ABSTRACT
We report the discovery and genome sequence of bacteriophage Aoka, an actinobacteriophage isolated from a soil sample in Pueblo, Colorado using Arthrobacter globiformis, B2880-SEA. Its genome length is 36,744 base pairs with 54 protein-coding genes. Based on gene content similarity to other actinobacteriophages, Aoka is assigned to cluster FO.
KEYWORDS: actinophage, bacteriophage, Arthrobacter globiformis, phage genome
ANNOUNCEMENT
Bacteriophage are viruses that infect bacteria (1, 2), resulting in bacterial-phage co-evolution (1, 3, 4). Knowledge of bacteriophage gene content and functions is essential to better understand the evolutionary and physiological impacts on bacteria of these infections, yet current findings indicate that the phage genetic space is mostly unknown (5, 6). Here, we present the genome of actinobacteriophage Aoka, sequenced in collaboration with the SEA-PHAGES program sponsored by Howard Hughes Medical Institute (HHMI) (7). Over a third of the open reading frames in Aoka’s genome code for unique proteins of unknown functions as of May 2023.
Aoka was isolated and purified from soil collected around a tree stump in a grassfield in Pueblo, Colorado (38.346111 N, 104.704722 W) in September 2022 using the host A. globiformis B2880-SEA and a host enrichment method with PYCa media at 30°C as outlined in the SEA-PHAGES manual (8), with two rounds of plaque purification. Aoka consistently forms large clear plaques and exhibits a Siphovirus morphology (9) with a capsid 45 ± 5 nm in diameter and tail 140 ± 5 nm in length (n = 3; Fig. 1).
Fig 1.
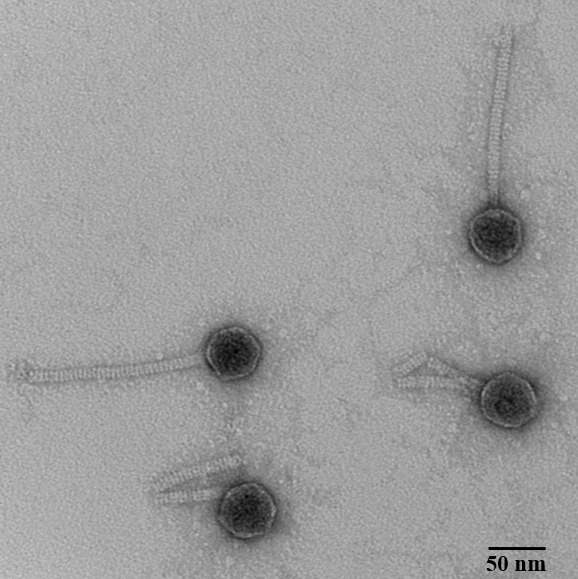
Transmission electron micrographs of phage Aoka. A high-titer (>1.0 × 105 pfu/mL) lysate was negatively stained with 1% uranyl acetate and visualized in a Thermo Fisher Tecnai G2 12 BioTwin electron microscope at 120.0 kV.
Genomic DNA extraction was performed from phage lysate using Promega’s Wizard DNA Extraction Kit (10). A sequencing library was prepared with a NEBNext Ultra II FS kit and sequenced using Illumina MiSeq platform (v3 reagents) yielding 414,384 150-base single-end reads. Raw reads were assembled using Newbler 2.9, with default settings, into a single-phage contig with approximate average coverage of 1,673-fold; the contig was checked for completeness, accuracy, and phage genomic termini with Consed 29 as described previously (10, 11). The genome is 36,744 bp long and has a G + C content of 68.6% with 11-bp 3′ single-stranded overhangs (5′-TTCGCCCGTTA-3′). Based on average nucleotide similarity of at least 35% to phages in the Actinobacteriophage database, Aoka is assigned to cluster FO (12, 13).
An automated annotation was generated using Glimmer (14) and GeneMark (15) and, subsequently, manually curated using DNA Master (16), Phamerator (17), and Starterator (18). Functions for each coding sequence were assigned based on top hits from searches using NCBI BLASTP (19), Phagesdb BLASTP (20), and HHpred (21). Membrane proteins were identified using TMHMM v2.0 (22) and SOSUI (23). Aragorn (24) and tRNAscan-SE (25) were used to identify tRNAs. All tools were run with default parameters.
Aoka is predicted to have 54 protein-coding genes; 24 of which (45%) have assigned functions, and 30 (55%) were annotated as hypothetical proteins. Currently, 19 (35%) of these hypotheticals are unique genes to Aoka (26, 27). No tRNA-coding genes were found. Most genes (n = 51) are predicted to be transcribed in a forward orientation and three in reverse orientation (Genes 27 and 32; both helix-turn-helix DNA binding domain, and Gene 28; hypothetical). No repressor or integrase coding genes were found suggesting Aoka is a lytic phage.
A BLASTn search using the nucleotide sequence of the entire Aoka genome as query against the phagesdb.org database (January 2022) returned phage Maja (84% identity; GenBank accession number MK279899) as most similar. Other similar phage retrieved belongs to clusters FB and AS (subcluster AS1) that also contain Arthrobacter-infecting phage.
ACKNOWLEDGMENTS
This project was generously supported by the Howard Hughes Medical Institute SEA-PHAGES program and by the Department of Biology and Discovery Scholars program at Colorado State University-Pueblo. Technical assistance for genome sequencing and electron microscopy was carried out, respectively, by the members of the Graham Hatfull laboratory at the University of Pittsburgh and by Anza Darehshouri in the Electron Microscopy Center at UC Denver-Anschutz Medical Campus.
Contributor Information
Amaya M. Garcia Costas, Email: amaya.garciacostas@csupueblo.edu.
Kenneth M. Stedman, Portland State University, Portland, Oregon, USA
DATA AVAILABILITY
The complete genome sequence of Aoka has been deposited in GenBank with accession number ON755180, Bioproject accession number PRJNA488469, and SRA accession number SRX14443516.
REFERENCES
- 1. Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, Cresawn SG, Jacobs Jr WR, Hendrix RW, Lawrence JG, Hatfull GF. 2015. science education alliance phage hunters advancing genomics and evolutionary science, phage hunters integrating research and education, mycobacterial genetics course.whole genome comparison of a large collection of Mycobacteriophages reveals a continuum of phage genetic diversity. eLife 4:e06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clokie MRJ, Millard AD, Letarov AV, Heaphy S. 2011. Phages in nature. Bacteriophage 1:31–45. doi: 10.4161/bact.1.1.14942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mavrich TN, Hatfull GF. 2017. Bacteriophage evolution differs by host, lifestyle and genome. Nat Microbiol 2:17112. doi: 10.1038/nmicrobiol.2017.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. 2017. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J 11:1511–1520. doi: 10.1038/ismej.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hatfull GF. 2015. Dark matter of the biosphere: the amazing world of bacteriophage diversity. J Virol 89:8107–8110. doi: 10.1128/JVI.01340-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fremin BJ, Bhatt AS, Kyrpides NC. n.d. Thousands of small, novel genes predicted in global phage genomes. Cell Rep 39:110984. doi: 10.1016/j.celrep.2022.110984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SCR, Findley AM, Gissendanner CR, Golebiewska UP, Guild N, Hartzog GA, Grillo WH, Hollowell GP, Hughes LE, Johnson A, King RA, Lewis LO, Li W, Rosenzweig F, Rubin MR, Saha MS, Sandoz J, Shaffer CD, Taylor B, Temple L, Vazquez E, Ware VC, Barker LP, Bradley KW, Jacobs-Sera D, Pope WH, Russell DA, Cresawn SG, Lopatto D, Bailey CP, Hatfull GF. 2014. A broadly Implementable research course in phage discovery and genomics for first-year undergraduate students. mBio 5:e01051-13. doi: 10.1128/mBio.01051-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poxleitner M, Pope W, Jacobs-Sera D, Sivanathan V, Hatfull G. 2016. Phage discovery guide. In Howard Hughes medical Institute. Chevy Chase. [Google Scholar]
- 9. Ackermann H-W. 1999. Tailed bacteriophages: the order Caudovirales. Adv Virus Res 51: 135–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Russell DA. 2018. Sequencing, assembling, and finishing complete bacteriophage genomes, p 109–125. In Clokie MRJ, Kropinski AM, Lavigne R (ed), Bacteriophages: methods and protocols, vol 3 Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 11. Gordon D, Green P. 2013. Consed: a graphical editor for next-generation sequencing. Bioinformatics 29:2936–2937. doi: 10.1093/bioinformatics/btt515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Russell DA, Hatfull GF. 2017. PhagesDB: the actinobacteriophage database. Bioinformatics 33:784–786. doi: 10.1093/bioinformatics/btw711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pope WH, Mavrich TN, Garlena RA, Guerrero-Bustamante CA, Jacobs-Sera D, Montgomery MT, Russell DA, Warner MH, Science (SEA-PHAGES) SEA-PHAG E, Hatfull GF. 2017. Bacteriophages of gordonia spp. display a spectrum of diversity and genetic relationships.. mBio. https://doi.org/10/gpf5g2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Besemer J, Borodovsky M. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res 33:W451–4. doi: 10.1093/nar/gki487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cobamide2.Bio.Pitt.Edu. Accessed on January 2022
- 17. Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2011. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics 12:395. doi: 10.1186/1471-2105-12-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Accessed on January-May 2022. http://phages.wustl.edu/starterator/
- 19. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 20. Russell, Daniel A, Hatfull, G, Wren J. 6 December 2016 Phagesdb: the actinobacteriophage database. Bioinformatics 33:784–786. doi: 10.1093/bioinformatics/btw711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Söding J, Biegert A, Lupas AN. 2005. The Hhpred interactive server for protein Homology detection and structure prediction. Nucleic Acids Res 33:W244–8. doi: 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Accessed on January-may 2022. n.d. Available from: http://www.cbs.dtu.dk/services/TMHMM
- 23. Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379. doi: 10.1093/bioinformatics/14.4.378 [DOI] [PubMed] [Google Scholar]
- 24. Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54–7. doi: 10.1093/nar/gkw413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hatfull GF, Pedulla ML, Jacobs-Sera D, Cichon PM, Foley A, Ford ME, Gonda RM, Houtz JM, Hryckowian AJ, Kelchner VA, Namburi S, Pajcini KV, Popovich MG, Schleicher DT, Simanek BZ, Smith AL, Zdanowicz GM, Kumar V, Peebles CL, Jacobs WJ, Lawrence JG, Hendrix RW. 2006. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet 2:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2011. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics 12:395. doi: 10.1186/1471-2105-12-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of Aoka has been deposited in GenBank with accession number ON755180, Bioproject accession number PRJNA488469, and SRA accession number SRX14443516.


