Abstract
Adipose tissues are closely related to physiological functions and pathological conditions in most organs. Although differentiated 3T3-L1 preadipocytes have been used for in vitro adipose studies, the difference in cellular characteristics of adipogenic differentiation in two-dimensional (2D) culture and three-dimensional (3D) culture remain unclear. In this study, we evaluated gene expression patterns using RNA sequencing and metabolic functions using an extracellular flux analyzer in 3T3-L1 preadipocytes with and without adipogenic induction in 2D culture and 3D culture. In 2D culture, 565 up-regulated genes and 391 down-regulated genes were identified as differentially expressed genes (DEGs) by adipogenic induction of 3T3-L1 preadipocytes, whereas only 69 up-regulated genes and 59 down-regulated genes were identified as DEGs in 3D culture. Ingenuity Pathway Analysis (IPA) revealed that genes associated with lipid metabolism were identified as 2 out of the top 3 causal networks related to diseases and function in 3D spheroids, whereas only one network related to lipid metabolism was identified within the top 9 of these causal networks in the 2D planar cells, suggesting that adipogenic induction in the 3D culture condition exhibits a more adipocyte-specific gene expression pattern in 3T3-L1 preadipocytes. Real-time metabolic analysis revealed that the metabolic capacity shifted from glycolysis to mitochondrial respiration in differentiated 3T3-L1 cells in the 3D culture condition but not in those in the 2D cultured condition, suggesting that adipogenic differentiation in 3D culture induces a metabolic phenotype of well-differentiated adipocytes. Consistently, expression levels of mitochondria-encoded genes including mt-Nd6, mt-Cytb, and mt-Co1 were significantly increased by adipogenic induction of 3T3-L1 preadipocytes in 3D culture compared with those in 2D culture. Taken together, the findings suggest that induction of adipogenesis in 3D culture provides a more adipocyte-specific gene expression pattern and enhances mitochondrial respiration, resulting in more adipocyte-like cellular properties.
Keywords: 3T3-L1 preadipocytes, 3D culture, 2D culture, RNA sequencing analysis, Gene ontology (GO), Enrichment analysis, Ingenuity pathway analysis (IPA), Seahorse Bioanalyzer
Graphical abstract
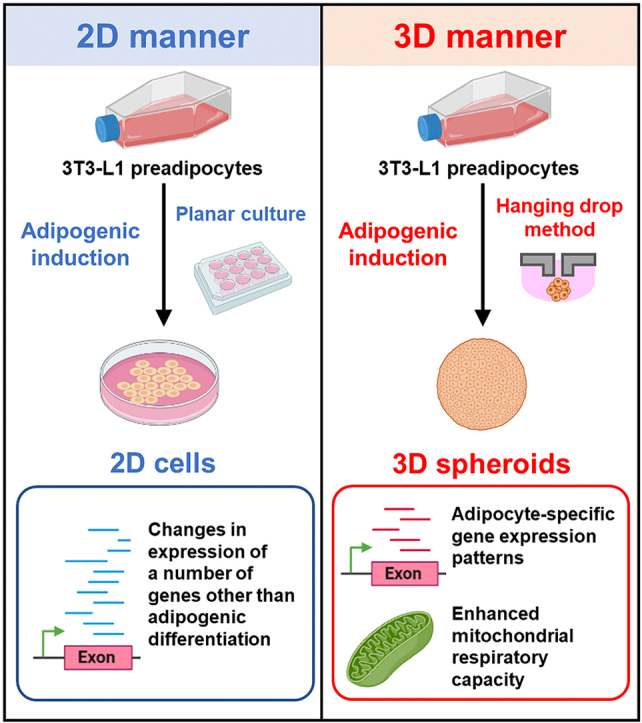
Highlights
-
•
Adipogenic induction in 2D and 3D cultures of 3T3-L1 cells was characterized.
-
•
The 3D culture condition induced more adipocyte-specific gene expression patterns.
-
•
The 3D culture condition enhanced mitochondrial respiration and inhibited glycolysis.
-
•
Adipogenic differentiation in 3D culture may induce more adipocyte-like properties.
1. Introduction
Adipose tissue is known to play pivotal roles in physiological functions and various pathological conditions in most organs [1]. In recent years, in addition to the conventional physiological and pathological roles of adipose tissue, novel functions of adipose tissue, such as aging [[2], [3], [4]], also have been identified, leading to an increasing need for advancing adipose tissue-related research. In this context, adipocytes, the primary cells that form adipose tissue, are characterized by their elasticity and plasticity, and these physiological structures are organized in three-dimensional (3D) space. Therefore, a more biological, accurate, and comprehensive environment is required to properly conduct adipocyte research. Indeed, various in vitro, in vivo, and ex vivo models have been developed using differentiated preadipocytes and primary mature adipocytes to study physiological and pathological aspects relevant to research on adipocytes [5]. Among these models, an important recent development has been the establishment of 3D culture methods that may mimic the physiological function and morphology of adipocytes [5]. 3D culture methods have been increasingly gaining importance in a variety of research areas in relation to extracellular matrix diversity, distinct gene expression patterns, cell-cell adhesion, and cellular metabolism [6,7].
Recently, we have demonstrated that cellular properties of 3D spheroids that are grown by using a drop culture method [8] are quite different from those of conventional 2D cultured cells not only for 3T3-L1 pre-adipocytes [[9], [10], [11], [12]] but also for other cells including trabecular meshwork cells [13], retinal pigment epithelium cells [14], conjunctival fibroblasts [15], orbital fibroblasts [16], and melanoma cells [17]. Notably, in 3T3-L1 preadipocytes, 3D culture significantly enhanced the expression of adipogenesis-related genes and promoted lipid content compared with those in conventional 2D culture, suggesting that a 3D culture condition promotes spontaneous adipogenesis [9,18]. However, it remains unclear whether responses to adipogenic induction affect gene expression patterns and whether cellular metabolic characteristics are different in 2D and 3D culture conditions. Novel insights into the adipogenic differentiation process in 2D and 3D cultures have the potential to advance the research field of adipocytes.
Therefore, the aim of the present study was to clarify the differences in cellular properties of 3T3-L1 preadipocytes and those that underwent adipogenic differentiation in 2D and 3D culture conditions. Cellular properties were analyzed by assessing the lipid content of differentiated cells, changes in gene expression patterns using an RNA sequencing method and gene function analyses, and cellular metabolism including mitochondrial respiration and glycolytic capacities, which are essential functional characteristics in adipocytes.
2. Materials and methods
2.1. Protocols for adipogenic differentiation of the 2D and 3D cultured 3T3-L1 preadipocytes
2D cell cultures and 3D spheroid cultures of 3T3-L1 preadipocytes (#EC86052701-G0, KAK) were performed as described in previous reports [[9], [10], [11], [12]]. In brief, 3T3-L1 preadipocytes were cultured in planar 2D culture dishes at 37 °C in a high-glucose (HG)-DMEM medium supplemented with 8 mg/L d-biotin, 4 mg/L calcium pantothenate, 100 U/mL penicillin, 100 μg/mL streptomycin (b.p. HG-DMEM), 10 % calf serum (CS) and methylcellulose (Methocel A4M). After reaching approximately 90 % confluence, the cells were divided into planar 2D cultures and 3D spheroid cultures using a hanging droplet spheroid 3D culture system [8]. Under these conditions, the 2D planar cultured 3T3-L1 cells were maintained with medium changes daily until Day 7. Alternatively, in the 3D 3T3-L1 spheroid cultures, each well of the drop culture plate (#HDP1385, Sigma-Aldrich) was seeded with approximately 20,000 cells in 28 μL of the medium (3D/Day 0) and thereafter maintained by exchanging half of the culture medium each day until Day 7. To obtain different levels of adipogenic differentiation in 2D planar cultured 3T3-L1 cells, the following three conditions were evaluated: 1) addition of 250 nM dexamethasone during the initial two days, and addition of 500 μM isobutylmethylxanthine (IBMX) and 1 μg/ml insulin each day for 7 or 10 days, 2) addition of 250 nM dexamethasone and 10 nM triiodothyronine (T3) during the initial two days, and addition of 10 μM troglitazone and 1 μg/ml insulin each day for 7 or 10 days, and 3) addition of 250 nM dexamethasone during the initial two days and addition of 500 μM IBMX, 10 μM troglitazone, and 1 μg/ml insulin each day for 7 or 10 days. After confirming the conditions for optimal induction of adipogenesis in 2D culture, induction of adipogenesis by conditions in the 3D cultures was performed by addition of 250 nM dexamethasone and 10 nM T3 during the initial two days, and addition of 10 μM troglitazone and 1 μg/ml insulin each day for 7 days.
2.2. Lipid staining
To assess lipid contents in the cells, Oil Red O lipid staining in the 2D cultured cells and BODIPY lipid staining in the 3D spheroids were performed as previously described [9].
2.3. RNA isolation, reverse transcription, and quantitative RT-PCR
RNA isolation, reverse transcription and quantitative RT-PCR in the 2D planar cells and 3D spheroids were performed as described in our previous report [10]. Briefly, total RNA was extracted using an RNeasy mini kit (Qiagen, Valencia, CA). Reverse transcription was performed with the SuperScript IV kit (Invitrogen) according to the manufacturer's instructions. Respective gene expression was quantified by RT-PCR with Universal Taqman Master mix using a StepOnePlus instrument (Applied Biosystems/Thermo Fisher Scientific). Gene expression levels were normalized to the expression of internal control 36B4 (Rplp0). All experiments in quantitative RT-PCR were performed in duplicate. Sequences of primers and Taqman probes used are shown in Supplementary Table 1.
2.4. RNA sequencing and gene function analyses
RNA sequencing and gene function analyses were performed as described in our previous report with slight modifications [10]. In brief, two independently prepared samples of 2D cultured 3T3-L1 cells or 3D cultured 3T3-L1 spheroids with or without adiopogenic induction were subjected to RNA isolation, and total RNA was initially extracted from each sample. After checking the RNA content and quality (more than 8.5 RIN; an RNA integrity number), cDNA libraries were prepared, and their quality and quantity were validated. After that, sequential steps including cluster generation, sequencing the multiplexed samples, image analysis, base calling, and quality filtering were carried out. The differentially expressed genes (DEGs, fold-change ≥2.0 and FDR adjusted P < 0.05 and q < 0.2) were statistically determined by using mapping of sequence data (Qiagen, Redwood City, CA, USA). To elucidate possible gene functions, gene ontology (GO) enrichment analysis and Ingenuity Pathway Analysis (IPA, Qiagen, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) were carried out. The datasets are available in the Gene Expression Omnibus (GEO) database (accession number: GSE233707).
2.5. Cellular metabolic function analyses using an extracellular flux analyzer
To assess cellular metabolic function in the 2D and 3D cultured 3T3-L1 preadipocytes with or without adipogenic induction, the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were determined by using an extracellular flux analyzer (Seahorse XFe96 Bioanalyzer, Agilent Technologies) as previously described with slight modification [13,14]. Briefly, 2.0 × 104 2D cultured cells were placed in each well of an XFe96 Cell Culture Microplate (Agilent Technologies, #103794-100). The culture medium was then replaced with 180 μL of XF DMEM Medium (Agilent Technologies, #103575-100) supplemented with 5.5 mM glucose, 1.0 mM sodium pyruvate, and 2.0 mM glutamine, with the pH of the assay buffer adjusted to 7.4. For 3D cultured cells, the spheroids were washed twice with phosphate buffered saline and three to five spheroids were then transferred into each well of an XFe96 Spheroid Microplate (Agilent Technologies, # 102978-100) containing 180 μL of the assay buffer. The assay plates were incubated in a CO2-free incubator at 37 °C for 1 h before the measurements. OCR and ECAR were simultaneously measured in a Seahorse XFe96 Bioanalyzer under a 3 min mix and a 3 min measure protocol at baseline and after following sequential injections of oligomycin (final concentration: 2.0 μM), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP, final concentration: 5.0 μM), rotenone/antimycin A mixture (R/A, final concentration: 1.0 μM), and 2-deoxyglucose (2DG, final concentration: 10 mM). Considering the difference in the effects of drug injection in 2D and 3D conditions, 3 cycles in each measurement were conducted for the 2D cells, and 8 cycles for measurement with oligomycin and 4 cycles for other measurements were used in the 3D spheroids. Each value was obtained from the measurement in a single well.
2.6. Statistical analysis
Statistical differences between groups were determined by Student's t-test for two group comparison or ANOVA followed by Tukey's multiple comparison test using Graph Pad Prism 9 (GraphPad Software, San Diego, CA).
3. Results
3.1. Determination of a method for inducing adipogenic differentiation of 3T3-L1 preadipocytes
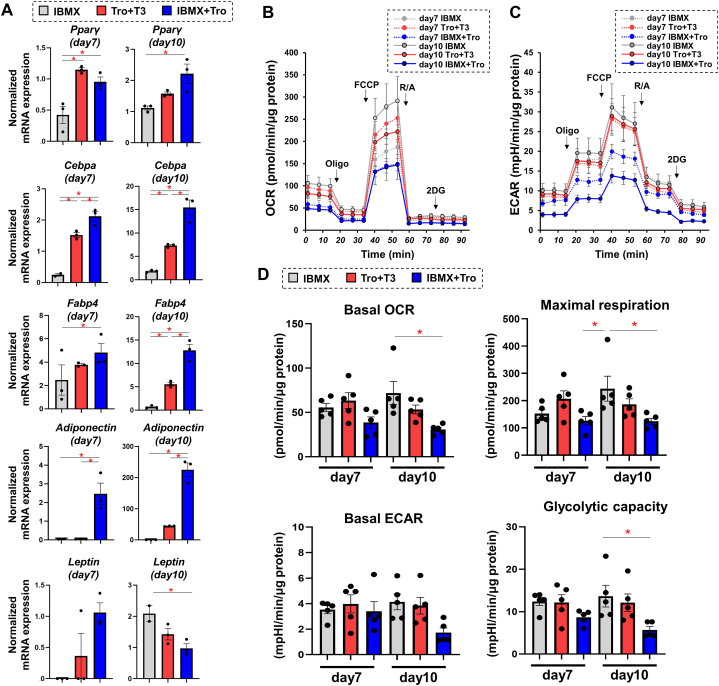
Before examining the characteristics of induction of adipogenic differentiation of 3D spheroids, we needed to evaluate an appropriate and effective way for inducing adipogenic differentiation in 3T3-L1 preadipocytes under 2D culture conditions, since spontaneous adipogenic differentiation was observed in 3T3-L1 preadipocytes in 3D culture conditions but not in 2D culture conditions in our previous study [9]. To determine suitable experimental conditions for differentiation for our research purpose, 2D-cultured 3T3-L1 preadipocytes were subjected to the following three different protocols for inducing adipogenic differentiation: 1) addition of 250 nM dexamethasone during the initial two days, and addition of 500 μM IBMX and 1 μg/ml insulin each day for 7 or 10 days, 2) addition of 250 nM dexamethasone and 10 nM triiodothyronine (T3) during the initial two days, and addition of 10 μM troglitazone and 1 μg/ml insulin each day for 7 or 10 days, and 3) addition of 250 nM dexamethasone during the initial two days, and addition of 500 μM IBMX, 10 μM troglitazone, and 1 μg/ml insulin each day for 7 or 10 days. Among the three different adipogenic induction protocols, the expression levels of five major adipogenesis-related genes, including peroxisome proliferator-activated receptor-γ (Pparγ), CCAAT-enhancer-binding protein α (Cebpa), fatty acid-binding protein 4 (Fabp4), adiponectin (Adiponectin), and leptin, and metabolic profiles were examined and compared at days 7 and 10 of adipogenic induction. As shown in Fig. 1A, expression levels of adipogenesis-related genes were significantly different among the three protocols and were generally more upregulated in the following order: 3) 500 μM and 10 mM troglitazone, 2) 10 μM T3 and 10 mM troglitazone, and 1) 500 μM IBMX. The difference of gene expression levels of PPARγ and leptin on days 7 and 10 were varied among three protocols, but expression levels of the other three genes were not different among the three protocols on days 7 and 10.
Fig. 1.
Determination of an optimal protocol for inducing adipogenic differentiation of 3T3L-1 preadipocytes by assessing adipogenesis-related gene expression and intracellular metabolic function in 2D culture.
Three different protocols for inducing adipogenic differentiation in 2D cultured 3T3-L1 cells were investigated: 1) addition of 250 nM dexamethasone during the initial two days, and addition of 500 μM IBMX and 1 μg/ml insulin each day for 7 or 10 days (=IBMX), 2) addition of 250 nM dexamethasone and 10 nM T3 during the initial two days, and addition of 10 μM troglitazone and 1 μg/ml insulin each day for 7 or 10 days (=Tro + T3), and 3) addition of 250 nM dexamethasone during the initial two days, and addition of 500 μM IBMX, 10 μM troglitazone, and 1 μg/ml insulin each day for 7 or 10 days (=IBMX + Tro). Each sample was subjected to quantitative RT-PCR analysis for adipogenesis-related genes (n = 3) and cellular metabolic function analyses using a Seahorse XFe96 Bioanalyzer (n = 5). Panel A: Expression levels of mRNA of Pparγ, Cebpa, Fabp4, Adiponectin, and Leptin in the three different protocols. Panel B: Measurement of the rate of oxygen consumption (OCR). Panel C: Measurement of the rate of extracellular acidification (ECAR). Panel D: Levels of major indices of mitochondrial functions and glycolytic functions. Basal OCR was determined as the average of OCR at baseline minus the average of OCR after a rotenone/antimycin injection. Maximal respiration was determined as the average of OCR after FCCP injection minus the average of OCR after a rotenone/antimycin injection. Basal ECAR was determined as the average of ECAR at baseline minus the final measurement of ECAR after 2-DG injection. Glycolytic capacity was determined as the average of ECAR after an oligomycin injection minus the final measurement of ECAR after injection of 2-DG. Data are presented as means ± the standard error of the mean (SEM). *P < 0.05, (ANOVA followed by Tukey's multiple comparison test). Tro: troglitazone, Oligo: oligomycin, FCCP: carbonyl cyanide p-trifluoromethoxyphenylhydrazone, R/A: rotenone/antimycin A, 2DG: 2-deoxyglucose.
The levels of both mitochondrial respiration and glycolytic capacities were also different among the three protocols for inducing adipogenic differentiation as shown in Fig. 1B–D. However, quite interestingly, those changes were in the reverse orders of the efficiencies of adipogenic induction assessed by levels of adipogenesis-related gene expression, suggesting that the level of differentiation and energy requirement are not proportional in 2D cultured cells. Based on these results, protocol 2) (addition of 250 nM dexamethasone and 10 nM T3 during the initial two days, and addition of 10 μM troglitazone and 1 μg/ml insulin each day for 7) was considered the most appropriate differentiation state for both expression levels of adipogenesis-related genes and energy status by adipogenesis induction among the three protocols. Thus, it was postulated that protocol 2 using T3 and troglitazone should be adopted to examine the characteristics of induction of adipogenic differentiation of 3T3-L1 preadipocytes in 2D and 3D culture conditions.
3.2. Responses to adipogenic induction are different in 2D and 3D cultured 3T3-L1 preadipocytes with different patterns of differentially expressed genes (DEGs)
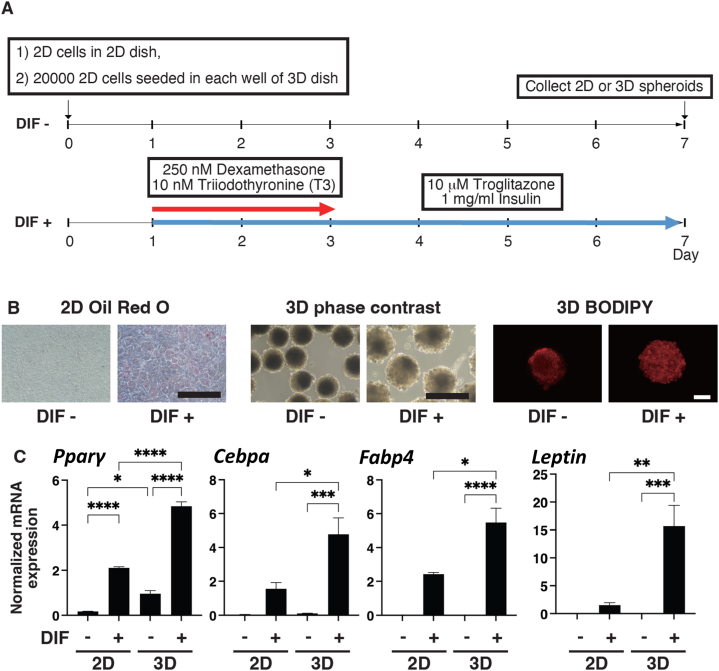
Next, we examined whether the responses to adipogenic induction are different in 2D and 3D cultured 3T3-L1 preadipocytes by performing lipid staining and RNA sequencing analysis. The protocol for induction of differentiation in 2D and 3D cultures in this study is shown in Fig. 2A. Upon adipogenic differentiation, lipid staining of the 2D cultured cells by Oil Red O was substantially increased, and the 3D spheroids were also associated with the numbers of oil droplets in phase contrast images and lipid staining by BODIPY was markedly enhanced (Fig. 2B), confirming that adipogenic differentiation was attained in both 2D and 3D culture conditions by adipogenic induction. However, the expression levels of adipogenesis-related genes including Pparγ, a master regulator gene of adipogenesis, were significantly higher in the 3D 3T3-L1 spheroids than in the 2D cultured cells (Fig. 2C), suggesting that the degrees of adipogenic differentiation induced by a mixture of dexamethasone, T3, troglitazone, and insulin are quite different in 2D and 3D cultures.
Fig. 2.
Diagram of the 2D and 3D cultures of 3T3-L1 preadipocytes and those differentiated by adipogenic induction. 2D cultured 3T3-L1 preadipocytes without induction of differentiation (DIF-) were further processed under 2D cell culture and 3D spheroid culture conditions for 7 days. In the 3D spheroid culture, approximately 20,000 cells were placed in each well of a drop culture plate for 3D spheroid generation (Day 0), and the 3D spheroids were then maintained as described in the Materials and Methods section (Panel A). To stimulate adipogenic differentiation (DIF+), the culture was supplemented with 250 nM dexamethasone and 10 nM T3 and with 10 mM troglitazone and 1 mg/ml insulin from Day 1 to Day 3 and from Day 1 to Day 7, respectively. Representative lipid Oil-red O (2D) and BODIPY (3D) stainings, and phase contrast (3D) images are shown in panel B. Scale bar: 100 μm.
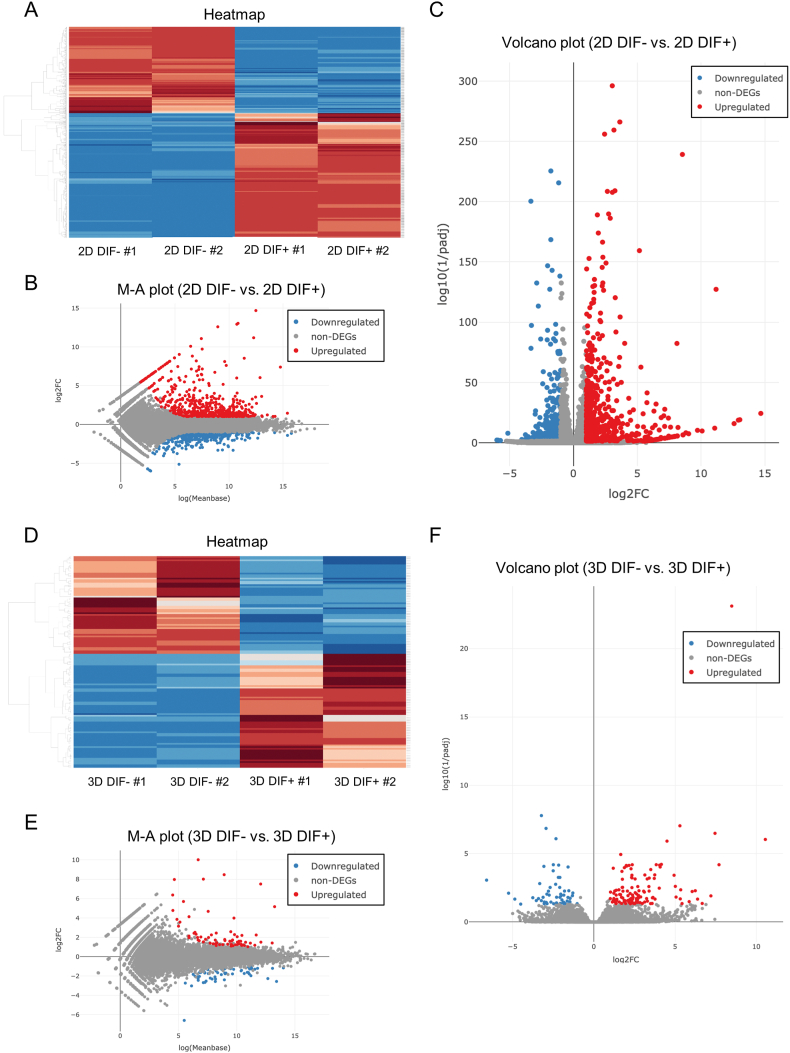
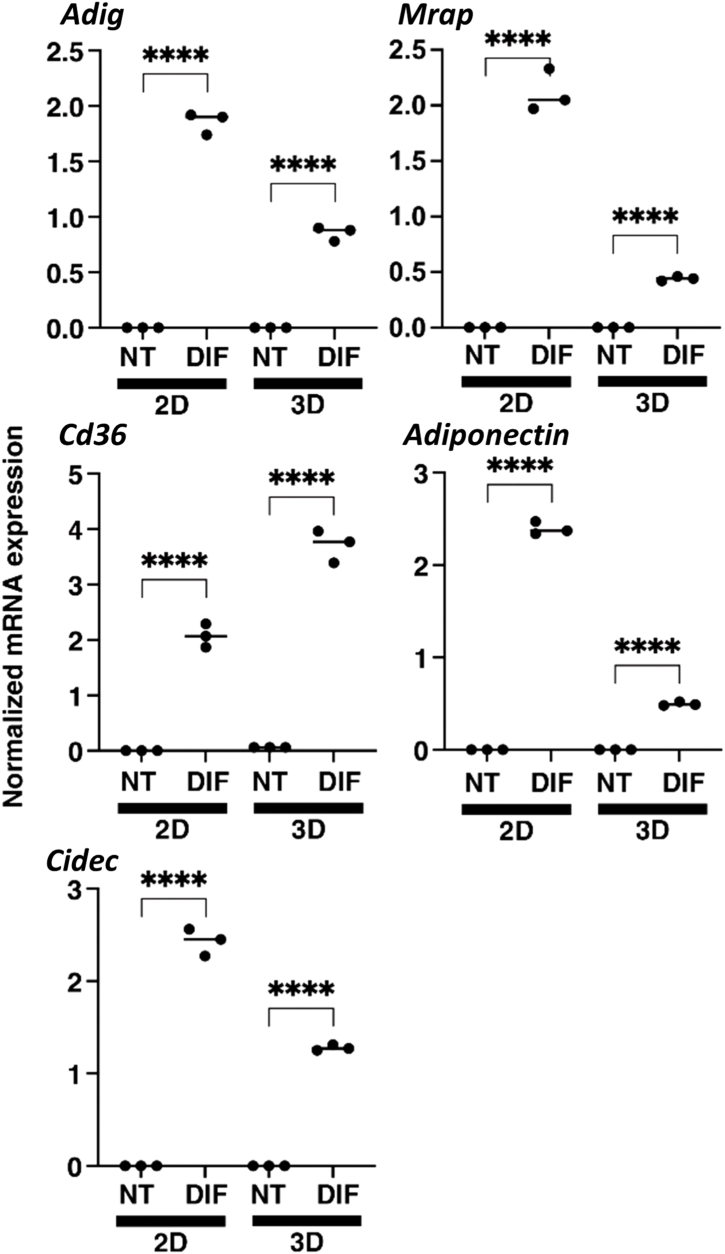
In order to further investigate the differences in gene expression patterns between 2D and 3D induction upon adipogenic differentiation, we comprehensively evaluated the patterns of gene expression changes by RNA sequencing analysis. Differentially expressed genes (DEGs) were determined with a false discovery rate (FDR) of <0.05 and an absolute fold-change ≥2. As shown in Fig. 3, 565 genes were up-regulated and 391 genes were down-regulated as DEGs upon differentiation in the 2D cultured 3T3-L1 preadipocytes (Fig. 3A–C), while in contrast, only 128 DEGs (69 up-regulated and 59 down-regulated genes) were detected in the 3D cultured 3T3-L1 spheroids (Fig. 3D–F). The top 10 up-regulated DEGs in the 2D and 3D cultured conditions are listed in Table 1, Table 2, respectively. Among them, two of the DEGs detected only in the 2D culture conditions including adipogenin (Adig) and Melanocortin 2 Receptor Accessory Protein (Mrap), and DEGs detected in both 2D and 3D culture conditions including cluster of differentiation 36 (Cd36), Adiponectin, and Cell Death Inducing DFFA Like Effector C (Cidec) were also evaluated by quantitative RT-PCR and consistency with the results of RNA sequencing was confirmed (Fig. 4). These results indicate that different responses in gene expression patterns are induced by adipogenic differentiation in 2D and 3D culture conditions.
Fig. 3.
Assessment of differentially expressed genes (DEGs) induced by adipogenic induction in 2D and 3D culture conditions in the 3T3-L1 preadipocytes using RNA sequencing.
DEGs induced by adipogenic induction in the 2D and 3D culture conditions of 3T3-L1 preadipocytes and are shown by hierarchical clustering heatmaps (2D: panel A, 3D: panel D), Minus-Average (M–A) plots of the relationship between the mean expression values [log (baseMean); x-axis] and the magnitude of change in gene expression (log2 of fold-change; y-axis) (2D: panel B, 3D: panel E) and volcano plots of the relationship between magnitude of gene expression change (log2 of fold-change; x-axis) and statistical significance of the change [−log10 of false discovery rate (FDR); y-axis] (2D: panel C, 3D: panel E). Red and blue points represent significantly over-expressed or under-expressed DEGs (cutoff FDR <0.05 and/or magnitude of change ≥2) upon adipogenic induction, respectively. DIF-: undifferentiated cells. DIF+: differentiated cells.
Table 1.
Top 10 genes that were up-regulated or down-regulated in 2D DIF + compared with those in 2D DIF-.
| 2D | Up-regulation (Total: 565 genes) |
Down-regulation (Total: 391 genes) |
||||
|---|---|---|---|---|---|---|
| log 2 Fold Change | Gene Symbol | Gene Biotype | log 2 Fold Change | Gene Symbol | Gene Biotype | |
| 1 | 14.68 | Cidec | protein_coding | −6.01 | Cxcl15 | protein_coding |
| 2 | 13.04 | Plin1 | protein_coding | −5.95 | Rtn4r | protein_coding |
| 3 | 12.92 | Adipoq | protein_coding | −5.91 | Tek | protein_coding |
| 4 | 12.59 | Retn | protein_coding | −5.75 | Acan | protein_coding |
| 5 | 11.17 | Cd36 | protein_coding | −5.73 | Adam5 | protein_coding |
| 6 | 11.07 | Mrap | protein_coding | −5.15 | GFgl2 | protein_coding |
| 7 | 10.08 | Slc2a4 | protein_coding | −4.05 | Atp6v0a4 | protein_coding |
| 8 | 9.72 | Fam83a | protein_coding | −3.81 | Prss16 | protein_coding |
| 9 | 9.02 | Ffar2 | protein_coding | −3.77 | 8020351A03Rik | Processed_transcript |
| 10 | 8.81 | Adig | protein_coding | −3.76 | Slc6a17 | protein_coding |
Table 2.
Top 10 genes that were up-regulated or down-regulated in 3D DIF + compared with those in 3D DIF-.
| 3D | Up-regulation (Total: 69 genes) |
Down-regulation (Total: 59 genes) |
||||
|---|---|---|---|---|---|---|
| log 2 Fold Change | Gene Symbol | Gene Biotype | log 2 Fold Change | Gene Symbol | Gene Biotype | |
| 1 | 10.01 | Adipoq | protein_coding | −6.60 | Aocl | protein_coding |
| 2 | 8.47 | Cidec | protein_coding | −3.03 | Akrlc18 | protein_coding |
| 3 | 8.01 | Aldh1a7 | protein_coding | −2.74 | U90926 | protein_coding |
| 4 | 7.97 | Atp1a2 | protein_coding | −2.69 | Mmp8 | protein_coding |
| 5 | 7.51 | Cd36 | protein_coding | −2.67 | Splnk5 | protein_coding |
| 6 | 6.37 | Thrsp | protein_coding | −2.58 | Cck | protein_coding |
| 7 | 5.70 | Sult1a1 | protein_coding | −2.57 | Saa3 | protein_coding |
| 8 | 5.16 | Fabp4 | protein_coding | −2.54 | Slco4a1 | protein_coding |
| 9 | 4.77 | Retn | protein_coding | −2.46 | Slpi | protein_coding |
| 10 | 4.68 | Tusc5 | protein_coding | −2.41 | C4b | protein_coding |
Fig. 4.
Quantitative RT-PCR analysis of selected genes among the top 10 up-regulated DEGs upon adipogenic differentiation in the 2D and 3D cultured 3T3-L1 preadipocytes. 2D and 3D cultures of 3T3-L1 preadipocyte (NT) and their adipogenesis forms (DIF) at Day 6 were subjected to qPCR analysis of Adig and Mrap, or Cd36, Adiponectin and Cidec which were identified as predominantly upregulated in the 2D culture condition or commonly upregulated in 2D and 3D culture conditions, respectively. All experiments were repeated twice using fresh preparations consisting of 16 spheroids for each qPCR analysis. Data are presented as means ± standard error of the mean (SEM). ****P < 0.001 (ANOVA followed by Tukey's multiple comparison test).
3.3. Gene ontology (GO) enrichment analysis and Ingenuity Pathway Analysis (IPA) of DEGs induced by adipogenic differentiation in 2D and 3D cultures
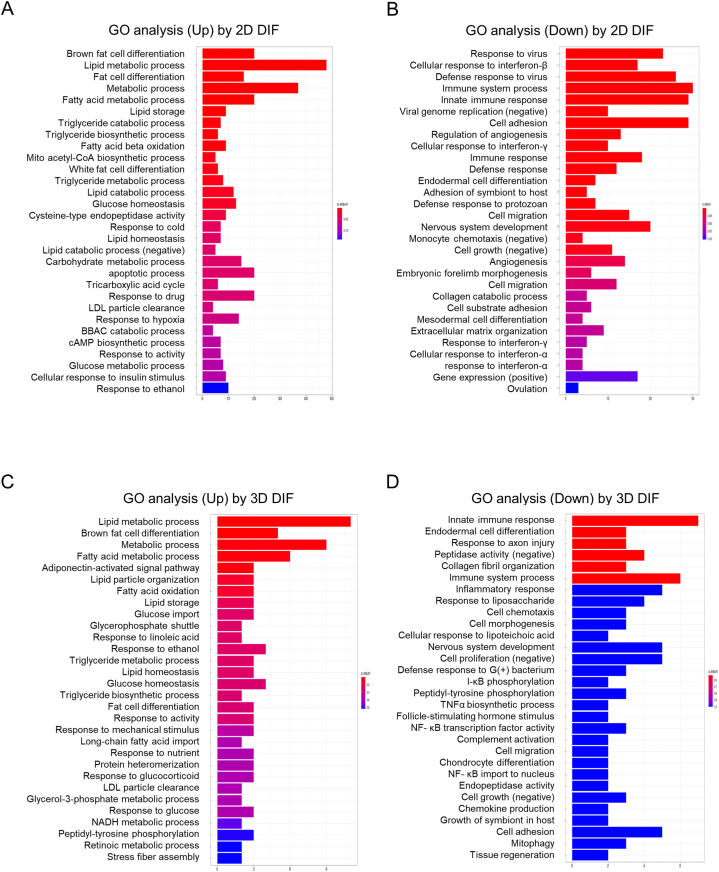
To estimate functional roles of the DEGs that were detected by adipogenic induction in 2D and 3D culture conditions of 3T3-L1 preadipocytes, multi-omics analysis including gene ontology (GO) enrichment analysis and Ingenuity Pathway Analysis (IPA) (Qiagen, Redwood City, CA) were employed. The results of GO enrichment analysis of DEGs that were up-regulated or down-regulated in the 2D and 3D culture conditions are shown in Fig. 5. Upon adipogenesis of 3T3-L1 preadipocytes in both 2D and 3D culture conditions, lipid metabolic processes, brown fat cell differentiation, and metabolic processes including fatty acid metabolic processes were significantly up-regulated, while immune system processes were significantly down-regulated (Fig. 5A and B). However, compared to the 2D culture condition, the 3D culture condition showed more prominent changes in a group of genes related to adipocyte function and metabolism without a decrease in important physiological functions such as angiogenesis and cell adhesion (Fig. 5C and D).
Fig. 5.
Gene ontology (GO) enrichment analysis of DEGs upon adipogenic differentiation in 2D and 3D 3T3-L1 preadipocytes. Up-regulated processes (2D: panel A, 3D: panel C) and down-regulated processes (2D: panel B, 3D: panel D) by adipogenic induction.
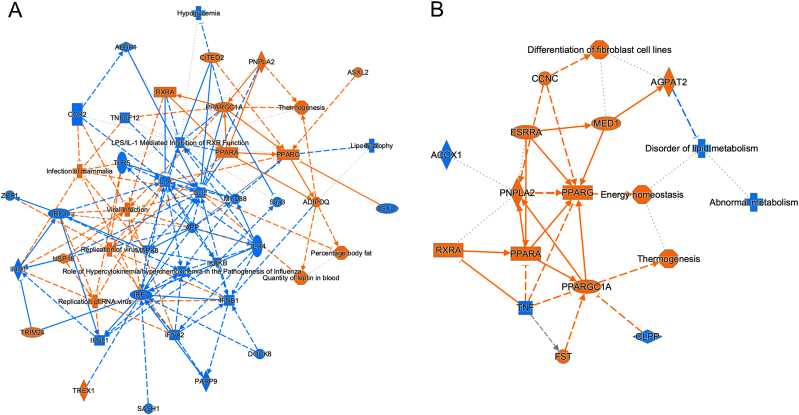
To compare the functional changes in DEGs by induction of adipogenesis in 2D and 3D culture conditions, graphical summaries of IPA in the 2D and 3D culture conditions are shown in Fig. 6. The 2D culture condition showed a more complex network compared to that in the 3D culture; however, PPARs, the master regulators of adipogenesis, were at the centers of the pathways in the 3D culture-based adipogenic induction, whereas pathways related to differentiation and proliferation were prominent in the 2D culture-based adipogenic induction, suggesting that induction of adipogenic differentiation in the 3D culture induces DEGs that are more specific to adipogenesis (Fig. 6A and B). In addition, energy homeostasis was conspicuously located next to the PPARγ pathway in adipogenic differentiation of 3D cultures (Fig. 6B). Furthermore, as shown in Table 3, the results of the IPA analysis showed that nine and three DEGs were significantly involved in the top networks related to disease and function with IPA network scores more than 30 in the 2D and 3D culture conditions upon adipogenic induction, respectively. Interestingly, two of the three networks in the 3D spheroids were associated with genes related to lipid metabolism, while only one of the nine network was found to be related to lipid metabolism in the 2D cells (Table 1). Thus, the results of the IPA analysis also suggest that 3D spheroid culture provides a more specific environment to study adipogenic differentiation of 3T3-L1 preadipocytes.
Fig. 6.
Graphical summary of Ingenuity Pathway Analysis (IPA) in the 2D and 3D culture conditions. Panel A: 2D culture condition. Panel B: 3D culture condition. Orange and blue indicate inhibition and activation, respectively.
Table 3.
Lists of the top 9 (2D) and 3 (3D) causal networks of diseases and functions with IPA network score ≥30.
| No. | Top Diseases and Functions | IPA Score |
Focus Molecules |
|
|---|---|---|---|---|
| 2D | 1 | Cancer, Organismal Injury and Abnormalities, Reproductive System Diseases | 54 | 34 |
| 2 | Carbohydrate Metabolism, Molecular Transport, Orgamismal Development | 46 | 31 | |
| 3 | Amino Acid Metabolism, Developmental Disorder, Small Molecular Biochemistry | 41 | 29 | |
| 4 | Antimicrobial Response, Digestive System Development and Function, Inflammatory Response | 37 | 27 | |
| 5 | Cellular Development, Digestive System Development and Function, Hepatic System Development and Function | 32 | 25 | |
| 6 | Embryonic Development, Nervous System Development and Function, Organ Development | 30 | 24 | |
| 7 | Lipid Metabolism, Molecular Transport, Small Molecular Biochemistry | 30 | 24 | |
| 8 | Cellular Assembly and Organization, Cellular Development, Cellular Function and Maintenance | 30 | 24 | |
| 9 | Amino Acid Metabolism, Molecular Transport, Protein Synthesis | 30 | 24 | |
| 3D | 1 | Energy Production, Lipid Metabolism, Small Molecules Biochemistry | 40 | 20 |
| 2 | Cancer, Hematological System Development and Function, Organismal Injury and Abnormalities | 30 | 16 | |
| 3 | Cellular Movement, Lipid Metabolism, Molecular Transport | 30 | 16 |
3.4. The 3D culture condition predominantly enhances mitochondrial spare respiratory capacity in adipogenic induction of 3T3-L1 cells with increased expression of mitochondria-encoded genes
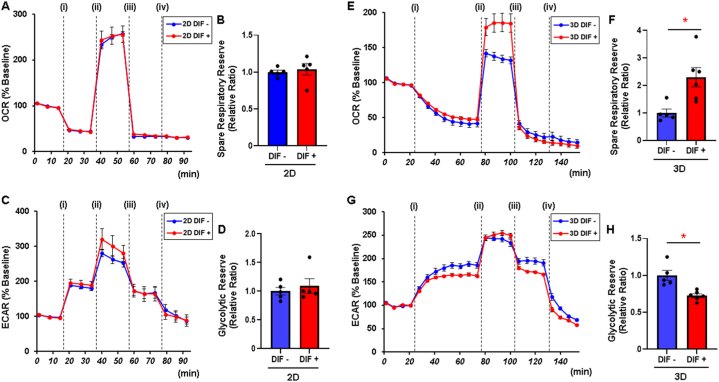
Finally, in order to assess the response in cellular metabolic function of 2D and 3D cultured 3T3-L1 preadipocytes to adipogenic induction, cells or spheroids were analyzed using a Seahorse XFe96 Bioanalyzer. To compare 2D planar cells and 3D spheroids, which are different in size and stress response, metabolic indices were corrected for %baseline in the present study. As shown in Fig. 7, no significant difference was observed between undifferentiated and differentiated cells in the 2D culture conditions in the oxygen consumption rate (OCR) in response to an ionophore FCCP injection, which reflects mitochondrial maximal respiratory capacity (Fig. 7A and B). The changes in extracellular acidification rate (ECAR) in response to an oligomycin injection, which reflects glycolytic reserve, were also comparable in undifferentiated and differentiated cells in the 2D culture conditions (Fig. 7C and D). However, the change in OCR in response to the FCCP injection, which means mitochondrial spare respiratory capacity, was significantly higher by 2.3-fold in differentiated spheroids than in undifferentiated spheroids (Fig. 7E and F). In addition, the change in ECAR in response to an oligomycin injection was significantly lower by 0.72-fold in differentiated spheroids than in undifferentiated spheroids (Fig. 7G and H). These results suggest that the intracellular metabolic ability is predominantly shifted from glycolysis to mitochondrial oxidative phosphorylation by induction of adipogenic differentiation in the 3D culture condition, whereas such metabolic adaptation is not observed in the 2D cultured condition.
Fig. 7.
Measurement of metabolic functions in the 2D and 3D culture conditions of 3T3-L1 preadipocytes with or without adipogenic differentiation.
2D and 3D cultures of the 3T3-L1 preadipocytes (DIF-) and those treated by adipogenic differentiation (DIF+) were subjected to mitochondrial and glycolysis function analyses using a Seahorse XFe96 Bioanalyzer. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) at baseline were calculated as 100 % and their changes were assessed by the following injections: (i) oligomycin, (ii) carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), (iii) rotenone/antimycin, and (iv) 2DG. Panel A: Measurements of OCR in 2D DIF- and DIF + cells. Panel B: Spare respiratory reserve defined as the difference between average of OCR in the presence FCCP and average of OCR at baseline in 2D DIF- and DIF + cells. Panel C: Measurements of ECAR in 2D DIF- and DIF + cells. Panel D: Glycolytic reserve defined as the difference between the final measurement of ECAR in the presence of oligomycin and average of ECAR at baseline in 2D DIF- and DIF + cells. Panel E: Measurements of OCR in 3D DIF- and DIF + spheroids. Panel F: Spare respiratory reserve in 3D DIF- and DIF + spheroids. Panel G: Measurements of ECAR in 3D DIF- and DIF + spheroids. Panel H: Glycolytic reserve in 3D DIF- and DIF + spheroids. Data are presented as means ± the standard error of the mean (SEM). *P < 0.05 (Student's t-test).
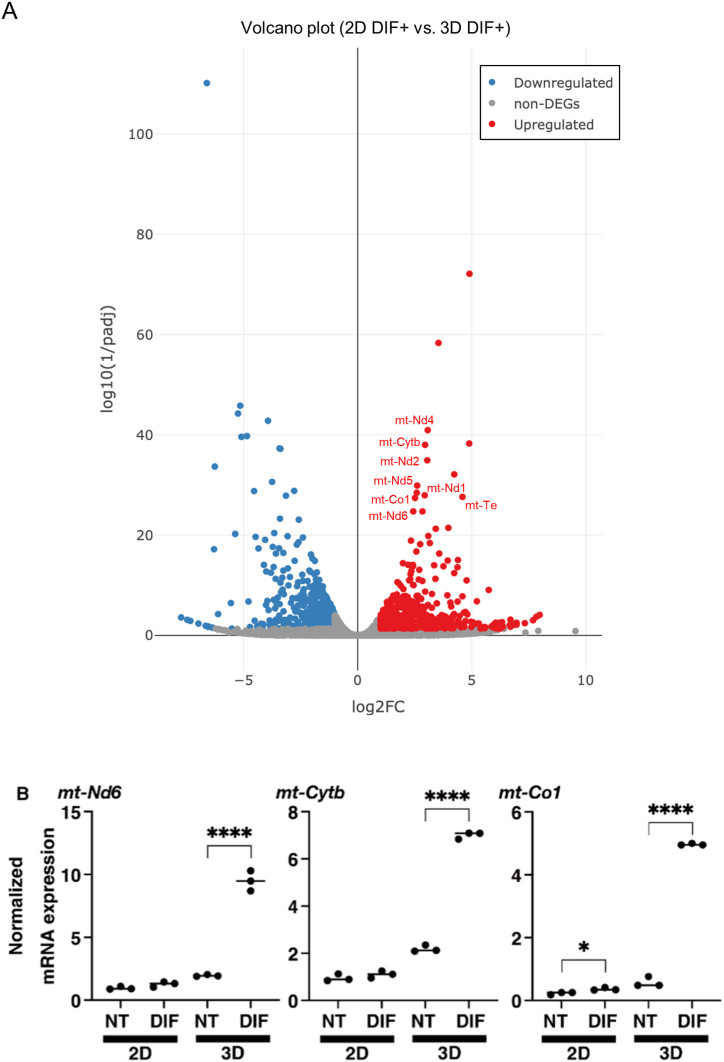
To address molecular mechanisms of different metabolic responses in 2D and 3D culture conditions upon adipogenic differentiation, we compared DEGs obtained by RNA sequencing in differentiated 3T3-L1 cells in 2D and 3D culture conditions. Quite interestingly, several mitochondria-encoded genes that are associated with oxidative phosphorylation (mt-Nd1, mt-Nd2, mt-Nd4, mt-Nd5, mt-Nd6, mt-Co1, and mt-Cytb) and amino-acid metabolism (mt-Te) were determined as significantly up-regulated DEGs in the 3D culture spheroids compared to those in 2D culture planar cells (Fig. 8A), suggesting that the shift of energy reserve to mitochondrial respiration in spheroids differentiated in 3D culture is associated with increased expression of mitochondrial electron transport chain complexes. Indeed, among these mitochondria-encoded genes, expression levels of mt-Nd6 (complex I), mt-Cytb (complex III) and mt-Co1 (complex IV) were also assessed by quantitative RT-PCR and were found to be markedly and significantly upregulated in differentiated 3D spheroids compared with other groups (Fig. 8B).
Fig. 8.
Differentially expressed genes (DEGs) for differentiated 3T3-L1 cells between 2D and 3D cultures.
Panel A: DEGs of DIF+ 3T3-L1 cells between 2D and 3D cultures are shown by a volcano plot. Among the significantly up-regulated DEGs, several mitochondria-related genes were detected. Panel B: 2D and 3D cultures of 3T3-L1 preadipocytes (NT) and their adipogenesis forms (DIF) at Day 7 were subjected to quantitative RT-PCR analysis of selected mitochondrial factors, in which there was a significant correlation with adipocyte metabolism: mt-Nd6 (complex I), mt-Cytb (complex III) and mt-Co1 (complex IV). All experiments were conducted using fresh preparations consisting of 16 spheroids each for qPCR analysis. Data are presented as means ± standard error of the mean (SEM). ****P < 0.001 (ANOVA followed by Tukey's multiple comparison test).
4. Discussion
Despite extensive studies on several biological aspects including gene expression patterns and cellular function in adipogenic differentiation of 3T3-L1 preadipocytes in 2D culture [19,20], knowledge of those in 3D culture has been limited. Even in this context, there have been several previous studies in which the differences between 2D and 3D cultures with respect to molecular expression were examined [21,22]. Turner et al. [21] showed by using transcriptome analysis that DEGs associated with adipogenesis, such as fatty acid metabolism, PPARγ signaling, and adipocytokine signaling, were increased in 3D spheroids of 3T3-L1 cells compared to those in 2D cells during adipogenesis, while DEGs associated with Rho-GTPase 3, matrix metalloproteinases, integrins, and actin-cytoskeletal molecules, which are associated with cell-cell interaction [[23], [24], [25], [26]], were decreased in the 3D condition. In the present study, despite the fact that the adipogenic induction protocol was different from that in the study by Turner et al. [21], similar gene expression patterns were observed. A graphical summary of IPA in the present study visually highlighted pathways specific to adipogenesis and lipid metabolism in the 3D culture conditions compared to the 2D culture conditions, in addition to the finding that the pathway of energy homeostasis was next to PPARγ, a master regulator of adipogenesis. These findings in the present study are in agreement with the results of proteomic analysis of 3D spheroids in adipose-differentiated 3T3-L1 cells in a previous study by Lee et al. [22] showing increased expression of proteins related to carbohydrate metabolism and mitochondrial fatty acid oxidation. These collective observations regarding molecular expression rationally suggest that a 3D spheroid environment may be more optimal for studies related to adipogenesis of 3T3-L1 cells regardless of the method of adipogenic induction. Although insulin signaling is one of the essential signals for lipid droplet synthesis, results of GO analysis in the present study revealed that the pathways involved in insulin signaling were not selected as highly characterized pathways under both 2D and 3D culture conditions, suggesting that differences in insulin signaling are, at least, not major regulators for differentiating 2D and 3D cells. However, it should be noted that gene expression levels do not necessarily correspond to protein expression levels. While our results of RNA sequencing analysis well explained the results of the metabolic function analysis, the recent multi-omics evaluation of proteins including proteome analysis and phosphoproteomics may lead to the elucidation of novel regulatory mechanisms for the differences between 2D and 3D cells.
While the precise mechanisms underlying these differences in adipogenesis between 2D and 3D cultures of 3T3-L1 preadipocytes remain undermined, the difference in cellular metabolism at least seems to be associated with the different phenotypes of 2D cells and 3D spheroids. In the present study, cellular metabolic capacity shifted from glycolysis to mitochondrial respiration in differentiated 3T3-L1 cells in the 3D culture condition but not in those in the 2D culture condition with increased expression of mitochondria-encoded genes in the 3D spheroids upon adipogenic induction. Indeed, there have been several reports showing that differentiated adipocytes exhibit higher oxygen consumption rates than those of undifferentiated cells [[27], [28], [29], [30]], although the difference in 2D and 3D culture conditions was not extensively assessed. Mitochondrial respiration is known to be regulated by not only simple molecular expression but also other multiple factors, including post-translational modification of molecules and supercomplex formation in the electron transport chain [31]. In the present study, since multiple mitochondria-encoded genes that are associated with oxidative phosphorylation were prominently increased in 3D cultured adipogenic spheroids, increased expression of mitochondrial electron transport chain complexes, at least partially, was associated with the increase in mitochondrial spare respiratory capacity observed in the 3D culture condition. PPARγ is a well-known regulator of mitochondrial biogenesis [32]. However, in the present study, PPARγ was upregulated by induction of adipogenesis in both 2D and 3D culture conditions, suggesting that increased PPARγ expression alone may not explain the differences in cellular metabolism between 2D and 3D cultures. Recently, the importance of mitochondria existing in the 3D condition, forming a reticular network contacting other cell organelles, has been proposed and studied [[33], [34], [35]]. Thus, the 3D environment itself, rather than molecular-based mechanisms, may be essential for increased mitochondria-encoded gene expression and altered metabolic characteristics in adipocytes. Further studies to address the detailed mechanisms of metabolic changes in the 3D spheroids of 3T3-L1 preadipocytes upon adipogenic induction are required.
The present study also showed that expression levels of adipogenesis-related genes and the energetic state of 3T3L-1 preadipocytes in 2D culture varied depending on the conditions used to induce adipogenic differentiation as shown in Fig. 1. Consistent with the fact that IBMX-based protocols have been commonly used for adipogenesis [36], our experiments showed that the adipogenesis-induction protocol using a combination of IBMX and thiazolidine in addition to dexamethasone and insulin derivatives increased expression levels of adipogenesis-related genes the most; however, this protocol resulted in the lowest levels of both mitochondrial spare respiratory capacity and glycolytic capacity, which is not consistent with the high metabolic capacity that is a well-known characteristic of differentiated cells. Recently, induction of adipogenic differentiation using both IBMX and thyroid hormones has been reported [37]. Nevertheless, the expression levels of adipogenesis-related genes may not match the metabolic state of the cells under 2D culture conditions regardless of the induction protocol, and from these points of view, induction of adipogenic differentiation in 3D culture, as obtained in the results of this study, may be optimal for mimicking the physiological phenotype of adipocytes.
We acknowledge that there are some limitations in the present study. First, the number of samples provided for RNA sequencing was small. Nevertheless, each sample was obtained in independent experiments, heatmaps (Fig. 3A and D) showed that there was little variation within groups, and gene expression levels of some DEGs were validated by quantitative RT-PCR in addition to RNA sequencing. Second, there are many different factors between 2D planar cells and 3D spheroids, making their comparison challenging. It remains unclear whether the results are due solely to culture conditions themselves or due to the differences in sensitivities of the drugs that induced adipogenesis. It should be noted that because of the different drug sensitivities between 2D and 3D culture conditions, different protocols for measuring real-time metabolic function had to be used for 2D planar cells and 3D spheroids [13,14]. Novel protocols need to be established through careful experiments to compare 2D and 3D culture conditions directly. In addition, lipidome analysis using recent mass spectrometry may be able to assess not only lipid content but also differences in lipid distribution and composition between 2D and 3D culture conditions [38]. Third, this study was conducted using cell lines, which may exhibit different properties compared to preadipocytes taken and grown directly from living organisms. Finally, no genetic modification or drug intervention for specific molecules was performed in the present study, since the purpose of this study was to evaluate the characteristics of induced adipogenic differentiation of 3T3-L1 preadipocytes in 2D and 3D cultures. Although all of the pathways highlighted by IPA in 3D spheroids in this study are important pathways for adipocytes, future studies, including genetic manipulation of molecules detected as DEGs only in 3D spheroids, may provide a precise understanding of the characteristics of 3D culture-based adipogenic induction of 3T3-L1 cells.
5. Conclusion
The findings in this study suggest that inducing adipogenesis in a 3D culture condition of 3T3-L1 preadipocytes leads to more adipocyte-specific gene expression patterns and metabolic phenotypes with increased expression of mitochondria-encoded genes, which may be associated with more differentiated adipocyte-like cellular properties compared with those in a 2D culture condition.
Funding
This research was partly supported by JSPS KAKENHI Grant Numbers 20K08913 (M.F.), 22K08210 (T.S.), and 23K07993 (M.F.)
Author contribution statement
Conceived and designed the experiments: Tatsuya Sato, Hiroshi Ohguro, Masato Furuhashi.
Performed the experiments: Keisuke Endo, Tatsuya Sato, Araya Umetsu, Fumihito Hikage, Megumi Watanabe, Yosuke Ida.
Analyzed and interpreted the data: Keisuke Endo, Tatsuya Sato, Araya Umetsu, Megumi Watanabe, Fumihito Hikage, Yosuke Ida, Hiroshi Ohguro, Masato Furuhashi.
Contributed reagents, materials, analysis tools or data: Keisuke Endo, Tatsuya Sato, Araya Umetsu, Megumi Watanabe, Fumihito Hikage, Yosuke Ida, Hiroshi Ohguro, Masato Furuhashi.
Wrote the paper: Keisuke Endo, Tatsuya Sato, Hiroshi Ohguro, Masato Furuhashi.
Funding statement
Masato Furuhashi was supported by Japan Society for the Promotion of Science {20K08913, 23K07993}.
Tatsuya Sato was supported by Japan Society for the Promotion of Science {22K08210}.
Data availability statement
Data associated with this study has been deposited at RNA-seq, GEO database under the accession number GSE233707.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The graphical abstract was partially created using licensed BioRender (T.S.). The RNA sequencing data set repository was made with the assistance of Rhelixa, Inc. (Tokyo, Japan). The authors acknowledge S.E.S. Translation and Proofreading Services for the language editing of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e20713.
Appendix ASupplementary data
The following is the Supplementary data to this article.
References
- 1.Parra-Peralbo E., Talamillo A., Barrio R. Origin and development of the adipose tissue, a key organ in physiology and disease. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.786129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ou M., Zhang H., Tan P., Zhou S.m, Li Q. Adipose tissue aging: mechanisms and therapeutic implications. Cell Death Dis. 2022;13(4):300. doi: 10.1038/s41419-022-04752-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silva G.N., Amato A.A. Thermogenic adipose tissue aging: mechanisms and implications. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.955612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai Z., He B. Adipose tissue aging: an update on mechanisms and therapeutic strategies. Metabolism. 2023;138 doi: 10.1016/j.metabol.2022.155328. [DOI] [PubMed] [Google Scholar]
- 5.Dufau J., Shen J.X., Couchet M., Barbosa T.D.C., Mejhert N., Massier L., Griseti E., Mouisel E., Amri E., Lauschke V.M., Rydén M., Langin D. In vitro and ex vivo models of adipocytes. Am J Physiol Cell Physiol. 2021;320(5):C822–C841. doi: 10.1152/ajpcell.00519.2020. [DOI] [PubMed] [Google Scholar]
- 6.Yamada K.M., Doyle A.D., Lu J. Cell-3D matrix interactions: recent advances and opportunities. Trends Cell Biol. 2022;32(10):883–895. doi: 10.1016/j.tcb.2022.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rybkowska P., Radoszkiewicz K., Kawalec M., Dymkowska D., Zabłocka B., Zabłocki K., Sarnowska A. The metabolic changes between monolayer (2D) and three-dimensional (3D) culture conditions in human mesenchymal stem/stromal cells derived from adipose tissue. Cells. 2023;12(1):178. doi: 10.3390/cells12010178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hikage F., Atkins S., Kahana A., Smith T.J., Chun T. HIF2A-LOX pathway promotes fibrotic tissue remodeling in thyroid-associated orbitopathy. Endocrinology. 2019;160(1):20–35. doi: 10.1210/en.2018-00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ida Y., Hikage F., Ohguro H. ROCK inhibitors enhance the production of large lipid-enriched 3D organoids of 3T3-L1 cells. Sci. Rep. 2021;11(1):5479. doi: 10.1038/s41598-021-84955-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohguro H., Ida Y., Hikage F., Umetsu A., Ichioka H., Watanabe M., Furuhashi M. STAT3 is the master regulator for the forming of 3D spheroids of 3T3-L1 preadipocytes. Cells. 2022;11(2):300. doi: 10.3390/cells11020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Umetsu A., Ida Y., Sato T., Watanabe M., Tsugeno Y., Furuhashi M., Hikage F., Ohguro H. Brimonidine modulates the ROCK1 signaling effects on adipogenic differentiation in 2D and 3D 3T3-L1 cells. Bioengineering (Basel) 2022;9(7):327. doi: 10.3390/bioengineering9070327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umetsu A., Sato T., Watanabe M., Ida Y., Furuhashi M., Tsugeno Y., Ohguro H. Unexpected crosslinking effects of a human thyroid stimulating monoclonal autoantibody, M22, with IGF1 on adipogenesis in 3t3l-1 cells. Int. J. Mol. Sci. 2023;24(2):1110. doi: 10.3390/ijms24021110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe M., Sato T., Tsugeno Y., Umetsu A., Suzuki S., Furuhashi M., Ida Y., Hikage F., Ohguro H. Human trabecular meshwork (HTM) cells treated with TGF-β2 or dexamethasone respond to compression stress in different manners. Biomedicines. 2022;10(6):1338. doi: 10.3390/biomedicines10061338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki S., Sato T., Watanabe M., Higashide M., Tsugeno Y., Umetsu A., Furuhashi M., Ida Y., Hikage F., Ohguro H. Hypoxia differently affects TGF-β2-induced epithelial mesenchymal transitions in the 2D and 3D culture of the human retinal pigment epithelium cells. Int. J. Mol. Sci. 2022;23(10):5473. doi: 10.3390/ijms23105473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsugeno Y., Furuhashi M., Sato T., Watanabe M., Umetsu A., Suzuki S., Ida Y., Hikage F., Ohguro H. FGF-2 enhances fibrogenetic changes in TGF-β2 treated human conjunctival fibroblasts. Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-20036-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hikage F., Ichioka H., Watanabe M., Umetsu A., Ohguro H., Ida Y. ROCK inhibitors modulate the physical properties and adipogenesis of 3D spheroids of human orbital fibroblasts in different manners. FASEB Bioadv. 2021;3(10):866–872. doi: 10.1096/fba.2021-00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohguro H., Watanabe M., Sato T., Hikage F., Furuhashi M., Okura M., Hida T., Uhara H. 3D spheroid configurations are possible indictors for evaluating the pathophysiology of melanoma cell lines. Cells. 2023;12(5):759. doi: 10.3390/cells12050759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Josan C., Kakar S., Raha S. Matrigel® enhances 3T3-L1 cell differentiation. Adipocyte. 2021;10(1):361–377. doi: 10.1080/21623945.2021.1951985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambele M.A., Dessels C., Durandt C., Pepper M.S. Genome-wide analysis of gene expression during adipogenesis in human adipose-derived stromal cells reveals novel patterns of gene expression during adipocyte differentiation. Stem Cell Res. 2016;16(3):725–734. doi: 10.1016/j.scr.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Sadowski H.B., Wheeler T.T., Young D.A. Gene expression during 3T3-L1 adipocyte differentiation. Characterization of initial responses to the inducing agents and changes during commitment to differentiation. J. Biol. Chem. 1992;267(7):4722–4731. [PubMed] [Google Scholar]
- 21.Turner P.A., Garrett M.R., Didion S.P., Janorkar A.V. Spheroid culture system confers differentiated transcriptome profile and functional advantage to 3T3-L1 adipocytes. Ann. Biomed. Eng. 2018;46(5):772–787. doi: 10.1007/s10439-018-1993-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S.Y., Park S.B., Kim Y.E., Yoo H.M., Hong J., Choi K., Kim K.Y., Kang D. iTRAQ-based quantitative proteomic comparison of 2D and 3D adipocyte cell models Co-cultured with macrophages using online 2D-nanoLC-ESI-MS/MS. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-53196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masri R.E., Delon J. RHO GTPases: from new partners to complex immune syndromes. Nat. Rev. Immunol. 2021;21(8):499–513. doi: 10.1038/s41577-021-00500-7. [DOI] [PubMed] [Google Scholar]
- 24.Kessenbrock K., Plaks V., Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gahmberg C.G., Grönholm M. How integrin phosphorylations regulate cell adhesion and signaling. Trends Biochem. Sci. 2022;47(3):265–278. doi: 10.1016/j.tibs.2021.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cronin N.M., DeMali K.A. Dynamics of the actin cytoskeleton at adhesion complexes. Biology. 2021;11(1):52. doi: 10.3390/biology11010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heimburg D., Hemmrich K., Zachariah S., Staiger H., Pallua N. Oxygen consumption in undifferentiated versus differentiated adipogenic mesenchymal precursor cells. Respir. Physiol. Neurobiol. 2005;146(2–3):107–116. doi: 10.1016/j.resp.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y., Marsboom G., Toth P.T., Rehman J. Mitochondrial respiration regulates adipogenic differentiation of human mesenchymal stem cells. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0077077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drehmer D.L., Aguiar A.M., Brandt A.P., Petiz L., Cadena S.M.S.C., Rebelatto C.K., Brofman P.R.S., Neto F.F., Dallagiovanna B., Abud A.P.L. Metabolic switches during the first steps of adipogenic stem cells differentiation. Stem Cell Res. 2016;17(2):413–421. doi: 10.1016/j.scr.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Moore T.M., Cheng L., Wolf D.M., Ngo J., Segawa M., Zhu X., Strumwasser A.R., Cao Y., Clifford B.L., Ma A., Scumpia P., Shirihai O.S., Vallim T.Q.A., Laakso M., Lusis A.J., Hevener A.L., Zhou Z. Parkin regulates adiposity by coordinating mitophagy with mitochondrial biogenesis in white adipocytes. Nat. Commun. 2022;13(1):6661. doi: 10.1038/s41467-022-34468-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sung A.Y., Floyd B.J., Pagliarini D.J. Systems biochemistry approaches to defining mitochondrial protein function. Cell Metab. 2020;31(4):669–678. doi: 10.1016/j.cmet.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan W., Evans R. PPARs and ERRs: molecular mediators of mitochondrial metabolism. Curr. Opin. Cell Biol. 2015;33:49–54. doi: 10.1016/j.ceb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fenton A.R., Jongens T.A., Holzbaur E.L.F. Mitochondrial dynamics: shaping and remodeling an organelle network. Curr. Opin. Cell Biol. 2021;68:28–36. doi: 10.1016/j.ceb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Q., An Y.A., Scherer P.E. Mitochondrial regulation and white adipose tissue homeostasis. Trends Cell Biol. 2022;32(4):351–364. doi: 10.1016/j.tcb.2021.10.008. Epub 2021 Nov 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lounas A., Lebrun A., Laflamme I., Vernoux N., Savage J., Tremblay M., Germain M., Richard F.J. A 3D analysis revealed complexe mitochondria morphologies in porcine cumulus cells. Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-19723-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott M.A., Nguyen V.T., Levi B., James A.W. Current methods of adipogenic differentiation of mesenchymal stem cells. Stem Cells Dev. 2011;20(10):1793–1804. doi: 10.1089/scd.2011.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu Y., Ding Q. Optimized protocol for gene editing in adipocytes using CRISPR-Cas9 technology. STAR Protoc. 2021;2(1) doi: 10.1016/j.xpro.2021.100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tobias F., Hummon A.B. Lipidomic comparison of 2D and 3D colon cancer cell culture models. J. Mass Spectrom. 2022;57(8) doi: 10.1002/jms.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at RNA-seq, GEO database under the accession number GSE233707.