Abstract
Net blotch (NB) and powdery mildew (PM) are major barley diseases with the potential to cause a dramatic loss in grain yield. Breeding for resistant barley genotypes in combination with identifying candidate resistant genes will accelerate the genetic improvement for resistance to NB and PM. To address this challenge, a set of 122 highly diverse barley genotypes from 34 countries were evaluated for NB and PM resistance under natural infection for in two growing seasons. Moreover, four yield traits; plant height (Ph), spike length (SL), spike weight (SW), and the number of spikelets per spike (NOS) were recorded. High genetic variation was found among genotypes in all traits scored in this study. No significant phenotypic correlation was found in the resistance between PM and NB. Immune genotypes for NB and PM were identified. A total of 21 genotypes were immune to both diseases. Of the 21 genotypes, the German genotype HOR_9570 was selected as the most promising genotype that can be used for future breeding programs. Furthermore, a genome-wide association study (GWAS) was used to identify resistant alleles to PM and NB. The results of GWAS revealed a set of 14 and 25 significant SNPs that were associated with increased resistance to PM and NB, respectively. This study provided very important genetic resources that are highly resistant to the Egyptian PM and NB pathotypes and revealed SNP markers that can be utilized to genetically improve resistance to PM and NB.
Keywords: Hordeum vulgare L., Gene models, Powdery mildew, Net blotch, GWAS, LD, Disease resistance
Graphical Abstract

1. Introduction
Barley is an important cereal crop that is used for human nutrition and animal feed [1]. Fungal infection is one of the most important biotic stresses affecting Barley's production and proactivity. Net blotch (NB) is a serious plant disease that is caused by Drechslera teres f. sp. teres [2]. The symptoms of NB spread quickly causing a significant loss in grain yield [2]. Another serious disease that is an important threat to barley production is powdery mildew (PM) which is caused by the pathogen Erysiphe graminis DC. f. sp. hordei Em Marchal (synamorph Blumeria graminis (DC.) Golovin ex Speer f. sp. hordei). Both diseases are wide-spread in Middle and Northern Egypt, while, PM disease is always observed in Upper Egypt [3], [4]. Climate change will have a great impact on increasing the spread of NB and PM. Moreover, climate change may help to produce new PM and NB pathotypes [5], [6], [7].
Treatment with fungicides to control the spread of PM and NB is a common practice to minimize the severity of both diseases on cereal crops [8], [9], [10], [11]. The high cost of fungicides and environmental concerns associated with the use of fungicides may lead to a gradual limitation of their use to control both diseases. Breeding for resistant genotypes to NB and PM is an alternative environmental-friendly solution to control the spread of NB and PM pathotypes. However, the resistance to the diseases in barley cultivars could be broken over time. Therefore, the continuous evaluation of the barley germplasms having highly diverse genotypes will help to select the most promising resistant genotypes to NB and/or PM pathotypes. Phenotypic screening for PM and NB resistance in large barley germplasm is necessary to have promising resistant genotypes for different pathotypes of PM and NB. Disease resistance is normally performed using a visual score which may be prone to human errors; hence, selection only based on the phenotypic variation could be misleading [8], [12].
The recent advances in DNA sequence technology led to the newborn of different DNA sequencing methods by which a lot of single-nucleotide polymorphisms (SNPs) can be detected [13], [14]. These SNPs are normally located within important genes. The SNPs can be utilized to identify genes associated with target traits (e.g. resistance to NB and PM) through a genome-wide association study (GWAS) [15]. GWAS provides very useful information in allele or/and gene identification. The identification of new genes and SNP markers will undoubtedly accelerate the breeding program and enhance marker-assisted selection (MAS) to genetically improve the resistance to PM and NB pathotypes. Many studies have successfully identified resistance genes associated with PM and NB in barley [16], [17], [18], [19], [20], [21], [22], [23], [24]. However, all the genes identified in the previous studies were resistant to pathotypes originating from Europe, Switzerland, Ethiopia, and the USA. To the best of our knowledge, there is no study on GWAS for resistance to the Egyptian NB and PM pathotype(s), and hence no information on the resistance genes to the Egyptian pathotypes.
Therefore, the objectives of this study were to (1) screen 122 highly diverse barley genotypes from different countries for resistance against Egyptian NB and PM pathotypes, (2) identify candidate genes associated with resistance to NP and PM, and (3) identify the most promising resistant genotypes to NB and PM combined with high-yielding traits to be integrated in future barley molecular breeding programs.
2. Material and methods
2.1. Plant material
A set of 122 highly diverse barley genotypes from 34 different countries was used in this study. All information on the population is presented in Supplementary Table 1. The plant materials were obtained from the gene bank at the Leibniz Institute of Plant Genetics and Crop Plant Research, Germany.
2.2. Evaluation of PM and NB resistance
Twenty seeds from each genotype were sown in a row 100 cm long and the distance between rows was 30 cm. The experimental layout was a randomized complete block design with three replications/year. All recommended cultural practices for barley crops in the commercial fields i.e., fertilization, irrigation, and other management were applied. Phenotyping of NB and PM resistance was carried out under a naturally infested unheated greenhouse which is located in the middle of Sakha Agriculture Research Station (31.094059° N, 30.933899°E), Plant Pathology Research Institute (PPRI), Agricultural Research Center (ARC), Egypt for two successive seasons, 2021 and 2022. The Sakha Agriculture Station is an infested open field with PM and NB diseases. For net blotch, a set of 12 Egyptian Pyrenophora teres f. sp. hordei pathotypes were characterized by Abdel-Fattah [25], while the pathotypes of Blumeria graminis f. sp. hordei (syn. Erysiphe graminis f. sp. Hordei which causes PM disease have not been characterized yet. The conditions of the unheated greenhouse were a temperature of ∼20–23 ± 5°C and relative humidity of ∼80–90% during the experiment (Supplementary Figure 1 and 2). According to Manadhar et al. [26], the infection of powdery mildew and net blotch were recorded on the whole plant and hinges on the value of 5 which has been defined as the midpoint. The scoring of adult plant infection was done when the powdery mildew and net blotch symptoms fully developed (around GS-75). A double-digit (D1D2) scale measured adult plant infection was used. The symptoms for each disease were visually scored on the plants (20 plants/accession) using a scale ranging from 0 to 9 [27]. The scale is based on infection types (ITs) as follows (Fig. 1):
Fig. 1.
Symptoms of powder mildew (a) and net blotch (b) appeared on barley leaves during the experiment.
0 = free from in (0).
1 = resistant: A few isolated lesions on only the lowest leaves (R).
2 = resistant: Scattered lesions on the second set of leaves (R).
3 = resistant: Light infection of the lower third of the plant (R, MR).
4 = moderately resistant: Moderate infection of lower leaves (MR).
5 = moderately susceptible: Severe infection of lower leaves; Moderate to light infection (MR, MS).
6 = moderately susceptible: Severe infection on the lower third of plant; Moderate on middle leaves (MS).
7 = susceptible: lesions Severe on lower and middle leaves (MS, S).
8 = susceptible: lesions Severe on lower and middle leaves; Moderate to Severe infection of the upper third of the plant, flag Leaf infected in amounts more than a trace (S).
9 = highly susceptible: Severe infection on all leaves; spike also infected to some degree (VS).
Four important yield traits were recorded on each genotype; plant height (Ph, cm), spike length (SPL, cm), spike weight (SW, g). and number of spikelets per spike (NOS, number).
Statistical analysis.
The analysis of variance (ANOVA) for all traits was performed using PLABSTAT software [28] according to the following statistical model,
| Yijk= μ + gi+ rj+ yk+ gyik+ εijk(error) |
where Yijk is the observation of a genotype i in a replication j tested in a year k. μ is the general average; gi, yk, and rj, refer to the effects of genotypes, years, and replications, respectively. gyik is genotype × year interaction. εijk is genotype × replications × year interaction (error). Genotypes, years, and replications were considered random effects. Also, interactions were also considered random effects.
The variance–covariance analysis was carried out using GENOT-command with PLABSTAT software to estimate the genetic correlation coefficient among the traits. The broad-sense heritability (H2) was estimated using HERTI command in PLABSTAT using the following equation
where g, r, and y are the number of genotypes, replications, and years, respectively. σ2g and σ2e are components of variance for genotypes and error, respectively.
2.3. Genome-wide association study (GWAS)
All genotypes used in this study were previously genotyped using the genotyping-by-sequencing method by Milner et al. [29]. The SNP calling was performed against the reference genome sequence of the barley cultivar Morex [29]. The SNP data of 122 genotypes extracted from Milner et al. [29] were used for the GWAS study. The SNP markers generated from the genotyping were filtered based on minor allele frequency (MAF) of > = 5% and a missing rate of < = 10%. As a result, a set of 18,525 SNPs were generated and used for GWAS.
Principal component analysis (PCA) was performed among genotypes using the 18,525 SNPs to examine the population structure. The GWAS analysis was carried out using nine models; MLM + kinship, GLM+kinship, FarmCPU+kinship, GLM+PCA, MLM+ PCA, FarmCPU+PCA, MLM+PCA+kinship, GLM+PCA+kinship, and FarmCPU+PCA+kinship by TASSEL v5.0 [30] and Genomic Association and Prediction Integrated Tool (GAPIT) in the R environment [31]. Five PCAs were included in the GWAS to account for the population. Marker-trait associations were detected a threshold P-value 0.001 equal to –log10(P)≥ 3 [33], [34]. The markers were finally selected if they were significant in at least five models. The best model for GWAS was determined based on the quantile-quantile (q-q) plot. For each significant marker, target alleles (resistant allele) were determined based on the allele effects. The gene annotation for all significant markers was performed against the recently released genome assembly: MorexV3_pseudomolecules_assembly using the ensemble plants genomic database (https://plants.ensembl.org/info/about/collaborations/barley.html). The candidate gene was selected if the significant SNP fell within the exon regions of that gene. Linkage disequilibrium was performed among markers located on the same chromosome using TASSEL 5.0 [30].
In this study, the population structure analysis for the set of 122 highly diverse barley genotypes from 34 countries and the 19 K set of molecular markers was performed. The analyses were conducted with ADMIXTURE v1.22 (Alexander et al., 2009). ADMIXTURE is a model-based clustering algorithm allowing the identification of the number of genetic clusters K. It assigns to each individual a membership probability to each one of the prespecified clusters under consideration. Here, successive values for the number of clusters K from 2 to 20 were considered. And for each K-value a ten-fold cross-validation was conducted with 30 replicates for each K-value thus a total of (19-values × 30 replicates) 570 unique combinations were considered. The most probably K-value is chosen based on the ADMIXTURE cross-validation trying to select the smallest number of clusters and the lowest cross-validation error. At each K-value, the CLUMPP software (Jakobsson and Rosenberg, 2007) was implemented to combine and align the results from the 30 replicates. And the membership proportions were averaged across runs using the permutation with the greatest similarity coefficient. At the end, the output from CLUMPP for the optimum K-value is considered to elaborate plots using the cluster visualization in R.
3. Results
3.1. Genetic variation in the NB resistance, PM resistance, and yield traits
The minimum, maximum, and average of all genotypes for all traits are presented in Table 1. The main average of all genotypes in PM and NB was approximately the same with a little bit higher in 2022 than in 2021. In addition, The average of all yield traits was higher in 2022 than in 2021.
Table 1.
Minimum, maximum, and average in each trait scored in the two growing seasons of 2021 and 2022.
| Season | Minimum | Maximum | Average |
|---|---|---|---|
| 2020–2021 | |||
| PM | 0 | 9 | 3.71 |
| NB | 0 | 8.66 | 2.21 |
| Ph | 60 | 110 | 90.03 |
| SW | 0.22 | 5.32 | 2.02 |
| SL | 4 | 15.67 | 8.92 |
| NOS | 7.33 | 19 | 12.46 |
| 2021–2022 | |||
| PM | 0 | 8.66 | 3.21 |
| NB | 0 | 9 | 2.24 |
| Ph | 61.33 | 111.67 | 91.48 |
| SW | 0.75 | 5.72 | 2.42 |
| SL | 5.33 | 16 | 9.65 |
| NOS | 8 | 19.33 | 13.23 |
PM, powdery mildew resistance; NB, bet blotch resistance; Ph, plant height; SW, seed weight; SL, spike length; NOS, number of spikelets
The analysis of variance for the net blotch resistance, powdery mildew resistance, and yield traits is presented in Table 2 and supplementary Table 2. High significant differences were found between the two years for all traits scored in this study. The differences among replications were significant in NB, PM, SL, and NOS. High significant genetic variation was found among genotypes for all traits. The G × Y was highly significant in Ph and SW.
Table 2.
F-values from analysis of variance (ANOVA) for all traits scored in the two growing seasons of 2021 and 2022.
| S.O.V | PM | NB | Ph | SW | SL | NOS |
|---|---|---|---|---|---|---|
| Years | 159.95 * * | 55.93 * * | 322.99 * * | 239.56 * * | 542.95 * * | 449.64 * * |
| Replications | 7.49 * * | 6.35 * * | 1.25 | 0.37 | 8.46 * * | 14.03 * * |
| Genotypes | 204.28 * * | 463.46 * * | 636.28 * * | 49.32 * * | 123.43 * * | 121.60 * * |
| G×Y | 0.80 | 0.48 | 2.52 * * | 1.88 * * | 0.37 | 0.21 |
* * refers tot he significant level at p > 0.01. PM, powdery mildew resistance; NB, bet blotch resistance; Ph, plant height; SW, seed weight; SL, spike length; NOS, number of spikelets
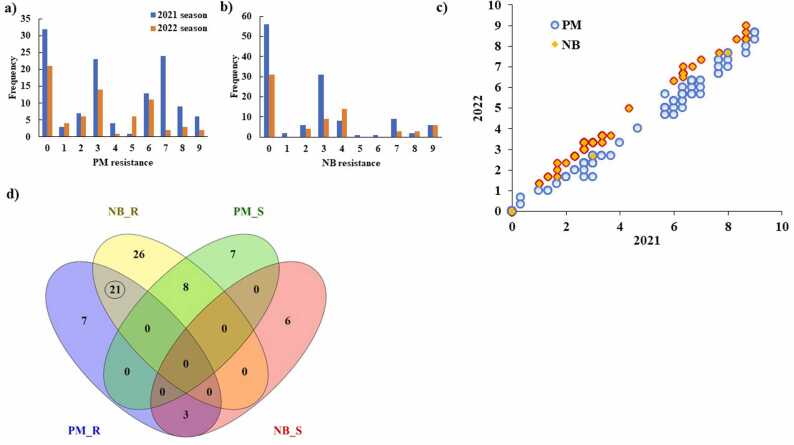
The distribution of all genotypes in PM and NB across the two years is presented in Fig. 2a and b. Most of the genotypes were immune to PM and NB and a wide range of resistance for each disease was observed. A highly significant positive correlation was found in PM between the two years with r = 0.99 * * (Fig. 2c). Likewise, the correlation in NB between the two years was highly and positively significant (r = 0.99 **). No significant correlation was found between NB and PM (r = 0.12). The phenotypic correlation among yield traits in each year and between the two years is presented in Supplementary Figure 3. No significant correlation was found among the four yield traits in each year. However, for each trait, highly significant correlations were found between the two years. The genetic correlation among all traits is presented in Supplementary Table 3. Significant genetic correlation was found between PM and NB.
Fig. 2.
Genotypes distribution for powdery mildew (PM) (a), genotype net blotch (NB) (b), phenotypic correlation in PM and NB between the two years (c), Venn diagram for the immune and susceptible genotypes to PM and NB (d). PM_R refers to the resistant genotypes in PM, NB_R refers to the resistant genotypes in NB, PM_S refers to PM_S refers to the susceptible genotypes in PM, and NB_S refers to PM_S refers to the susceptible genotypes in NB. In Fig. 2c: some circles (PM) and squares (NB) represent many genotypes having the same phenotypic score.
In each disease, all immune genotypes with 0 infections were selected (supplementary Table 4, Fig. 2d). Moreover, the susceptible genotypes with a visual score ranging from 7 to 9 in each disease were determined (Fig. 2d). A total of 28 and 47 genotypes were found to be immune to PM and NB resistance, respectively (Fig. 2d). A set of 21 genotypes were found to be immune to both diseases. Eight genotypes were resistant to NB and susceptible to PM, and vice versa for three genotypes. No common genotypes were susceptible in both diseases. Eight genotypes were immune to NB and susceptible to PM, while, three genotypes were immune to PM and susceptible to NB (Fig. 2b). The 21 immune genotypes were from 13 different countries. The highest number of immune genotypes (five) were from Egypt. The yield traits (Ph, SW, SL, and NOS) for the 21 genotypes are presented in Table 3, while, the yield traits for all immune genotypes to both diseases are presented in Supplementary Table 4. A wide range was found in all yield traits among the immune genotypes of PM and NB. The Ph ranged from 71 (HOR_14100) to 105.4 (HOR_4023) cm. The SW extended from 0.785 (HOR_2252) to 4.79 (HOR_9570) g. Spike length (SL) ranged from 6.33 ( HOR_16102) to 15.33 (HOR_1938) cm. The NOS extended from 9.835 (HOR_16102) to 19.165 (HOR_1938).
Table 3.
The LD (r2) structure between e SNP-marker pair located on the same chromosome.
| Chro. | r2 | No. sig. LD | Average Sig. LD | No. of non-sig. LD | Average non-sig. LD |
|---|---|---|---|---|---|
| 1 H | 0.025965 | 55,949 | 0.280547 | 2208,615 | 0.0195156 |
| 2 H | 0.026534 | 107,148 | 0.266738 | 5223,133 | 0.0216067 |
| 3 H | 0.034789 | 6,7277 | 0.2787828 | 1721,590 | 0.0252542 |
| 4 H | 0.025813 | 8,0271 | 0.26629 | 4059,505 | 0.0210577 |
| 5 H | 0.02621 | 87241 | 0.2837086 | 4822,135 | 0.0215513 |
| 6 H | 0.023678 | 16,7096 | 0.2762896 | 12,975,253 | 0.0204249 |
3.2. Genome-wide association mapping
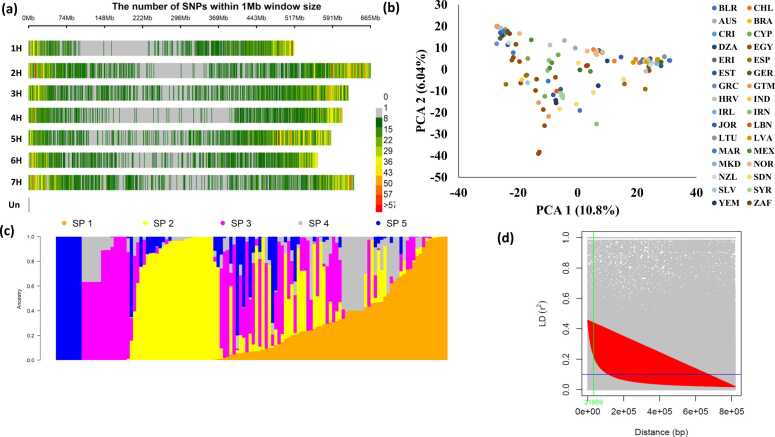
The PCA based on genetic distance was performed among all genotypes using 18, 525 SNP markers (Fig. 3a). A clear population structure can be observed from the analysis of the genotypes. Therefore, population structure correction was considered in the GWAS analysis. The SNP markers were distributed on all chromosomes. The 4 H chromosome had the lowest number of markers (1918), while the highest number of SNPs was found on the 2 H chromosome (Fig. 3b). The results of ADMIXTURE divided the population into five subpopulations (Fig. 3d, supplementary Figure 4). The number of subpopulations or clusters was determined to be 5 based on the cross-validation error and the computed delta value between successive number of subpopulations.
Fig. 3.
the distribution of SNP markers on barley chromosome (a) principle component analysis (PCA) based on the genetic distance among the genotypes (b), analysis of population structure (c), LD decay (d).
The structure of LD is presented in Table 3. The mean of r2 between SNP pairs located on the same chromosome ranged from 0.0223 (1 H) to 0.347 (3 H). Chromosome 3 H had the highest percentage (3.77%) of significance with an average of r2 of 0.278, while, chromone 6 H had the lowest percentage (1.27%) of the significant SNPs with an r2 average of 0.278. The r2 between each pair of markers was plotted against the physical distance (bp) in order to determine LD decay (Fig. 3c). The LD declined below r2 = 0.2 at about 31,959 pb.
3.2.1. GWAS for PM and NB resistance
Due to the non-significant Y × G interaction, the GWAS for PM and NB was performed using the average of the two years for each genotype. The analysis of GWAS is presented in Supplementary Table 5.
A total of 12 SNP markers were found to be significantly associated with decreased NB symptoms in barley. These SNPs were located on 2 H, 3 H, 5 H, and 7 H (Fig. 4). The phenotypic variation explained by markers (R2) ranged from 9.03 (chr2H:625802290:C:T) to 13.93% (SNP6H-30133310). Of the 12 SNP markers, eight had major effects with R2 > 10%. The target (resistant) allele effects extended from − 2.0412 (SNP5H-483460389) to − 4.091 (chr2H:625802290:C:T). Chromosome 3 H had 7 markers, while, only two significant SNP markers were found on 2 H. Significant SNP clusters were noted for NB resistance. The seven significant SNPs on the 3 H chromosome were found in two clusters representing two genomic regions. A cluster of two significant markers was observed on the 6 H chromosome (Fig. 4). The linkage disequilibrium (r2) was calculated among the significant markers located on the same chromosome. two high LD genomic regions distributed on 3 H (two), 5 H (one), and 7 H (two) were identified (Fig. 4). The analysis of gene annotation revealed that nine significant markers were found to be located within eight gene models that encode six proteins. Out of the 12 SNP markers, two (3 H and 6 H) were located within HORVU.MOREX.r3.3HG0295830 and HORVU.MOREX.r3.6HG0546030 gene models that encode leucine-rich repeat domain superfamily.
Fig. 4.
The position of significant SNPs associated with PM (green) and NB (red) on barley chromosomes and the LD among SNPs located on the same chromosome.
For PM resistance, the GWAS highlighted nine significant markers associated with decreased PM symptoms. The R2 ranged from 9.649 (chr5H:566275755:G:C & chr5H:566275753:T:C) to 11.97% (SNP7H-588256863). Of the nine significant markers, seven had major effects (R2 > 10%). The resistant allele effect extended from − 1.99 (G, SNP5H-19353755) to − 3.57 (T, SNP7H-589537963). The nine markers were located on 5 H (three), and 7 H (six) (Fig. 4). Three high LD genomic regions were found with one on chromosome 5 H and two on chromosome 7 H. Seven significant markers were located within five gene models that encode three proteins. three markers on 7 H; SNP7H-589537804, SNP7H-589537934, and SNP7H-589537963 were located within HORVU.MOREX.r3.7HG0734260 gene model that encodes P-loop containing nucleoside triphosphate hydrolase.
3.2.2. GWAS for yield traits scored under NP and PM diseases
The GWAS was performed using the mean average of each genotype for SL and NOS due to the non-significant Y x G interaction, while the GWAS was performed for each year for SW and Ph. The analysis of GWAS for yield traits is presented in Supplementary Table 6.
For NOS, the GWAS revealed five significant markers with R2 ranging from 6.38% to 8.72%. A total of 23 significant SNPs were found to be significantly associated with SL. The majority of these SNPs were located on the 3 H chromosome (12 SNPs). Out of the 23 SNPs, six had R2 > 10%. For SW, 23 and 21 significant SNP markers were detected in 2021 and 2022, respectively. Interestingly, 14 SNPs were significantly associated with SW in both years (supplementary Figure 4). Six markers (chr3H:531671175:T:A, chr3H:531873168:T:, chr3H:532237118:C:T, chr3H:532439487:C:T, chr3H:532439628:G:A, and chr6H:536036292:T:G) had major effects on increasing spike weight. One marker chr2H:24431549:C:T (2 H) was found to be associated with Ph in 2021 and 2022. This marker had R2 of 12.434% in both years.
3.3. Number of different resistance alleles and genetic diversity among the immune genotypes to PM and NB
The number of target alleles associated with the resistance to PM (supplementary Table 7) and NB (Supplementary Table 8) were examined in each immune genotype (Fig. 5a). In PM resistance, the number of resistant alleles ranged from 4 (two genotypes) to nine (nine genotypes). For NB resistance, the number of resistant alleles extended from six (one genotype) to 22 (12 genotypes). HOR_85 did not have any resistant allele. Given the 21 resistant genotypes to PM and NB (Fig. 5a), the number of resistant alleles to both diseases ranged from 14 in HOR_9570 to 21 in BCC_809 (Brazil), HOR_1556 (Cyprus), HOR_2252 (Egypt), and HOR_14100 (Syria).
Fig. 5.
Number of resistant allele to PM and NB in each immune genotype (a) and genetic distance among the immune genotypes calculated based on the resistant allele of NB and PM (b).
It is worth investigating the genetic distance among the immune genotypes to both diseases (21 genotypes) based on the resistance target allele (Fig. 5a&b, Supplementary Table 9a). HOR_9570 from Germany has separated away from all immune genotypes in a specific cluster. The genetic distance ranged from 0 to 0.69 between HOR_9570 (Germany) and HOR_899 (Chile). It was noted that six genotypes’ pairs were found to share the same resistant alleles. The HOR_9570 had high genetic distance based on resistant allele with genotype with a range extended from 0.48 to 0.69. The genetic distance based on all markers used for GWAS among the 21 genotypes is presented in supplementary Table 9b.
4. Discussion
4.1. Genetic variation in NB and PM resistance
The high genetic variation found among all genotypes in NB and PM allows barley breeders to select the most promising resistant genotypes to PM and NB pathotypes under Egyptian conditions. Such high genetic variation was expected as the tested genotypes represented 34 different countries. Using highly diverse germplasm is very useful to capture the possible genetic variation in the target traits [9], [35], [36], [37]. The G × Y interaction was non-significant indicating that the performance of genotypes for NB and PM was approximately the same in the two years. This result was supported by the high correlation found between the two years in each trait. No phenotypic significant correlation was found between NB and PM, but both traits had a low significant genotypic correlation. The negative association between the NB resistance and PM resistance was previously reported [38].
The 122 genotypes were evaluated under unheated greenhouse conditions in which humidity ranged from 80% to 90% at the Sakha Experimental Field Station, a disease-infested research station. It was reported that the optimum temperature and humidity for NB and PM are 20–25 ⁰C and 80–100%, respectively [1]. The temperature and humidity records from January to April across the two growing seasons are presented in supplementary Figures 1 and 2. Therefore, all genotypes in the two years were exposed to the natural infection with PM and NB races. Natural infection evaluation was used also in earlier studies to assess the variation in PM and NB resistance [39], [40], [41]. A set of nine and 15 genotypes were found to be very susceptible (IT score of 7–9) to NB and PM. The presence of highly susceptible genotypes indicated the success of natural infection for all genotypes. No common genotypes were susceptible to both diseases. HOR_3737, HOR_97, and BCC_1550 genotypes can be used as susceptible checks for PM, while HOR_8212 and BCC_1710 can be used as susceptible checks for NB in resistance evaluation for NB and PM Egyptian populations.
For PM resistance, a total of 32 (26%) genotypes were found to be immune to the Egyptian PM populations. These genotypes were from 18 countries with six genotypes from Egypt. Pogoda et al. [1] examined the genetic variation in the severity of powdery mildew infection at the seedlings stage of 267 barley accessions against the Blumeria graminis (DC.) E. O. Speer f. sp. hordei (Bgh) isolates (European pathotypes). Seven genotypes HOR_8658 and HOR_8212 (Egypt), BCC_1468 and BCC_1469 (Kazakhstan), HOR_2589 and HOR_3045 (Sudan), and BCC_1467 (Berlaus) were common between our study and the study of Pogoda et al. [1] (Supplementary Table 10). Interestingly, HOR_3045 (Sudan) had a score of 0.3% disease severity (Ø) at the seedling stage and an IT value of 0 at the adult growth stage in this study. Moreover, BCC_1469 (Kazakhstan) had a score of 20% disease severity (Ø) at the seedling stage and an IT value of 2.3 at the adult growth stage in this study. BCC_1468 (Kazakhstan) was susceptible to PM in the current study (IT=6) and also in the study of Pogoda et al. [1] (Ø= 60.2). Bearing in mind that the seven genotypes were exposed to the different origins of PM populations and at two different growth stages. Therefore, HOR_3045 (Sudan) could be a good resistant genotype to broad PM populations at different growth stages.
For net blotch resistance, 56 (46%) immune genotypes were found in the two years, while, nine genotypes were very susceptible to NB (IT =8–9) and can be used for susceptible checks to the Egyptian NB populations in future experiments.
A set of 21 accessions was found to be immune (no symptoms) for both NB and PM and had a high variation among yield traits. Therefore, this study provided very useful new immune genotypes to the Egyptian NB and PM populations individually or in combination. Such immune genotypes are very useful for future breeding programs not only to produce new barley cultivars having high resistance to a wide range of the Egyptian NB and PM pathotype(s) combined with high-yielding attributes. Selection to improve target traits should be combined with high-yielding attributes [42], [43], [44]. The resistance of 21 genotypes was deeply investigated in more detail in the genetic analyses to select the best candidate genotypes for future crossing in molecular breeding programs.
4.2. SNP markers and candidate genes for NB and PM resistance revealed by GWAS
The analysis of GBS results using a set of 18,525 SNP markers that were distributed on the seven barley chromosomes allowing the detection of candidate genes associated with PM and NB resistance. The PCA and kinship analyses were included in GWAS to correct the effect of population structure which could cause spurious associations [13], [14], [33], [45]. The result of q-q plot indicated that the best GWAS model for most of traits was GLM+PCA. High significant LDs were found among significant markers located on the same chromosome indicating that these markers tend to be co-inherited. Such high LD genomic regions that are associated with NB and PM resistance could be useful for marker-assisted selection and further genetic validation [8], [46]. From each high LD genomic region, one marker can be selected and converted to Kompetitive allele-specific PCR (KASP) marker for further validation in a different genetic background [47], [48], [49].
4.2.1. GWAS for NB resistance
12 significant markers were found to be associated with decreased NB symptoms in barley. These markers were located on chromosomes 2 H, 3 H, 5 H, and 6 H. Most of the identified SNPs were located on the 3 H chromosome a total of seven markers. Three net blotch resistance genes were mapped in cM on 3 H (Pt,a) and 6 H (rpt.k and rpt.r) chromosomes [50]. The GWAS results highlighted that 3 H could carry important genes against the Egyptian NB pathotype(s). Different LD degrees were observed among the 13 markers that clustered in two groups on the 3 H chromosome. Some of these 12 markers had complete LD and tend to be co-inherited, while the rest of the markers represented individual and independent QTLs (e.g. NB-4, NB-5, NB-9, NB10, NB-11 and NB-12).
A set of 22 SNP markers associated with NB resistance were detected in 234 diverse barley genotypes that were evaluated in Ethiopia and USA [51]. The SNPs were distributed on all barley chromosomes. Seven SNPs were located on 6 H chromosomes. The physical positions of the 22 SNPs were completely different from those reported in this study. Maurer et al. [52] reported 24 QTL for resistance against net blotch (German pathotypes) in a barley population HEB-25 comprising 1420 lines. The 24 QTL were distributed on all barley chromosomes. Many earlier studies reported significant SNPs associated with a net blotch in barley. Moreover, most of these studies shared the evaluation of the same NB pathotypes and successfully reported the same significant SNPs [16], [17], [18], [19], [20], [51], [53], [54], [55], [56], [57]. However, all SNPs detected in this study were in physical positions that were not previously reported, indicating that resistance to the Egyptian NB pathotypes is controlled by different genes. Unfortunately, no GWAS experiment was conducted to identify alleles and/or genes against the Egyptian NB pathotype(s). In the current study, a set of eight gene models were identified from GWAS that encodes eight different proteins. Notably, two different gene models HORVU.MOREX.r3.3HG0295830 (3 H) and HORVU.MOREX.r3.6HG0546030 (6 H) were found to encode the same protein leucine-rich repeat domain superfamily. Leucine-rich repeat protein is a resistance protein encoded by the nucleotide-binding leucine-rich repeat sequence (NBS-LRR) gene in plants which plays a vital role in plant defense against the invasion of many various pathogens. Specifically, the LRR protein was found to be involved in the barley defense mechanism to spot blot resistance [58]. Therefore, the two makers that were located within the two genes could be useful for marker-assisted selection after further validation in a different genetic background. Also, other proteins that play a vital role in disease resistance in crops were reported in this study such as BTB/POZ and MATH domain-containing protein 1–6 [60], protein egg apparatus-1 [61], Osmotin/thaumatin-like superfamily [62], metal-dependent hydrolase [63], and amino acid/polyamine transporter I [64]. The analysis of gene annotation confirmed the power of GWAS conducted in this study to identify alleles associated with NB resistance.
4.2.2. GWAS for PM resistance
The GWAS revealed a set of nine significant resistant SNP markers to the Egyptian PM pathotype(s) under natural field conditions. Unfortunately, very few studies reported significant markers for PM resistance in barley. The nine significant markers were located on chromosomes 5 H, and 7 H. Pogoda et al. [24] performed GWAS for PM resistance against the European pathotypes in a set of 267 barley accessions at the seeding stage and they reported 214 significantly associated SNPs. The positions of already known significant markers associated with NBwere compared with those detected in this study and no common significant markers were found. However, strong peaks of the significant markers were found on the 5 H chromosome [24]. In the current study, a cluster of five significant markers associated with PM was located on the same chromosome. A mapping population was evaluated against the Swiss powdery mildew field isolate CH4.8 by Hoseinzadeh et al. [22]. These authors mapped a dominant resistance locus (Mlhb.A42) on the 2 H chromosome. In our study, no significant markers associated with PM resistance were found on 2 H. Four QTLs on 4 H (one) and 6 H (three) were identified using GWAS in 169 highly diverse barley genotypes at natural field conditions [23]. The position of these SNPs was in cM which is different from the position of SNP markers reported in this study. Gupta et al. [21] reported associated markers with PM at the adult growth stage on chromosome 5 H and these markers are located between 619.7 and 627.3 Mbp on the reference sequence of Morex. In this regard, the marker cluster located on 5 H in this study was located between 514.8 and 566.2Mbp which was far from those that were previously published. This result indicated that our GWAS identified putative novel markers and genomic regions resistant to the Egyptian PM pathotype(s). Moreover, the promising novel seven significant markers with major effects were located on the 7 H chromosome. The gene annotation for the significant SNPs revealed seven gene models. Interestingly, two gene models HORVU.MOREX.r3.5HG0427780 (5 H) and HORVU.MOREX.r3.7HG0734260 (7 H) encode to P-loop containing nucleoside triphosphate hydrolase which was highly expressed under PM infection [66]. These results further supported the importance of these two chromosomes for carrying very valuable genes that control PM resistance.
Interestingly, no common significant SNP was found between PM and NB resistance. This indicates that the genetic control of the two disease resistances is different. The non-significant correlation between NB and PM resistance further supported the absence of the common markers controlling both diseases. Moreover, none of the significant markers for PM and NB detected in this study were previously reported in earlier studies. This could be due to the races that may differ by graphical region [67]. Also, race for the same region could differ by year due to the emergence of new races by mutations [67]. As each race is probably controlled by different genes, it is expected that the genes that are resistant to the Egyptian races of NB and PM could be different from those that were resistant to other races reported in earlier studies. Therefore, marker-assisted selection should be performed for each disease resistance individually. In wheat and barley, many makers were found to be associated with resistance to many plant diseases [8], [68], [69].
4.3. Genetic selection for the most promising and highly resistant genotypes to NB and PM resistance for the future breeding program
To genetically improve the resistance to NB and PM in barley, candidate genotypes should be selected to produce barley cultivars with high resistance to the NB and PM populations. Here, we utilized the features of the GWAS that identified resistant alleles and then we examined the number of resistant alleles in the immune genotypes for PM (supplementary Table 7) and NB (Supplementary Table 8). It was reported that phenotypic selection alone can be misleading due to human errors [43], [70], [71]. Selecting the promising genotypes based on the phenotype and number of target alleles will help to accurately produce cultivars with target traits (e.g. disease resistance) and make the results of crossing in breeding programs fruitful. In this study, we identified several resistant alleles in each immune genotype to NB (55 genotypes) and PM (32 genotypes). We focused on the 21 immune genotypes for both diseases to improve the resistance to PM and NB in parallel through molecular barley breeding programs.
The 21 immune genotypes to NB and PM resistance provide very useful genetic resources for breeding to create cultivars that have resistance to both diseases. The number of resistance alleles for NB and PM diseases varied among the immune genotypes. However, the resistance alleles could be similar when two parents are selected for crossing, hence improvement will not be fruitful and the F1 of this crossing will inherit the same resistance allele [42], [47], [72]. Therefore, the genetic distance among immune genotypes using only the resistance alleles was estimated to investigate the number of different resistance alleles between each two gentoypes. This will help to precisely select the highly divergent genotypes having different resistance alleles. For example, HOR_2252 (Egypt) and HOR_1566 (Cyprus) had the same resistance alleles for NB and PM with 21 resistance alleles in each genotype. Therefore, crossing between these two genotypes may be not the most useful crossing and the same 21 alleles will be passed to the F1. Bearing in mind that both genotypes are from different countries, therefore the diversity among these genotypes is expected and could be useful for the breeding of other target traits. Notably, the German genotype HOR_9570, which had the lowest number of resistant genes to NB and PM, had a high genetic distance with all genotypes with a range extended from 0.487 to 0.690 (BCC_899, Chile), indicating that this genotype had specific resistance alleles that did not exist in the other genotypes. The German genotype HOR_9570 (German) had eight resistance alleles for NB, while, BCC_899 (Chile) had 11 resistance alleles. The number of different alleles between these two genotypes was five resistance alleles. For PM, HOR_9570 had six resistance alleles, while BCC_899 had seven resistance alleles. The number of different alleles was five. So, crossing between these two genotypes will result in inheriting 9 resistance alleles to PM (same four alleles and five different alleles) and 12 resistance alleles to NB (same seven alleles and five different alleles). In total, the F1 will have the same 11 alleles that are common and 10 different alleles between the two parents (HOR_9570 and BCC_899). As the immune genotypes were resistant to the Egyptian NB and PM pathotype(s), it is worth investigating the genetic distance between the German genotype HOR_9570 and the immune Egyptian genotypes. High genetic distance was also found. Crossing between HOR_9570 with the Egyptian genotypes should be considered to produce cultivars with high adaptability to Egyptian conditions. Another important selection criterion that HOR_9570 had the highest SW, high NOS, high Ph, and high SL. Therefore, including this genotype will undoubtedly improve the yield attributes combined with high resistance to NB and PM.
The GWAS analysis is still the most powerful statistical method for identifying the resistant alleles to plant diseases [16], [18], [73], [74]. It results in identifying the number of target alleles in the highly resistant genotypes, leading to the precise selection of the candidate genotypes that can be used as parents in molecular breeding programs.
In conclusion, potential genes and SNP markers associated with the Egyptian PM and NB resistance were reported in this study for the first time. The germplasm used in this study was very useful for selection and breeding to improve NB and PM resistance. Among all immune genotypes for NB and PM, the German HOR_9570 genotype had a high genetic distance based on resistance alleles with all genotypes and it was also characterized as a high-yielding genotype. Important candidate parents for future crossing were highlighted in this study and it is highly recommended to include the HOR_9570 in future breeding programs to improve NB and PM resistance and yield traits. All markers detected in this study can be converted to KASP markers for validation in different genetic backgrounds before using them in MAS. Finally, the 21 immune genotypes are very important genetic resources not only for improving NB and PM resistance but also for increasing the circle of genetic diversity of barley genotypes grown in Egypt.
Authorship contribution statement
SME phenotyped all genotypes to NB and PM, and revised the manuscript; DJ performed the analysis of population structure and edited the manuscript. AB provided the plant material of this study and edited the manuscript; AS designed the study, performed all phenotypic and genetic analyses, and wrote the manuscript.
Funding
Costs for open access publishing were partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, grant 491250510).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported on this page.
Acknowledgment
The authors would like to express their grateful thanks to Prof. Dr. Peter Stephen Baenziger, University of Nebraska-Lincoln, for his useful discussion and comments on the results of this study. Also, the authors would like to thank the reviewers for their valuable comments, time, and efforts to improve the quality of this research article.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.10.014.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Backes A., Guerriero G., Ait Barka E., Jacquard C. Pyrenophora teres: taxonomy, morphology, interaction with barley, and mode of control. Front Plant Sci. 2021;12:509. doi: 10.3389/fpls.2021.614951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hafez Y., Abdelfatah A., El-Nashar F., Badr M., Elkady S. Management of barley net blotch using Trichoderma asperellum (T34), eugenol, non-traditional compounds and fungicides. Egypt J Biol Pest Control. 2019;29 [Google Scholar]
- 3.El-Nashar F. Cairo University; 1983. Net blotch disease of barley caused by Drechslera teres (Sacc.) Shoem. [Google Scholar]
- 4.Abdelrhim, A., Abd-Alla, H.M., Abdou, E.-S. & Ismail, M.E. Virulence of Egyptian Blumeria graminis f. sp. tritici Population and Response of Egyptian Wheat Cultivars. doi: 10.1094/PDIS-07-17-0975-RE. [DOI] [PubMed]
- 5.Dawson I.K., et al. Barley: a translational model for adaptation to climate change. N Phytol. 2015;206:913–931. doi: 10.1111/nph.13266. [DOI] [PubMed] [Google Scholar]
- 6.Mourad A.M.I., Hamdy R.M., Esmail S.M. Novel genomic regions on chromosome 5B controlling wheat powdery mildew seedling resistance under Egyptian conditions. Front Plant Sci. 2023;14:1160657. doi: 10.3389/fpls.2023.1160657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.New hope for powdery mildew resistant barley -- ScienceDaily. 〈https://www.sciencedaily.com/releases/2014/07/140725080154.htm〉.
- 8.Eltaher S., et al. Identification and validation of high LD hotspot genomic regions harboring stem rust resistant genes on 1B, 2A (Sr38), and 7B chromosomes in wheat. Front Genet. 2021;12:1875. doi: 10.3389/fgene.2021.749675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhavani S., et al. Globally important wheat diseases: status, challenges, breeding and genomic tools to enhance resistance durability. Genom Des Biot Stress Resist Cereal Crops. 2021:59–128. doi: 10.1007/978-3-030-75879-0_2. [DOI] [Google Scholar]
- 10.Mourad A.M.I., et al. Molecular marker dissection of stem rust resistance in Nebraska bread wheat germplasm. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-47986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czembor J.H., Hodowli I., Roslin A., Czembor J.H. Resistance to powdery mildew in barley (Hordeum vulgare L.) landraces from Egypt. Plant Genet Resour Newsl. 2000 〈https://www.researchgate.net/publication/264037257〉 [Google Scholar]
- 12.Sallam A., Mourad A.M.I., Hussain W. & Stephen Baenziger, P. Genetic variation in drought tolerance at seedling stage and grain yield in low rainfall environments in wheat (Triticum aestivum L.) Euphytica. 2018;214:1–18. [Google Scholar]
- 13.Mourad A.M.I., Belamkar V., Baenziger P.S. Molecular genetic analysis of spring wheat core collection using genetic diversity, population structure, and linkage disequilibrium. BMC Genom. 2020;21:434. doi: 10.1186/s12864-020-06835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eltaher S., et al. Genetic diversity and population structure of F3:6 Nebraska Winter wheat genotypes using genotyping-by-sequencing. Front Genet. 2018;9:76. doi: 10.3389/fgene.2018.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alqudah A.M., Sallam A., Stephen Baenziger P., Börner A. GWAS: fast-forwarding gene identification and characterization in temperate Cereals: lessons from Barley – A review. J Adv Res. 2020;22:119–135. doi: 10.1016/j.jare.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vatter T., et al. A nested association mapping population identifies multiple small effect QTL conferring resistance against net blotch (Pyrenophora teres f. teres) in wild barley. PLoS One. 2017;12 doi: 10.1371/journal.pone.0186803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manninen O.M., et al. Mapping of major spot-type and net-type net- blotch resistance genes in the Ethiopian barley line CI 9819. Genome. 2006;49:1564–1571. doi: 10.1139/g06-119. [DOI] [PubMed] [Google Scholar]
- 18.Richards J.K., Friesen T.L., Brueggeman R.S. Association mapping utilizing diverse barley lines reveals net form net blotch seedling resistance / susceptibility loci. Theor Appl Genet. 2017;130:915–927. doi: 10.1007/s00122-017-2860-1. [DOI] [PubMed] [Google Scholar]
- 19.Rozanova I.V., et al. SNPs associated with barley resistance to isolates of Pyrenophora teres f. teres. BMC Genom. 2019;20 doi: 10.1186/s12864-019-5623-3. 1DUMMMY. 1DUMMMY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amezrou R., et al. Genome-wide association studies of net form of net blotch resistance at seedling and adult plant stages in spring barley collection. Mol Breed. 2018;38:58. [Google Scholar]
- 21.Gupta S., et al. A locus on barley chromosome 5H affects adult plant resistance to powdery mildew. Mol Breed. 2018;38 doi: 10.1007/s11032-018-0858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoseinzadeh P., Ruge-Wehling B., Schweizer P., Stein N., Pidon H. High resolution mapping of a hordeum bulbosum-derived powdery mildew resistance locus in barley using distinct homologous introgression lines. Front Plant Sci. 2020;11:225. doi: 10.3389/fpls.2020.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bengtsson T., et al. A novel QTL for powdery mildew resistance in nordic spring barley (Hordeum vulgare L. ssp. vulgare) revealed by genome-wide association study. Front Plant Sci. 2017;8:1954. doi: 10.3389/fpls.2017.01954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pogoda M., et al. Identification of novel genetic factors underlying the host-pathogen interaction between barley (Hordeum vulgare L.) and powdery mildew (Blumeria graminis f. sp. hordei) PLoS One. 2020;15 doi: 10.1371/journal.pone.0235565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atef, A. & Fattah, A.- Studies on net blotch disease of barley in Egypt. (2015).
- 26.Kaji Manandhar, H. et al. A field guide for identification and scoring methods of diseases in the mountain crops of Nepal.
- 27.Saari E., Prescott J.M. A scale for appraising the foliar intensity of wheat diseases. Plant Dis Report. 1975 [Google Scholar]
- 28.Utz H.F. Institute of Plant Breeding, Seed Science and Population Genetics. University of Hohenheim; Stuttgart, Germany: 1997. A computer program for statistical analysis of plant breeding experiments. Version 2N. [Google Scholar]
- 29.Milner S.G., et al. Genebank genomics highlights the diversity of a global barley collection. Nat Genet. 2019;51:319–326. doi: 10.1038/s41588-018-0266-x. [DOI] [PubMed] [Google Scholar]
- 30.Bradbury P.J., et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 31.AE L., et al. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 33.Alqudah A.M., Sallam A., Stephen Baenziger P., Börner A. GWAS: fast-forwarding gene identification and characterization in temperate cereals: lessons from Barley – A review. J Adv Res. 2020;22:119–135. doi: 10.1016/j.jare.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang, X. & Huang, B. Natural variation and genome- wide Association studies in crop plants. Annual Review of Plant Biology. 2014;65(1):531–551. 65, (2014). [DOI] [PubMed]
- 35.Mourad A.M.I., Alomari D.Z., Alqudah A.M., Sallam A., Salem K.F.M. Recent advances in wheat ( spp.) breeding. Adv Plant Breed Strateg: Cereals. 2019;5:559–593. [Google Scholar]
- 36.Dawood M.F.A., Moursi Y.S., Amro A., Baenziger P.S., Sallam A. Investigation of heat-induced changes in the grain yield and grains metabolites, with molecular insights on the candidate genes in barley. Agronomy. 2020;10:1730. [Google Scholar]
- 37.Mondal S., et al. Advances in breeding for abiotic stress tolerance in wheat. Genom Des Abiotic Stress Resist Cereal Crops. 2021:71–103. doi: 10.1007/978-3-030-75875-2_2. [DOI] [Google Scholar]
- 38.Saghai Maroof M.A., Webster R.K., Allard R.W. Evolution of resistance to scald, powdery mildew, and net blotch in barley composite cross II populations. Theor Appl Genet. 1983;66:279–283. doi: 10.1007/BF00251159. [DOI] [PubMed] [Google Scholar]
- 39.Hovmøller, M.S., Caffier, V., Jalli, M. & Andersen, Ø.M. The European barley powdery mildew virulence survey and disease nursery Barley Genetic Resources: Advancing Conservation and Application for Breeding View project Picea abies View project. (1993) doi:. [DOI]
- 40.WHITE E.M. The effects of mixing barley cultivars on incidence of powdery mildew (Erysighe graminis) and on yield in Northern Ireland. Ann Appl Biol. 1982;101:539–545. [Google Scholar]
- 41.Hafez Y., Abdelfatah A., El-Nashar F., Badr M., Elkady S. Management of barley net blotch using Trichoderma asperellum (T34), eugenol, non-traditional compounds and fungicides. Egypt J Biol Pest Control. 2019;29:1–12. [Google Scholar]
- 42.Sallam A., et al. Marker–trait association for grain weight of spring barley in well-watered and drought environments. Mol Biol Rep. 2019 doi: 10.1007/s11033-019-04750-6. [DOI] [PubMed] [Google Scholar]
- 43.Ahmed A.A.M., Mohamed E.A., Hussein M.Y., Sallam A. Genomic regions associated with leaf wilting traits under drought stress in spring wheat at the seedling stage revealed by GWAS. Environ Exp Bot. 2021;184 [Google Scholar]
- 44.Sallam A., Dhanapal A.P., Liu S. Association mapping of winter hardiness and yield traits in faba bean (Vicia faba L.) Crop Pasture Sci. 2016;67:55–68. [Google Scholar]
- 45.Mourad A.M.I., et al. Genome-wide association study for identification and validation of novel SNP markers for Sr6 stem rust resistance gene in bread wheat. Front Plant Sci. 2018;9:1–12. doi: 10.3389/fpls.2018.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sallam A., Alqudah A.M., Baenziger P.S., Rasheed A. Editorial: genetic validation and its role in crop improvement. Front Genet. 2023;13:3705. doi: 10.3389/fgene.2022.1078246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abo-Elyousr K.A.M., et al. Identification of putative SNP markers associated with resistance to egyptian loose smut race(s) in spring barley. Genes. 2022;13:1075. doi: 10.3390/genes13061075. 2022, Vol. 13, Page 1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasheed A., et al. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor Appl Genet. 2016;129:1843–1860. doi: 10.1007/s00122-016-2743-x. [DOI] [PubMed] [Google Scholar]
- 49.Kaur B., Mavi G.S., Gill M.S., Saini D.K. Utilization of KASP technology for wheat improvement. Cereal Res Commun. 2020:1–13. [Google Scholar]
- 50.Abu Qamar M., et al. A region of barley chromosome 6H harbors multiple major genes associated with net type net blotch resistance. Theor Appl Genet. 2008;117:1261–1270. doi: 10.1007/s00122-008-0860-x. [DOI] [PubMed] [Google Scholar]
- 51.Daba S.D., Horsley R., Brueggeman R., Chao S., Mohammadi M. Genome-wide association studies and candidate gene identification for leaf scald and net blotch in barley (Hordeum vulgare L.) Plant Dis. 2019;103:880–889. doi: 10.1094/PDIS-07-18-1190-RE. [DOI] [PubMed] [Google Scholar]
- 52.Vatter T., et al. A nested association mapping population identifies multiple small effect QTL conferring resistance against net blotch (Pyrenophora teres f. teres) in wild barley. PLoS One. 2017;12 doi: 10.1371/journal.pone.0186803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma Z., Lapitan N.L.V., Steffenson B. QTL mapping of net blotch resistance genes in a doubled-haploid population of six-rowed barley. Euphytica. 2004;137:291–296. [Google Scholar]
- 54.Steffenson B.J., Hayes P.M., Kleinhofs A. Genetics of seedling and adult plant resistance to net blotch (Pyrenophora teres f. teres) and spot blotch (Cochliobolus sativus) in barley. Theor Appl Genet. 1996;92:552–558. doi: 10.1007/BF00224557. [DOI] [PubMed] [Google Scholar]
- 55.Wonneberger R., Ficke A., Lillemo M. Mapping of quantitative trait loci associated with resistance to net form net blotch (Pyrenophora teres f. teres) in a doubled haploid Norwegian barley population. PLoS One. 2017;12 doi: 10.1371/journal.pone.0175773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamang P., Neupane A., Mamidi S., Friesen T., Brueggeman R. Association mapping of seedling resistance to spot form net blotch in a worldwide collection of barley. Phytopathology. 2015;105:500–508. doi: 10.1094/PHYTO-04-14-0106-R. [DOI] [PubMed] [Google Scholar]
- 57.Grewal T.S., Rossnagel B.G., Scoles G.J. Mapping quantitative trait loci associated with spot blotch and net blotch resistance in a doubled-haploid barley population. Mol Breed. 2012;30:267–279. [Google Scholar]
- 58.Ameen G., et al. rcs5-mediated spot blotch resistance in barley is conferred by wall-associated kinases that resist pathogen manipulation. bioRxiv. 2020 doi: 10.1101/2020.04.13.040238. 2020.04.13.040238. [DOI] [Google Scholar]
- 60.Madala N.E., Leone M.R., Molinaro A., Dubery I.A. Deciphering the structural and biological properties of the lipid A moiety of lipopolysaccharides from Burkholderia cepacia strain ASP B 2D, in Arabidopsis thaliana. Glycobiology. 2011;21:184–194. doi: 10.1093/glycob/cwq146. [DOI] [PubMed] [Google Scholar]
- 61.Gardiner S.A., et al. Transcriptome analysis of the barley-deoxynivalenol interaction: Evidence for a role of glutathione in deoxynivalenol detoxification. Mol Plant-Microbe Interact. 2010;23:962–976. doi: 10.1094/MPMI-23-7-0962. [DOI] [PubMed] [Google Scholar]
- 62.Hohnjec N., Vieweg M.F., Pühler A., Becker A., Küster H. Overlaps in the transcriptional profiles of medicago truncatula roots inoculated with two different glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol. 2005;137:1283–1301. doi: 10.1104/pp.104.056572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aragunde H., Biarnés X., Planas A. Substrate recognition and specificity of chitin deacetylases and related family 4 carbohydrate esterases. Int J Mol Sci. 2018;19:412. doi: 10.3390/ijms19020412. 2018, Vol. 19, Page 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu G., et al. Amino acid homeostasis modulates salicylic acid–associated redox status and defense responses in arabidopsis. Plant Cell. 2010;22:3845–3863. doi: 10.1105/tpc.110.079392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu X., et al. Identification of MicroRNAs and their targets that respond to powdery mildew infection in cucumber by small RNA and degradome sequencing. Front Genet. 2020;11 doi: 10.3389/fgene.2020.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shahin A., et al. Geographical correlation and genetic diversity of newly emerged races within the Ug99 lineage of stem rust pathogen, puccinia graminis f. sp. tritici, in different wheat-producing areas. J Fungi. 2022;8:1041. doi: 10.3390/jof8101041. 2022, Vol. 8, Page 1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mourad A.M.I., Draz I.S., Omar G.E., Börner A., Esmail S.M. Genome-wide screening of broad-spectrum resistance to leaf rust (Puccinia triticina Eriks) in spring wheat (Triticum aestivum L.) Front Plant Sci. 2022;13 doi: 10.3389/fpls.2022.921230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abou-Zeid M.A., Mourad A.M.I. Genomic regions associated with stripe rust resistance against the Egyptian race revealed by genome-wide association study. BMC Plant Biol. 2021;21:1–14. doi: 10.1186/s12870-020-02813-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sallam A., Martsch R. Association mapping for frost tolerance using multi-parent advanced generation inter-cross (MAGIC) population in faba bean (Vicia faba L.) Genetica. 2015;143:501–514. doi: 10.1007/s10709-015-9848-z. [DOI] [PubMed] [Google Scholar]
- 71.Mourad A.M.I., et al. Molecular marker dissection of stem rust resistance in Nebraska bread wheat germplasm. Sci Rep. 2019;9 doi: 10.1038/s41598-019-47986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eltaher S., et al. GWAS revealed effect of genotype × environment interactions for grain yield of Nebraska winter wheat. BMC Genom. 2021;22:1–14. doi: 10.1186/s12864-020-07308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Esmail S.M., Omar G.E., Mourad A.M.I. In-Depth Understanding of the Genetic Control of Stripe Rust Resistance (Puccinia striiformis f. sp. tritici) Induced in Wheat (Triticum aestivum) by Trichoderma asperellum T34. 2023;107:457–472. doi: 10.1094/PDIS-07-22-1593-RE. [DOI] [PubMed] [Google Scholar]
- 74.Mourad, A.M.I., Hamdy, R.M. & Esmail, S.M. Novel genomic regions on chromosome 5B controlling wheat powdery mildew seedling resistance under Egyptian conditions. Front Plant Sci14, 1293. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material