Abstract
With the advances in endoscopic technologies and artificial intelligence, a large number of endoscopic imaging datasets have been made public to researchers around the world. This study aims to review and introduce these datasets. An extensive literature search was conducted to identify appropriate datasets in PubMed, and other targeted searches were conducted in GitHub, Kaggle, and Simula to identify datasets directly. We provided a brief introduction to each dataset and evaluated the characteristics of the datasets included. Moreover, two national datasets in progress were discussed. A total of 40 datasets of endoscopic images were included, of which 34 were accessible for use. Basic and detailed information on each dataset was reported. Of all the datasets, 16 focus on polyps, and 6 focus on small bowel lesions. Most datasets (n = 16) were constructed by colonoscopy only, followed by normal gastrointestinal endoscopy and capsule endoscopy (n = 9). This review may facilitate the usage of public dataset resources in endoscopic research.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10278-023-00844-7.
Keywords: Datasets, Endoscopy, Artificial intelligence, Review
Introduction
In recent years, gastrointestinal (GI) endoscopy has developed rapidly. Due to its minimal invasiveness, endoscopy has become a primary diagnostic tool for early GI lesions [1]. However, the diagnostic rate of various diseases has not increased significantly with the development of endoscopy. The miss rate for colorectal polyps is reportedly as high as 25% [2]. The endoscopic miss rate of upper gastrointestinal cancers is more than 5% [3]. With the increasing number of GI endoscopies, imaging interpretation makes endoscopists tired and indirectly influences diagnostic accuracy and efficiency. Taking capsule endoscopy (CE) as an example, although CE is considered the standard criterion for investigating small bowel (SB) lesions [4], SB-CE reading is tedious (30 to 60 min per video) and time-consuming (approximately 50,000 frames per video) [5]. This increases the risk of missed diagnosis during the reading process by endoscopists. The limitation of endoscopy alone provides an opportunity for objective diagnostic techniques to improve the detection rate of gastrointestinal lesions.
In the past decade, the development of artificial intelligence (AI)-based technologies in medicine has been advancing rapidly [6]. In the field of GI endoscopy, various AI applications have been proposed, especially with the use of deep learning (DL) technology, including convolutional neural networks (CNNs). Rapid advances in AI-based technology have improved diagnosis accuracy, but they have also increased the need for endoscopists to be familiar with AI as well as high-quality and great-quantity endoscopic images. However, obtaining a large sample size can be time-consuming and expensive. Moreover, due to the lack of manually labeled data and legal restrictions, it is difficult to create a dataset. Therefore, publicly available datasets have gained increasing popularity and may overcome the difficulties described above. Compared with the clinical dataset, the endoscopy dataset is defined as a dataset of high-definition endoscopic videos or images from the esophagus to the cecum used in endoscopic research in a peer-reviewed journal, and is not restricted by visit duration or encounter setting. The images or videos from the endoscopy dataset are used for different tasks, such as segmentation, classification and detection. However, because of barriers to access and usability, such as governance and cost barriers, endoscopy datasets need specialized websites, which are hard to find. Currently, there is no centralized directory of endoscopy datasets and, therefore, little knowledge regarding available endoscopic imaging data.
With the significant advances in endoscopic technologies and AI, a large number of endoscopic imaging datasets have been made public to researchers worldwide. This review aims to collect these datasets and provide a guide for researchers using them.
Method
Eligibility Criteria
The search was restricted to humans and English. Datasets containing endoscopic videos and images were eligible for inclusion since 2010. No datasets were excluded due to the age, sex, or ethnicity of the patients. Datasets containing nonendoscopic images, videos, or numerical-only data were excluded.
Retrieval of GI Endoscopy Dataset
PubMed was initially searched for relevant publications. Two independent authors used the following search terms to perform a systematic search of the endoscopy dataset: “endoscop*,” “gastrointestinal endoscop*,” “colonoscop*,” “capsule endoscop*,” “database*,” and “dataset*,” and then we attempted to access these datasets at the source. After the initial search, we performed a second search of Kaggle (https://www.kaggle.com/), GitHub (https://github.com/), and Simula (https://datasets.simula.no/). We also manually reviewed the references of the articles identified from the initial search. The last search was performed on December 31, 2022. This systematic search was performed under the supervision of a medical doctor. Endnote, which is a specialized software for managing bibliographies, was used for recording.
Dataset Selection
Two authors independently screened search results in duplicate to identify the name and source of any relevant study. Where the status of availability was unclear, we checked these datasets and attempted to access their source. Duplicates were excluded, and obviously irrelevant studies were removed based on title and abstract. Then, full-text screening was performed. Search results from Kaggle, GitHub, and Simula were also screened by two authors to identify relevant datasets directly. Discrepancies were resolved through negotiation. In our reviews, dataset accessibility was defined as three types: (1) not available anymore because of unpredictable reasons, such as no response to request; (2) available publicly: there are no requirements or restrictions; and (3) available only by request or registration: there are minimum requirements (submission for personal information) or formal agreement.
Extraction of Dataset Characteristics
Two independent authors recorded the characteristics of each dataset, including the direct link to the dataset, publish year, accessibility, country, content (imaging number, data type, and endoscopic type), and other features. Two other authors reviewed the characteristics for correctness and resolved inconsistencies.
Results
Datasets Identified from the Literature and Targeted Search
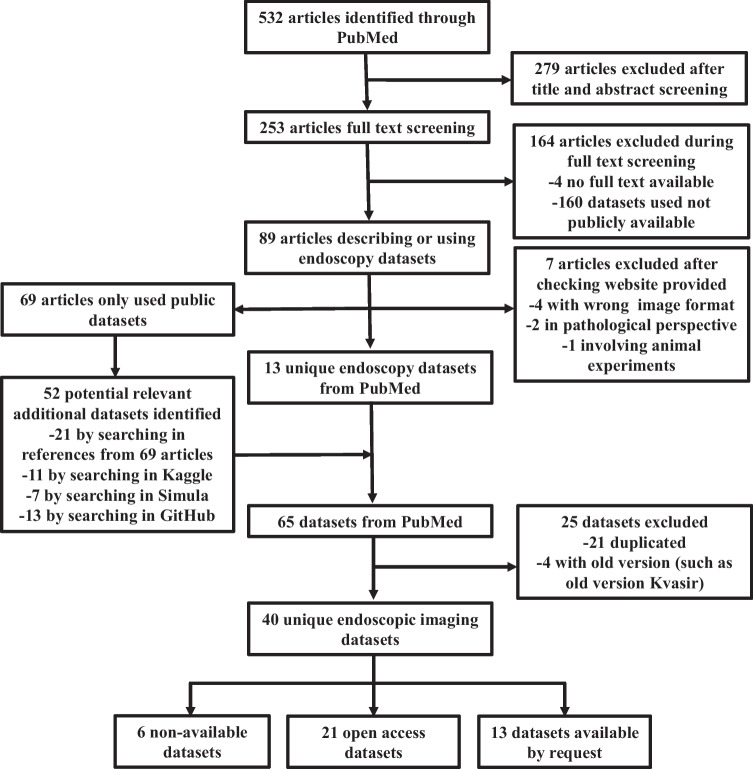
The dataset search and selection process flowchart is shown in Fig. 1. A total of 532 articles were identified from the initial PubMed search, of which 279 were excluded after screening the title and abstract. A total of 253 were assessed to be eligible for full-text review. Of these, 13 unique datasets were identified, and 69 potential articles were chosen for further review. The same datasets were often referenced by multiple articles. The second search from Kaggle, Simula, GitHub, and references identified 52 potential endoscopic imaging datasets. After combining the results, 21 duplicate datasets were excluded, and 4 datasets with old versions were excluded. Finally, 40 endoscopic imaging datasets were identified and included for further data extraction. The basic information of these datasets is summarized in Table 1. The detailed characteristics are summarized in Table 2. Moreover, the application of these datasets in the establishment of the AI model is presented in Supplementary Table 2.
Fig. 1.
Flowchart of literature search
Table 1.
Basic information of endoscopic imaging datasets
| Name | Publish year | Geographical distribution | Accessibility | Data type | Endoscopic type | Refs |
|---|---|---|---|---|---|---|
| Endoscopy datasets of polyps (n = 16) | ||||||
| CVC-ColonDB | 2013 | Spain | Unavailablea | Colon polyps | Colonoscopy | [7] |
| CVC-ClinicDB (CVC-612) | 2015 | Spain | https://www.kaggle.com/datasets/balraj98/cvcclinicdb?select=metadata.csv; https://polyp.grand-challenge.org/CVCClinicDB/b | Colon polyps | Colonoscopy | [8] |
| ETIS-Larib Polyp DB | 2014 | France | Unavailablea | Polyps | Capsule endoscopy | [9] |
| ASU-Mayo | 2016 | America | https://polyp.grand-challenge.org/AsuMayo/c | Colon polyps | Colonoscopy | [10] |
| GI lesions in Regular Colonoscopy Dataset | 2016 | France | http://www.depeca.uah.es/colonoscopy_dataset/b | Colon polyps | Colonoscopy | [11] |
| EndoScene | 2016 | Secondary dataset | Unavailablea | Colon polyps | Colonoscopy | [12] |
| Kvasir SEG | 2019 | Secondary dataset | https://datasets.simula.no/kvasir-seg/b | Gastrointestinal polyps | Gastrointestinal endoscopy | [13] |
| NBIPolyp-Ucdb | 2019 | Portugal | http://www.mat.uc.pt/~isabelf/Polyp-UCdb/NBIPolyp-UCdb.htmlc | Colon polyps | Colonoscopy | [14] |
| WLPolyp-UCdb | 2019 | Portugal | http://www.mat.uc.pt/~isabelf/Polyp-UCdb/WLPolyp-UCdb.htmlc | Colon polyps | Colonoscopy | [15] |
| KUMC | 2020 | Secondary dataset | https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/FCBUORb | Colon polyps | Colonoscopy | [16] |
| SUN | 2020 | Japan | http://amed8k.sundatabase.org/c | Colon polyps | Colonoscopy | [17] |
| PICCOLO | 2020 | Spain | https://www.biobancovasco.org/en/Sample-and-data-catalog/Databases/PD178-PICCOLO-EN.htmlc | Colon polyps | Colonoscopy | [18] |
| CP-CHILD | 2020 | China | https://figshare.com/articles/dataset/CP-CHILD_zip/12554042b | Colon polyps | Colonoscopy | [19] |
| LD Polyp Video | 2021 | China | https://github.com/dashishi/LDPolypVideo-Benchmark; https://pan.baidu.com/s/1ri3lDBDl3v00TAXc6euzvb | Colon polyps | Colonoscopy | [20] |
| SUN-SEG | 2022 | Secondary dataset | https://github.com/GewelsJI/VPSb | Colon polyps | Colonoscopy | [21] |
| PolypGen | 2022 | Multi-sites | https://www.synapse.org/#!Synapse:syn26376615/wiki/613312b | Colon polyps | Colonoscopy | [22] |
| Endoscopy datasets of small bowel lesions (n = 6) | ||||||
| KID | 2017 | Online | https://mdss.uth.gr/datasets/endoscopy/kid/a | Small bowel lesions | Capsule endoscopy | [23] |
| CAD-CAP | 2020 | France | Unavailablea | Small bowel lesions | Capsule endoscopy | [24] |
| Kvasir-Capsule | 2021 | Norway | https://www.kaggle.com/datasets/manishkc06/the-kvasircapsule-dataset; https://osf.io/dv2ag/; https://github.com/simula/kvasir-capsuleb | Small bowel anatomical landmarks and lesions | Capsule endoscopy | [25] |
| Endoscopy Crohn’s Disease dataset | 2021 | China | https://www.frontiersin.org/articles/10.3389/fmolb.2021.614277/fullc | Crohn’s disease | Capsule endoscopy | [26] |
| CrohnIPI | 2021 | France | https://crohnipi.ls2n.fr/en/crohn-ipi-project/c | Crohn’s disease | Capsule endoscopy | [27] |
| The AICE Project | 2022 | Japan | https://www.kaggle.com/datasets/capsuleyolo/kyucapsule; https://osf.io/pf5nm/b | Small bowel lesions | Capsule endoscopy | d |
| Endoscopy datasets of gastro-esophageal lesions (n = 2) | ||||||
| IPCL | 2020 | Taiwan, China | https://github.com/luiscarlosgph/ipcl; https://www.synapse.org/#!Synapse:syn21636566/wiki/601346c | IPCL in esophageal | Esophageal endoscopy | [28] |
| IM and GA Benchmark | 2022 | China | https://github.com/fengcherenxi/LAGc | Stomach lesions | Gastroscopy | [29] |
| Endoscopy datasets of comprehensive GI detection (n = 5) | ||||||
| Kvasir | 2017 | Norway | https://www.kaggle.com/datasets/yasserhessein/the-kvasir-datasetb | Findings in gastrointestinal tract | Gastrointestinal endoscopy | [30] |
| Hyper Kvasir | 2020 | Norway | https://www.kaggle.com/datasets/kelkalot/the-hyper-kvasir-dataset; https://datasets.simula.no/hyper-kvasir; https://github.com/simula/hyper-kvasirb | Findings in gastrointestinal tract | Gastrointestinal endoscopy | [31] |
| Rhode Island | 2022 | America | https://springernature.figshare.com/collections/Rhode_island_gastroenterology_video_capsule_endoscopy_data_set/6071216/1; https://github.com/acharoen/Rhode-Island-GI-VCE-Technical-Validationb | Findings in gastrointestinal tract | Capsule endoscopy | [32] |
| WCE Curated Colon Disease | 2022 | Secondary dataset | https://www.kaggle.com/datasets/francismon/curated-colon-dataset-for-deep-learning; https://github.com/francismontalbo/mfurecnnb | Findings in gastrointestinal tract | Capsule endoscopy | [33] |
| ERS | 2022 | Poland | https://cvlab.eti.pg.gda.pl/publications/endoscopy-datasetc | Findings in gastrointestinal tract | Multi-tissue endoscopy | [34] |
| Atlases of GI endoscopy (n = 4)e | ||||||
| Gastrolab | NA | Online | http://www.gastrolab.net/index.htmb | Gastrointestinal atlas | Gastrointestinal endoscopy | [35] |
| WEO Clinical Endoscopy Atlas | NA | Online | http://www.endoatlas.org/index.phpb | Gastrointestinal atlas | Gastrointestinal endoscopy | [36] |
| Atlas of Gastrointestinal Endoscopy | NA | Online | http://www.endoatlas.com/atlas_1.htmlb | Gastrointestinal atlas | Gastrointestinal endoscopy | [37] |
| El Salvador atlas | NA | Online | http://www.gastrointestinalatlas.com/index.htmlb | Gastrointestinal atlas | Gastrointestinal endoscopy | [38] |
| Others (n = 7) | ||||||
| GIANA 2017 | 2017 | Multi-sites | https://endovissub2017-giana.grand-challenge.org/c | Angiodysplasia and polyps | Multi-tissue endoscopy (Colonoscopy and Capsule endoscopy) | [39] |
| Nerthus | 2017 | Norway | https://www.kaggle.com/datasets/waltervanhuissteden/the-nerthus-dataset; https://datasets.simula.no/nerthus/b | Bowel cleanliness | Colonoscopy | [40] |
| GIANA 2018 | 2018 | Multi-sites | Unavailablea | Colon polyps and small bowel lesions | Multi-tissue endoscopy (colonoscopy and capsule endoscopy) | [41] |
| EAD 2019 | 2019 | Multi-sites | https://ead2019.grand-challenge.org/; https://github.com/sharib-vision/EAD2019c | Endoscopy artifacts | Multi-tissue endoscopy | [42] |
| Cho et al. 2019 | 2019 | Korea | https://figshare.com/articles/dataset/Colonoscopy_images/7937336/1b | Colon polyps | Colonoscopy | [43] |
| EDD 2020 | 2021 | Multi-sites | https://edd2020.grand-challenge.org/Home/, https://github.com/sharib-vision/EDD2020c | Five lesions in gastrointestinal tract | Gastrointestinal endoscopy | [44] |
| Kvasir-Instrument | 2021 | Norway | https://datasets.simula.no/kvasir-instrument/; https://github.com/DebeshJha/Kvasir-Instrumentb | Diagnostic and therapeutic tool | Gastrointestinal endoscopy | [45] |
GI gastrointestinal, IPCL intrapapillary capillary loops
aNot available anymore
bAvailable publicly from the web address provided
cAvailable only by request or registration
dPractical Artificial Intelligence Model and Data Repository of Small intestine Capsule Endoscopy: The AICE Project, the article is under submission
eIt is more of a medical atlas or database for education with several findings in the GI tract than a dataset for machine learning or deep learning
Table 2.
Detailed characteristics of endoscopic imaging datasets
| Name | Findings | Clinical detailsa |
|---|---|---|
| Endoscopy datasets of polyps (n = 16) | ||
| CVC-ColonDB | Polyps | No |
| CVC-ClinicDB (CVC-612) | Polyps | Yes |
| ETIS-Larib Polyp DB | Polyps and normal mucosa | No |
| ASU-Mayo | Polyps and normal mucosa | No |
| GI lesions in Regular Colonoscopy Dataset | Serrated adenomas, hyperplastic polyps and adenomas | Yes |
| EndoScene | Polyps | No |
| Kvasir SEG | Polyps | No |
| NBIPolyp-Ucdb | Adenomas and hyperplastic polyps | No |
| WLPolyp-UCdb | Polyps and normal mucosa | No |
| KUMC | Adenomas and hyperplastic polyps | No |
| SUN | Polyps | Yes |
| PICCOLO | Polyps | Yes |
| CP-CHILD | Polyps and normal mucosa | No |
| LD Polyp Video | Polyps | No |
| SUN-SEG | Polyps | Yes |
| PolypGen | Polyps and normal mucosa | Yes |
| Endoscopy datasets of small bowel lesions (n = 6) | ||
| KID | SB lesions (vascular lesions, inflammatory lesions, lymphangiectasias, polypoid lesions) and normal mucosa | No |
| CAD-CAP | SB lesions (vascular lesions, fresh blood, ulcer-inflammatory lesions, and other lesions) and normal mucosa | No |
| Kvasir-Capsule | Anatomy (pylorus, ampulla of Vater, ileocecal valve), SB lesions (mucosal atrophy, lymphangiectasia, erythema, angiectasia, blood-fresh, blood-hematin, erosion, ulcer, polyp, foreign body) and normal mucosa | No |
| Endoscopy Crohn’s Disease dataset | Clearness degree classification in Crohn’s tract (clearness, blur and invisible) | Yes |
| CrohnIPI | Crohn’s lesions (erythema, edema, aphthoid ulceration, 3–10 mm ulceration, > 10 mm ulceration, stenosis) and normal mucosa | No |
| The AICE Project | SB lesions (angiodysplasia, erosion, stenosis, lymphangiectasia, lymph follicle, polyp-like lesions, submucosal tumor, bleeding, diverticulum, erythema, foreign body, vein lesions) and normal mucosa | No |
| Endoscopy datasets of gastro-esophageal lesions (n = 2) | ||
| IPCL | Four classes of abnormal esophageal IPCL (A, B1, B2, B3) and normal mucosa | No |
| IM and GA Benchmark | GA, IM and normal images in five gastric lesion location (antrum, angle, cardia, fundus and body) by WLI and LCI | No |
| Endoscopy datasets of comprehensive GI detection (n = 5) | ||
| Kvasir | 8 classes in GI tract (Z-line, pylorus, cecum, esophagitis, polyps, ulcerative colitis, “dyed and lifted polyp” and “dyed resection margins”) | No |
| Hyper Kvasir | 23 classes of GI findings including anatomical landmarks, quality of mucosal views, pathological findings, therapeutic interventions and so onc | No |
| Rhode Island | Findings in four anatomical organs (esophagus, stomach, small bowel, colon) | No |
| WCE Curated Colon Disease | 3 classes of GI lesions (ulcer, polyps, esophagitis) and normal mucosa | No |
| ERS | 34 classes of colonoscopy findings, 70 classes of upper endoscopy findings and 7 classes of healthy tissues, 2 classes of blood and 10 classes of imaging qualityc | Yes |
| Atlases of GI endoscopy (n = 4) | ||
| Gastrolab | Findings in GI tract (14 anatomical structures, 7 classes of GI lesions and 4 others)c | No |
| WEO Clinical Endoscopy Atlas | Findings in 6 classes of GI tract (lumen, contents, mucosa, flat lesions, protruding lesions, excavated lesions) | No |
| Atlas of Gastrointestinal Endoscopy | Findings in GI tract (esophagus, stomach, capsule endoscopy, duodenum and ampulla, inflammatory bowel disease, colon and ileum, miscellaneous) with several rare diseases | No |
| El Salvador atlas | Findings in GI tract | No |
| Others (n = 7) | ||
| GIANA 2017 | Polyps and angiodysplasia | No |
| Nerthus | 4 degrees of bowel preparation (BBPS 0–3) | No |
| GIANA 2018 | Polyps and lesions in WCE | No |
| EAD 2019 | 7 classes of artifacts during GI endoscopy process (imaging artefacts, pixel contrast, specular reflections, motion blur, bubbles, pixel saturation and instrument) | No |
| Cho et al. 2019 | Cecal landmarks in insertion, withdrawal and stopping point | No |
| EDD 2020 | 5 classes of GI lesions (Barrett’s esophagus, suspicious area, high-grade dysplasia; adenocarcinoma and polyps) | No |
| Kvasir-Instrument | Diagnostic and therapeutic tools in endoscopic images (such as biopsy forceps, metallic clip, probe, snares) | No |
| Name | Source of ground truth annotations | Endoscopy system brand | ||
|---|---|---|---|---|
| Ground truthb | Content | Performed by | ||
| Endoscopy datasets of polyps (n = 16) | ||||
| CVC-ColonDB | Binary mask | Polyps | Expert endoscopists | NR |
| CVC-ClinicDB (CVC-612) | Binary mask | Polyps and specular highlights | Expert endoscopists | NR |
| ETIS-Larib Polyp DB | Bounding box | Polyps | Expert endoscopists | NR |
| ASU-Mayo | Binary mask and bounding box | Polyps | Expert endoscopists | NR |
| GI lesions in Regular Colonoscopy Dataset | Annotated file and bounding box in videos | Polys | Histopathology, and the human operators’ opinion (4 expert endoscopists and 3 beginners) | Olympus |
| EndoScene | Binary mask | Polyps, specular highlights, and lumen | Expert endoscopists | NR |
| Kvasir SEG | Binary mask and bounding box | Polyps | Expert endoscopists | Olympus |
| NBIPolyp-Ucdb | Binary mask | Polyps | Endoscopists (experience not specified) | Olympus |
| WLPolyp-UCdb | Annotated file | Polyps | NR | Olympus |
| KUMC | Bounding box | Polyps in dataset 3 and 4 | Endoscopists (experience not specified) | NR |
| SUN | Bounding box | Polyps | Expert endoscopists and research assistants | Olympus |
| PICCOLO | Binary mask | Polyps | Expert endoscopists | Olympus |
| CP-CHILD | Annotated file | Polyps | Expert endoscopists | Olympus for A; Fujifilm for B |
| LD Polyp Video | Bounding box | Polyps | Annotation tool | NR |
| SUN-SEG | Binary mask, bounding box, scribble, and polygon | Polyps | 10 expert endoscopists and 2 researchers | Olympus |
| PolypGen | Binary mask and bounding box | Polyps | 6 expert endoscopists, 2 post-doctoral researchers and 1 student | Multi-brands |
| Endoscopy datasets of small bowel lesions (n = 6) | ||||
| KID | Binary mask and graphical annotation | Small bowel lesions | Expert endoscopists | NR |
| CAD-CAP | Binary mask | Small bowel lesions | Expert endoscopists | Medtronic |
| Kvasir-Capsule | Bounding box | Small bowel lesions | Expert endoscopists, master students and junior endoscopists | Olympus |
| Endoscopy Crohn’s Disease dataset | Annotated file | Clearness degrees in Crohn’s tract | Expert endoscopists and junior endoscopists | Jinshan Groupd |
| CrohnIPI | Annotated file | 6 Crohn’s lesions | Expert endoscopists and one reader | Medtronic |
| The AICE Project | Annotated file | Small bowel lesions | NR | Medtronic |
| Endoscopy datasets of gastro-esophageal lesions (n = 2) | ||||
| IPCL | Annotated file | IPCL in esophageal | Histopathology and expert endoscopists | Olympus |
| IM and GA Benchmark | Annotated file | Intestinal metaplasia and gastritis atrophy | Biopsy and endoscopists (experience not specified) | Fujifilm |
| Endoscopy datasets of comprehensive GI detection (n = 5) | ||||
| Kvasir | Annotated file | 8 classes in GI tract | Expert endoscopists | Olympus |
| Hyper Kvasir | Annotated file, binary mask, and bounding box | Annotated file for 23 classes; bounding box and binary mask only for polyps | Expert endoscopists and junior endoscopists | Olympus and Pentax |
| Rhode Island | Annotated file | Findings in four anatomical organs | Endoscopists (experience not specified) | Medtronic |
| WCE Curated Colon Disease | Annotated file | 3 classes of GI lesions | Expert endoscopists | NR |
| ERS | Binary mask and bounding box | 3600 precise and 22,600 approximate segmentation masks | Expert endoscopists | NR |
| Atlases of GI endoscopy (n = 4) | ||||
| Gastrolab | Annotated file | Findings in the GI tract | NR | NR |
| WEO Clinical Endoscopy Atlas | Annotated file | Findings in the GI tract | Expert endoscopists | NR |
| Atlas of Gastrointestinal Endoscopy | Annotated file | Findings in the GI tract | Expert endoscopists | NR |
| El Salvador atlas | Annotated file | Findings in the GI tract | NR | NR |
| Others (n = 7) | ||||
| GIANA 2017 | Binary mask and bounding box | Binary mask for polyps and bounding box for angiodysplasia | NR | NR |
| Nerthus | Annotated videos | Bowel preparation quality | Expert endoscopists | Olympus |
| GIANA 2018 | Binary mask and bounding box | Polys | NR | NR |
| EAD 2019 | Bounding box | Endoscopy artifact | Expert endoscopists and postdoctoral fellows | Multi-brands |
| Cho et al. 2019 | Annotated file | Cecal landmarks | Expert endoscopists | Olympus |
| EDD 2020 | Binary mask and bounding box | GI lesions | Expert endoscopists and post-doctoral researchers | NR |
| Kvasir-Instrument | Binary mask, bounding box, and image annotation | Diagnostic and therapeutic tools in gastrointestinal endoscopy | Research assistants and expert endoscopists | Olympus and Pentax |
| Name | No. of patients | No. of images | ||
|---|---|---|---|---|
| Total | Annotated/Not annotated | Classification of annotated images | ||
| Endoscopy datasets of polyps (n = 16) | ||||
| CVC-ColonDB | 13 | 0 | 0 | 0 |
| CVC-ClinicDB (CVC-612) | 23 | 0 | 0 | 0 |
| ETIS-Larib Polyp DB | NR | 1500 | 300/1200 | 300 of polyp |
| ASU-Mayo | 20 | 19,400 | 19,400/0 | 5200 of polyp; 14,200 of normal mucosa |
| GI lesions in Regular Colonoscopy Dataset | NR | 0 | 0 | 0 |
| EndoScene | 36 | 0 | 0 | 0 |
| Kvasir SEG | NR | 1000 | 1000/0 | 1000 of polyp |
| NBIPolyp-Ucdb | 10 | 0 | 0 | 0 |
| WLPolyp-UCdb | 42 | 3040 | 3040/0 | 1680 of polyp; 1360 of normal colon mucosa |
| KUMC | NR | 0 | 0 | 0 |
| SUN | 99 | 0 | 0 | 0 |
| PICCOLO | 40 | 3433 | 3433/0 | 3433 of polyp (2131 by WLI, 1302 by NBI) |
| CP-CHILD | NR | 9500 | 9500/0 | 1400 of polyp (1000 by Olympus, 400 by Fujifilm); 8100 of non-polyp (7000 by Olympus, 1100 by Fujifilm) |
| LD Polyp Video | NR | 0 | 0 | 0 |
| SUN-SEG | 99 | 0 | 0 | 0 |
| PolypGen | > 300 | 6282 | 6282/0 | 3762 of polyp; 2520 of normal colon mucosa |
| Endoscopy datasets of small bowel lesions (n = 6) | ||||
| KID | NR | 2500 | 2500/0 | 2500 of SB lesions |
| CAD-CAP | NR | 0 | 0 | 0 |
| Kvasir-Capsule | NR | 0 | 0 | 0 |
| Endoscopy Crohn’s disease dataset | 15 | 466 | 466/0 | 323 of clearness; 101 of blur; 42 of invisible |
| CrohnIPI | ≥ 200 | 3498 | 3498/0 | 2124 of normal SB; 1360 of lesions; 14 of inconclusive findings |
| The AICE Project | NR | 18,481 | 18,481/0 | 12,320 images with 19,459 annotations (931 of angiodysplasia, 5988 of erosion, 477 of stenosis, 612 of lymphangiectasia, 6792 of lymph follicle, 547 of SMT, 3236 of polyp, 875 of bleeding); 6161 of normal SB |
| Endoscopy datasets of gastro-esophageal lesions (n = 2) | ||||
| IPCL | 114 | 67,740 | 67,740/0 | 39,662 of lesion; 28,078 of normal mucosa |
| IM and GA Benchmark | 630 | 21,420 | 21,240/0 | 2438 of WLI-IM; 3381 of WLI-GA; 5854 of WLI-Normal; 2549 of LCI-IM; 3270 of LCI-GA; 3928 of LCI-Normal |
| Endoscopy datasets of comprehensive GI detection (n = 5) | ||||
| Kvasir | NR | 8000 | 8000/0 | 1000 of Z-line; 1000 of pylorus; 1000 of cecum; 1000 of esophagitis; 1000 of polyps; 1000 of ulcerative colitis; 1000 of “dyed and lifted polyp”; 1000 of “dyed resection margins” |
| Hyper Kvasir | NR | 110,079 | 10,662/99,417 | 3452 of 7 classes from upper GI tract; 7210 of 16 classes from lower GI tractc |
| Rhode Island | 424 | 0 | 0 | 0 |
| WCE Curated Colon Disease | NR | 6000 | 6000/0 | 1500 of polyp; 1500 of ulcerative colitis; 1500 of esophagitis; 1500 of normal mucosa |
| ERS | 1135 | 0 | 0 | 0 |
| Atlases of GI endoscopy (n = 4) | ||||
| Gastrolab | NR | ≥ 1498 | ≥ 1498/0 | ≥ 1498 of GI findingsc |
| WEO Clinical Endoscopy Atlas | NR | 148 | 148/0 | 31 of lumen; 5 of content; 25 of mucosa; 14 of flat lesions; 49 of protruding lesions; 24 of excavated lesions |
| Atlas of Gastrointestinal Endoscopy | NR | 1259 | 1259/0 | 1259 of GI findings |
| El Salvador atlas | NR | 0 | 0 | 0 |
| Others (n = 7) | ||||
| GIANA 2017 | NR | ≥ 1500 | ≥ 1500/0 | 600 of angiodysplasia; ≥ 900 of poly |
| Nerthus | 21 | 5525 | 5525/0 | 5525 of four classes of bowel cleanliness |
| GIANA 2018 | NR | 8262 | 8262/0 | 8262 of polyp and small bowel lesion |
| EAD 2019 | NR | ≥ 2500 | ≥ 2500/0 | ≥ 2500 of endoscopy artifacts |
| Cho et al. 2019 | 112 | 0 | 0 | 0 |
| EDD 2020 | NR | 385 | 385/0 | 385 images with 502 ground truth annotations (160 of non-dysplastic Barrett’s; 88 of precancerous lesion; 74 of high-grade dysplasia; 53 of cancer; 127 of cancer) |
| Kvasir-Instrument | NR | 590 | 590/0 | 590 of diagnostic and therapeutic tools |
| Name | No. of videos (frames) | Image/ frame resolution (pixels) | Imaging modality | ||
|---|---|---|---|---|---|
| Total | Annotated/Not annotated | Classification of annotated videos(frames) | |||
| Endoscopy datasets of polyps (n = 16) | |||||
| CVC-ColonDB | 13 (300) | 13 (300)/0 | 13 (300) of polyp | 500 × 574 | WLI |
| CVC-ClinicDB (CVC-612) | 31 (612) | 31 (612)/0 | 31 (612) of polyp | 576 × 768 | WLI |
| ETIS-Larib Polyp DB | 0 | 0 | 0 | 1225 × 966 | NR |
| ASU-Mayo | 0 | 0 | 0 | 512 × 512 | NR |
| GI lesions in Regular Colonoscopy Dataset | 76 (NR) | 76 (NR)/0 | 15 of serrated adenoma; 21 of hyperplastic polyp; 40 of adenoma | 30 frames of 768 × 576 pixels every second | NBI and WLI |
| EndoScene | 44 (912) | 44 (912)/0 | 44 (912) of polyp (300 from CVC-ColonDB and 612 from CVC-ClinicDB) | 224 × 224 | WLI |
| Kvasir SEG | 0 | 0 | 0 | 320 × 320 | WLI |
| NBIPolyp-Ucdb | 11 (86) | 11 (86)/0 | 10 videos of adenoma and 1 video of hyperplastic polyp | 726 × 576 | NBI |
| WLPolyp-UCdb | 0 | 0 | 0 | 726 × 576 | WLI |
| KUMC | 157 (35,981) | 76 (4955)/81 (31,026) | 76 (4955) of polyp (38 videos of adenoma and 38 videos of hyperplastic polyp) | 224 × 224 | WLI |
| SUN | 113(158,690) | 113 (158,690)/0 | 100 (49,136) of polyp; 13 (109,554) of non-polyp | 416 × 416 | WLI |
| PICCOLO | 0 | 0 | 0 | 854 × 480 or 1920 × 1080 | NBI and WLI |
| CP-CHILD | 0 | 0 | 0 | 256 × 256 | NR |
| LD Polyp Video | 263 (901,666) | 160 (40,266)/103 (861,400) | 33,884 frames of polyp; 6382 frames of non-polyp | 560 × 480 | WLI |
| SUN-SEG | 1106 (158,690) | 1106 (158,690)/0 | 378 (49,136) of polyp; 728 (109,554) of non-polyp | 416 × 416 | WLI |
| PolypGen | 0 | 0 | 0 | 384 × 288 to 1920 × 1080 | NR |
| Endoscopy datasets of small bowel lesions (n = 6) | |||||
| KID | 47 | 47/ 0 | 47 videos of SBesions | NR | NR |
| CAD-CAP | 1686 (25,124) | 1686 (25,124)/0 | 1480 (5124) of abnormal SB findings (3103 of vascular lesion, 651 of fresh blood, 1370 of ulcer-inflammatory lesion); 206 (20,000) of normal SB | NR | WLI |
| Kvasir-Capsule | 117 (4,741,504) | 43 (47,238)/74 (4,694,266) | 1529 frames of pylorus; 10 frames of ampulla of Vater; 4189 of ileocecal valve; 2906 of mucosal atrophy; 592 of lymphangiectasia; 159 of erythema; 866 of angiectasia; 466 of fresh blood; 12 of blood-hematin; 506 of erosion; 854 of ulcer; 55 of polyp; 776 of foreign body; 4189 of normal mucosa | 256 × 256 to 512 × 512 | WLI |
| Endoscopy Crohn’s disease dataset | 0 | 0 | 0 | 240 × 240 | WLI |
| CrohnIPI | 0 | 0 | 0 | NR | WLI |
| The AICE Project | 0 | 0 | 0 | NR | WLI |
| Endoscopy datasets of gastro-esophageal lesions (n = 2) | |||||
| IPCL | 0 | 0 | 0 | 256 × 256 | ME-NBI |
| IM and GA Benchmark | 0 | 0 | 0 | 1280 × 1024 | WLI and LCI |
| Endoscopy datasets of comprehensive GI detection (n = 5) | |||||
| Kvasir | 0 | 0 | 0 | 720 × 574 to 1920 × 1072 | WLI |
| Hyper Kvasir | 374 | 374/ 0 | 60 videos of 14 classes from upper GI tract; 314 videos of 16 classes from lower GI tractc | 332 × 352 to 1921 × 1073 | WLI |
| Rhode Island | 424 (5,247,588) | 424 (5,247,588)/0 | 13,715 frames of esophagus; 557,049 frames of stomach; 4,111,865 frames of small bowel; 564,959 frames of colon | 320 × 320 | WLI |
| WCE Curated Colon Disease | 0 | 0 | 0 | 224 × 224 | NR |
| ERS | 1520 (≥ 1,230,000) | 121,000/1230000 (frames) | 6000 frames of precisely labeled; 115,000 frames of approximately labeledc | NR | NR |
| Atlases of GI endoscopy (n = 4) | |||||
| Gastrolab | 0 | 0 | 0 | NR | NR |
| WEO Clinical Endoscopy Atlas | 0 | 0 | 0 | NR | NR |
| Atlas of Gastrointestinal Endoscopy | 0 | 0 | 0 | NR | NR |
| El Salvador atlas | 5138 (NR) | 5138 (NR)/0 | 5138 videos of GI findings | NR | NR |
| Others (n = 7) | |||||
| GIANA 2017 | ≥ 38 | ≥ 38/0 | ≥ 38 videos of polyp | NR | NR |
| Nerthus | 0 | 0 | 0 | 720 × 576 | WL |
| GIANA 2018 | 38 | 38/0 | 38 videos of polyp and SB lesions | NR | NR |
| EAD 2019 | 0 | 0 | 0 | NR | Multi-modal (WLI, BNI and fluorescence) |
| Cho et al. 2019 | 112 (328,927) | 2 (100) available | 2 (100) of cecal landmarks | 850 × 750 | WLI |
| EDD 2020 | 0 | 0 | 0 | NR | NR |
| Kvasir-Instrument | 0 | 0 | 0 | 720 × 576 to 1280 × 1024 | NR |
NR not reported, SB small bowel, GA gastric atrophy, IM intestinal metaplasia, WLI white light imaging, LCI linked color imaging, BBPS Boston bowel preparation scale, WCE wireless capsule endoscopy, NBI narrow band imaging
aClinical details includes clinical metadata (patients’ information, location of polyp, type of polyp and so on) or annotation details (annotators’ number, annotation process and so on)
bAnnotated file for classification task, binary mask for segmentation task, and bounding box for detection task
cComplete information of classification was shown in Supplementary Table 1
dChongqing Jinshan Science & Technology (Jinshan Group), a national high-tech enterprise that integrates research and development, manufacturing, marketing, and service of digital medical devices
According to the data type, we divided 40 endoscopic imaging datasets into 6 groups: polyp datasets (n = 16), small bowel lesions datasets (n = 6), gastro-esophageal lesions datasets (n = 2), comprehensive GI detection datasets (n = 5), atlases of GI endoscopy (n = 4), and others (n = 7).
Polyp Datasets
Most endoscopy datasets are polyp related (n = 16). One dataset collected GI polyp images by wireless capsule endoscopy (WCE). The others collected colon-polyp images by colonoscopy.
CVC-ColonDB, the first endoscopy colon-polyp dataset constructed in 2013, contained 300 polyp images with associated masks from 13 polyp videos. In addition to polyp images, the dataset reported three key characteristics of each video: length, number of frames, and polyp shape (flat or peduncular). CVC-ColonDB was used to train and test automatic polyp detection models from 2013 to 2015 [7, 46–48]. However, rough boundary outlines and a small number of polyp images cannot be ignored. Moreover, the dataset is unavailable from the official website.
CVC-ClinicDB, a dataset from Spain in 2015, contained 612 polyp images from 31 polyp videos. Compared with CVC-ColonDB, more polyp images were included. Moreover, it provided binary masks for both polyps and specular highlights. Clinical metadata associated with each polyp were also included, such as polyp size and classification according to Paris criteria [49]. However, complete boundary information was still lacking in the CVC-ClinicDB, which might lose the advantage over the gradient information accumulation process.
ETIS-Larib Polyp DB contained 300 colon-polyp images with corresponding bounding boxes and 1200 non-polyp images by WCE. However, the dataset is unavailable.
ASU-Mayo, built in 2016 by Arizona State University in America, contained 10 positive shots and 10 negative shots from colonoscopy. A total of 5200 polyp frames were extracted from positive shots, while 14,200 normal frames were extracted from negative shots. More than 3500 frames came with corresponding masks and boxes. To avoid overfitting, images varied from different levels of colon preparation, colonoscopy events, and artifacts, which maintained a large degree of variability and complexity. The copyrighted dataset is only available through direct contact with the administrations.
GI lesions in the Regular Colonoscopy Dataset included 76 colonoscopy videos from 15 serrated adenomas, 21 hyperplastic lesions, and 40 adenomas. Lesions were recorded from five modalities: WL frame, NBI frame, WL video less than 30 s, NBI video less than 30 s, and camera calibration. The calibration was associated with ground truth from histopathology and the human operators’ diagnosis (4 experts and 3 beginners). This was the first endoscopy dataset to provide a detailed ground truth for videos and differentiate between serrated adenomas, hyperplastic polyps, and adenomas.
The LD PolypVideo dataset, the largest polyp-related dataset to date, was constructed at the First Affiliated Hospital of Anhui Medical University, Hefei, China. The dataset contained 160 colonoscopy videos and 40,266 frames with annotation in total. Moreover, there were 33,884 frames of colon polyps, which were more than 11 times that of CVC-ClinicVideoDB. Due to diverse morphologies and data, the LD PolypVideo dataset can be used in unsupervised and semi-supervised tasks.
The Kvasir-SEG, EndoScene, and KUMC datasets are secondary datasets that were constructed based on original datasets. The Kvasir-SEG dataset was based on the polyp class of Kvasir. Researchers replaced 13 original polyp images with new high-quality images and added corresponding segmentation masks and bounding boxes for 1000 images of polyps. The EndoScene dataset was composed of 912 images (300 from CVC-ColonDB and 612 from CVC-ClinicDB). Each frame came with three corresponding masks: poly, specular highlights, and lumen masks. However, the dataset is unavailable online. The KUMC dataset contained 4955 images with corresponding bounding boxes from 38 adenomatous and 38 hyperplastic polyps. The KUMC dataset was based on the CVC-ColonDB, ASU-Mayo, and Colonoscopic and KUMC Colonoscopy datasets. It provided two sets of supporting information, in which set 1 contained the extracted polyp patches, while set 2 contained not only the extracted polyp patches but also the background around the polyps.
The NBIPolyp-Ucdb and WLPolyp-UCdb datasets came from the same author and organization in Portugal. NBIPolyp-Ucdb contained 86 colon-polyp images from 10 adenomas and 1 hyperplastic polyp, recorded with NBI colonoscopy. A corresponding mask of each image was also provided. WLPolyp-UCdb contained 1680 images of polyps and 1360 of normal colon mucosa with WL colonoscopy. A form with personal information needs to be completed to download both datasets.
The SUN dataset was collected from the Showa University and Nagoya University databases in Japan and updated in December 2022. The dataset included 49,136 polyp frames annotated with bounding boxes and 109,554 non-polyp frames. Moreover, detailed characteristics of each polyp, including shape, median size, location, and pathological diagnosis, were provided. The data can be requested by email.
The SUN-SEG dataset, a secondary dataset based on the SUN dataset, contained 1106 short video clips with 158,690 frames in total. Researchers manually separated 113 original colonoscopy videos into 378 positive and 728 negative sequences. Moreover, in addition to primary bounding boxes and detailed information, more annotations and ground truth were provided, including visual attributes, masks, scribbles, and polygons in SUN-SEG.
The PICCOLO dataset was collected from October 2017 to December 2019 in Spain. This dataset included 2131 WL and 1302 NBI polyp images from 76 different lesions. For each image, a binary mask was created manually, indicating that there was a polyp. Moreover, clinical metadata were provided as follows: the number of polyps, polyp identification, polyp size (in millimeters), Paris classification, NICE rating, preliminary and literal diagnoses, and histological stratification. Although the PICCOLO dataset is publicly available, a dedicated form to request a download must be completed.
The CP-CHILD dataset recorded the colonoscopy data of children from Hunan Children’s Hospital in China. It was divided into CP-CHILD-A and CP-CHILD-B datasets. The CP-CHILD-A dataset contained 8000 RGB images, including 1000 colonic polyp images and 7000 normal or other pathological images by Olympus PCF-H290DI; the CP-CHILD-B dataset contained 1500 RGB images taken by FUJIFLIM EC-530wm, including 400 colon-polyp images and 1100 normal or other pathological images.
PolypGen, a multicenter polyp detection and segmentation dataset, was composed of 3672 positive frames and 2520 negative frames. For each polyp frame, a mask was created by expert endoscopists. Moreover, detailed information containing size, location, artifacts, and visibility was provided.
Small Bowel Lesion Dataset
Capsule endoscopy has been used as a complementary test for patients with GI bleeding since early 2020, with great potential to become an authoritative diagnostic tool for the small bowel [50, 51]. The development of CE contributes several datasets to small bowel lesions.
The Kid dataset, the earliest open-source CE dataset in 2017, aimed to provide a reference for research on the development of medical decision support systems for CE. More than 2500 annotated CE images and 47 videos were provided through the Kid dataset. Images included normal CE and vascular lesions. Detailed information on the classification can be seen in Table 2. Researchers need to register for access to the Kid database. However, precise definitions are lacking for several diseases in the small intestine, and a certain number of images are difficult to annotate and set.
CAD-CAP, a national multicenter dataset from twelve French endoscopic units, provided 1685 SB-CE videos, 5124 images with abnormal findings, and 20,000 normal images. Abnormal findings are divided into three categories: vascular lesions (n = 3103), fresh blood (n = 651), ulcer-inflammatory lesions (n = 1370). However, bowel preparation quality and polyps were not considered in this version of the CAD-CAP. The dataset is unavailable now.
Kvasir-Capsule, a large VCE dataset collected from Norwegian Hospital in 2021, contained 117 videos and 4,741,504 extracted frames. A total of 47,238 frames were labeled and divided into 2 classes: anatomy and small bowel findings. There were also 74 unlabeled videos and 4,694,266 unlabeled images for further research. Detailed information on the classification can be seen in Table 2.
The AICE Project dataset was created at Kaggle in 2022. The dataset contained 18,481 images. It consisted of 12,320 lesion images and 6161 normal images. Small bowel lesions included angiodysplasia, erosion, stenosis, lymphangiectasia, lymph follicle, polyp-like lesions, submucosal tumor, bleeding, diverticulum, erythema, foreign body, and vein lesions. The data are available by the author.
CrohnIPI dataset, a multicentric dataset of pathological and nonpathological images of Crohn’s disease, included 3498 annotated images extracted from 66 video capsules of diagnosed patients. A total of 1630 images contained 6 Crohn’s lesions (erythema, edema, aphthoid ulceration, 3–10 mm ulceration, > 10 mm ulceration, stenosis), 2124 images were labeled as nonpathological, and 14 images were inconclusive findings.
Endoscopy Crohn’s Disease dataset, a large-scale Crohn’s gastrointestinal image dataset for lesions and challenges faced in CE, covered 466 images from 15 patients. The content involved in Crohn’s lesions includes various complex challenges, including motion blur, excreta occlusion, and reflection. Moreover, the clinical and demographic characteristics of 15 patients were provided by the dataset. To obtain the data, a message needs to be sent.
Gastroesophageal lesion Datasets
IPCL, the first dataset of normal and abnormal intrapapillary capillary loops (IPCL), was constructed from magnification endoscopy (ME) in Taiwan. IPCL is a clinical microvascular feature considered an endoscopic marker for early squamous cell neoplasia (ESCN). A total of 67,740 frames from 114 videos were classified into four types of IPCL, A, B1, B2, and B3, based on the Japanese Endoscopic Society (JES) IPCL system correlated with histopathology. The IPCL dataset can serve as a benchmark for future work on the detection of ESCNs and has great potential in the endoscopic diagnosis of early esophageal neoplasia, which remains in its infancy [52].
IM and GA Benchmark, a dataset to detect intestinal metaplasia (IM) and gastritis atrophy (GA), was built in China by traditional WLI and LCI endoscopy. The dataset included GA, IM, and normal images in five gastric lesion locations (antrum, angle, cardia, fundus, and body). There were 21,420 annotated LCI and WLI images that were annotated by four radiologists and validated by biopsy examination results. The advantages of this dataset are as follows: (1) more than 20,000 images covering all five key locations in the stomach, (2) detailing and reserving the original image resolution, and (3) few studies of LCI-related GI and IM.
Comprehensive GI Detection Datasets
Conventional endoscopy examination is currently the gold-standard procedure for investigating the GI tract, including gastroscopy and colonoscopy. Gastroscopy covers the upper GI tract from the esophagus to the duodenum, while colonoscopy covers the colon and rectum [53, 54]. Comprehensive GI detection datasets are considered multiclass image or video datasets in GI endoscopy. It covers several anatomical structures or lesions and has abundant data for automatic algorithmic detection of many aspects of the GI tract, which is not limited to a specific lesion, a specific part or a specific endoscopic technology.
Kvasir, the first comprehensive GI detection dataset containing multiclass images, was created in 2017 from the Vestre Viken Health Trust in Norway. Images were classified into three important anatomical landmarks, three clinically pathological findings, and two endoscopic procedures. Eight detailed classes included Z-line, pylorus, cecum, esophagitis, polyps, ulcerative colitis, “dyed and lifted polyp,” and “dyed resection margins.” Each class contained 1000 images, which was sufficient for different tasks. Two sets of images related to the removal of polyps, the “dyed and lifted polyp” and “dyed resection margins,” were provided for automatic recognition of the site of polyp removal. The dataset was used for the Multimedia for Medicine Challenge (the Medico Task) in 2017 [55] and 2018 [56] at the MediaEval Benchmarking Initiative for Multimedia Evaluation. However, due to only frame-wise annotations, the dataset is limited to frame classification only.
HyperKvasir, a comprehensive multiclass image and video dataset of the GI tract available today, was collected during gastroscopy and colonoscopy at Bærum Hospital in Norway from 2008 to 2016. It contained 110,079 images and 374 videos, including anatomical landmarks and pathological & normal findings. A total of 10,662 labeled images showed 23 different classes, including anatomical landmarks, quality of mucosal views, pathological findings, and therapeutic interventions. One thousand mask images of corresponding polyps were provided for the segmentation task. Thirty classes of findings were identified in 374 videos from the GI tract. Moreover, HyperKvasir provided 99,417 unlabeled images that can be used for semi-supervised and unsupervised tasks. Chang et al. combined 10,417 images from a local hospital and 3157 from the HyperKvasir dataset to develop a quality assurance algorithm for colonoscopy [57]. To the best of our knowledge, HyperKvasir is the most diverse dataset of GI endoscopy, which enables researchers not only to analyze, classify, segment, and retrieve various GI findings but also to differentiate between the severity of the findings.
The Rhode Island Gastroenterology VCE dataset, the latest and largest public dataset available, included 424 videos and 5,247,588 labeled images from the VCE procedures. Images were divided into four classes by anatomical organ: esophagus (n = 13,715), stomach (n = 557,049), small bowel (n = 4,111,865), and colon (n = 564,959).
WCE Curated Colon Disease, a secondary dataset constructed in 2022, relied on two readily available datasets: Kvasir and ETIS-Larib Polyp DB. After intensive data collection and evaluation, a total of 6000 images were chosen for this dataset. Images were divided into four classes: normal, ulcer, polys, and esophagitis. Each class had equal numbers for training, validation, and testing, which contained 800, 500, and 200 images, respectively.
The ERS dataset, a multitissue comprehensive imaging dataset from flexible endoscopy, colonoscopy, and capsule endoscopy, described all possible findings in the GI tract. It contained 6000 precisely and 115,000 approximately labeled images from endoscopy videos and 3600 precisely and 22,600 approximately labeled images with segmentation masks. Images were annotated and divided into 27 different types of colonoscopic findings and 54 different upper endoscopy findings. In addition to traditional findings, three categories of terms were included in the dataset: healthy GI tract tissues, image quality attributes (such as sharp, blur, motion, and stool), and images with blood. Detailed information is provided in Supplementary Table 1. Researchers can fill in a form and obtain a link to download the ERS dataset.
Atlases of GI Endoscopy
In the next section, there are four atlases of GI endoscopy with several findings in the GI tract. It is more of a medical atlas or database for education than a dataset for traditional ML or DL. With the appearance of few-shot learning [58], the atlas of GI endoscopy might be used in this field.
The WEO Clinical Endoscopy Atlas, an atlas of GI from the World Endoscopy Organization (WTO), was compiled from personal contributions to endoscopists throughout the web from 2009 to 2022. The atlas contained hundreds of images divided into 6 classes: lumen (n = 31), contents (n = 5), mucosa (n = 25), flat lesions (n = 14), protruding lesions (n = 49), and excavated lesions (n = 24). The website provides a description, source, and data below every image. Researchers can search for images of interest using the minimal standard terminology (MST) term.
The Atlas of Gastrointestinal Endoscopy was constructed by Atlanta South Gastroenterology from 1996 to 2016. It provided 1259 images from the esophagus to the colon/ileum. To our surprise, the atlas provided several rare lesions in the GI tract, such as gastrointestinal syphilis and gastrostomy tube ulcers. It can be used for educational and general informational purposes with a brief case report and description for every lesion.
The El Salvador atlas, a video atlas of GI endoscopy, contains 5138 video clips of the GI tract. Although covering almost all areas of GI pathology is detectable, low-quality and low-resolution videos cannot be avoided.
Gastrolab contains more than 1498 images of GI anatomical structure, lesions, infectious diseases, and GI devices. Detailed classification information is provided in Supplementary Table 1. Moreover, hundreds of videos can be downloaded through this website.
Others
A number of detection and segmentation challenges posted online contribute endoscopy datasets to GI lesions. However, these datasets are restricted to being available to the registers of these challenges only. Here, we find three challenges and corresponding datasets: GIANA 2017, GIANA 2018, and EDD 2020.
GINAN 2017 defined four different tasks: polyp detection, polyp segmentation, angiodysplasia detection, and angiodysplasia localization. It contained 600 images of angiodysplasia, 38 videos captured by WCE, and more than 900 images of polys captured by colonoscopy. Organizers provided ground-truth segmentation masks with each image for detection, segmentation, and classification.
GINAN 2018 defined three types of tasks: polyp detection and localization in video colonoscopy, polyp segmentation in colonoscopy images, and lesion detection and localization in WCE images. It contained 8262 images and 38 videos with corresponding ground-truth segmentation masks.
Moreover, due to successful iterations of the Endoscopic Vision Challenge (GIANA 2017 and GIANA 2018), a new challenge was released in 2021 that was associated with colonoscopy image analysis: lesion detection, segmentation, and classification. More information about GIANA 2021 can be found at https://giana.grand-challenge.org/.
EDD 2020, a multiclass, multiorgan, and multipopulation disease detection and segmentation challenge in clinical endoscopy, provided a comprehensive dataset to benchmark algorithms for disease detection. The dataset incorporated multiple populations with 4 different international centers and 3 GI organs: the colon, esophagus, and stomach. There were 385 images from GI videos and 503 ground-truth annotations consisting of five types of GI lesions: normal dysplastic Barrett’s esophagus, suspicious area, high-grade dysplasia, adenocarcinoma, and polyps.
The final section, with databases not easily lying within the earlier categories, contains the unusual content of the GI tract.
The Cho et al. 2019 dataset is a single-center colon-polyp dataset from Seoul National University Hospital in Korea. It recorded the complete process in colonoscopy, containing 328,927 frames of cecal landmarks in insertion, withdrawal and stopping points, but data were only available for 100 polyp images from 2 videos online. The complete raw data may be available through direct contact with the author.
EAD (endoscopy artifact detection) 2019, a multiclass artifact detection dataset, was constructed from 6 different institutions in 2019. Artifacts are considered heavy imaging interference during the GI endoscopy process, such as motion blur and bubbles, which remain a challenge and problem in the diagnosis and treatment of disease in hollow organs through GI endoscopy. The dataset contained 7 classes of artifacts: imaging artifacts, pixel contrast, specular reflections, motion blur, bubbles, pixel saturation, and instruments from multiple organs (the esophagus, stomach, liver, colon, and bladder) and multiple modalities (white light, narrow band, and fluorescence light). EAD 2019 was used to solve three tasks: multiartifact detection, region segmentation, and generalization.
For detection, 2147 annotated frames over all 7-artifact classes were provided. For semantic segmentation, 475 annotated frames for 5 classes (pixel saturation, specular reflections, imaging artifacts, bubbles, and instrument) were provided. For generalization, 53 images were provided.
Kvasir-Instrument, the first diagnostic and therapeutic tool segmentation dataset in GI endoscopy, was partly collected from endoscopic examinations performed at the Bærum Hospital and partly extracted from the HyperKvasir and Kvasir-SEG datasets. There were 590 annotated frames, including GI procedure tools such as snares, balloons, and biopsy forceps. In addition to the images, ground-truth masks and bounding boxes were provided. Kvasir-Instrument helped to set up an automated system algorithm for the segmentation of GI tract diagnostic and therapeutic endoscopy tools to locate and guide GI tract biopsies and surgeries.
Nerthus showed different degrees of bowel preparation. It contained a total number of 5525 annotated frames. The frames were divided into four classes (ranging from 0 to 3) by the Boston bowel preparation scale (BBPS) [59, 60] within each section according to a defined numeric scale. Most frames were provided with the location inside the bowel due to different values in different positions. Thanks to the Nerthus dataset, automatic systems would be made for evaluating the quality of bowel cleansing to achieve high-quality colonoscopy examinations.
Dataset Characteristics
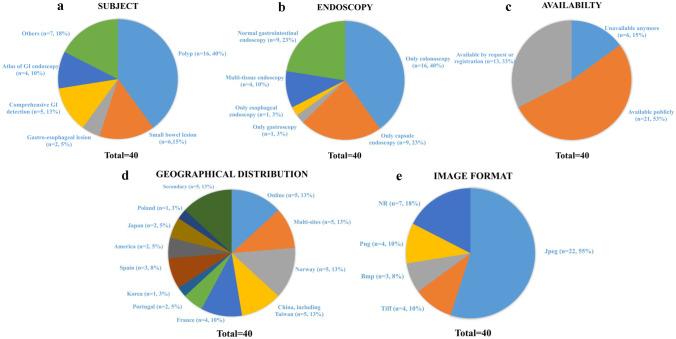
The study includes 40 endoscopy datasets from 2010 to 2022. The subject of the datasets included is summarized in Fig. 2a. A total of 36 (90%) datasets are endoscopy datasets, including a large number of images for ML or DL, and 4 (10%) are atlases of GI endoscopy with several samples of various findings in the GI tract. Polyp-related datasets were the most numerous (n = 16, 40%), followed by small bowel lesions (n = 6, 15%), comprehensive GI detection (n = 5, 13%), and gastroesophageal lesions (n = 2, 5%). Of the remaining 7 datasets, 3 (8%) are challenge-related datasets, and 4 (10%) are related to unusual findings such as instruments and artifacts.
Fig. 2.
Characteristics of datasets involved
The endoscopic type of the datasets included is shown in Fig. 2b. Most datasets (n = 16, 40%) were constructed by colonoscopy only, followed by normal gastrointestinal endoscopy consisting of gastroscopy and colonoscopy and capsule endoscopy (n = 9, 23%).
Dataset access is evaluated in Fig. 2c. Twenty-one (53%) datasets can be viewed and downloaded freely and publicly, while 6 (15%) are unavailable from official websites and 13 (33%) are available by request or registration with minimum requirements for access (such as creating an account or a form for personal information).
The geographical distribution of the datasets included is shown in Fig. 2d. Five (13%) were secondary datasets based on original datasets, 5 (13%) were collected online, and 5 (13%) were constructed from multiple endoscopic centers from different countries. Other studies (n = 25, 63%) were conducted in a single country. Of these, Norway and China (including Taiwan) developed the most endoscopy datasets (n = 5, 13%, respectively), followed by France (n = 4, 10%), Spain (n = 3, 8%), Japan, America, and Portugal (n = 2, 5%, respectively). One (3%) dataset was retrieved in Poland and Korea.
Most datasets (22 of 40, 55%) stored images in joint photographic experts portable network graphics (JEPG/JPG), 4 (10%) in tagged image file format (TIFF), 4 (10%) in portable network graphics (PNG), and 3 (8%) in bitmap image file (BMP). The image format is unreported in 7 datasets (18%). The image format of the datasets included is summarized in Fig. 2e.
National Endoscopy Datasets
When constructing a public dataset, processes such as encryption and de-identification are required to remove data related to patient identification [61]. This protects patients’ privacy, but restricts certain areas of clinical metadata. This issue can be resolved by a large-scale, high-quality dataset. Here, we introduce two potential national datasets in process: Japan Endoscopy Database (JED) [62] and the United Kingdom National Endoscopy Database (NED) [63].
JED, a multicenter endoscopy dataset launched by the Japan Gastroenterological Endoscopy Society, was proposed in 2015 [64]. The purposes of JED were as follows: (1) to construct the largest endoscopic practice database worldwide; (2) to store diagnostic information for therapeutic procedures and examinations; (3) to standardize the terminology and fundamental items for endoscopy; and (4) to provide adequate data for clinical and basic research. JED consisted of both structured and unstructured information. Structured information was obtained for all endoscopic procedures including patients’ fundamental information and the duration of the procedure. Unstructured information was obtained from four endoscopic examinations: upper GI endoscopy, small bowel endoscopy, lower GI endoscopy, and endoscopic retrograde cholangiopancreatography-related procedures (ERCP). Taking upper GI endoscopy as an example, endoscopic images of upper GI findings and Helicobacter pylori (Hp) infection status were collected.
There were several reports and researches based on JED. Kodashima et al. published the first JED project status report in 2017, describing over 60,000 endoscopic procedures and identifying several problems that need to be addressed [65]. Based on JED, Saito et al. reported the current status of diagnostic and therapeutic colonoscopy, while Oda et al. reported the current status of Hp infection and gastric mucosal atrophy in patients with gastric cancer [66, 67].
Due to combination with structured and unstructured data, JED will contribute to the future development of GI endoscopy AI technology. More multi-model algorithms will be mined based on JED.
The United Kingdom National Endoscopy Database (NED), a centralized dataset launched by the Joint Advisory Group, comprehensively represents the entire UK endoscopy practice. It was used to capture near-real-time data from GI endoscopy procedure. Standardization of endoscopic data and key performance indicators made for the implementation of NED, such as the modified Aronchick classification for bowel preparation quality. At present, 411 of 520 UK endoscopic units, both public and private, are actively uploading endoscopic data to NED, which has collected more than 2.5 million endoscopic procedures. The initial purpose of NED was to generate personalized trainee metrics and learning curves to evaluate competency progression and quantify endoscopy quality. With the development of NED, the NED IT team promised that they would provide procedure-level data and images of the GI tract. Rutter et al. utilized NED data from January 2020 to May 2020 to evaluate the impact of the COVID-19 pandemic on endoscopic activity in UK [68]. NED is likely to become a high-quality endoscopic dataset for future.
Discussion
From the broad search of medical literature and targeted search engines, we found 40 unique endoscopy datasets. The most common subject is polyp-related lesions. Colorectal polyp detection and characterization have been most likely representative of the application of DL for GI endoscopy [69, 70].
Across all datasets, colonoscopy is the most common endoscopy type, probably because of its widespread availability and common use of colonic lesions. The second most common endoscopy is capsule endoscopy. Capsule endoscopy has been developed rapidly not only for the small bowel but also for the whole GI tract. Moreover, AI with capsule endoscopy has demonstrated better diagnostic accuracy with a shorter reading time [71].
The availability of the current endoscopy datasets appears to be an issue. Of 40 datasets, 53% of datasets (n = 21) are available publicly, while 33% of datasets (n = 13) are available by request or registration. Although a number of studies used endoscopy datasets, they did not open access to datasets. Moreover, discoverability also appears to be an issue. Although a few datasets have been used multiple times, such as CVC-ClinicDB, Kvasir-SEG, and HyperKvasir, many are not. This difference in use might lead to a loss of research opportunities and selection bias due to an overuse of several potential nonrepresentative datasets. In this regard, it is necessary to improve their discoverability. Therefore, our study provides an initial point of access that will improve their discoverability. We encourage journals and authors to improve data accessibility for the future.
Most datasets (n = 33, 82%) provided images in a common and portable file, which is generally accepted in various algorithms in ML or DL.
Strengths and Weaknesses
To the best of our knowledge, this is the first review to curate a comprehensive list of endoscopic imaging datasets. One of the strengths is the broad search strategy, including scientific and online search engines. Moreover, two public colonoscopy image datasets are included for reference [72, 73].
Furthermore, we attempt to verify the statements and availabilities of all datasets involved because several datasets that could be obtained publicly are unavailable today, such as CVC-ColonDB and ETIS-Larib Polyp DB. This process helps us determine the extent to which datasets are truly accessible and provide users with the latest guidance.
Finally, whether image labels, ground masks, and clinical metadata are provided is included in the brief introduction of each dataset involved. Therefore, users can select an appropriate dataset according to the introduction, which greatly saves time and energy.
There are several limitations in our review. First, only one medical search engine, PubMed, was used to screen endoscopy datasets. In addition, we failed to report details regarding the selection and annotation processes which have an effect on the establishment of the dataset [74]. Finally, we excluded specific datasets for the purpose of Visual Simultaneous Localization and Mapping, such as Endomapper and VR-Caps.
Implications
To reduce tedious work and accomplish complicated tasks, the need for AI-assisted tools in clinical practice is on the rise. Publicly available imaging datasets can be a powerful and essential benchmark for AI-assisted tools; however, the implications and limitations of these datasets must be considered. In this section, we discuss three implications of these datasets: accessibility, details, and adequate samples.
The first implication is accessibility. It is amazing that we identified 34 datasets that have open access; however, there are two issues remaining. One is that unbalanced use and reference might result in bias or neglect of the latest but high-quality datasets. Another is that users’ desire for datasets with regulated access might not be strong. Although these datasets might have higher quality and reflect stronger attention to governance and metadata reporting, users still prefer to select datasets that have immediate, unregulated access but low quality.
The second implication is the details. Most endoscopy datasets provide original images with corresponding masks or bounding boxes to outline the subjects. However, clinical and annotation details are not reported in most datasets. Clinical data includes what type of patients and lesions were involved and categories & locations of lesions. Taking the PICCOLO dataset as an example, in addition to original images, it provides a bounding box, binary mask, polyp identification, polyp size, Paris classification, NICE rating, preliminary and literal diagnoses, and histological stratification for each polyp. These clinical metadata help distinguish the diverse population of humans and their diseases to launch more accurate algorithms and make assumptions on the generalization of the real world. Annotation details regarding annotators’ number, annotators’ expertise, and annotation processes are reported in a few datasets. There is no doubt that several assumptions and definitions were made during the annotation process, which could influence the ultimate performance of an algorithm. Incomplete annotation details may result in inappropriate use of data and even biased results. However, there are acknowledged challenges associated with clinical and annotation details. In addition to ethical supervision during curation, storage, and access, the curation of metadata is demanding, costly, time-consuming, and requires careful treatment to ensure accuracy and completeness. Therefore, such a dataset is difficult to build.
The last key implication is adequate samples. A small sample size of images may make models overfitted, which limits the development of AI systems in the diagnosis of GI diseases. Fortunately, there have been an adequate number of endoscopic images in datasets in recent years. Additionally, it is of great significance to ensure adequate high-quality samples. Specifically, multicenter, authoritative datasets are needed; therefore, we introduced two national endoscopy datasets. Meanwhile, more video-related datasets are required to improve model verification by simulating the real setting in clinical practice [75]. AI has been applied to most GI lesions, especially in intestinal polyps and inflammatory bowel disease; however, esophageal polyps, gastric cysts, and other lesions remain apparent exceptions [76]. In addition, esophageal, stomach, and colorectal cancers continue to pose major challenges to public health; however, we find that there are few endoscopy datasets regarding GI cancers [77]. Moreover, there are relatively few datasets that focus on the change between diagnosis and prognosis due to the difficulty of follow-up.
Conclusions
Publicly available endoscopy datasets, as prerequisites for computer vision-based algorithms, can be used both as training datasets or validation datasets. Endoscopy datasets can assist in the development of state-of-the-art solutions for lesion images captured by GI endoscopy, and decrease the morbidity and mortality of GI diseases. However, poor accessibility and visibility, absence of details, and inadequate samples dissuade researchers from further usage. This brief review of the comprehensive list of endoscopic imaging datasets provides potential value to promoting AI’s application in gastroenterology. We hope that more researchers can use these datasets through this review and more large, high-quality, comprehensive, annotated endoscopic imaging datasets can be made accessible.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
All authors contributed to the study conception and design. Zhu JZ conception and design; Zhu SQ drafting of the article; Zhu SQ and Yin MY literature research; Gao JW and Lin JX data extraction; Xu C and Liu L quality assessment; Zhu JZ and Xu CF critical revision of the article; Xu CF and Zhu JZ final approval of the article.
Funding
This work was supported by the National Natural Science Foundation of China (82000540), Science and Technology Plan of Suzhou City (SKY2021038), Suzhou Clinical Center of Digestive Diseases (Szlcyxzx202101), and Youth Program of Suzhou Health Committee (KJXW2019001).
Data Availability
The endoscopic imaging data supporting the findings of the review are available within the article. The websites of available datasets are provided in Table 1.
Declarations
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chunfang Xu, Email: xuchunfang@suda.edu.cn.
Jinzhou Zhu, Email: jzzhu@zju.edu.cn.
References
- 1.Nishiyama S, et al. Clinical usefulness of endocytoscopy in the remission stage of ulcerative colitis: a pilot study. J Gastroenterol. 2015;50:1087–1093. doi: 10.1007/s00535-015-1059-y. [DOI] [PubMed] [Google Scholar]
- 2.Corley DA, Levin TR, Doubeni CA. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370:2541. doi: 10.1056/NEJMc1405329. [DOI] [PubMed] [Google Scholar]
- 3.Telford JJ, Enns RA. Endoscopic missed rates of upper gastrointestinal cancers: parallels with colonoscopy. Am J Gastroenterol. 2010;105:1298–1300. doi: 10.1038/ajg.2009.739. [DOI] [PubMed] [Google Scholar]
- 4.Iddan G, Meron G, Glukhovsky A, Swain P. Wireless capsule endoscopy. Nature. 2000;405:417. doi: 10.1038/35013140. [DOI] [PubMed] [Google Scholar]
- 5.McAlindon ME, Ching HL, Yung D, Sidhu R, Koulaouzidis A: Capsule endoscopy of the small bowel. Ann Transl Med 4:369, 2016. 10.21037/atm.2016.09.18 [DOI] [PMC free article] [PubMed]
- 6.He J, Baxter SL, Xu J, Xu J, Zhou X, Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med. 2019;25:30–36. doi: 10.1038/s41591-018-0307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernal J, Sánchez J, Vilariño F. Towards automatic polyp detection with a polyp appearance model. Pattern Recognition. 2012;45:3166–3182. [Google Scholar]
- 8.Bernal J, Sánchez FJ, Fernández-Esparrach G, Gil D, Rodríguez C, Vilariño F: WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput Med Imaging Graph 43:99–111, 2015 [DOI] [PubMed]
- 9.Silva J, Histace A, Romain O, Dray X, Granado B. Toward embedded detection of polyps in WCE images for early diagnosis of colorectal cancer. Int J Comput Assist Radiol Surg. 2014;9:283–293. doi: 10.1007/s11548-013-0926-3. [DOI] [PubMed] [Google Scholar]
- 10.Tajbakhsh N, Gurudu SR, Liang J. Automated Polyp Detection in Colonoscopy Videos Using Shape and Context Information. IEEE Trans Med Imaging. 2016;35:630–644. doi: 10.1109/TMI.2015.2487997. [DOI] [PubMed] [Google Scholar]
- 11.Mesejo P, et al. Computer-Aided Classification of Gastrointestinal Lesions in Regular Colonoscopy. IEEE Trans Med Imaging. 2016;35:2051–2063. doi: 10.1109/TMI.2016.2547947. [DOI] [PubMed] [Google Scholar]
- 12.Vázquez D, et al. A Benchmark for Endoluminal Scene Segmentation of Colonoscopy Images. J Healthc Eng. 2017;2017:4037190. doi: 10.1155/2017/4037190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jha D, Smedsrud PH, Riegler MA et al.: Kvasir-seg: A segmented polyp dataset. In: International Conference on MultiMedia Modeling (MMM), pp 451–462, 2020. 10.1007/978-3-030-37734-2_37
- 14.Figueiredo I, Pinto L, Figueiredo P, Tsai R: Unsupervised segmentation of colonic polyps in narrow-band imaging data based on manifold representation of images and Wasserstein distance. Biomedical Signal Processing and Control 53:101577, 2019. 10.1016/j.bspc.2019.101577
- 15.Figueiredo P, Figueiredo I, Pinto L, Kumar S, Tsai R, Mamonov A. Polyp detection with computer-aided diagnosis in white light colonoscopy: comparison of three different methods. Endoscopy International Open. 2019;07:E209–E215. doi: 10.1055/a-0808-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel K, et al.: A comparative study on polyp classification using convolutional neural networks. PLoS One 15:e0236452, 2020. 10.1371/journal.pone.0236452 [DOI] [PMC free article] [PubMed]
- 17.Misawa M, et al. Development of a computer-aided detection system for colonoscopy and a publicly accessible large colonoscopy video database (with video) Gastrointest Endosc. 2021;93:960–967.e963. doi: 10.1016/j.gie.2020.07.060. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Peralta LF, et al. PICCOLO White-Light and Narrow-Band Imaging Colonoscopic Dataset: A Performance Comparative of Models and Datasets. Applied Sciences. 2020;10:8501. doi: 10.3390/app10238501. [DOI] [Google Scholar]
- 19.Wang W, Tian J, Zhang C, Luo Y, Wang X, Li J. An improved deep learning approach and its applications on colonic polyp images detection. BMC Med Imaging. 2020;20:83. doi: 10.1186/s12880-020-00482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y, Chen X, Cheng K, Li Y, Sun B: LDPolypVideo Benchmark: A Large-Scale Colonoscopy Video Dataset of Diverse Polyps. In: Medical Image Computing and Computer Assisted Intervention (MICCAI), pp 387–396, 2021. 10.1007/978-3-030-87240-3_37
- 21.Ji GP, et al. Video Polyp Segmentation: A Deep Learning Perspective. Machine Intelligence Research. 2022;19:1–19. [Google Scholar]
- 22.Ali S, et al. A multi-centre polyp detection and segmentation dataset for generalisability assessment. Sci Data. 2022;10:75. doi: 10.1038/s41597-023-01981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koulaouzidis A, et al. KID Project: an internet-based digital video atlas of capsule endoscopy for research purposes. Endosc Int Open. 2017;5:E477–e483. doi: 10.1055/s-0043-105488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leenhardt R, et al. CAD-CAP: a 25,000-image database serving the development of artificial intelligence for capsule endoscopy. Endosc Int Open. 2020;8:E415–e420. doi: 10.1055/a-1035-9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smedsrud PH, et al. Kvasir-Capsule, a video capsule endoscopy dataset. Sci Data. 2021;8:142. doi: 10.1038/s41597-021-00920-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong Z, et al.: Multi-Task Classification and Segmentation for Explicable Capsule Endoscopy Diagnostics. Front Mol Biosci 8:614277, 2021. 10.3389/fmolb.2021.614277 [DOI] [PMC free article] [PubMed]
- 27.de Maissin A, et al. Multi-expert annotation of Crohn's disease images of the small bowel for automatic detection using a convolutional recurrent attention neural network. Endosc Int Open. 2021;9:E1136–e1144. doi: 10.1055/a-1468-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García-Peraza-Herrera LC, et al. Intrapapillary capillary loop classification in magnification endoscopy: open dataset and baseline methodology. Int J Comput Assist Radiol Surg. 2020;15:651–659. doi: 10.1007/s11548-020-02127-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, et al. A benchmark dataset of endoscopic images and novel deep learning method to detect intestinal metaplasia and gastritis atrophy. IEEE Journal of Biomedical and Health Informatics. 2023;27:7–16. doi: 10.1109/JBHI.2022.3217944. [DOI] [PubMed] [Google Scholar]
- 30.Pogorelov K, Randel KR, Griwodz C, Lange TD, Halvorsen P: KVASIR: A Multi-Class Image Dataset for Computer Aided Gastrointestinal Disease Detection. In: the 8th Acm on Multimedia Systems Conference, pp 164–169, 2017. 10.1145/3083187.3083212
- 31.Borgli H, et al. HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci Data. 2020;7:283. doi: 10.1038/s41597-020-00622-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charoen A, et al. Rhode Island gastroenterology video capsule endoscopy data set. Sci Data. 2022;9:602. doi: 10.1038/s41597-022-01726-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montalbo F: Diagnosing gastrointestinal diseases from endoscopy images through a multi-fused CNN with auxiliary layers, alpha dropouts, and a fusion residual block. Biomedical signal processing and control 76:103683, 2022. 10.1016/j.bspc.2022.103683
- 34.Cychnerski J, Dziubich T, Brzeski A: ERS: a novel comprehensive endoscopy image dataset for machine learning, compliant with the MST 3.0 specification. arXiv e-prints, 2022. 10.48550/arXiv.2201.08746
- 35.Gastrolab. Available at: http://www.gastrolab.net/index.htm
- 36.WEO Clinical Endoscopy Atlas. Available at: http://www.endoatlas.org/index.php
- 37.Atlas of Gastrointestinal Endoscopy. Available at: http://www.endoatlas.com/atlas_1.html.
- 38.EI salvador atlas. Available at: http://www.gastrointestinalatlas.com/index.html.
- 39.Gastrointestinal Image Analysis (GIANA) Angiodysplasia D&L challenge. [Online] https://endovissub2017-giana.grand-challenge.org/home/. Accessed 20 Nov 2017
- 40.Pogorelov K, et al.: Nerthus: A Bowel Preparation Quality Video Dataset. In: the 8th Acm on Multimedia Systems Conference, pp 170–174, 2017. 10.1145/3083187.3083216
- 41.Angermann Q, et al.: Towards Real-Time Polyp Detection in Colonoscopy Videos: Adapting Still Frame-Based Methodologies for Video Sequences Analysis. In: Computer Assisted and Robotic Endoscopy and Clinical Image-Based Procedures, pp 29–41, 2017. 10.1007/978-3-319-67543-5_3
- 42.Endoscopy Artefact Detection (EAD) Dataset. [Online] 10.17632/c7fjbxcgj9.2. Accessed 30 Aug 2019
- 43.Cho M, Kim JH, Hong KS, Kim JS, Kong HJ, Kim S: Identification of cecum time-location in a colonoscopy video by deep learning analysis of colonoscope movement. PeerJ 7:e7256, 2019. 10.7717/peerj.7256 [DOI] [PMC free article] [PubMed]
- 44.Endoscopy Disease Detection and Segmentation (EDD2020). [Online] https://edd2020.grand-challenge.org/Home/
- 45.Jha D, et al.: Kvasir-Instrument: Diagnostic and Therapeutic Tool Segmentation Dataset in Gastrointestinal Endoscopy. In: International Conference on MultiMedia Modeling (MMM), pp 218–229, 2020. 10.1007/978-3-030-67835-7_19
- 46.Bae S-H, Yoon K-J: Polyp Detection via Imbalanced Learning and Discriminative Feature Learning. IEEE transactions on medical imaging 34, 2015. 10.1109/TMI.2015.2434398 [DOI] [PubMed]
- 47.Bernal J, Sanchez J, Vilariño F: Impact of image preprocessing methods on polyp localization in colonoscopy frames. In: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Conference, pp 7350–7354, 2013. 10.1109/EMBC.2013.6611256 [DOI] [PubMed]
- 48.Tajbakhsh N, Gurudu S, Liang J. A Classification-Enhanced Vote Accumulation Scheme for Detecting Colonic Polyps. Computation and Clinical Applications. 2013;8198:53–62. [Google Scholar]
- 49.Inoue HKH, et al. The Paris endoscopic classification of superficial neoplastic lesions: esophagus, stomach, and colon: November 30 to December 1, 2002. Gastrointest Endosc. 2003;58:S3–43. doi: 10.1016/s0016-5107(03)02159-x. [DOI] [PubMed] [Google Scholar]
- 50.Enns RA, et al. Clinical Practice Guidelines for the Use of Video Capsule Endoscopy. Gastroenterology. 2017;152:497–514. doi: 10.1053/j.gastro.2016.12.032. [DOI] [PubMed] [Google Scholar]
- 51.Hale M, McAlindon ME. Capsule endoscopy as a panenteric diagnostic tool. Br J Surg. 2014;101:148–149. doi: 10.1002/bjs.9321. [DOI] [PubMed] [Google Scholar]
- 52.Everson M, et al. Artificial intelligence for the real-time classification of intrapapillary capillary loop patterns in the endoscopic diagnosis of early oesophageal squamous cell carcinoma: A proof-of-concept study. United European Gastroenterol J. 2019;7:297–306. doi: 10.1177/2050640618821800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishihara R, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095–1105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norwood DA, Montalvan EE, Dominguez RL, Morgan DR. Gastric Cancer: Emerging Trends in Prevention, Diagnosis, and Treatment. Gastroenterol Clin North Am. 2022;51:501–518. doi: 10.1016/j.gtc.2022.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Riegler M, et al.: Multimedia for Medicine: The Medico Task at MediaEval. In: MediaEval Benchmarking Initiative for Multimedia Evaluation 2017, pp 13–15, 2017
- 56.Pogorelov K, et al.: Medico Multimedia Task at MediaEval 2018. In: MediaEval 2018, pp 29–31, 2018
- 57.Chang YY, et al. Development and validation of a deep learning-based algorithm for colonoscopy quality assessment. Surg Endosc. 2022;36:6446–6455. doi: 10.1007/s00464-021-08993-y. [DOI] [PubMed] [Google Scholar]
- 58.Das D, Lee CSG. A Two-Stage Approach to Few-Shot Learning for Image Recognition. IEEE Trans Image Process. 2020;29:3336–3350. doi: 10.1109/TIP.2019.2959254. [DOI] [PubMed] [Google Scholar]
- 59.Calderwood AH, Jacobson BC. Comprehensive validation of the Boston Bowel Preparation Scale. Gastrointest Endosc. 2010;72:686–692. doi: 10.1016/j.gie.2010.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai EJ, Calderwood AH, Doros G, Fix OK, Jacobson BC. The Boston bowel preparation scale: a valid and reliable instrument for colonoscopy-oriented research. Gastrointest Endosc. 2009;69:620–625. doi: 10.1016/j.gie.2008.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang CB, Kim SH, Lim YJ. Preparation of image databases for artificial intelligence algorithm development in gastrointestinal endoscopy. Clin Endosc. 2022;55:594–604. doi: 10.5946/ce.2021.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka K. Japan Endoscopy Database project. Dig Endosc. 2022;34(Suppl 2):20–22. doi: 10.1111/den.14122. [DOI] [PubMed] [Google Scholar]
- 63.Lee TJ, et al. Development of a national automated endoscopy database: The United Kingdom National Endoscopy Database (NED) United European Gastroenterol J. 2019;7:798–806. doi: 10.1177/2050640619841539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuda K, et al. Design paper: Japan Endoscopy Database (JED): A prospective, large database project related to gastroenterological endoscopy in Japan. Dig Endosc. 2018;30:5–19. doi: 10.1111/den.12964. [DOI] [PubMed] [Google Scholar]
- 65.Kodashima S, et al. First progress report on the Japan Endoscopy Database project. Dig Endosc. 2018;30:20–28. doi: 10.1111/den.12963. [DOI] [PubMed] [Google Scholar]
- 66.Oda I, Hoteya S, Fujishiro M. Status of Helicobacter pylori infection and gastric mucosal atrophy in patients with gastric cancer: Analysis based on the Japan Endoscopy Database. Dig Endosc. 2019;31:103. doi: 10.1111/den.13287. [DOI] [PubMed] [Google Scholar]
- 67.Saito Y, et al. Current status of diagnostic and therapeutic colonoscopy in Japan: The Japan Endoscopic Database Project. Dig Endosc. 2022;34:144–152. doi: 10.1111/den.13980. [DOI] [PubMed] [Google Scholar]
- 68.Rutter MD, Brookes M, Lee TJ, Rogers P, Sharp L. Impact of the COVID-19 pandemic on UK endoscopic activity and cancer detection: a National Endoscopy Database Analysis. Gut. 2021;70:537–543. doi: 10.1136/gutjnl-2020-322179. [DOI] [PubMed] [Google Scholar]
- 69.Hann A, Troya J, Fitting D. Current status and limitations of artificial intelligence in colonoscopy. United European Gastroenterol J. 2021;9:527–533. doi: 10.1002/ueg2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nogueira-Rodríguez A, et al. Deep Neural Networks approaches for detecting and classifying colorectal polyps. Neurocomputing. 2021;423:721–734. [Google Scholar]
- 71.Chetcuti Zammit S, Sidhu R. Capsule endoscopy - Recent developments and future directions. Expert Rev Gastroenterol Hepatol. 2021;15:127–137. doi: 10.1080/17474124.2021.1840351. [DOI] [PubMed] [Google Scholar]
- 72.Houwen B, Nass KJ, Vleugels JLA, Fockens P, Hazewinkel Y, Dekker E. Comprehensive review of publicly available colonoscopic imaging databases for artificial intelligence research: availability, accessibility, and usability. Gastrointest Endosc. 2023;97:184–199.e116. doi: 10.1016/j.gie.2022.08.043. [DOI] [PubMed] [Google Scholar]
- 73.Nogueira-Rodríguez A, Reboiro-Jato M, Glez-Peña D, López-Fernández H: Performance of Convolutional Neural Networks for Polyp Localization on Public Colonoscopy Image Datasets. Diagnostics (Basel) 12, 2022. 10.3390/diagnostics12040898 [DOI] [PMC free article] [PubMed]
- 74.Krause J, et al. Grader Variability and the Importance of Reference Standards for Evaluating Machine Learning Models for Diabetic Retinopathy. Ophthalmology. 2018;125:1264–1272. doi: 10.1016/j.ophtha.2018.01.034. [DOI] [PubMed] [Google Scholar]
- 75.Luo H, et al. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645–1654. doi: 10.1016/S1470-2045(19)30637-0. [DOI] [PubMed] [Google Scholar]
- 76.Zhou J, et al.: Application of artificial intelligence in gastrointestinal disease: a narrative review. Ann Transl Med 9:1188, 2021. 10.21037/atm-21-3001 [DOI] [PMC free article] [PubMed]
- 77.Arnold M, et al. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology. 2020;159:335–349.e15. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The endoscopic imaging data supporting the findings of the review are available within the article. The websites of available datasets are provided in Table 1.