured annotations for a large volume of radiographic images are difficult to obtain because the manual annotation process requires tremendous amounts of experts’ attention and is very costly.
Deep learning (DL) solutions are typically developed using large numbers of labeled data. Specifically, image classification is an important supervised DL task to achieve various objectives in radiology, including disease detection, characterization, and monitoring [4–7]. An important example approached in this paper is classification of knee radiographs, which are commonly utilized for clinical evaluation of knee abnormalities [8–13] and knee arthroplasty [14–16]. Multiple studies incorporated DL-based image classification approaches for automating such evaluations [17–25]. These studies demonstrated the feasibility of leveraging large datasets for DL-based image classification in knee radiology, but a significant amount of effort in annotating knee radiographs has already been made to curate annotations for the existing datasets.
Alternatively, a label of a knee radiographic image can be directly interpreted and extracted from the corresponding radiology report. However, this is challenging for the following reasons. First, precise information extraction from unstructured knee radiology reports is difficult due to lack of standardization reporting [26]. Second, radiology reports are complex given the intricacy of knee pathologies in general, and reporting discrepancies or even errors across different radiologists [27]. Third, there is a lack of automated systems to interpret unstructured reports in knee radiology with high accuracy and sufficiency.
To overcome these limitations, this study aims to develop a hybrid DL framework, combining image classification and natural language processing (NLP) approaches, that leverages a large volume of unlabeled data to improve the performance of a multiclass classification model using only a limited number of manually labeled data. We decided on three classification labels, namely normal, abnormal, and arthroplasty. These labels represent visible conditions in the bilateral posterior to anterior standing (BLPA) knee weightbearing views, which are commonly utilized to assess radiographic changes in the knee [8, 28, 29].
To the best of our knowledge, our proposed approach is the first to simultaneously utilize radiographs and their corresponding radiology reports in a DL framework for knee radiograph classification. The inherent association between a radiograph and its corresponding report ensures the same label for these two data sources. Our proposed framework consists of two main DL components. First, we developed an NLP-based automated labeler on labeled radiology reports and applied the labeler to a large volume of unlabeled reports to generate pseudo labels. Second, to validate the improvement when trained with additional pseudo-labeled data, we developed two pretrained image classification models, one with only manually labeled data and the other with additional pseudo-labeled data, and evaluated their performance on a hold-out test set.
Methods
Data Collection
This study was HIPAA-compliant and approved by the institutional review board (IRB) of the Duke University Health System (DUHS). In the initial cohort, we retrospectively collected a dataset of 25,657 patients who had knee X-ray imaging studies finalized in 2019 within our large institutional health system. For each study, we downloaded radiology reports and knee radiographs in Digital Imaging and Communications in Medicine (DICOM) format from our electronic medical record (EMR) database.
We identified the initial cohort and obtained the dataset in two steps. First, we utilized a reporting workbench tool to search for radiology reports of knee X-ray imaging studies in our health system. The searching query was built based on the 10 most common knee procedure names in 2019 across our institution. The main searching results included (1) unique patient identifiers, (2) unique imaging study accession numbers, and (3) radiology reports. Second, we queried our large institutional picture archiving and communication system (PACS) to retrieve the DICOM objects for each imaging study by using the study accession numbers obtained from the main search.
In the final dataset, we only included patients with BLPA X-rays by filtering two DICOM attributes (1) modality = {CR, DX}, (2) series description = {PA axial, PA weight bearing, PA tunnel}. The final dataset consisted of 8140 patients with 8659 imaging studies. Each imaging study contained one radiology report and one BLPA radiograph.
Annotation
BLPA radiographs were annotated with three labels: normal, abnormal, and arthroplasty, by using the handcraft rules, created by a team including four personnel: a 4-year experience of MSK sub-specialist and three non-expert researchers. The annotation rules consisted of descriptions for one category of the presence of arthroplasty, and eleven categories of knee abnormalities. Eleven abnormal categories included degenerative changes, postoperative changes or presence of non-arthroplasty orthopedic hardware, fractures, lesions, fragmentation, bone lucency, malalignment, osseous abnormalities, soft tissue abnormalities, developmental abnormalities, and trauma. The annotation team determined categories of images by applying the annotation rules to the associated radiology reports of BLPA radiographic images. A radiographic image was labeled abnormal if it met with at least one abnormal category, arthroplasty if it met with the arthroplasty category, and normal if none of abnormal or arthroplasty rules were met. The final annotation rule was approved by the MSK sub-specialist.
Dataset Split
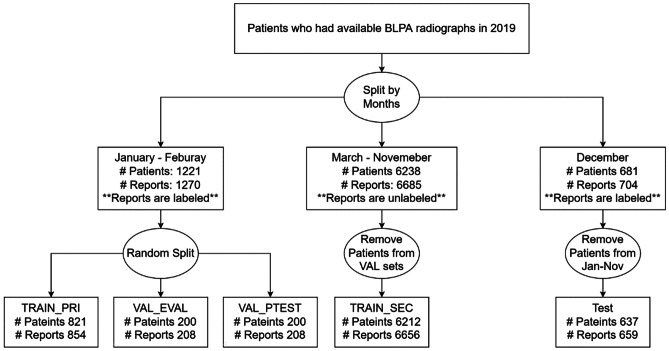
All patients (N = 8140) who had BLPA radiographs in 2019 were split into three groups based on the dates of radiology studies: (1) January to February, (2) March to November, and (3) December (Fig. 1). Patients from January to February, with reports being manually labeled, were randomly split into the primary training set (TRAIN_PRI), the validation set for tuning hyperparameters and saving the best checkpoints (VAL_EVAL), and a held-out validation set for determining the final model (VAL_PTEST). Patients from March to November served as the secondary training set (TRAIN_SEC). Reports in TRAIN_SEC were not manually labeled, but were pseudo labeled by the NLP model. We removed 26 overlapping patients with those in the two validation sets to prevent “data leakage” issues when the same patient appeared in both training and validation or test set. We called the above four dataset as the “development set.” Patients from December were served as the test set (TEST) with all reports being manually labeled. Likewise, to prevent data leakage, we removed 44 overlapping patients from January to November, which had already been used in training and developing models.
Fig. 1.
Dataset split plan. TRAIN_PRI: used for training both image/NLP. VAL_EVAL: used for saving checkpoints/tuning hyperparameters during training. VAL_PTEST: used for last evaluations before applying models to TEST. TRAIN_SEC: used for automatic pseudo labeling. We called the above 4 datasets as the “development set.” TEST: a completely untouched hold-out set for final evaluation
Automated Labeling of Reports Using an NLP model
The NLP model took a preprocessed report as an input and returned predictive probabilities of the three labels as an output. The final label had the highest predictive probability. Reports were preprocessed in two steps: (1) punctuations and numbers were removed and (2) only findings and impressions sections of the radiology reports were extracted. The model structure consisted of a feature extraction backbone and a classification module. An input unstructured report was tokenized and fed into the feature extraction backbone. We picked Bidirectional Encoder Representations from Transformers for Biomedical Text Mining (BioBERT) [30] model as the multiclass text classification backbone to output feature vectors, which were then fed into a linear layer with three output units for classification.
We trained candidate NLP models on TRAIN_PRI. The maximum token length of each report input to the model was fixed at 512. Batch size was set as 16, which is the optimal size for fitting the GPU RAM. Patience was set as 15 for early stopping purposes. Models were trained with the ADAM optimizer and tuned the following learning rates: {1e−5, 5e−5}. The model with the best performance on VAL_EVAL was used to annotate unlabeled reports in TRAIN_SEC. The NLP models were trained with deep learning libraries in Python, namely PyTorch and HuggingFace [31].
Image Classification Model
The image classification model took pixel arrays of an image as an input and returned predictive probabilities of the three labels. The final label had the highest predictive probability. We extracted pixel arrays from each corresponding DICOM file of an image and normalized them into the [0, 255] range. The range of image width was [1309, 3056], and height was [1287, 3424]. Similar to the NLP model structure, the imaging model structure consisted of a feature extraction backbone and a classification module. In this work, we picked EfficientNet-b4 [32] as the multiclass image classification backbone output feature vectors, which were then fed into a linear layer with three output units for classification.
The baseline image classification model used manual labels and was trained on TRAIN_PRI. Our proposed model leveraged additional pseudo-labeled data in TRAIN_SEC. The input image size for training was 380 × 380, the same as the size used to train a full EfficientNet-b4 model in the original paper. Batch size was set as 16, which is the optimal size given the GPU RAM. Patience was set as 10 for early stopping purposes. We trained image classification models using the ADAM optimizer and tuned the following learning rates: {1e−5, 5e−5}. We determined the final learning rate configuration by selecting the model with the best performance on VAL_EVAL. Using this configuration, we evaluated performance of models with five random experiments on VAL_PTEST and selected the best seed to be tested on TEST. Image classification models were trained using PyTorch with the “EfficientNet PyTorch” package.
Transfer Learning
Training feature extraction backbones, namely BioBERT and EfficientNet-b4, from scratch required significant amount of data and computational resources. Thus, to expedite the training while maintaining high accuracy, we utilized “transfer learning” approach for training our classification models. To do so, we initialized weights of feature extraction backbones using pretrained weights and further fine-tuned such model on the downstream classification task. Weights of BioBERT were initialized using pretrained BioBERT-Base v1.1 weights optimized on English Wikipedia, BooksCorpus, and PubMed abstracts, and fine-tuned in downstream training; weights of EfficientNet-b4 were initialized using pretrained EfficientNet weights optimized on ImageNet, a large visual database consisting of high-resolution natural images with human annotations.
Statistical Analysis
Each model was trained with five different seeds which control randomness. We collected mean, standard deviation, and median of weighted average area under receiver operating characteristic (ROC) curve (WA-AUC) based on five experiments of each model. WA-AUC is calculated as following:
where and is the frequency percentage and area under ROC curve (AUC) of label in the dataset, respectively. When we compared two models, the model with larger mean WA-AUC was deemed as a better model and selected as the representative for further evaluations. Once we obtained the final model, we reported the AUC for each class and WA-AUC, and plotted ROC curves for each class accordingly. The DeLong test [33] was performed on each class to evaluate if there was a significant difference in AUC when adding pseudo-labeled data in training. For WA-AUC, we collected 10,000 paired bootstrap samples in TEST and performed a two-sided normal test to evaluate if there was a significant difference.
Results
Dataset Analysis
A total of 8019 patients (7382 in the development set that includes TRAIN_PRI, VAL_EVAL, VAL_PTEST, TRAIN_SEC, 637 in the test set) and 8585 images were included in the final analysis. In the development set, the mean age was 53 years old with a standard deviation (SD) of 19 years old. The youngest patient was 2 years old, and the oldest patient was 96 years old. Two thousand nine hundred eighty-eight (40%) patients were male, and 4394 (60%) patients were female. Four thousand eight hundred ninety-nine (66%) patients were white, 1694 (23%) patients were black, and 182 (3%) patients were Asian. In the test set, the mean age was 54 years old with SD of 19 years old. The youngest patient was 6 years old, and the oldest patient was 97 years old. Two hundred seventy-six (43%) patients were male, and 361 (57%) patients were female. Four hundred twenty-eight (67%) patients were white, 133 (21%) patients were white, and 24 (3%) patients were Asian (Table 1). There was no significant difference of age, sex, and race between the development and test set.
Table 1.
Demographic characteristics of the patients at baseline
| Characteristic | Development set (N = 7382) | Test set (N = 637) | p value |
|---|---|---|---|
| Mean age ± SDa year (range) | 53 ± 19 (2 – 96) | 54 ± 19 (6 – 97) | 0.200 |
| Male no. (%) − female no. (%) | 2988 (40%) – 4394 (60%) | 276 (43%) – 361 (57%) | 0.173 |
| Race or ethnic group no. (%) | 0.326 | ||
| White | 4899 (66%) | 428 (67%) | |
| Black | 1694 (23%) | 133 (21%) | |
| Asian | 182 (3%) | 22 (3%) | |
| Otherb | 607 (8%) | 54 (9%) |
aSD standard deviation
bCategory “Other” includes American Indian or Alaska Native, Native Hawaiian or other Pacific Islander, not reported/unavailable, not Hispanic or Latino
TRAIN_PRI contained 854 cases, with 198 (23%) normal cases. VAL_EVAL and VAL_PTEST contained 208 cases each, with 41 (20%) and 44 (21%) normal cases, respectively. TRAIN_SEC contained 6656 cases, with 1867 (28%) cases being pseudo-labeled normal. TEST contained 659 cases, with 151 (23%) normal cases (Table 2). The ratio of manually labeled data to pseudo-labeled data used in the development set was approximately 1:5.2.
Table 2.
Label distributions of images in January to February, including TRAIN_PRI, VAL_EVAL, and VAL_PTEST
| Dataset | Normal no. (%) | Abnormal no. (%) | Arthroplasty no. (%) | Total no. |
|---|---|---|---|---|
| TRAIN_PRI | 198 (23%) | 589 (69%) | 67 (8%) | 854 |
| VAL_EVAL | 41 (20%) | 152 (73%) | 15 (7%) | 208 |
| VAL_PTEST | 44 (21%) | 142 (68%) | 22 (11%) | 208 |
| TRAIN_SECa | 1867 (28%) | 4282 (64%) | 507 (8%) | 6656 |
| TEST | 151 (23%) | 457 (69%) | 51 (8%) | 659 |
aLabels in TRAIN_SEC were pseudo-labeled by the NLP model
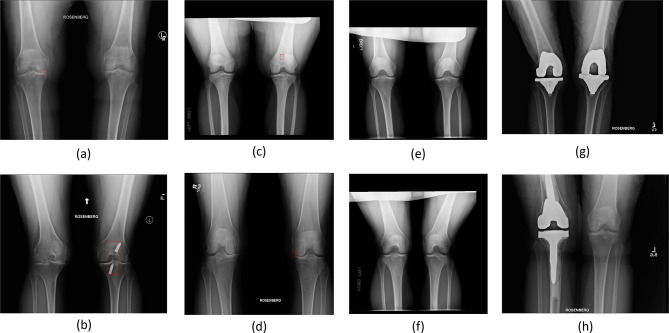
We provided typical examples of normal, abnormal, and arthroplasty radiographs in Fig. 2. We showed two examples of abnormal images with clear visibility of degenerative changes (Fig. 2a) and orthopedic hardware (Fig. 2b). We also recognized that some of the abnormal categories, such as fracture (Fig. 2c) and lucency (Fig. 2d), were visually closer to normal on radiographs. Figure 2e, f show two examples of normal images. Figure 2g, h show two examples of arthroplasty images.
Fig. 2.
Two examples of “easy abnormal” images. a, b An example of degenerative changes in the joint space. c, d The clear evidence of non-arthroplasty orthopedic hardware (nails) implanted in the knee. Our expert labeled the regions of abnormal categories in a–d on the images in a red bounding box. e, f Two examples of normal images. g, h Two examples of images with clear evidence of arthroplasty hardware
Test Results of BLPA Image Classification
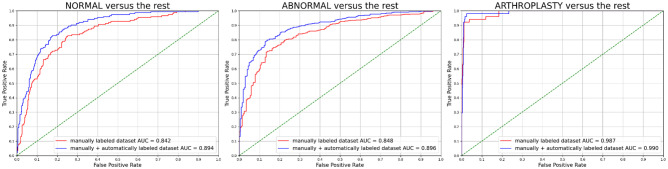
The NLP model had the best performance of WA-AUC = 0.995 (AUC_normal = 0.993, AUC_abnormal = 0.996, AUC_arthroplasty = 1.000) on VAL_EVAL, indicating near-perfect quality of pseudo labels. Table 3 shows the results on TEST using only manually labeled data vs. manually and automatically labeled data. When training with additional pseudo-labeled data, all reported metrics were higher (normal AUC normal + 0.052, abnormal AUC + 0.048, arthroplasty AUC + 0.003, WA-AUC + 0.046) than training with only manual-labeled data. The model almost perfectly predicted images with evidence of arthroplasty hardware in both specifications (manual-labeled arthroplasty AUC = 0.987, manual + pseudo-labeled arthroplasty AUC = 0.990). For each class, we plotted one class versus rest ROC curves in Fig. 3. Statistical tests showed that there is significant improvement in normal AUC (p value < 0.002), abnormal AUC (p value < 0.001), and WA-AUC (p value = 0.001).
Table 3.
Comparison of performance on TEST using different data based on the configuration selection on VAL_PTEST. AUC for each class versus rest and WA-AUC are provided
| Only manually labeled data (TRAIN_PRI) | Manually and pseudo- labeled data (TRAIN_PRI + pseudo-labeled TRAIN_SEC) | p value | |
|---|---|---|---|
| AUC normala | 0.842 | 0.894 | < 0.002 |
| AUC abnormala | 0.848 | 0.896 | < 0.001 |
| AUC arthroplasty | 0.987 | 0.990 | 0.4006 |
| WA-AUCa | 0.857 | 0.903 | 0.001 |
aSignificantly improved when training with manually and pseudo-labeled data compared to manually labeled data only
Fig. 3.
ROC plots on TEST of the best specification in each tested model using different data: red: only manually labeled data (only TRAIN_PRI, WA-AUC = 0.857) and blue: manually and automatically labeled data (TRAIN_PRI + pseudo-labeled TRAIN_SEC, WA-AUC = 0.903). DeLong tests provide p values for each class: p value_normal < 0.002, p value_abnormal < 0.001, p value_arthroplasty = 0.4006
Discussion
In this study, we proposed a DL framework that improves the baseline image classification performance by introducing a large volume of unlabeled data. A state-of-the-art NLP model, trained on a small set of labeled reports, served as an automated labeler to provide accurate pseudo-labels of unlabeled reports and their corresponding images. By augmenting the training size by approximately eight times, we trained image classification models with additional pseudo-labeled images and achieved significantly better classification results on normal (AUC + 0.052, p value_normal < 0.002) and abnormal (AUC + 0.048, p value_abnormal < 0.001) images, and overall performance was improved (WA-AUC + 0.046, p value = 0.001). Although no significant improvements were found for arthroplasty images, models had already achieved high performance in both settings (AUC = 0.987 using only manually labeled cases; AUC = 0.990 using additional pseudo-labeled cases) and improved only marginally when we added pseudo-labeled cases (AUC + 0.003).
An alternative approach to increase the training size for better performance is to adopt traditional data augmentation technique, such as affine augmentation. However, as opposed to our proposed NLP approach, using such method generates artificial imaging data based on the training samples, such as rotation or scaling, and thus did not expand the breadth of the training data. To improve the classification performance, our approach adopted the idea of additive augmentation when training with additional pseudo-labeled images. Different from online augmentation methods in which the transformations were randomly applied to the images in the mini-batches during training, we prepared the augmented pseudo-labeled images beforehand and then curated the expanded dataset for final training. Similarly in literature, Fabi et al. has also shown the benefits of additive augmentation in improving the classification of abnormal knee radiographs by adding synthetic images to the baseline [34]. Therefore, considering a wide range of data augmentation methods available in literature, we recommend future work to thoroughly explore the effects of data augmentation by integrating various transformation techniques into our current experiments and designing appropriate online or offline training pipelines.
Our findings demonstrate important practical values. First, by applying the DL framework, our approach requires only 16% of the development set to be manually annotated. Such low percentage of required annotations significantly reduced the burden of human annotations and improved the inherit limitations of human interpretation, such as observer variability, time constraints, cost, and bias. Second, similar to the literature in which BioBERT and EfficientNet models have been proven to be effective feature extraction backbones in general radiology text [35, 36] and image classification [37–39] tasks, our results demonstrated the power and potentials of DL tools in knee radiology. When our framework is adopted to other datasets in future research, the DL backbones can be easily substituted with other tools for optimal usage. Third, our dataset split plan reflected a real-world scenario where DL developers trained models using retrospective data and validated performance on prospective data. Hence, we believe that our approach can be adopted to provide reliable assistance in the clinical applications related to the identification and diagnosis of knee abnormalities using knee radiographs.
Pseudo-labeling is an important feature of our framework. The idea of pseudo-labeling is fundamental in conventional semi-supervised learning (SSL) tasks [40–43]. Such tasks have also been proven to be effective in predicting knee abnormalities [44, 45]. SSL-based algorithms require a small amount of labeled input, train the whole model with a joint input of labeled and unlabeled cases with, and learn the pseudo labels for unlabeled cases accordingly. Our approach shares the same goal with SSL tasks: improve the classification performance by utilizing additional pseudo-labeled cases generated from a large unlabeled dataset. However, unlike SSL tasks, first, we utilized NLP to learn knowledge from the manually labeled reports to automatically pseudo-label the unlabeled reports. Thanks to the inherent association between radiology reports and images, we treated report labels as image labels. Second, in a separate model, we trained both labeled and pseudo-labeled images for final prediction. We acknowledge that such association might be vulnerable in practice. Thus, future work can involve SSL-based models to learn pseudo-labels on the same input source, without considering the inherent link between different types of input sources.
Another important feature of our framework is to exploit both radiology reports and images in one workflow for image classification using a large volume of unlabeled radiology data. In the literature, multiple studies have shown the effectiveness of including this feature for curating large datasets of radiographs. For example, Jeremy et al. curated the CheXpert dataset and investigated different approaches of incorporating uncertainty labels in training to predict lung pathologies [46]. Xiaosong et al. curated the ChestX-ray8 dataset aided by a concept detection tool and proposed an image classification model to detect and locate thoracic disease [47]. In both studies, curation of large datasets was aided by an automated NLP labeler. The curated data were then tested feasible for downstream image classification tasks. While our proposed framework shared such feature, a key difference is that we utilized a state-of-the-art DL-based NLP model as the automated labeler to generate high-quality pseudo-labels for knee radiology reports. This demonstrates great potentials in applications of our framework to curate large datasets of knee radiographs. Future work can focus on applying our framework to significantly larger datasets of unlabeled knee radiographs and further validated for downstream tasks.
Limitations
We acknowledge several limitations in this study. First, our proposed framework was trained and validated using data in one health system without being externally validated. Future work could apply our framework to multicenter data and test the generalizability of our approach. Second, annotation rules were developed based on the experience of a single expert. As a result, our defined rules may not comprehensively reflect all characteristics on knee radiographs. Third, label noises may exist in pseudo-labeled cases that were generated by the trained NLP model. However, we believe that the image classification model can tolerate such label noise because of the high-quality pseudo-labels. Fourth, we determined the selection criteria of BLPA view by manually reviewing a small number of knee radiographs in the dataset. Future work could involve a more rigorous process in consolidating such criteria. Fifth, our study has inherent biases in that a significantly leading proportion of patients in the study are white. While our study design was focused on evaluations of knee abnormalities, future work can investigate the biases of algorithms based on race. Lastly, our utilization of BLPA view for this study does not account for the differences in joint space loss that can be seen with variable flexion of the knee which has been shown in other studies [8].
Conclusion
By harnessing DL powers for annotating a large volume of unlabeled reports using only a small number of labeled data, we have shown the feasibility of our proposed approach to improve image classification performance for knee radiographic diagnosis, without the labor of interpreting an overwhelming number of images. As a result, the proposed approach minimizes the inherent limitations of human annotations and can be potentially useful for curating large knee datasets. Our focus on a commonly used imaging modality of the knee will allow for wide utilization of the application in knee radiology to improve patient care for knee abnormality diagnosis.
Data Availability
Portions of data may be shared on reasonable request to the corresponding author, though IRB restrictions may limit sharing in some instances. We shared our code on GitHub: https://github.com/mazurowski-lab/knee_clf.
Declarations
Ethics Approval
This study was HIPAA-compliant and approved by the institutional review board (IRB) of Duke University.
Consent to Participate/For Publication
Our IRB waived the requirement for informed consent given the retrospective nature of the research and the minimal risk to subjects.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kansagra AP, Yu JPJ, Chatterjee AR, et al. Big Data and the Future of Radiology Informatics. Acad Radiol. 2016;23(1):30–42. doi: 10.1016/j.acra.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Aiello M, Cavaliere C, D’Albore A, Salvatore M. The Challenges of Diagnostic Imaging in the Era of Big Data. J Clin Med. 2019;8(3). 10.3390/jcm8030316 [DOI] [PMC free article] [PubMed]
- 3.Murdoch TB, Detsky AS. The Inevitable Application of Big Data to Health Care. JAMA. 2013;309(13):1351–1352. doi: 10.1001/jama.2013.393. [DOI] [PubMed] [Google Scholar]
- 4.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18(8):500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ueda D, Shimazaki A, Miki Y. Technical and clinical overview of deep learning in radiology. Jpn J Radiol. 2019;37(1):15–33. doi: 10.1007/s11604-018-0795-3. [DOI] [PubMed] [Google Scholar]
- 6.McBee MP, Awan OA, Colucci AT, et al. Deep Learning in Radiology. Acad Radiol. 2018;25(11):1472–1480. doi: 10.1016/j.acra.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 7.Saba L, Biswas M, Kuppili V, et al. The present and future of deep learning in radiology. Eur J Radiol. 2019;114:14–24. doi: 10.1016/j.ejrad.2019.02.038. [DOI] [PubMed] [Google Scholar]
- 8.Roemer FW, Demehri S, Omoumi P, et al. State of the Art: Imaging of Osteoarthritis—Revisited 2020. Radiology. 2020;296(1):5–21. doi: 10.1148/radiol.2020192498. [DOI] [PubMed] [Google Scholar]
- 9.Prieto-Alhambra D, Judge A, Javaid MK, Cooper C, Diez-Perez A, Arden NK. Incidence and risk factors for clinically diagnosed knee, hip and hand osteoarthritis: influences of age, gender and osteoarthritis affecting other joints. Ann Rheum Dis. 2014;73(9):1659–1664. doi: 10.1136/annrheumdis-2013-203355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turkiewicz A, Petersson IF, Björk J, et al. Current and future impact of osteoarthritis on health care: a population-based study with projections to year 2032. Osteoarthritis Cartilage. 2014;22(11):1826–1832. doi: 10.1016/j.joca.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Dunn R, Greenhouse J, James D, Ohlssen D, Mesenbrink P. Risk Scoring for Time to End-Stage Knee Osteoarthritis: Data from the Osteoarthritis Initiative. Osteoarthritis Cartilage. 2020;28. 10.1016/j.joca.2019.12.013 [DOI] [PMC free article] [PubMed]
- 12.Koplas M, Schils J, Sundaram M. The painful knee: Choosing the right imaging test. Cleve Clin J Med. 2008;75(5):377. http://www.ccjm.org/content/75/5/377.abstract [DOI] [PubMed]
- 13.Wang K, Kim HA, Felson DT, et al. Radiographic Knee Osteoarthritis and Knee Pain: Cross-sectional study from Five Different Racial/Ethnic Populations. Sci Rep. 2018;8(1):1364. doi: 10.1038/s41598-018-19470-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen TL, Engh GA. Radiographic assessment of knee alignment after total knee arthroplasty. J Arthroplasty. 1988;3(1):67–72. doi: 10.1016/S0883-5403(88)80054-8. [DOI] [PubMed] [Google Scholar]
- 15.Gu S, Kuriyama S, Nakamura S, Nishitani K, Ito H, Matsuda S. Underhang of the tibial component increases tibial bone resorption after total knee arthroplasty. Knee Surgery, Sports Traumatology, Arthroscopy. 2019;27(4):1270–1279. doi: 10.1007/s00167-018-5309-4. [DOI] [PubMed] [Google Scholar]
- 16.Meneghini RM, Mont MA, Backstein DB, Bourne RB, Dennis DA, Scuderi GR. Development of a Modern Knee Society Radiographic Evaluation System and Methodology for Total Knee Arthroplasty. J Arthroplasty. 2015;30(12):2311–2314. doi: 10.1016/j.arth.2015.05.049. [DOI] [PubMed] [Google Scholar]
- 17.Tiulpin A, Thevenot J, Rahtu E, Lehenkari P, Saarakkala S. Automatic Knee Osteoarthritis Diagnosis from Plain Radiographs: A Deep Learning-Based Approach. Sci Rep. 2018;8(1):1727. doi: 10.1038/s41598-018-20132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung K, Zhang B, Tan J, et al. Prediction of Total Knee Replacement and Diagnosis of Osteoarthritis by Using Deep Learning on Knee Radiographs: Data from the Osteoarthritis Initiative. Radiology. 2020;296(3):584–593. doi: 10.1148/radiol.2020192091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antony J, McGuinness K, O’Connor N, Moran K. Quantifying Radiographic Knee Osteoarthritis Severity using Deep Convolutional Neural Networks. In: ; 2016. 10.1109/ICPR.2016.7899799
- 20.Górriz M, Antony J, McGuinness K, Giró-i-Nieto X, O’Connor NE. Assessing Knee OA Severity with CNN attention-based end-to-end architectures. In: Cardoso MJ, Feragen A, Glocker B, et al., eds. Proceedings of The 2nd International Conference on Medical Imaging with Deep Learning. Vol 102. Proceedings of Machine Learning Research. PMLR; 2019:197–214. https://proceedings.mlr.press/v102/gorriz19a.html
- 21.Wahyuningrum RT, Anifah L, Eddy Purnama IK, Hery Purnomo M. A New Approach to Classify Knee Osteoarthritis Severity from Radiographic Images based on CNN-LSTM Method. In: 2019 IEEE 10th International Conference on Awareness Science and Technology (ICAST). ; 2019:1–6. 10.1109/ICAwST.2019.8923284
- 22.Chen P, Gao L, Shi X, Allen K, Yang L. Fully automatic knee osteoarthritis severity grading using deep neural networks with a novel ordinal loss. Computerized Medical Imaging and Graphics. 2019;75:84–92. doi: 10.1016/j.compmedimag.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bien N, Rajpurkar P, Ball RL, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLoS Med. 2018;15(11):e1002699-. 10.1371/journal.pmed.1002699 [DOI] [PMC free article] [PubMed]
- 24.Ramkumar PN, Karnuta JM, Navarro SM, et al. Deep Learning Preoperatively Predicts Value Metrics for Primary Total Knee Arthroplasty: Development and Validation of an Artificial Neural Network Model. J Arthroplasty. 2019;34(10):2220–2227.e1. doi: 10.1016/j.arth.2019.05.034. [DOI] [PubMed] [Google Scholar]
- 25.Yi PH, Wei J, Kim TK, et al. Automated detection & classification of knee arthroplasty using deep learning. Knee. 2020;27(2):535–542. doi: 10.1016/j.knee.2019.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Marcovici PA, Taylor GA. JOURNAL CLUB: Structured Radiology Reports Are More Complete and More Effective Than Unstructured Reports. American Journal of Roentgenology. 2014;203(6):1265–1271. doi: 10.2214/AJR.14.12636. [DOI] [PubMed] [Google Scholar]
- 27.Brady AP. Error and discrepancy in radiology: inevitable or avoidable? Insights Imaging. 2017;8(1):171–182. doi: 10.1007/s13244-016-0534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braun HJ, Gold GE. Diagnosis of osteoarthritis: Imaging. Bone. 2012;51(2):278–288. doi: 10.1016/j.bone.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohn MD, Sassoon AA, Fernando ND. Classifications in Brief: Kellgren-Lawrence Classification of Osteoarthritis. Clin Orthop Relat Res. 2016;474(8). https://journals.lww.com/clinorthop/Fulltext/2016/08000/Classifications_in_Brief__Kellgren_Lawrence.28.aspx [DOI] [PMC free article] [PubMed]
- 30.Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234–1240. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolf T, Debut L, Sanh V, et al. HuggingFace’s Transformers: State-of-the-art Natural Language Processing. ArXiv. 2019;abs/1910.03771.
- 32.Tan M, Le Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In: Chaudhuri K, Salakhutdinov R, eds. Proceedings of the 36th International Conference on Machine Learning. Vol 97. Proceedings of Machine Learning Research. PMLR; 2019:6105–6114. https://proceedings.mlr.press/v97/tan19a.html
- 33.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics. 1988;44(3):837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 34.Prezja F, Paloneva J, Pölönen I, Niinimäki E, Äyrämö S. DeepFake knee osteoarthritis X-rays from generative adversarial neural networks deceive medical experts and offer augmentation potential to automatic classification. Sci Rep. 2022;12(1):18573. doi: 10.1038/s41598-022-23081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood DA, Lynch J, Kafiabadi S, et al. Automated Labelling using an Attention model for Radiology reports of MRI scans (ALARM). In: Arbel T, ben Ayed I, de Bruijne M, Descoteaux M, Lombaert H, Pal C, eds. Proceedings of the Third Conference on Medical Imaging with Deep Learning. Vol 121. Proceedings of Machine Learning Research. PMLR; 2020:811–826. https://proceedings.mlr.press/v121/wood20a.html
- 36.Smit A, Jain S, Rajpurkar P, Pareek A, Ng A, Lungren M. Combining Automatic Labelers and Expert Annotations for Accurate Radiology Report Labeling Using BERT. In: Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP). Association for Computational Linguistics; 2020:1500–1519. 10.18653/v1/2020.emnlp-main.117
- 37.Marques G, Agarwal D, de la Torre Díez I. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl Soft Comput. 2020;96:106691. 10.1016/j.asoc.2020.106691 [DOI] [PMC free article] [PubMed]
- 38.Nayak DR, Padhy N, Mallick PK, Zymbler M, Kumar S. Brain Tumor Classification Using Dense Efficient-Net. Axioms. 2022;11(1). 10.3390/axioms11010034
- 39.Oloko-Oba M, Viriri S. Ensemble of EfficientNets for the Diagnosis of Tuberculosis. lo Bosco G, ed. Comput Intell Neurosci. 2021;2021:9790894. 10.1155/2021/9790894 [DOI] [PMC free article] [PubMed]
- 40.Lee DH. Pseudo-Label : The Simple and Efficient Semi-Supervised Learning Method for Deep Neural Networks. ICML 2013 Workshop : Challenges in Representation Learning (WREPL). Published online July 10, 2013.
- 41.Berthelot D, Carlini N, Goodfellow I, Papernot N, Oliver A, Raffel CA. MixMatch: A Holistic Approach to Semi-Supervised Learning. In: Wallach H, Larochelle H, Beygelzimer A, d Alché-Buc F, Fox E, Garnett R, eds. Advances in Neural Information Processing Systems. Vol 32. Curran Associates, Inc.; 2019. https://proceedings.neurips.cc/paper/2019/file/1cd138d0499a68f4bb72bee04bbec2d7-Paper.pdf
- 42.Laine S, Aila T. Temporal Ensembling for Semi-Supervised Learning. ArXiv. 2016;abs/1610.02242.
- 43.Verma V, Lamb A, Kannala J, Bengio Y, Lopez-Paz D. Interpolation Consistency Training for Semi-Supervised Learning. Neural Netw. 2019;145:90–106. doi: 10.1016/j.neunet.2021.10.008. [DOI] [PubMed] [Google Scholar]
- 44.Huo J, Ouyang X, Si L, et al. Automatic Grading Assessments for Knee MRI Cartilage Defects via Self-ensembling Semi-supervised Learning with Dual-Consistency. Med Image Anal. 2022;80:102508. 10.1016/j.media.2022.102508 [DOI] [PubMed]
- 45.Nguyen HH, Saarakkala S, Blaschko MB, Tiulpin A. Semixup: In- and Out-of-Manifold Regularization for Deep Semi-Supervised Knee Osteoarthritis Severity Grading From Plain Radiographs. IEEE Trans Med Imaging. 2020;39(12):4346–4356. doi: 10.1109/TMI.2020.3017007. [DOI] [PubMed] [Google Scholar]
- 46.Irvin J, Rajpurkar P, Ko M, et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. Proceedings of the AAAI Conference on Artificial Intelligence. 2019;33:590–597. doi: 10.1609/aaai.v33i01.3301590. [DOI] [Google Scholar]
- 47.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Published online 2017:3462–3471.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Portions of data may be shared on reasonable request to the corresponding author, though IRB restrictions may limit sharing in some instances. We shared our code on GitHub: https://github.com/mazurowski-lab/knee_clf.