Abstract
Background
Epicardial adipose tissue (EAT) is a key aspect in the investigation of cardiac pathophysiology. We sought to develop a deep learning (DL) model for fully automatic extraction and quantification of EAT through pulmonary computed tomography venography (PCTV) images.
Methods
In this retrospective study, we included 128 patients with atrial fibrillation and PCTV from 2 hospitals. A DL model for automated EAT segmentation was developed from a training set of 51 patients and a validation set of 13 patients from hospital A. The algorithm was further validated using an internal test set of 16 patients from hospital A and an external test set of 48 patients from hospital B. The consistency and measurement agreement of EAT quantification were compared between the DL model and the conventional manual protocol using the Dice score coefficient (DSC), Hausdorff distance (HD95), Pearson correlation coefficient, and Bland-Altman plot.
Results
In the internal and external test set, automated segmentation with DL was successful in all cases. The total analysis time was shorter for DL than for manual reconstruction (5.43±2.52 vs. 106.20±15.90 min; P<0.001). The EAT segmented with the DL model had good consistency with manual segmentation (the DSC of the internal and external test sets were 0.92±0.02 and 0.88±0.03, respectively). The quantification of EAT evaluated with the 2 methods showed excellent correlation (all correlation coefficients >0.9; all P values <0.001) and minimal measurement difference.
Conclusions
The proposed DL model achieved fully automatic quantification of EAT from PCTV images. The yielded results were highly consistent with those of manual quantification.
Keywords: Epicardial adipose tissue (EAT), deep learning (DL), pulmonary computed tomography venography (PCTV)
Introduction
Epicardial adipose tissue (EAT) stores fat between the myocardium and the epicardium and is derived from the splanchnopleuric mesoderm (1). EAT affects local myocardial electrophysiology via direct infiltration and intermyocyte disruption, tissue fibrosis, and gap junction remodeling (2). In addition, the thickness, volume, radiodensity, and radiomic features of EAT are associated with atrial fibrillation (AF) and other adverse cardiovascular events such as atherosclerosis (3-6). Therefore, accurate EAT quantification is fundamental for cardiac adipose tissue-related research, in which EAT usually serves as an imaging biomarker.
Coronary artery calcium score (CACS) computed tomography (CT) imaging is the most widely applied data source for EAT quantification (7,8), owing to its high accessibility and ease of generation. However, CACS cannot depict the full spectrum of EAT characteristics, especially for EAT vascularization, which can be additionally provided using a contrast-enhanced scan. Nevertheless, using contrast-enhanced CT for manual EAT quantification is labor-intensive because of the need for slice-by-slice editing of the border between the EAT and pericardium.
Recently, deep neural networks have been widely used in the field of medical image segmentation (9). Their application to the fully automatic segmentation of EAT has enabled EAT quantification for large-cohort studies (3,10-12). Currently, most deep learning (DL) algorithms with automatic EAT segmentation set the pericardium as the detection target and use threshold segmentation to separate EAT from the pericardium. Commandeur et al. used 2 multitask convolutional neural networks (CNNs) to segment EAT. The first CNN detected the heart’s boundaries and performed segmentation, and the second CNN used a statistical shape model to detect the pericardium; the median Dice score coefficient (DSC) was 0.823 (13). Hoori et al. applied a novel look ahead slab-of-slices with bisection, in which they split the heart into halves, retaining the increasing curvature of the sac and presenting similar images to the training network. The average DSC of this method was 0.89 (14).
Pulmonary CT venography (PCTV) is commonly used before radiofrequency ablation in patients with AF to define the left atrium (LA) and pulmonary vein anatomy. PCTV is ideal for studying the EAT and prognosis in patients with AF. However, no prior studies have used PCTV for EAT quantification. To fully explore the potential risk factors for AF (15-17), it is necessary to develop a model for the fully automatic segmentation of EAT based on PCTV scans.
U-net is one of the most successful network architectures for DL in medical image segmentation (18). One of its derivatives, no new U-net (nnU-Net), demonstrates remarkable performance across multiple tasks and has been adopted by researchers for diverse segmentation challenges (19). Its outstanding efficacy and reliability were demonstrated by its exceptional performance in the Medical Segmentation Decathlon Challenge (20). We hypothesized that nnU-Net is a suitable DL algorithm for automatic EAT quantification. Thus, this study aimed to develop and validate an nnU-Net-based DL model for automatic segmentation and EAT quantification based on PCTV data.
Methods
Study population
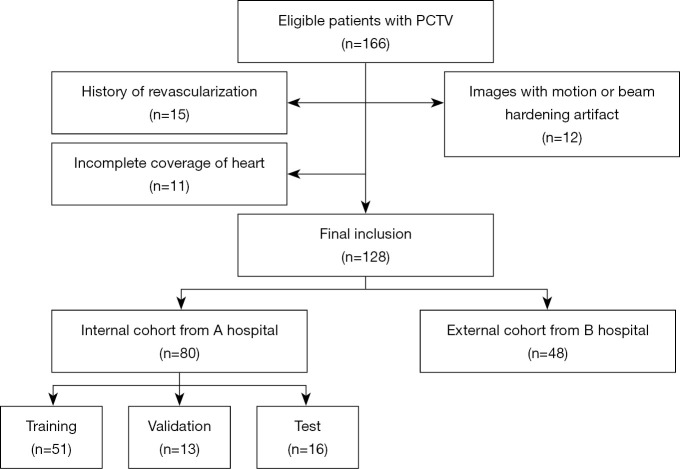
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional review boards of Shanghai General Hospital and Dongtai People’s Hospital approved this retrospective study, and the requirement for written informed consent from patients was waived due to the retrospective nature of the study. For model development, PCTV images of patients with AF who were candidates for ablation therapy between June 2018 and August 2018, were retrospectively included from 1 tertiary hospital. We randomly selected 20% of cases as the test cohort. The remaining data were divided into training and validation cohorts in a 4:1 ratio. The external validation cohort comprised 48 randomly selected cases undergoing PCTV from another tertiary hospital. We excluded (I) patients with significant motion artifacts due to failed breath holding, (II) patients with histories of prior percutaneous coronary interventions, (III) patients with histories of prior chest surgeries, and (IV) patients in whom the scanning range did not cover the whole heart. Figure 1 shows the patient inclusion and exclusion criteria flowchart.
Figure 1.
Inclusion and exclusion criteria for the internal and external cohort. PCTV, pulmonary computed tomography venography.
Pulmonary CT venography scan protocol
For the internal cohort, all patients were subjected to scanning with a 256-slice wide detector CT scanner (Revolution HD, GE HealthCare, Chicago, IL, USA). PCTV images were acquired using a nongated helical mode, with the region of interest placed in the LA. A 50- to 60-mL contrast media bolus [Iomeron (iomeprol), 400 mg iodine/m; Bracco Medical Technologies, Milan, Italy] was injected into the antecubital vein at a rate of 4–5 mL/s, which was followed by flushing with 40 mL of saline. The detailed scan parameters were as follows: collimation =256×0.625 mm, reconstructed slice thickness =0.625 mm, reconstructed slice interval =0.5 mm, rotation time =280 ms, and matrix =512×512 pixels. In addition, we applied automated tube voltage and current modulation (KV Assist, Smart mA, GE HealthCare).
For the external test cohort, PCTV data were acquired from a second-generation dual-source CT scanner (SOMATOM Definition Flash, Siemens Healthineers, Erlangen, Germany) through a nongated helical mode. We employed contrast injection and triggering techniques similar to those used with the internal training cohort. The detailed scan parameters were as follows: collimation =64×0.6 mm, reconstructed slice thickness =0.75 mm, reconstructed slice interval =0.5 mm, and rotation time =280 ms. Moreover, we applied automated tube voltage and current modulation (CAREKv, CAREDose 4D, Siemens Healthineers).
Development of a DL model for EAT quantification
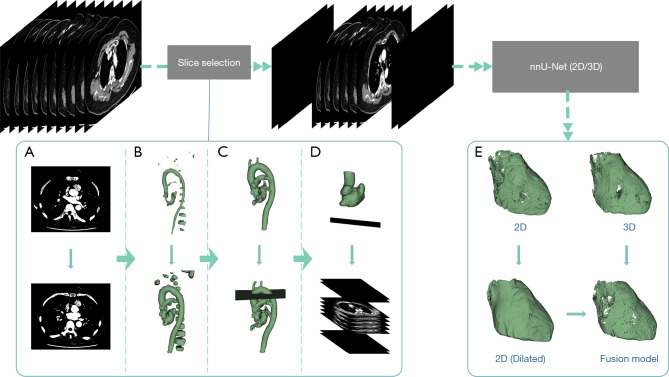
Our proposed fully automatic processing flow based on the nnU-Net framework is shown in Figure 2. This process was implemented according to the 3 steps described below.
Figure 2.
Automatic EAT segmentation workflow. (A-D) Steps for the preprocessing pipeline: (A) binarize and fill the images, (B) perform the opening operation, (C) keep the largest connected component and delete the image slice by slice, and (D) keep the largest connected component and determine the lower bound. (E) The postprocessing procedure. nnU-Net, no new U-net; EAT, epicardial adipose tissue.
In the first step, to avoid missegmentation of the neck and abdominal fat with nnU-Net, the input volume was preprocessed using morphological processing to detect the upper and lower ranges of the slices, and the slices out of this range were set to 0. All morphological processing was performed using the Python toolkit SimpleITK (https://simpleitk.org/). Details of morphological processing are included in the online appendix.
For the second step, we used 2-dimensional (2D) and 3-dimensional (3D) nnU-Net to train the EAT segmentation models based on the preprocessed training set described above. nnU-Net is a DL-based segmentation method that automatically configures itself for any new task, including preprocessing, network architecture, training, and postprocessing (19). The source code for nnU-Net is publicly available on GitHub (https://github.com/MIC-DKFZ/nnunet). The preprocessed internal validation and test sets were sent to the 2 networks for obtaining their segmentation results during inference. We did not use the postprocessing method of selecting the largest connected domain in nnU-Net, as this processing is unsuitable for EAT segmentation. Instead, we trained the model using Python 3.9 and PyTorch 1.12.1 on a PC configured with an Intel Core i9-9900K CPU and a GeForce RTX 3090Ti GPU. The epoch was set at 1000.
Upon analyzing the segmentation results of the validation set, we observed distinct strengths and weaknesses in both the 2D nnU-Net and 3D nnU-Net models. The 2D nnU-Net accurately determined the spatial position of the heart but misidentified the fat around the vessels as EAT. Conversely, the 3D nnU-Net correctly segmented EAT along the pericardium contour but misidentified some fat located far from the heart.
Finally, we decided to merge the segmentation outcomes of the 2D and 3D models through the intersection of the 3D results with the expanded 2D results. The fused result was then postprocessed via thresholding; thus, voxels in the fat Hounsfield range (−190/−30 HU) were selected. We calculated the volume and radiodensity of the EAT with SimpleITK 2.2.0 using Python 3.9.12. The automatic segmentation model of EAT is available online (http://www.huyifan.top:8000).
Manual segmentation and quantification of EAT
We used the 3D slicer 4.11.20210226 (21) for EAT segmentation and calculated the volume and radiodensity in Python, as described above. The pulmonary trunk bifurcation was considered the superior limit of the pericardium. We further manually corrected the pericardial boundary slice by slice. Once the manual editing was finished, EAT was identified according to the preset Hounsfield thresholds (−190/−30 HU). The above analysis was performed by a radiologist with 7 years of relevant experience.
We obtained the EAT volume by multiplying the volume per unit in a voxel-by-voxel count (n), which was defined as follows:
| [1] |
Then, we accumulated the radiodensity-per-unit voxel (Densityi) in EAT and calculated the average (DensityEAT), which was defined as follows:
| [2] |
The masks segmented by the automatic and manual segmentation models are binary images, where 0 indicates background and 1 indicates EAT. The number of voxels for EAT is obtained by counting the number of occurrences of 1 in the binary image. In this study, the slice and voxel spacings were obtained from Digital Imaging and Communications in Medicine (DICOM) images, and the volume per voxel was obtained by multiplying the 3D voxel spacings. The radiodensity of each EAT voxel was obtained from the corresponding DICOM voxel radiodensity.
Statistical analysis
We used SegMetrics 1.0.36 and Scipy 1.7.3 in Python 3.9.12 for all statistical analyses. Continuous variables are presented as the mean ± standard deviation (SD) if normally distributed and otherwise as the median with interquartile range. We evaluated the performance of automatic segmentation models through DSC and Hausdorff distance (HD95). DSC is a measure of the similarity of 2 samples and is defined as follows:
| [3] |
HD(dH(X,Y)) measures how far 2 subsets of a metric space are from each other and is defined as follows:
| [4] |
where d(x,Y) finds the shortest distance for each point x to set Y, and obtains the largest value from the calculated shortest distances of all points; d(y,X) and act similarly but in a different set order. HD95 is based on the calculation of the 95th percentile of the distances between boundary points in X and Y.
The correlation of the EAT characteristics between automatic and manual quantification was evaluated with Pearson correlation coefficient. The Bland-Altman plot was used to illustrate the consistency and difference in EAT quantification results between the automatic and manual approaches. Statistical significance was defined as a 2-sided P value less than 0.05.
Results
Clinical characteristics
A total of 166 patients with AF who were referred for PCTV were initially screened. Of these patients, 15 and 11 patients were excluded due to their clinical history of revascularization or due to incomplete imaging coverage of the whole pericardium, respectively. Another 12 patients with severe motion or metal artifacts were also excluded. Finally, 128 patients {68 males; median age 65 [interquartile range (IQR), 59–73] years} were included in the analysis. Their detailed demographic characteristics are shown in Table 1.
Table 1. Clinical characteristics of the internal and external cohorts.
| Clinical characteristics | Internal cohort | External cohort | |||
|---|---|---|---|---|---|
| Training (n=51) | Validation (n=13) | Test (n=16) | Test (n=48) | ||
| Age (years)* | 64±11 | 61±10 | 66±11 | 66±12 | |
| Sex (male), n (%) | 29 (56.9) | 6 (46.2) | 9 (56.3) | 24 (50.0) | |
| AF type (PAF), n (%) | 30 (58.8) | 11 (84.6) | 9 (56.3) | 28 (58.3) | |
| (PeAF), n (%) | 21 (41.2) | 2 (15.4) | 7 (43.8) | 20 (41.7) | |
| Diabetes, n (%) | 12 (23.5) | 2 (15.4) | 3 (18.8) | 8 (16.7) | |
| Hyperlipidemia, n (%) | 22 (43.1) | 3 (23.1) | 8 (50.0) | 26 (54.2) | |
| Hypertension, n (%) | 32 (62.7) | 6 (46.2) | 9 (56.3) | 32 (66.7) | |
| CAD, n (%) | 13 (25.5) | 2 (15.4) | 1 (6.3) | 7 (14.6) | |
*, values are presented as the mean ± SD. AF, atrial fibrillation; PAF, paroxysmal atrial fibrillation; PeAF, persistent atrial fibrillation; CAD, coronary artery disease; SD, standard deviation.
Efficacy of DL-based automatic EAT quantification
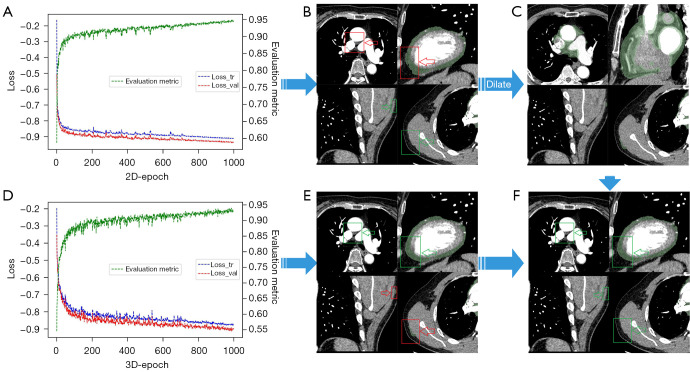
Before preprocessing, there were 15 cases of missegmentation outside of the heart. After morphological processing, the heart slices were correctly identified in all 128 patients. The fused results are presented in Figure 3. The performance of the fusion model for each dataset is shown in Table 2.
Figure 3.
The fused results of the 2D and 3D models. (A) The 2D and (D) 3D model training loss curves. The 3 curves in the figure are the training (blue), validation loss (red), and results of the evaluation (green). The segmentation results of the (B) 2D and (E) 3D models. (C) The process of dilating the 2D model segmentation results. (F) The results of the fusion model. The red box represents the missegmentation, and the green box represents the correct segmentation.
Table 2. Automated segmentation performance of EAT with the DL model.
| Segmentation index | Internal set | External set | |||
|---|---|---|---|---|---|
| Training | Validation | Test | Test | ||
| DSC | 0.94±0.02 | 0.93±0.03 | 0.92±0.02 | 0.88±0.03 | |
| HD95 (mm) | 0.66±0.13 | 0.93±0.31 | 0.88±0.28 | 0.99±0.52 | |
Data are presented as the mean ± SD. EAT, epicardial adipose tissue; DL, deep learning; DSC, Dice score coefficient; HD95, Hausdorff distance 95%; SD, standard deviation.
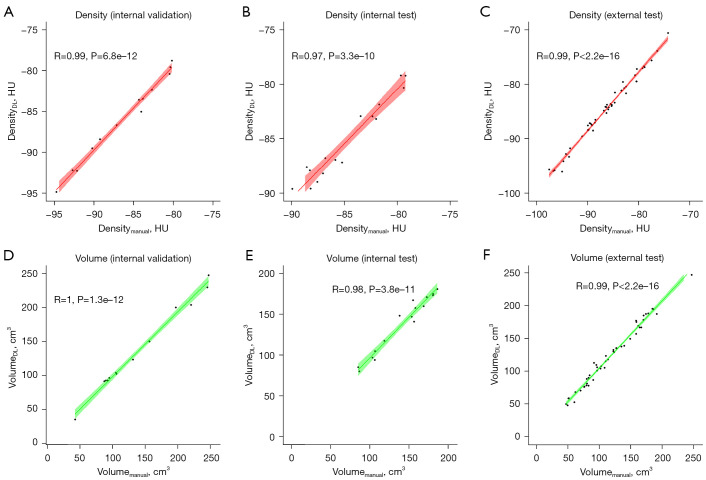
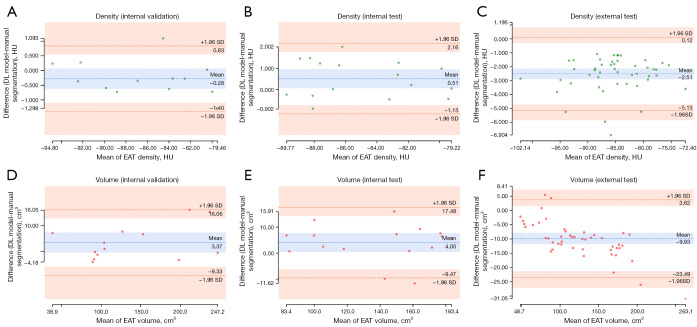
Table 3 shows the volume and radiodensity of the manually and automatically segmented EATs for each dataset. Compared with manual segmentation, Pearson correlation coefficient for the EAT volume and radiodensity reached 0.95 (P<0.01), indicating an excellent correlation between the 2 approaches (Figure 4). The Bland-Altman plots indicate that the 2 have good consistency (Figure 5).
Table 3. Quantification results of EAT with 2 methods.
| EAT parameters | DL model | Manual | Correlation coefficient | 95% CI | P value |
|---|---|---|---|---|---|
| EAT density (HU) | |||||
| Internal validation | −86.82±5.36 | −86.26±5.04 | 0.99 | 0.98–1.00 | <0.001 |
| Internal test | −85.18±3.70 | −84.68±3.59 | 0.97 | 0.92–0.99 | <0.001 |
| External test | −85.24±6.63 | −86.81±6.09 | 0.99 | 0.99–1.00 | <0.001 |
| EAT volume (cm3) | |||||
| Internal validation | 136.07±64.44 | 139.44±67.03 | 1 | 0.98–1.00 | <0.001 |
| Internal test | 137.15±35.26 | 141.15±34.78 | 0.98 | 0.94–0.99 | <0.001 |
| External test | 123.13±47.57 | 118.53±45.92 | 0.99 | 0.98–1.00 | <0.001 |
Data are presented as the mean ± SD. EAT, epicardial adipose tissue; DL, deep learning; CI, confidence interval; HU, Hounsfield unit; SD, standard deviation.
Figure 4.
Pearson correlation coefficients between the manual and deep learning models for EAT volumetric and radiodensity measurements. (A-C) Scatterplots representing the radiation density. (D-F) Scatterplots representing the volume. HU, Hounsfield unit; DL, deep learning; EAT, epicardial adipose tissue.
Figure 5.
Bland-Altman plots for the measurement difference of EAT between the manual and DL model segmentation. (A) The variability in radiodensity measurement of EAT between the manual and DL model segmentations in the internal validation datasets. (B) The variability in the radiodensity measurement of EAT between the manual and DL model segmentations in the internal test datasets. (C) The variability in the radiodensity measurement of EAT between the manual and DL model segmentations in the external test datasets. (D) The variability in the volume measurement of EAT between the manual and DL model segmentations in the internal validation datasets. (E) The variability in the volume measurement of EAT between the manual and DL model segmentations in the internal test datasets. (F) The variability in the volume measurement of EAT between the manual and DL model segmentations in the external test datasets. DL, deep learning; HU, Hounsfield unit; SD, standard deviation; EAT, epicardial adipose tissue.
Efficiency of DL-based automatic EAT quantification
In the internal test cohort, the average EAT segmentation and quantification processing time was 5.43±2.52 and 106.20±15.90 min for the DL and manual approaches, respectively. Similarly, with the external validation set considered, the average time for the DL model was 4.26±1.21 min, whereas the average time for manual segmentation was 100.80±12.20 min.
Discussion
We developed and evaluated a DL model based on nnU-Net for fully automatic EAT extraction from PCTV scans. Our model accurately and efficiently determined the volume and radiodensity of EAT in clinical studies. Moreover, we found that the predicted and manual segmentation methods were highly consistent.
EAT is a unique fat deposition located between the myocardium and the visceral layer of the epicardium. EAT can be highly protective for the adjacent myocardium through its dynamic brown fat-like thermogenic function but also deeply harmful due to the paracrine or vasocrine secretion of pro-inflammatory and profibrotic cytokines (22). Therefore, EAT has been considered to play a pivotal role in the pathogenesis of AF, heart failure, and coronary artery diseases (23). Quantitative data obtained from body composition imaging analysis has been shown to be associated with the development, risk, and clinical outcomes of diverse diseases, encompassing cardiovascular and oncological conditions (24). Recent evidence supports the crucial involvement of EAT accumulation in the pathogenesis of AF and coronary artery disease (15,25). The emergence of automatic segmentation models based on CT angiography and CACS scans has significantly advanced research on the relationship between EAT and cardiovascular adverse events, such as myocardial infarction (10,11,26,27). However, the investigation into the association between EAT and AF still lacks validation from large-scale cohorts (16,28,29). This discrepancy can be attributed to the limitations of existing EAT segmentation models when applied to scans from PCTV. The comprehensive coverage of the chest in PCTV scans poses challenges, as existing segmentation models may missegment the neck root and midabdominal structures. To address this issue, our model offers a fully automated approach to accurately segmenting EAT from PCTV scans, eliminating the instability associated with manual segmentation. This innovative method provides an efficient tool for conducting large-scale multicenter studies and enhances the reliability of research investigating the relationship between EAT and AF.
Setting the pericardium as the detection target requires the detection of an intact pericardium; however, the pericardium is difficult to identify on slices from the bottom of the heart. The pericardium is disconnected by the vasculum on the top slices of the heart. Consequently, we established EAT as the direct learning target. For network structure, we selected nnU-Net due to its ability to self-configure (19). This method avoids tedious tuning work and allows for a greater focus on the segmentation results. In this study, we combined the segmentation results of the 2 networks by observing the segmentation in the validation set and mutually complementing each network’s advantages, making our segmentation results more suitable for clinical practice.
The postprocessing of the original nnU-Net network includes selecting the largest connected component. Given that EAT is scattered over a wide physical area, it is unsuitable for such postprocessing. We found that the missegmentation differed between the 2D and 3D nnU-Nets. The 2D model could pinpoint the location of the heart, likely because training the 2D slices requires less memory. This allows the model to identify the spatial location of the heart in each slice. Conversely, the 3D model could not determine the position of the heart relative to the whole. This led to the fat under the diaphragm being misidentified as EAT. When the contour of the pericardium is difficult to identify, the 2D model likely exhibits missegmentations on some slices, which can lead to a slight decrease in DSC; however, in the multiplanar reconstruction (MPR), the segmentation boundary is highly uneven. The 3D model connects the epicardium at the upper and lower slices and also fits (almost exactly) the boundary of the pericardium. Therefore, we use the segmentation of the 3D model as the cornerstone when fusing the segmentation of the 2 models.
Our model allows for the quantitative analysis of EAT in large-scale cohorts to study and validate its association with AF. For example, our model can determine the relationship between EAT volume and recurrence after radiofrequency ablation of AF and can stably segment EAT from PCTV scans. The extracted radiomic features have strong consistency, and the resultant markers will produce more accurate measures than those from studies based on manual segmentation.
Despite the promising results described above, our study has some limitations. First, we used a nonelectrocardiography-gated acquisition protocol for PCTV. This inevitably resulted in mild motion artifacts in a small number of slices, which might have affected EAT quantification. In addition, our model employs morphological processing, which increases processing time and requires the scan range to encompass most of the aortic arch; otherwise, some slices containing the heart would be missed. Therefore, the current model cannot process source data directly acquired from different scanning protocols. Further investigations with data from various CT scanners and acquisition protocols are needed to improve the generalizability of our proposed model.
Conclusions
Our DL model uses PCTV data to achieve faster EAT segmentation than do manual methods. In addition, the EAT volume and density, quantified using the DL model, were consistent and correlated well with the manual quantification results.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by Shanghai Jiao Tong University “Star Project” of Biomedical Multi-discipline Research Program (No. YG2022ZD015 to J Zhang).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional review boards of Shanghai General Hospital and Dongtai People’s Hospital approved this study, and the requirement for written informed consent from patients was waived due to the retrospective study design.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-233/coif). JZ reports a grant from the Shanghai Jiao Tong University “Star Project” of Biomedical Multi-discipline Research Program (No. YG2022ZD015). SJ, SH and XZ are employees of Philips Healthcare. The other authors have no conflicts of interest to declare.
References
- 1.Sacks HS, Fain JN. Human epicardial adipose tissue: a review. Am Heart J 2007;153:907-17. 10.1016/j.ahj.2007.03.019 [DOI] [PubMed] [Google Scholar]
- 2.Nalliah CJ, Bell JR, Raaijmakers AJA, et al. Epicardial Adipose Tissue Accumulation Confers Atrial Conduction Abnormality. J Am Coll Cardiol 2020;76:1197-211. 10.1016/j.jacc.2020.07.017 [DOI] [PubMed] [Google Scholar]
- 3.Eisenberg E, McElhinney PA, Commandeur F, et al. Deep Learning-Based Quantification of Epicardial Adipose Tissue Volume and Attenuation Predicts Major Adverse Cardiovascular Events in Asymptomatic Subjects. Circ Cardiovasc Imaging 2020;13:e009829. 10.1161/CIRCIMAGING.119.009829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mancio J, Azevedo D, Fragao-Marques M, et al. Meta-Analysis of Relation of Epicardial Adipose Tissue Volume to Left Atrial Dilation and to Left Ventricular Hypertrophy and Functions. Am J Cardiol 2019;123:523-31. 10.1016/j.amjcard.2018.10.020 [DOI] [PubMed] [Google Scholar]
- 5.Sepehri Shamloo A, Schoene K, Stauber A, et al. Epicardial adipose tissue thickness as an independent predictor of ventricular tachycardia recurrence following ablation. Heart Rhythm 2019;16:1492-8. 10.1016/j.hrthm.2019.06.009 [DOI] [PubMed] [Google Scholar]
- 6.Hammache N, Pegorer-Sfes H, Benali K, et al. Is There an Association between Epicardial Adipose Tissue and Outcomes after Paroxysmal Atrial Fibrillation Catheter Ablation? J Clin Med 2021;10:3037. 10.3390/jcm10143037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangili LC, Mangili OC, Bittencourt MS, et al. Epicardial fat is associated with severity of subclinical coronary atherosclerosis in familial hypercholesterolemia. Atherosclerosis 2016;254:73-7. 10.1016/j.atherosclerosis.2016.09.010 [DOI] [PubMed] [Google Scholar]
- 8.Mahabadi AA, Balcer B, Dykun I, et al. Cardiac computed tomography-derived epicardial fat volume and attenuation independently distinguish patients with and without myocardial infarction. PLoS One 2017;12:e0183514. 10.1371/journal.pone.0183514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malhotra P, Gupta S, Koundal D, et al. Deep Neural Networks for Medical Image Segmentation. J Healthc Eng 2022;2022:9580991. 10.1155/2022/9580991 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Commandeur F, Goeller M, Razipour A, et al. Fully Automated CT Quantification of Epicardial Adipose Tissue by Deep Learning: A Multicenter Study. Radiol Artif Intell 2019;1:e190045. 10.1148/ryai.2019190045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Commandeur F, Slomka PJ, Goeller M, et al. Machine learning to predict the long-term risk of myocardial infarction and cardiac death based on clinical risk, coronary calcium, and epicardial adipose tissue: a prospective study. Cardiovasc Res 2020;116:2216-25. 10.1093/cvr/cvz321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroll L, Nassenstein K, Jochims M, et al. Assessing the Role of Pericardial Fat as a Biomarker Connected to Coronary Calcification-A Deep Learning Based Approach Using Fully Automated Body Composition Analysis. J Clin Med 2021;10:356. 10.3390/jcm10020356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Commandeur F, Goeller M, Betancur J, et al. Deep Learning for Quantification of Epicardial and Thoracic Adipose Tissue From Non-Contrast CT. IEEE Trans Med Imaging 2018;37:1835-46. 10.1109/TMI.2018.2804799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoori A, Hu T, Lee J, et al. Deep learning segmentation and quantification method for assessing epicardial adipose tissue in CT calcium score scans. Sci Rep 2022;12:2276. 10.1038/s41598-022-06351-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couselo-Seijas M, Rodríguez-Mañero M, González-Juanatey JR, et al. Updates on epicardial adipose tissue mechanisms on atrial fibrillation. Obes Rev 2021;22:e13277. 10.1111/obr.13277 [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Xu Z, Jiang B, et al. Machine-learning-based radiomics identifies atrial fibrillation on the epicardial fat in contrast-enhanced and non-enhanced chest CT. Br J Radiol 2022;95:20211274. 10.1259/bjr.20211274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber AT, Fankhauser S, Chollet L, et al. The Relationship between Enhancing Left Atrial Adipose Tissue at CT and Recurrent Atrial Fibrillation. Radiology 2022;305:56-65. 10.1148/radiol.212644 [DOI] [PubMed] [Google Scholar]
- 18.Kelly BS, Judge C, Bollard SM, et al. Radiology artificial intelligence: a systematic review and evaluation of methods (RAISE). Eur Radiol 2022;32:7998-8007. 10.1007/s00330-022-08784-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isensee F, Jaeger PF, Kohl SAA, et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18:203-11. 10.1038/s41592-020-01008-z [DOI] [PubMed] [Google Scholar]
- 20.Antonelli M, Reinke A, Bakas S, et al. The Medical Segmentation Decathlon. Nat Commun 2022;13:4128. 10.1038/s41467-022-30695-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012;30:1323-41. 10.1016/j.mri.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iacobellis G. Local and systemic effects of the multifaceted epicardial adipose tissue depot. Nat Rev Endocrinol 2015;11:363-71. [DOI] [PubMed] [Google Scholar]
- 23.Iacobellis G. Epicardial adipose tissue in contemporary cardiology. Nat Rev Cardiol 2022;19:593-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greco F, Mallio CA. Artificial intelligence and abdominal adipose tissue analysis: a literature review. Quant Imaging Med Surg 2021;11:4461-74. 10.21037/qims-21-370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mancio J, Azevedo D, Saraiva F, et al. Epicardial adipose tissue volume assessed by computed tomography and coronary artery disease: a systematic review and meta-analysis. Eur Heart J Cardiovasc Imaging 2018;19:490-7. 10.1093/ehjci/jex314 [DOI] [PubMed] [Google Scholar]
- 26.Lin A, Wong ND, Razipour A, et al. Metabolic syndrome, fatty liver, and artificial intelligence-based epicardial adipose tissue measures predict long-term risk of cardiac events: a prospective study. Cardiovasc Diabetol 2021;20:27. 10.1186/s12933-021-01220-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greco F, Salgado R, Van Hecke W, et al. Epicardial and pericardial fat analysis on CT images and artificial intelligence: a literature review. Quant Imaging Med Surg 2022;12:2075-89. 10.21037/qims-21-945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang M, Cao Q, Xu Z, et al. Development and Validation of a Machine Learning-Based Radiomics Model on Cardiac Computed Tomography of Epicardial Adipose Tissue in Predicting Characteristics and Recurrence of Atrial Fibrillation. Front Cardiovasc Med 2022;9:813085. 10.3389/fcvm.2022.813085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu J, Zhuo K, Zhang B, et al. Sex Differences in Epicardial Adipose Tissue: Association With Atrial Fibrillation Ablation Outcomes. Front Cardiovasc Med 2022;9:905351. 10.3389/fcvm.2022.905351 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as