ABSTRACT
Here, we announce the complete coding sequence of two strains of feline panleukopenia virus (FPLV) that were obtained from deceased domestic cats in animal shelters in Tennessee. The provided sequence data will contribute to a deeper comprehension of the genetic characteristics and evolutionary patterns of FPLV in the USA.
KEYWORDS: feline panleukopenia virus, parvoviridae, carnivore protoparvovirus 1, feline parvovirus, feline parvoviral enteritis, virology, veterinary microbiology
ANNOUNCEMENT
Feline panleukopenia virus (FPLV), belonging to the Parvovirus genus, is a non-enveloped, single-stranded DNA virus that infects a wide range of animals beyond domestic cats, including raccoons, tigers, lions, and minks, making it a significant concern for both domestic and wild feline populations (1–5). It causes acute gastroenteritis and leukopenia, particularly in cats that have not been vaccinated (6, 7). FPLV is highly contagious and spreads easily between cats, especially in shelters and catteries (7, 8).
Two loops of small intestinal tissues were collected from deceased cats in 2022. Subsequently, one gram of the collected tissue was homogenized in 200 µL of phosphate-buffered saline using sterile glass beads for 15 minutes and subsequently filtered using a sterile 0.22-µM syringe filter. The resulting filtrate was then used to inoculate a monolayer of Crandell-Rees feline kidney (CRFK) cells (ATCC, CCL-94) (9). The total DNA was then extracted from infected CRFK cells using MagMAX Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific, USA). The NanoDrop 2000 spectrophotometer and Qubit fluorometer were used to assess the quality and quantity of extracted DNA. Using the complete genome of feline parvovirus (FPV) (MT614366) as the reference, the forward primer (5′-GAATGATAGGCGGTTTGTGTGT-3′) and reverse primer (5′-CCTACGCGGTCTGGTTGATT-3′) were designed to amplify the coding sequence of FPVL using Geneious Prime (10).
The coding sequence of FPV was amplified using the LongAmp Taq 2× Master Mix (New England Biolabs, USA), following the manufacturer’s instructions. The amplicons were purified using GenCleanTurbo kit (MP Biomedicals, USA)
PCR amplicon libraries were prepared using the Nextera DNA Flex kit (Illumina, Inc., USA) following the manufacturer’s instructions. Two FPLV strains were sequenced using Illumina MiniSeq (150 bp × 2) (Illumina, Inc.) resulting in 621,221 and 604,347 high-quality reads for each VI2882 and VI2534 strains, respectively. The raw reads for each sequence were trimmed using BBDuk followed by mapping to reference by Geneious Prime version 2023.0.1. Default parameters were used for all software unless otherwise specified (10). The National Center for Biotechnology Information open reading frame (ORF) finder (https://www.ncbi.nlm.nih.gov/orffinder/) was used to predict each ORF using default parameters.
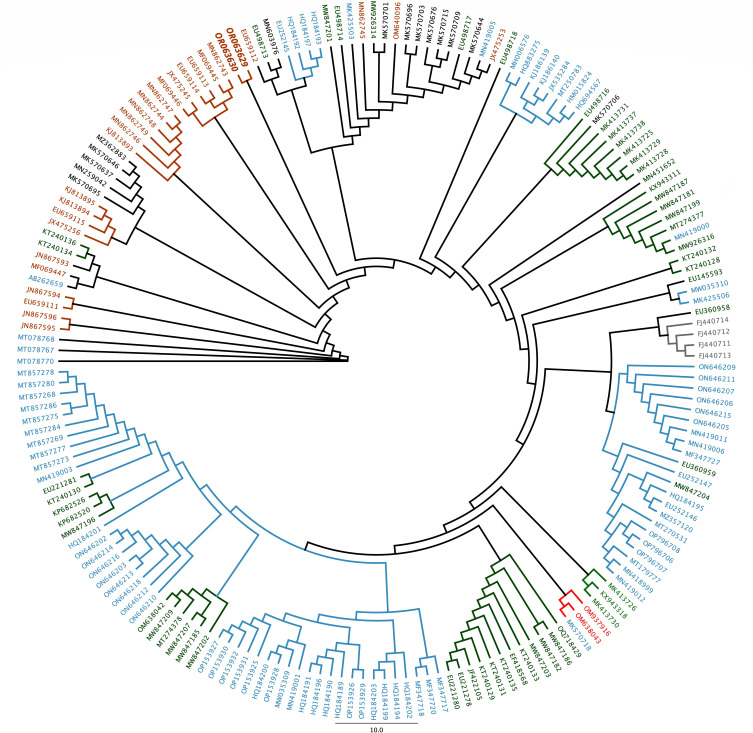
The complete coding sequence of strains VI2882 and VI2534, spanning nearly the entire genome, consists of 4,269 nucleotides. Moreover, the mean read coverage for VI2882 and VI2534 isolates was 34,117 and 28,327, respectively. The guanine-cytosine (GC) contents for VI2882 and VI2534 were 36.2% and 35.9%, respectively. The full-length VP2 gene of VI 2882 and VI 2534 FPVL strains, along with feline parvovirus strains from various continents, was retrieved from the National Center for Biotechnology Information virus (11) (Fig. 1). The sequences were aligned using Clustal Omega version 1.2.3 (12, 13). Phylogenetic relationships among FPVL strains based on VP2 sequences were analyzed using maximum-likelihood method through PHYML version 3.2.20180621 (14). The strains subjected to sequencing in this study clustered together with FPVL strains isolated from domestic cats in North America.
Fig 1.
Maximum-likelihood tree showing the genetic relationship of the full-length VP2 gene of feline panleukopenia viruses. The strains that were sequenced in this study, which are denoted in bold and italic, formed a cluster within the FPVL strains isolated from domestic cats in North America. The sequences shown in orange correspond to strains isolated from North America, while blue koi indicates strains from Asia; dark green represents strains from Europe; black designates strains from Oceania; red from Africa; and gray from South America.
ACKNOWLEDGMENTS
We thank the University of Tennessee College of Veterinary Medicine, Virology and Immunology Labs, for technical assistance and the CVM Department of Biomedical & Diagnostic Sciences at the University of Tennessee for their support of this project.
This work used Bridges-2 at Pittsburgh Supercomputing Center through allocation AGR220002 from the Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support (ACCESS) program, which is supported by National Science Foundation grants #2138259, #2138286, #2138307, #2137603, and #2138296. This work was funded by the University of Tennessee, Knoxville, Student/Faculty Research Award in 2023.
Contributor Information
Mohamed A. Abouelkhair, Email: mabouelk@utk.edu.
Jelle Matthijnssens Matthijnssens, Katholieke Universiteit Leuven, Leuven, Belgium.
DATA AVAILABILITY
The complete coding sequence of the strains has been deposited in GenBank under accession numbers OR063629 and OR063630. The National Center for Biotechnology Information's (NCBI's) Sequence Read Archive (SRA) accession numbers are available under SRS17591485 and SRS17591484.
REFERENCES
- 1. Duarte MD, Barros SC, Henriques M, Fernandes TL, Bernardino R, Monteiro M, Fevereiro M. 2009. Fatal infection with feline panleukopenia virus in two captive wild carnivores (Panthera tigris and Panthera leo). J Zoo Wildl Med 40:354–359. doi: 10.1638/2008-0015.1 [DOI] [PubMed] [Google Scholar]
- 2. Liu C, Liu Y, Liu D, Qiu Z, Tian J, Guo D, Li Z, Liu M, Li Y, Qu L. 2015. Complete genome sequence of feline panleukopenia virus strain HRB-CS1, isolated from a domestic cat in northeastern China. Genome Announc 3:e01556-14. doi: 10.1128/genomeA.01556-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Isaya R, Ciccarelli S, Enache D, Specchi S, Pesaresi M, Ferri F, Porporato F, Auriemma E, Contiero B, Coppola LM, Zini E. 2021. Gastrointestinal ultrasonographic findings in cats with Feline panleukopenia: a case series. BMC Vet Res 17:143. doi: 10.1186/s12917-021-02856-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miller L, Janeczko S, Hurley KF. 2021. Feline panleukopenia, p 337–366. In Infectious disease management in animal shelters. doi: 10.1002/9781119294382 [DOI] [Google Scholar]
- 5. Kolangath SM, Upadhye SV, Dhoot VM, Pawshe MD, Bhadane BK, Gawande AP, Kolangath RM. 2023. Molecular investigation of Feline panleukopenia in an endangered leopard (Panthera pardus) - a case report. BMC Vet Res 19:56. doi: 10.1186/s12917-023-03612-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ohshima T, Mochizuki M. 2009. Evidence for recombination between Feline panleukopenia virus and canine parvovirus type 2. J Vet Med Sci 71:403–408. doi: 10.1292/jvms.71.403 [DOI] [PubMed] [Google Scholar]
- 7. Diakoudi G, Desario C, Lanave G, Salucci S, Ndiana LA, Zarea AAK, Fouad EA, Lorusso A, Alfano F, Cavalli A, Buonavoglia C, Martella V, Decaro N. 2022. Feline panleukopenia virus in dogs from Italy and Egypt. Emerg Infect Dis 28:1933–1935. doi: 10.3201/eid2809.220388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rehme T, Hartmann K, Truyen U, Zablotski Y, Bergmann M. 2022. Feline panleukopenia outbreaks and risk factors in cats in animal shelters. Viruses 14:1248. doi: 10.3390/v14061248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Puentes R, Eliopulos N, Pérez R, Franco G, Sosa K, Bianchi P, Furtado A, Hübner SO, Esteves PA. 2012. Isolation and characterization of canine parvovirus type 2c (CPV-2c) from symptomatic puppies. Braz J Microbiol 43:1005–1009. doi: 10.1590/S1517-838220120003000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kearse M, Sturrock S, Meintjes P. 2012. The Geneious 6.0. 3 read mapper. Auckland, New Zealand: Biomatters, Ltd [Google Scholar]
- 11. NCBI Virus . 2004. Bethesda (MD): National library of medicine (US), national center for biotechnology information; 2004 –2023 08 08. Internet. Available from: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/
- 12. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res 38:W695–W699. doi: 10.1093/nar/gkq313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete coding sequence of the strains has been deposited in GenBank under accession numbers OR063629 and OR063630. The National Center for Biotechnology Information's (NCBI's) Sequence Read Archive (SRA) accession numbers are available under SRS17591485 and SRS17591484.