Abstract
Granulomas are important immunological structures in the host defense against the fungus Paracoccidioides brasiliensis, the main etiologic agent of Paracoccidioidomycosis (PCM), a granulomatous systemic mycosis endemic in Latin America. We have performed transcriptional and proteomic studies of yeasts present in the pulmonary granulomas of PCM aiming to identify relevant genes and proteins that act under stressing conditions. C57BL/6 mice were infected with 1x106 yeasts and after 8- and 12-weeks of infection, granulomatous lesions were obtained for extraction of fungal and murine RNAs and fungal proteins. Dual transcriptional profiling was done comparing lung cells and P. brasiliensis yeasts from granulomas with uninfected lung cells and the original yeast suspension used in the infection, respectively. Mouse transcripts indicated a lung malfunction, with low expression of genes related to muscle contraction and organization. In addition, an increased expression of transcripts related to the activity of neutrophils, eosinophils, macrophages, lymphocytes as well as an elevated expression of IL-1β, TNF-α, IFN-γ, IL-17 transcripts were observed. The increased expression of transcripts for CTLA-4, PD-1 and arginase-1, provided evidence of immune regulatory mechanisms within the granulomatous lesions. Also, our results indicate iron as a key element for the granuloma to function, where a high number of transcripts related to fungal siderophores for iron uptake was observed, a mechanism of fungal virulence not previously described in granulomas. Furthermore, transcriptomics and proteomics analyzes indicated a low fungal activity within the granuloma, as demonstrated by the decreased expression of genes and proteins related to energy metabolism and cell cycle.
Keywords: paracoccidioidomycosis, granuloma, transcriptomic, proteomic, fungal infection
Introduction
Paracoccidioidomycosis (PCM) is a systemic mycosis characterized by granulomatous lesions caused by the thermically dimorphic fungus Paracoccidioides brasiliensis, P. lutzii and the recently described P. restrepiensis, P. venezuelensis, and P. americana. PCM is restricted to Latin America countries and has a high incidence in Brazil, Venezuela, and Colombia (Montenegro and Franco, 1994; Turissini et al., 2017). In Brazil, PCM ranges from 3,360 to 5,600 cases yearly, and represents the eighth cause of death among infectious and parasitic diseases. The infection occurs after the inhalation of mycelia fragments or conidia that remains as a localized benign lesion or evolve as an overt disease. The disease mainly affects rural works and can progress from a primary lesion or reactivation of a latent pulmonary focus (McEwen et al., 1987; Coutinho et al., 2015; de Oliveira et al., 2016; Martinez, 2017).
Granulomas are important immunological structures in the host defense against P. brasiliensis (Montenegro and Franco, 1994). The interaction with the host tissue initially triggers a congestive-exudative inflammatory reaction with a predominant influx of neutrophils. Progressively, these cells are replaced by macrophages, which are arranged in nodules and multinucleated giant cells, many of which contain fungi (Montenegro and Franco, 1994). With the evolution of the process, epithelioid cells are found and, concomitantly, lymphoplasmacytic halo forms. The presence of a central area of suppuration is a common finding as well as inflammatory exudate rich in lymphocytes, plasma cells, and eosinophils permeating or surrounding the granulomatous lesions. Fibrosis of varying intensity is usually found around the granuloma or areas of necrosis, which are gradually replaced by fibrous scar tissue (San-Blas, 1992; Egen et al., 2008).
Granuloma formation depends on the genetics of the host and the virulence factors of the fungus (San-Blas, 1992; Calich and Kashino, 1998; Bocca et al., 2013). Studies with granulomas from tuberculosis showed a reduced replication of Mycobacterium tuberculosis, and the induced expression of genes related to low pH, oxygen depletion, iron limitation, nitrosative stress, and nutrient starvation, revealing new stress-adaptive genes (Timm et al., 2003; Talaat et al., 2007; Garton et al., 2008). In the context of PCM, a change of environment associated with infection requires an adaptation of the fungal pathogen aiming to survive from the host immune response. This adaptation process involves changes in the pattern of gene expression, mainly of genes related to virulence and pathogenicity (San-Blas, 1992; Bocca et al., 2013). However, there is no information about which pathways are modulated in the host and in the fungus after the development of granulomatous pulmonary lesions. In contrast, previous P. brasiliensis transcriptomics and proteomics studies revealed potential virulence factors when yeasts were recovered from hepatic lesions of mice (Costa et al., 2007). In these studies, a high degree of virulence was correlated with an increase in the expression of genes involved in the glyoxylate cycle, nitrogen metabolism, and lipid biosynthesis (Costa et al., 2007). In another in vivo assay, after 6 hours of infection, yeasts had increased gene and protein expression for β-oxidation, glyoxylate cycle, pentose-phosphate pathway, and oxidative stress defense (Lacerda Pigosso et al., 2017).
Here, a transcriptomic approach was performed by using a dual-RNAseq assay from RNA extracted from pulmonary granulomatous lesions of mice 8- and 12-weeks post-infection with P. brasiliensis. We also carry out proteomic analysis of yeasts isolated from these lesions and from yeasts that were cultivated after recovery from lesions. Our data suggested an active and regulated host immune innate response, with elevated expression of genes that were not previously described in PCM, such as Ptx3, Lcn2, Clec4a2, when compared with healthy, uninfected lungs. Furthermore, our data indicated prominent Th1 and Th17 responses tightly regulated by checkpoint inhibition molecules. Additionally, there is an indication that iron is a major player in the chronic murine PCM granuloma, where the repression of the fungus energy metabolism occurs along with an increase in siderophore synthesis for iron uptake, revealing virulence mechanisms that have never been described in fungal granulomas.
Materials and methods
Ethics statement
All the experiments were performed in strict accordance with the Brazilian Federal Law 11,794 establishing procedures for the scientific use of animals, and the State Law establishing the Animal Protection Code of the State of São Paulo. All efforts were made to minimize animal suffering. The procedures were approved by the Ethics Committee on Animal Experiments of the Federal University of São Paulo (N° 9596270919).
Animals
8 to 12 weeks old male C57BL/6 mice obtained from the Center for the Development of Experimental Models for Biology and Medicine (CEDEME-UNIFESP) and bred as specific pathogen-free mice in the animal facility of the Institute of Science and Technology at UNIFESP were used throughout this study.
Paracoccidioides brasiliensis
The virulent strain Pb18 of the fungus P. brasiliensis was used and maintained by weekly sub-cultivation in a semi-solid medium of Fava Netto (Netto, 1955) at 37°C. The fungus was collected and washed with phosphate-buffered saline (PBS, pH 7.4). The number of yeasts was evaluated using a hemocytometer and their viability was determined using the Janus-Green vital dye. All procedures were performed with fungal suspension with viability greater than 95%.
Infection of mice
Groups of C57BL/6 WT mice were anesthetized with intraperitoneal (ip.) injection of ketamine (90 mg/kg) and xylazine (10 mg/kg) and subjected to intratracheal (IT.) infection as previously described (Cano et al., 1995). Briefly, after anesthesia the animals were infected with 1 x 106 yeasts of P. brasiliensis contained in 50 μL of PBS by surgical (IT.) inoculation, allowing the direct dispense of the yeasts into the lungs. After the infection the experiments were performed as showed in Figure 1 .
Figure 1.
Schematic figure of the experiments.
Histological analysis and colony-forming unit assays
To analyze the histopathology of the infected lungs, groups of 3-4 animals with two, six, eight and twelve weeks of infection were euthanized with xylazine (30 mg/kg) and ketamine (180 mg/kg) and the removed lungs were stored in 10% formaldehyde at 4 °C. Sections of 5 µm were stained with Hematoxylin-Eosin (H&E) for analysis of lesions and stained with Grocott for fungal evaluation. The anatomy of the lesion was analyzed according to size, morphology, and presence of fungi. The measurement of the area of the lesions was performed using the Leica DM750 microscope and the Leica Application Suite V4.8 software, thus allowing the observation of the establishment of the granuloma through time. To evaluate the fungal load a colony-forming unit (CFU) assay was performed. Briefly, the granulomatous lesions were separated from the apparently healthy regions with the help of a scalpel, and the tissue was macerated with a Doucer. Then, the cells were centrifuged at 1,200 x g for 10 minutes and the supernatant was discarded. The pellet was resuspended in 1 mL before cultivation of the yeasts in BHI. The colonies were counted daily.
Granulomatous lesions extraction and yeast recovery
After eight and twelve weeks of infection, 3 independent groups with 2-4 animals were euthanized, the lungs were removed, and the granulomatous lesions were separated from the apparently healthy lung regions with the help of a scalpel. For the transcriptomics analysis the entire granulomas were frozen before the RNA extraction. For the proteomic analyzes of yeasts present in the granulomas, the lesions were enzymatically digested with collagenase (2 mg/mL) for 40 minutes at 37°C, as previously described (Lindell et al., 2006). The digested tissue was gently macerated with a Doucer and then centrifuged at 1,200 x g for 10 minutes. The supernatant was discarded, and the pellet resuspended in 2 mL of distilled water. Then 1 mL of this suspension was dispensed in two different Petri dishes, and the volume completed with 4 mL of distilled water. The dishes were incubated at 37 °C for 1 h to lyse the mouse cells. The suspension was then centrifuged (1,200 x g, 10 minutes) and the supernatant discarded, remaining only fungi cells in the tube. To evaluate the virulence and the proteins expressed by yeasts recovered from granulomas, these lesions were macerated with a Doucer, centrifuged (1,200 x g, 10 minutes) and then pellet resuspended in Phosphate-buffered saline (PBS). The recovered yeasts were cultivated in BHI for 5 days before cultivation in Fava Netto medium for 7 days (recovered yeast). The cultivation of the yeasts in BHI before cultivation in Fava Netto medium is necessary because there is growth factor in the BHI composition, which is essential for fungal growth after recovery from the lesions.
Mortality rate
To evaluate the virulence of cultivated yeasts recovered from granulomas (experimental group) in comparison to the yeasts used in the original inoculation (control group), a group of six C57BL/6 WT mice IT. infected with the recovered fungus and another group of 6 mice was infected with the control fungus. Deaths were recorded daily. The Log-rank test (Mantel-Cox) was used for the survival curve.
Total RNA extraction
Frozen granulomatous lesions were first macerated with the help of a pestle and pot. To proceed with total RNA extration, TRIzol LS (Thermo Scientific) was used according to the protocol described by Pisu et al., 2020. For the control lung was used uninfected healthy lungs, while for the control yeasts was used the original yeast suspension used in the infection. The control lung and control fungus were mechanical lysed with pestle and pot and zirconium beads, respectively, followed by RNA extraction with TRIzol LS (Thermo Scientific) according to the manufacturer’s protocol. After RNA extraction, the DNA was digested with Rnase-free Dnase (RQ1 Rnase-Free Dnase, Promega Corporation) in the presence of the ribonuclease inhibitor Rnasin® (Promega Corporation). The purification was done with the addition of 1 volume of phenol:chloroform (1:1), followed by 100% ethanol:3M sodium acetate (10:1) RNA precipitation. RNA integrity was verified with a 2100 Bioanalyzer (Agilent) and RNA 6000 Nano Kit (Agilent).
cDNA preparation, sequencing, and alignment
First, rRNA was depleted using the RiboMinus™ Eukaryote Kit for RNA-Seq (Thermo Scientific). Then, cDNA library was synthesized using the NEBNext® Ultra II Directional RNA Library Prep Kit for Illumina® (New England BioLabs) and the NEBNext Multiplex oligos E7335 and E7500 (New England BioLabs). The cDNA library was sequenced in the NGS equipment Brazilian Biorenewables National Laboratory (LNBR), part of the Brazilian Centre for Research in Energy and Materials (CNPEM), a private non-profit organization under the supervision of the Brazilian Ministry for Science, Technology, and Innovations (MCTI). Because there is genetic material from two different organisms in the sample, these reads refer to two different genomes. Therefore, we chose to align against a chimeric genome of P. brasiliensis (Refseq accession number GCF_000150735.1) and Mus musculus (Refseq accession number GCF_000001635. 27). Bowtie2 was used in the default configuration for paired reads, but as it only returns the best pairing by default, the output mode was changed to report the best 5 alignments.
Yeast protein extraction and trypsin digestion
Proteins from the yeasts were extracted using the Yeast Buster Protein Extraction Reagent (MerckMillipore) according to the manufacturer’s specifications. Benzonase Nuclease (MerckMillipore), protease inhibitors (Protease Inhibitor Cocktail, Sigma Aldrich) and phosphatase inhibitors (PhoSTOP, Sigma Aldrich) were also added to the buffer. In-solution trypsin digestion was performed according to Kleifeld et al., 2011 with modifications (Kleifeld et al., 2011). Briefly, a solution of 6 M guanidine hydrochloride (GuHCl) was added to a sample of 100 μg of protein from each cell lysate to a final concentration of 3 M GuHCl, followed by the addition of 5 mM dithiothreitol (DTT) (final concentration). The mixture was incubated at 37°C for 1 h. Iodoacetamide (IAA) was then added to a final concentration of 15 mM and the samples were incubated for 30 min at room temperature, in the dark. To quench the excess of IAA, DTT was added to a final concentration of 15 mM followed by the incubation of samples for 20 min at room temperature. Clean-up of samples was performed by the addition of ice-cold acetone (8 volumes) and methanol (1 volume), followed by the incubation of samples for 3 h at -80° C. After centrifugation at 14,000 x g for 10 min, protein pellets were washed twice with one volume of ice cold methanol and then resolubilized with NaOH solution (final concentration of 10 mM), followed by the addition of HEPES buffer (final concentration of 50 mM), pH 7.5, to a final volume of 100 μL. Trypsin (Proteomics grade; Sigma, USA) was added at 1:100 ratio (enzyme/substrate) and protein samples were incubated at 37°C for 18 h.
Tryptic peptides were desalted using C-18 cartridges SPE Extraction disks (3M EmporeTM), resuspended in 50 μL of 0.1% formic acid, quantified with Pierce™ Quantitative Colorimetric Peptide Assay (Thermo Scientific) and stored at -80° C.
LC-MS/MS and protein identification
LC-MS/MS was made using the Thermo Fisher Q-Exactive mass spectrometer coupled to a Dionex RSLC nano. LC gradients ran from 4% to 35% B over 2 h, and data were collected using a Top15 method for MS/MS scans (O'Keeffe et al., 2014; Dolan et al., 2017). Mass spectrometric (RAW) data were analyzed with MaxQuant software (version 2.0.3.0). A False Discovery Rate (FDR) of 1% was required for both protein and peptide-to-spectrum match identifications. Raw data were searched against a target database restricted to the taxonomy ‘Paracoccidioides brasiliensis’ (UniProt/Proteomes - UP000001628; 8,399 entries). This database was also combined with the sequences of 245 common contaminants and concatenated with the reversed versions of all sequences. Enzyme specificity was set to trypsin and up to two missed cleavages were allowed; cysteine carbamidomethylation was selected as fixed modification whereas methionine oxidation, glutamine/asparagine deamidation and protein N-terminal acetylation were selected as variable modifications. Peptide identification was based on a search with an initial mass deviation of the precursor ion of 4.5 ppm and the fragment mass tolerance was set to 20 ppm. Label-free quantitation was performed using the MaxLFQ algorithm (Cox and Mann, 2008; Cox et al., 2014) with the ‘re-quantify’ function of MaxQuant software enabled. As is observed from complex proteomes such as those of vertebrates, peptides can be shared between homologous proteins or splice variants, leading to “protein groups”. For each protein group in the MaxQuant’s ‘proteinGroups.txt’ file, the first protein entry was selected as representative.
Differentially expressed genes and proteins analysis
Transcriptomic data were normalized by log2 and Z-score. For proteomics data, all the protein intensity values were log2-transformed and quantile-normalized using the ‘preprocessCore’ library in R scripting and statistical environment (Ihaka and Gentleman, 1996; Bolstad et al., 2003). Statistical analyses were performed using the ‘limma’ package in R/Bioconductor, using eBayes for granuloma data and treat for the recovered yeast (Gentleman et al., 2004; Smyth, 2004; Ritchie et al., 2015). Differentially expressed genes (DEG) and proteins analyzed by eBayes were delimited by an adjusted p-value < 0,05 and a cutoff of |fold change| > 1, where the p-values were adjusted for multiple testing with the Benjamini-Hochberg method. The data analyzed by treat don’t need a delimitation, as specified in the limma package user’s guide. Correlations between replicates were also calculated in R, using Pearson correlation coefficient. Genes and proteins that were differentially expressed in both infection groups in comparison to the control group were reanalyzed by pooling the expression values for 8 and 12 weeks, generating a new log (Fold change) and adjusted P-value for the disease for both experimental time points (disease).
Functional analysis
For function prediction, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) annotations were taken, as well as enrichment analyzes (Ashburner et al., 2000; Kanehisa and Goto, 2000; Kanehisa, 2019; Gene Ontology Consortium, 2021; Kanehisa et al., 2023). The P. brasiliensis Gene Onthology functions were taken from FungiDB. Gene Onthology enrichment was performed by Enrichr for Mus musculus, using default settings (Chen et al., 2013; Kuleshov et al., 2016; Xie et al., 2021). The P. brasiliensis metabolic pathway was obtained from KEGG.
Staining of neutral lipids in yeast
Fungi obtained from 8 weeks granulomatous lesions, fungi recovered from granulomas and control fungi were stained with Oil Red O (Merck Millipore) using the manufacturer’s protocol with minor modifications. Briefly, a 0.5% solution of Oil Red O in isopropanol was diluted in water at a 3:2 ratio and incubated at room temperature (~ 25°C) for 1 hour. After that, the diluted solution was filtered using a 0.22 μm filter. The cells, previously fixed in 10% paraformaldehyde, were centrifuged and the supernatant discarded; then, the filtered Oil Red O solution was added in sufficient quantity to cover the fungi. Staining took place for 2 hours at room temperature. Subsequently, the cells were washed twice with ultrapure water and then resuspended in PBS for analysis on a microscope slide. The photos were taken using the Leica DM750 microscope and the Leica Application Suite V4.8 software, thus allowing the observation of stained neutral lipids inside the cells.
Secondary metabolites extraction and analysis
Lung granuloma, control lung, control yeasts and recovered yeasts were frozen, and 100 mg of frozen material was macerated with the help of a pestle and pot. The secondary metabolites extraction was performed with 1 mL of methanol (100%; HPLC grade), followed by sonication bath for 40 minutes. For sample preparation, 500 L of each sample extract were filtered in a 0.22 μm PTFE and diluted to 1 mL volume with HPLC-grade methanol. LC-HRMS/MS analysis were performed in positive ionization mode in a Thermo Scientific QExactive Hybrid Quadrupole-Orbitrap Mass Spectrometer, with m/z range of 100–1500, capillary voltage at 3.5 kV, source temperature at 300°C and S-lens 50 V. The stationary phase was a Thermo Scientific Accucore C18 2.6 μm (2.1 mm x 100 mm) column. Mobile phase composition was a gradient of formic acid (A) and acetonitrile (B). Eluent profile (A/B %): 95/5 for 5 minutes, up to 60/40 within 5 minutes, up to 55/45 within 2 minutes, up to 2/98 for 6 minutes, maintaning 2/98 for 2 minutes, down to 95/5 within 2 minutes, maintaning 95/5 for 2 minutes. Total run time was 24 minutes for each sample run and flow rate was 400 µL/min. Injection volume was 5 μL. MS spectra were acquired with m/z ranges from 100 to 1500, with 70000 mass resolution at 200 Da. MS2 spectra were acquired in data-dependent acquisition (DDA) mode. Normalized collision energy was applied stepwise (20, 30 and 40), and the 5 most intense precursors per cycle were fragmented with 17500 resolutions at 200 Da. Spectra data were processed with Xcalibur software (version 3.0.63) developed by Thermo Fisher Scientific. Compound Ferricrocin was annotated based on its exact mass and MS/MS fragmentation pattern.
RT-qPCR
The total RNA extracted from granulomas was subjected to reverse transcriptase using ImProm-II (Promega) according to the manufacturer’s instructions. The synthesized cDNA was used for real-time PCR analysis using the SYBR green PCR master mix kit (Applied Biosystems) on the ABI 7500 Fast real-time PCR system (Applied Biosystems, Foster City, CA, USA). For the M. musculus, Usp9x was used as normalizer, while for the P. brasiliensis, pyruvate decarboxylase (PADG_00714) was employed. The genes and primers used for RNA-seq validation are listed in Supplementary Table 1 .
Statistical analysis
For statistical analyses, the mean ± standard deviation/standard error was compared. For comparison between two groups, an unpaired Student’s t-test was performed, while for multiple analyses, a one-way ANOVA was conducted with Bonferroni correction. The level of significance was set at p < 0.05. The Log-rank test (Mantel-Cox) was employed for survival curve analysis, for comparison between two groups.
Results
The granuloma is well-established after 8 weeks of infection
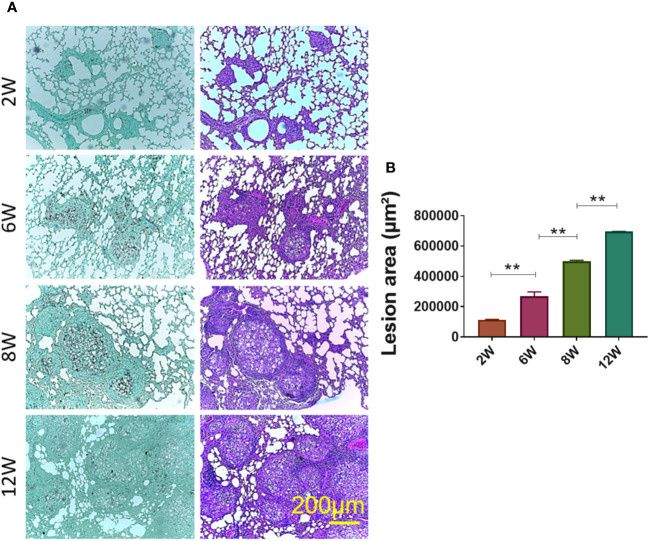
C57BL/6 mice were infected with 1x106 P. brasiliensis yeasts via IT. route. Histological sections of two, six, eight and, twelve weeks of infection indicated that eight and twelve weeks of infection are the most adequate time points to obtain chronic granulomatous lesions considering the lesions organization and lung area occupied ( Figures 2A, B ).
Figure 2.
Establishment of granuloma. Groups of four C57BL/6 mice were infected by the IT. route with 1 × 106 P. brasiliensis yeasts contained in 50 µL of PBS. After two (2W), six (6W), eight (8W) and twelve (12W) weeks of infection, the animals were euthanized and the lungs removed, which were stored in 10% formaldehyde at 4 °C. Sections of 5 µm were stained with Hematoxylin-Eosin (H&E; pink) for analysis of lesions and stained with Grocott (green) for fungal evaluation (A). The anatomy of the lesion was analyzed according to size, morphology and presence of fungi. The image and the lesion area calculation were taken using the Leica DM750 microscope and the Leica Application Suite V4.8 software. Statistical analysis was performed using one-way ANOVA. Bars represent means ± standard error of lesion area (µm²) of groups of three mice (B) **p<0.01.
To evaluate the fungal load a CFU assay was performed with the granulomatous lesions and the apparently healthy lung tissue. As it can be seen, approximately 90% of the yeasts present in the lung are within the granulomatous lesions, whereas 10% are present in the apparently healthy lung tissues ( Supplementary Figure 1 ). Thus, all results from the granulomatous lesions comprise 90% of yeasts present in the lungs.
Murine transcriptional profiling of pulmonary granulomatous lesions reveals new players in PCM host immune response
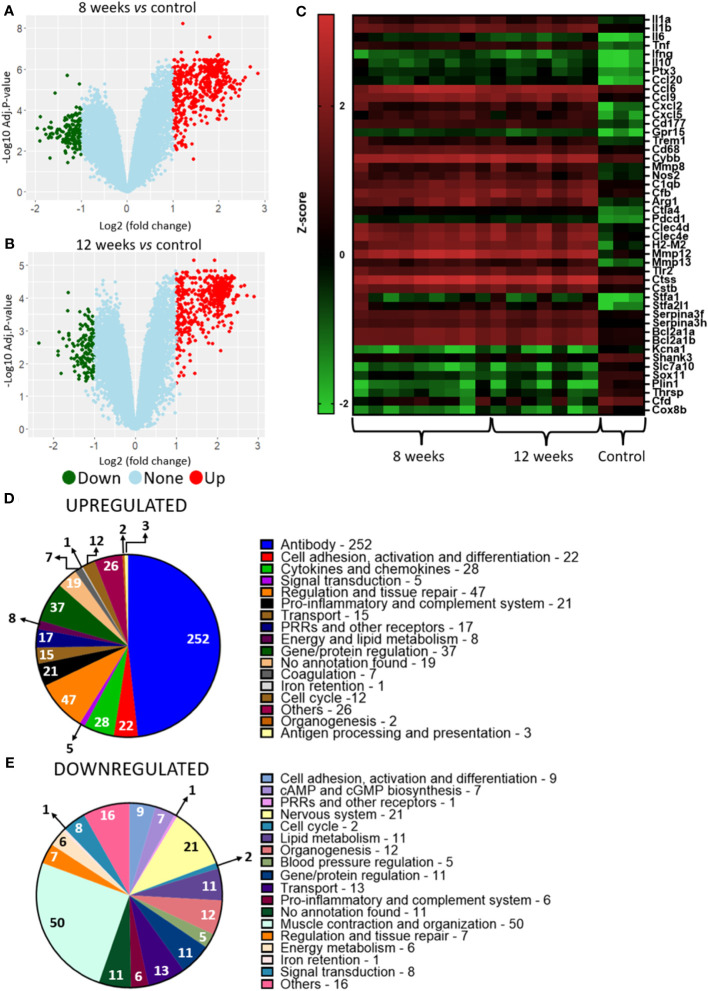
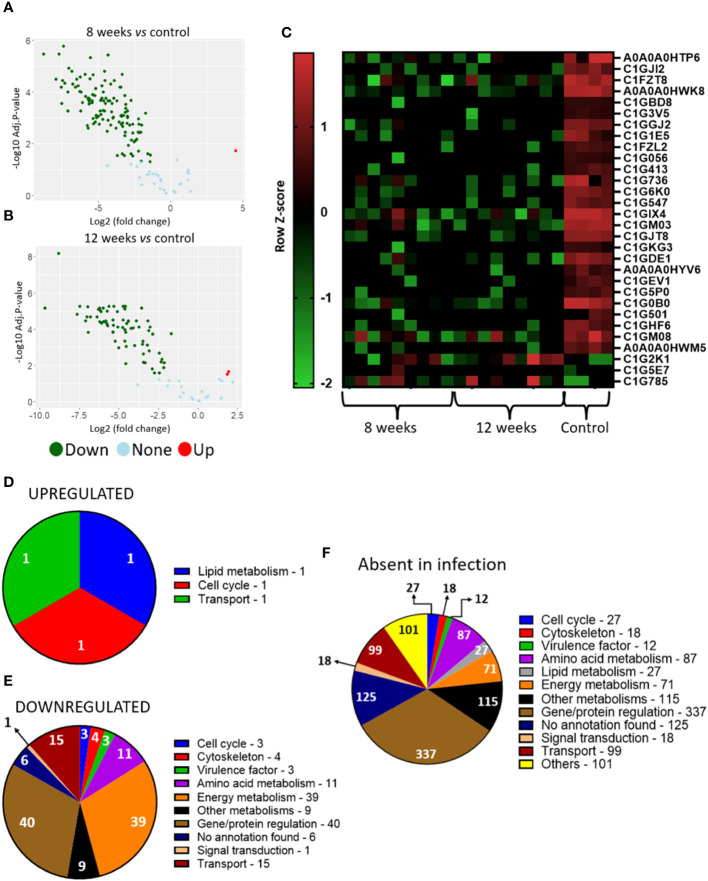
Analysis of hosts’ lung transcriptional profile comparing 8-weeks infected lungs to non-infected lungs show 563 transcripts upregulated and 131 with downregulated, while lungs from 12-weeks infection display 509 up- and 136 downregulated genes ( Figures 3A–C ). Functional analysis of the host’s transcripts whose abundance increased during P. brasiliensis infection using KEGG, GO and Panther databases identified transcripts encondig Pattern Recognition Receptors (PRRs) and other receptors that are related to antigen presentation, such as CD68, Ptx3 (Pentraxin-related protein PTX3), Clec4a2 (C-type lectin domain family 4 member A2), Clec4d (C-type lectin domain family 4 member D), Clec4n (C-type lectin domain family 4 member N), Clec7a (C-type lectin domain family 7 member A), TLR13 (toll-like receptor 13), TLR2 (toll-like receptor 2), and H2-M2 gene (Histocompatibility 2, M region locus 2) ( Figure 3D ; Table 1 ). Likewise, transcripts of some cytokines such as IL-1α, IL-1β, TNF and IFN-γ and chemokines (CXCL2, CXCL9, CXCL5, CCL8, CCL7, CCL9, CCL20, CXCL1, CC20, CXCL10, CCL6, CCL4 and CXCL13) were also increased in granulomatous host cells. The abundance of transcripts related to cell activation and chemotaxis functions (which are not chemokines) were also found in the disease, such as Cd177 and Trem1 (triggering receptor expressed on myeloid cells 1), Trem3 (triggering receptor expressed on myeloid cells 3) and Gpr15 (G protein-coupled receptor 15). Transcripts for pro-inflammatory proteins were also identified as more abundant in infected lungs, such as Nos2 (nitric-oxide synthase), Gbp2b (Guanylate-binding protein 2b), Cd300ld (CMRF35-like molecule 5), Cfb, C1qb, C3ar1, and C1qc. Transcripts for immune regulatory proteins, such as Arg1 (arginase-1), Il1rn (IL-1 receptor antagonist), CTLA4 (cytotoxic T-lymphocyte-associated protein 4) and, Pdcd1 (programmed cell death protein 1), as well as genes related to tissue repair (matrix metalloproteinase-12 (Mmp12) and collagenase 3 (Mmp13) were also found in higher levels when compared to transcripts in naïve lung control. Additionally, a high number of transcripts related to antibody chains were also identified as more expressed in the fungal granuloma. Taken together, these data point to a prominent and highly regulated Th1 response in addition to the production of antibodies by the host immune system.
Figure 3.
Murine transcriptional profiling of pulmonary granulomatous lesions reveals new players in PCM host immune response. Granulomatous lesions were extracted from two to four mice from three independent infections at eight weeks (8W) and twelve weeks (12W) after being infected with 1 x 106 P. brasiliensis yeasts. Then, extraction of total RNA, removal of rRNA, and assembly of cDNA libraries were performed. The samples were sequenced and the fragments aligned with the reference genome for M. musculus using bowtie2. The FeatureCounts software was used to count the amount of each identified gene, which were normalized. Then, a differentially expressed genes analysis was made using limma and a functional analysis was carried out. To illustrate the differentially expressed genes, a volcano plot of the 8-weeks infection (8W) group in relation to the uninfected control (A) and a volcano plot of the 12-weeks infection group (12W) in relation to the uninfected control (B) were made. Additionally, a heatmap was created with selected genes (C). The genes in red represent a higher expression and the green ones a lower expression for a given sample. The functions of the upregulated genes (D) and downregulated genes (E) were taken using GO, KEGG and Panther.
Table 1.
Differential expression of mice genes.
| Acession number | Protein | Expression status | Log (Fold Change) | Adjusted p-value |
|---|---|---|---|---|
| UPs | ||||
| PRRs and other receptors | ||||
| Clec4n | C-type lectin domain family 6 member A | UP(D) | 2,078333 | 3,78E-05 |
| Clec7a | C-type lectin domain family 7 member A | UP(D) | 2,020814 | 3,01E-05 |
| Tlr2 | toll-like receptor 2 | UP(D) | 1,960263 | 2,92E-05 |
| Tlr13 | toll-like receptor 13 | UP(D) | 1,903147 | 3,03E-05 |
| Clec4d | C-type lectin domain family 4 member D | UP(D) | 1,868473 | 0,000104 |
| Ptx3 | Pentraxin-related protein PTX3 | UP(D) | 1,851374 | 2,55E-05 |
| Clec4e | C-type lectin domain family 4 member E | UP(D) | 1,715215 | 5,88E-05 |
| Cd68 | CD68 antigen | UP(D) | 1,338493 | 0,001332 |
| Cell adhesion, activation and differentiation | ||||
| Gpr15 | G protein-coupled receptor 15 | UP(D) | 2,086638 | 3,19E-05 |
| Trem3 | Triggering receptor expressed on myeloid cells 3 | UP(D) | 1,611529 | 8,33E-06 |
| Trem1 | triggering receptor expressed on myeloid cells 1 | UP(D) | 1,408704 | 0,002771 |
| Cd177 | CD177 antigen | UP(D) | 1,202699 | 0,000194 |
| Pro-inflammatory and complement system | ||||
| Nos2 | nitric-oxide synthase, inducible [EC:1.14.13.39] | UP(D) | 2,033614 | 3,01E-05 |
| C1qa | Complement C1q subcomponent subunit A | UP(D) | 2,025756 | 1,64E-05 |
| Cfb | complement factor B [EC:3.4.21.47] | UP(D) | 1,935486 | 2,77E-05 |
| Cybb | NADPH oxidase 2 [EC:1.-.-.-] | UP(D) | 1,775452 | 6,73E-05 |
| C1qc | complement C1q subcomponent subunit C | UP(D) | 1,729555 | 0,000174 |
| C3ar1 | C3a anaphylatoxin chemotactic receptor | UP(D) | 1,615371 | 8,47E-05 |
| C1qb | complement C1q subcomponent subunit B | UP(D) | 1,346731 | 0,000138 |
| Cd300ld | CMRF35-like molecule 5 | UP(8) | 1,049027 | 7,73E-07 |
| Cytokines and chemokines | ||||
| Cxcl9 | C-X-C motif chemokine 9 | UP(D) | 2,2741 | 1,45E-05 |
| Ccl20 | C-C motif chemokine 20 | UP(D) | 2,103374 | 4,42E-05 |
| Cxcl1 | C-X-C motif chemokine 1/2/3 | UP(D) | 2,073216 | 0,000421 |
| Ccl7 | C-C motif chemokine 7 | UP(D) | 2,057447 | 2,19E-05 |
| Cxcl10 | C-X-C motif chemokine 10 | UP(D) | 2,011544 | 8,14E-05 |
| Cxcl13 | C-X-C motif chemokine 13 | UP(D) | 1,95105 | 1,27E-05 |
| Tnf | tumor necrosis factor superfamily, member 2 | UP(D) | 1,930536 | 1,23E-05 |
| Il1b | interleukin 1 beta | UP(D) | 1,895732 | 7,86E-05 |
| Ccl8 | C-C motif chemokine 8 | UP(D) | 1,886362 | 2,85E-05 |
| Il1a | interleukin 1 alpha | UP(D) | 1,850909 | 7,78E-06 |
| Cxcl2 | C-X-C motif chemokine 1/2/3 | UP(D) | 1,208573 | 0,000461 |
| Ccl6 | C-C motif chemokine 6 | UP(D) | 1,161484 | 1,39E-05 |
| Cxcl5 | C-X-C motif chemokine 5/6 | UP(D) | 1,114359 | 2,2E-05 |
| Ifng | interferon gamma | UP(8) | 1,105272 | 0,001062 |
| Ccl4 | C-C motif chemokine 4 | UP(D) | 1,103517 | 3,92E-05 |
| Ccl9 | C-C motif chemokine 9 | UP(D) | 1,070775 | 0,002911 |
| Antigen processing and presentation | ||||
| H2-M2 | Histocompatibility 2, M region locus 2 | UP(D) | 1,964557 | 3,04E-05 |
| Antibody | ||||
| Ighv1-56 | Immunoglobulin heavy variable 1-56 | UP(D) | 2,417073 | 2,54E-05 |
| Igha | Immunoglobulin heavy constant alpha | UP(D) | 2,233271 | 2,44E-05 |
| Ighv1-22 | Immunoglobulin heavy variable 1-22 | UP(D) | 2,007573 | 1,24E-05 |
| Ighv1-21-1 | Immunoglobulin heavy variable 1-21-1 | UP(D) | 2,002292 | 5,28E-05 |
| Regulation and tissue repair | ||||
| Pdcd1 | programmed cell death protein 1 | UP(D) | 2,073949 | 4,75E-05 |
| Arg1 | Arginase-1 | UP(D) | 2,059942 | 4,73E-05 |
| Mmp12 | matrix metalloproteinase-12 (macrophage elastase) [EC:3.4.24.65] | UP(D) | 1,548912 | 8,8E-05 |
| Mmp13 | Collagenase 3 | UP(D) | 1,522556 | 2,12E-05 |
| Ctla4 | cytotoxic T-lymphocyte-associated protein 4 | UP(D) | 1,368578 | 0,000165 |
| Il1rn | interleukin 1 receptor antagonist | UP(D) | 1,244346 | 0,000409 |
| Gene/protein regulation | ||||
| Serpina3f | Serine protease inhibitor A3F | UP(D) | 2,06118 | 7,26E-05 |
| Cstb | cystatin-A/B | UP(D) | 1,830246 | 1,44E-05 |
| Cstdc4 | Cystatin domain-containing 4 | UP(D) | 1,791522 | 2,81E-05 |
| Serpina3g | Serine protease inhibitor A3G | UP(D) | 1,695611 | 0,001231 |
| Ctss | cathepsin S [EC:3.4.22.27] | UP(D) | 1,520351 | 3,51E-05 |
| Stfa2l1 | stefin A2 like 1 | UP(D) | 1,390768 | 0,000282 |
| Stfa2 | cystatin-A/B | UP(D) | 1,205505 | 4,08E-05 |
| Csta2 | cystatin-A/B | UP(D) | 1,088477 | 0,00027 |
| Serpina3h | Serine protease inhibitor A3H | UP(D) | 1,082125 | 0,000453 |
| Ctsk | cathepsin K [EC:3.4.22.38] | UP(D) | 1,064556 | 3,06E-05 |
| Energy and lipid metabolism | ||||
| Gla | Growth factor independent protein 1 | UP(D) | 1,303367 | 0,001139 |
| Pla1a | phosphatidylserine sn-1 acylhydrolase [EC:3.1.1.111] | UP(D) | 1,285157 | 1,13E-05 |
| Pla2g7 | platelet-activating factor acetylhydrolase [EC:3.1.1.47] | UP(D) | 2,167659 | 0,000343 |
| Cell cycle | ||||
| Bcl2a1d | hematopoietic Bcl-2-related protein A1 | UP(D) | 1,946559 | 1,99E-05 |
| Bcl2a1c | hematopoietic Bcl-2-related protein A1 | UP(D) | 1,878795 | 2,29E-05 |
| Bcl2a1a | hematopoietic Bcl-2-related protein A1 | UP(D) | 1,806497 | 1,6E-05 |
| Bcl2a1b | hematopoietic Bcl-2-related protein A1 | UP(D) | 1,722721 | 0,0003 |
| Iron retention | ||||
| Lcn2 | lipocalin 2 | UP(D) | 1,294766 | 0,000263 |
| DOWNs | ||||
| Muscle contraction and organization | ||||
| Tnnc1 | troponin C, slow skeletal and cardiac muscles | DOWN(D) | -1,77016 | 0,000138 |
| Myh4 | Myosin-4 | DOWN(8) | -1,7189 | 0,002297 |
| Myh13 | myosin heavy chain 1/2/3/4/8/13/7B/15 | DOWN(8) | -1,61232 | 0,00143 |
| Tnnt2 | troponin T, cardiac muscle | DOWN(D) | -1,45826 | 0,011979 |
| Tnni3 | troponin I, cardiac muscle | DOWN(D) | -1,22069 | 0,003786 |
| Myh3 | Myosin-3 | DOWN(8) | -1,18001 | 0,007459 |
| Myh6 | myosin heavy chain 6/7 | DOWN(D) | -1,17542 | 0,000529 |
| Myl1 | Myosin light chain 1/3, skeletal muscle isoform | DOWN(8) | -1,12704 | 0,006081 |
| Nervous system | ||||
| Kcna1 | potassium voltage-gated channel Shaker-related subfamily A member 1 | DOWN(D) | -1,43254 | 0,005246 |
| Slc17a7 | MFS transporter, ACS family, solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6/7/8 | DOWN(8) | -1,32168 | 0,000511 |
| Kcnh2 | potassium voltage-gated channel Eag-related subfamily H member 2 | DOWN(D) | -1,3173 | 0,013463 |
| Shank3 | SH3 and multiple ankyrin repeat domains protein | DOWN(D) | -1,25195 | 0,005262 |
| Slc7a10 | solute carrier family 7 (D/L-type amino acid transporter), member 10 | DOWN(D) | -1,13547 | 0,00345 |
| Organogenesis | ||||
| Bmp6 | bone morphogenetic protein 6 | DOWN(D) | -1,4241 | 0,008604 |
| Sox11 | transcription factor SOX11/12 (SOX group C) | DOWN(D) | -1,13558 | 0,002878 |
| Lipid metabolism | ||||
| Cyp1a1 | cytochrome P450 family 1 subfamily A1 [EC:1.14.14.1] | DOWN(D) | -1,23017 | 0,001407 |
| Thrsp | thyroid hormone responsive | DOWN(D) | -1,14802 | 0,001858 |
| Plin1 | perilipin-1 | DOWN(D) | -1,13331 | 0,00144 |
| Energy metabolism | ||||
| Cox8b | cytochrome c oxidase subunit 8 | DOWN(D) | -1,3867 | 0,004553 |
| Cox7a1 | cytochrome c oxidase subunit 7a | DOWN(12) | -1,12561 | 0,008107 |
| Pro-inflammatory | ||||
| Cfd | complement factor D [EC:3.4.21.46] | DOWN(D) | -1,04996 | 0,001562 |
| Iron retention | ||||
| Hamp | hepcidin | DOWN(D) | -1,50399 | 0,002563 |
Granulomatous lesions were extracted from two to four mice from three independent infections at eight weeks and twelve weeks after being infected with 1x106 P. brasiliensis yeasts. Then, extraction of total RNA, removal of rRNA, and assembly of cDNA libraries were performed. The samples were sequenced and the fragments aligned with the reference genome for M. musculus using bowtie2. The FeatureCounts software was used to count the amount of each identified gene, which were normalized. Then, a differentially expressed gene was made using the limma and a functional analysis was carried out using GO, KEGG and Panther. The Table shows the differentially expressed genes with the values of Log (Fold change) and adjusted p-value. The expression status column represents the upregulated or downregulated genes for the eight-week infection group (8), the twelve-week infection group (12), and the genes that were differentially expressed in both infection groups were combined and reanalyzed, resulting in an expression status designation for the disease (D).
Transcripts to IL-17 and IL-21 were found exclusively in granulomas from infected animals, as well as some transcripts related to PRRs (the C-type lectin domain family 6 member A and theCD209 antigen-like protein E), several variable portions of T cell receptors, myeloperoxidase and genes related to the regulation of the immune response and tissue repair (Matrix metalloproteinase-1, Matrix metalloproteinase-7) ( Supplementary Figure 2 ), indicating a Th17 response in the granuloma. Together with the enrichment analysis where a Th17-related response was present in higher abundance in the lung granuloma when compared to uninfected lungs, the transcripts profile exclusively found in our analysis ( Supplementary Table 5 ) support that the susceptibility of the mice linage used in this work might be a Th17 response. Not only transcripts for immune response proteins were upregulated within the granuloma, but also transcripts for enzymes related to lipid metabolism such as Pla2g7 (platelet-activating factor acetylhydrolase), Gla (alpha-galactosidase) and Pla1a (phosphatidylserine sn-1 acylhydrolase), as well as genes related to protein regulation (Ctss, Ctsk, Stlfa2l1, stfa2, stfa1, Cstdc6, Csta2, Cstb, Serpina3g, Serpina3h and Serpina3f) and cell cycle/cell death regulation (Bcl2a1b, Bcl2a1c, Bcl2a1d and Bcl2a1a).
In contrast, genes involved in muscle organization and contraction, such as myosin and troponin, nervous system, action potential and organogenesis (Shank3, Slc7a10, Slc17a7, Kcna1, Kcnh2 and Sox11) were down-regulated ( Figure 3E ; Table 1 ). The repression of these genes may be related to impaired lung function within the granuloma since immune cells are expected to be located there, and as seen from Figure 2A there are less normal lung tissue present. Interestingly, lipid metabolism was also identified among repressed genes, as for example, [Cyp1a1 (cytochrome P450 family 1 subfamily A1), Plin1 (perilipin-1), Thrsp (thyroid hormone responsive)]. Curiously, Hepcidin (Hamp), a gene related to the negative regulation of iron availability was also downregulated in the granulomatous lesions.
Transcriptional profiling of granulomatous Paracoccidioides brasiliensis yeasts shows transcripts associated with siderophores biosynthesis and downregulation of metabolism and cell cycle associated genes
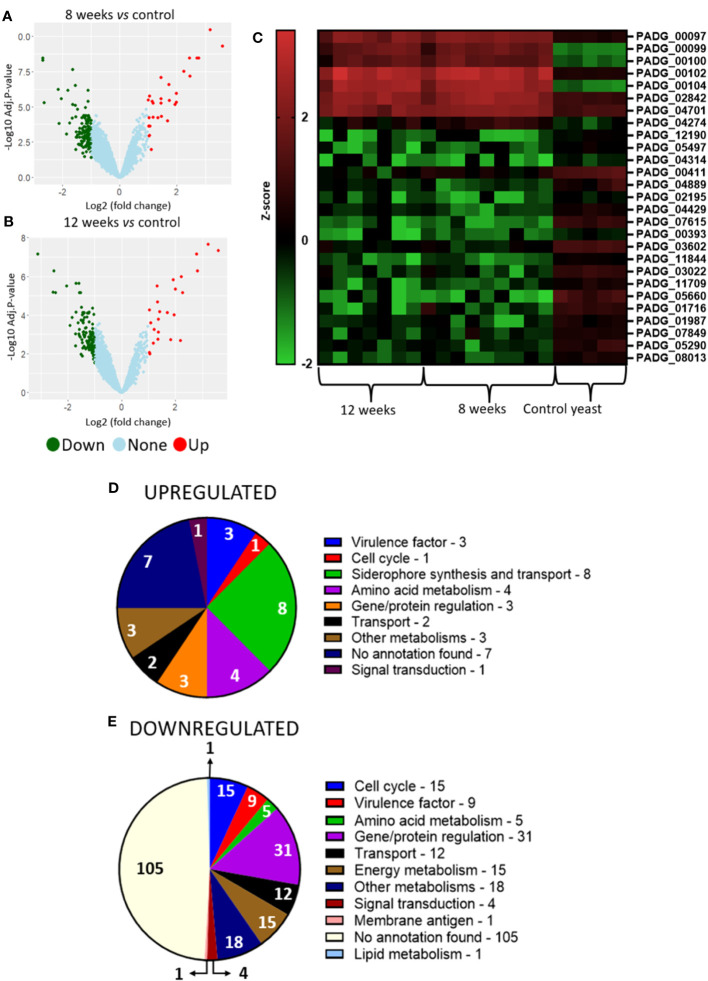
Analysis of granuloma yeasts DEGs comparing 8- and 12-weeks infected lungs to control yeasts show 37 up- and 139 downregulated genes and 28 up- and 157 downregulated genes, respectively ( Figures 4A–C ). This large number of repressed genes when compared to induced genes may indicate a possible fungal metabolism repression within the granuloma. Functional analysis of the corresponding gene products was also carried out by using the KEGG, GO and FungiDB databases. A large number of the transcripts augmented in the fungus within the granuloma are related to the synthesis and transport of siderophores, such as: sidA (L-ornithine-N5-oxygenase; PADG_00097), sidI (Acyl-CoA ligase; PADG_00099), sidF (Hydroxyornithine transacylase; PADG_00100), fusarinin C synthase (sidD; PADG_00102), and OXR1 oxidoreductase (PADG_00104) ( Figure 4D ; Table 2 ). Siderophores are molecules responsible for the capture and storage of iron, being considered an important virulence factor in several pathogens (Schrettl et al., 2007; Blatzer et al., 2011; Silva et al., 2020).
Figure 4.
Transcriptome of granulomatous yeasts indicates upregulation of siderophores biosynthesis and downregulation of metabolism and cell cycle. Granulomatous lesions were extracted from two to four mice from three independent infections at eight weeks (8W) and twelve weeks (12W) after being infected with 1 x 106 P. brasiliensis yeasts. Then, extraction of total RNA, removal of rRNA, and assembly of cDNA libraries were performed. The samples were sequenced and the fragments aligned with the reference genome for P. brasiliensis using bowtie2. The FeatureCounts software was used to count the amount of each identified gene, which were normalized. Then, a differentially expressed genes analysis was made using limma and a functional analysis was carried out. To illustrate the differentially expressed genes, a volcano plot of the 8-weeks infection (8W) group in relation to the control yeasts (A) and a volcano plot of the 12-weeks infection group (12W) in relation to the control yeasts (B) were made. Additionally, a heatmap was created with selected genes (C). The genes in red represent a higher expression and the green ones a lower expression for a given sample. The functions of the upregulated genes (D) and downregulated genes (E) were taken using GO, KEGG and FungiDB.
Table 2.
Differential expression of yeasts genes.
| Acession number | Protein | Expression status | Log (Fold Change) | Adjusted p-value |
|---|---|---|---|---|
| UPs | ||||
| Siderophore synthesis and transport | ||||
| PADG_00097 | L-ornithine N5-oxygenase SidA | UP(D) | 3,594664 | 2,32E-08 |
| PADG_00095 | MFS siderochrome iron transporter 1 | UP(D) | 3,188131 | 1,12E-08 |
| PADG_00100 | Hydroxyornithine transacylase SIDF | UP(D) | 2,354155 | 3,4E-06 |
| PADG_00102 | fusarinine C synthase | UP(D) | 2,227774 | 5,26E-07 |
| PADG_00104 | Oxidoreductase OXR1 | UP(D) | 1,957033 | 4,87E-05 |
| PADG_00099 | Acyl-CoA ligase SID4 | UP(D) | 1,406269 | 8,21E-05 |
| Virulence factors | ||||
| PADG_02842 | Superoxide dismutase, Cu-Zn | UP(D) | 1,424825 | 3,48E-05 |
| PADG_04701 | alcohol dehydrogenase [EC:1.1.1.-] | UP(8) | 1,042112 | 1,75E-06 |
| PADG_04274 | polysaccharide synthase Cps1 | UP(8) | 1,027865 | 0,001103 |
| Amino acid metabolis | ||||
| PADG_07440 | Amino acid permease | UP(D) | 1,812242 | 8,64E-07 |
| PADG_08406 | O-acetylhomoserine/O-acetylserine sulfhydrylase [EC:2.5.1.49 2.5.1.47] | UP(D) | 1,637297 | 0,000873 |
| PADG_00215 | aromatic-L-amino-acid/L-tryptophan decarboxylase [EC:4.1.1.28 4.1.1.105] | UP(D) | 1,514618 | 0,000419 |
| PADG_02214 | 4-aminobutyrate aminotransferase/(S)-3-amino-2-methylpropionate transaminase [EC:2.6.1.19 2.6.1.22] | UP(8) | 1,140801 | 4,12E-06 |
| Other metabolisms | ||||
| PADG_06196 | Flavin oxidoreductase hxnT | UP(D) | 1,98727 | 4,73E-06 |
| PADG_03085 | Mannose-P-dolichol utilization defect 1 protein homolog | UP(12) | 1,076101 | 0,000244 |
| PADG_05433 | pyridoxamine 5’-phosphate oxidase [EC:1.4.3.5] | UP(8) | 1,032033 | 5,7E-05 |
| DOWNs | ||||
| Gene/protein regulation | ||||
| PADG_04314 | Zn(2)-C6 fungal-type domain-containing protein | DOWN(8) | -1,24281 | 4,13E-05 |
| PADG_04889 | Zn(2)-C6 fungal-type domain-containing protein | DOWN(D) | -1,23166 | 0,002813 |
| PADG_05497 | GATA-binding protein, other eukaryote | DOWN(D) | -1,22538 | 0,002127 |
| PADG_02195 | DNA repair protein Swi5/Sae3 | DOWN(8) | -1,21794 | 0,00134 |
| PADG_04429 | Structure-specific endonuclease subunit SLX4 | DOWN(8) | -1,14817 | 0,00099 |
| PADG_00411 | C6 transcription factor (Ctf1B) | DOWN(D) | -1,05195 | 0,00555 |
| PADG_06413 | C6 finger domain transcription factor nscR | DOWN(8) | -1,00975 | 0,008026 |
| Energy metabolism | ||||
| PADG_11844 | aconitate hydratase | DOWN(D) | -1,52072 | 0,01042 |
| PADG_00393 | hexokinase [EC:2.7.1.1] | DOWN(12) | -1,38879 | 0,000134 |
| PADG_03602 | acyl-CoA dehydrogenase | DOWN(D) | -1,32085 | 0,000697 |
| PADG_05290 | Cytochrome b5 heme-binding domain-containing protein | DOWN(D) | -1,07453 | 0,005483 |
| Cell cylce | ||||
| PADG_03022 | PLK/PLK1 protein kinase | DOWN(D) | -1,25446 | 0,002207 |
| PADG_11709 | serine/threonine-protein kinase TTK/MPS1 [EC:2.7.12.1] | DOWN(D) | -1,23479 | 5,54E-05 |
| Virulence factors | ||||
| PADG_01954 | superoxide dismutase, Fe-Mn family [EC:1.15.1.1] | DOWN(D) | -2,55152 | 5,85E-06 |
| PADG_05660 | Fungal nitric oxide reductase | DOWN(D) | -1,38287 | 0,007157 |
| PADG_01716 | alcohol dehydrogenase zinc-binding | DOWN(D) | -1,35725 | 0,000574 |
| Other metabolisms | ||||
| PADG_02384 | porphobilinogen synthase [EC:4.2.1.24] | DOWN(D) | -1,38467 | 0,000981 |
| PADG_07849 | N5-hydroxyornithine acetyltransferase [EC:2.3.1.-] | DOWN(12) | -1,12162 | 0,005081 |
| PADG_04432 | alpha-amylase [EC:3.2.1.1] | DOWN(D) | -1,06127 | 0,002921 |
Granulomatous lesions were extracted from two to four mice from three independent infections at eight weeks and twelve weeks after being infected with 1x106 P. brasiliensis yeasts. Then, extraction of total RNA, removal of rRNA, and assembly of cDNA libraries were performed. The samples were sequenced and the fragments aligned with the reference genome for P. brasiliensis using bowtie2. The FeatureCounts software was used to count the amount of each identified gene, which were normalized. Then, a differentially expressed gene was made using the limma and a functional analysis was carried out using GO, KEGG and FungiDB. The Table shows the differentially expressed genes with the values of Log (Fold change) and adjusted p-value. The expression status column represents the upregulated or downregulated genes for the eight-week infection group (8), the twelve-week infection group (12), and the genes that were differentially expressed in both infection groups were combined and reanalyzed, resulting in an expression status designation for the disease (D).
In addition, genes that have already been described as virulence factors such as superoxide dismutase (PADG_02842), alcohol dehydrogenase (PADG_04701) (two non-iron enzymes that also manage antioxidant defenses) and polysaccharide synthase Cps1 (PADG_04274) were also found as upregulated. Superoxide dismutase was identified as upregulated in both 8- and 12-weeks infection groups. Transcripts which products are related to amino acid metabolism such as amino acid permease (PADG_07440), O-acetylhomoserine/O-acetylserine sulfhydrylase (PADG_08406) and aromatic-L-amino-acid/L-tryptophan decarboxylase (PADG_00215) were induced in both infection groups, while 4-aminobutyrate aminotransferase/(S)-3-amino-2-methylpropionate transaminase (PADG_02214) was induced only in the 8-weeks infection group.
Transcripts for gene and protein expression/degradation regulators are the most downregulated in the transcriptome of the fungus within the granuloma, in addition to transcripts related to the control of cell cycle, energy metabolism and virulence factors ( Figure 4E ; Table 2 ). Among the repressed genes, we identified the transcription factors (TFs) C6 finger domain TF nscR (PADG_06413), GATA-binding protein (PADG_05497), Zn (2)-C6 fungal-type domain-containing protein (PADG_04314), C6 TF (Ctf1B) (PADG_00411) and Zn (2)-C6 fungal-type domain-containing protein (PADG_04889). In addition to the repressed TFs, there are also two genes related to DNA repair that are downregulated, the DNA repair protein Swi5/Sae3 (PADG_02195) and the structure-specific endonuclease subunit SLX4 (PADG_04429). Furthermore, transcripts which products are related to energy metabolism were also repressed in the granulomatous yeasts, such as glucan 1,3-beta-glucosidase (PADG_07615), hexokinase (PADG_00393), acyl-CoA dehydrogenase (PADG_03602) and aconitate hydratase (PADG_11844). We also identified the downregulation of genes related to oxidative phosphorylation, such as cytochrome b5 (PADG_05290) and NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 2 (PADG_04699). Furthermore, some genes related to fungal virulence were also found at lower expression levels when compared to the control fungus, such as fungal nitric oxide reductase (PADG_05660), alcohol dehydrogenase (PADG_01716 and PADG_01987), superoxide dismutase (PADG_01954) and sidL (N5- hydroxyornithine acetyltransferase (PADG_07849).
Proteome of the Paracoccidioides brasiliensis present in the granuloma indicate low protein production, low protein turnover and reprograming of the primary metabolism
We identified 157 and 97 proteins after 8- and 12-weeks of infection, respectively, and 1199 in the control group. When comparing the yeasts present in the granulomatous lesions at 8-weeks of infection with the control group, 1 upregulated protein and 124 downregulated proteins were identified, while the comparison of the 12-weeks infection group with the control group showed a total of 2 more abundant and 74 less abundant proteins ( Figures 5A–C ).
Figure 5.
P. brasiliensis proteins present in the granuloma indicate low protein production and a metabolic repression. Granulomatous lesions were extracted from two to four mice from three independent infections at eight weeks (8W) and twelve weeks (12W) after being infected with 1 x 106 P. brasiliensis yeasts. After lysing the animal cells, the fungal proteins were extracted and digested with trypsin and resolubilized in 0.1% formic acid. Peptides were analyzed by LC-MS/MS and proteins identified by MaxQuant software. Proteins containing more than two unique peptides had their intensity normalized by log2 followed by quantile normalization within each experimental repeat. Then, a differentially abundant proteins analysis was made using limma and a functional analysis was carried out. To illustrate the differentially abundant proteins, a volcano plot of the 8-weeks infection (8W) group in relation to the control yeasts (A) and a volcano plot of the 12-weeks infection group (12W) in relation to the control yeasts (B) were made. Additionally, a heatmap was created with selected proteins (C). The proteins in red represent a higher abundance and the green ones a lower abundance for a given sample. The functions of the upregulated proteins (D), downregulated proteins (E) and the proteins absent in the infection (F) were taken using GO, KEGG and FungiDB.
Increased abundance was observed in only three proteins, glycerol-3-phosphate dehydrogenase (C1G2K1), which is related to lipid metabolism; replication factor C subunit 3 (C1G5E7), involved in DNA replication, DNA repair and regulation of cell cycle progression in yeast; and GTP-binding protein ypt5 (C1G785), involved in vesicles transport ( Figure 5D ; Table 3 ). The six different proteins related to siderophore biosynthesis and transport ( Table 2 ) which transcripts have accumulated in granulomatous yeasts, have not been identified among the proteins revealed by the proteome, nor any other iron-capture protein have been identified among the accumulated proteins.
Table 3.
Differential abundance of yeasts proteins.
| Acession number | Protein | Expression status | Log (Fold Change) | Adjusted p-value |
|---|---|---|---|---|
| UPs | ||||
| Lipid metabolism | ||||
| C1G2K1 | Glycerol-3-phosphate dehydrogenase | UP(12) | 1,924377 | 0,02212 |
| Cell cycle | ||||
| C1G5E7 | Replication factor C subunit 3 | UP(8) | 4,474333 | 0,018531 |
| Transport | ||||
| C1G785 | GTP-binding protein ypt5 | UP(12) | 1,864712 | 0,030992 |
| DOWNs | ||||
| Gene/protein regulation | ||||
| C1G0E5 | 40S ribosomal protein S14 | DOWN(8) | -5,76095 | 0,001957 |
| C1G5P0 | pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13] | DOWN(D) | -4,64777 | 2,39E-05 |
| C1G0P0 | molecular chaperone DnaK | DOWN(12) | -2,42964 | 0,026103 |
| Energy metabolism | ||||
| C1G294 | succinyl-CoA synthetase alpha subunit [EC:6.2.1.4 6.2.1.5] | DOWN(D) | -3,29572 | 0,003605 |
| C1G2P3 | enoyl-CoA hydratase [EC:4.2.1.17] | DOWN(D) | -4,37849 | 2,82E-05 |
| C1G2W2 | Pyruvate kinase (EC 2.7.1.40) | DOWN(D) | -5,32367 | 0,00092 |
| C1GM03 | Cytochrome b-c1 complex subunit 2 | DOWN(D) | -4,10832 | 0,000135 |
| Transport | ||||
| C1FZ88 | importin subunit alpha-6/7 | DOWN(8) | -4,36192 | 0,003272 |
| C1GAF5 | Coatomer subunit alpha | DOWN(8) | -2,60008 | 0,001756 |
| C1GJS2 | Phosphatidylinositol transfer protein SFH5 (PITP SFH5) | DOWN(D) | -7,21769 | 1,52E-05 |
| Amino acid metabolism | ||||
| C1G3V5 | Aspartate aminotransferase (EC 2.6.1.1) | DOWN(12) | -5,05193 | 7,6E-05 |
| C1GBD8 | 3-oxoacid CoA-transferase [EC:2.8.3.5] | DOWN(8) | -6,39235 | 5,76E-05 |
| C1GBT4 | 1-pyrroline-5-carboxylate dehydrogenase [EC:1.2.1.88] | DOWN(8) | -6,07164 | 0,00025 |
| Virulence factor | ||||
| C1GKT9 | Thioredoxin domain-containing protein | DOWN(8) | -6,48413 | 3,21E-05 |
| C1G7K8 | cytochrome c peroxidase [EC:1.11.1.5] | DOWN(D) | -3,10035 | 0,000272 |
| C1GJI2 | Superoxide dismutase [Cu-Zn] | DOWN(D) | -3,5651 | 0,00071 |
| Other metabolisms | ||||
| C1GG77 | Carboxypeptidase Y homolog A (EC 3.4.16.5) | DOWN(D) | -2,92665 | 0,00822 |
| C1GJM4 | Vacuolar aminopeptidase I | DOWN(D) | -4,32296 | 0,000825 |
Granulomatous lesions were extracted from two to four mice from three independent infections at eight weeks and twelve weeks after being infected with 1x106 P. brasiliensis yeasts. After lysing the animal cells, the fungal proteins were extracted and digested with trypsin and resolubilized in 0.1% formic acid. Peptides were analyzed by LC-MS/MS and proteins identified by MaxQuant software. Proteins containing more than two unique peptides had their intensity normalized by log2, then quantile normalized within each experimental repeat. The limma package was used for differentially abundant proteins analysis and GO, KEGG and FungiDB were used for functional analysis. The Table shows the differentially abundant proteins with the values of Log (Fold change) and adjusted p-value. The expression status column represents the upregulated or downregulated proteins for the eight-week infection group (8), the twelve-week infection group (12), and the proteins that were differentially expressed in both infection groups were combined and reanalyzed, resulting in an expression status designation for the disease (D).
Instead, yeast cells sharply down regulate two key iron-dependent enzymes required for energy production by mitochondria and for antioxidant defence, respectively, the Cytochrome b-c1 complex subunit 2 (C1GM03), decreased by 4,11-fold; and cytochrome c peroxidase [EC:1.11.1.5], decreased by 3,10035 fold; that will be further discussed in this work.
Along with these iron-dependent enzymes, a large number of reduced abundance proteins were found ( Figure 5E ; Table 3 ), which mainly are related to redox potential, energy production, protein/amino acid and lipid metabolism have also been significantly down regulated in the imprisoned yeasts. Those are Superoxide dismutase [Cu-Zn], that converts superoxide radicals (O2-) into molecular oxygen (O2) and hydrogen peroxide (H2O2), which is further converted to water (H2O) by catalase, an enzyme that contains four iron-containing heme groups; the allosteric enzyme Pyruvate kinase (C1G2W2), that provide pyruvate to be converted to acetyl-CoA in mitochondria, to gluconeogenesis and to amino acid anabolism; succinyl-CoA synthetase alpha subunit (C1G294), which is part of an enzyme that regulates citric acid cycle by controlling the interconversion between succinyl CoA and succinate and the GTP/ATP production at substrate level, and to provide precursors for porphyrin and heme biosynthesis; enoyl-CoA hydratase (C1G2P3) for beta-oxidation, that provide acetyl-CoA to citric acid cycle, but also intermediates for the metabolism of lipids, cholesterol and ketone body; and, Aspartate aminotransferase (C1G3V5), that catalyses the interconversion of aspartate and α-ketoglutarate to oxaloacetate and glutamate, important for both amino acid synthesis and for providing intermediates for the citric acid cycle/energy production; and enzymes in amino acid metabolism, such as aspartate aminotransferase, 3-oxoacid CoA-transferase (C1GBD8) and 1 -pyrroline-5-carboxylate dehydrogenase (C1GBT4). Protein synthesis, degradation and folding have also been sharply affected in granulomatous yeasts as seen by the down regulation of two different peptidases, the Carboxypeptidase Y homolog A (EC 3.4.16.5) and the Vacuolar aminopeptidase I; the molecular chaperone DNAK (C1G0P0); the 40S ribosomal protein S14 (C1G0E5); and the pre-mRNA-splicing helicase BRR2 (C1G5P0). Finally, proteins involved in vesicle transport, such as coatomer subunit alpha (C1GAF5), importin subunit alpha-6/7 (C1FZ88) and phosphatidylinositol transfer protein SFH5 (C1GJS2) have also been decrease in abundance.
We hypothesize that the fungi inside the granulomas avoid energy loss by lowering protein synthesis, since these are one of the most consuming energy processes in a cell. At the same time there is a decrease of protein degradation in an attempt to tune the protein content to adapt the general metabolism to the constraint of the granulomatous environment.
We should keep in mind that although granulomatous yeasts have lowered their protein content in general, with 1037 proteins that have not been detected (either because they were not present or were below the detection threshold of the proteome analysis), the protein that have been detected were those that the yeast maintained because somehow they are important to cope with the constraint during the host-parasite interaction, are required to resist to/modulate of immune response, to keep yeast viability and competence for proliferation and dissemination within host. These assumptions make the proteins identified in this work as promising target for drug design and screening for inhibitors.
Additionally, the absent proteins in the infection groups were also related to gene/protein regulation, amino acid metabolism, energy metabolism, and cell cycle and transport ( Figure 5F ), which support a reprogramming of these biochemical processes by the imprisoned yeasts, indicating a possible fungal low activity within the granuloma, but with their virulence preserved, as shown in the next session.
Proteome of Paracoccidioides brasiliensis yeasts recovered from the granuloma indicates an enhanced metabolism concomitant with the expression of virulence factors after cultivation
The analysis of differentially expressed proteins shows that there is a large number of upregulated proteins. When yeasts from 8-week granulomatous lesions were cultivated and compared with control yeasts, 382 proteins were found to be induced and only 1 repressed. Similarly, when comparing the fungal proteins from granulomatous lesions at 12 weeks of infection, 319 proteins were found to be more abundant ( Supplementary Figures 3A–C ).
Opposing to what was observed for RNAseq and the proteome of the yeasts present in the lesions, a greater number of differentially abundant proteins were observed in the post-infection groups when compared to control group. Those abundant proteins have functions related to gene/protein regulation, energy metabolism, lipid and amino acid metabolism ( Supplementary Figure 3D ; Supplementary Table 11 ).
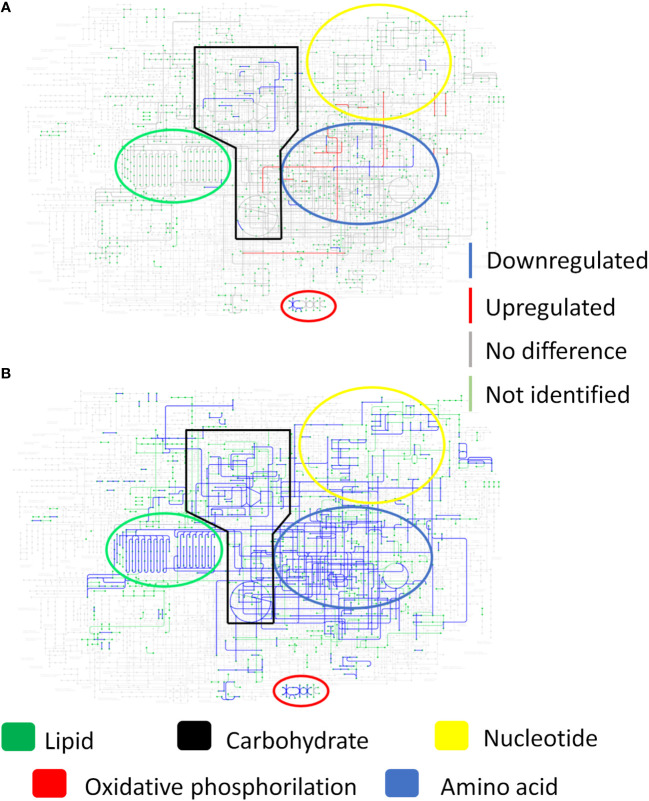
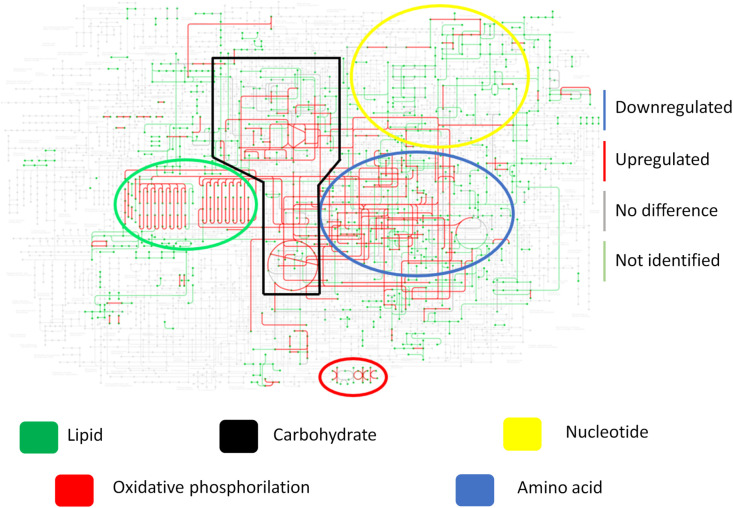
To better illustrate the yeast metabolism present in the granuloma when compared to the recovered yeasts (cultivated in Fava Netto medium), the Figure 6A shows the KEGG metabolic pathway of the P. brasiliensis in a transcriptional level, while Figure 6B shows the metabolic pathway at a proteomic level. This data indicates that there is a repression of the metabolism, especially the energy metabolism, both at transcriptional and proteomic levels in granulomatous yeasts. Instead, when we analyze the metabolic pathway of the recovered yeasts ( Figure 7 ) we can see that the metabolism that was once repressed it is now induced, evidenced by the upregulation of lipid, nucleotide, energy, carbohydrate, and amino acid metabolisms after yeast cultivation.
Figure 6.
Yeast metabolism in transcriptional and proteomic levels show a repressed metabolism within the granuloma. Granulomatous lesions were extracted from two to four mice from three independent infections at eight weeks and twelve weeks after being infected with 1 x 106 P. brasiliensis yeasts. After extracting total RNA and proteins, alignment and protein identification were carried out, respectively. The limma software was used to evaluate differentially abundant genes and proteins, and then the metabolic map of P. brasiliensis obtained from KEGG was used to evaluate the induction and repression of metabolism. (A) Metabolic map at the transcriptional level. (B) Metabolic map at protein level. Genes and proteins marked in blue are downregulated, those marked in red are upregulated, those marked in gray are not at statistically different levels, while those marked in green were not identified in the samples.
Figure 7.
Fungal metabolism of recovered yeasts shows a recovered and enhanced metabolism after cultivation. Granulomatous lesions were extracted from two to four mice from three independent infections at eight weeks and twelve weeks after being infected with 1x106 P. brasiliensis yeasts. After recovering the fungi present in the granulomatous lesions, cultivation was carried out in Fava Netto culture medium for 14 days. Then, extraction of fungal proteins was performed, which were digested with trypsin and resolubilized in 0.1% formic acid. Peptides were analyzed by LC-MS/MS and proteins identified by MaxQuant software. Proteins containing more than two unique peptides had their intensity normalized by log2, then quantile normalized within each experimental repeat. The limma software was used to evaluate differentially abundant proteins, and then use the metabolic map of P. brasiliensis obtained from KEGG to evaluate the induction and repression of metabolism as a whole. Proteins marked in blue are downregulated, those marked in red are upregulated, those marked in gray are not at statistically different levels, while those marked in green were not identified in the samples.
RT-qPCR analysis validates host-pathogen transcriptomics
RT-qPCR analysis was performed to validate the dual-RNAseq results for both mouse ( Supplementary Figure 4 ) and P. brasiliensis genes ( Supplementary Figure 5 ). For the host validation, we used Cxcl9, Tnf, Gbp2b, Pdcd1, Tnnc1, Lcn2, and Hamp. For the pathogen validation, we used PADG_00104 (Oxidoreductase OXR1), PADG_00097 (L-ornithine N5-oxygenase SidA) and PADG_00099 (Acyl-CoA ligase SID4), PADG_05660 (fungal nitric oxide reductase), and PADG_01954 (superoxide dismutase, Fe-Mn family). Both datasets were validated by RT-qPCR data, supporting the findings in the RNAseq with a Pearson statistical correlation between 0.8981 and 0.9679 for the mice dataset and between 0.8906 and 0.906 for the P. brasiliensis dataset.
Paracoccidioides brasiliensis yeasts present in the granulomatous lesions produces lower levels of intracellular siderophore ferricrocin and has low lipid content.
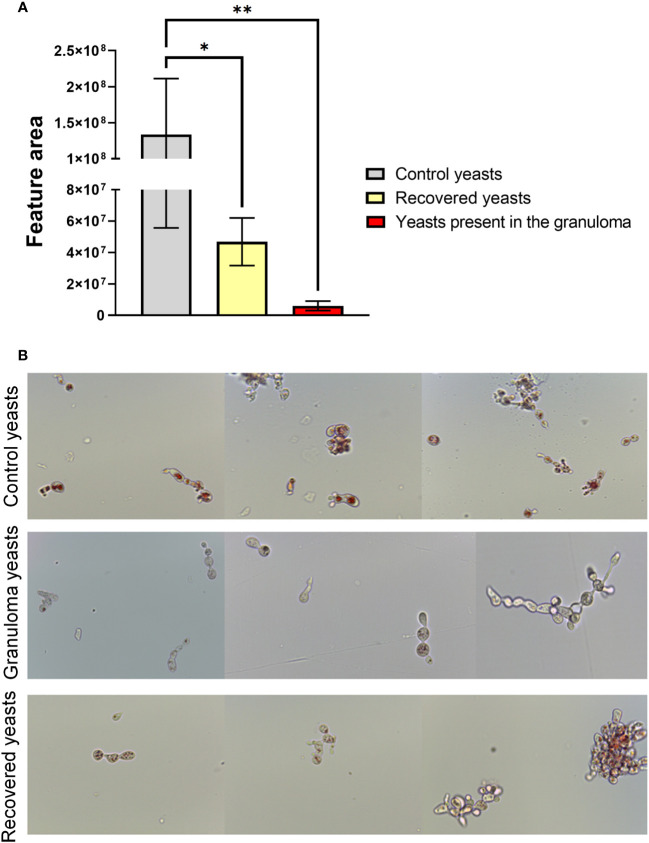
To verify the expression of siderophores by P. brasiliensis, LC-HRMS/MS analyses were performed with the granuloma fungus extracts, control fungus and recovered fungus. The siderophore Ferricrocin (m/z 718.3370) was annotated based on its exact mass and MS/MS fragmentation pattern typical of this siderophore class. Ferricrocin levels were lower in the yeasts present in the granuloma and the recovered ones when compared to the control yeasts ( Figure 8 ). This result corroborates the RNAseq data considering a lower expression of SidL observed in the granuloma yeasts ( Figure 4E ; Table 2 ).
Figure 8.
Yeasts present in the granuloma produces low levels of intracellular siderophore ferricrocin and has low lipid availability. For the ferricrocin levels analysis (A) a quadruplicate of lung granuloma, control lung, control yeasts and recovered yeasts were frozen, and 100 mg of frozen material was macerated with the help of a pestle and pot. The secondary metabolites extraction was performed with 1 mL of methanol (100%; HPLC grade), followed by sonication bath for 40 minutes. For sample preparation, 500 μL of each sample extract were filtered in a 0.22 μm PTFE and diluted to 1 mL volume with HPLC-grade methanol. Then LC-HRMS/MS analysis were performed and data were processed with Xcalibur software (version 3.0.63). Statistical analysis was performed using one-way ANOVA. Bars represent means ± standard error of the feature area of groups of 4 to 5 samples (*p<0.05; **p < 0.01). For lipid availability analysis (B) control yeasts, granuloma yeasts and recovered yeasts were stained with filtered Oil Red O for 2 hours at room temperature. Subsequently, the cells were washed twice with ultrapure water and then resuspended in PBS for analysis on a microscope slide. The photos were taken using the Leica DM750 microscope and the Leica Application Suite V4.8 software, thus allowing the observation of stained neutral lipids inside the cells.
Furthermore, a OilRed O lipid staining was performed aiming to evaluate the neutral lipid content in the three yeasts groups (control, recovered and present in the lesions). Oil Red O dye binds to neutral lipids, as those found in lipid droplets, but not to phospholipids in membranes, revealing the amount and size of lipid droplets by staining them with a red colour. As showed in the Figure 8B , the lipid droplets in the infection yeasts are less abundant when compared to control yeasts, supporting both the transcript and proteomic data, where genes and proteins related to beta-oxidation were significantly less abundant in the fungus inside the granuloma. Because lipid droplets are in greater number and size in the recovered and cultured yeasts compared to the yeasts present in the lesions, our results indicate 1) neutral lipid stocks decrease during infection and 2) that lipid biosynthesis is resumed by subsequent cultivation of yeast isolated from the lesions in rich medium.
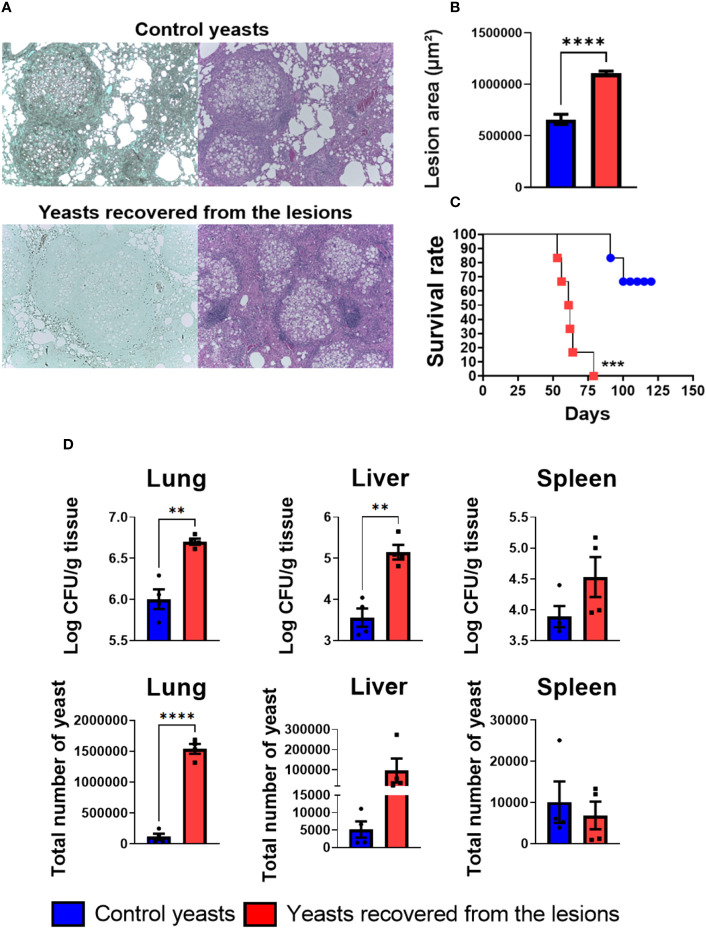
Yeasts isolated from granulomas are more virulent than those of non-granuloma yeasts
Aiming to compare the virulence of the yeasts obtained from the granuloma after 8 weeks of infection with the non-granuloma yeasts (control yeasts, i.e, yeasts used for mice infection), a histopathology analysis of the lung was made. The mice infected with the yeasts isolated from granulomatous lesions had a less compact granuloma with a larger lesion area when compared to the granulomas size formed in mice infected with the control yeasts ( Figures 9A, B ). Furthermore, a survival analysis of infected C57BL/6 wild-type (WT) mice was performed. All mice infected with granulomatous lesion-yeasts died within 79 days; in contrast, mice infected with control yeasts start to die on day 91 and 100 days after infection only two mice were dead ( Figure 9C ). These results demonstrate that yeasts recovered from granulomatous lesions are more virulent than the non-granuloma control yeasts. To evaluate the fungal load a CFU assay was performed with lungs, liver, and spleen of mice. As it can be seen in the Figure 9D , the mice infected with the recovered yeast had a higher fungal load in the lungs and liver when compared to animals infected with control yeasts, showing that the yeasts from granulomatous lesions can disseminate more easily than control yeasts.
Figure 9.
Recovered yeasts induces looser granulomas, is more virulent and disseminate more than control yeasts. Groups of four C57BL/6 mice were infected by i.t. route with 1 × 106 control P. brasiliensis yeasts or recovered yeasts contained in 50 µL of PBS. After eight and weeks of infection, the animals were euthanized and the lungs removed, which were stored in 10% formaldehyde at 4 °C. Sections of 5 µm were stained with Hematoxylin-Eosin (H&E; pink) for analysis of lesions and stained with Grocott (green) for fungal evaluation (A). The anatomy of the lesion was analyzed according to size, morphology, and presence of fungal cells. The image and the lesion area calculation were taken using the Leica DM750 microscope and the Leica Application Suite V4.8 software. Statistical analysis was performed using one-way ANOVA. Bars represent means ± standard error of lesion area (µm²) of groups of three mice (B). For the survival analysis (C) groups of six C57Bl/6 WT mice were infected via i.t. with 1x106 yeasts extracted from granulomatous lesions or control yeasts to verify the virulence of freshly extracted granuloma yeasts. Deaths were recorded daily. The survival curve used the Log-rank test (Mantel-Cox) to compare the two groups. For the CFU assay (D) groups of three to four C57BL/6 mice were infected by the i.t. route with 1 × 106 P. brasiliensis yeasts contained in 50 µL of PBS. After eight weeks of infection, the lungs, liver, and spleen were removed and macerated. After that, the samples were centrifuged to obtain the fungus and inoculated in plates containing BHI medium for subsequent counting of CFU. Bars represent means ± standard deviation of log10 or total yeast number. Values were considered significant when **p < 0.01; ***p<0.001; ****p<0.0001.
Discussion
The granuloma is a host defense mechanism that aims to contain the pathogen and prevent the spread of infection. However, at the same time that the granuloma keeps the infection under control, allows the pathogen to survive inside, since it does not eliminate it completely (Rubin, 2009; Hams et al., 2013).
The functional analysis of the DEGs of the infected host tissue indicates the expression of genes associated with innate and adaptive immunity. To our knowledge, it is the first time that both PRRs Clec4d and Clec4a2 were observed in chronic PCM, although Dectin-3 (Clec4d) expression was demonstrated in vitro in human plasmacytoid dendritic cells (pDCs), where it has been associated to type I IFN and IL-1β secretion (Preite et al., 2018). Besides, Dectin-3 has been described to initiate a host defense against Candida albicans and Cryptococcus ssp (Zhu et al., 2013; Huang et al., 2018). Clec4a2 belongs to the Dectin-2 family, and it is also named DC immunoreceptor (DCIR) (Bates et al., 1999; Lambert et al., 2008; Kerscher et al., 2013; Bloem et al., 2014; Nagae et al., 2016) In tuberculosis, DCIR was reported to be protective by modulating the immune response through IL-12 expression and Th1 expansion (Troegeler et al., 2017). The expression of Pentraxin 3 (Ptx3), a PRR related gene stimulated by tumor necrosis factor and interleukin-1beta, which remains underexploited in PCM, has already been reported to be involved in the P. brasiliensis opsonization by macrophages (Diniz et al., 2004). Additionally, Dectin-1 (Clec7a) and TLR2 important PRRs in PCM and others mycosis were also observed with a transcript expression in the granuloma. These receptors play an important role in the immune response of human and murine phagocytic cells when exposed to P. brasiliensis infection (Brown, 2006; Bonfim et al., 2009; Loures et al., 2009; Loures et al., 2014; Feriotti et al., 2015; Calich et al., 2019; de Quaglia E Silva et al., 2019; Li et al., 2019; Rodriguez-Echeverri et al., 2021).
The transcripts to chemokines Ccl3 (MIP-1α), and Ccl7 (MCP3), that have never been described in a PCM context, were also expressed in the granuloma. Those chemokines are responsible for recruiting monocytes, neutrophils, DCs, and eosinophils, which reflects the presence of these cells within the lesions (Luster, 1998). In fact, Ccl3 and Ccl7 were associated to a milder Cryptococcus neoformans and Histoplasma capstulatum infection, since they promote a protective profile by inhibiting a Th2 response (Huffnagle et al., 1997; Olszewski et al., 2000; Szymczak and Deepe, 2009; Qiu et al., 2012). Additionally, the expression of Ccl3 and Ccl4 (MIP-1β), a chemokine responsible for monocytes, neutrophils, DCs, and eosinophils migration has already been related to neutrophil accumulation in the lungs in Aspergillus fumigatus, C. neoformans, H. capsulatum and P. brasiliensis infection, indicating an important neutrophil participation in the granuloma (Huffnagle et al., 1997; Mehrad et al., 2000; Olszewski et al., 2000; Souto et al., 2003; Kroetz and Deepe, 2010; Puerta-Arias et al., 2016). Since Ccl3, Ccl4, and Ccl7 attract eosinophils, these cell populations are also expected in the granulomatous lesions. Although eosinophils are common in the acute PCM form, they have already been identified in the lesions by the presence of the eosinophil granule major basic protein, but their function in the chronic form of PCM remain unclear (Wagner et al., 1998; Braga et al., 2017).
The presence of Ifng, Tnf, Tnfrsf9, and Nos2 transcripts (iNOS) corroborates with the fact that macrophages and a Th1 response play an essential role in the granuloma function. TNF-α is responsible for macrophages accumulation and differentiation in epithelioid cells, resulting in a well-organized granuloma (Kindler et al., 1989; Figueiredo et al., 1993). Therefore, the expression of TNF-α can explain why the granulomas were still tight in our histology analysis. Furthermore, IFN-γ can activate infected macrophages to produce TNF-α and iNOS, leading to a reduced P. brasiliensis replication (Brummer et al., 1988; Gonzalez et al., 2000), which may explain why yeasts transcripts and proteins related to cell cycle are in a lower level inside the granuloma when compared to control fungi.
Concomitant to the presence of macrophages and Th1 lymphocytes, there is an indication that neutrophils and Th17 lymphocytes also play an important role in the granuloma activity. The Th17 response, although described as a crucial T cell subset in granuloma formation, it has never been described inside a chronic PCM granulomatous lesion (Tristão et al., 2017). However, IL-17 expressing cells were found in the skin and mucosal lesions of patients (Pagliari et al., 2011) besides a protective role of Th17 cells that has been described by our group (Galdino et al., 2018; Preite et al., 2023). The depletion of myeloid-derived suppressor cells (MDSCs) as well as Treg cells leads to robust Th1/Th17 lymphocyte responses associated to a regressive disease with reduced fungal burden, lung pathology and mortality ratios in mice (Galdino et al., 2018; Preite et al., 2023).
As reported to several disease, controlling the immune response is crucial, aiming for a homeostasis of the inflammatory response, as an exaggerated immune reaction can result in host damage and an inhibited immune response as well (Marques et al., 2016). The expression of Arginase-1 and collagenase 3 (Mmp13) indicates a well-regulated immune response with tissue repair activation and the participation of M2 polarized macrophages which are related to an anti-inflammatory profile (Elkington et al., 2005; Yunna et al., 2020). M2 macrophages have already been linked to a less severe PCM, since a M2 profile was found in the immunoregulatory mechanism developed by resistant mice (Feriotti et al., 2013). Mmp13 was described to cleave some chemokines, such as Ccl2 and Ccl7, leading to a negative control of chemotaxis and inhibiting the fibrosis formation in the PCM granuloma (Arango et al., 2017; Giannandrea and Parks, 2014). Pdcd1 (PD-1) and CTLA4 induction in the granuloma may suggest that these immune checkpoint inhibition molecules play a role in the lymphocyte’s homeostasis, controlling the Th1/Th17 responses (Sansom, 2000; Keir et al., 2008). In fact, those two receptors have been explored as a drug target in some respiratory infections (Hamashima et al., 2020). In a murine model of C. neoformans infection, the administration of anti PD-1 and anti CTLA-4 antibodies resulted in a better survival of the mice (McGaha and Murphy, 2000; Roussey et al., 2017).
Although the host develops a highly active and regulated immune response inside the granuloma, an impaired function of the lung was here detected, since transcripts related to the normal function of lung were found in lower levels within the lesions when compared to naïve control lung, as revealed by the enrichment analysis of the downregulated genes. The low abundance of these genes can also be explained because the granuloma is mostly composed of immune and yeast cells, so there are less healthy lung and consequently, less transcripts. However, since the immunological structure is occupying almost the whole lung, it is safe to assume an impaired lung function. In fact, in chronic obstructive pulmonary diseases, the expression of myosin is impaired, due to the shortening of the telomere (Thériault et al., 2012; Thériault et al., 2014; Pomiès et al., 2015).
To our knowledge, this is the first time a transcriptional study has been conducted within the murine granuloma in PCM. For this reason, we presented several genes and mechanisms that can be explored in the future. The presence of transcripts within the lesions does not guarantee that these responses are active, as post-translational modifications and regulatory mechanisms can hinder the translation of these transcripts into proteins. Therefore, the results presented so far serve to guide future studies on the immunology of PCM.
The functional analysis of the differentially expressed genes of the fungus shows an upregulation of genes related to the synthesis and transport of the siderophores fusarinine C and triacetylfusarinine C. This can be considered a very interesting data, considering that it has already been described that a greater supply of iron to the fungus is related to its higher virulence (Parente et al., 2011; Silva et al., 2020). The upregulation of these genes related to extracellular siderophores may indicate a lack of iron within the granuloma (Timm et al., 2003; Hilty et al., 2011; Haas, 2014). In fact, previous studies have shown that P. brasiliensis cultivated in iron deficient medium expressed transcripts related to extracellular siderophores (Parente et al., 2011). The sidL analogue (N5-hydroxyornithine acetyltransferase (PADG_07849) was found downregulated in the infection. This gene is part of the synthesis pathway of the intracellular siderophores ferricrocin and hydroxyferricrocin, responsible for intracellular storage and distribution of iron (Schrettl et al., 2007; Blatzer et al., 2011). This repression of SidL may indicate that there is no iron available for storage by the fungus within the granuloma (Miethke and Marahiel, 2007; Schrettl et al., 2007). Interestingly, the phenomenon of increased synthesis of siderophores for iron uptake and repression of synthesis of siderophores for maintenance within the granuloma was previously described for M. tuberculosis Timm et al., 2003). However, the expression of siderophores by P. brasiliensis present in the granuloma have never been described in PCM. The identification of ferricrocin by LC-HRMS/MS in lower levels within the granuloma strengthen the fact there is an iron deficiency by the pathogen inside the immunological structure (Haas, 2014).
Another fact that indicates there is an iron deficiency within the granuloma, is that the host induces the expression of Lipocalin 2 (Lcn2), a protein that binds to siderophores chelated to iron, helping the defense against siderophores expressing pathogens, acting as an iron sequestrator and as a bacteriostatic agent (Goetz et al., 2002; Holmes et al., 2005). Lipocalin 2 has also been described as an important player in the tuberculosis granuloma formation. LcnKO mice infected with M. tuberculosis had increased granulomatous inflammation when compared to WT infected mice, because Lcn2 has the ability to induce neutrophil migration and lymphocytes regulation (Guglani et al., 2012). High levels of Lcn2 transcripts were also reported in C. neoformans and A. fumigatus infection (Gessner et al., 2012; Wozniak et al., 2014). Inhibiting the fungal intracellular iron is crucial for host survival. Previous reports in which the authors used chloroquine to restrict intracellular iron in P. brasiliensis yeasts, was reported a lower fungal load inside human and mouse monocytes when compared to cells that did not receive chloroquine treatment (Dias-Melicio et al., 2006). These results, along with our data, indicate that iron is crucial for fungal survival and replication in the host.
The repressed expression of Hamp can indicate that there is a lack of iron even for host, because this hormone binds to ferroportin, leading to its internalization and degradation, causing an inhibition of iron efflux (Nemeth et al., 2004; Nemeth and Ganz, 2021). Because this hormone cause host cell to trap the iron inside and iron is a major player in the course of several diseases, an upregulation was observed in many infections, such as those caused by M. tuberculosis, Vibrio vulnificus, A. fumigatus and Fusarium oxysporum (Leal et al., 2013; Arezes et al., 2015; Harrington-Kandt et al., 2018). Therefore, the repressed expression of hepcidin in a chronic PCM context, may indicate that there is an iron deficiency for both the host and the pathogen, revealing iron to be a crucial element for the granuloma function. In fact, A recent work showed that patients with chronic PCM developed anemia due to iron deficiency and due to inflammation process (de Brito et al., 2023).
An induction of some genes that have been described as P. brasiliensis virulence factors in other contexts were also found. Superoxide dismutase Cu-Zn (PADG_02842) has been described to help against the oxidative stress caused by the synthesis of reactive oxygen species by the host. Likewise, alcohol dehydrogenase (PADG_04701) synthesis has been reported to help against the increase in pyruvate released by amino acid degradation (Castilho et al., 2014; Camacho and Niño-Vega, 2017). Another transcript that is a possible virulence factor is CPS1 (polysaccharide synthase; PADG_04274), which has been described in C. neoformans and contributes to the pathology, since it is associated with the rigidity of the cell wall (Chang et al., 2006). These transcripts related to virulence factors that were found upregulated do not use iron as a cofactor, while some described in lower transcriptional and proteomic levels use iron as a cofactor, strengthening the hypothesis of iron deficiency in the granuloma.
The analyzes of repressed genes and proteins by the yeasts inside the granulomatous lesions revealed a large number related to transcriptional factors. The reduced expression of these genes and proteins favors the idea that the fungus is repressing a good part of its gene expression within the granuloma, which can explain the low protein identification in the lesions yeasts when compared to the control group. It is important to emphasize that these transcripts that have been modulated to increase or decrease in abundance may not be directly involved in the maintenance of the low metabolic activity state of the fungus or in coping with the host-imposed constrains because their protein products were not identified in the proteome. Instead, keeping these transcripts may be related to the fungus ability to recover from the constrain and to prepare them to resume full metabolism once free.
The energy production of the fungus within the granuloma is also repressed since there are genes and proteins related to energy metabolism downregulated, such as those related to glycolysis, beta oxidation, Krebs cycle, and oxidative phosphorylation, including Cytochrome b5 heme-binding domain-containing protein. Iron is essential for the survival of the fungus, so siderophores synthesis is a way to circumvent iron deficiency. Another way to control iron availability is by altering their metabolism, limiting the synthesis of proteins that use iron as a cofactor, as is the case of some proteins related to oxidative phosphorylation and Krebs cycle (do Amaral et al., 2019). In previous studies, P. brasiliensis cultivated in iron deficient medium had also repressed proteins related to Krebs Cycle, glyoxylate cycle and oxidative phosphorylation (Parente et al., 2011). This report supports the hypothesis of iron deficiency inside the granuloma.
In addition, multiple proteins, including the PLK1 (PADG_03022) and serine/threonine-protein gene TTK/MPS1 kinase (PADG_11709), which are essential for the continuation of mitosis were also detected in low levels. Another fact that corroborates with the repression of fungus reproduction is that energy production and amino acid metabolism are repressed, which are essential for reproduction (Zhu and Thompson, 2019). Additionally, a limitation in pathogen growth within the murine lung and the granuloma was previously described for M. tuberculosis, which enters a latent state after a long period of infection, mostly in more resistant mice (Wayne and Sohaskey, 2001; Shi et al., 2003). The latent state in M. tuberculosis was associated to hypoxia conditions and nutrient starvation, which lead to a low energy and lipid production, corroborating with our findings and indicating a possible nutrient starvation by the fungus (Betts et al., 2002). Although there is a repression of the fungus reproduction, the pathogen is still able to grow inside the granuloma, as seen from the histological analysis, revealing a fight between the pathogen and host, in which the pathogen succeeds.
Interestingly, a low expression of 15 cell cycle genes associated with fungal reproduction were here detected. Therefore, our findings lead us to speculate that fungi are with low metabolic activity, when metabolism must be reprogrammed in order to fit yeast cells to such a macro and micro-nutrient restrictive environment. We even hypothesized that the fungus is suppressing the protein production to save energy loss, only expressing proteins that are essential for survival. Nevertheless, the fungus succeeds in infection: it disseminates within the lung and to other tissues/organs after 12 weeks of infection and eventually kill the host. Importantly, the low activity state should not be interpreted as dormancy, but as a delay of this fungus to reach widespread dissemination when compared to its behaviour in a fully susceptible host, in which PCM causes death much faster. We also speculate that the metabolic fitness may further protect yeasts against the host immune response, albeit we cannot rule out the possibility that host immune response also contribute to shape yeast adaptation within granulomas of susceptible mice. The fact is that the host-pathogen relationship somehow enables yeast cells to successfully recover from the restrictive condition, and even burst, their virulence once released from the constrains inflicted by the host, as seen from the proteome of the recovered yeasts. This data agrees with previous reports where it was shown that after several animal passages the antioxidant repertoire of the recovered P. brasileinsis-yeasts had changed in comparison with control avirulent yeasts (Castilho et al., 2014; Castilho et al., 2018). Looking from the parasite perspective, survival within granulomas by Paracoccidioides sp. and M. tuberculosis require adaptation to iron deficiency, nutrient starvation, and hypoxia. Instead, adaptation to low pH and nitrosative stress reported in the bacterial granuloma may not be relevant in PCM granuloma (Timm et al., 2003; Talaat et al., 2007; Garton et al., 2008).
The results obtained from the basic lipids staining support the analyzes of the fungal transcriptomics and proteomics data that show a wide reprogramming of the lipid metabolism. Because a reduction in abundance of genes and proteins related to beta-oxidation within the granuloma was seen and because some of the enzymes can act either way to catabolism or anabolism depending on carbon source availability, the decrease of lipid droplets abundance in yeast inside the granuloma clearly show that the basic lipid catabolism rates were actually higher than lipids’ synthesis rate at this site. Indeed, after 6 hours of infection there is an increase in beta-oxidation by the yeasts, as well as an increase in the glyoxylate cycle, causing consumption of the yeast’s lipid stock (Lacerda Pigosso et al., 2017).
Taken together our data demonstrate an important iron deficiency for both the host and the pathogen inside the fungal granuloma, concomitant with the activity of genes related to the innate and adaptive immunity of hosts. Furthermore, our data indicate that the fungus represses its metabolism and proliferation inside the granuloma, but still grow, and once it is recovered from the lesions and in vitro cultivated a more active metabolism is established concomitant with an elevated expression of virulence molecules. In fact, the recovered yeasts induce a looser granuloma with a larger lesion area when compared to control yeasts, concomitant with an uncontrolled dissemination as observed in the CFU analysis. Additionally, the premature death of animals infected with yeasts recovered from granulomatous lesions demonstrates the enhanced virulent profile of the recovered yeasts.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA945862 http://www.proteomexchange.org/ PXD040826 http://www.proteomexchange.org/, PXD040829.
Ethics statement
The animal study was approved by Ethics Committee on Animal Experiments (CEUA) of the Federal University of São Paulo (UNIFESP), No. 9596270919. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
BB: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. RR: Investigation, Methodology, Writing – review & editing, Data curation, Formal Analysis, Visualization. NP: Formal Analysis, Investigation, Methodology, Visualization, Writing – review & editing. VK: Formal Analysis, Investigation, Methodology, Visualization, Writing – review & editing. PA: Data curation, Formal Analysis, Investigation, Methodology, Visualization, Writing – review & editing. MC: Data curation, Methodology, Writing – review & editing, Formal Analysis. MM: Formal Analysis, Investigation, Writing – review & editing. TF: Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. VC: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. AT: Data curation, Methodology, Writing – review & editing. OS-B: Funding acquisition, Writing – review & editing, Formal Analysis. SD: Funding acquisition, Writing – review & editing. ÖB: Funding acquisition, Methodology, Resources, Writing – review & editing. CC: Conceptualization, Formal Analysis, Investigation, Writing – original draft, Writing – review & editing. AZ: Data curation, Investigation, Methodology, Writing – review & editing. GG: Conceptualization, Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. FL: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Acknowledgments
We also want to thank the Brazilian Biorenewables National Laboratory (LNBR), part of the Brazilian Centre for Research in Energy and Materials (CNPEM), a private non-profit organization under the supervision of the Brazilian Ministry for Science, Technology, and Innovations (MCTI) for their assistance.
Funding Statement
We thank the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) grant number 2018-14762-3 (FL), 2019/17324-0 (BB), 2021/04977-5 (GG) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) grant numbers 301058/2019-9 and 404735/2018-5 (GG), both from Brazil, and the National Institutes of Health/National Institute of Allergy and Infectious Diseases grants R01AI153356 (GG). AT was funded by a John and Pat Hume PhD Scholarship from Maynooth University. The Science Foundation Ireland provided grants SFI-21/FFP-P/10146 to ÖB and SFI-21/PATH-S/9444 to ÖS-B. Protein mass spectrometry facilities were funded by a competitive award from Science Foundation Ireland (12/RI/2346 (3)) to SD.
Conflict of interest
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1268959/full#supplementary-material
References
- Arango J. C., Puerta-Arias J. D., Pino-Tamayo P. A., Salazar-Peláez L. M., Rojas M., González Á. (2017). Impaired anti-fibrotic effect of bone marrow-derived mesenchymal stem cell in a mouse model of pulmonary paracoccidioidomycosis. PloS Negl. Trop. Dis. 11 (10), e0006006. doi: 10.1371/journal.pntd.0006006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arezes J., Jung G., Gabayan V., Valore E., Ruchala P., Gulig P. A., et al. (2015). Hepcidin-induced hypoferremia is a critical host defense mechanism against the siderophilic bacterium Vibrio vulnificus . Cell Host Microbe 17 (1), 47–57. doi: 10.1016/j.chom.2014.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25 (1), 25–29. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates E. E., Fournier N., Garcia E., Valladeau J., Durand I., Pin J. J., et al. (1999). APCs express DCIR, a novel C-type lectin surface receptor containing an immunoreceptor tyrosine-based inhibitory motif. J. Immunol. 163 (4), 1973–1983. doi: 10.4049/jimmunol.163.4.1973 [DOI] [PubMed] [Google Scholar]
- Betts J. C., Lukey P. T., Robb L. C., McAdam R. A., Duncan K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43 (3), 717–731. doi: 10.1046/j.1365-2958.2002.02779.x [DOI] [PubMed] [Google Scholar]
- Blatzer M., Schrettl M., Sarg B., Lindner H. H., Pfaller K., Haas H. (2011). SidL, an Aspergillus fumigatus transacetylase involved in biosynthesis of the siderophores ferricrocin and hydroxyferricrocin. Appl. Environ. Microbiol. 77 (14), 4959–4966. doi: 10.1128/AEM.00182-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloem K., Vuist I. M., van den Berk M., Klaver E. J., van Die I., Knippels L. M., et al. (2014). DCIR interacts with ligands from both endogenous and pathogenic origin. Immunol. Lett. 158 (1-2), 33–41. doi: 10.1016/j.imlet.2013.11.007 [DOI] [PubMed] [Google Scholar]
- Bocca A. L., Amaral A. C., Teixeira M. M., Sato P. K., Shikanai-Yasuda M. A., Soares Felipe M. S. (2013). Paracoccidioidomycosis: eco-epidemiology, taxonomy and clinical and therapeutic issues. Future Microbiol. 8 (9), 1177–1191. doi: 10.2217/fmb.13.68 [DOI] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 (2), 185–193. doi: 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
- Bonfim C. V., Mamoni R. L., Blotta M. H. (2009). TLR-2, TLR-4 and dectin-1 expression in human monocytes and neutrophils stimulated by Paracoccidioides brasiliensis. Med. Mycol. 47 (7), 722–733. doi: 10.3109/13693780802641425 [DOI] [PubMed] [Google Scholar]
- Braga F. G., Ruas L. P., Pereira R. M., Lima X. T., Antunes E., Mamoni R. L., et al. (2017). Functional and phenotypic evaluation of eosinophils from patients with the acute form of paracoccidioidomycosis. PLoS. Negl. Trop. Dis. 11 (5), e0005601. doi: 10.1371/journal.pntd.0005601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G. D. (2006). Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat. Rev. Immunol. 6 (1), 33–43. doi: 10.1038/nri1745 [DOI] [PubMed] [Google Scholar]
- Brummer E., Hanson L. H., Restrepo A., Stevens D. A. (1988). In vivo and in vitro activation of pulmonary macrophages by IFN-gamma for enhanced killing of Paracoccidioides brasiliensis or Blastomyces dermatitidis . J. Immunol. 140 (8), 2786–2789. doi: 10.4049/jimmunol.140.8.2786 [DOI] [PubMed] [Google Scholar]
- Calich V. L., Kashino S. S. (1998). Cytokines produced by susceptible and resistant mice in the course of Paracoccidioides brasiliensis infection. Braz. J. Med. Biol. Res. 31 (5), 615–623. doi: 10.1590/s0100-879x1998000500003 [DOI] [PubMed] [Google Scholar]
- Calich V. L. G., Mamoni R. L., Loures F. V. (2019). Regulatory T cells in paracoccidioidomycosis. Virulence. 10 (1), 810–821. doi: 10.1080/21505594.2018.1483674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho E., Niño-Vega G. A. (2017). Paracoccidioides spp.: virulence factors and immune-evasion strategies. Mediators Inflamm. 2017, 5313691. doi: 10.1155/2017/5313691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano L. E., Singer-Vermes L. M., Vaz C. A., Russo M., Calich V. L. (1995). Pulmonary paracoccidioidomycosis in resistant and susceptible mice: relationship among progression of infection, bronchoalveolar cell activation, cellular immune response, and specific isotype patterns. Infect. Immun. 63 (5), 1777–1783. doi: 10.1128/iai.63.5.1777-1783.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho D. G., Chaves A. F., Xander P., Zelanis A., Kitano E. S., Serrano S. M., et al. (2014). Exploring potential virulence regulators in Paracoccidioides brasiliensis isolates of varying virulence through quantitative proteomics. J. Proteome Res. 13 (10), 4259–4271. doi: 10.1021/pr5002274 [DOI] [PubMed] [Google Scholar]
- Castilho D. G., Navarro M. V., Chaves A. F. A., Xander P., Batista W. L. (2018). Recovery of the Paracoccidioides brasiliensis virulence after animal passage promotes changes in the antioxidant repertoire of the fungus. FEMS Yeast Res. 18 (2). doi: 10.1093/femsyr/foy007 [DOI] [PubMed] [Google Scholar]
- Chang Y. C., Jong A., Huang S., Zerfas P., Kwon-Chung K. J. (2006). CPS1, a homolog of the Streptococcus pneumoniae type 3 polysaccharide synthase gene, is important for the pathobiology of Cryptococcus neoformans . Infect. Immun. 74 (7), 3930–3938. doi: 10.1128/IAI.00089-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., et al. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 14, 128. doi: 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Borges C. L., Bailão A. M., Meirelles G. V., Mendonça Y. A., Dantas S. F. I. M., et al. (2007). Transcriptome profiling of Paracoccidioides brasiliensis yeast-phase cells recovered from infected mice brings new insights into fungal response upon host interaction. Microbiol. (Reading) 153 (Pt 12), 4194–4207. doi: 10.1099/mic.0.2007/009332-0 [DOI] [PubMed] [Google Scholar]
- Coutinho Z. F., Wanke B., Travassos C., Oliveira R. M., Xavier D. R., Coimbra C. E., Jr. (2015). Hospital morbidity due to paracoccidioidomycosis in Brazil, (1998-2006). Trop. Med. Int. Health 20 (5), 673–680. doi: 10.1111/tmi.12472 [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., Mann M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics. 13 (9), 2513–2526. doi: 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26 (12), 1367–1372. doi: 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- de Brito E. D. C. A., Siqueira I. V., Venturini J., Félix V. L. T., Dos Santos A. O. G. M., Mendes R. P., et al. (2023). Iron metabolism disorders of patients with chronic paracoccidioidomycosis. PloS One 18 (6), e0282218. doi: 10.1371/journal.pone.0282218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira H. C., Michaloski J. S., da Silva J. F., Scorzoni L., de Paula E Silva A. C., Marcos C. M., et al. (2016). Peptides Derived from a Phage Display Library Inhibit Adhesion and Protect the Host against Infection by Paracoccidioides brasiliensis and Paracoccidioides lutzii . Front. Pharmacol. 7. doi: 10.3389/fphar.2016.00509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Quaglia E Silva J. C., Della Coletta A. M., Gardizani T. P., Romagnoli G. G., Kaneno R., Dias-Melicio L. A. (2019). Involvement of the Dectin-1 Receptor upon the Effector Mechanisms of Human Phagocytic Cells against Paracoccidioides brasiliensis . J. Immunol. Res. 2019, 1529189. doi: 10.1155/2019/1529189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias-Melicio L. A., Moreira A. P., Calvi S. A., Soares A. M. (2006). Chloroquine inhibits Paracoccidioides brasiliensis survival within human monocytes by limiting the availability of intracellular iron. Microbiol. Immunol. 50 (4), 307–314. doi: 10.1111/j.1348-0421.2006.tb03798.x [DOI] [PubMed] [Google Scholar]
- Diniz S. N., Nomizo R., Cisalpino P. S., Teixeira M. M., Brown G. D., Mantovani A., et al. (2004). PTX3 function as an opsonin for the dectin-1-dependent internalization of zymosan by macrophages. J. Leukoc. Biol. 75 (4), 649–656. doi: 10.1189/jlb.0803371 [DOI] [PubMed] [Google Scholar]
- do Amaral C. C., Fernandes G. F., Rodrigues A. M., Burger E., de Camargo Z. P. (2019). Proteomic analysis of Paracoccidioides brasiliensis complex isolates: Correlation of the levels of differentially expressed proteins with in vivo virulence. PloS One 14 (7), e0218013. doi: 10.1371/journal.pone.0218013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan S. K., Bock T., Hering V., Owens R. A., Jones G. W., Blankenfeldt W., et al. (2017). Structural, mechanistic and functional insight into gliotoxin bis-thiomethylation in Aspergillus fumigatus . Open Biol. 7 (2), 160292. doi: 10.1098/rsob.160292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egen J. G., Rothfuchs A. G., Feng C. G., Winter N., Sher A., Germain R. N. (2008). Macrophage and T cell dynamics during the development and disintegration of mycobacterial granulomas. Immunity. 28 (2), 271–284. doi: 10.1016/j.immuni.2007.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkington P. T., O'Kane C. M., Friedland J. S. (2005). The paradox of matrix metalloproteinases in infectious disease. Clin. Exp. Immunol. 142 (1), 12–20. doi: 10.1111/j.1365-2249.2005.02840.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feriotti C., Bazan S. B., Loures F. V., Araújo E. F., Costa T. A., Calich V. L. (2015). Expression of dectin-1 and enhanced activation of NALP3 inflammasome are associated with resistance to paracoccidioidomycosis. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feriotti C., Loures F. V., Frank de Araújo E., da Costa T. A., Calich V. L. (2013). Mannosyl-recognizing receptors induce an M1-like phenotype in macrophages of susceptible mice but an M2-like phenotype in mice resistant to a fungal infection. PloS One 8 (1), e54845. doi: 10.1371/journal.pone.0054845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo F., Alves L. M., Silva C. L. (1993). Tumour necrosis factor production in vivo and in vitro in response to Paracoccidioides brasiliensis and the cell wall fractions thereof. Clin. Exp. Immunol. 93 (2), 189–194. doi: 10.1111/j.1365-2249.1993.tb07964.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdino N. A. L., Loures F. V., de Araújo E. F., da Costa T. A., Preite N. W., Calich V. L. G. (2018). Depletion of regulatory T cells in ongoing paracoccidioidomycosis rescues protective Th1/Th17 immunity and prevents fatal disease outcome. Sci. Rep. 8 (1), 16544. doi: 10.1038/s41598-018-35037-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton N. J., Waddell S. J., Sherratt A. L., Lee S. M., Smith R. J., Senner C., et al. (2008). Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PloS Med. 5 (4), e75. doi: 10.1371/journal.pmed.0050075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2021). The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 49 (D1), D325–D334. doi: 10.1093/nar/gkaa1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5 (10), R80. doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessner M. A., Werner J. L., Lilly L. M., Nelson M. P., Metz A. E., Dunaway C. W., et al. (2012). Dectin-1-dependent interleukin-22 contributes to early innate lung defense against Aspergillus fumigatus . Infect. Immun. 80 (1), 410–417. doi: 10.1128/IAI.05939-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannandrea M., Parks W. C. (2014). Diverse functions of matrix metalloproteinases during fibrosis. Model. Mech. 7 (2), 193–203. doi: 10.1242/dmm.012062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz D. H., Holmes M. A., Borregaard N., Bluhm M. E., Raymond K. N., Strong R. K. (2002). The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell. 10 (5), 1033–1043. doi: 10.1016/s1097-2765(02)00708-6 [DOI] [PubMed] [Google Scholar]
- Gonzalez A., de Gregori W., Velez D., Restrepo A., Cano L. E. (2000). Nitric oxide participation in the fungicidal mechanism of gamma interferon-activated murine macrophages against Paracoccidioides brasiliensis conidia. Infect. Immun. 68 (5), 2546–2552. doi: 10.1128/IAI.68.5.2546-2552.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglani L., Gopal R., Rangel-Moreno J., Junecko B. F., Lin Y., Berger T., et al. (2012). Lipocalin 2 regulates inflammation during pulmonary mycobacterial infections. PloS One 7 (11), e50052. doi: 10.1371/journal.pone.0050052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H. (2014). Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat. Prod. Rep. 31 (10), 1266–1276. doi: 10.1039/c4np00071d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamashima R., Uchino J., Morimoto Y., Iwasaku M., Kaneko Y., Yamada T., et al. (2020). Association of immune checkpoint inhibitors with respiratory infections: A review. Cancer. Treat. Rev. 90, 102109. doi: 10.1016/j.ctrv.2020.102109 [DOI] [PubMed] [Google Scholar]
- Hams E., Aviello G., Fallon P. G. (2013). The schistosoma granuloma: friend or foe? Front. Immunol. 4. doi: 10.3389/fimmu.2013.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington-Kandt R., Stylianou E., Eddowes L. A., Lim P. J., Stockdale L., Pinpathomrat N., et al. (2018). Hepcidin deficiency and iron deficiency do not alter tuberculosis susceptibility in a murine M.tb infection model. PloS One 13 (1), e0191038. doi: 10.1371/journal.pone.0191038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilty J., George Smulian A., Newman S. L. (2011). Histoplasma capsulatum utilizes siderophores for intracellular iron acquisition in macrophages. Med. Mycol. 49 (6), 633–642. doi: 10.3109/13693786.2011.558930 [DOI] [PubMed] [Google Scholar]
- Holmes M. A., Paulsene W., Jide X., Ratledge C., Strong R. K. (2005). Siderocalin (Lcn 2) also binds carboxymycobactins, potentially defending against mycobacterial infections through iron sequestration. Structure. 13 (1), 29–41. doi: 10.1016/j.str.2004.10.009 [DOI] [PubMed] [Google Scholar]
- Huang H. R., Li F., Han H., Xu X., Li N., Wang S., et al. (2018). Dectin-3 Recognizes Glucuronoxylomannan of Cryptococcus neoformans Serotype AD and Cryptococcus gattii Serotype B to Initiate Host Defense Against Cryptococcosis. Front. Immunol. 9. doi: 10.3389/fimmu.2018.01781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle G. B., Strieter R. M., McNeil L. K., McDonald R. A., Burdick M. D., Kunkel S. L., et al. (1997). Macrophage inflammatory protein-1alpha (MIP-1alpha) is required for the efferent phase of pulmonary cell-mediated immunity to a Cryptococcus neoformans infection. J. Immunol. 159 (1), 318–327. doi: 10.4049/jimmunol.159.1.318 [DOI] [PubMed] [Google Scholar]
- Ihaka R., Gentleman R. (1996). R: A language for data analysis and graphics. J. Comput. Graph. Stat. 5 (3), 299–314. doi: 10.2307/1390807 [DOI] [Google Scholar]
- Kanehisa M. (2019). Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28 (11), 1947–1951. doi: 10.1002/pro.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Sato Y., Kawashima M., Ishiguro-Watanabe M. (2023). KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51 (D1), D587–D592. doi: 10.1093/nar/gkac963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir M. E., Butte M. J., Freeman G. J., Sharpe A. H. (2008). PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 26, 677–704. doi: 10.1146/annurev.immunol.26.021607.090331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher B., Willment J. A., Brown G. D. (2013). The Dectin-2 family of C-type lectin-like receptors: an update. Int. Immunol. 25 (5), 271–277. doi: 10.1093/intimm/dxt006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler V., Sappino A. P., Grau G. E., Piguet P. F., Vassalli P. (1989). The inducing role of tumor necrosis factor in the development of bactericidal granulomas during BCG infection. Cell. 56 (5), 731–740. doi: 10.1016/0092-8674(89)90676-4 [DOI] [PubMed] [Google Scholar]
- Kleifeld O., Doucet A., Prudova A., auf dem Keller U., Gioia M., Kizhakkedathu J. N., et al. (2011). Identifying and quantifying proteolytic events and the natural N terminome by terminal amine isotopic labeling of substrates. Nat. Protoc. 6 (10), 1578–1611. doi: 10.1038/nprot.2011.382 [DOI] [PubMed] [Google Scholar]
- Kroetz D. N., Deepe G. S., Jr. (2010). CCR5 dictates the equilibrium of proinflammatory IL-17+ and regulatory Foxp3+ T cells in fungal infection. J. Immunol. 184 (9), 5224–5231. doi: 10.4049/jimmunol.1000032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44 (W1), W90–W97. doi: 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda Pigosso L., Baeza L. C., Vieira Tomazett M., Batista Rodrigues Faleiro M., Brianezi Dignani de Moura V. M., Melo Bailão A., et al. (2017). Paracoccidioides brasiliensis presents metabolic reprogramming and secretes a serine proteinase during murine infection. Virulence. 8 (7), 1417–1434. doi: 10.1080/21505594.2017.1355660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A. A., Gilbert C., Richard M., Beaulieu A. D., Tremblay M. J. (2008). The C-type lectin surface receptor DCIR acts as a new attachment factor for HIV-1 in dendritic cells and contributes to trans- and cis-infection pathways. Blood. 112 (4), 1299–1307. doi: 10.1182/blood-2008-01-136473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal S. M., Jr, Roy S., Vareechon C., Carrion S.D., Clark H., Lopez-Berges M. S., et al. (2013). Targeting iron acquisition blocks infection with the fungal pathogens Aspergillus fumigatus and Fusarium oxysporum . PloS Pathog. 9 (7), e1003436. doi: 10.1371/journal.ppat.1003436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Lu G., Meng G. (2019). Pathogenic fungal infection in the lung. Front. Immunol. 10. doi: 10.3389/fimmu.2019.01524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell D. M., Moore T. A., McDonald R. A., Toews G. B., Huffnagle G. B. (2006). Distinct compartmentalization of CD4+ T-cell effector function versus proliferative capacity during pulmonary cryptococcosis. Am. J. Pathol. 168 (3), 847–855. doi: 10.2353/ajpath.2006.050522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loures F. V., Araújo E. F., Feriotti C., Bazan S. B., Costa T. A., Brown G. D., et al. (2014). Dectin-1 induces M1 macrophages and prominent expansion of CD8+IL-17+ cells in pulmonary Paracoccidioidomycosis. J. Infect. Dis. 210 (5), 762–773. doi: 10.1093/infdis/jiu136 [DOI] [PubMed] [Google Scholar]
- Loures F. V., Pina A., Felonato M., Calich V. L. (2009). TLR2 is a negative regulator of Th17 cells and tissue pathology in a pulmonary model of fungal infection. J. Immunol. 183 (2), 1279–1290. doi: 10.4049/jimmunol.0801599 [DOI] [PubMed] [Google Scholar]
- Luster A. D. (1998). Chemokines–chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 338 (7), 436–445. doi: 10.1056/NEJM199802123380706 [DOI] [PubMed] [Google Scholar]
- Marques R. E., Marques P. E., Guabiraba R., Teixeira M. M. (2016). Exploring the homeostatic and sensory roles of the immune system. Front. Immunol. 7, 125. doi: 10.3389/fimmu.2016.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R. (2017). New trends in paracoccidioidomycosis epidemiology. J. fungi. 3 (1), 1. doi: 10.3390/jof3010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen J. G., Bedoya V., Patiño M. M., Salazar M. E., Restrepo A. (1987). Experimental murine paracoccidiodomycosis induced by the inhalation of conidia. J. Med. Vet. Mycol. 25 (3), 165–175. doi: 10.1080/02681218780000231 [DOI] [PubMed] [Google Scholar]
- McGaha T., Murphy J. W. (2000). CTLA-4 down-regulates the protective anticryptococcal cell-mediated immune response. Infect. Immun. 68 (8), 4624–4630. doi: 10.1128/IAI.68.8.4624-4630.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrad B., Moore T. A., Standiford T. J. (2000). Macrophage inflammatory protein-1 alpha is a critical mediator of host defense against invasive pulmonary aspergillosis in neutropenic hosts. J. Immunol. 165 (2), 962–968. doi: 10.4049/jimmunol.165.2.962 [DOI] [PubMed] [Google Scholar]
- Miethke M., Marahiel M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71 (3), 413–451. doi: 10.1128/MMBR.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro M. R., Franco C. S. (1994). ““Pathology”,” in Paracoccidioidomycosis. Eds. Franco M., Lacaz C. S., Restrepo-Moreno A., Negro G.D. (Boca Raton, FL: CRC Press; ), 131–147. [Google Scholar]
- Nagae M., Ikeda A., Hanashima S., Kojima T., Matsumoto N., Yamamoto K., et al. (2016). Crystal structure of human dendritic cell inhibitory receptor C-type lectin domain reveals the binding mode with N-glycan. FEBS Lett. 590 (8), 1280–1288. doi: 10.1002/1873-3468.12162 [DOI] [PubMed] [Google Scholar]
- Nemeth E., Ganz T. (2021). Hepcidin-ferroportin interaction controls systemic iron homeostasis. Int. J. Mol. Sci. 22 (12), 6493. doi: 10.3390/ijms22126493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth E., Tuttle M. S., Powelson J., Vaughn M. B., Donovan A., Ward D. M., et al. (2004). Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 306 (5704), 2090–2093. doi: 10.1126/science.1104742 [DOI] [PubMed] [Google Scholar]
- Netto C. F. (1955). Estudos quantitativos sôbre a fixaçào do complemento na blastomicose sul-americana, com antígeno polissacarídico [Quantitative studies on fixation of complement in South American blastomycosis with polysaccharide antigen]. Arq. Cir. Clin. Exp. 18 (5-6), 197–254. [PubMed] [Google Scholar]
- O'Keeffe G., Hammel S., Owens R. A., Keane T. M., Fitzpatrick D. A., Jones G. W., et al. (2014). RNA-seq reveals the pan-transcriptomic impact of attenuating the gliotoxin self-protection mechanism in Aspergillus fumigatus . BMC Genomics 15 (1), 894. doi: 10.1186/1471-2164-15-894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski M. A., Huffnagle G. B., McDonald R. A., Lindell D. M., Moore B. B., Cook D. N., et al. (2000). The role of macrophage inflammatory protein-1 alpha/CCL3 in regulation of T cell-mediated immunity to Cryptococcus neoformans infection. J. Immunol. 165 (11), 6429–6436. doi: 10.4049/jimmunol.165.11.6429 [DOI] [PubMed] [Google Scholar]
- Pagliari C., Fernandes E. R., Stegun F. W., da Silva W. L., Seixas Duarte M. I., Sotto M. N. (2011). Paracoccidioidomycosis: cells expressing IL17 and Foxp3 in cutaneous and mucosal lesions. Microb. Pathog. 50 (5), 263–267. doi: 10.1016/j.micpath.2010.12.008 [DOI] [PubMed] [Google Scholar]
- Parente A. F., Bailão A. M., Borges C. L., Parente J. A., Magalhães A. D., Ricart C. A., et al. (2011). Proteomic analysis reveals that iron availability alters the metabolic status of the pathogenic fungus Paracoccidioides brasiliensis . PloS One 6 (7), e22810. doi: 10.1371/journal.pone.0022810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisu D., Huang L., Rin Lee B. N., Grenier J. K., Russell D. G. (2020). Dual RNA-sequencing of Mycobacterium tuberculosis-infected cells from a murine infection model. STAR Protoc. 1 (3), 100123. doi: 10.1016/j.xpro.2020.100123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomiès P., Rodriguez J., Blaquière M., Sedraoui S., Gouzi F., Carnac G., et al. (2015). Reduced myotube diameter, atrophic signalling and elevated oxidative stress in cultured satellite cells from COPD patients. J. Cell. Mol. Med. 19 (1), 175–186. doi: 10.1111/jcmm.12390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preite N. W., Feriotti C., Souza de Lima D., da Silva B. B., Condino-Neto A., Pontillo A., et al. (2018). The syk-coupled C-type lectin receptors dectin-2 and dectin-3 are involved in Paracoccidioides brasiliensis recognition by human plasmacytoid dendritic cells. Front. Immunol. 9. doi: 10.3389/fimmu.2018.00464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preite N. W., Kaminski V. L., Borges B. M., Calich V. L. G., Loures F. V. (2023). Myeloid-derived suppressor cells are associated with impaired Th1 and Th17 responses and severe pulmonary paracoccidioidomycosis which is reversed by anti-Gr1 therapy. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1039244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerta-Arias J. D., Pino-Tamayo P. A., Arango J. C., González Á. (2016). Depletion of neutrophils promotes the resolution of pulmonary inflammation and fibrosis in mice infected with Paracoccidioides brasiliensis . PloS One 11 (9), e0163985. doi: 10.1371/journal.pone.0163985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Zeltzer S., Zhang Y., Wang F., Chen G. H., Dayrit J., et al. (2012). Early induction of CCL7 downstream of TLR9 signaling promotes the development of robust immunity to cryptococcal infection. J. Immunol. 188 (8), 3940–3948. doi: 10.4049/jimmunol.1103053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Echeverri C., Puerta-Arias J. D., González Á. (2021). Paracoccidioides brasiliensis activates mesenchymal stem cells through TLR2, TLR4, and Dectin-1. Med. Mycol. 59 (2), 149–157. doi: 10.1093/mmy/myaa039 [DOI] [PubMed] [Google Scholar]
- Roussey J. A., Viglianti S. P., Teitz-Tennenbaum S., Olszewski M. A., Osterholzer J. J. (2017). Anti-PD-1 antibody treatment promotes clearance of persistent cryptococcal lung infection in mice. J. Immunol. 199 (10), 3535–3546. doi: 10.4049/jimmunol.1700840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E. J. (2009). The granuloma in tuberculosis–friend or foe? N. Engl. J. Med. 360 (23), 2471–2473. doi: 10.1056/NEJMcibr0902539 [DOI] [PubMed] [Google Scholar]
- San-Blas G. (1992). Regulaciones bioquímicas en el dimorfismo y la virulencia de hongos patógenos para humanos [Biochemical regulations in the dimorphism and virulence of pathogenic fungi for humans]. Acta Cient. Venez. 43 (1), 3–10. [PubMed] [Google Scholar]
- Sansom D. M. (2000). CD28, CTLA-4 and their ligands: who does what and to whom? Immunology. 101 (2), 169–177. doi: 10.1046/j.1365-2567.2000.00121.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Bignell E., Kragl C., Sabiha Y., Loss O., Eisendle M., et al. (2007). Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PloS Pathog. 3 (9), 1195–1207. doi: 10.1371/journal.ppat.0030128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Jung Y. J., Tyagi S., Gennaro M. L., North R. J. (2003). Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc. Natl. Acad. Sci. U. S. A. 100 (1), 241–246. doi: 10.1073/pnas.0136863100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M. G., de Curcio J. S., Silva-Bailão M. G., Lima R. M., Tomazett M. V., de Souza A. F., et al. (2020). Molecular characterization of siderophore biosynthesis in Paracoccidioides brasiliensis . IMA Fungus 11, 11. doi: 10.1186/s43008-020-00035-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3. doi: 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Souto J. T., Aliberti J. C., Campanelli A. P., Livonesi M. C., Maffei C. M., Ferreira B. R., et al. (2003). Chemokine production and leukocyte recruitment to the lungs of Paracoccidioides brasiliensis-infected mice is modulated by interferon-gamma. Am. J. Pathol. 163 (2), 583–590. doi: 10.1016/s0002-9440(10)63686-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak W. A., Deepe G. S., Jr. (2009). The CCL7-CCL2-CCR2 axis regulates IL-4 production in lungs and fungal immunity. J. Immunol. 183 (3), 1964–1974. doi: 10.4049/jimmunol.0901316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaat A. M., Ward S. K., Wu C. W., Rondon E., Tavano C., Bannantine J. P., et al. (2007). Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J. Bacteriol. 189 (11), 4265–4274. doi: 10.1128/JB.00011-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thériault M. E., Paré M.È., Lemire B. B., Maltais F., Debigaré R. (2014). Regenerative defect in vastus lateralis muscle of patients with chronic obstructive pulmonary disease. Respir. Res. 15 (1), 35. doi: 10.1186/1465-9921-15-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thériault M. E., Paré M. È., Maltais F., Debigaré R. (2012). Satellite cells senescence in limb muscle of severe patients with COPD. PloS One 7 (6), e39124. doi: 10.1371/journal.pone.0039124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm J., Post F. A., Bekker L. G., Walther G. B., Wainwright H. C., Manganelli R., et al. (2003). Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. U. S. A. 100 (24), 14321–14326. doi: 10.1073/pnas.2436197100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristão F. S. M., Rocha F. A., Carlos D., Ketelut-Carneiro N., Souza C. O. S., Milanezi C. M., et al. (2017). Th17-inducing cytokines IL-6 and IL-23 are crucial for granuloma formation during experimental paracoccidioidomycosis. Front. Immunol. 8. doi: 10.3389/fimmu.2017.00949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troegeler A., Mercier I., Cougoule C., Pietretti D., Colom A., Duval C., et al. (2017). C-type lectin receptor DCIR modulates immunity to tuberculosis by sustaining type I interferon signaling in dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 114 (4), E540–E549. doi: 10.1073/pnas.1613254114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turissini D. A., Gomez O. M., Teixeira M. M., McEwen J. G., Matute D. R. (2017). Species boundaries in the human pathogen Paracoccidioides. Fungal Genet. Biol. 106, 9–25. doi: 10.1016/j.fgb.2017.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. M., Franco M., Kephart G. M., Gleich G. J. (1998). Localization of eosinophil granule major basic protein in paracoccidioidomycosis lesions. Am. J. Trop. Med. Hyg. 59 (1), 66–72. doi: 10.4269/ajtmh.1998.59.66 [DOI] [PubMed] [Google Scholar]
- Wayne L. G., Sohaskey C. D. (2001). Nonreplicating persistence of mycobacterium tuberculosis. Annu. Rev. Microbiol. 55, 139–163. doi: 10.1146/annurev.micro.55.1.139 [DOI] [PubMed] [Google Scholar]
- Wozniak K. L., Hole C. R., Yano J., Fidel P. L., Wormley F. L. (2014). Characterization of IL-22 and antimicrobial peptide production in mice protected against pulmonary Cryptococcus neoformans infection. Microbiol. (Reading). 160 (Pt 7), 1440–1452. doi: 10.1099/mic.0.073445-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Bailey A., Kuleshov M. V., Clarke D. J. B., Evangelista J. E., Jenkins S. L., et al. (2021). Gene set knowledge discovery with enrichr. Curr. Protoc. 1 (3), e90. doi: 10.1002/cpz1.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunna C., Mengru H., Lei W., Weidong C. (2020). Macrophage M1/M2 polarization. Eur. J. Pharmacol. 877, 173090. doi: 10.1016/j.ejphar.2020.173090 [DOI] [PubMed] [Google Scholar]
- Zhu J., Thompson C. B. (2019). Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell. Biol. 20 (7), 436–450. doi: 10.1038/s41580-019-0123-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. L., Zhao X. Q., Jiang C., You Y., Chen X. P., Jiang Y. Y., et al. (2013). C-type lectin receptors Dectin-3 and Dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection. Immunity 39 (2), 324–334. doi: 10.1016/j.immuni.2013.05.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA945862 http://www.proteomexchange.org/ PXD040826 http://www.proteomexchange.org/, PXD040829.