Abstract
Objective
Lung cancer is the second most frequent cancer type and the most common cause of cancer-related deaths worldwide. Alteration of gene copy numbers are associated with lung cancer and the determination of copy number variations (CNV) is appropriate for the discrimination between tumor and non-tumor tissue in lung cancer. As telomerase reverse transcriptase (TERT) and v-myc avian myelocytomatosis viral oncogene homolog (MYC) play a role in lung cancer the aims of this study were the verification of our recent results analyzing MYC CNV in tumor and non-tumor tissue of lung cancer patients using an independent study group and the assessment of TERT CNV as an additional marker.
Results
TERT and MYC status was analyzed using digital PCR (dPCR) in tumor and adjacent non-tumor tissue samples of 114 lung cancer patients. The difference between tumor and non-tumor samples were statistically significant (p < 0.0001) for TERT and MYC. Using a predefined specificity of 99% a sensitivity of 41% and 51% was observed for TERT and MYC, respectively. For the combination of TERT and MYC the overall sensitivity increased to 60% at 99% specificity. We demonstrated that a combination of markers increases the performance in comparison to individual markers. Additionally, the determination of CNV using dPCR might be an appropriate tool in precision medicine.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-023-06566-x.
Keywords: Marker combination, Copy number variations, Digital polymerase chain reaction, Diagnosis, Non-small cell lung cancer, Frozen tissue
Introduction
Lung cancer constitutes a major health burden. Worldwide, 2,206,771 new cases and 1,796,144 deaths were assigned to lung cancer in 2020, representing the second most frequent cancer type (11.4%) and the most common cause of cancer-related deaths (18.0%) [1].
The major histological lung cancer group (~ 85%) is non-small cell lung cancer (NSCLC). NSCLC shows a 5-year survival of 10% for patients at advanced stages in comparison to 68% for patients at early stages [2, 3].
Many cancers exhibit chromosomal instability resulting in amplification/gain or loss/deletion of genomic DNA. Copy number variation (CNV) is characterized by a change of DNA sequence number in comparison to the normal (diploid) genome [4]. Lung cancer as well as several other cancers are associated with CNV [5, 6].
Recently, we have shown that the determination of v-myc avian myelocytomatosis viral oncogene homolog (MYC) CNV is an appropriate tool to discriminate between lung tumor and non-tumor tissues with a sensitivity of 43% and 99% specificity [7]. MYC is located on chromosome 8q24.21 and encodes a transcription factor playing a role in cell cycle, apoptosis, and cellular transformation. Notably, MYC is one of the most frequently amplified genes in lung cancer [8].
Similarly, telomerase reverse transcriptase (TERT) is frequent amplified in early-stage lung cancer [9–11]. Barthel et al. observed a TERT amplification in 13% and 14% of lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LSCC) cases, respectively [12]. TERT is located on chromosome 5p15.33 and is a major component of the telomerase complex. Up to 90% of all tumors are marked by telomerase activation [13, 14]. Cao et al. proposed that an increase of the TERT copy number is an important mechanism for upregulation of telomerase activity in human cancer [15]. TERT is a key player supporting immortality of cancer cells and the amplification of TERT with other chromosomal aberrations plays a role in tumor development and progression [16, 17].
Due to the roles of MYC and TERT in lung cancer, the aims of this study were the verification of our recent results analyzing MYC CNV in tumor and non-tumor tissue of lung cancer patients using an independent study group and the assessment of TERT CNV as an additional marker.
Main text
Methods
Study group
Between 08/2017 and 06/2020, 423 subjects were recruited in the Malteser Krankenhaus Seliger Gerhard Bonn/Rhein-Sieg. Of those, within the context of surgical interventions, lung tissue samples were collected from 144 lung cancer patients, immediately cooled to 4 °C and washed with isotonic saline solution. Pathological evaluation was performed and finally, 114 primary tumor and adjacent non-tumor tissues without signs of inflammation were selected as appropriate for CNV analyses. Samples were stored at − 80 °C until analyses.
All participants of the study provided informed consent. The study was designed according to the rules guarding patient privacy and with the approval from the ethics committee of the Faculty of Medicine, Ruhr University Bochum (registration number 4552-12) and performed in accordance with the Declaration of Helsinki.
Detection of CNV
Isolation of genomic DNA (gDNA) was performed from two 40 µm sections of frozen tissue using the QIAamp Fast DNA Tissue Kit (Qiagen, Hilden, Germany) as recently described [7]. Notably, isolated gDNA was not pretreated, i.e., fragmented, according to Brik et al. [7]. Digital PCR (dPCR) was carried out using the QuantStudio 3D Digital PCR 20 K Chip Kits v2 (AppliedBiosystems, Pleasanton, CA, USA) with 35 ng gDNA as template according to the manufacturer’s instructions using FAM-labeled TERT (Hs06005815_cn; Thermo Fisher Scientific) and MYC (Hs02758348_cn; Thermo Fisher Scientific) as marker, and VIC-labeled RNase P (4403326; Thermo Fisher Scientific) as reference. All assays were performed using duplex assays, i.e., RNase P and MYC and TERT, respectively, were determined in parallel within a single reaction. CNV was calculated as 2 × ratio of the MYC and TERT copy number, respectively, to the RNase P copy number.
Statistical analyses
Statistical analyses were performed using SAS software, version 9.4 (SAS Institute Inc., Cary, NC, USA). Dot plots with median and inter-quartile range (IQR) were used to depict the distribution of single markers. Mann–Whitney U tests were applied to examine group differences and Wilcoxon signed-rank tests to compare two related samples. P < 0.05 were considered as statistically significant. Receiver operating characteristic (ROC) curves were used to quantify classification performance and cut-offs of single markers. The accuracy of the diagnostic tests was depicted by the area under curve (AUC) and its 95% confidence interval (CI). Marker cut-offs were determined by the highest sensitivity at fixed specificities of 99%. The chi-squared test was used to compare assays status and clinicopathological parameters.
Combinations of MYC and TERT were evaluated using the two Boolean operators AND and OR to combine both markers. Applying the AND operator, the two-marker combination was defined as positive if both markers were positive, whereas applying the OR operator the two-marker combination was defined as positive if at least one of the two markers was positive. For both operators, the cut-off values correspond to the highest sensitivity at 99% specificity of all potential cut-off combinations.
Results
Study population
In Table 1 the clinicopathologic parameters of the included 114 lung cancer patients are presented. Median age of the patients was 70 years. Most of the participants were current (47.4%) or former smokers (35.9%), whereas 14.9% were never smokers. Mostly, lung cancer patients were diagnosed with LUAD (64.9%) or LSCC (23.7%) and the majority of patients (71.9%) were diagnosed with tumor stage T1 or T2.
Table 1.
Patients´ characteristics and clinicopathological parameters in the study group of 114 lung cancer patients
| Characteristic | Group | N (%) |
|---|---|---|
| Age (years), median (range) | 70 (50–85) | |
| Sex | Male | 57 (50.0) |
| Female | 57 (50.0) | |
| Smoking status | Never | 17 (14.9) |
| Former | 41 (35.9) | |
| Current | 54 (47.4) | |
| Missing | 2 (1.8) | |
| Histological subtypes | Lung adenocarcinoma | 74 (64.9) |
| Lung squamous cell carcinoma | 27 (23.7) | |
| Other# | 13 (11.4) | |
| Grade§ | G1 | 5 (4.4) |
| G2 | 50 (43.9) | |
| G3 | 52 (45.6) | |
| GX | 7 (6.1) | |
| T Stage | T1 | 43 (37.7) |
| T2 | 39 (34.2) | |
| T3 | 16 (14.0) | |
| T4 | 11 (9.7) | |
| TX | 5 (4.4) |
#four carcinoid, three large cell neuroendocrine carcinomas, two non-small cell lung cancer-not otherwise specified, two adenosquamous carcinoma, one large cell carcinoma-not otherwise specified and one carcinosarcoma of the lung
§Grade 1: well differentiated, low grade; Grade 2: moderately differentiated, intermediate grade; Grade 3: poorly differentiated, high grade; Grade X: grade cannot be assessed (undetermined grade)
Assessing of TERT and MYC copy number
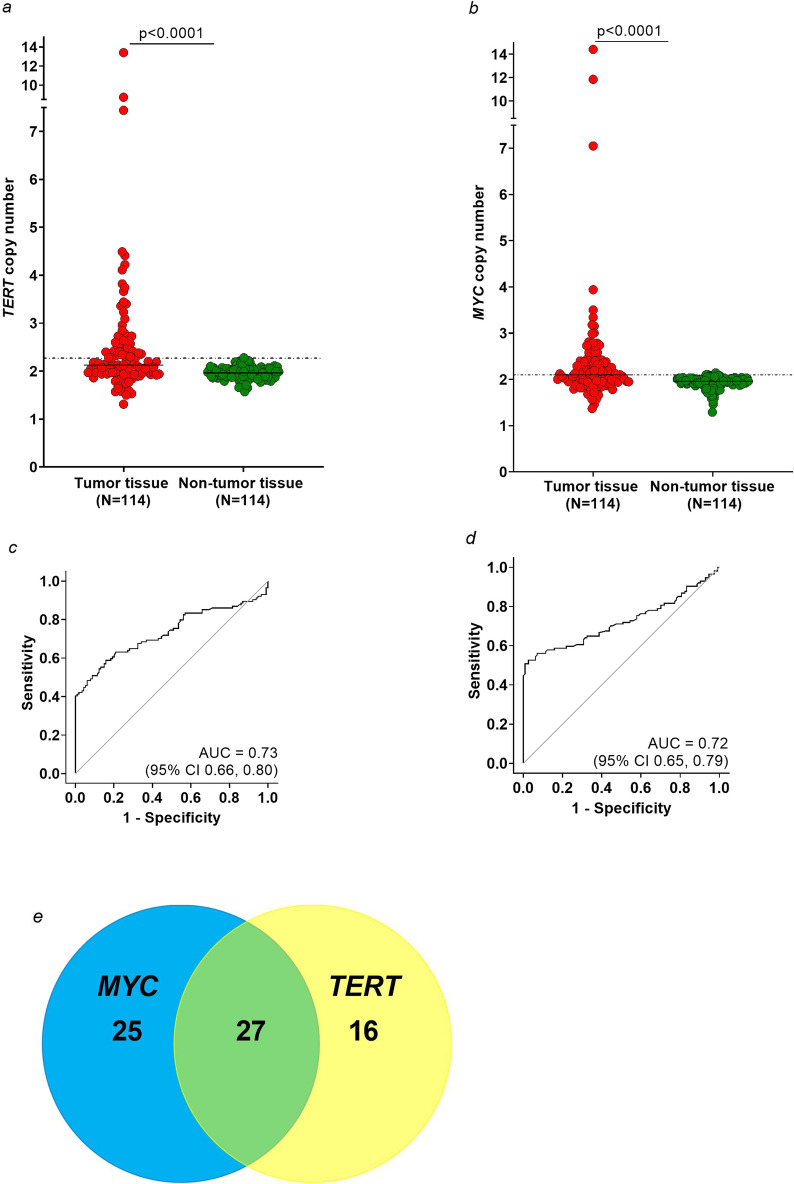
The median TERT copy number in tumor samples was 2.12 (IQR 1.96–2.56) and in non-tumor samples 1.96 (IQR 1.88–2.04). The difference between tumor and non-tumor samples was statistically significant (p < 0.0001), (Fig. 1a). The highest TERT copy number was 13.40 in tumor tissue and 2.28 in non-tumor tissue. Regarding the histological subtype, differences between LUAD and LSCC samples and their corresponding non-tumor samples were also statistically significant (p < 0.0001 and p = 0.0002, respectively), (Additional file 1: Figure S1a–b).
Fig. 1.
a–b: Distribution of copy number variation (CNV) of TERT (a) and MYC (b) in tumor (red) and non-tumor (green) tissue samples from 114 lung cancer patients. The dotted lines indicate the cut-offs for TERT (2.27) and MYC (2.10) and the horizontal lines the median. c–d: Receiver operating characteristic (ROC) analysis for TERT (c) and MYC (d) based on tumor and adjacent non-tumor tissues from the 114 lung cancer patients. e: Venn diagram of the number of true positive tests in lung cancer patients using MYC (blue) and TERT (yellow)
For MYC the median copy number was 2.10 (IQR 1.94–2.38) in tumor tissues and 1.96 (IQR 1.89–2.03) in non-tumor tissues and the difference between tumor and non-tumor was statistically significant (p < 0.0001), (Fig. 1b). The highest MYC copy number in tumor samples was 14.41 in contrast to 2.14 in non-tumor samples. Differences between LUAD and LSCC tissues and their corresponding non-tumor tissues were statistically significant (p < 0.0001 and p = 0.0010, respectively), (Additional file 1: Figure S1c–d).
Differences of copy numbers between the two histological subtypes LUAD and LSCC were not statistically significant, neither for MYC nor for TERT.
For the differentiation between tumor and non-tumor tissue the ROC analysis revealed an AUC of 0.73 (95% CI 0.66, 0.80) for TERT (Fig. 1c) and 0.72 (95% CI 0.65, 0.79) for MYC (Fig. 1d). Using a predefined specificity of 99% (cut-off 2.27) a sensitivity of 41% was observed for TERT. Accordingly, TERT was amplified in 47 of 114 tumor tissues of lung cancer patients. Regarding the histological subtype, TERT was amplified in 30 of 74 LUAD (40.5%) and 13 of 27 LSCC cases (48.1%), (Table 2).
Table 2.
Distribution of clinicopathological characteristics regarding different CNV cut-off values in tumor tissue
| Characteristic | Group | N | MYC ≥ 2.10A (N = 58, 50.9%) | MYC < 2.10A (N = 56, 49.1%) | p-value (chi2 test) | TERT ≥ 2.27B (N = 47, 41.2%) | TERT < 2.27B (N = 67, 58.8%) | p-value (chi2 test) |
|---|---|---|---|---|---|---|---|---|
| Age | ≤ 70 years | 62 | 31 (53.5) | 31 (55.4) | 0.8379 | 24 (51.1) | 38 (56.7) | 0.5509 |
| > 70 years | 52 | 27 (46.6) | 25 (44.6) | 23 (48.9) | 29 (43.3) | |||
| Sex | Female | 57 | 22 (37.9) | 35 (62.5) | 0.0087 | 21 (44.7) | 36 (53.7) | 0.3414 |
| Male | 57 | 36 (62.1) | 21 (37.5) | 26 (55.3) | 31 (46.3) | |||
| Smoking status | Never | 17 | 6 (10.3) | 11 (19.6) | 0.3230* | 6 (12.8) | 11 (16.4) | 0.0590* |
| Former | 41 | 21 (36.2) | 20 (35.7) | 12 (25.5) | 29 (43.9) | |||
| Current | 54 | 29 (50.0) | 25 (44.6) | 27 (57.5) | 27 (40.3) | |||
| Missing | 2 | 2 (3.5) | 0 (0.0) | 2 (4.3) | 0 (0.0) | |||
| Histological subtypes | Lung adenocarcinoma | 74 | 36 (62.1) | 38 (67.9) | 0.2827 | 30 (63.8) | 44 (65.7) | 0.5670 |
| Lung squamous cell carcinoma | 27 | 17 (29.3) | 10 (17.9) | 13 (27.7) | 14 (20.9) | |||
| Other | 13 | 5 (8.6) | 8 (14.3) | 4 (8.5) | 9 (13.4) | |||
| Grade | G1 + G2 | 55 | 24 (41.4) | 31 (55.4) | 0.0399 | 23 (48.9) | 32 (47.8) | 0.6515 |
| G3 | 52 | 33 (56.9) | 19 (33.9) | 24 (51.1) | 28 (41.8) | |||
| GX§ | 7 | 1 (1.7) | 6 (10.7) | 0 (0.0) | 7 (10.5) | |||
| T Stage | T1 | 43 | 22 (37.9) | 21 (37.5) | 0.6470 | 15 (31.9) | 28 (41.8) | 0.3371 |
| T2 | 39 | 23 (39.7) | 16 (28.6) | 18 (38.3) | 21 (31.3) | |||
| T3 + T4 | 27 | 13 (22.4) | 14 (25.0) | 14 (29.8) | 13 (19.4) | |||
| TX$ | 5 | 0 (0.0) | 5 (8.9) | 0 (0.0) | 5 (7.5) |
Acut-off value for MYC at 99% specificity and 51% sensitivity
Bcut-off value for TERT at 99% specificity and 41% sensitivity
§GX was excluded from tests
$TX was excluded from tests
*p-value from Fisher’s exact test
For MYC 51% sensitivity was observed using the predefined specificity of 99% (cut-off 2.10). Thus, MYC was amplified in 58 of 114 tumor tissues of lung cancer patients. Regarding the histological subtype MYC was amplified in 36 of 74 LUAD (48.6%) and in 17 of 27 LSCC cases (63.0%), (Table 2).
For the combination of TERT (cut-off 2.29) and MYC (cut-off 2.14) the overall sensitivity increased to 60% at the predefined specificity of 99% using the OR operator, whereas a sensitivity of 39% at 99% specificity was revealed using the AND operator. Thus, using the OR combination for TERT and MYC, a total of 68 lung cancer tissues were identified correctly, 16 samples by TERT, 25 by MYC, and 27 by both markers (Fig. 1e). In particular, the OR combination detected 44 of 74 LUAD (59.5%) and 18 of 27 LSCC cases (66.7%) correctly.
Influence of clinicopathological parameters on TERT and MYC
No significant group difference between tumor tissue and non-tumor tissue with regard to age, smoking status, tumor histological subtype, grading, and T stage could be observed for TERT, whereas MYC showed statistically significant differences between males and females (p = 0.0217) and regarding grading, i.e., G1 + G2 vs. G3 (p = 0.0399) (Table 2).
Discussion
Amplification of MYC and TERT is a common event in lung cancer. Thus, the assessment of TERT and MYC CNV might be meaningful for the application in clinical diagnostics. In this study, we observed a TERT amplification in 41% of the lung cancer samples, confirming recent studies showing an amplification of TERT in lung cancer cases between 38% [18] and 57% [9]. In contrast to TERT the published percentages for amplification of MYC in lung cancer vary more, ranging from 11% [19] to 88% [20]. In this study, MYC was amplified in 51% of the lung cancer cases, verifying our former results (43% sensitivity and 99% specificity) in an independent study group [7]. Both study groups are comparable regarding most of the clinicopathological parameters. Differences between our two study groups exist only regarding sex, but this difference may be due to the relatively small number of samples in both study groups. However, Li et al. have showed that amplification of MYC is more frequent in males (48%) than in females (37%) [21]. Thus, a detailed analysis regarding CNV and sex should be performed in the future.
Generally, the combination of markers within a panel has the potential to increase the performance of the single markers. For example, in lung tumors a panel of the four markers MYC, TP63, CEP3, and CEP6 showed an improved performance in comparison to each individual marker [22]. In accordance, in this study the combination of MYC and TERT lead to an increase of sensitivity to 60% to detect lung cancer, suggesting that the combination of both markers could be useful for the differentiation between lung cancer and normal tissue.
Regarding the histological subtypes, TERT amplification was detected in 41% of the LUAD and 48% of the LSCC cases. Barthel et al. observed TERT amplification in 13% of LUAD and 14% of LSCC cases [12], whereas other studies detected a TERT amplification in 83.8% [23] and 74.8% [24] of the analysed LUAD patients [23, 24]. Similarly, MYC is amplificated in 45% of the LUAD and 59% of the LSCC cases in this study. This is in accordance with Han et al. observing a MYC amplification in 68.1% of LUAD [24].
Despite the general discrimination between lung cancer and normal tissue MYC and TERT CNV are appropriate to distinguish between each histological subtype and normal tissue, but not to discriminate LUAD from LSCC.
Besides their use as diagnostic markers, the assessment of MYC and TERT in tissue might also be meaningful for the application as prognostic markers in lung tumor patients. MYC is a promising marker candidate for LUAD and associated with poor prognosis [25] and Flacco et al. have shown that an increase of MYC copy number is a predictor of worse survival in NSCLC [26]. Amplification of TERT may be a marker for poorer prognosis in early-stage NSCLC [9] and Liu et al. demonstrated prognostic significance for TERT CNV, showing that TERT was associated with a 35% risk reduction of LUAD progression [23]. Additionally, for ovarian cancer it was shown that TERT and MYC, as well as PIC3CA, CCNE1, and KRAS, might be useful to tailor therapeutics in precision medicine [27], and the same might be true for lung cancer. Alidousty et al. have shown that patients with amplification of both MYC [28] and TERT [29] are prone to fast development of resistance to ALK inhibitors after treatment of lung tumor. This should facilitate further assessment of TERT and MYC for prognostic or predictive purposes.
We are currently performing a follow-up of our patient group in order to update information on progression, recurrence, and survival. This should facilitate further assessment of TERT and MYC for prognostic purposes.
Diagnostic and prognostic use of CNV analysis of amplified genes would greatly benefit from the development of blood-based tests. A high dilution of circulating tumor DNA by normal DNA, however, still poses a challenge. The development of appropriate methods to selectively enrich tumor DNA may be a possible approach.
It was already shown, that dPCR is a valuable tool for CNV analysis [7, 30, 31] and the used method in this study confirms that the application of dPCR enables high accuracy in the detection of CNV in tumor tissues without high levels of false-positive test results in normal tissues. Thus, dPCR may be an appropriate tool in precision medicine [32–34].
Conclusion
We demonstrated that amplification of MYC and TERT is a common event in lung cancer patients. The combination of TERT and MYC leads to an increased performance in comparison to the individual markers. For the detection of CNV, dPCR appears to be a reliable method. In the future, the assessment of MYC and TERT CNV using dPCR might be a reliable tool in precision medicine.
Limitations
The analyzed study group is relatively small and differences of MYC and TERT detection rates in comparison to other studies are based on different group sizes, ranging from 21 [18] to 2,032 lung cancer cases [35], and different methods for the detection of CNV, dPCR [7, 32], quantitative PCR (qPCR) [9, 19], and whole genome sequencing (WGS) [2, 23, 36]. Additionally, discrepancies of RNase P values during determination of MYC and TERT in identical samples makes it necessary to develop appropriate verification methods. However, measurement of references in parallel to the marker using duplex assays already compensate differences during dPCR in comparison to simplex assays. However, consistency between simplex and duplex assays should be assessed in detail. Thus, larger studies with consistent methods for the assessment of CNV are needed.
Additionally, no follow-up data of the patients are available at the moment. Thus, the feasibility of MYC and TERT as prognostic markers could not be evaluated yet.
Supplementary Information
Additional file 1: Figure S1. Distribution of copy number variation (CNV) of TERT in LUAD, TERT in LSCC, MYC in LUAD and MYC in LSCC in tumor and non-tumor tissue samples from 74 LUAD and 27 LSCC patients, respectively.
Additional file 2: Table S1. Raw Data: Levels of RNaseP, MYC and TERT (copies/µL) by dPCR.
Acknowledgements
The authors gratefully acknowledge Bettina Dumont and Simone Naumann for excellent technical assistance. We acknowledge support by the DFG Open Access Publication Funds of the Ruhr University Bochum.
Abbreviations
- AUC
Area under the curve
- Cl
Confidence interval
- CNV
Copy number variations
- dPCR
Digital PCR
- IQR
Inter-quartile range
- LSCC
Lung squamous cell carcinoma
- LUAD
Lung adenocarcinomas
- MYC
V-myc avian myelocytomatosis viral oncogene homolog
- NSCLC
Non-small cell lung cancer
- qPCR
Quantitative PCR
- ROC
Receiver operating characteristic
- TERT
Telomerase reverse transcriptase
- WGS
Whole genome sequencing
Author contributions
AB conceived the study, participated in its design and coordination, performed the experiments and drafted the manuscript. KW performed the statistical analyses and helped to draft the manuscript. DGW conceived of the study, participated in study design and coordination and helped to draft the manuscript. KS performed the experiments. PR and SM participated in study design and helped to draft the manuscript. Y-DK, RB, KG participated in study design and coordination. TBe coordinated field work, participated in study design and helped to draft the manuscript. TBr participated in study design and coordination. GJ conceived of the study, participated in study design and coordination, and helped to draft the manuscript. All authors have read, commented, and approved the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. Not applicable.
Availability of data and materials
The raw data supporting the conclusions of this article are included as Additional file 2.
Declarations
Ethics approval and consent to participate
All participants of the study provided informed consent. The study was designed according to the rules guarding patient privacy and with the approval from the ethics committee of the Faculty of Medicine, Ruhr University Bochum (registration number 4552-12) and performed in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Chen X, Chang C-W, Spoerke JM, Yoh KE, Kapoor V, Baudo C, et al. Low-pass whole-genome sequencing of circulating cell-free DNA demonstrates dynamic changes in genomic copy number in a squamous lung cancer clinical cohort. Clin Cancer Res. 2019;25(7):2254–2263. doi: 10.1158/1078-0432.CCR-18-1593. [DOI] [PubMed] [Google Scholar]
- 3.Fan Y, Sun R, Wang Z, Zhang Y, Xiao X, Liu Y, et al. Detection of MET amplification by droplet digital PCR in peripheral blood samples of non-small cell lung cancer. J Cancer Res Clin Oncol. 2023;149(5):1667–1677. doi: 10.1007/s00432-022-04048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishra S, Whetstine JR. Different facets of copy number changes: permanent, transient, and adaptive. Mol Cell Biol. 2016;36(7):1050–1063. doi: 10.1128/MCB.00652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albertson DG. Gene amplification in cancer. Trends Genetics. 2006;22(8):447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Bowcock AM. Invited review DNA copy number changes as diagnostic tools for lung cancer. Thorax. 2014;69(5):495–496. doi: 10.1136/thoraxjnl-2013-204681. [DOI] [PubMed] [Google Scholar]
- 7.Brik A, Weber DG, Casjens S, Rozynek P, Meier S, Behrens T, et al. Digital PCR for the analysis of MYC copy number variation in lung cancer. Dis Markers. 2020;2020:4176376. doi: 10.1155/2020/4176376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lockwood WW, Chari R, Coe BP, Girard L, MacAulay C, Lam S, et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008;27(33):4615–4624. doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu C-Q, Cutz J-C, Liu N, Lau D, Shepherd FA, Squire JA, et al. Amplification of telomerase (hTERT) gene is a poor prognostic marker in non-small-cell lung cancer. Br J Cancer. 2006;94(10):1452–1459. doi: 10.1038/sj.bjc.6603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eldholm V, Haugen A, Zienolddiny S. CTCF mediates the TERT enhancer-promoter interactions in lung cancer cells: identification of a novel enhancer region involved in the regulation of TERT gene. Int J Cancer. 2014;134(10):2305–2313. doi: 10.1002/ijc.28570. [DOI] [PubMed] [Google Scholar]
- 11.Gaspar TB, Sá A, Lopes JM, Sobrinho-Simões M, Soares P, Vinagre J. Telomere maintenance mechanisms in cancer. Genes (Basel) 2018;9(5):241. doi: 10.3390/genes9050241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barthel FP, Wei W, Tang M, Martinez-Ledesma E, Hu X, Amin SB, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;49(3):349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W, Duan X, Zhang Z, Yang Z, Zhao C, Liang C, et al. Combination of CT and telomerase+ circulating tumor cells improves diagnosis of small pulmonary nodules. JCI Insight. 2021;6(11):e148182. doi: 10.1172/jci.insight.148182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shay JW, Wright WE. Telomeres and telomerase: three decades of progress. Nat Rev Genet. 2019;20(5):299–309. doi: 10.1038/s41576-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 15.Cao Y, Bryan TM, Reddel RR. Increased copy number of the TERT and TERC telomerase subunit genes in cancer cells. Cancer Sci. 2008;99(6):1092–1099. doi: 10.1111/j.1349-7006.2008.00815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan X, Larsson C, Xu D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene. 2019;38(34):6172–6183. doi: 10.1038/s41388-019-0872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dratwa M, Wysoczańska B, Łacina P, Kubik T, Bogunia-Kubik K. TERT-regulation and roles in cancer formation. Front Immunol. 2020;11:589929. doi: 10.3389/fimmu.2020.589929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang A, Zheng C, Lindvall C, Hou M, Ekedahl J, Lewensohn R, et al. Frequent amplification of the telomerase reverse transcriptase gene in human tumors. Cancer Res. 2000;60(22):6230–6235. [PubMed] [Google Scholar]
- 19.Iwakawa R, Kohno T, Kato M, Shiraishi K, Tsuta K, Noguchi M, et al. MYC amplification as a prognostic marker of early-stage lung adenocarcinoma identified by whole genome copy number analysis. Clin Cancer Res. 2011;17(6):1481–1489. doi: 10.1158/1078-0432.CCR-10-2484. [DOI] [PubMed] [Google Scholar]
- 20.Kubokura H, Tenjin T, Akiyama H, Koizumi K, Nishimura H, Yamamoto M, et al. Relations of the c-myc gene and chromosome 8 in non-small cell lung cancer: analysis by fluorescence in situ hybridization. Ann Thorac Cardiovasc Surg. 2001;7(4):197–203. [PubMed] [Google Scholar]
- 21.Li CH, Haider S, Shiah Y-J, Thai K, Boutros PC. Sex differences in cancer driver genes and biomarkers. Cancer Res. 2018;78(19):5527–5537. doi: 10.1158/0008-5472.CAN-18-0362. [DOI] [PubMed] [Google Scholar]
- 22.Massion PP, Zou Y, Uner H, Kiatsimkul P, Wolf HJ, Baron AE, et al. Recurrent genomic gains in preinvasive lesions as a biomarker of risk for lung cancer. PLoS ONE. 2009;4(6):e5611. doi: 10.1371/journal.pone.0005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, Huang J, Wang K, Li L, Li Y, Yuan J, et al. Identification of hallmarks of lung adenocarcinoma prognosis using whole genome sequencing. Oncotarget. 2015;6(35):38016–38028. doi: 10.18632/oncotarget.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han X, Tan Q, Yang S, Li J, Xu J, Hao X, et al. Comprehensive profiling of gene copy number alterations predicts patient prognosis in resected stages i-iii lung adenocarcinoma. Front Oncol. 2019;9:556. doi: 10.3389/fonc.2019.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seo AN, Yang JM, Kim H, Jheon S, Kim K, Lee CT, et al. Clinicopathologic and prognostic significance of c-MYC copy number gain in lung adenocarcinomas. Br J Cancer. 2014;110(11):2688–2699. doi: 10.1038/bjc.2014.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flacco A, Ludovini V, Bianconi F, Ragusa M, Bellezza G, Tofanetti FR, et al. MYC and human telomerase gene (TERC) copy number gain in early-stage non-small cell lung cancer. Am J Clin Oncol. 2015;38(2):152–158. doi: 10.1097/COC.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martins FC, Couturier D-L, de Santiago I, Sauer CM, Vias M, Angelova M, et al. Clonal somatic copy number altered driver events inform drug sensitivity in high-grade serous ovarian cancer. Nat Commun. 2022;13(1):6360. doi: 10.1038/s41467-022-33870-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alidousty C, Baar T, Martelotto LG, Heydt C, Wagener S, Fassunke J, et al. Genetic instability and recurrent MYC amplification in ALK-translocated NSCLC: a central role of TP53 mutations. J Pathol. 2018;246(1):67–76. doi: 10.1002/path.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alidousty C, Duerbaum N, Wagener-Ryczek S, Baar T, Martelotto LG, Heydt C, et al. Prevalence and potential biological role of TERT amplifications in ALK translocated adenocarcinoma of the lung. Histopathology. 2021;78(4):578–585. doi: 10.1111/his.14256. [DOI] [PubMed] [Google Scholar]
- 30.Cai Z, Chen H, Bai J, Zheng Y, Ma J, Cai X, et al. Copy Number variations of CEP63, FOSL2 and PAQR6 serve as novel signatures for the prognosis of bladder cancer. Front Oncol. 2021;11:674933. doi: 10.3389/fonc.2021.674933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kockerols CC, Valk PJ, Levin M-D, Pallisgaard N, Cornelissen JJ, Westerweel PE. Digital PCR for BCR-ABL1 quantification in CML: current applications in clinical practice. HemaSphere. 2020;4:1–9. doi: 10.1097/HS9.0000000000000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee KS, Nam SK, Seo SH, Park KU, Oh H-K, Kim D-W, et al. Digital polymerase chain reaction for detecting c-MYC copy number gain in tissue and cell-free plasma samples of colorectal cancer patients. Sci Rep. 2019;9(1):1611. doi: 10.1038/s41598-018-38415-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min S, Shin S, Chung Y-J. Detection of KRAS mutations in plasma cell-free DNA of colorectal cancer patients and comparison with cancer panel data for tissue samples of the same cancers. Genomics Inform. 2019;17(4):e42. doi: 10.5808/GI.2019.17.4.e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whale AS, Jones GM, Pavšič J, Dreo T, Redshaw N, Akyürek S, et al. Assessment of digital PCR as a primary reference measurement procedure to support advances in precision medicine. Clin Chem. 2018;64(9):1296–1307. doi: 10.1373/clinchem.2017.285478. [DOI] [PubMed] [Google Scholar]
- 35.Qiu Z-W, Bi J-H, Gazdar AF, Song K. Genome-wide copy number variation pattern analysis and a classification signature for non-small cell lung cancer. Genes Chromosomes Cancer. 2017;56(7):559–569. doi: 10.1002/gcc.22460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simbolo M, Mafficini A, Sikora KO, Fassan M, Barbi S, Corbo V, et al. Lung neuroendocrine tumours: deep sequencing of the four World Health Organization histotypes reveals chromatin-remodelling genes as major players and a prognostic role for TERT, RB1, MEN1 and KMT2D. J Pathol. 2017;241(4):488–500. doi: 10.1002/path.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Distribution of copy number variation (CNV) of TERT in LUAD, TERT in LSCC, MYC in LUAD and MYC in LSCC in tumor and non-tumor tissue samples from 74 LUAD and 27 LSCC patients, respectively.
Additional file 2: Table S1. Raw Data: Levels of RNaseP, MYC and TERT (copies/µL) by dPCR.
Data Availability Statement
The raw data supporting the conclusions of this article are included as Additional file 2.