Abstract
Multidrug-resistant fungal infections have become much more common in recent years, especially in immune-compromised patients. Therefore, researchers and pharmaceutical professionals have focused on the development of novel antifungal agents that can tackle the problem of resistance. In continuation to this, a novel series of pyrazole-bearing pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione derivatives (4a–4o) have been developed. These compounds have been screened against Candida albicans, Aspergillus niger, and Aspergillus clavatus. The synthesized compounds were characterized by well-known spectroscopic techniques, i.e., IR, 1H NMR, 13C NMR, and mass spectrometry. In vitro antifungal results revealed that compound 4n showed activity against C. albicans having MIC value of 200 μg/mL. To know the plausible mode of action, the active derivatives were screened for anti-biofilm and ergosterol biosynthesis inhibition activities. The compounds 4h, 4j, 4k, and 4n showed greater ergosterol biosynthesis inhibition than the control DMSO. To comprehend how molecules interact with the receptor, studies of molecular docking of 4k and 4n have been performed on the homology-modeled protein of β-tubulin. The molecular docking revealed that the active compounds 4h, 4j, 4k, 4l, and 4n interacting with the active site amino acid of sterol 14-alpha demethylase (PDB ID: 5v5z) indicate one of the possible modes of action of ergosterol inhibition activity. The synthesized compounds 4c, 4e, 4h, 4i, 4j, 4k, 4l, and 4n inhibited biofilm formation and possessed the potential for anti-biofilm activity. DFT-based quantum mechanical calculations were carried out to optimize, predict, and compare the vibration modes of the molecule 4a.
1. Introduction
Invasive fungal infections are becoming more common as a result of stem cell transplantation, organ transplantation, chemotherapy, and the human immunodeficiency virus.1,2 The most common pathogenic strains that cause systemic fungal infections are Candida albicans, Cryptococcus neoformans, and Aspergillus fumigatus.3 Fluconazole, voriconazole, itraconazole, and miconazole are azole-based broad-spectrum antifungal drugs that are widely used (Figure 1).4 They have broad-spectrum antifungal activity against the majority of filamentous fungi, but several of them are ineffective against invasive aspergillosis and have substantial drug resistance.5 Furthermore, widespread usage of existing antifungal medications has resulted in significant drug resistance.6 As a result, novel antifungal agents with outstanding action against a wide range of clinical fungal species are urgently needed.
Figure 1.
Commercially available azole-based antifungal drugs.
Heterocycles are suggestively a wealthy target for the development of bioactive agents with a wide range of biological activities acting as oxygen carriers (hemoglobin), constituents of DNA, energy storage (adenosine triphosphate), and natural antimicrobial agents (diketopiperazines).7 Thus, these compounds with various functional moieties have been explored for their antimicrobial characteristics.8,9 Compounds with nitrogen-rich five- and six-membered heterocyclic structures, such as pyrazoles, pyridine, and pyrimidine, are a vital source of biologically active components for a variety of applications.10
Pyrazole contains two nitrogen atoms in its five-membered ring and is chemically 1,2-diazoles. Pyrazole-containing compounds have been effectively synthesized as they possess various biological activities like antimicrobial,11 anti-inflammatory,12 anticancer,13 and antitubercular.14 Moreover, pyridine is a is a six-membered heterocyclic entity containing nitrogen atoms while pyrimidine has two nitrogen atoms at its adjacent carbon. Pyridine and pyrimidine derivatives have a wide range of biological activities, i.e., antimicrobial,15,16 anti-inflammatory,17 antitumor,18 anticancer,19 antitubercular,20,21 etc.
We have used heterocycles to invent our novel hybrid heterocyclic compounds focusing on different fused pyridine–pyrimidines with pyrazole motifs, as noted in the present study. The synthesized compounds were uniformly auspicious to the medicinal chemist for correlation of the similarities with commercially available medicine consisting of pyridine scaffolds. The pyridine and pyrimidine scaffolds clubbed with the pyrazole showed potential antimicrobial activities. The pyrazole ring-containing compounds showed antifungal activity via ergosterol biosynthesis inhibition22 and biofilm inhibition. The structural modification of the ciprofloxacin to 7-phenylpyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione core highlighted in Figure 2 indicates that the pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione pharmacophore matches with the quinolin-4(1H)-one core of the ciprofloxacin and the phenyl ring at 7-position of the target scaffold matches with the piperazine ring. Also, the 5-position of pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione was modified by substituting the pyrazole for enhancing the activity. These things enlightened us the synthesis of 5-(1,3-diphenyl-1H-pyrazol-4-yl)-7-phenylpyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione as an antimicrobial agent. The possible mode of action of antifungal activity was also evaluated by ergosterol biosynthesis inhibition and biofilm inhibition activities.
Figure 2.
Design of target molecule based on the commercially available drug ciprofloxacin.
2. Results and Discussion
2.1. Chemistry
The titled molecules (4a–4o) were synthesized by multi-step reactions. Chalcone derivatives (3a–3o) were prepared by using an aldol condensation of 1,3-diphenyl-1H-pyrazole-4-carbaldehyde (1) and substituted acetophenones (2a–2o). Final compounds (4a–4o) were prepared from chalcone derivatives, which was reacted with 6-aminouracil using acetic acid as a catalyst. The synthetic root adopted for the synthesis of compounds (4a–4o) is illustrated in Scheme 1. IR spectra of compound 4a, at frequencies of 3178 and 2811 cm–1, confirmed the presence of −CH– and −CH=CH– groups, respectively. The presence of >C=O was confirmed at 1755 cm–1. Stretching vibrations at 1689, 1659, and 1276 cm–1 confirmed the presence of −C=N, −C=C–, and −C–N groups, respectively. The proton of secondary amine in the pyrimidine ring was confirmed at δ = 12.09–10.39 ppm while the proton of pyrazole was confirmed at 8.55 ppm, and the signal observed at 8.36 ppm confirmed the proton of the pyridine ring. Moreover, the signal appeared in the range of δ = 8.04–7.45 ppm confirming 15 aromatic protons. In 13C NMR, chemical shifts at 159, 153.5, and 151.3 ppm agreed with the carbon of the pyrimidine ring. Furthermore, carbons of the pyridine ring were confirmed at 150.5, 149.0, 112.4, and 110.3 ppm. Signals that appeared at 145.8, 131.6, and 101.3 ppm confirmed the presence of a heterocyclic ring of pyrazole. Molecular weight at m/z = 457.17 of mass spectra is evidence of compound 4a.
Scheme 1. Synthetic Pathways of Synthesized Compounds 4a–4o.
2.2. Biological Activity
2.2.1. Discussion on Antifungal Activity
The main scaffold of newly synthesized molecules (4a–4o) consists of the pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione moiety. There are many reports wherein pyrido/pyrimido-pyrimidines were tested for antifungal activity. Abdelhameed et al. have reported arylidene hydrazinylpyrido[2,3-d]pyrimidin-4-ones as potent anti-microbial agents targeting C. albicans.23 Pyrimido[4,5-d][1,2,4]triazolo[4,3-a]pyrimidines were studied for their antifungal activity by Mangoud et al. against C. albicans.24 Sharma et al. have synthesized a pyrimido[4,5-d]pyrimidine-2,5-dione derivative that showed 8–100 ppm MIC values against C. albicans.25 Taking a cue from these literature reports, as well as our previous work26 wherein we have studied fused pyridine–pyrimidine hybrids for their antifungal activity, we have evaluated novel synthesized 4a–4o for in vitro antifungal activity using the Broth dilution method. The results of antifungal activity revealed that some of the compounds have inhibition effects toward screened fungal strains. Antifungal activity data revealed that compound 4n showed to be most potent against C. albicans having an MIC value of 200 μg/mL. Moreover, compound 4k showed activity against C. albicans having an MIC value of 250 μg/mL. Compounds 4n and 4k exhibited significant inhibition than the standard drug griseofulvin. Additionally, compounds 4c, 4e, 4h, 4i, 4j, and 4l showed comparable potency with reference drug griseofulvin against C. albicans with MIC 500 μg/mL. The in vitro antifungal activity data of synthesized compounds 4a–4o are listed in Table 1.
Table 1. Antifungal Screening of Compounds 4a–4o Having Different Fungal Strainsa.

SD = standard deviation; C. A. = Candida albicans; A. n. = Aspergillus niger; A. c. = Aspergillus clavatus.
p ≤ 0.0001.
2.3. Computational Study
2.3.1. Molecular Docking Study
The molecular interaction between the receptor and the ligand is vital information obtained from docking studies. Griseofulvin, which is used as the standard for antifungal activity, is considered to be a tubulin polymerization inhibitor.27,28 β-Tubulin was used as a receptor for molecular docking studies. Since the experimental three-dimensional structure of Candida albicans tubulin is not available in the Protein Data Bank, the structure was modeled by homology modeling using Yeast tubulin (PDB ID: 5w3f) as a template protein. The sequence coverage was around 96% with 87% identities and 96% positives. The modeled protein structure was verified by verifying 3D, Errat, and Procheck using protein structure validation server SAVES v6.0 of UCLA-DOE Lab and ProSA-web (Z-score: −9.84).
The modeled protein was prepared and used for molecular docking studies. Rathinasamy et al.28 have reported that griseofulvin binds at a site distinct from the colchicine-binding site in tubulin; hence, docking studies were carried out at a site comprising amino acid residues Glu 27, Leu 215, Gln 224, Asn 227, Leu 228, Ser 231, Tyr 270, Leu 273, Thr 274, Arg 318, Lys 359, Asp 360, and Leu 361. Compound 4g showed the highest XP dock score of −5.933 kcal/mol, and the standard compound griseofulvin showed a dock score of −4.064 kcal/mol. All the compounds showed similar dock scores within a range of −5.993 to −3.982 kcal/mol. The prime MM-GBSA binding energy (MMGBSA – ΔGbind) values of all the compounds were better than those of griseofulvin (Table 2). The dock scores and ΔGbind values showed a similar trend as the experimental antifungal activity.
Table 2. Glide XP Dock Scores and Prime MMGBSA−ΔGbind Scores.
| entry | XP GScore (kcal/mol) | MMGBSA ΔGbind (kcal/mol) |
|---|---|---|
| 4a | –4.340 | –45.07 |
| 4b | –4.838 | –62.30 |
| 4c | –4.128 | –38.55 |
| 4d | –4.699 | –42.64 |
| 4e | –4.169 | –42.75 |
| 4f | –3.997 | –41.98 |
| 4g | –5.933 | –68.57 |
| 4h | –4.628 | –42.99 |
| 4i | –4.151 | –43.61 |
| 4j | –5.052 | –48.61 |
| 4k | –4.427 | –63.31 |
| 4l | –4.295 | –45.27 |
| 4m | –3.982 | –47.09 |
| 4n | –4.079 | –44.75 |
| 4o | –4.574 | –45.36 |
| griseofulvin | –4.064 | –41.41 |
A dock pose analysis of all the compounds was carried out, and the molecules showed a majorly hydrophobic interaction with protein amino acid residues. Griseofulvin showed a hydrogen bond interaction with Ser 231 and Tyr 270 (Figure 3). Molecules 4k and 4n showed better antifungal activity having similar binding energy values, but they had an additional hydrogen bond interaction with Arg 318 and Lys 359. Molecule 4k showed an additional hydrophobic π–π interaction with Tyr 270 and a π-cation interaction with Lys 359, which corresponds to the higher binding value and better antifungal activity when compared to griseofulvin (Figure 4). Molecule 4n also showed a similar hydrogen bond interaction to molecule 4k along with an additional halogen interaction with Lys 359 (Figure 5).
Figure 3.
Ligand interaction diagram griseofulvin docked into tubulin showing a hydrogen bond interaction with residues Ser 231 and Tyr 270, and a halogen bond with Asn 227.
Figure 4.
Ligand interaction diagram 4k docked into tubulin showing a hydrogen bond interaction with residues Tyr 270 and Arg 318, a hydrophobic π–π interaction with Tyr 270, and a π–cation interaction with Lys 359.
Figure 5.
Ligand interaction diagram 4n docked into tubulin showing a hydrogen bond interaction with residues Ser 231 Tyr 270 and Lys 359 and a halogen bond with Lys 359.
2.3.1.1. Evaluation of ADME-T Properties
ADME properties and toxicity were calculated for synthesized molecules to evaluate their drug-like properties. A heat map table of the ADME properties and toxicity parameters is provided in Table 3. Physicochemical properties like partition coefficient (logPoctanol/water), cell membrane permeability (PCaco and PMDCK), and oral absorption of all molecules are in the acceptable range. Only molecule 4l showed lower MDCK cell permeability and oral absorption value than the acceptable range. However, all the molecules showed lesser water solubility (LogS) compared to standard griseofulvin. Furthermore, the toxicity profiling using the pkCSM server revealed that all the molecules were showing a nontoxic nature.
Table 3. ADME-T Properties of Synthesized Molecules 4a–4oa.
| molecule | MW | logPo/w | logS | PCaco | PMDCK | #metab | % human oral absorption | AMES toxicity | max. tolerated dose (human) | Minnow toxicity |
|---|---|---|---|---|---|---|---|---|---|---|
| 4a | 457.49 | 5.173 | –7.847 | 422.086 | 194.739 | 0 | 91.265 | no | 0.458 | 1.452 |
| 4b | 536.386 | 5.704 | –8.619 | 387.788 | 469.626 | 0 | 80.758 | no | 0.446 | 0.919 |
| 4c | 536.386 | 5.696 | –8.551 | 431.878 | 530.023 | 0 | 81.551 | no | 0.46 | 0.875 |
| 4d | 491.935 | 5.554 | –8.237 | 396.664 | 377.95 | 0 | 93.012 | no | 0.457 | 1.138 |
| 4e | 491.935 | 5.61 | –8.483 | 436.948 | 499.41 | 0 | 94.092 | no | 0.46 | 1.021 |
| 4f | 475.481 | 5.37 | –8.035 | 445.758 | 373.901 | 0 | 92.841 | no | 0.456 | 1.765 |
| 4g | 473.49 | 4.469 | –7.055 | 213.81 | 93.366 | 1 | 94.818 | no | 0.438 | 1.761 |
| 4h | 473.49 | 4.354 | –7.275 | 147.736 | 62.613 | 1 | 91.268 | no | 0.442 | 2.007 |
| 4i | 487.517 | 5.179 | –7.59 | 447.053 | 207.219 | 1 | 91.75 | no | 0.456 | 1.472 |
| 4j | 487.517 | 5.177 | –7.785 | 411.643 | 189.536 | 1 | 91.096 | no | 0.457 | 1.671 |
| 4k | 471.517 | 5.437 | –8.312 | 420.486 | 193.941 | 1 | 92.783 | no | 0.447 | 1.093 |
| 4l | 502.488 | 4.432 | –7.946 | 54.14 | 21.156 | 1 | 70.965 | no | 0.443 | 0.504 |
| 4m | 526.381 | 5.835 | –8.457 | 430.176 | 768.652 | 0 | 82.331 | no | 0.464 | 0.503 |
| 4n | 526.381 | 6.056 | –9.04 | 446.201 | 1057.187 | 0 | 83.911 | no | 0.462 | 0.67 |
| 4o | 493.471 | 5.57 | –8.418 | 415.555 | 518.208 | 0 | 93.472 | no | 0.453 | 1.541 |
| griseofulvin | 352.771 | 2.185 | –4.759 | 1553.757 | 1853.409 | 3 | 96.857 | no | 1.223 | –0.274 |
Acceptable range for properties: 350≤ MW ≥650, −2.0 ≤ logPo/w ≥6.5, −6.5 ≤ logS ≥0.5. PCaco and PMDCK cell permeability: <25 poor, >500 good. Percent human oral absorption >80. #metab: 1 to 8. Max. tolerated dose (MRTD) ≥0.47. Minnow toxicity: log LC50 >−0.3.
2.3.2. Vibrational Analysis
The structure of molecule 4a was optimized with C1 symmetry; the predicted IR spectra for molecule 4a at the B3LYP/6-31G* level in the gas phase is provided in the supplementary figures, and the comparison with experimental IR data is provided in Table 4 along with the assignment of wavenumbers to corresponding stretching modes. The calculations predicted the CH stretching modes corresponding to the phenyl rings in the expected region of ∼3200 cm–1; >C=O stretching was predicted at ∼1700 cm–1, and >C=N stretching was predicted at ∼1400 cm–1.
Table 4. Experimental, Calculated IR Frequency and Assignment for Molecule 4a.
| IR experimental frequency (wavenumber cm–1) | IR calculated (B3LYP/6-31G*) (wavenumber cm–1) | assignment |
|---|---|---|
| 519.65 | 527.86 | ν C–H |
| 632.55 | 642.54 | ν C–H |
| 706.39 | 718.36 | ν N–H |
| 802.65 | 807.04 | ν C–H |
| 904.65 | 947.40 | ν C–H |
| 1276.49 | 1274.85 | ν C–H |
| 1432.54 | 1422.39 | ν C=N |
| 1479.98 | 1496.53 | ν C–H |
| 1543.76 | 1566.16 | ν C–N |
| 1609.18 | 1599.23 | ν C=C |
| 1659.98 | 1636.42 | ν C=C |
| 1689.78 | 1717.16 | ν C=O |
| 1755.21 | 1798.15 | ν C=O |
| 3001.34 | 3213.37 | ν C–H aromatic |
| 3178.76 | 3220.99 | ν C–H aromatic |
2.3.3. Ergosterol Inhibition Activity
The antifungal mechanism of action of azole drugs is to disrupt the sterol biosynthetic pathway that leads to ergosterol biosynthesis inhibition.29 Ergosterol is one of the components of the fungal plasma membrane. Inhibition of synthesis of this major sterol is one of the common modes of action for many antifungal drugs. The quantitative estimation of ergosterol biosynthesis was done using the protocol of Breivik et al. in 1957.30,31 Ergosterol quantitation was represented in terms of % ergosterol/g wet weight (Figure 6). From the ergosterol biosynthesis inhibition activity, it is clear from the graph that the % of ergosterol was decreased in compounds 4h, 4j, 4k, and 4n as compared to DMSO control in all conditions except compound 4l (Figure 6). This clearly indicates that the plausible mode of action of these compounds is ergosterol biosynthesis inhibition and Hasan antifungal potential.
Figure 6.
Ergosterol quantitation in terms of % ergosterol/g wet weight.
The ergosterol inhibition activity represented by compounds 4h, 4j, 4k, 4l, and 4n was further confirmed by molecular docking studies. Table 5 shows molecular docking data represented in terms of binding energy (ΔG) in kcal/mol for sterol 14-alpha demethylase (PDB ID: 5v5z), the key enzyme from the ergosterol biosynthetic pathway from C. albicans (5v5z).32 The results indicate that all compounds bind to this enzyme with less binding energy indicating stable interactions, as shown in Table 5. Compound 4l shows the lowest binding energy of −12.8 kcal/mol. It is observed that all five compounds bind in the active site of the enzyme, which is represented in Figure 7. Figure 7A shows the active site of 5v5z, and Figure 7B represents the binding poses of ligands 4h, 4j, 4k, 4l, and 4n. The docking analysis of these poses was performed to check the interaction of these compounds with amino acids present in the active site (Figure 8). It was observed that all compounds were interacting with active site amino acids, suggesting its strong and stable interaction with the enzyme. These newly synthesized derivatives were subjected to the ADME study33 for pharmacological analysis, and the data is represented in Table 5. Lipinski’s rule gives us a good approach to predicting drug-likeness. All five compounds are following Lipinski’s rule and therefore have the potential to be developed as lead candidates.
Table 5. ADME Analysis of Active Compounds.
| 4h | 4j | 4k | 4l | 4n | |
|---|---|---|---|---|---|
| molecular weight | 473.49 | 487.52 | 471.52 | 502.49 | 526.38 |
| A log P | 4.5 | 4.81 | 5.11 | 4.71 | 6.1 |
| H-bond acceptor | 6 | 6 | 5 | 7 | 5 |
| H-bond donor | 3 | 2 | 2 | 2 | 2 |
| rotatable bonds | 4 | 5 | 4 | 5 | 4 |
Figure 7.

(A) Active site of 5v5z. (B) The two-dimensional interacting mode of the hybrid derivative in the active region of sterol 14-alpha demethylase from Candida albicans (5v5z).
Figure 8.
The two-dimensional interacting mode of the hybrid derivative in the active region of sterol 14-alpha demethylase from Candida albicans (5v5z).
2.3.4. Anti-Biofilm Activity
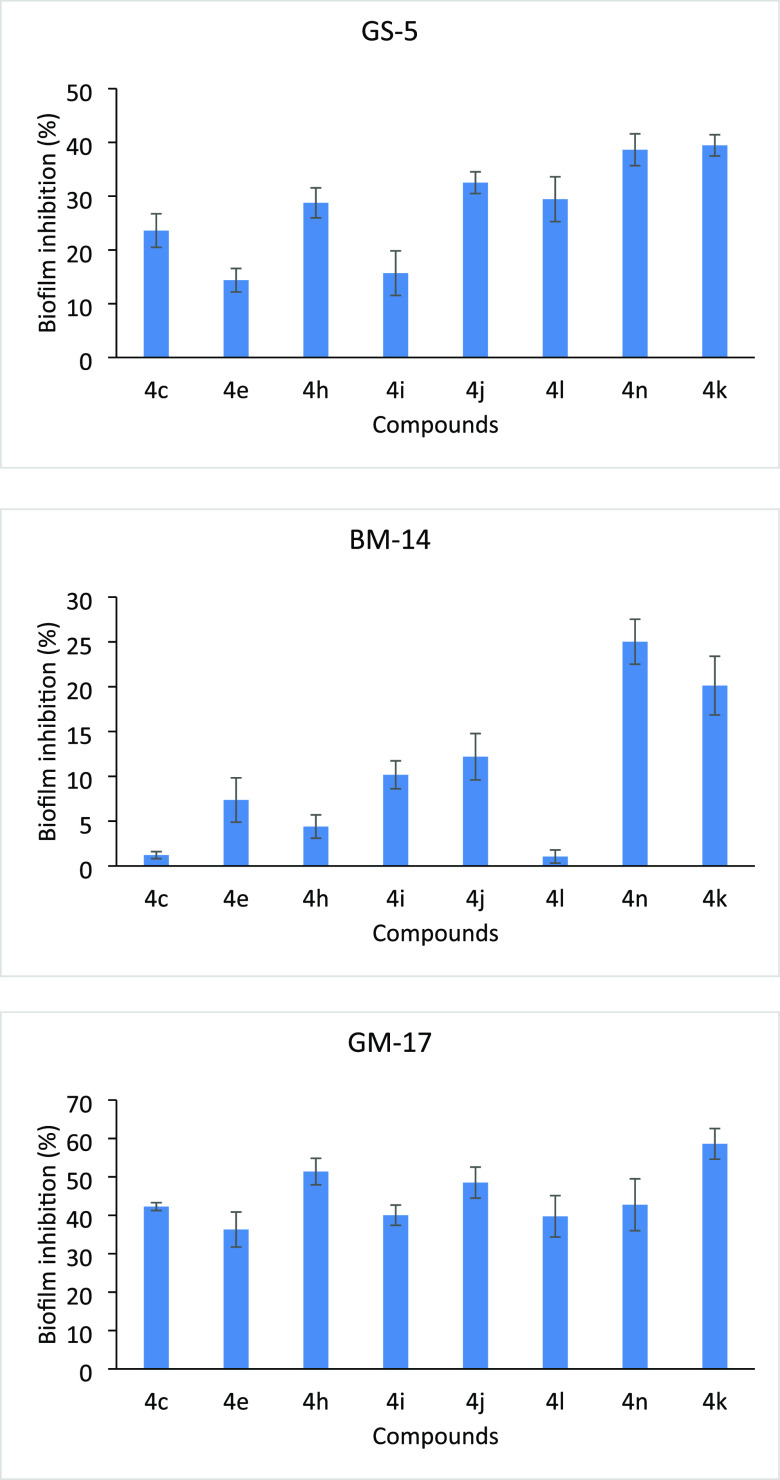
Bacterial biofilms are highly organized bacterial complexes enclosed in a self-produced matrix (extracellular polymeric substance, EPS) protected from the host defense. EPS plays an important role in bacterial cell aggregation, cell adhesion, and biofilm formation and protects bacterial cells from a hostile environment. Biofilms are known to have high adaptive resistance to antibiotics and other disinfectants, which makes it very difficult to treat infections.34 The bacterial isolates used in the present study are of marine origin and known for biofilm formation.35 The results of the biofilm inhibition of different compounds are presented in Figure 9. The inhibitory actions for biofilm formation activities were shown by all compounds at varying levels. In general, compounds highly inhibited the biofilm formation by Gram-negative bacteria (GS-5 and GM-17) compared to Gram-positive (BM-14)). This may be due to the differences in the structure between Gram-positive and Gram-negative cell walls and showed different modes of action of the tested compounds.36 The tested compounds showed 14.38% (4e) to 39.45% (4k) inhibition against Vibrio sp. (GS-5), similarly 36.30% (4e) to 58.59% (4k) against Gallaecimonas sp. (GM-17). Contrarily, these compounds showed less inhibitory action for biofilm formation in Staphylococcus sp. (BM-14), which varied from 1.05% (4l) to 25.02% (4n). The reduced inhibition of biofilm development exhibited that the bacterial cell in a biofilm is more resistant to antimicrobial agents.37 Altogether, our study demonstrated that the compounds inhibit biofilm formation and had the potential for anti-biofilm activity. The disc diffusion method was used to enumerate the antibacterial activities of the synthesized compounds; no zones of inhibition were observed around the discs impregnated with chemical compounds (Figure S1). The results indicated that the inhibition of biofilm formation by bacteria is not due to the toxic effect of the chemical compounds but because chemical compounds inhibited the initial attachment of bacterial cells to the surface as well as reducing cell-to-cell surface interactions.
Figure 9.
Biofilm formation inhibitory effects of different compounds on bacterial isolates (GS-5, Vibrio sp.; BM-14, Staphylococcus sp.; and GM-17, Gallaecimonas sp.).
2.3.5. The Effect of Various-Synthesized Derivatization on Antifungal Activity
The hybrids of pyrazole-bearing pyridine–pyrimidine (4a–4o) were synthesized and evaluated for their antifungal potency. The effect of various functional groups on antifungal activity is shown in Figure 10. Compound 4n having an electron-withdrawing chloro group at the third and fourth positions possessed MIC of 200 μg/mL, while compound 4k having an electron-donating methyl group at the fourth position possessed MIC of 250 μg/mL against C. albicans. Compounds 4c, 4e, and 4l having electron-withdrawing bromo, chloro, and nitro groups on the fourth position possessed comparable antifungal activity against C. albicans. Compounds with electron-donating functional groups on the second (4i-2-OCH3), third (4j-3-OCH3), and fourth (4h-4-OH) positions were also found to have similar MIC values in comparison to griseofulvin.
Figure 10.
The effect of various functional groups on antifungal activity of pyrazole bearing pyrido[2,3-d]pyrimidine-2,4-dione analogues.
The effect of various functional groups on ergosterol biosynthesis is shown in Figure 11. Compound 4k having an electron-donating methyl group at the fourth position on the phenyl ring possesses an ergosterol content of 0.0018%. Similarly, compound 4h having an electron-donating hydroxyl group at the fourth position on the phenyl ring possesses an ergosterol content of 0.0108%. Compound 4n having an electron-withdrawing chloro group at the third and fourth positions on the phenyl ring possesses an ergosterol content of 0.0064%, and 4j with an electron-donating methoxy group at the third position on the phenyl ring has ergosterol content 0.0079%. Compounds having different election-donating and -withdrawing functional groups on the third and fourth positions on the phenyl ring inhibited ergosterol biosynthesis in comparison to DMSO (0.0182%).
Figure 11.
The effect of compound derivatization on antifungal activity of synthetic hybrids against ergosterol biosynthesis of compounds 4h, 4j, 4k, and 4n.
The effect of various functional groups on biofilm formation by Gram-positive and Gram-negative bacteria is shown in Figure 12. The potent antifungal compounds having various electron-donating and -withdrawing functional groups on the phenyl ring highly inhibited biofilm formation by Gram-negative bacteria (GS-5 and GM-17). The tested compounds showed 14.38 to 39.45% inhibition against Gram-negative Vibrio sp. (GS-5) and 36.30 to 58.59% against Gram-negative Gallaecimonas sp. (GM-17). Compounds having various electron-donating functional groups on the second (4i-2-OCH3), third (4j-3-OCH3), and fourth (4k-4-CH3) positions on the phenyl ring inhibited biofilm formation by Gram-positive bacteria (BM-14) with 10.17 to 20.12%. Compound 4k with the electron-negative chloro group on the third and fourth positions inhibited biofilm formation by Gram-positive bacteria (BM-14) with 25.02%.
Figure 12.

The effect of various functional groups on biofilm formation by Gram-positive and Gram-negative bacteria.
3. Conclusions
The aldol condensation of 1,3-diphenyl-1H-pyrazole-4-carbaldehyde and appropriate acetophenone derivatives in methanolic KOH afforded the chalcone derivatives. The reaction of substituted chalcones with 6-aminouracil in the presence of acetic acid yielded the corresponding pyrazole-containing pyridine–pyrimidine derivatives. The synthesized compounds (4a–4o) were screened for their in vitro antifungal activity against three fungi. The −3,4-Cl2 group in compound 4n (MIC 200 μg/mL) and the 4-CH3 group in compound 4k (MIC 250 μg/mL) increased the potency for C. albicans, making them more active than the standard drug griseofulvin. The antifungal active derivatives were screened for ergosterol biosynthesis inhibition activity, and the compounds 4h, 4j, 4k, and 4n showed ergosterol biosynthesis inhibition. The molecular docking analysis showed that the active compounds 4h, 4j, 4k, 4l, and 4n interacted with the active site amino acid of sterol 14-alpha demethylase (PDB ID: 5v5z), indicating that one of the possible modes of action is ergosterol biosynthesis inhibition. The potent antifungal agents were highly inhibitory to the biofilm formation by Gram-negative bacteria (GS-5 and GM-17) and Gram-positive bacteria (BM-14) and possessed anti-biofilm activity. A stable homologous model of β-tubulin was constructed for molecular docking studies. Molecules with better antifungal activity than standard griseofulvin showed higher binding affinity, which may be attributed to an additional hydrogen bond interaction with Arg 318 and Lys 359.
4. Experimental Procedure
4.1. Materials and Instruments
“Completion of the reaction and purity of compounds was checked on Aluminium coated TLC plates [60 F254 (E. Merck)]. n-hexane: ethyl acetate (3:2 V/V) was used as a mobile phase and was visualized in an iodine chamber. An electrothermal melting point apparatus was used to determine melting points and was uncorrected. Elemental analysis (% C, H, N) was confirmed by a Perkin-Elmer 2400 CHN analyzer. A Perkin-Elmer FT-IR spectrophotometer was used to record IR spectra by using KBr. 1H NMR and 13C NMR spectra were recorded on a Bruker Avance III 400 MHz in CDCl3 as a solvent and tetramethylsilane (TMS) as an internal standard using a 5 mm tube. Mass spectra were obtained on SHIMADZU MS 2010 spectrometer.”38
4.1.1. Synthesis of (3-(1,3-Diphenyl-1H-pyrazol-4-yl)-1-arylprop-2-en-1-ones (3a–3o)
(3-(1,3-Diphenyl-1H-pyrazol-4-yl)-1-arylprop-2-en-1-ones (3a–3o) were prepared according to the method as per the literature method.39
4.1.2. Synthesis of 5-(1,3-Diphenyl-1H-Pyrazol-4-yl)-7-arylpyrido[2,3-d]pyrimidine-2,4(1H,3H)-diones (4a–4o)
A mixture of chalcone derivatives (3a–3o) (10 mmol), EtOH (40 mL), and 6-aminouracil (10 mmol) was refluxed in a round-bottom flask fitted with a reflux condenser. The refluxing was continued for 4 h and poured into ice-cold water. The solid precipitate was filtered and washed with hot water and dried overnight in a vacuum oven. The crude product was recrystallized from ethanol.
4.1.3. 5-(1,3-Diphenyl-1H-pyrazol-4-yl)-7-phenylpyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4a)
Yield: 75%; solid; M.P. 211–213 °C; IR (KBr, cm–1): 3178, 2811 (C–H, −CH=CH−), 1755 (>C=O), 1689, 1659, 1276 (C=N, C=C, C–N); 1H NMR (400 MHz, CDCl3): δ = 9.45 (s, 1H, Ar-NH), 9.24 (s, 1H, Ar-NH), 8.27 (s, 1H, -CH-CN), 7.83–7.76 (m, 5H, Ar-H), 7.72–7.57 (m, 5H, Ar-H), 7.30–7.24 (m, 6H, Ar-H); 13C NMR (100 MHz, CDCl3): δ = 163.0 (>C=O), 160.6 (>C=O), 155.0 (Ar-C), 151.3 (Ar-C), 150.8 (Ar-C), 140.5 (C3 of pyrazole), 139.2 (Ar-C), 137.9 (Ar-C), 135.3 (Ar-C), 133.6 (Ar-C), 132.5 (Ar-C), 131.2 (C5 of pyrazole), 130.9 (Ar-C), 130.4 (Ar-C), 130.1 (Ar-C), 129.4 (Ar-C), 128.7 (Ar-C), 128.5 (Ar-C), 127.1 (Ar-C), 126.5 (Ar-C), 126.3 (Ar-C), 126.0 (Ar-C), 124.9 (Ar-C), 120.5 (Ar-C), 118.9 (Ar-C), 118.3 (Ar-C), 111.4 (Ar-C), 105.1 (C4 of pyrazole); MS (m/z): 457.17 (M+); element analysis calculated (%) for C28H19N5O2: C 73.51, H 4.19, N 15.31; found: C 73.57, H 4.29, N 15.42.
4.1.4. 7-(3-Bromophenyl)-5-(1,3-diphenyl-1H-pyrazol-4-yl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4b)
Yield: 69%; solid; M.P. 225–227 °C; IR (KBr, cm–1): 3184, 2845 (C–H, −CH=CH−), 1746 (>C=O), 1679, 1661, 1267 (C=N, C=C, C–N), 679 (C-Br); 1H NMR (400 MHz, CDCl3): δ = 9.56 (s, 1H, Ar-NH), 9.47 (s, 1H, Ar-NH), 8.03 (s, 1H), 7.99–7.88 (m, 4H), 7.73–7.64 (m, 4H), 7.26–7.21 (m, 7H); 13C NMR (100 MHz, CDCl3): δ = 164.5 (>C=O), 160.4 (>C=O), 154.5 (Ar-C), 149.5 (Ar-C), 149.1 (Ar-C), 146.5 (C3 of pyrazole), 140.3 (Ar-C), 136.9 (Ar-C), 135.4 (Ar-C), 134.4 (Ar-C), 134.1 (Ar-C), 132.9 (C5 of pyrazole), 130.4 (Ar-C), 129.9 (Ar-C), 129.6 (Ar-C), 128.6 (Ar-C), 128.4 (Ar-C), 127.5 (Ar-C), 125.8 (Ar-C), 122.3 (Ar-C), 119.5 (Ar-C), 117.4 (Ar-C), 115.5 (Ar-C), 110.8 (Ar-C), 102.4 (C4 of pyrazole); MS (m/z): 535.06 (M+); element analysis calculated (%) for C28H18BrN5O2: C 62.70, H 3.38, N 13.06; found: C 62.87, H 3.42, N 13.15.
4.1.5. 7-(4-Bromophenyl)-5-(1,3-diphenyl-1H-pyrazol-4-yl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4c)
Yield: 67%; solid; M.P. 228–230 °C; IR (KBr, cm–1): 3173, 2831 (C–H, −CH=CH−), 1744 (>C=O), 1689, 1676, 1254 (C=N, C=C, C–N), 689 (C-Br); 1H NMR (400 MHz, CDCl3): δ = 9.53 (s, 1H, Ar-NH), 9.41 (s, 1H, Ar-NH), 8.06 (s, 1H), 7.94–7.86 (m, 4H), 7.71–7.60 (m, 4H), 7.28–7.22 (m, 7H); 13C NMR (100 MHz, CDCl3): δ = 164.7 (>C=O), 160.0 (>C=O), 154.8 (Ar-C), 149.8 (Ar-C), 149.0 (Ar-C), 146.8 (C3 of pyrazole), 140.6 (Ar-C), 136.7 (Ar-C), 135.3 (Ar-C), 134.5 (Ar-C), 134.2 (Ar-C), 132.5 (C5 of pyrazole), 130.1 (Ar-C), 129.8 (Ar-C), 129.3 (Ar-C), 128.7 (Ar-C), 128.2 (Ar-C), 127.6 (Ar-C), 125.2 (Ar-C), 122.8 (Ar-C), 119.7 (Ar-C), 117.3 (Ar-C), 115.0 (Ar-C), 110.6 (Ar-C), 102.1 (C4 of pyrazole); MS (m/z): 535.06 (M+); element analysis calculated (%) for C28H18BrN5O2: C 62.70, H 3.38, N 13.06; found: C 62.85, H 3.47, N 13.16.
4.1.6. 7-(2-Chlorophenyl)-5-(1,3-diphenyl-1H-pyrazol-4-yl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4d)
Yield: 65%; solid; M.P. 192–193 °C; IR (KBr, cm–1): 3185, 2817 (C–H, −CH=CH−), 1759 (>C=O), 1689, 1662, 1282 (C=N, C=C, C–N), 743 (C-Cl); 1H NMR (400 MHz, CDCl3): δ = 9.34 (s, 1H, Ar-NH), 9.21 (s, 1H, Ar-NH), 8.46 (s, 1H, Ar-H), 7.89–7.69 (m, 5H, Ar-H), 7.58–7.51 (m, 5H, Ar-H), 7.23–7.16 (m, 5H, Ar-H); 13C NMR (100 MHz, CDCl3): δ = 163.6 (>C=O), 158.3 (>C=O), 152.6 (Ar-C), 149.4 (Ar-C), 147.4 (C3 of pyrazole), 140.8 (Ar-C), 139.5 (Ar-C), 136.3 (Ar-C), 135.7 (Ar-C), 134.4 (Ar-C), 132.8 (Ar-C), 131.6 (Ar-C), 131.4 (Ar-C), 131.1 (C5 of pyrazole), 129.6 (Ar-C), 129.4 (Ar-C), 129.1 (Ar-C), 128.4 (Ar-C), 128.2 (Ar-C), 127.6 (Ar-C), 126.3 (Ar-C), 122.4 (Ar-C), 119.7 (Ar-C), 118.5 (Ar-C), 112.7 (Ar-C), 106.4 (C4 of pyrazole); MS (m/z): 491.11 (M+); element analysis calculated (%) for C28H18ClN5O2: C 68.36, H 3.69, N 14.24; found: C 68.43, H,3.75, N 14.32.
4.1.7. 7-(4-Chlorophenyl)-5-(1,3-diphenyl-1H-pyrazol-4-yl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4e)
Yield: 69%; solid; M.P. 202–204 °C; IR (KBr, cm–1): 3182, 2837 (C–H, −CH=CH−), 1754 (>C=O), 1694, 1689, 1265 (C=N, C=C, C–N), 753 (C-Cl); 1H NMR (400 MHz, CDCl3): δ = 9.24 (s, 1H, Ar-NH), 9.18 (s, 1H, Ar-NH), 8.40 (s, 1H, Ar-H), 7.86–7.78 (m, 5H, Ar-H), 7.55–7.50 (m, 5H, Ar-H), 7.28–7.20 (m, 5H, Ar-H); 13C NMR (100 MHz, CDCl3): δ = 163.3 (>C=O), 158.6 (>C=O), 152.3 (Ar-C), 149.8 (Ar-C), 147.4 (C3 of pyrazole), 140.5 (Ar-C), 139.7 (Ar-C), 136.7 (Ar-C), 135.3 (Ar-C), 134.8 (Ar-C), 132.5 (Ar-C), 131.7 (Ar-C), 131.5 (Ar-C), 131.2 (C5 of pyrazole), 129.8 (Ar-C), 129.2 (Ar-C), 129.0 (Ar-C), 128.8 (Ar-C), 128.5 (Ar-C), 127.1 (Ar-C), 126.0 (Ar-C), 122.4 (Ar-C), 119.4 (Ar-C), 118.3 (Ar-C), 112.0 (Ar-C), 106.5 (C4 of pyrazole); MS (m/z): 491.11 (M+); element analysis calculated (%) for C28H18ClN5O2: C 68.36, H 3.69, N 14.24; found: C 68.49, H 3.78, N 14.34.
4.1.8. 5-(1,3-Diphenyl-1H-pyrazol-4-yl)-7-(4-fluorophenyl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4f)
Yield: 65%; solid; M.P. 199–201 °C; IR (KBr, cm–1): 3188, 2816 (C–H, −CH=CH−), 1759 (>C=O), 1679, 1652, 1275 (C=N, C=C, C–N), 1269 (C-F); 1H NMR (400 MHz, CDCl3): δ = 9.29 (s, 1H, Ar-NH), 9.22 (s, 1H, Ar-NH), 8.37 (s, 1H, Ar-H), 7.84–7.76 (m, 5H, Ar-H), 7.51–7.46 (m, 5H, Ar-H), 7.32–7.27 (m, 5H, Ar-H); 13C NMR (100 MHz, CDCl3): δ = 164.3 (>C=O), 158.3 (>C=O), 152.7 (Ar-C), 149.3 (Ar-C), 147.7 (C3 of pyrazole), 140.3 (Ar-C), 139.8 (Ar-C), 136.3 (Ar-C), 135.5 (Ar-C), 134.5 (Ar-C), 132.4 (Ar-C), 131.4 (Ar-C), 131.2 (Ar-C), 131.1 (C5 of pyrazole), 129.5 (Ar-C), 129.3 (Ar-C), 129.1 (Ar-C), 128.9 (Ar-C), 128.4 (Ar-C), 127.5 (Ar-C), 126.2 (Ar-C), 122.7 (Ar-C), 119.2 (Ar-C), 118.8 (Ar-C), 112.4 (Ar-C), 106.3 (C4 of pyrazole); MS (m/z): 475.14 (M+); element analysis calculated (%) for C28H18FN5O2: C 70.73, H 3.82, N 14.73; found: C 70.85, H 3.96, N 14.79.
4.1.9. 5-(1,3-Diphenyl-1H-pyrazol-4-yl)-7-(2-hydroxyphenyl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4g)
Yield: 73%; solid; M.P. 206–208 °C; IR (KBr, cm–1): 3545 (−OH), 3181, 2819 (C–H, −CH=CH−), 1752 (>C=O), 1685, 1665, 1279 (C=N, C=C, C–N); 1H NMR (400 MHz, CDCl3): δ 9.41 (s, 1H, Ar-NH), 9.12 (s, 1H, Ar-NH), 8.53 (s, 1H, Ar-H), 8.11–7.89 (m, 4H, Ar-H), 7.75–7.60 (m, 3H, Ar-H), 7.51–7.42 (m, 4H, Ar-H), 7.26–7.17 (m, 4H, Ar-H), 7.04 (s, 1H, Ar-OH); 13C NMR (100 MHz, CDCl3): δ = 165.8 (>C=O), 161.3 (>C=O), 156.4 (Ar-C), 153.6 (Ar-C), 151.7 (Ar-C), 149.3 (Ar-C), 142.6 (C3 of pyrazole), 140.3 (Ar-C), 135.7 (Ar-C), 133.4 (Ar-C), 132.4 (Ar-C), 131.6 (C5 of pyrazole), 129.6 (Ar-C), 129.5 (Ar-C), 129.2 (Ar-C), 129.1 (Ar-C), 128.5 (Ar-C), 126.7 (Ar-C), 126.8 (Ar-C), 122.2 (Ar-C), 122.1 (Ar-C), 120.7 (Ar-C), 118.3 (Ar-C), 116.6 (Ar-C), 113.6 (Ar-C), 107.4 (C4 of pyrazole); MS (m/z): 473.15 (M+); element analysis calculated (%) for C28H19N5O3: C 71.03, H 4.04, N 14.79; found: C 71.2, H 4.23, N 14.98.
4.1.10. 5-(1,3-Diphenyl-1H-pyrazol-4-yl)-7-(4-hydroxyphenyl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4h)
Yield: 77%; solid; M.P. 201–203 °C; IR (KBr, cm–1): 3554 (−OH), 3179, 2814 (C–H, −CH=CH−), 1758 (>C=O), 1679, 1661, 1277 (C=N, C=C, C–N); 1H NMR (400 MHz, CDCl3): δ = 9.34 (s, 1H, Ar-NH), 9.09 (s, 1H, Ar-NH), 8.44 (s, 1H, Ar-H), 8.05–7.99 (m, 4H, Ar-H), 7.70–7.65 (m, 3H, Ar-H), 7.56–7.44 (m, 4H, Ar-H), 7.27–7.20 (m, 4H, Ar-H), 7.08 (s, 1H, Ar-OH); 13C NMR (100 MHz, CDCl3): δ = 163.3 (>C=O), 161.0 (>C=O), 156.7 (Ar-C), 153.6 (Ar-C), 151.3 (Ar-C), 149.1 (Ar-C), 142.5 (C3 of pyrazole), 140.3 (Ar-C), 135.2 (Ar-C), 133.7 (Ar-C), 132.7 (Ar-C), 131.2 (C5 of pyrazole), 129.8 (Ar-C), 129.7 (Ar-C), 129.5 (Ar-C), 129.2 (Ar-C), 128.5 (Ar-C), 126.6 (Ar-C), 126.4 (Ar-C), 122.8 (Ar-C), 122.3 (Ar-C), 120.5 (Ar-C), 118.6 (Ar-C), 116.3 (Ar-C), 113.3 (Ar-C), 107.9 (C4 of pyrazole); MS (m/z): 473.15 (M+); element analysis calculated (%) for C28H19N5O3: C 71.03, H 4.04, N 14.79; found: C 71.19, H 4.22, N 14.96.
4.1.11. 5-(1,3-Diphenyl-1H-pyrazol-4-yl)-7-(2-methoxyphenyl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4i)
Yield: 81%; solid; M.P. 207–209 °C; IR (KBr, cm–1): 3182, 2816 (C–H, −CH=CH−), 1754 (>C=O), 1686, 1651, 1275 (C–N, C=C, C–N), 1141 (C-OCH3); 1H NMR (400 MHz, CDCl3): δ = 9.66 (s, 1H, Ar-NH), 9.42 (s, 1H, Ar-NH), 8.74 (s, 1H, Ar-H), 7.70–7.67 (m, 4H, Ar-H), 7.49–7.44 (m, 1H, Ar-H), 7.51–7.39 (m, 6H, Ar-H), 7.34–7.26 (m, 4H, Ar-H), 3.88 (s, 3H, Ar-OCH3); 13C NMR (100 MHz, CDCl3): δ = 163.4 (>C=O), 162.4 (>C=O), 158.6 (Ar-C), 154.8 (Ar-C), 153.6 (Ar-C), 149.7 (Ar-C), 143.4 (C3 of pyrazole), 140.5 (Ar-C), 139.6 (Ar-C), 138.3 (Ar-C), 132.7 (Ar-C), 132.4 (Ar-C), 131.4 (Ar-C), 131.1 (C5 of pyrazole), 129.9 (Ar-C), 129.7 (Ar-C), 129.1 (Ar-C), 127.6 (Ar-C), 126.3 (Ar-C), 125.2 (Ar-C), 121.7 (Ar-C), 120.4 (Ar-C), 118.4 (Ar-C), 111.6 (Ar-C), 110.3 (Ar-C), 105.6 (C4 of pyrazole), 50.9 (Ar-OCH3); 487.16 (M+); element analysis calculated (%) for C29H21N5O3: C 71.45, H 4.34, N 14.37; found: C 71.55, H 4.49, N 14.47.
4.1.12. 5-(1,3-Diphenyl-1H-pyrazol-4-yl)-7-(3-methoxyphenyl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4j)
Yield: 75%; solid; M.P. 210–212 °C; IR (KBr, cm–1): 3182, 2818 (C–H, −CH=CH−), 1756 (>C=O), 1684, 1658, 1278 (C=N, C=C, C–N), 1148 (C-OCH3); 1H NMR (400 MHz, CDCl3): δ = 9.50 (s, 1H, Ar-NH), 9.37 (s, 1H, Ar-NH), 8.71 (s, 1H, Ar-H), 7.76–7.70 (m, 4H, Ar-H), 7.57–7.53 (m, 1H, Ar-H), 7.51–7.44 (m, 6H, Ar-H), 7.28–7.23 (m, 4H, Ar-H), 3.91 (s, 3H, Ar-OCH3); 13C NMR (100 MHz, CDCl3): δ = 163.9 (>C=O), 162.8 (>C=O), 158.9 (Ar-C), 154.5 (Ar-C), 153.7 (Ar-C), 149.6 (Ar-C), 143.8 (C3 of pyrazole), 140.3 (Ar-C), 139.3 (Ar-C), 138.9 (Ar-C), 132.8 (Ar-C), 132.3 (Ar-C), 131.7 (Ar-C), 131.2 (C5 of pyrazole), 129.8 (Ar-C), 129.6 (Ar-C), 129.3 (Ar-C), 127.7 (Ar-C), 126.8 (Ar-C), 125.4 (Ar-C), 121.1 (Ar-C), 120.5 (Ar-C), 118.3 (Ar-C), 111.2 (Ar-C), 110.5 (Ar-C), 105.7 (C4 of pyrazole), 50.1 (Ar-OCH3); MS (m/z): 487.16 (M+); element analysis calculated (%) for C29H21N5O3: C 71.45, H 4.34, N 14.37; found: C 71.61, H 4.51, N 14.54.
4.1.13. 5-(1,3-Diphenyl-1H-pyrazol-4-yl)-7-(p-tolyl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4k)
Yield: 77%; solid; M.P. 198–200 °C; IR (KBr, cm–1): 3179, 2819 (C–H, −CH=CH−), 1758 (>C=O), 1685, 1667, 1278 (C=N, C=C, C–N); 1H NMR (400 MHz, CDCl3): δ = 9.02 (s, 1H, Ar-NH), 8.94 (s, 1H, Ar-NH), 8.17 (s, 1H, Ar-H), 7.75–7.68 (m, 6H, Ar-H), 7.56–7.39 (m, 6H, Ar-H), 7.28–7.22 (m, 3H, Ar-H), 2.53 (s, 3H, , Ar-CH3); 13C NMR (100 MHz, CDCl3): δ = 158.5 (>C=O), 154.7 (>C=O), 149.7 (Ar-C), 148.4 (Ar-C), 147.7 (Ar-C), 142.9 (C3 of pyrazole), 139.2 (Ar-C), 138.0 (Ar-C), 137.6 (Ar-C), 133.5 (Ar-C), 133.3 (Ar-C), 131.5 (C5 of pyrazole), 131.0 (Ar-C), 129.8 (Ar-C), 129.3 (Ar-C), 129.2 (Ar-C), 128.5 (Ar-C), 127.7 (Ar-C), 127.3 (Ar-C), 126.2 (Ar-C), 124.2 (Ar-C), 121.7 (Ar-C), 118.7 (Ar-C), 115.1 (Ar-C), 113.1 (Ar-C), 106.3 (C4 of pyrazole), 25.3 (Ar-CH3); MS (m/z): 471.17 (M+); element analysis calculated (%) for C29H21N5O2: C 73.87, H 4.49, N 14.85; found: C 73.97, H 4.57, N 14.92.
4.1.14. 5-(1,3-Diphenyl-1H-pyrazol-4-yl)-7-(4-nitrophenyl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4l)
Yield: 67%; solid; M.P. 228–230 °C; IR (KBr, cm–1): 3180, 2824 (C–H, −CH=CH−), 1761 (>C=O), 1679, 1669, 1545 (C-NO2), 1279 (C=N, C=C, C–N); 1H NMR (400 MHz, CDCl3): δ = 9.34 (s, 1H, Ar-NH), 9.27 (s, 1H, Ar-NH), 8.34 (s, 1H, Ar-H), 7.83–7.78 (m, 5H, Ar-H), 7.56–7.48 (m, 5H, Ar-H), 7.33–7.25 (m, 5H, Ar-H); 13C NMR (100 MHz, CDCl3): δ = 164.5 (>C=O), 158.3 (>C=O), 152.4 (Ar-C), 149.3 (Ar-C), 147.7 (C3 of pyrazole), 140.6 (Ar-C), 139.3 (Ar-C), 136.4 (Ar-C), 135.8 (Ar-C), 134.4 (Ar-C), 132.2 (Ar-C), 131.8 (Ar-C), 131.4 (Ar-C), 131.3 (C5 of pyrazole), 129.7 (Ar-C), 129.3 (Ar-C), 129.2 (Ar-C), 128.6 (Ar-C), 128.3 (Ar-C), 127.5 (Ar-C), 126.3 (Ar-C), 122.8 (Ar-C), 119.6 (Ar-C), 118.2 (Ar-C), 112.6 (Ar-C), 106.3 (C4 of pyrazole); MS (m/z): 502.14 (M+); element analysis calculated (%) for C28H18N6O4: C 66.93, H 3.61, N 16.73; found: C 67.12, H 3.75, N 16.77.
4.1.15. 7-(2,4-Dichlorophenyl)-5-(1,3-diphenyl-1H-pyrazol-4-yl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4m)
Yield: 77%; solid; M.P. 213–215 °C; IR (KBr, cm–1): 3182, 2818 (C–H, −CH=CH−), 1759 (>C=O), 1684, 1657, 1271 (C=N, C=C, C–N), 758 (C-Cl); 1H NMR (400 MHz, CDCl3): δ = 9.46 (s, 1H, Ar-NH), 9.37 (s, 1H, Ar-NH), 8.62 (s, 1H, Ar-H), 7.89–7.86 (m, 4H, Ar-H), 7.64–7.57 (m, 8H, Ar-H), 7.30–7.19 (m, 2H, Ar-H); 13C NMR (100 MHz, CDCl3): δ = 163.4 (>C=O), 161.3 (>C=O), 157.8 (Ar-C), 155.3 (Ar-C), 151.8 (Ar-C), 144.4 (Ar-C), 141.2 (C3 of pyrazole), 138.7 (Ar-C), 137.2 (Ar-C), 135.6 (Ar-C), 134.2 (Ar-C), 132.9 (Ar-C), 132.5 (C5 of pyrazole), 130.3 (Ar-C), 129.7 (Ar-C), 129.5 (Ar-C), 128.8 (Ar-C), 127.3 (Ar-C), 127.4 (Ar-C), 126.3 (Ar-C), 123.5 (Ar-C), 118.3 (Ar-C), 116.7 (Ar-C), 112.3 (Ar-C), 101.9 (C4 of pyrazole); MS (m/z): 525.08 (M+); element analysis calculated (%) for C28H17Cl2N5O2: C 63.89, H 3.26, N 13.31; found: C 63.98, H 3.34, N 13.45.
4.1.16. 7-(3,4-Dichlorophenyl)-5-(1,3-diphenyl-1H-pyrazol-4-yl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4n)
Yield: 77%; solid; M.P. 213–215 °C; IR (KBr, cm–1): 3182, 2818 (C–H, −CH=CH−), 1759 (>C=O), 1684, 1657, 1271 (C=N, C=C, C–N), 758 (C-Cl); 1H NMR (400 MHz, CDCl3): δ = 9.39 (s, 1H, Ar-NH), 9.32 (s, 1H, Ar-NH), 8.67 (s, 1H, Ar-H), 7.98–7.90 (m, 4H, Ar-H), 7.57–7.42 (m, 8H, Ar-H), 7.32–7.22 (m, 2H, Ar-H); 13C NMR (100 MHz, CDCl3): δ = 163.9 (>C=O), 161.7 (>C=O), 157.5 (Ar-C), 155.6 (Ar-C), 151.5 (Ar-C), 144.7 (Ar-C), 141.9 (C3 of pyrazole), 138.6 (Ar-C), 137.5 (Ar-C), 135.0 (Ar-C), 134.7 (Ar-C), 132.8 (Ar-C), 132.0 (C5 of pyrazole), 130.4 (Ar-C), 129.9 (Ar-C), 129.3 (Ar-C), 128.2 (Ar-C), 127.9 (Ar-C), 127.7 (Ar-C), 126.8 (Ar-C), 123.5 (Ar-C), 118.3 (Ar-C), 116.1 (Ar-C), 112.5 (Ar-C), 101.6 (C4 of pyrazole); MS (m/z): 525.08 (M+); element analysis calculated (%) for C28H17Cl2N5O2: C 63.89, H 3.26, N 13.31; found: C 63.98, H 3.34, N 13.45.
4.1.17. 7-(2,4-Difluorophenyl)-5-(1,3-diphenyl-1H-pyrazol-4-yl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione (4o)
Yield: 75%; solid; M.P. 204–206 °C; IR (KBr, cm–1): 3174, 2814 (C–H, −CH=CH−), 1751 (>C=O), 1684, 1664, 1277 (C=N, C=C, C–N), 1287 (C-F); 1H NMR (400 MHz, CDCl3): δ = 9.43 (s, 1H, Ar-NH), 9.40 (s, 1H, Ar-NH), 8.76 (s, 1H, Ar-H), 7.87–7.76 (m, 4H, Ar-H), 7.49–7.39 (m, 8H, Ar-H), 7.27–7.15 (m, 2H, Ar-H); 13C NMR (100 MHz, CDCl3): δ = 165.2 (>C=O), 161.5 (>C=O), 157.6 (Ar-C), 155.3 (Ar-C), 151.8 (Ar-C), 144.5 (Ar-C), 141.6 (C3 of pyrazole), 138.3 (Ar-C), 137.7 (Ar-C), 135.4 (Ar-C), 134.8 (Ar-C), 132.4 (Ar-C), 132.1 (C5 of pyrazole), 130.7 (Ar-C), 129.8 (Ar-C), 129.5 (Ar-C), 128.1 (Ar-C), 127.6 (Ar-C), 127.3 (Ar-C), 126.3 (Ar-C), 123.7 (Ar-C), 118.4 (Ar-C), 116.6 (Ar-C), 112.3 (Ar-C), 101.6 (C4 of pyrazole); MS (m/z): 493.14 (M+); element analysis calculated (%) for C28H17F2N5O2: C 68.15, H 3.47, N 14.19; found: C 68.45, H 3.54, N 14.56.
5. Materials and Methods
5.1. Antifungal Bioassay
“The synthesized compounds (4a–4o) were screened for their antifungal activity against Candida albicans (MTCC-227), Aspergillus niger (MTCC-282), Aspergillus clavatus (MTCC-1323) at concentrations of 1000, 500, and 250 μg/mL. The experiments were conducted in triplicates. The active compounds obtained as above in primary screening were similarly diluted to obtain 200, 125, 100, 62.5, 50, 25, and 12.5 μg/mL concentrations for secondary screening to test in the second set of dilutions against all microorganisms. ‘Griseofulvin’ was used as a standard drug for antifungal activity and was considered a positive control. For the growth of fungi, Sabouraud dextrose broth was used and incubated at 28 °C in aerobic conditions for 48 h. DMSO and sterilized distilled water were used as negative controls.”40 (Figures S2–S4).
5.2. Molecular Docking and Computational Methodology for Target β-Tubulin
The β-tubulin protein sequence of C. albicans was retrieved from the NCBI protein database. A protein–protein blast (BLASTP)41,42 was carried out to obtain homologous proteins having an experimental structure retrieved from the Protein Data Bank (PDB). The sequences of target and template proteins were aligned using clustalX, and the protein structure of C. albicans β-tubulin protein was constructed using the template protein by homology modeling using Modeller 10.43−45 The protein structure was validated by protein structure validation server SAVES v 6.0 of UCLA-DOE Lab.46−48 The constructed protein of C. albicans β-tubulin was prepared using the protein preparation wizard in Schrödinger Suite using the default parameter. A grid was constructed at the Taxol binding site, and molecules 4a–4o were docked into the Taxol binding site by the GLIDE module using the extra precision (XP) mode. The docked complexes were further subjected to binding energy calculation using PRIME-MMGBSA. Molecule 4a was optimized, and vibrational frequency calculations were performed in Gaussian applying the DFT method using B3LYP/6-31G* level theory. ADME properties were calculated using the QikProp module of Schrödinger suite, and toxicity parameters were calculated by the online pkCSM server.49
5.3. Ergosterol Inhibition Activity
5.3.1. Materials and Methods
Ergosterols were extracted by using the standard protocol.50 The sterols were quantitated by Breivik and Owades in 1957.30 Overnight-grown Candida culture was inoculated in 50 mL of the potato dextrose broth containing the MIC concentration of compounds. All the solutions were prepared in 2 mL of DMSO according to their MIC (500 μg/mL for compounds 4h, 4j, and 4l; 250 μg/mL for compounds 4k; and 200 μg/mL for compounds 4n), 50 μg/mL concentration of fluconazole, and 2 mL concentration of DMSO. The cultures were incubated on a shaker at 37 °C for 72 h. The cultures were centrifuged at 5000 rpm for 5 min and washed once with sterile distilled water for harvesting the fungal cells. The net wet weight of the cell pellet was measured. 3 mL of 25% alcoholic KOH was added to each pellet and mixed by vortexing for 1 min. This was incubated at 85 °C water bath for 1 h. A mixture of 1 mL of sterile distilled water and 3 mL of n-heptane was added to each tube after cooling for extraction of sterols. The upper heptane layer was transferred to a clean tube and diluted 10 times with n-heptane:ethanol (1:1) and canned spectrophotometrically from 240 and 300 nm. Ergosterol content was calculated as a percentage of the wet weight of the cell by the following equations:
| 1 |
| 2 |
| 3 |
| 4 |
where F is the factor for dilution in ethanol and 290 and 518 are the E values (in percentages per centimeter) determined for crystalline ergosterol and 24(28)DHE, respectively.
5.3.2. Molecular Docking Analysis of Target Sterol 14-Alpha Demethylase Proteins/Macromolecules
Sterol 14-demethylase is one of the important enzymes in ergosterol biosynthesis in fungi and is used as a target for many antifungal agents.32,33 This enzyme was selected to find the mechanism of action of these compounds as an antifungal candidate. The 3D crystal structure of this enzyme from C. albicans (PDB ID: 5v5z) was downloaded from the Protein Data Bank (https://www.rcsb. org/), in .pdb format.51 The protein molecule was cleaned by removing bound complexes. The protein structure was prepared by removing water molecules using PyMOL version 2.3.3.
5.3.3. Preparation of Ligands
The 3D structures of the ligands were drawn, and energy was minimized by Open Babel, an open-source platform.
5.3.4. Determination of Active Sites
The Computed Atlas for Surface Topography of Proteins (CASTp)52 and BIOVIA Discovery Studio 4.553 were used to determine the amino acids in the active site of a protein (http://sts.bioe.uic.edu/castp/index.html?201l).
5.3.5. Molecular Docking
Ligand and protein optimization was done using PyMOL version 2.3.3 [The PyMOL Molecular Graphics System, Version 2.0 Schrödinger LLC]. For ligand optimization, the geometry of ligands was cleaned, whereas, for protein, the water was removed. The docking was performed by using PyRx 0.8.54 The docking analyses were performed using both PyMOL as well as BIOVIA Discovery Studio 4.5.51
5.4. Anti-Biofilm Activity
5.4.1. Materials and Methods
The ability of the compounds (4c, 4e, 4h, 4i, 4j, 4l, 4n, and 4k) to inhibit biofilm formation was measured using the microplate-based assay, as described in ref (34) with three different marine biofilm-forming bacterial strains (GS-5, Vibrio sp. (Gram-negative); BM-14, Staphylococcus sp. (Gram-positive); and GM-17, Gallaecimonas sp. (Gram-negative)).35 Bacterial isolates were cultured in ZMB (Zobell Marine Broth) overnight and diluted in sterile ZMB broth containing 1% glucose in 1:10 ratio. 10 μL of each chemical compound in the concentration of 400 μg/mL dissolved in DMSO was transferred into the wells of a microtiter plate containing 90 μL of diluted bacterial culture and replicated six times. Thereafter, the plate was kept on a stirrer for proper mixing of culture and compounds and kept in a static incubator at 30 °C for 24 h. After incubation, the wells were washed three times with distilled water and dripped with 1% crystal violet solution (100 μL), incubated at room temperature for 30 min, and again, washed with distilled water three times and dried at room temperature. Next, 100 μL of 70% ethanol was added to each well, and optical density was measured at 590 nm using a microplate spectrophotometer. Bacterial culture without compound was used as control. The percentage biofilm inhibition was calculated with the following formula:
| 5 |
For the antimicrobial susceptibility assay, the disc diffusion method was used. With the help of a sterile cell spreader, the bacterial colony adjusted to 0.6 OD, spread on Zobell marine agar, and dried for 15 min. Discs were impregnated in the chemical compounds (400 μg/mL) and then placed on a Zobell Marine Agar plate. Each plate contained five disc, four chemical-compound-treated discs, and one as a control followed by incubation at 37 °C for 48 h, and the zone of inhibition was examined.
Acknowledgments
N.C.D. is appreciative to the UGC, New Delhi, for grant BSR Faculty Fellowship 2019 [No. F. 18-1/2011(BSR)] and financial help. J.D.M. is grateful to the Department of Science and Technology, INSPIRE PROGRAM, for the award of INSPIRE Fellowship [No. IF180817]. A.M.J. is thankful to SHODH (ScHeme Of Developing High-quality research) for providing a research fellowship, KCG, Government of Gujarat. The authors are thankful to Priyanka Desai, founder of iScribblers for the linguistic editing of the manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c01722.
Antibacterial activities of chemical compounds; antifungal activity against C. albicans; antifungal activity against A. niger; antifungal activity against A. clavatus; Procheck–Ramachandran plot; Verify3D plot (Verify3D—Pass); Errat—overall quality factor 77.6722; ProSa; ligand interaction diagram of 4a; ligand interaction diagram of 4k; ligand interaction diagram of 4n; ligand interaction diagram of griseofulvin; IR frequency data obtained from DFT; IR frequency data; 1H NMR spectra of compound 4a; 13C NMR spectra of compound 4a; mass spectra of compound 4a; IR spectra of compound 4a; 1H NMR spectra of compound 4c; 13C NMR spectra of compound 4c; 1H NMR spectra of compound 4e; 13C NMR spectra of compound 4e; mass spectra of compound 4e; 1H NMR spectra of compound 4h; 13C NMR spectra of compound 4h; mass spectra of compound 4h; 1H NMR spectra of compound 4j; 13C NMR spectra of compound 4j; mass spectra of compound 4j; 1H NMR spectra of compound 4k; 13C NMR spectra of compound 4k; mass spectra of compound 4k; 1H NMR spectra of compound 4n; 13C NMR spectra of compound 4n; mass spectra of compound 4n (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Vandeputte P.; Ferrari S.; Coste A. T. Antifungal Resistance and New Strategies to Control Fungal Infections. Int. J. Microbiol. 2012, 2012, 1–26. 10.1155/2012/713687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch D. A.; Ludlam H. A.; Brown N. M. Invasive Fungal Infections: A Review of Epidemiology and Management Options. J. Med. Microbiol. 2006, 55, 809–818. 10.1099/jmm.0.46548-0. [DOI] [PubMed] [Google Scholar]
- Steenbergen J. N.; Casadevall A. Prevalence of Cryptococcus Neoformans Var. Neoformans (Serotype D) and Cryptococcus Neoformans Var. Grubii (Serotype A) Isolates in New York City. J. Clin. Microbiol. 2000, 38, 1974–1976. 10.1128/JCM.38.5.1974-1976.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan D. J.; Hitchcock C. A.; Sibley C. M. Current and Emerging Azole Antifungal Agents. Clin. Microbiol. Rev. 1999, 12, 40–79. 10.1128/CMR.12.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert R.; Sedlacek L.; Kahl B.; Hogardt M.; Hamprecht A.; Haase G.; Gunzer F.; Haas A.; Grauling-Halama S.; MacKenzie C. R.; Essig A.; Stehling F.; Sutharsan S.; DIttmer S.; Killengray D.; Schmidt D.; Eskandarian N.; Steinmann E.; Buer J.; Hagen F.; Meis J. F.; Rath P. M.; Steinmann J. Prevalence and Characterization of Azole-Resistant Aspergillus Fumigatus in Patients with Cystic Fibrosis: A Prospective Multicentre Study in Germany. J. Antimicrob. Chemother. 2018, 73, 2047–2053. 10.1093/jac/dky147. [DOI] [PubMed] [Google Scholar]
- Desai N.; Monapara J.; Jethawa A.; Khedkar V.; Shingate B. Oxadiazole: A highly versatile scaffold in drug discovery. Arch. Pharm. 2022, 355, e2200123 10.1002/ardp.202200123. [DOI] [PubMed] [Google Scholar]
- Almulla A. F.; Pharma D.; Al-Mulla A. ISSN 0975-413X CODEN (USA): PCHHAX A Review: Biological Importance of Heterocyclic Compounds Biomarkers of Treatment Resistant (Deficit) Schizophrenia View Project Immunopsychiatry View Project ISSN 0975-413X CODEN (USA): PCHHAX A Review. Biological Importance of Heterocyclic Compounds. 2017, 9, 141–147. [Google Scholar]
- Mermer A.; Keles T.; Sirin Y. Recent Studies of Nitrogen Containing Heterocyclic Compounds as Novel Antiviral Agents: A Review. Bioorg. Chem. 2021, 114, 105076 10.1016/j.bioorg.2021.105076. [DOI] [PubMed] [Google Scholar]
- Desai N.; Shihory N.; Khasiya A.; Pandit U.; Khedkar V. Quinazoline Clubbed Thiazole and 1,3,4-Oxadiazole Heterocycles: Synthesis, Characterization, Antibacterial Evaluation, and Molecular Docking Studies. Phosphorus, Sulfur Silicon Relat. Elem. 2021, 196, 569–577. 10.1080/10426507.2021.1871732. [DOI] [Google Scholar]
- Thakral S.; Singh V. Recent Development on Importance of Heterocyclic Amides as Potential Bioactive Molecules: A Review. Curr. Bioact. Compd. 2018, 15, 316–336. [Google Scholar]
- Desai N. C.; Pandya D. D.; Jadeja D. J.; Panda S. K.; Rana M. K. Design, Synthesis, Biological Evaluation and Molecular Docking Study of Novel Hybrid of Pyrazole and Benzimidazoles. Chem. Data Collect. 2021, 33, 100703 10.1016/j.cdc.2021.100703. [DOI] [Google Scholar]
- Surendra Kumar R.; Arif I. A.; Ahamed A.; Idhayadhulla A. Anti-Inflammatory and Antimicrobial Activities of Novel Pyrazole Analogues. Saudi J. Biol. Sci. 2016, 23, 614–620. 10.1016/j.sjbs.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R.; Mary Y. S.; Resmi K. S.; Narayana B.; Sarojini B. K.; Vijayakumar G.; Van Alsenoy C. Two Neoteric Pyrazole Compounds as Potential Anti-Cancer Agents: Synthesis, Electronic Structure, Physico-Chemical Properties and Docking Analysis. J. Mol. Struct. 2019, 1181, 455–466. 10.1016/j.molstruc.2019.01.003. [DOI] [Google Scholar]
- Desai N. C.; Bhatt K.; Monapara J.; Pandit U.; Khedkar V. M. Conventional and Microwave-Assisted Synthesis, Antitubercular Activity, and Molecular Docking Studies of Pyrazole and Oxadiazole Hybrids. ACS Omega 2021, 6, 28270–28284. 10.1021/acsomega.1c04411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N. C.; Monapara J. D.; Jethawa A. M.; Pandit U.. Contemporary Development in the Synthesis and Biological Applications of Pyridine-Based Heterocyclic Motifs. Recent Developments in the Synthesis and Applications of Pyridines; Elsevier.2023, 253–298, 10.1016/B978-0-323-91221-1.00007-5. [DOI] [Google Scholar]
- Malani A.; Makwana A.; Monapara J.; Ahmad I.; Patel H.; Desai N. Synthesis, Molecular Docking, DFT Study, and in Vitro Antimicrobial Activity of Some 4-(Biphenyl-4-Yl)-1,4-Dihydropyridine and 4-(Biphenyl-4-Yl)Pyridine Derivatives. J. Biochem. Mol. Toxicol. 2021, 35, e22903 10.1002/jbt.22903. [DOI] [PubMed] [Google Scholar]
- El-Sayed N. N. E.; Abdelaziz M. A.; Wardakhan W. W.; Mohareb R. M. The Knoevenagel Reaction of Cyanoacetylhydrazine with Pregnenolone: Synthesis of Thiophene, Thieno[2,3-d]Pyrimidine, 1,2,4-Triazole, Pyran and Pyridine Derivatives with Anti-Inflammatory and Anti-Ulcer Activities. Steroids 2016, 2016, 98–111. 10.1016/j.steroids.2015.12.023. [DOI] [PubMed] [Google Scholar]
- Abdel-latif E.; Abdel-fattah S.; Gaffer H. E.; Etman H. A. Synthesis and Antitumor Activity of Some New Pyrazolo[3,4-d]Pyrimidine and Pyrazolo[3,4-b]Pyridine Derivatives. Egypt. J. Basic Appl. Sci. 2016, 3, 118–124. 10.1016/j.ejbas.2015.11.001. [DOI] [Google Scholar]
- Prachayasittikul S.; Pingaew R.; Worachartcheewan A.; Sinthupoom N.; Prachayasittikul V.; Ruchirawat S.; Prachayasittikul V. Roles of Pyridine and Pyrimidine Derivatives as Privileged Scaffolds in Anticancer Agents. Mini-Rev. Med. Chem. 2016, 17, 869–901. 10.2174/1389557516666160923125801. [DOI] [PubMed] [Google Scholar]
- Desai N. C.; Trivedi A. R.; Khedkar V. M. Preparation, Biological Evaluation and Molecular Docking Study of Imidazolyl Dihydropyrimidines as Potential Mycobacterium Tuberculosis Dihydrofolate Reductase Inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 4030–4035. 10.1016/j.bmcl.2016.06.082. [DOI] [PubMed] [Google Scholar]
- Desai N. C.; Somani H. C.; Mehta H. K.; Jadeja D. J.; Khasiya A. G.; Khedkar V. M. Microwave-Assisted Organic Synthesis, Antimycobacterial Activity, Structure–Activity Relationship and Molecular Docking Studies of Some Novel Indole-Oxadiazole Hybrids. SAR QSAR Environ. Res. 2022, 33, 89–109. 10.1080/1062936X.2022.2032333. [DOI] [PubMed] [Google Scholar]
- Walunj Y.; Nandurkar Y.; Shinde A.; Jagadale S.; Shaikh A. L. N.; Modak M.; Mhaske P. C. Synthesis, antimicrobial and ergosterol biosynthesis inhibition activity of clubbed 1,1′-biphenyl-pyrazole derivatives. New J. Chem. 2023, 47, 3810–3824. 10.1039/D2NJ04449H. [DOI] [Google Scholar]
- Abdelhameed R. M.; Darwesh O. M.; El-Shahat M. Synthesis of Arylidene Hydrazinylpyrido [2, 3-d] pyrimidin-4-ones as Potent Anti-microbial Agents. Heliyon 2020, 6, e04956 10.1016/j.heliyon.2020.e04956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelhamid A. O.; El Sayed I. E.; Hussein M. Z.; Mangoud M. M. Synthesis and Antimicrobial Activity of some New Thiadiazoles, Thioamides, 5-Arylazothiazoles and Pyrimido [4, 5-d][1, 2, 4] triazolo [4, 3-a] pyrimidines. Molecules 2016, 21, 1072. 10.3390/molecules21081072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P.; Rane N.; Gurram V. K. Synthesis and QSAR Studies of Pyrimido [4, 5-d] pyrimidine-2, 5-dione Derivatives as Potential Antimicrobial Agents. Bioorg. Med. Chem. Lett. 2004, 14, 4185–4190. 10.1016/j.bmcl.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Desai N. C.; Khasiya A. G. Design, Synthesis and in vitro Antimicrobial Activity of Fused Pyridine-pyrimidine Hybrids. Indian J. Chem. 2022, 61, 582–590. 10.3390/scipharm89040049. [DOI] [Google Scholar]
- Keates R. A. B. Griseofulvin at low concentration inhibits the rate of microtubule polymerization in vitro. Biochem. Biophys. Res. Commun. 1981, 102, 746–752. 10.1016/S0006-291X(81)80195-7. [DOI] [PubMed] [Google Scholar]
- Rathinasamy K.; Jindal B.; Asthana J.; Singh P.; Balaji P. V.; Panda D. Griseofulvin Stabilizes Microtubule Dynamics, Activates P53 and Inhibits the Proliferation of MCF-7 Cells Synergistically with Vinblastine. BMC Cancer 2010, 10, 213. 10.1186/1471-2407-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthington-Skaggs B. A.; Jradi H.; Desai T.; Morrison C. J. Quantitation of Ergosterol Content : Novel Method for Determination of Fluconazole Susceptibility of Candida Albicans. J. Clin. Microbiol. 1999, 37, 3332–3337. 10.1128/JCM.37.10.3332-3337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breivik O. N.; Owades J. L. Yeast Analysis, Spectrophotometric Semimicrodetermination of Ergosterol in Yeast. J. Agric. Food Chem. 1957, 5, 360–363. 10.1021/jf60075a005. [DOI] [Google Scholar]
- Hargrove T. Y.; Friggeri L.; Wawrzak Z.; Qi A.; Hoekstra W. J.; Schotzinger J.; York J. D.; Guengerich F. P.; Lepesheva G. I. Structural Analyses of Candida Albicans Sterol 14 α -Demethylase Complexed with Azole Drugs Address the Molecular Basis of Azole-Mediated Inhibition of Fungal Sterol Biosynthesis. J. Biol. Chem. 2017, 292, 6728–6743. 10.1074/jbc.M117.778308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove T. Y.; Friggeri L.; Wawrzak Z.; Qi A.; Hoekstra W. J.; Schotzinger R. J.; York J. D.; Guengerich F. P.; Lepesheva G. I. Structural analyses of Candida albicans sterol 14α-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis. J. Biol. Chem. 2017, 292, 6728–6743. 10.1074/jbc.M117.778308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A.; Michielin O.; Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amengor C. D. K.; Kekessie F. K.; Brobbey A.; Addotey J. N.; Peprah P.; Amaning-Danquah C.; Adu J.; Tetteh M.; Adusei E. B. A. Anti-biofilm Formation Activities of 4-hydroxyindole Azo Compounds against Pseudomonas aeruginosa and Staphylococcus aureus. J. Sci. Res. Rep. 2022, 28, 77–88. 10.9734/jsrr/2022/v28i830541. [DOI] [Google Scholar]
- Kumar M.; Kumar R.; Chaudhary D. R.; Jha B. An appraisal of early stage biofilm-forming bacterial community assemblage and diversity in the Arabian Sea, India. Mar. Pollut. Bull. 2022, 180, 113732 10.1016/j.marpolbul.2022.113732. [DOI] [PubMed] [Google Scholar]
- Lotlikar S. R.; Gallaway E.; Grant T.; Popis S.; Whited M.; Guragain M.; Rogers R.; Hamilton S.; Gerasimchuk N. G.; Patrauchan M. A. Polymeric composites with silver (I) cyanoximates inhibit biofilm formation of gram-positive and gram-negative bacteria. Polymer 2019, 11, 1018. 10.3390/polym11061018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourkhosravani E.; Dehghan Nayeri F.; Mohammadi Bazargani M. Decoding antibacterial and antibiofilm properties of cinnamon and cardamom essential oils: a combined molecular docking and experimental study. AMB Express 2021, 11, 143. 10.1186/s13568-021-01305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N. C.; Joshi S. B.; Jadeja K. A. A One-Pot Multicomponent Biginelli Reaction for the Preparation of Novel Pyrimidinthione Derivatives as Antimicrobial Agents. J. Heterocycl. Chem. 2020, 57, 791–795. 10.1002/jhet.3821. [DOI] [Google Scholar]
- Desai N. C.; Joshi S. B.; Khedkar V. M. Synthesis, Antimicrobial Activity and Molecular Docking of Pyrazole Bearing the Benzodiazepine Moiety. Anal. Chem. Lett. 2020, 10, 307–320. 10.1080/22297928.2020.1785325. [DOI] [Google Scholar]
- Desai N. C.; Rupala Y. M.; Khasiya A. G.; Shah K. N.; Pandit U. P.; Khedkar V. M. Synthesis, Biological Evaluation, and Molecular Docking Study of Thiophene-, Piperazine-, and Thiazolidinone-Based Hybrids as Potential Antimicrobial Agents. J. Heterocycl. Chem. 2022, 59, 75–87. 10.1002/jhet.4366. [DOI] [Google Scholar]
- Altschul S. F.; Madden T. L.; Schäffer A. A.; Zhang J.; Zhang Z.; Miller W.; Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F.; Wootton J. C.; Gertz E. M.; Agarwala R.; Morgulis A.; Schäffer A. A.; Yu Y.-K. Protein Database Searches Using Compositionally Adjusted Substitution Matrices. FEBS J. 2005, 272, 5101–5109. 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí-Renom M. A.; Stuart A. C.; Fiser A.; Sänchez R.; Melo F.; Šali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 291–325. 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Šali A.; Blundell T. L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 779–815. 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Fiser A.; Do R. K. G.; Šali A. Modeling of Loops in Protein Structures. Protein Sci. 2000, 9, 1753–1773. 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos C.; Yeates T. O. Verification of Protein Structures: Patterns of Nonbonded Atomic Interactions. Protein Sci. 1993, 2, 1511–1519. 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie J. U.; Lüthy R.; Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991, 253, 164–170. 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Laskowski R. A.; MacArthur M. W.; Moss D. S.; Thornton J. M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Pires D. E. V.; Blundell T. L.; Ascher D. B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalawade J.; Shinde A.; Chavan A.; Patil S.; Suryavanshi M.; Modak M.; Choudhari P.; Bobade V. D.; Mhaske P. C. Synthesis of New Thiazolyl-Pyrazolyl-1 , 2 , 3-Triazole Derivatives as Potential Antimicrobial Agents. Eur. J. Med. Chem. 2019, 179, 649–659. 10.1016/j.ejmech.2019.06.074. [DOI] [PubMed] [Google Scholar]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W.; Chen C.; Lei X.; Zhao J.; Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biovia D. S.Discovery Studio Visualizer; BIOVIA: San Diego, 2019. [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 3, 1455–1461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.