Abstract
Oxaliplatin (cyclohexane-1,2-diamine; oxalate; platinum [2+]) is a third-generation chemotherapeutic drug with anticancer effects. Oxaliplatin has a role in the treatment of several cancers. It is one of the few drugs which can eliminate the neoplastic cells of colorectal cancer. Also, it has an influential role in breast cancer, lung cancer, bladder cancer, prostate cancer, and gastric cancer. Although oxaliplatin has many beneficial effects in cancer treatment, resistance to this drug is in the way to cure neoplastic cells and reduce treatment efficacy. microRNAs are a subtype of small noncoding RNAs with ∼22 nucleotides that exist among species. They have diverse roles in physiological processes, including cellular proliferation and cell death. Moreover, miRNAs have essential roles in resistance to cancer treatment and can strengthen sensitivity to chemotherapeutic drugs and regimens. In colorectal cancer, the co-treatment of oxaliplatin with anti-miR-19a can partially reverse the oxaliplatin resistance through the upregulation of phosphatase and tensin homolog (PTEN). Moreover, by preventing the spread of gastric cancer cells and downregulating glypican-3 (GPC3), MiR-4510 may modify immunosuppressive signals in the tumor microenvironment. Treatment with oxaliplatin may develop into a specialized therapeutic drug for patients with miR-4510 inhibition and glypican-3-expressing gastric cancer. Eventually, miR-122 upregulation or Wnt/β-catenin signaling suppression boosted the death of HCC cells and made them more sensitive to oxaliplatin. Herein, we have reviewed the role of microRNAs in regulating cancer cells’ response to oxaliplatin, with particular attention to gastrointestinal cancers. We also discussed the role of these noncoding RNAs in the pathophysiology of oxaliplatin-induced neuropathic pain.
Keywords: cancer, chemoresistance, oxaliplatin, microRNAs, gastrointestinal
Introduction
Oxaliplatin (cyclohexane-1,2-diamine; oxalate; platinum [2+]) is a third-generation chemotherapeutic drug with anticancer effects. Oxaliplatin benefits were approved 20 years after identifying it in 1996. It can be used in a chemotherapeutic regimen with either cisplatin or carboplatin and has no contraindications with them. Also, it has lower toxicity and more efficacy than them.1–3 Unfortunately, using oxaliplatin can cause peripheral sensory neuropathy.4,5 Other side effects of oxaliplatin are allodynia, hyperalgesia, dysesthesia, and paresthesia.6,7
Oxaliplatin has a role in the treatment of several cancers. It is one of few drugs that can eliminate the neoplastic cells of colorectal cancer. 8 Also, it has an influential role in bladder cancer, prostate cancer, breast cancer, lung cancer, and gastric cancer.9–12
There are some suggested mechanisms for oxaliplatin. First, it binds to DNA and forms intra-strand cross-links. 13 This act reduces cell capability to transcription, and replacing DNA also arrests the cell cycle, leads to the death of neoplastic cells, and treats cancers with a high turnover rate. 13 Another mechanism is when we treat a colorectal tumor, treated C26 cells release an immunogenic signal that activates an immune response, which leads to an anticancer effect by the immune system. 14
Although oxaliplatin has many beneficial effects in cancer treatment, resistance to this drug is in the way to cure neoplastic cells and reduce treatment efficacy. Some suggested reasons for this obstacle are Forkhead box proteins, g-glutamyl transpeptidase, and miRNAs. There are several mechanisms for resistance to oxaliplatin. First is resistance imposed by FOX (Forkhead box) proteins, a family of proteins with essential roles in biological processes and cancer. They have roles in the development of the immune system, nervous system, kidney, lung, and hair. 15 They can enhance the action of other regulators by binding to chromatin. 16 FOXC2 is a member of this family that evokes MAPK/ERK signaling pathway to induce epithelial-to-mesenchymal transition resulting in more progression, malignancy of cancer cells, and resistance to oxaliplatin in colorectal cancer. 17 Second, another mechanism suggested by researchers is the promotion of glutathione intervened by g-glutamyl transpeptidase. 18 Lastly, some miRNAs have a role in this resistance, too. miR-107 is an onco-suppressor and reduces migration and invasion of neoplastic cells in oral squamous carcinoma cells. 19 This miRNA also has an oncogenic role and elevates the expression of mTOR by reducing the expression of CAB39 and AMPK, which leads to oxaliplatin resistance. 20 miR-744 overexpression reduces colony formation and proliferation of neoplastic cells. 21 However, it also imposes resistance to oxaliplatin by downregulating BIN1. 22 MiR-46146 inhibits PDCD10, and it causes resistance to oxaliplatin. 23 At last, miR-19b-3p enhances resistance to oxaliplatin by downregulating SMAD4. 24
Oxaliplatin is a third-generation platinum-based drug with an effect for treating several malignancies like colorectal, pancreatic, and gastric cancer. Unfortunately, resistance to treatment with platinum-based drugs is a significant problem. Studies suggest epigenetic mechanisms as a cause of this problem. There are three epigenetic regulation classes: DNA methylation, histone modification, and noncoding RNA action. 25 DNA methylation is an important mechanism that raises the resistance to oxaliplatin in different forms.26–28 Also, histone modification increases resistance to oxaliplatin as a treatment method. 29 The variety and role of noncoding RNAs are extensive, which are explained below:
Noncoding RNAs (ncRNAs) are a part of the transcriptome that do not have a prominent role in protein coding but participate in many biological processes. Noncoding RNAs are made of two major branches: structural noncoding RNAs and regulatory noncoding RNAs. Also, regulatory noncoding RNAs are divided into long noncoding RNAs, medium noncoding RNAs, and small noncoding RNAs. 30
microRNAs are a subtype of small noncoding RNAs with ∼22 nucleotides that exist among species. They have diverse roles in physiological processes. In cellular proliferation and cell death, they have crucial roles. They are responsible for immune system maintenance and immune cell development. In the neurological aspect, they have crucial roles in central nervous system development and physiological functions.31,32 They also play essential roles in cardiac development and related diseases.33–35
miRNAs can alter the sensitivity to chemotherapeutic drugs and cancer treatment regimens. For example, in lung cancer, normalizing the regulation of miR-155 removes resistance to chemotherapeutic drugs. 36 Another study concluded that in breast cancer, miR-181 a/b elevates chemoresistance and metastasis. 37 Another example is the miR-106/b cluster. They have roles in drug resistance, metastasis, invasion, and immune evasion. 38 In another study, it is suggested that downregulation of miR-205 will increase resistance to docetaxel in prostate cancer.39,40 They also play essential roles in resistance to chemotherapy in gastric cancer. BCL-2 is a gene that regulates cell apoptosis. Downregulation of the miR-200bc/429 cluster has been observed despite the overexpression of BCL-2 and XIAP (X-linked inhibitor of apoptosis protein) in multidrug resistance. 41 In liver cancer, changes in the regulation of miRNA-130a, miRNA-340, miRNA-182, miRNA-96, and miRNA-199a-5p could elevate or reduce the sensitivity of hepatocellular carcinoma (HCC) to the chemotherapy drug.42–46
1. microRNAs Regulating Cancer Cell Response to Oxaliplatin in Colorectal Cancer
Colorectal cancer is the third most common type of cancer in the world. Approximately 1.9 million patients were newly diagnosed with colorectal cancer, and 935,000 related deaths occurred in 2020. 47 Family history, diet, obesity, and smoking are the most crucial risk factors for colorectal cancer. 48 Early colorectal cancer may not present noticeable symptoms, but changes in bowel habits, stool characteristics, abdominal discomfort, and weight loss may occur at a particular stage. A combination of imaging tests, endoscopy methods, and laboratory examinations are currently used to detect colorectal cancer. After diagnosis, surgery, radiation therapy, targeted therapy, and chemotherapy are commonly used for palliative or curative treatment.49–51
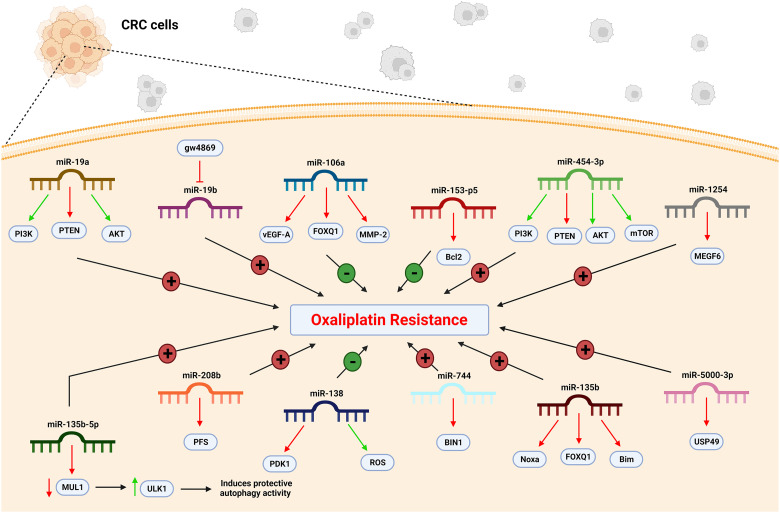
MiR-19a can induce oxaliplatin resistance in colorectal cancer cell lines (Table 1). These cell lines express a significantly higher level of miR-19a, while the knockdown of miR-19a increases their sensitivity to oxaliplatin and oxaliplatin-induced apoptosis. In 2020, Zhang et al investigated how miR-19a can affect cell resistance. They found PTEN as the target of miR-19a and found a negative correlation between them (Figure 1). In colorectal cancer oxaliplatin-resistant cells, upregulation of miR-19a reduces the expression of PTEN. In these cells, co-treatment of oxaliplatin with anti-miR-19a can partially reverse the oxaliplatin resistance through the upregulation of PTEN. They also studied the role of PI3K and AKT and indicated that anti-miR-19a targets the PTEN/PI3K/AKT pathway. 52
Table 1.
miRNAs and Their Induced Effect on Cancer Cell Response to Oxaliplatin-Containing Regimens.
| miRNA | Target | Overexpression effect | Reference |
|---|---|---|---|
| Colorectal cancer | |||
| miR-19a | PTEN | Resistance | 52 |
| miR-19b | gw4869 | Sensitivity | 53 |
| miR-106a | FOXQ1 | Sensitivity | 54 |
| miR-135b | FOXO1 | Resistance | 55 |
| miR-135b-5p | MUL1 | Resistance | 56 |
| miR-138 | PDK1 | Sensitivity | 57 |
| miR-153-5p | Bcl-2 | Sensitivity | 58 |
| miR-208b | PDCD4 | Resistance | 59 |
| miR-454-3p | PTEN | Resistance | 60 |
| miR-744 | BIN1 | Resistance | 22 |
| miR-1254 | MEGF6 | Resistance | 61 |
| miR-5000-3p | USP49 | Resistance | 62 |
| Gastric cancer | |||
| miR-4510 | GPC3 | Sensitivity | 63 |
| miR-130a | FOSL1, RAB5A | Sensitivity | 64 |
| miR-15a-5p | PHLPP2 | Resistance | 65 |
| miR188-3p | EIF3J-DT | Resistance | 66 |
| miR-421 | ATM | Resistance | 67 |
| miR-22-3p | MALAT1 | Sensitivity | 68 |
| miR-211-5p | lncSLCO1C1 | Sensitivity | 69 |
| miR-204-5p | |||
| Hepatocellular carcinoma | |||
| miR-31-5p | PARP1 | Resistance | 70 |
| miR-122 | Wnt/β-catenin | Sensitivity | 71,72 |
| Ovarian cancer | |||
| miR-199b-5p | JAG1-Notch1 | Sensitivity | 73 |
| miR-506 | RAD51 | Sensitivity | 74 |
| Breast cancer | |||
| miR-320c | Chk1 | Sensitivity | 75 |
| miR-525-5p | CircFAT1 | Sensitivity | 76 |
| Esophageal cancer | |||
| miR-141-3p | PTEN | Resistance | 77 |
Figure 1.
miRNA change and CRC cell resistance to the oxaliplatin. In CRC cells, the level of miR-106a, miR-153-5p, and miR-138 decrease. Conversely, the level of miR-19a, miR-19b, miR-454-3p, miR-1254, miR-135-5p, miR-208b, miR-744, miR-135b, and miR-5000-3p increase. Thus, the CRC cells become resistant to oxaliplatin. CRC, colorectal cancer; miR, microRNA.
Exosomal miR-19b participates in oxaliplatin sensitivity of colorectal cancer cell lines. Gu et al in 2018 indicated that miR-19b overexpression decreases colorectal cancer cell lines’ oxaliplatin sensitivity. Suppressing the secretion of exosomal miR-19b in these cells is a role of gw4869, which is a nSMase2 (neutral sphingomyelinase 2) inhibitor. Altogether, gw4869 regulates colorectal cancer cell oxaliplatin sensitivity by suppressing exosomal miR-19b release. 53
MiR-106a is a critical factor in responding to oxaliplatin-containing treatment in colorectal cancer cells and tissues. In these cells, the level of miR-106a is elevated, which can increase oxaliplatin sensitivity (Table 1). Forkhead box Q1, also known as FOXQ1, is the direct target of miR-106a in these cells (Figure 1). Liu et al in 2020 showed that miR-106a could bind to FOXQ1 mRNA and directly inhibit its expression in vitro and in vivo. In addition, miR-106a can suppress the transcription of VEGFA and MMP-2, which are target genes for FOXQ1. 54
MiR-135b functions as a tumor promoter in colorectal cancer. The miR-135b expression is remarkably elevated in colorectal cancer cell lines. In the patients’ serum also, a significant increase in miR-135b level in colorectal cancer patients can be observed. In contrast, the downregulation of miR-135b can sensitize colorectal cancer cells to chemotherapy drugs, including oxaliplatin, cisplatin, and 5-fluorouracil, and their related cytotoxicity. It can also increase the antitumor effect of oxaliplatin on colorectal cancer in vivo. 55
FOXO1 is a tumor suppressor and a target of miR-135b. According to Qin et al, the protein level of FOXO1 is negatively correlated with miR-135b in colorectal cancer cells, and a reduction in miR-135b expression can increase FOXO1 expression in colorectal cancer. FOXO1 expression regulates chemosensitivity to oxaliplatin in colorectal cancer, and downregulation of miR-135b is found to sensitize colorectal cancer cells to oxaliplatin-induced cytotoxicity through the increase of FOXO1. Downregulated miR-135b and FOXO1 play a crucial role in promoting Bim and Noxa proteins’ function and mitochondrial apoptosis in oxaliplatin-treated colorectal cancer cells (Figure 1). 55
MiR-135b-5p escalates the resistance of colorectal cancer cells to chemotherapy drugs like oxaliplatin, augments malignant proliferation, and significantly increases tumor volume, weight, and growth rate. The miR-135b-5p expression level is also elevated in colorectal cancer cell lines and the patient's serum. In 2021, Wang et al indicated that this overexpression is due to the activation of heat shock factor 1 (HSF1). In these cells, autophagy plays a vital role in inducing oxaliplatin resistance. MiR-135b-5p can induce protective autophagy and, by that, enhance oxaliplatin resistance. 56
Mitochondrial ubiquitin ligase 1, also known as MUL1, is the miR-135b-5p direct target in colorectal cancer, and they are negatively correlated. In oxaliplatin-resistant cells, a decrease in MUL1 expression leads to an increase in unc-51-like kinase 1 (ULK1) expression, an autophagy-related gene. Thus, the miR-135b-5p/MUL1 axis induces protective autophagy activity by regulating ULK1. 56
Prolonged exposure to oxaliplatin can stimulate resistance in colorectal tumor cells. These resistant cells produce more lactate, consume more glucose, and show higher lactic dehydrogenase (LDH) activity. Altogether, they present an elevated glycolysis rate and a decreased OCR (oxygen consumption rate). Resistant cells also elevate the level of PDK1, a key enzyme in mitochondrial metabolism. In 2020, Wang et al found that PDK1 overexpression significantly decreases the colorectal cancer cells’ sensitivity to oxaliplatin. 57
MiR-138 negatively targets PDK1 and regulates the colorectal cancer cells’ resistance to oxaliplatin. In these cells, the expression of miR-138 is downregulated, leading to overexpression of PDK1. Suppressing the expression of PDK1 cannot influence the glycolysis in resistant cells, but it can augment the OCR and lead to partially reversing the oxaliplatin resistance. Increasing ROS level is a key mechanism for miR-138 in reversing oxaliplatin resistance (Figure 1). Wang et al also showed that miR-138 promotes apoptosis due to oxaliplatin in resistant cells through the PDK1/ROS/ASK1/JNK/Bcl-2 pathway. 57
MiR-153-5p correlates with chemoresistance in colorectal cancer cells. In oxaliplatin-resistant colorectal cancer cell lines, miR-153-5p expression is significantly downregulated. As reported in 2020, the upregulation of miR-153-5p efficiently increases cell apoptosis and inhibits the cell viability and growth of resistant cells under oxaliplatin treatment. In these cells, miR-153-5p potentially targets Bcl-2 and reduces its mRNA and protein levels (Table 1 and Figure 1). Through targeting Bcl-2-mediated autophagy, miR-153-5p enhances the sensitivity of colorectal cancer cell lines to oxaliplatin. 58
MiR-208b is a predictive marker of oxaliplatin sensitivity in colorectal cancer. The level of miR-208b expression in oxaliplatin-sensitive patients is significantly reduced (P < 0.001), but it is increased in resistant patients. The expression of miR-208b is connected to PFS (progression-free survival) in colorectal cancer patients (Figure 1). Patients with a lower miR-208b have a significantly longer PFS (P < 0.001). Moreover, overexpression of miR-208b can promote tumor growth in vivo by inducing increased regulatory T cells. Exosomal miR-208b is secreted by colorectal tumor cells. According to Ning et al, this miRNA can promote regulatory T cell expansion by directly targeting programmed cell death protein 4 (PDCD4) in CD4+ T cells. PDCD4 is a crucial gene that negatively regulates the differentiation of regulatory T cells, which is related to the chemosensitivity of colorectal cancer cells to oxaliplatin. 59
MiR-454-3p is a crucial factor in oxaliplatin-resistant colorectal cancer cells. The expression of this miRNA in resistant cells is approximately 7-fold higher than in nonresistant cells. This overexpression significantly induces resistance to oxaliplatin in colorectal cancer cells, and a positive association can be found between miR-454-3p expression and oxaliplatin resistance. Moreover, colorectal cancer patients with overexpressed miR-454-3p exhibit shorter PFS versus patients with low miR-454-3p expression. 60
PTEN is a tumor suppressor that is directly targeted by miR-454-3p (Figure 1). Qian et al in 2021 demonstrated that the miR-454-3p overexpression significantly downregulates the expression of PTEN, while the inhibition of miR-454-3p significantly upregulates the expression of PTEN. This tumor suppressor downregulates the PI3K/AKT pathway. Altogether, miR-454-3p lowers the oxaliplatin-induced apoptosis by activating the PTEN/AKT/mTOR pathway. 60
MiR-744 interferes with oxaliplatin-induced chemoresistance in colorectal cancer cells. Oxaliplatin treatment induces an elevated level of miR-744 in colorectal cancer tissues and cell lines. It demonstrates that long-term oxaliplatin administration promotes the chemoresistance of colorectal cancer cells and upregulates the expression of miR-744. Inhibition of miR-744 sensitizes these cells to oxaliplatin. Bridging integrator 1 or BIN1, a tumor suppressor, is directly targeted by miR-744, and a negative correlation can be found between them. Zhou et al in 2019 confirmed that the level of BIN1, unlike the level of miR-744 in oxaliplatin-resistant cells, is decreased. They mentioned that miR-744 mediates oxaliplatin resistance via downregulating BIN1. 22
MiR-1254 is related to chemoresistance in colorectal cancer. In oxaliplatin-resistant colorectal cancer cells, the expression level of miR-1254 is upregulated. This overexpression leads to reduced cell apoptosis and escalated oxaliplatin resistance. In contrast, inhibiting miR-1254 can restore oxaliplatin responsiveness in colorectal-resistant cells by increasing apoptosis. As Mou et al mentioned in 2021, miR-1254 negatively targets multiple EGF-like domains 6 (MEGF6) to regulate oxaliplatin resistance (Figure 1). Inhibiting the expression of MEGF6 decreases apoptosis and promotes resistance in colorectal cancer cells. Altogether, miR-1254 modulates oxaliplatin resistance in colorectal cancer cells by MEGF6. 61
MiR-5000-3p assists the malignant characteristics of colorectal cancer cells. The expression level of miR-5000-3p is significantly increased in these cell lines. By promoting cell growth and metastasis and suppressing cell apoptosis, miR-5000-3p contributes to chemoresistance in colorectal cancer both in vivo and in vitro. Zhuang et al in 2021 showed USP49 as a target of miR-5000-3p and found a significantly negative association between them. mRNA and protein levels of USP49 were lower in oxaliplatin-resistant cells. Additionally, overexpression of USP49 remarkably inhibits cell proliferation and regulates the PI3K/AKT signal pathway. Taken together, the downregulation of miR-5000-3p expression improves the sensitivity of colorectal cancer cells to oxaliplatin through binding to USP49 and regulating the PI3K/AKT signaling pathway. 62
2. microRNAs Regulating Cancer Cell Response to Oxaliplatin in Gastric Cancer
One of the most common malignant tumors of the gastrointestinal system is gastric cancer (GC), which is the third cause of cancer-related deaths. 78 Every year, around 1 million people worldwide receive a new diagnosis of stomach cancer, with China accounting for 47% of those cases. When they are diagnosed, the majority of them already have metastases. Surgery combined with chemotherapy is a primary treatment for GC. 79 In chemotherapy for gastric cancer, platinum medicines—primarily cisplatin and oxaliplatin—are among the most frequently utilized therapeutic agents. 80
Small, single-stranded, noncoding RNAs are known as mRNAs. The formation and progression of various cancer types, including gastric cancer, ovarian cancer, T-cell lymphoma, and melanoma, have been linked to critical molecules participating in miRNA production, such as RNase III enzymes DROSHA, DICER, and RAN-GTPase (RAN).81–84
In cell growth and malignant transformation, glypican-3 (GPC3) is essential. 85 MiR-4510 is a tumor suppressor in some cancer types. In gastrointestinal stromal tumors, miR-4510 expression was lower in the tumor tissue than in the matched adjacent tissue. 86 MiR-4510 was found to be negatively linked with GPC3 mRNA and protein levels in samples of hepatocellular carcinoma (HCC). 87 By preventing the spread of gastric cancer cells and downregulating GPC3, MiR-4510 may modify immunosuppressive signals in the tumor microenvironment. Treatment with oxaliplatin may develop into a specialized therapeutic drug for patients with miR-4510 inhibition and glypican-3-expressing gastric cancer (Table 1). 63
It has been demonstrated that miR-130a can play a role in the pathogenesis of several malignancies, such as leukemia, ovarian cancer, glioblastoma, and cervical cancer.88–91 MiR-130a, which blocks GC cells from migrating and invading when overexpressed, targets the 3′-UTR of RAB5A. 92 Reduced miR-130a expression promotes cancer cell proliferation via overexpression of FOSL1 and RAB5A. Direct targets of miR-130a, which suppresses protein production and cell division, are FOSL1 and RAB5A. Together, oxaliplatinand miR-130a significantly inhibit cell invasion. 64
Among different forms of carcinoma, PHLPP2 increases apoptosis and slows the cell cycle. 93 MiR-15a-5p interacts with PHLPP2 and subsequently phosphorylates AKT to increase cell survival and decrease apoptosis, reducing the susceptibility of MGC803 cells to cisplatin. Additionally, there is a strong association between blood miR-15a-5p levels and patients’ responses to oxaliplatin-based chemotherapy. 65
When cells are exposed to a hostile environment, such as hypoxia, nutritional shortage, or drug toxicity, autophagy is a process that cells use to protect themselves.94,95 Autophagy can alter autophagy genes, microRNAs, TP53, the PI3K-AKT-mTOR and MAPK signaling pathways, or other mechanisms that control drug resistance.96,97 In gastric cancer cells resistant to oxaliplatin and 5-fluorouracil (5-Fu), the lncRNA EIF3J-DT gene increased the expression of ATG14. Chemotherapy resistance resulted from the sequestration of MIR188-3p by EIF3J-DT, which maintained ATG14 mRNA and prevented its degradation. 66
A decreased circ 0001546 in the GC is associated with a bad prognosis. Circ 0001546 targets miR-421, which targets ATM (ataxia telangiectasia mutated). Additionally, miR-421 overexpression inverts the circ 0001546 overexpression effect. Circ 0001546 reduces HGC-27 cells’ chemoresistance to oxaliplatin, possibly through activating the ATM/checkpoint kinase. 67
lncRNA MALAT1 is elevated in GC and oxaliplatin-resistant GC tissues and cells, indicating that MALAT1 was likely linked to drug resistance in GC. MiR-22-3p and zinc finger protein 91 (ZFP91) were possible targets for miR-22-3p, respectively. MALAT1 may control ZFP91 expression by competitively interacting with miR-22-3p to influence GC cell oxaliplatin resistance. 68
LncSLCO1C1 increases the tumor resistance to treatment with oxaliplatin by reducing GC DNA damage and tumor cell proliferation and migration, while lncSLCO1C1 enhances SSRP1 expression in GC cells by acting as a sponge to absorb both miR-211-5p and miR-204-5p in the cytoplasm. 69
3. microRNAs Regulating Cancer Cell Response to Oxaliplatin in Hepatocellular Carcinoma
Hepatocellular carcinomas (HCC) constitute ninety percent of primary liver tumors, one of the worst cancers in the world.98,99 Asian patients make up more than half of all cases of HCC worldwide. HCC has the world's second most common cancer fatality. 100 The main risk factors for HCC are hepatitis caused by infection with the hepatitis B and/or hepatitis C virus, aflatoxin B1 ingestion, high alcohol consumption, and smoking.99,101 Because of inadequate chemotherapy or a multidrug resistance (MDR) mechanism, patients with advanced stages of HCC frequently have poor prognoses and unfavorable clinical outcomes. 102 Some malignancies, including HCC, are undoubtedly influenced by miRNA dysfunction.103–105
MiR-31-5p has recently gained a lot of attention as a potential inhibitor or inducer of tumor growth in various cancers, including HCC. 106 Many studies report that miR-31-5p is the most repeatedly deleted miRNA in HCC. 107 The DNA repair mechanisms base excision repair (BER), homology-directed repair (HDR), and nonhomologous end-joining (NHEJ) all require poly ADP-ribose polymerase-1 (PARP1). 108 Multiple miR-31 binding sites in the 3′-UTR region of PARP1, a possible transcriptional negative regulator, may encourage the increase of lysosomal drug transport through ABCB9, suggesting the sequestration of oxaliplatin into intracellular compartments. 109 Through a protein-protein interaction between PARP1 and ABCB9, miR-31-5p mediates PARP1, which results in oxaliplatin resistance in HCC (Table 1). MiR-31-5p re-expression unexpectedly raised the relative intracellular aggregation of oxaliplatin even while it increased chemoresistance. 70
It has been demonstrated that miR-122, a liver-specific miRNA, is essential for controlling hepatocyte growth, differentiation, and cholesterol metabolism. 110 MiR-122 expression was revealed to be significantly downregulated in HCC tissues, and HCC formation and progression have both been linked to miR-122 downregulation.111,112 However, miR-122 is observed to be decreased in HCC. Its role in HCC chemosensitivity is yet unknown. The Wnt/β-catenin pathway is negatively regulated by miR122. MiR-122 upregulation or Wnt/β-catenin signaling suppression boosted the death of HCC cells and made them more sensitive to oxaliplatin.71,72
4. microRNAs Regulating Cancer Cell Response to Oxaliplatin in Ovarian Cancer
Ovarian cancer is one of the gynecologic malignancies and causes about 140,000 deaths per year among women worldwide. 113 This cancer often is diagnosed at advanced stages, which provides a slight chance of a cure. 114 Standard treatment for ovarian cancer is platinum-based first-line chemotherapies such as carboplatin, cisplatin, and oxaliplatin.114,115 Although mucinous ovarian cancer (MOC) treatment is known for its poor outcomes when treated with platinum-based drugs, the antitumor effect of oxaliplatin plus siPRKRA has shown notable results in MOC treatment. 115 Novel studies have shown that some miRNAs, including miR-199b-5p and miR-506, are clinical markers used in sensitizing the tumor cells toward ovarian cancer chemotherapy (Table 1).73,74 Drug resistance in patients toward chemotherapy is a multifactorial issue, and no single miRNA can help us predict the patient's outcome in treatment with chemotherapy. 116
5. microRNAs Regulating Cancer Cell Response to Oxaliplatin in Breast Cancer
Breast cancer is the first common cancer type worldwide. Approximately 2.3 million new cases of female breast cancer were diagnosed in 2020, with 685,000 deaths. 47 Various factors contribute to the development of breast cancer. Having a family history, late menopause onset, and using oral contraceptives are some factors. 117 Patients with breast cancer usually experience breast lumps, nipple discharge, skin changes, and enlarged lymph nodes. Mammography is the most important breast cancer detection tool, but other methods, such as breast ultrasound and MRI, can also be used. After diagnosis, patients often undergo surgery. Other treatment methods like radiation therapy and chemotherapy can be used before or after surgery depending on cancer's characteristics. 118
MiR-320c is a potential indicator of oxaliplatin responsiveness in patients with triple-negative breast cancer. In triple-negative breast cancer cells, miR-320c expression is downregulated, and this expression is strongly associated with poor overall survival rates. This association could be due to checkpoint kinase 1 (Chk1) overexpression. MiR-320c inhibits Chk1 expression in tissue and cell lines (P < 0.0001). MiR-320c mimic injection decreases Chk1 expression in triple-negative breast cancer cell lines and improves their responsiveness to oxaliplatin. Additionally, in mice, upregulating miR-320c can reduce tumor volume and weight by downregulating Chk1 expression. Reduced Chk1 expression can lead to increased apoptosis and DNA damage accumulation. In 2020, Lim et al treated triple-negative breast cancer cell lines with oxaliplatin and miR-320c mimics. They demonstrated that increased miR-320c expression caused oxaliplatin-induced apoptosis and confirmed that the effects of miR-320c on apoptosis were due to reduced Chk1 expression (Table 1). They also discovered that Chk1 expression was suppressed, and DNA damage was increased in triple-negative breast cancer cell lines treated with overexpressed miR-320c and oxaliplatin. They reported that induced DNA damage successfully activated Chk1 in triple-negative breast cancer cells after oxaliplatin treatment. Chk1 activation prevented RAD51 binding to the DNA damage site for DNA repair and damage-induced apoptosis. However, miR-320c injection prevented the synthesis of sufficient amounts of Chk1. Therefore, miR-320c-upregulated triple-negative breast cancer cells showed increased apoptosis and reduced RAD51 recruitment, which caused a reduction in DNA repair and an escalation in DNA damage. Taken together, the upregulation of miR-320c can improve the sensitivity of triple-negative breast cancer cells to oxaliplatin in vitro and in vivo. 75
MiR-525-5p is a key element in oxaliplatin resistance in breast cancer. Breast cancer cell lines exhibit downregulated miR-525-5p, which is linked to circFAT1. Circular RNA FAT atypical cadherin 1, also known as circFAT1, is essential for developing chemoresistance to platinum-based drugs. The expression of circFAT1 is elevated in oxaliplatin-resistant breast cancer tissues and cell lines. While inhibiting circFAT1 expression, it declines the resistance of breast cancer cells to oxaliplatin. Other effects of circFAT1 silencing include enhanced apoptosis and inhibited metastatic characteristics such as cell migration and invasion in oxaliplatin-resistant breast cancer tissues. 76
According to Yao et al in 2021, circFAT1 directly targets miR-525-5p in oxaliplatin-resistant breast cancer cells to promote metastasis, reduce apoptosis, and worsen oxaliplatin resistance. Their findings showed a negative association between the expressions of circFAT1 and miR-525-5p from oxaliplatin-resistant breast cancer cells. They verified that SKA1 was directly targeted by miR-525-5p and that the expression of SKA1 in breast cancer cells was inversely linked with miR-525-5p. They showed that circFAT1 inhibition reduced SKA1 abundance while miR-525-5p knockdown significantly raised the SKA1 level. Moreover, they discovered that SKA1 silencing reduced the expression of the Notch2, GSK-3, and β-catenin proteins in breast cancer cells, demonstrating that SKA1 expression in breast cancer is positively correlated with the Notch and Wnt signaling pathways. 76
6. microRNAs Regulating Cancer Cell Response to Oxaliplatin in Esophageal Cancer
Sixth among all cancers that cause death globally is esophageal cancer.119,120 Esophageal cancer is responsible for over than 500,000 deaths globally each year, which accounts for 5.3% of all cancer fatalities worldwide. 121 In the USA, esophageal adenocarcinoma (EAC) is one of the most common kind of cancer. 122 As esophageal cancer incidence rises with age, and many diagnosed patients are over 60. 123
The only known precursor for esophageal adenocarcinoma is Barrett's esophagus, which undergoes a metaplastic transition from the typical squamous mucosa of the esophagus to a columnar lining. Patients with Barrett's esophagus have a 30- to 40-fold more significant risk of developing esophageal adenocarcinoma.124,125 The consumption of polyunsaturated fat, omega-3 fatty acids, total fiber, dietary vitamin C, fiber from fruits and vegetables, and beta-carotene, along with vitamin E, has all been reported to be related to a lessened risk of being diagnosed with Barrett's esophagus. Smoking, GERD, and ethanol are risk factors associated with Barrett's esophagus and esophageal adenocarcinoma.126–130 Early detection of EC (EC restricted to the mucosa or superficial submucosa) and endoscopic or surgical treatment can significantly increase the 5-year survival rate to more than 90%.131,132
In order to treat esophageal cancer, multimodality neoadjuvant concurrent chemoradiotherapy (CCRT) is used. 133 Despite the widespread use of adjuvant chemotherapy, such as 5-FU and oxaliplatin, patient survival rates have not improved over time, and recurring patients are found to have acquired drug resistance. 134 Unfortunately, advanced esophageal cancer is typically discovered when surgery alone cannot cure it. 135
To understand how MiR-141-3p modulates acquired chemoresistance, MiR-141-3p inhibitor or mimic was transfected into EC9706R cells. A miR-141-3p inhibitor transfection of EC9706 significantly decreased the level of miR-141-3p. In line with this, suppression of miR141-3p resulted in a larger proportion of apoptosis when compared to control cells, suggesting that miR-141-3p is an essential mediator of chemoresistance in these cells. The cells that had been transfected with the miR-141-3p inhibitor were much more sensitive to oxaliplatin, with a drop in IC50 of roughly 4-fold, respectively. 77
7. The Role of microRNAs in Oxaliplatin-Induced Neuropathic Pain
Many diseases can damage the central or peripheral nervous system and lead to neuropathic pain. One of the influential generative factors that causes chronic neuropathic pain is cancer and the side effects of related drugs. According to research, the severity of neuropathic pain differs based on the type and site of the cancer. A study considered cancer patients in 5 years window (2006 to 2011). About 35% of patients had moderate pain, and 46% had severe neuropathic pain. In a neoplastic disease like hematologic cancer, pain intensity was between 20 to 80%.136,137 In another study, on 2301 patients, 8.8% presented severe neuropathic pain in their 3-month examinations. 138
Oxaliplatin is one of the cancer-related drugs used in treating colon and rectal cancer that can lead to neuropathic pain. 139 The main mechanism of neuropathic pain is under debate. In vivo studies have suggested several mechanisms for neuropathic pain, but these mechanisms in humans may differ completely. Anterior neurons have a significant role in these mechanisms. These neurons may increase the number of neurotransmitters and sodium channels that can lead to spontaneous and movement-related pain. On the other hand, inhibitory circuits’ disorder in the dorsal horn or brain stem can lead to neuropathic pain. Generally, three primary mechanisms have been suggested for neuropathic pain: (1) peripheral, (2) central sensitization, and (3) denervation. Structural and functional damage to peripheral and central nerves can lead to neuropathic pain. 140
microRNAs have a significant role in the pain system. Noxious stimuli correlated with chemical stimuli affected miRNA expressions and pain behaviors acutely. In the following, we will examine the number of these examples. microRNAs were detected in nearly all dorsal root ganglions. 141 Other trials proved that miRNA-1 and miRNA-16 were decreased after Freund's adjuvant (CFA) injection and partial sciatic nerve ligation. Other trials showed that several genes related to pain are predicted to be targets of miR-143. 142
According to recent studies, the use of microRNA is practical in the treatment of neuropathic pain. In clinical practice, dorsal root ganglion (DRG) and trigeminal ganglion (TG) neurons are the principal origins of neuropathic pain. 143 However, the miRNAs and deep-sequencing analyses showed significant changes in miRNA expressions in the DRG after nerve injuries.144,145 Nerve injury also changes the miRNA expressions in the proximal and distal sciatic nerves, which leads to DRG microRNA expressions. Next to microRNA-143, microRNA-7a has a significant effect on neuropathic pain. In DRG neurons, this microRNA regulates neuronal excitability. 146 According to the studies, miR-96 and miR-183 suppress neuropathic pain induced by traumatic peripheral nerve injury.147,148 Autophagy was found in the spinal cord caused by nerve injury. MiRNA-195 acts as a mediator in neuroinflammation by regulating autophagy, which happens due to nerve injury in the spinal cord and leads to neuropathic pain. 149 MiRNAs amount and expression changes in cancer. As confirmed by Bali KK et al’s (2013) study, the number of related miRNAs to neuropathic pain in dorsal root ganglion neurons changed more than 2.5-fold. It showed a significant effect of neuropathic pain on the expression of several miRNAs. 150
Neuronal damage significantly affects transmitters, ionic channels, and proteins. These molecular changes can lead to neuropathic pain. 151 According to animal trials, neuropathic pain leads to changes in specific microRNAs. MiR-21 expression increases during different types of peripheral nerve injury.144,152 In Sakai and Suzuki's study, mechanical allodynia and thermal hyperalgesia were reduced after miR-21 inhibitor injection. miR-21 is specifically upregulated in the injured DRG. 149 The destruction of micRNA-124a leads to nociceptive behavior in mice. On the other hand, an increment of miR-124a can lead to nociceptive behavior reduction. 153 MiR-143 is expressed in nociceptive neurons. It could selectively participate in miRNA regulation in specific populations of nociceptors. 154 miR-103 has a significant role in pain relief by regulating the L-type calcium channel in the spinal dorsal horn. 155 MiR-15b increased in chronic neuropathic pain. In the research of Sakai and colleagues, in the first 5 days of oxaliplatin injection, the level of mir-15b expression was raised. After 28 days of this test, there were significant changes in this mir-15b expression. Overexpression of mir-15b can lead to allodynia (a type of neuropathic pain). In further research, beta-site amyloid precursor protein-cleaving enzyme 1 (BACE1) has an essential effect on neuropathic pain. After oxaliplatin injection, miRNA-15b upregulated. This event can affect BACE1 genes and lead to the downregulation of BACE1. BACE1 participates in the myelination and nociception process. Conversely, BACE1 modulates the expression of TNF receptor 1. This receptor is a critical mediator of the inflammatory process. Downregulation of BACE1 by miRNA-15b significantly affects BACE1 tasks like the neuropathy process. This study can demonstrate that the proper dose of inhibitors and enzymes can be used as neuropathic pain drugs. 156
One of the essential miRNAs in neuropathic pain is miRNA-30b. According to several studies, it can inhibit the overexpression of miRNAs caused by oxaliplatin. It can reduce neuropathic pain by stopping its mechanism and pain-related RNA expression. On the other hand, miRNA-30b can relieve oxaliplatin-induced neuropathic pain behaviors in rats. 157
MiRNA-155 has a significant effect on the regulation of inflammation-related disease. According to animal trials, injection of oxaliplatin can lead to cold hypersensitivity in rats. It can affect specific protein expressions like transient receptor potential ankyrin 1 (TRAP1). In another way, oxaliplatin can have an oxidative stress-TRAP1 pathway disorder effect. Inhibiting miRNA-155 can lead to this pathway improvement and reduction of neuropathic pain. Conversely, inhibiting NADPH oxidants like NADPH oxidative 4 can reduce products of oxidative stress in the dorsal horn and reduce neuropathic pain. 158 Similar to the inhibition of miRNA-155, inhibiting miRNA-195 can have the same effect. MiRNA-195 contributes to neuropathic pain by increasing neuro-inflammation. This process occurs by affecting neuro-inflammation factors like ATG14. This factor has an essential effect on the neuroinflammatory process.159,160
MiRNA treatment as new potential therapy can have a significant role in future neuropathic treatment. Although many studies proved that microRNA regulation has an important effect on nociceptive stimulators, further studies needed to make these small RNAs into practical treatment tools.
8. Conclusion
In the current paper, we have reviewed the role of microRNAs in regulating cancer cells’ response to oxaliplatin, with particular attention to GI cancers. We also discussed the role of these noncoding RNAs in the pathophysiology of oxaliplatin-induced neuropathic pain. Although cancerous cells and tissues use perplexing molecular mechanisms to suppress the antitumor effect of chemotherapeutics, we hope that our work can lead clinicians to a better understanding and efficient chemotherapy.
Acknowledgments
All figures are created using BioRender.com.
Abbreviations
- ABCB9
ATP binding cassette subfamily B member 9
- AKT
protein kinase B
- AMPK
Adenosine monophosphate-activated protein kinase
- ASK1
Apoptosis signal-regulating kinase 1
- ATG14
Autophagy-related Gene 14
- ATM
Ataxia Telangiectasia Mutated
- BACE1
Beta-site amyloid precursor protein cleaving enzyme 1
- BCL-2
B-cell lymphoma 2
- BER
Base excision repair
- BIN1
Myc boxdependent-interacting protein 1
- BIM
Bcl-2-like protein 11
- CAB39
Calcium-binding protein 39
- CCRT
Concurrent chemoradiotherapy
- CD4
Cluster of differentiation 4
- CFA
Complete Freund's Adjuvant
- Chk1
Checkpoint kinase 1
- CircRNAs
Circular RNAs
- DICER
endoribonuclease Dicer
- DNA
Deoxyribonucleic acid
- DRG
Dorsal root ganglion neurons
- DROSHA
Drosha Ribonuclease III
- EAC
Esophageal adenocarcinoma
- EC
Esophageal cancer
- EC9706
Esophageal cancer cell line
- EC9706R
5-fluorouracil and oxaliplatin resistant subline cells
- EGF
Epidermal growth factor
- ERK
Extracellular signal-regulated kinase
- FAT1
FAT atypical cadherin 1
- FOSL1
Fos-related antigen 1
- FOX
Forkhead box proteins family
- FOXC2
Forkhead transcription factor C2
- FOXQ1
Forkhead box Q1
- GC
Gastric cancer
- GERD
Gastroesophageal reflux disease
- GI
Gastrointestinal
- GPC3
glypican-3
- GSK-3
Glycogen synthase kinase 3
- gw4869
A neutral, noncompetitive inhibitor of sphingomyelinase
- HCC
Hepatocellular carcinoma
- HDR
Homology-directed repair
- HSF1
Heat shock factor 1
- JNK
Jun N-terminal kinase
- LDH
Lactate dehydrogenase
- lncRNA
Long noncoding RNAs
- MALAT1
Metastasis Associated Lung Adenocarcinoma Transcript 1
- MAPK
Mitogen-activated protein kinase
- MDR
Multidrug Resistance
- MEGF6
Multiple EGF Like Domains 6
- miR
microRNA
- miRNAs
microRNAs
- MOC
Mucinous ovarian cancer
- MMP-2
Matrix metalloproteinase-2
- MRI
Magnetic resonance imaging
- mRNA
messenger ribonucleic acid
- mTOR
mammalian target of rapamycin
- MUL1
Mitochondrial E3 ubiquitin protein ligase 1
- NADPH
Nicotinamide Adenine Dinucleotide Phosphate Hydrogen
- ncRNAs
non-coding RNAs
- NHEJ
Nonhomologous end joining
- Notch2
Neurogenic locus notch homolog protein 2
- Noxa
Phorbol-12-myristate-13-acetate-induced protein 1
- nSMase2
Neutral sphingomyelinase-2
- OCR
Oxygen Consumption Rate
- PARP1
Poly (ADP-ribose) polymerase 1
- PDCD4
Programmed cell death protein 4
- PDCD10
Programmed cell death protein 10
- PDK1
3-Phosphoinositide-dependent kinase 1
- PFS
Progression-free survival
- PHLPP2
PH domain leucine-rich repeat protein phosphatase 2
- PI3K
Phosphoinositide 3-kinases
- PTEN
Phosphatase and tensin homologue deleted on chromosome ten
- RAB5A
Ras-related protein Rab-5A
- RAD51
RAD51 recombinase
- RAN
Ras-related nuclear protein
- RNA
Ribonucleic acid
- ROS
Reactive oxygen species
- siPRKRA
PRKRA Small Interfering RNA
- SKA1
Spindle and kinetochore associated protein 1
- SMAD4
SMAD family member 4
- SSRP1
Structure specific recognition protein 1
- TG
Trigeminal ganglion
- TP53
Tumor protein p53
- TRAP1
Transient receptor potential ankyrin 1
- ULK1
Unc-51-like autophagy-activating kinases 1
- US
United States of America
- USP49
Ubiquitin specific peptidase 49
- VEGFA
Vascular endothelial growth factor A
- Wnt
Wingless-related integration site
- XIAP
X-Linked Inhibitor of Apoptosis
- ZFP91
Zinc finger protein 91
- 3′-UTR
3'-untranslated region
- 5-FU
5-fluorouracil
Footnotes
Data Availability Statement: The data that supports the findings of this study are available from the corresponding author upon reasonable request.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iDs: Ali Tavakoli Pirzaman https://orcid.org/0000-0002-9426-7034
Sohrab Kazemi https://orcid.org/0000-0002-8068-5745
References
- 1.Dolan ME, El Charif O, Wheeler HE, et al. Clinical and genome-wide analysis of cisplatin-induced peripheral neuropathy in survivors of adult-onset cancer. Clin Cancer Res. 2017;23(19):5757-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang J, Zhao Y, Xu Y, et al. Comparative effectiveness and safety between oxaliplatin-based and cisplatin-based therapy in advanced gastric cancer: A meta-analysis of randomized controlled trials. Oncotarget. 2016;7(23):34824-34831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raymond E, Faivre S, Chaney S, Woynarowski J, Cvitkovic E. Cellular and molecular pharmacology of oxaliplatin. Mol Cancer Ther. 2002;1(3):227-235. [PubMed] [Google Scholar]
- 4.Cavaletti G, Marmiroli P. Management of oxaliplatin-induced peripheral sensory neuropathy. Cancers (Basel). 2020;12(6):1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang L, Tian Y, Xu S, Chen H. Oxaliplatin-induced peripheral neuropathy: Clinical features, mechanisms, prevention and treatment. J Neurol. 2021;268(9):3269-3282. [DOI] [PubMed] [Google Scholar]
- 6.Wolf S, Barton D, Kottschade L, Grothey A, Loprinzi C. Chemotherapy-induced peripheral neuropathy: Prevention and treatment strategies. Eur J Cancer. 2008;44(11):1507-1515. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C, Xu C, Gao X, Yao Q. Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics. 2022;12(5):2115-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothenberg ML. Efficacy of oxaliplatin in the treatment of colorectal cancer. Oncology (Williston Park). 2000;14(12 Suppl 11):9-14. [PubMed] [Google Scholar]
- 9.Misset JL, Bleiberg H, Sutherland W, Bekradda M, Cvitkovic E. Oxaliplatin clinical activity: A review. Crit Rev Oncol Hematol. 2000;35(2):75-93. [DOI] [PubMed] [Google Scholar]
- 10.Pouessel D, Huguet H, Iborra F, et al. A pilot study of gemcitabine in combination with oxaliplatin and vinorelbine in patients with metastatic bladder cancer. Anticancer Res. 2010;30(11):4711-4715. [PubMed] [Google Scholar]
- 11.Takahashi H, Morizane C, Nomura S, et al. 731PPhase II clinical trial of gemcitabine plus oxaliplatin combination therapy (GEMOX) in patients with advanced pancreatic adenocarcinoma with a family history of pancreatic/breast/ovarian/prostate cancer or personal history of breast/ovarian/prostate cancer (FABRIC study). Ann Oncol. 2018;29(10):viii248-viviii9. [Google Scholar]
- 12.Zhang F, Zhang Y, Jia Z, Wu H, Gu K. Oxaliplatin-based regimen is superior to cisplatin-based regimen in tumour remission as first-line chemotherapy for advanced gastric cancer: A meta-analysis. J Cancer. 2019;10(8):1923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alcindor T, Beauger N. Oxaliplatin: A review in the era of molecularly targeted therapy. Curr Oncol. 2011;18(1):18-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tesniere A, Schlemmer F, Boige V, et al. Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene. 2010;29(4):482-491. [DOI] [PubMed] [Google Scholar]
- 15.Jonsson H, Peng SL. Forkhead transcription factors in immunology. Cell Mol Life Sci. 2005;62(4):397-409. [DOI] [PubMed] [Google Scholar]
- 16.Bach DH, Long NP, Luu TT, Anh NH, Kwon SW, Lee SK. The Dominant Role of Forkhead Box Proteins in Cancer. Int J Mol Sci. 2018;19(10):3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Deng G, Fu Y, et al. FOXC2 promotes oxaliplatin resistance by inducing epithelial-mesenchymal transition via MAPK/ERK signaling in colorectal cancer. Onco Targets Ther. 2020;13:1625-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hector S, Bolanowska-Higdon W, Zdanowicz J, Hitt S, Pendyala L. In vitro studies on the mechanisms of oxaliplatin resistance. Cancer Chemother Pharmacol. 2001;48(5):398-406. [DOI] [PubMed] [Google Scholar]
- 19.Na C, Li X, Zhang J, Han L, Li Y, Zhang H. miR-107 targets TRIAP1 to regulate oral squamous cell carcinoma proliferation and migration. Int J Clin Exp Pathol. 2019;12(5):1820-1825. [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Y, Zhu D, Hou L, et al. MiR-107 confers chemoresistance to colorectal cancer by targeting calcium-binding protein 39. Br J Cancer. 2020;122(5):705-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng Y, Li Y, Fang Q, Luo H, Zhu G. microRNA-744 is downregulated in glioblastoma and inhibits the aggressive behaviors by directly targeting NOB1. Am J Cancer Res. 2018;8(11):2238-2253. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y, He A, Zhang L, Yi G. MiR-744 mediates the Oxaliplatin chemoresistance in colorectal cancer through inhibiting BIN1. Neoplasma. 2020;67(2):296-303. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Zhu M. Novel exosomal miR-46146 transfer oxaliplatin chemoresistance in colorectal cancer. Clin Transl Oncol. 2020;22(7):1105-1116. [DOI] [PubMed] [Google Scholar]
- 24.Jiang T, Ye L, Han Z, et al. miR-19b-3p promotes colon cancer proliferation and oxaliplatin-based chemoresistance by targeting SMAD4: Validation by bioinformatics and experimental analyses. J Exp Clin Cancer Res. 2017;36(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Balibrea E, Martínez-Cardús A, Ginés A, et al. Tumor-Related molecular mechanisms of oxaliplatin resistance. Mol Cancer Ther. 2015;14(8):1767-1776. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Gan Y, Liu J, et al. Downregulation of MEIS1 mediated by ELFN1-AS1/EZH2/DNMT3a axis promotes tumorigenesis and oxaliplatin resistance in colorectal cancer. Signal Transduct Target Ther. 2022;7(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohitesh K, Chowdhury R, Mukherjee S. Resistance a major hindrance to chemotherapy in hepatocellular carcinoma: An insight. Cancer Cell Int. 2018;18:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo M, Yang X, Chen HN, Nice EC, Huang C. Drug resistance in colorectal cancer: An epigenetic overview. Biochim Biophys Acta Rev Cancer. 2021;1876(2):188623. [DOI] [PubMed] [Google Scholar]
- 29.Yan D, Tu L, Yuan H, et al. WBSCR22 confers oxaliplatin resistance in human colorectal cancer. Sci Rep. 2017;7(1):15443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dahariya S, Paddibhatla I, Kumar S, Raghuwanshi S, Pallepati A, Gutti RK. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol Immunol. 2019;112:82-92. [DOI] [PubMed] [Google Scholar]
- 31.Ngo C, Kothary R. MicroRNAs in oligodendrocyte development and remyelination. J Neurochem. 2022;162(4):310-321. [DOI] [PubMed] [Google Scholar]
- 32.Vieira MS, Santos AK, Vasconcellos R, et al. Neural stem cell differentiation into mature neurons: Mechanisms of regulation and biotechnological applications. Biotechnol Adv. 2018;36(7):1946-1970. [DOI] [PubMed] [Google Scholar]
- 33.Kalayinia S, Arjmand F, Maleki M, Malakootian M, Singh CP. MicroRNAs: Roles in cardiovascular development and disease. Cardiovasc Pathol. 2021;50:107296. [DOI] [PubMed] [Google Scholar]
- 34.Porrello ER. microRNAs in cardiac development and regeneration. Clin Sci (Lond). 2013;125(4):151-166. [DOI] [PubMed] [Google Scholar]
- 35.Tüfekci KU, Oner MG, Meuwissen RL, Genç S. The role of microRNAs in human diseases. Methods Mol Biol. 2014;1107:33-50. [DOI] [PubMed] [Google Scholar]
- 36.Van Roosbroeck K, Fanini F, Setoyama T, et al. Combining anti-mir-155 with chemotherapy for the treatment of lung cancers. Clin Cancer Res. 2017;23(11):2891-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123(1):150-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mehlich D, Garbicz F, Włodarski PK. The emerging roles of the polycistronic miR-106b∼25 cluster in cancer - A comprehensive review. Biomed Pharmacother. 2018;107:1183-1195. [DOI] [PubMed] [Google Scholar]
- 39.Puhr M, Hoefer J, Schäfer G, et al. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. Am J Pathol. 2012;181(6):2188-2201. [DOI] [PubMed] [Google Scholar]
- 40.Qin D, Li H, Xie H. Ultrasound-targeted microbubble destruction-mediated miR-205 enhances cisplatin cytotoxicity in prostate cancer cells. Mol Med Rep. 2018;18(3):3242-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riquelme I, Letelier P, Riffo-Campos AL, Brebi P, Roa JC. Emerging role of miRNAs in the drug resistance of gastric cancer. Int J Mol Sci. 2016;17(3):424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu N, Shen C, Luo Y, et al. Upregulated miR-130a increases drug resistance by regulating RUNX3 and Wnt signaling in cisplatin-treated HCC cell. Biochem Biophys Res Commun. 2012;425(2):468-472. [DOI] [PubMed] [Google Scholar]
- 43.Shi L, Chen Z-G, Wu L-L, et al. miR-340 reverses cisplatin resistance of hepatocellular carcinoma cell lines by targeting Nrf2-dependent antioxidant pathway. Asian Pac J Cancer Prev. 2015;15(23):10439-10444. [DOI] [PubMed] [Google Scholar]
- 44.Qin J, Luo M, Qian H, Chen W. Upregulated miR-182 increases drug resistance in cisplatin-treated HCC cell by regulating TP53INP1. Gene. 2014;538(2):342-347. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Huang J-W, Calses P, Kemp CJ, Taniguchi T. MiR-96 downregulates REV1 and RAD51 to promote cellular sensitivity to cisplatin and PARP InhibitionMiR-96 in chemosensitivity. Cancer Res. 2012;72(16):4037-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu N, Zhang J, Shen C, et al. Cisplatin-induced downregulation of miR-199a-5p increases drug resistance by activating autophagy in HCC cell. Biochem Biophys Res Commun. 2012;423(4):826-831. [DOI] [PubMed] [Google Scholar]
- 47.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249. [DOI] [PubMed] [Google Scholar]
- 48.Hossain MS, Karuniawati H, Jairoun AA, et al. Colorectal cancer: A review of carcinogenesis, global epidemiology, current challenges, risk factors, preventive and treatment strategies. Cancers (Basel). 2022;14(7):1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: A review. Jama. 2021;325(7):669-685. [DOI] [PubMed] [Google Scholar]
- 50.Buccafusca G, Proserpio I, Tralongo AC, Rametta Giuliano S, Tralongo P. Early colorectal cancer: Diagnosis, treatment and survivorship care. Crit Rev Oncol Hematol. 2019;136:20-30. [DOI] [PubMed] [Google Scholar]
- 51.National Health Commission Of The People's Republic Of C. National guidelines for diagnosis and treatment of colorectal cancer 2020 in China (English version). Chin J Cancer Res. 2020;32(4):415-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Liu X, Zhang J, et al. Inhibition of miR-19a partially reversed the resistance of colorectal cancer to oxaliplatin via PTEN/PI3K/AKT pathway. Aging (Albany NY). 2020;12(7):5640-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu YY, Yu J, Zhang JF, Wang C. Suppressing the secretion of exosomal miR-19b by gw4869 could regulate oxaliplatin sensitivity in colorectal cancer. Neoplasma. 2019;66(1):39-45. [DOI] [PubMed] [Google Scholar]
- 54.Liu Z, Qin Y, Dong S, Chen X, Huo Z, Zhen Z. Overexpression of miR-106a enhances oxaliplatin sensitivity of colorectal cancer through regulation of FOXQ1. Oncol Lett. 2020;19(1):663-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin Y, Li L, Wang F, et al. Knockdown of Mir-135b sensitizes colorectal cancer cells to oxaliplatin-induced apoptosis through increase of FOXO1. Cell Physiol Biochem. 2018;48(4):1628-1637. [DOI] [PubMed] [Google Scholar]
- 56.Wang H, Wang X, Zhang H, et al. The HSF1/miR-135b-5p axis induces protective autophagy to promote oxaliplatin resistance through the MUL1/ULK1 pathway in colorectal cancer. Oncogene. 2021;40(28):4695-4708. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Zhang D, Li Y, Fang F. MiR-138 suppresses the PDK1 expression to decrease the oxaliplatin resistance of colorectal cancer. Onco Targets Ther. 2020;13:3607-3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He Y, Zhang L, Tan F, et al. MiR-153-5p promotes sensibility of colorectal cancer cells to oxaliplatin via targeting Bcl-2-mediated autophagy pathway. Biosci Biotechnol Biochem. 2020;84(8):1645-1651. [DOI] [PubMed] [Google Scholar]
- 59.Ning T, Li J, He Y, et al. Exosomal miR-208b related with oxaliplatin resistance promotes Treg expansion in colorectal cancer. Mol Ther. 2021;29(9):2723-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qian XL, Zhou F, Xu S, et al. MiR-454-3p promotes oxaliplatin resistance by targeting PTEN in colorectal cancer. Front Oncol. 2021;11:638537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mou Y, He N, Su M, et al. MiR-1254 and MEGF6 regulates oxaliplatin resistance in human colorectal cancer cells. Am J Transl Res. 2021;13(1):183-196. [PMC free article] [PubMed] [Google Scholar]
- 62.Zhuang YY, Zhong W, Xia ZS, et al. miR-5000-3p confers oxaliplatin resistance by targeting ubiquitin-specific peptidase 49 in colorectal cancer. Cell Death Discov. 2021;7(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma H-F, Shu P, Shi X-H, Wang M, Jiang M-F. Identification of miR-4510 as a metastasis suppressor of gastric cancer through regulation of tumor microenvironment via targeting GPC3. Clin Exp Metastasis. 2022;39(2):363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang F, Sun Y, Shi J. Programmed death-ligand 1 monoclonal antibody-linked immunoliposomes for synergistic efficacy of miR-130a and oxaliplatin in gastric cancers. Nanomedicine. 2019;14(13):1729-1744. [DOI] [PubMed] [Google Scholar]
- 65.Pang K, Song J, Bai Z, Zhang Z. miR-15a-5p targets PHLPP2 in gastric cancer cells to modulate platinum resistance and is a suitable serum biomarker for oxaliplatin resistance. Neoplasma. 2020;67(5):1114-1121. [DOI] [PubMed] [Google Scholar]
- 66.Luo Y, Zheng S, Wu Q, et al. Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy. 2021;17(12):4083-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu Q, Wang H, Liu L, Zhu K, Yu W, Guo J. Hsa_circ_0001546 acts as a miRNA-421 sponge to inhibit the chemoresistance of gastric cancer cells via ATM/Chk2/p53-dependent pathway. Biochem Biophys Res Commun. 2020;521(2):303-309. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Z, Li M, Zhang Z. lncRNA MALAT1 modulates oxaliplatin resistance of gastric cancer via sponging miR-22-3p. Onco Targets Ther. 2020;13:1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao YF, Li BS, Liu JJ, et al. Role of lncSLCO1C1 in gastric cancer progression and resistance to oxaliplatin therapy. Clin Transl Med. 2022;12(4):e691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Que KT, Zhou Y, You Y, et al. Retraction note: MicroRNA-31-5p regulates chemosensitivity by preventing the nuclear location of PARP1 in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu J, Zhu X, Wu L, et al. Micro RNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting wnt/β-catenin pathway. Liver Int. 2012;32(5):752-760. [DOI] [PubMed] [Google Scholar]
- 72.Cao F, Yin L-X. miR-122 enhances sensitivity of hepatocellular carcinoma to oxaliplatin via inhibiting MDR1 by targeting Wnt/β-catenin pathway. Exp Mol Pathol. 2019;106:34-43. [DOI] [PubMed] [Google Scholar]
- 73.Liu MX, Siu MK, Liu SS, Yam JW, Ngan HY, Chan DW. Epigenetic silencing of microRNA-199b-5p is associated with acquired chemoresistance via activation of JAG1-Notch1 signaling in ovarian cancer. Oncotarget. 2014;5(4):944-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu G, Yang D, Rupaimoole R, et al. Augmentation of response to chemotherapy by microRNA-506 through regulation of RAD51 in serous ovarian cancers. J Natl Cancer Inst. 2015;107(7):djv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lim S, Kim Y, Lee SB, et al. Inhibition of Chk1 by miR-320c increases oxaliplatin responsiveness in triple-negative breast cancer. Oncogenesis. 2020;9(10):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao Y, Li X, Cheng L, Wu X, Wu B. Circular RNA FAT atypical cadherin 1 (circFAT1)/microRNA-525-5p/spindle and kinetochore-associated complex subunit 1 (SKA1) axis regulates oxaliplatin resistance in breast cancer by activating the notch and Wnt signaling pathway. Bioengineered. 2021;12(1):4032-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin Y-Y, Chen Q-J, Xu K, et al. Involvement of microRNA-141-3p in 5-fluorouracil and oxaliplatin chemo-resistance in esophageal cancer cells via regulation of PTEN. Mol Cell Biochem. 2016;422(1):161-170. [DOI] [PubMed] [Google Scholar]
- 78.Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumor Biol. 2017;39(7):1010428317714626. [DOI] [PubMed] [Google Scholar]
- 79.Räihä MR, Puolakkainen PA. Tumor-associated macrophages (TAMs) as biomarkers for gastric cancer: A review. Chron Dis Transl Med. 2018;4(3):156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33(1):9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma Z, Swede H, Cassarino D, Fleming E, Fire A, Dadras SS. Up-regulated dicer expression in patients with cutaneous melanoma. PloS one. 2011;6(6):e20494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359(25):2641-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valencak J, Schmid K, Trautinger F, et al. High expression of dicer reveals a negative prognostic influence in certain subtypes of primary cutaneous T cell lymphomas. J Dermatol Sci. 2011;64(3):185-190. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J, Zhang X-H, Wang C-X, et al. Dysregulation of microRNA biosynthesis enzyme dicer plays an important role in gastric cancer progression. Int J Clin Exp Pathol. 2014;7(4):1702. [PMC free article] [PubMed] [Google Scholar]
- 85.Wei X, Zheng W, Tian P, et al. Oncogenic hsa_circ_0091581 promotes the malignancy of HCC cell through blocking miR-526b from degrading c-MYC mRNA. Cell Cycle. 2020;19(7):817-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen Y, Qin C, Cui X, Geng W, Xian G, Wang Z. Mir-4510 acts as a tumor suppressor in gastrointestinal stromal tumor by targeting APOC2. J Cell Physiol. 2020;235(7–8):5711-5721. [DOI] [PubMed] [Google Scholar]
- 87.Cartier F, Indersie E, Lesjean S, et al. New tumor suppressor microRNAs target glypican-3 in human liver cancer. Oncotarget. 2017;8(25):41211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang J, Wu H, Li P, Zhao Y, Liu M, Tang H. NF-κB-modulated miR-130a targets TNF-α in cervical cancer cells. J Transl Med. 2014;12(1):1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xa Z, Huang L, Zhao Y, Tan W. Downregulation of miR-130a contributes to cisplatin resistance in ovarian cancer cells by targeting X-linked inhibitor of apoptosis (XIAP) directly. Acta Biochim Biophys Sin. 2013;45(12):995-1001. [DOI] [PubMed] [Google Scholar]
- 90.Qiu S, Lin S, Hu D, Feng Y, Tan Y, Peng Y. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J Transl Med. 2013;11(1):1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kovaleva V, Mora R, Park YJ, et al. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012;72(7):1763-1772. [DOI] [PubMed] [Google Scholar]
- 92.Chen X, Zhao M, Huang J, et al. MicroRNA-130a suppresses breast cancer cell migration and invasion by targeting FOSL1 and upregulating ZO-1. J Cell Biochem. 2018;119(6):4945-4956. [DOI] [PubMed] [Google Scholar]
- 93.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP And a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25(6):917-931. [DOI] [PubMed] [Google Scholar]
- 94.Singletary K, Milner J. Diet, autophagy, and cancer: A review. Cancer Epidemiol Biomark Prev. 2008;17(7):1596-1610. [DOI] [PubMed] [Google Scholar]
- 95.Ouyang L, Shi Z, Zhao S, et al. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45(6):487-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kumar P, Zhang D-M, Degenhardt K, Chen Z-S. Autophagy and transporter-based multi-drug resistance. Cells. 2012;1(3):558-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu Q, Yang Z, Nie Y, Shi Y, Fan D. Multi-drug resistance in cancer chemotherapeutics: Mechanisms and lab approaches. Cancer Lett. 2014;347(2):159-166. [DOI] [PubMed] [Google Scholar]
- 98.Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941-1953. [DOI] [PubMed] [Google Scholar]
- 99.Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. [DOI] [PubMed] [Google Scholar]
- 100.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359-EE86. [DOI] [PubMed] [Google Scholar]
- 101.Craig P, Young S, Golzarian J. Current trends in the treatment of hepatocellular carcinoma with transarterial embolization: Variability in technical aspects. Cardiovasc Intervent Radiol. 2019;42(9):1322-1328. [DOI] [PubMed] [Google Scholar]
- 102.Ma J, Zeng S, Zhang Y, et al. BMP4 Promotes oxaliplatin resistance by an induction of epithelial-mesenchymal transition via MEK1/ERK/ELK1 signaling in hepatocellular carcinoma. Cancer Lett. 2017;411:117-129. [DOI] [PubMed] [Google Scholar]
- 103.Callegari E, Gramantieri L, Domenicali M, D'abundo L, Sabbioni S, Negrini M. MicroRNAs in liver cancer: A model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 2015;22(1):46-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang W, Wang Y, Liu W, van Wijnen AJ. Regulation and biological roles of the multifaceted miRNA-23b (MIR23B). Gene. 2018;642:103-109. [DOI] [PubMed] [Google Scholar]
- 105.Qiu X, Dong S, Qiao F, et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene. 2013;32(27):3296-3305. [DOI] [PubMed] [Google Scholar]
- 106.Kim HS, Lee KS, Bae HJ, et al. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget. 2015;6(10):8089-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52(4):297-303. [DOI] [PubMed] [Google Scholar]
- 108.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moody HL, Lind MJ, Maher SG. MicroRNA-31 regulates chemosensitivity in malignant pleural mesothelioma. Molecular Therapy-Nucleic Acids. 2017;8:317-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122–a key factor and therapeutic target in liver disease. J Hepatol. 2015;62(2):448-457. [DOI] [PubMed] [Google Scholar]
- 111.Morita K, Taketomi A, Shirabe K, et al. Clinical significance and potential of hepatic microRNA-122 expression in hepatitis C. Liver Int. 2011;31(4):474-484. [DOI] [PubMed] [Google Scholar]
- 112.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28(40):3526-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Penny SM. Ovarian cancer: An overview. Radiol Technol. 2020;91(6):561-575. [PubMed] [Google Scholar]
- 114.Kujawa KA, Lisowska KM. [Ovarian cancer--from biology to clinic]. Postepy Hig Med Dosw (Online). 2015;69:1275-1290. [DOI] [PubMed] [Google Scholar]
- 115.Hisamatsu T, McGuire M, Wu SY, et al. PRKRA/PACT expression promotes chemoresistance of mucinous ovarian cancer. Mol Cancer Ther. 2019;18(1):162-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Samuel P, Pink RC, Brooks SA, Carter DR. miRNAs and ovarian cancer: A miRiad of mechanisms to induce cisplatin drug resistance. Expert Rev Anticancer Ther. 2016;16(1):57-70. [DOI] [PubMed] [Google Scholar]
- 117.Liu H, Shi S, Gao J, Guo J, Li M, Wang L. Analysis of risk factors associated with breast cancer in women: A systematic review and meta-analysis. Transl Cancer Res. 2022;11(5):1344-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Health Commission Of The People's Republic Of China N. National guidelines for diagnosis and treatment of breast cancer 2022 in China (English version). Chin J Cancer Res. 2022;34(3):151-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pakzad R, Mohammadian-Hafshejani A, Khosravi B, et al. The incidence and mortality of esophageal cancer and their relationship to development in Asia. Ann Transl Med. 2016;4(2):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268-274. [DOI] [PubMed] [Google Scholar]
- 121.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. [DOI] [PubMed] [Google Scholar]
- 122.Willett CG, Chang DT, Czito BG, Meyer J, Wo J. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012(5), Int J Radiat Oncol Biol Phys. 2013;86(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381(9864):400-412. [DOI] [PubMed] [Google Scholar]
- 124.Sharma P, McQuaid K, Dent J, et al. A critical review of the diagnosis and management of barrett’s esophagus: The AGA Chicago workshop. Gastroenterology. 2004;127(1):310-330. [DOI] [PubMed] [Google Scholar]
- 125.Reid B. Barrett’s esophagus: Ordering the events that. Eur J Cancer Prev. 1996;5(2):57. [DOI] [PubMed] [Google Scholar]
- 126.Cook MB, Kamangar F, Whiteman DC, et al. Cigarette smoking and adenocarcinomas of the esophagus and esophagogastric junction: A pooled analysis from the international BEACON consortium. J Natl Cancer Inst. 2010;102(17):1344-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Toh Y, Oki E, Ohgaki K, et al. Alcohol drinking, cigarette smoking, and the development of squamous cell carcinoma of the esophagus: Molecular mechanisms of carcinogenesis. Int J Clin Oncol. 2010;15(2):135-144. [DOI] [PubMed] [Google Scholar]
- 128.Fennerty MB. Barrett's-related esophageal cancer: Has the final hurdle been cleared, now paving the way for human chemoprevention trials? Gastroenterology. 2002;122(4):1172-1175. [DOI] [PubMed] [Google Scholar]
- 129.Kubo A, Block G, Quesenberry Jr CP, Buffler P, Corley DA. Effects of dietary fiber, fats, and meat intakes on the risk of Barrett's esophagus. Nutr Cancer. 2009;61(5):607-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kubo A, Levin TR, Block G, et al. Dietary antioxidants, fruits and vegetables, and the risk of Barrett’s esophagus. Am J Gastroenterol. 2008;103(7):1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lopes AB, Fagundes RB. Esophageal squamous cell carcinoma-precursor lesions and early diagnosis. World J Gastrointest Endosc. 2012;4(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sun F, Yuan P, Chen T, Hu J. Efficacy and complication of endoscopic submucosal dissection for superficial esophageal carcinoma: A systematic review and meta-analysis. J Cardiothorac Surg. 2014;9(1):1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Merkow RP, Bilimoria KY, McCarter MD, Chow WB, Ko CY, Bentrem DJ. Use of multimodality neoadjuvant therapy for esophageal cancer in the United States: Assessment of 987 hospitals. Ann Surg Oncol. 2012;19(2):357-364. [DOI] [PubMed] [Google Scholar]
- 134.Ilson DH. Esophageal cancer chemotherapy: Recent advances. Gastrointestinal Cancer Research: GCR. 2008;2(2):85. [PMC free article] [PubMed] [Google Scholar]
- 135.Watanabe M, Otake R, Kozuki R, et al. Recent progress in multidisciplinary treatment for patients with esophageal cancer. Surg Today. 2020;50(1):12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Russo MM, Sundaramurthi T. An overview of cancer pain: Epidemiology and pathophysiology. Semin Oncol Nurs. 2019;35(3):223-228. [DOI] [PubMed] [Google Scholar]
- 137.Higginson IJ MF, Osborne TR. Epidemiology of pain in cancer. In: Cancer pain. London: Springer; 2013:5-24. [Google Scholar]
- 138.Yawn BP, Wollan PC, Weingarten TN, Watson JC, Hooten WM, Melton L3. The prevalence of neuropathic pain: Clinical evaluation compared with screening tools in a community population. Pain Med. 2009;10(3):586-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li M, Li Z, Ma X, et al. Huangqi guizhi wuwu decoction can prevent and treat oxaliplatin-induced neuropathic pain by TNFα/IL-1β/IL-6/MAPK/NF-kB pathway. Aging (Albany NY). 2022;14(12):5013-5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sommer C, Leinders M, Üçeyler N. Inflammation in the pathophysiology of neuropathic pain. Pain. 2018;159(3):595-602. [DOI] [PubMed] [Google Scholar]
- 141.Bastian I, Tam Tam S, Zhou XF, et al. Differential expression of microRNA-1 in dorsal root ganglion neurons. Histochem Cell Biol. 2011;135(1):37-45. [DOI] [PubMed] [Google Scholar]
- 142.Kusuda R, Cadetti F, Ravanelli MI, et al. Differential expression of microRNAs in mouse pain models. Mol Pain. 2011;7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu M, Wood JN. The roles of sodium channels in nociception: Implications for mechanisms of neuropathic pain. Pain Med. 2011;12(Suppl 3):S93-S99. [DOI] [PubMed] [Google Scholar]
- 144.Strickland IT, Richards L, Holmes FE, Wynick D, Uney JB, Wong LF. Axotomy-induced miR-21 promotes axon growth in adult dorsal root ganglion neurons. PLoS One. 2011;6(8):e23423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lu A, Huang Z, Zhang C, et al. Differential expression of microRNAs in dorsal root ganglia after sciatic nerve injury. Neural Regen Res. 2014;9(10):1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sakai A, Saitow F, Miyake N, Miyake K, Shimada T, Suzuki H. miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain. 2013;136(Pt 9):2738-2750. [DOI] [PubMed] [Google Scholar]
- 147.Chen HP, Zhou W, Kang LM, et al. Intrathecal miR-96 inhibits Nav1.3 expression and alleviates neuropathic pain in rat following chronic construction injury. Neurochem Res. 2014;39(1):76-83. [DOI] [PubMed] [Google Scholar]
- 148.Lin CR, Chen KH, Yang CH, Huang HW, Sheen-Chen SM. Intrathecal miR-183 delivery suppresses mechanical allodynia in mononeuropathic rats. Eur J Neurosci. 2014;39(10):1682-1689. [DOI] [PubMed] [Google Scholar]
- 149.Sakai A, Suzuki H. Nerve injury-induced upregulation of miR-21 in the primary sensory neurons contributes to neuropathic pain in rats. Biochem Biophys Res Commun. 2013;435(2):176-181. [DOI] [PubMed] [Google Scholar]
- 150.Bali KK, Selvaraj D, Satagopam VP, Lu J, Schneider R, Kuner R. Genome-wide identification and functional analyses of microRNA signatures associated with cancer pain. EMBO Mol Med. 2013;5(11):1740-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ji RR, Kohno T, Moore KA, Woolf CJ. Central sensitization and LTP: Do pain and memory share similar mechanisms? Trends Neurosci. 2003;26(12):696-705. [DOI] [PubMed] [Google Scholar]
- 152.Wu D, Raafat M, Pak E, Hammond S, Murashov AK. MicroRNA machinery responds to peripheral nerve lesion in an injury-regulated pattern. Neuroscience. 2011;190:386-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kynast KL, Russe OQ, Möser CV, Geisslinger G, Niederberger E. Modulation of central nervous system-specific microRNA-124a alters the inflammatory response in the formalin test in mice. Pain. 2013;154(3):368-376. [DOI] [PubMed] [Google Scholar]
- 154.Tam S T, Bastian I, Zhou XF, et al. MicroRNA-143 expression in dorsal root ganglion neurons. Cell Tissue Res. 2011;346(2):163-173. [DOI] [PubMed] [Google Scholar]
- 155.Favereaux A, Thoumine O, Bouali-Benazzouz R, et al. Bidirectional integrative regulation of Cav1.2 calcium channel by microRNA miR-103: Role in pain. Embo j. 2011;30(18):3830-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ito N, Sakai A, Miyake N, et al. miR-15b mediates oxaliplatin-induced chronic neuropathic pain through BACE1 down-regulation. Br J Pharmacol. 2017;174(5):386-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li L, Shao J, Wang J, et al. MiR-30b-5p attenuates oxaliplatin-induced peripheral neuropathic pain through the voltage-gated sodium channel Na(v)1.6 in rats. Neuropharmacology. 2019;153:111-120. [DOI] [PubMed] [Google Scholar]
- 158.Arroyo DS, Gaviglio EA, Peralta Ramos JM, Bussi C, Rodriguez-Galan MC, Iribarren P. Autophagy in inflammation, infection, neurodegeneration and cancer. Int Immunopharmacol. 2014;18(1):55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li H, Shen L, Ma C, Huang Y. Differential expression of miRNAs in the nervous system of a rat model of bilateral sciatic nerve chronic constriction injury. Int J Mol Med. 2013;32(1):219-226. [DOI] [PubMed] [Google Scholar]
- 160.Li H, Huang Y, Ma C, Yu X, Zhang Z, Shen L. MiR-203 involves in neuropathic pain development and represses Rap1a expression in nerve growth factor differentiated neuronal PC12 cells. Clin J Pain. 2015;31(1):36-43. [DOI] [PubMed] [Google Scholar]