ABSTRACT
We report the complete genome sequences of seven virulent Newcastle disease viruses (NDVs) that were isolated from chickens from live bird markets in the Arusha, Iringa, Mbeya, and Tanga regions of Tanzania in 2012. Phylogenetic analysis revealed that all isolates belong to sub-genotype XIII.1.1.
KEYWORDS: Newcastle disease virus, NDV, Avian avulavirus 1, Avian orthoavulavirus 1, orthoavulavirus javaense, XIII.1.1, complete genome, next-generation sequencing, live bird market, Tanzania
ANNOUNCEMENT
Newcastle disease virus (NDV) is a single-stranded, non-segmented RNA virus that belongs to the family Paramyxoviridae (1). It has a single serotype and at least 22 different genotypes that can be divided into two classes (2–4). Outbreaks of velogenic NDV (vNDV) occur worldwide, and the disease is endemic in many countries in Africa, Asia, the Middle East, and Central and South America (5–7).
In this study, 36 vNDVs were isolated from 796 oropharyngeal swabs collected from chickens at live bird markets in six regions of Tanzania in 2012, as described previously (8). Out of these 36 vNDVs, seven isolates were chosen for whole-genome sequencing (Fig. 1A). The viruses were propagated in 9-day-old specific-pathogen-free embryonating chicken eggs, and the intracerebral pathogenicity index (ICPI) was estimated following standard procedures (9). Viral RNA was isolated from the allantoic fluid using the Trizol LS Reagent (Invitrogen, USA). The Illumina libraries were prepared using the KAPA Stranded RNA-Seq Library Preparation Kit (Kapa Biosystems, USA) as per the manufacturer’s instructions. The distribution size and concentration of the prepared libraries were checked on a Bioanalyzer 2100, using the Agilent High Sensitivity DNA Kit (Agilent Technologies, Germany) and a Qubit fluorometer, using the dsDNA HS Assay Kit (Life Technologies, USA), respectively. Next-generation paired-end sequencing was performed on an Illumina MiSeq instrument using the 500-cycle MiSeq reagent kit v.2 (Illumina, USA). Sequence data were assembled using a de novo approach and utilizing MIRA version 3.4.1 (10) within a customized workflow on the Galaxy platform (11), as described previously (12, 13).
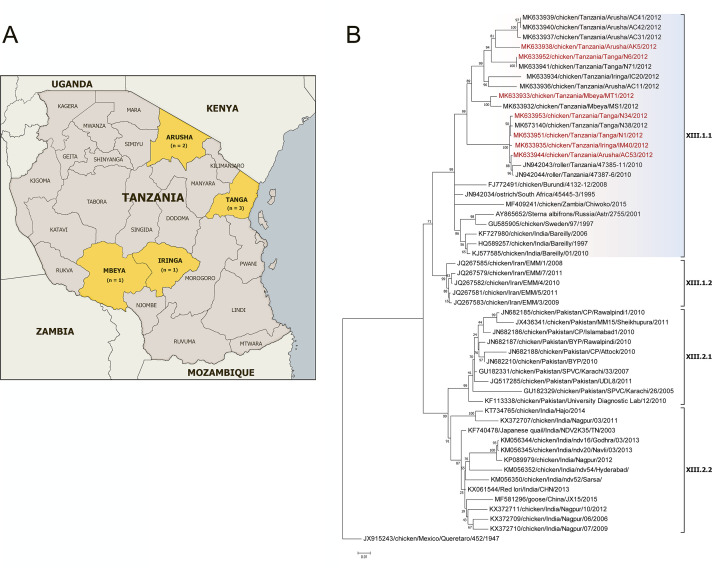
Fig 1.
(A) Locations for the live bird market surveillance. (B) Phylogenetic analysis of NDV isolates of genotype XIII based on the complete fusion gene sequences constructed with the Maximum Likelihood method based on the General Time Reversible model in MEGA version 7.0.26. The tree with the highest log likelihood (−9896.03) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 0. 4735)]. The rate variation model allowed for some sites to be evolutionarily invariable [(+I), 38.62% sites]. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 54 nucleotide sequences (sequence from genotype XVI is included as an outgroup). All positions containing gaps and missing data were eliminated. There were a total of 1,662 positions in the final dataset. The isolates used in this study are shown in red.
The MiSeq run generated from 671,067 to 2,263,648 total paired-end reads per sample (Table 1). All final consensuses were called from the raw reads that were aligned to the de novo-generated contigs using BWA-MEM (14) and were 15,192 nucleotides long with 100% genome coverage. Phylogenetic analysis in MEGA 7.0.26 revealed that all isolates had a 0.003 to 0.065% pairwise distance value (P-distance) compared to each other, which indicates a high level of nucleotide identity (93.5 to 99.7%) among them. The initial NCBI BLASTn (15) comparison to the currently available full-length NDV genome sequences showed that four isolates (AC53, N1, IM40, and N34) had the highest nucleotide identity (98.93 to 99.58%) to a previously published vNDV strain chicken/Tanzania/Tanga/N38/2012 (GenBank accession number MK673140) (8), while the other three isolates (MT1, N6, and AK5) showed the highest nucleotide identity (94.46 to 95.2%) to the vNDV strain chicken/India/Bareilly/01/2010 (KJ577585) (16). Detailed phylogenetic analysis based on the complete fusion gene classified all seven isolates as members of sub-genotype XIII.1.1 (Fig. 1B). The phylogenetic tree revealed that all publicly available fusion gene sequences of sub-genotype XIII.1.1 isolates from Tanzania cluster into a distinct branch from XIII.1.1 isolates detected in other countries (8, 16–21).
TABLE 1.
Isolates, sampling locations, dates, sequencing metrics, ICPI, and accession numbers of genomes of the virulent Newcastle diseases viruses in this report
| Isolate name | Collection date | Location (region) | Total no. of raw read pairs | No. of mapped read pairs | Median coverage depth (reads) | ICPIa | GenBank accession no. | SRA accession no. |
|---|---|---|---|---|---|---|---|---|
| Chicken/Tanzania/Mbeya/MT1/2012 | 05/30/2012 | Sokomatola (Mbeya) | 2,263,648 | 687,157 | 7,300 | 1.88 | MK633933 | SRR24142067 |
| Chicken/Tanzania/Iringa/IM40/2012 | 06/04/2012 | Mashine tatu (Iringa) | 671,067 | 449,556 | 5,366 | 1.88 | MK633935 | SRR24142063 |
| Chicken/Tanzania/Arusha/AK5/2012 | 05/27/2012 | Kilombero (Arusha) | 1,006,401 | 454,248 | 5,287 | 1.78 | MK633938 | SRR24142066 |
| Chicken/Tanzania/Arusha/AC53/2012 | 05/27/2012 | Arusha Central (Arusha) | 1,325,590 | 917,398 | 10,854 | 1.83 | MK633944 | SRR24142065 |
| Chicken/Tanzania/Tanga/N1/2012 | 05/21/2012 | Ngamiani (Tanga) | 813,624 | 316,250 | 3,292 | 1.95 | MK633951 | SRR24142064 |
| Chicken/Tanzania/Tanga/N34/2012 | 05/21/2012 | Ngamiani (Tanga) | 881,511 | 437,253 | 5,182 | 1.88 | MK633953 | SRR24142062 |
| Chicken/Tanzania/Tanga/N6/2012 | 05/21/2012 | Ngamiani (Tanga) | 844,044 | 484,424 | 5,873 | 1.88 | MK633952 | SRR24142061 |
ICPI = intracerebral pathogenicity index.
Analysis of the deduced amino acid sequence of the fusion protein cleavage sites (9, 22) of isolates from this study showed a polybasic amino acid motif 112RRQKR↓F117, which is typical for vNDV. This result was consistent with the ICPI values ranging between 1.78 and 1.95 (9). Overall, this study provides valuable sequence information on vNDVs from Tanzania and sheds light on their genetic diversity and virulence.
ACKNOWLEDGMENTS
The authors acknowledge the Arusha, Iringa, Mbeya, and Tanga regions of Tanzania’s local government authorities for facilitating sample collection from chickens in the live bird market owned by traders. We acknowledge Timothy Olivier and Suzanne DeBlois for their technical assistance in characterizing these viruses.
The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply a recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
This study was supported by USDA CRIS project 6040-32000-082 and by a visiting scientist appointment to the Oak Ridge Institute for Science and Education (ORISE).
Contributor Information
David L. Suarez, Email: david.suarez@usda.gov.
Jelle Matthijnssens, Katholieke Universiteit Leuven, Leuven, Belgium.
DATA AVAILABILITY
The complete genome sequence of seven isolates has been deposited in GenBank under the accession numbers MK633933, MK633935, MK633938, MK633944, MK633951, MK633952, and MK633953. Raw data were deposited in the NCBI Sequence Read Archive (SRA) under accession numbers SRR24142061, SRR24142062, SRR24142063, SRR24142064, SRR24142065, SRR24142066, and SRR24142067 under the BioProject number PRJNA543308.
REFERENCES
- 1. Rima B. 2019. ICTV virus taxonomy profile: Paramyxoviridae. J Gen Virol 100:1593–1594. doi: 10.1099/jgv.0.001328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dimitrov KM, Ramey AM, Qiu X, Bahl J, Afonso CL. 2016. Temporal, geographic, and host distribution of avian paramyxovirus 1 (Newcastle disease virus). Infect Genet Evol 39:22–34. doi: 10.1016/j.meegid.2016.01.008 [DOI] [PubMed] [Google Scholar]
- 3. Snoeck CJ. 2013. High genetic diversity of Newcastle disease virus in poultry in West and central Africa: cocirculation of genotype XIV and newly defined genotypes XVII and XVIII. J Clin Microbiol 51:2250–2260. doi: 10.1128/JCM.00684-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dimitrov KM. 2019. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect Genet Evol 74:103917. doi: 10.1016/j.meegid.2019.103917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. da Silva AP. 2020. Molecular characterization of Newcastle disease viruses isolated from chickens in Tanzania and Ghana. Viruses 12. doi: 10.3390/v12090916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dimitrov KM. 2016. Repeated isolation of virulent Newcastle disease viruses of sub-genotype VIId from backyard chickens in Bulgaria and Ukraine between 2002 and 2013. Arch Virol 161:3345–3353. doi: 10.1007/s00705-016-3033-2 [DOI] [PubMed] [Google Scholar]
- 7. Alexander DJ, Senne DA. 2008. Edited by Dufour-Zavala L.. A laboratory manual for the isolation, identification and characterization of avian pathogens, p 135–141. American association of avian pathologists, Athens, GA. [Google Scholar]
- 8. Msoffe PLM, Chiwanga GH, Cardona CJ, Miller PJ, Suarez DL. 2019. Isolation and characterization of Newcastle disease virus from live bird markets in Tanzania. Avian Dis 63:634–640. doi: 10.1637/aviandiseases-D-19-00089 [DOI] [PubMed] [Google Scholar]
- 9. OIE . 2012. Biological standards commission, manual of diagnostic tests and vaccines for terrestrial animals, p 964–983. World Organization for Animal Health, Paris, France. [Google Scholar]
- 10. Chevreux B, Wetter T, Suhai S. 1999. Computer science and biology, p 45–56. Hanover, Germany. [Google Scholar]
- 11. Afgan E. 2016. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44:W3–W10. doi: 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dimitrov KM. 2017. A robust and cost-effective approach to sequence and analyze complete genomes of small RNA viruses. Virol J 14:72. doi: 10.1186/s12985-017-0741-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goraichuk IV. 2019. First complete genome sequence of a subgenotype Vd Newcastle disease virus isolate. Microbiol Resour Announc 8. doi: 10.1128/MRA.00436-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H, Durbin R. 2009. Fast and accurate short read alignment with burrows-Wheeler transform. Bioinf 25:1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 16. Morla S, Kumar Tiwari A, Joshi V, Kumar S. 2014. Complete genome sequence of a Newcastle disease virus isolate from an outbreak in northern India. Genome Announc 2. doi: 10.1128/genomeA.00342-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cattoli G. 2010. Emergence of a new genetic lineage of Newcastle disease virus in West and central Africa--implications for diagnosis and control. Vet Microbiol 142:168–176. doi: 10.1016/j.vetmic.2009.09.063 [DOI] [PubMed] [Google Scholar]
- 18. Abolnik C, Mubamba C, Dautu G, Gummow B. 2017. Complete genome sequence of a Newcastle disease genotype XIII virus isolated from indigenous chickens in Zambia. Genome Announc 5. doi: 10.1128/genomeA.00841-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Linde A-M. 2010. Complete genome Characterisation of a Newcastle disease virus isolated during an outbreak in Sweden in 1997. Virus Genes 41:165–173. doi: 10.1007/s11262-010-0498-z [DOI] [PubMed] [Google Scholar]
- 20. Sawant PM. 2011. Immunomodulation of bivalent Newcastle disease DNA vaccine induced immune response by co-delivery of chicken IFN-γ and IL-4 genes. Vet Immunol Immunopathol 144:36–44. doi: 10.1016/j.vetimm.2011.07.006 [DOI] [PubMed] [Google Scholar]
- 21. Ebrahimi MM, Shahsavandi S, Moazenijula G, Shamsara M. 2012. Phylogeny and evolution of Newcastle disease virus genotypes isolated in Asia during 2008-2011. Virus Genes 45:63–68. doi: 10.1007/s11262-012-0738-5 [DOI] [PubMed] [Google Scholar]
- 22. Peeters BP, de Leeuw OS, Koch G, Gielkens AL. 1999. Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J Virol 73:5001–5009. doi: 10.1128/JVI.73.6.5001-5009.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of seven isolates has been deposited in GenBank under the accession numbers MK633933, MK633935, MK633938, MK633944, MK633951, MK633952, and MK633953. Raw data were deposited in the NCBI Sequence Read Archive (SRA) under accession numbers SRR24142061, SRR24142062, SRR24142063, SRR24142064, SRR24142065, SRR24142066, and SRR24142067 under the BioProject number PRJNA543308.