Abstract
Recessive mutations in the DNAJB2 gene, encoding the J-domain co-chaperones DNAJB2a and DNAJB2b, have previously been reported as the genetic cause of progressive peripheral neuropathies, rarely involving pyramidal signs, parkinsonism and myopathy. We describe here a family with the first dominantly acting DNAJB2 mutation resulting in a late-onset neuromyopathy phenotype. The c.832 T > G p.(*278Glyext*83) mutation abolishes the stop codon of the DNAJB2a isoform resulting in a C-terminal extension of the protein, with no direct effect predicted on the DNAJB2b isoform of the protein. Analysis of the muscle biopsy showed reduction of both protein isoforms. In functional studies, the mutant protein mislocalized to the endoplasmic reticulum due to a transmembrane helix in the C-terminal extension. The mutant protein underwent rapid proteasomal degradation and also increased the turnover of co-expressed wild-type DNAJB2a, potentially explaining the reduced protein amount in the patient muscle tissue. In line with this dominant negative effect, both wild-type and mutant DNAJB2a were shown to form polydisperse oligomers.
Introduction
The J-domain proteins (JDPs, also known as DNAJ proteins, or Hsp40) are a large and diverse group of cochaperones essential for cellular protein quality control (PQC). The defining feature of all JDPs is the J domain, which stimulates the ATPase activity of HSPA (Hsp70) chaperones, enabling the chaperone cycle, i.e. alternating binding and release of client (substrate) proteins. JDPs may also recognize and present clients to the HSPA machinery and affect their triage to refolding and/or degradative pathways. Some JDPs also possess HSPA-independent anti-aggregation activity (1,2).
The DNAJB2 (HSJ1) gene encodes two JDP cochaperones with alternative C-terminal parts generated through an alternative splice site within the last exon (3). The shorter DNAJB2a (HSJ1a) isoform is localized to the nucleus and cytoplasm, while the longer DNAJB2b (HSJ1b) isoform has a geranylgeranyl anchor attaching it to the cytoplasmic face of the endoplasmic reticulum (4). Common to both isoforms are the N-terminal J domain (JD), which is followed by glycine/phenylalanine-rich (G/F) domain and a C-terminal domain (CTD) containing a short serine-rich (SR) region and two ubiquitin-interacting motifs (UIMs) (4,5). DNAJB2 shows highest expression in neurons, particularly in the neocortex, and lower levels have been reported in various cells and tissues (3,4,6). In most tissues, DNAJB2b is the predominant isoform (4,6–8). A notable exception is human skeletal muscle, where the levels of the protein isoforms have been reported to be roughly equal (6).
The principal function of DNAJB2 is to mediate degradation of HSPA client proteins through the ubiquitin–proteasome system (UPS). DNAJB2 interacts with polyubiquitinated proteins and the proteasome through its UIMs, and cooperates with STUB1 (CHIP) to promote client ubiquitination and sorting to UPS (5,9). The membrane-anchored DNAJB2b isoform has been shown to promote the proteasomal degradation of endoplasmic reticulum (ER)-located proteins via the ER-associated degradation pathway (5). Studies using a variety of model systems and clients have demonstrated that DNAJB2 also has intrinsic chaperone activity and PQC functions independent of the HSPA system and/or UIMs (reviewed in Sarparanta et al. 10). DNAJB2a has, for instance, been shown to efficiently counteract TARDBP (TDP-43) aggregation by promoting its HSPA-mediated refolding (11). For a more detailed discussion on the reported functions of DNAJB2, we refer to our recent review (10).
Recessive mutations in DNAJB2 cause a spectrum of progressive peripheral axonal neuropathies, ranging from a pure motor phenotype [distal hereditary motor neuropathy (dHMN), a.k.a. distal spinal muscular atrophy] to sensorimotor axonal Charcot–Marie–Tooth disease (CMT2) (7,8,12–21). Pyramidal tract signs and Parkinson’s disease (PD) have been described in some patients, and hearing loss in one family (14–17,21). Recently, rimmed-vacuolar myopathy in combination with dHMN was described in a patient homozygous for a DNAJB2 missense mutation (20). The clinical course of DNAJB2-related neuropathies is typically moderate to severe, with onset in the second to third decades and progressing to severe muscle weakness over the course of the disease (7,8,13,15–21).
All the hitherto described DNAJB2 mutations (Table 1) are recessive and apparently act through a loss-of-function mechanism. Most of the mutations are demonstrated or expected to completely abolish DNAJB2 expression (7,8,21). In addition, a few missense mutations have been described, most of them affecting the J domain (8,12,17,20). On the basis of their recessive mode of action and their clinical consequences being comparable with the null alleles, the missense changes are likely to act by inactivating or destabilizing the protein. The downstream mechanisms leading from DNAJB2 loss of function to neuropathy are largely unknown, but accumulation of phosphorylated TARDBP in patient skin biopsies suggests that TARDBP aggregation has a potential role in the pathomechanism (21). Moreover, axonal accumulation of phosphorylated α-synuclein was specifically observed in a skin biopsy from a patient with PD-associated CMT2 due to a DNAJB2 mutation (21).
Table 1.
Published DNAJB2 mutations
| DNA change | Effect | Clinical phenotype | Ref. |
|---|---|---|---|
| c.14A > G | p.Tyr5Cys | CMT2 | (8,12) |
| c.125C > A | p.Ala42Asp | dHMN, YOPD, pyramidal | (17) |
| c.145delG | p.Val49Trpfs*25 | CTM2, PD, hearing loss | (21) |
| c.176-3C > G | Splice change | dHMN, pyramidal | (16) |
| c.184C > T | p.Arg62Trp | dHMN, myopathy | (20) |
| c.229 + 1G > A | Splice change | dHMN | (8,18) |
| c.310delC | p.R104Gfs*97 | CMT2 | (13) |
| c.352 + 1G > A | Splice change | dHMN, YOPD | (7,14,17) |
| c.620-1G > Aa | Splice change | CMT2 | (13) |
| c.700A > Tb | p.Thr234Ser | dHMN | (19) |
| g.219277938_219281781del | Likely null | dHMN, YOPD | (15) |
Reference sequences: NM_001039550.1 (cDNA), NP_001034639.1 (protein), NC_000002.12 (genomic)
dHMN: distal hereditary motor neuropathy; CMT2: axonal Charcot–Marie–Tooth disease; (YO)PD: (young onset) Parkinson’s disease
aAnnotated as c.619-1G > A in the original publication
bClassified as a variant of unknown significance (VUS) in the original publication
Results
Patients
Our proband is a German male first examined at the age of 66 years. He had suffered from balance problems and leg weakness, worsened by exercise, since the fifth decade, walking difficulties since the sixth decade and stocking-like paraesthesias in later years. The proband’s younger brother had no subjective symptoms, but neuromuscular examination revealed muscle hypotrophy of lower legs. A further brother had died in his 60s of cardiac causes without known neurological complaints. Their mother was reported to have suffered from walking difficulties since her early 60s, with a broad-based, slow gait. The proband’s maternal half-sister suffered from an unrelated paternally inherited spinocerebellar ataxia since her 50s (Fig. 1A).
Figure 1.
Patients and clinical findings. (A) Pedigree of our family. The DNAJB2 genotype (+, wild-type; ext, c.832 T > G p.*278Glyext*83) is shown for the family members available for genetic analysis. (B) Muscle MRIs of the proband (left) show diffuse neurogenic degenerative change in the soleus and gastrocnemius lateralis muscles of the lower legs but also myopathic-dystrophic fatty replacement focal changes in anterior parts of gluteus minimus and left soleus (arrows). Degenerative changes in the younger brother (right) are similar but milder with more neurogenic but also spots of myopathic replacement in the left adductor magnus (arrow) and the outer part of both peroneus longus muscles. (C) Gastrocnemius muscle biopsy of the proband (1 and 2 haematoxylin/eosin, 3 NADH and 4 ATPase pH 4.6) showing classical neurogenic changes such as fibre type grouping and groups of atrophic fibres together with clear myopathic changes such as rimmed vacuoles (arrowhead in 1), fibre splitting (arrowhead in 2) and heavily increased number of internalized myonuclei. Furthermore, in NADH staining (3), some small dark angulated fibres (black arrowhead in 3) and few moth-eaten fibres (white arrowhead in 3) are found. Scale bars 25 μm for 1 and 2, 100 μm for 3 and 4.
Clinical and laboratory findings
Neuromuscular examination of the proband at the age of 66 years showed diminished knee reflexes and absent ankle reflexes. There was bilateral foot dorsiflexion weakness Medical Research Council (MRC) grade 4 and plantar flexion grade 3–4. The patient had severe ataxia in tandem gait. No central motor signs, myoclonus or dystonia were present. He reported paraesthesia in the soles of the feet and showed loss of vibration sensation at the ankles, while reduced at the knee level. He reported reduced sense of position of the toes and had mild claw toes. Serum creatine kinase (CK) was in excess of 7× the upper limit of normal (ULN), and laboratory screening revealed increased rheumatoid factor and low vitamin B12. Supplementation was started for the latter, and vitamin B12 levels normalized over 6 weeks; no macrocytosis or anaemia was observed. His medical history included arterial hypertension and sleep apnoea under CPAP treatment. Chronic renal failure (stage G3a according to Kidney Disease Improving Global Outcomes) due to chronic mesangioproliferative glomerulonephritis type IgA had recently been diagnosed, but creatine values did not worsen over the time of observation, and steroid medication was not used.
Muscle MRI showed both neurogenic and myopathic degenerative changes (Fig. 1B), while head and spinal MRI failed to detect pathology of the brain or spinal cord.
Neurographic examination (Supplementary Material, Table S1) showed clear sensorimotor polyneuropathy with minor signs suggesting demyelination (proximal decreased compound muscle action potential, prolonged F-wave and distal motor latencies), but without slowing of nerve conduction velocities. Transcranial magnetic stimulation (TMS) showed increased total and central motor conduction times to all four limbs. Electromyography (EMG) detected chronic neurogenic changes (increased motor unit potential durations) but no spontaneous activity in upper limb muscles.
Muscle biopsy from the gastrocnemius revealed severe chronic neurogenic changes with signs of extensive reinnervation (broad, double-peaked fibre diameter distribution, fibre type grouping, grouped atrophy) together with myopathic changes (fibre splitting, internalized myonuclei, rimmed vacuoles) (Fig. 1C), whereas biopsy of the sural nerve showed mixed axonal and demyelinizing changes. The highly elevated CK, focal fatty replacement changes on MRI, even in gluteus minimus, and the muscle biopsy findings nevertheless indicate a partly myogenic origin to the disease.
Upon follow-up, the proband’s symptoms and neurophysiological findings had worsened. At the age to 70, examinations showed bilateral foot dorsiflexion weakness MRC grade 3–4 and plantar flexion grade 2, knee flexion and extension now weak at grade 4. There was Trendelenburg gait and mild atrophy of the thighs, in particular the posterior compartment. Atrophy of the anterior and posterior distal leg muscles on the left and normal calf size on the right with few fasciculations were evident. No central motor signs, myoclonus or dystonia were present. He complained of stocking-like hypaesthesia with hyperalgesia of the soles. There was loss of vibration sense at the ankles, and marked claw toes had developed. His serum CK was 2.5× ULN.
Repeat neurophysiological examinations show some progression of the neuropathy, sparing the right peroneal nerve. In TMS, potentials could no longer be recorded from the tibialis anterior muscles. In EMG, all tested limb muscles except right tibialis anterior and paravertebrals showed chronic neurogenic changes, and some spontaneous activity could be detected in the right vastus lateralis and the left tibialis anterior.
The proband’s brother was examined at the age of 66 years showing diminished knee reflexes and absent ankle reflexes. He had mild foot dorsiflexion weakness on the right, grade 4 on the left and weak big toe extension grade 2–3. Anterior and posterior compartments of the legs showed asymmetric (right > left) atrophy. No central motor signs, myoclonus or dystonia were present. He had mild ataxia in tandem gait and reduced vibration sense at the ankles with otherwise normal sensory examination. His serum CK was normal. Muscle MRI of pelvis and lower extremities showed milder neurogenic degenerative changes distally more on the right side (Fig. 1B), and neurography revealed a mild sensorimotor polyneuropathy.
Molecular genetics
Targeted high-throughput sequencing of the proband with the MyoCap panel identified 20 rare variants (minor allele frequency < 1% in gnomAD_ALL). Of these, the only likely pathogenic variant shared between the proband and his affected brother was the heterozygous stop-loss variant NM_001039550.2:c.832 T > G p.(*278Glyext*83) in the DNAJB2 gene. The variant was not previously reported in the gnomAD or ClinVar databases. Sanger sequencing confirmed that the variant was present in the proband and his brother, and absent from their unaffected half-sister (Fig. 1A). The identified variant abolishes the termination codon of the DNAJB2a isoform (NP_001034639.1), causing the extension of the protein product by 83 amino acids (Fig. 2A). The variant sequence is also present deep in the 3′ UTR of the DNAJB2b transcript (NM_006736.6:c.1983 T > G), with no effects expected on the protein product. RNA sequencing (RNAseq) analysis of the proband’s muscle biopsy showed expression of the wild-type and mutant transcripts at the expected ratio (Fig. 2B), indicating that the mutant allele is normally expressed and does not affect RNA stability. RNAseq detected transcript variants 1 (DNAJB2a) and 2 (DNAJB2b) at a ~5:1 ratio, with no indication of altered isoform ratio or other DNAJB2 splicing changes in the proband (Fig. 2B).
Figure 2.
The DNAJB2 mutation. (A) The T > G change identified in the proband affects the termination codon of the DNAJB2 transcript variant 1 (NM_001039550.2) and is predicted to cause a C-terminal extension of the DNAJB2a protein isoform (NP_001034639.1:p.*278Glyext*83). In variant 2 (NM_006736.6) encoding the DNAJB2b isoform, the altered nucleotide lies within the 3′ UTR and does not affect the protein product. In the diagram of the DNAJB2 transcript, the non-coding regions are shown in grey, the normal coding regions in black and the extended open reading frame caused by the mutation in magenta. Both the DNAJB2a (top) and DNAJB2b (bottom) proteins contain an N-terminal J domain (JD; orange) followed by a glycine/phenylalanine-rich region (G/F; blue), and a C-terminal domain (CTD; yellow) containing a serine-rich region (SR) and two ubiquitin-interacting motifs (UIMs). The two isoforms differ in their C-terminal parts (green), and DNAJB2b has a C-terminal geranylgeranyl moiety (GG) anchoring it to the endoplasmic reticulum. The extended DNAJB2a protein (p.*278Glyext*83) produced from the mutant allele has a C-terminal extension of 83 amino acids (magenta; sequence shown in the box). (B) RNA sequencing (RNAseq) coverage graphs covering the last exon(s) of the DNAJB2 transcripts. RNAseq of the proband (P) muscle sample showed equal expression of the wild-type and mutant alleles (arrow) and did not indicate splicing changes or altered isoform ratio compared with other samples run in the same batch (C1–5).
To exclude causative variants outside the MyoCap panel, whole-exome sequencing was performed for all three available family members (Supplementary Material, Fig. S1). Apart from the DNAJB2 variant, the only other variant predicted as likely pathogenic and segregating with the disease was a heterozygous deletion abolishing the start codon of the FIBP gene. As recessive loss-of-function mutations in FIBP cause Thauvin–Robinet–Faivre syndrome, a congenital syndrome characterized by tall stature, intellectual disability and renal anomalies (22,23), the identified variant is unlikely relevant for the neuromyopathy phenotype in our family.
The SGCE variant NM_003919.3:c.1046_1047del:p.(Arg349Lysfs*29), predicted by VarSome as likely pathogenic, was identified by MyoCap and by exome sequencing in the proband but not in his brother or sister. Dominant SGCE mutations are a reported cause of myoclonus–dystonia, a phenotype not compatible with that seen in our patients (24). The identified frameshift variant affects an alternatively spliced exon and was according to RNAseq only present in ~25% of SGCE transcripts in the proband’s muscle biopsy.
Western blotting and immunofluorescence analysis of patient biopsy
Western blotting was performed to evaluate the effects of the DNAJB2 mutation in a muscle biopsy from the proband. The extended protein product was not present at a detectable level in total muscle lysate. Interestingly, however, the protein levels of both DNAJB2a and DNAJB2b isoforms appeared reduced by >50% compared with pooled control muscle (Fig. 3A). Prompted by the membrane localization of mutant DNAJB2 (see below), we performed subcellular fractionation of patient and control muscles. Although the extended protein was not detectable in the membrane fraction, the results supported the decrease of both DNAJB2a and DNAJB2b in the soluble and membrane fractions (Fig. 3B and C).
Figure 3.

Western blotting of patient biopsy. (A) Western blotting of a muscle biopsy from the proband (P) revealed a reduced amount of both DNAJB2a and DNAJB2b proteins compared with pooled control (C), and no detectable mutant protein. P1 and P2 are independently prepared samples from the same biopsy. Post-blotting Coomassie staining of the myosin heavy chain (MYHC CBB) is shown as loading control. (B) Control (C1, C2) and proband (P) muscle biopsies were fractionated with the ProteoExtract Subcellular Proteome Extraction Kit to cytosolic (F1), membrane/organelle (F2), nuclear (F3) and cytoskeletal/insoluble (F4) fractions and analysed by western blotting. Total protein, tubulin, calnexin and histone 3 are shown as loading and fractionation controls. (C) The levels of DNAJB2 relative to total protein were quantified from the F1 and F2 fractions in (B) and represented normalized to the mean of control samples. Both DNAJB2 isoforms showed a ~50% reduction in the biopsy of the proband.
In immunofluorescence microscopy (not shown), the biopsy of the proband showed myopathic features including fibre size variation and internal nuclei. Occasional rimmed-vacuolar fibres were encountered, however, without major SQSTM1/p62 or TARDBP accumulation pathology. Immunofluorescent analysis did not show significant DNAJB2 or DNAJB6 accumulation pathology.
Characterization of the mutant protein
To understand the effects of the p.*278Glyext*83 extension, we analysed the properties of the mutant protein in silico and in cell culture experiments. Sequence analysis with the TMPRED and TMHMM algorithms predicted a transmembrane helix in the C-terminal extension (Fig. 4A). In line with this, subcellular fractionation of stably transfected C2C12 myotubes revealed increased localization of DNAJB2a p.*278Glyext*83 to the membrane fraction, whereas overexpressed and endogenous wild-type DNAJB2a were mostly found in the soluble cytosolic fraction (Fig. 4B). Similarly, the overexpressed p.*278Glyext*83 protein showed enrichment to the microsomal fraction in T-REx 293 cells (Fig. 4C) and partially colocalized with the ER in HeLa cells (Fig. 4D).
Figure 4.
Membrane localization of mutant DNAJB2. (A) In silico prediction. The amino acid sequence of DNAJB2a p.*278Glyext*83 was analysed with transmembrane helix prediction algorithms. The graph shows the scores from TMHMM (orange trace) and TMPRED (blue solid trace, in–out orientation, dashed trace, out–in) for the 83 amino acid extension. (B) Stably transfected C2C12 myotubes induced to express wild-type or p.*278Glyext*83 (ext) DNAJB2a, and non-induced control cells, were fractionated with the ProteoExtract Subcellular Proteome Extraction Kit to cytosolic (F1), membrane/organelle (F2), nuclear (F3) and cytoskeletal/insoluble (F4) fractions. Endogenous and overexpressed DNAJB2a were predominantly cytosolic, whereas p.*278Glyext*83 was enriched in the membrane fraction similarly to endogenous DNAJB2b. (C) T-REx 293 cells were transfected with a combination of wild-type DNAJB2a and DNAJB2b (a + b wt) or DNAJB2a p.*278Glyext*83 (ext) and fractionated. Wild-type DNAJB2a was mostly found in the cytosolic (CYT) fraction, whereas DNAJB2b and p.*278Glyext*83 were enriched in the microsomal (MIC) fraction, similarly to the ER marker calnexin. PNS, post-nuclear supernatant. (D) In transfected HeLa cells, wild-type (wt) V5-DNAJB2a showed diffuse nuclear and cytoplasmic localization, whereas p.*278Glyext*83 (ext) partially colocalized with the endoplasmic reticulum (ER) visualized with the Cytopainter ER staining kit.
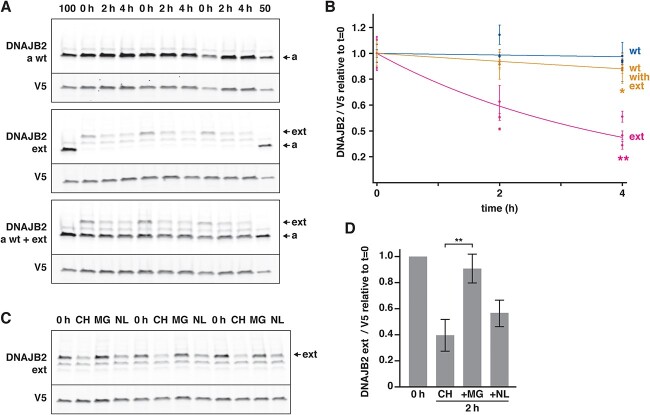
The absence of detectable p.*278Glyext*83 in patient muscle and the lower steady-state levels of the mutant constructs in transfection experiments suggested increased turnover of the mutant protein. In line with this, cycloheximide chase experiments indicated dramatically increased turnover rate of mutant DNAJB2a p.*278Glyext*83 (Fig. 5A and B). Moreover, DNAJB2a p.*278Glyext*83 slightly accelerated the turnover of cotransfected wild-type DNAJB2a. The turnover of the mutant protein was efficiently blocked with the proteasomal inhibitor MG132, whereas lysosomal inhibitors showed little effect (Fig. 5C and D). Altogether, these findings indicate that DNAJB2a p.*278Glyext*83 undergoes rapid degradation through the ubiquitin–proteasome pathway and may also destabilize the wild-type protein, consistently with the decreased amount of DNAJB2 in patient muscle.
Figure 5.
Turnover studies. (A–B) Wild-type (wt) or p.*278Glyext*83 (ext) DNAJB2a were expressed in T-REx 293 cells alone or in combination, and their levels were assayed at 0, 2 and 4 h of cycloheximide treatment. (A) A representative experiment performed in triplicate. 100 and 50 indicate a normalization sample at 100% and 50% loading, common for all three blots. (B) Quantification of three replicate experiments. The level of DNAJB2 was normalized to the transfection marker (GFP-V5) and represented relative to the initial level (t = 0). Each data point represents the mean ± SD of one experiment performed in triplicate. Asterisks indicate significant differences in remaining protein amount at t = 4 compared with wild-type (2-tailed t-test; *P = 0.029, **P = 0.001). DNAJB2 p.*278Glyext*83 showed an increased turnover rate and also increased the turnover of the co-expressed wild-type DNAJB2a. (C–D) T-REx 293 cells expressing DNAJB2 p.*278Glyext*83 were treated with cycloheximide alone (CH) or in combination with the proteasome inhibitor MG132 (MG) or lysosomal inhibitors (NH4Cl/leupeptin; NL) for 2 h. (D) The level of DNAJB2 was normalized to the transfection marker (GFP-V5) and represented relative to the initial level (t = 0). The graph shows means ± SD from three replicate experiments, each performed in triplicate. MG132 efficiently blocked the turnover of mutant DNAJB2 (**P = 0.006, 2-tailed t-test).
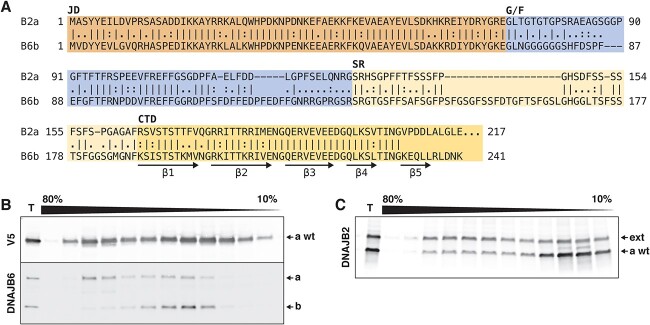
A possible explanation for the dominant negative effect of DNAJB2a p.*278Glyext*83 could be co-oligomerization with the wild-type protein. Although oligomerization of DNAJB2 has not been reported, the related cochaperone DNAJB6 forms polydisperse oligomers through its CTD (25–27), which shows a high degree of similarity to DNAJB2 (Fig. 6A). Indeed, DNAJB2a constructs expressed in T-REx 293 cells showed polydispersity in sucrose density gradient centrifugation, supporting potential oligomerization of DNAJB2a (Fig. 6B). Although the fractionation profiles of wild-type and p.*278Glyext*83 DNAJB2a were somewhat dissimilar, both showed a polydisperse distribution in sucrose gradients, consistently with a possibility of co-oligomerization (Fig. 6C).
Figure 6.
Oligomerization of DNAJB2. (A) Local sequence alignment of DNAJB2a (B2a) and DNAJB6b (B6b), with the coloured shadings indicating the J domain (JD), the glycine/phenylalanine-rich domain (G/F), the serine-rich region (SR) and the C-terminal domain (CTD) as defined for DNAJB6b by Karamanos et al. (25). Most of the region mediating DNAJB6b oligomerization (CTD β strands β1–β5) is highly similar between the two proteins. The C-terminal part of DNAJB2a (amino acids 218–277), not homologous to DNAJB6, is not shown. (B–C) Density gradient centrifugation of T-REx 293 cell lysates in 10–80% sucrose gradients. Fractionation profile of wild-type V5-DNAJB2a (a wt) suggests its oligomerization into polydisperse oligomers, similarly to endogenous DNAJB6 (a and b isoforms indicated) in the same samples (B). Co-expressed untagged wild-type (wt) and p.*278Glyext*83 (ext) DNAJB2a are both distributed throughout the gradient, albeit with somewhat different profiles (C). T, total samples.
Discussion
Our functional studies provide strong evidence for the abnormal behaviour of the DNAJB2a p.*278Glyext*83 protein, consistent with the idea that the identified DNAJB2 variant is the molecular cause of dominantly inherited neuromyopathy in our family. The causative role of DNAJB2 is further supported by the DNAJB2 protein deficiency observed in patient muscle. However, due to the small family size and unavailability of the parents’ DNA samples, contribution of other genetic factors to the phenotypic diversity in the family cannot be excluded.
Our proband presented with a combination of sensorimotor polyneuropathy and myopathy. The initial vitamin B12 deficiency may have aggravated the neurogenic component in the proband. However, considering the progressive course of the disease despite a timely and effective vitamin B12 supplementation, the absence of corresponding spinal cord changes in MRI and the presence of neuropathy also in the brother, the role of B12 deficiency is not major. The neuropathy symptoms seen in the proband are similar to those caused by recessive loss-of-function mutations in DNAJB2, albeit with later onset. The subjectively almost asymptomatic clinical phenotype in the proband’s brother defines the mild end of the spectrum, and it is likely that the walking difficulties in their mother were related to the same disease. In contrast to previous report of pyramidal tract signs in some dHMN patients with homozygous DNAJB2 variants (16), our proband’s neurophysiological examinations point to central motor affection but without a clinical correlate. Again, vitamin B12 deficiency seems an unlikely cause of this in the absence of MRI abnormalities correlating to this measurement and with the deficiency corrected promptly.
The myopathological features and the prominent hyperCKemia in our proband suggest a concomitant myopathic process, not usually associated with DNAJB2 mutations, although elevated CK has been previously reported in individual patients with the c.14A > G p.(Tyr5Cys) (12) and c.184C > T p.(Arg62Trp) mutations (20). In the latter case, the patient showed neurogenic myopathology (angulated atrophy, fibre type grouping) with infrequent rimmed-vacuolar changes (20), suggesting a possible myopathic component. Additional studies will be needed to clarify the frequency and mechanisms of myopathy associated with DNAJB2 loss of function.
The recessive manifestation of DNAJB2 null mutations suggests that ~50% reduction in DNAJB2 level is not alone sufficient to cause disease. Indeed, Saveri et al. showed reduced transcript and protein expression in lymphoblasts from a non-symptomatic heterozygous carrier of the c.145delG p.Val49Trpfs*25 variant (21). We therefore hypothesize that the DNAJB2 c.832 T > G p.(*278Glyext*83) mutation may cause disease through a combination of mechanisms. On one hand, the loss of functional DNAJB2a expression from the mutant allele combined with accelerated turnover of the wild-type protein via a dominant negative effect would lead to the DNAJB2 protein deficiency observed in our patient. On the other hand, the extended protein—notwithstanding its low steady-state level—could have an additional toxic gain-of-function effect. In a similar fashion, C-terminal extension mutations in HSPB8 cause disease through a gain-of-function mechanism even at an undetectable protein level (10,28,29).
Interestingly, dominant mutations in other JDP cochaperones, DNAJB6 (26,30,31) and DNAJB4 (32), also lead to muscle disease. These mutations likely disrupt the interaction between the G/F and J domains, leading to toxic gain-of-function by stalling the HSPA chaperone complex, and are associated with reduced turnover and/or accumulation of the mutant JDP (10,26,32–34). Given the accelerated turnover of mutant DNAJB2, the dominant toxicity in our case appears mechanistically distinct, and could possibly depend on mislocalized DNAJB2 interfering with PQC or calcium handling at the ER. Although our attempts to determine the membrane topology of the mutant protein on microsomal preparations remained inconclusive (not shown), the TMHMM prediction places the N-terminal major part on the protein ‘outside’, i.e. facing the ER lumen. That said, the mutant protein can be envisioned to exert a harmful effect in either orientation.
The apparent primary myopathic component and the relatively benign though progressive clinical course compared with DNAJB2 loss-of-function mutations likely reflect the fact that the identified variant is predicted to specifically affect DNAJB2a. We found this isoform, in line with previous work (6), to be the predominant one in skeletal muscle both at RNA and protein levels. In muscle, both the amount of the toxic protein and the dominant negative effect leading to loss of wild-type DNAJB2 would be more pronounced compared with neurons and other cell types where DNAJB2b is the predominant isoform.
In summary, the robust in vitro effects on protein localization and stability, and altered DNAJB2 level in patient muscle support classification of the DNAJB2 variant NM_001039550.2:c.832 T > G p.(*278Glyext*83) as likely pathogenic. Although additional genetic or environmental factors may contribute to the disease phenotypes, the DNAJB2 variant can be concluded to be the probable main cause of dominantly inherited neuromyopathy in our family.
Materials and Methods
Targeted high-throughput sequencing
Targeted high-throughput sequencing of genomic DNA was performed with the MyoCap panel (35) versions 5 (for the proband) and 6 (for his brother). The panels cover, respectively, 328 and 344 reported or candidate genes for myopathy (Supplementary Material, Table S2). Raw data were analysed by an in-house pipeline (35). VarSome (36) was used for variant interpretation.
Whole-exome sequencing
Whole-exome sequencing was performed using the Twist Human Core Exome + RefSeq + Mitochondrial Panel (Twist Bioscience) at CeGaT (Tübingen, Germany) on Illumina NovaSeq 6000 (PE 2×100). Short variant (SNPs and indels) discovery was done on the Euformatics (Espoo, Finland) Orchestrator implementing the GATK Best Practices per sample pipeline. In brief, alignment of the raw reads was done using Burrows–Wheeler alignment tool on the University of California, Santa Cruz hg38 reference genome and variant calling was done using the Haplotype Caller. Variant annotation and automated classification were performed on the Euformatics omnomicsNGS system, which integrates data from multiple annotation sources and uses tools such as the Variant Effect Predictor (VEP v.107) for enriching every variant with annotations covering ~150 ‘dimensions’. Variant filtering was performed on omnomicsNGS (Supplementary Material, Fig. S1). VarSome (36) was used for variant interpretation.
RNA sequencing
For RNAseq, total RNA was isolated from a gastrocnemius muscle biopsy of the proband with Qiagen RNeasy Plus Universal Mini Kit (Qiagen, Hilden, Germany). Total RNAseq library was prepared using the Illumina Ribo-Zero Plus rRNA Depletion kit (Illumina, Palo Alto, CA, USA) and run on a NovaSeq 6000 (Illumina) at the Oxford Genomics Centre, Wellcome Trust Institute, Oxford, UK, generating approximately 133 million 150 bp long reads (37). Sequences were mapped to Gencode.v39 human reference genome (based on ENSEMBL GRCh38.p13), using the STAR (v 2.7.0d) (38) two-pass method.
Protein sequence analysis
The primary sequence of mutant DNAJB2 was analysed for transmembrane domains by TMPRED (http://embnet.vital-it.ch/software/TMPRED_form.html) and TMHMM (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0). Local pairwise sequence alignment of DNAJB2a and DNAJB6b was performed with EMBOSS Matcher (https://www.ebi.ac.uk/Tools/psa/emboss_matcher/) using default parameters.
SDS-PAGE and western blotting
For western blotting analyses, protein samples were separated in TGX minigels (Bio-Rad, Hercules, CA, USA) and transferred on nitrocellulose membranes with the Trans-Blot Turbo system (Bio-Rad). Total protein was stained with the Revert 700 Total Protein Stain (Li-Cor Biosciences, Lincoln, NE, USA) and scanned with the Odyssey scanner (Li-Cor). For muscle lysates, gels were stained post-blotting with Coomassie blue. Blots were stained with the following primary antibodies: calnexin Rb mAb C5C9 (Cell Signaling Technology, 2679, RRID:AB_2228381); DNAJB2 Rb pAb (Proteintech, 10 838-1-AP, RRID:AB_2277491); DNAJB6 Rb mAb [EPR17122] (Abcam, ab198995, RRID:AB_2924896); histone 3 Rb pAb (Abcam, ab1791, RRID:AB_302613); tubulin Rt mAb YL1/2 (Abcam, ab6160, RRID:AB_305328); V5 Ms mAb (Thermo Fisher, R960-25, RRID:AB_2556564); V5 Rb mAb D3H8Q (Cell Signaling Technology, 13202, RRID:AB_2687461). Detection was performed with fluorescent secondary antibodies using an Odyssey scanner or with HRP-conjugated secondary antibodies visualized by enhanced chemiluminescence. Image analysis was done in Fiji (39).
Plasmid constructs
Tetracycline-inducible untagged and N-terminally V5-tagged DNAJB2a and DNAJB2b constructs in pCDNA5/FRT/TO (40) were a kind gift from Harm Kampinga. The sequence encoding the C-terminal extension was synthesized at Eurofins Genomics (Ebersberg, Germany) and added to the DNAJB2a constructs to create the p.*278Glyext*83 constructs. The inserts were transferred to pSBtet-Hyg (41) (a kind gift from Eric Kowarz; Addgene plasmid # 60508; RRID: Addgene_60 508) to create pSBtet-Hyg-DNAJB2a wild-type and p.*278Glyext*83. pEGFP-V5, encoding C-terminally V5-tagged green fluorescent protein, was constructed in pEGFP-C1.
Histological analyses
The proband underwent gastrocnemius muscle biopsy at the age of 66 years. Conventional muscle biopsy procedure and histological analyses were performed, including haematoxylin/eosin, NADH diaforase and ATPase pH 4.6 stainings.
For immunofluorescence microscopy, cryosections prepared from the snap-frozen muscle biopsy were fixed using 4% paraformaldehyde (PFA) in phosphate buffered saline and were then immunostained. Primary antibodies were incubated overnight at +8°C and secondary antibodies 1 h at room temperature. Primary antibodies: DNAJB2 Rb pAb (Proteintech, 10838-1-AP, RRID:AB_2277491); DNAJB6 Rb mAb [EPR17122] (Abcam, ab198995, RRID:AB_2924896); TDP-43 Ms mAb (Sigma-Aldrich, WH00234, clone 2E2-D3, RRID:AB_1843869); p62 Rb pAb (Sigma-Aldrich, P0067, RRID:AB_1841064). Secondary antibodies: Alexa Fluor-488 and -546 conjugated secondary antirabbit and antimouse antibodies.
Fractionation and western blotting of patient biopsy
For a total protein lysate, biopsied gastrocnemius muscle from the proband was homogenized in sample buffer (50 mm Tris–HCl pH 6.8; 4% sodium dodecyl sulphate, 8.7% glycerol, 10% 2-mercaptoethanol, bromophenol blue). DNAJB2 was analysed by western blotting, using post-blotting myosin heavy chain as loading control. For subcellular fractions, the biopsy was fractionated with the ProteoExtract Subcellular Proteome Extraction kit (Merck Millipore) as per the kit instructions. Tubulin, calnexin and histone 3 were used as markers for soluble, membrane and nuclear fractions, respectively.
Immunofluorescence microscopy of HeLa cells
For comparing the localization of wild-type and mutant DNAJB2, HeLa cells seeded on coverslips were transfected with V5-tagged or untagged pCDNA5/FRT/TO–DNAJB2 constructs and fixed with 4% PFA the next day. The cells were stained with V5 or DNAJB2 antibodies, and counterstained with the Cytopainter ER Staining Kit—Red Fluorescence (ab139482; Abcam plc, Cambridge, UK). Images were acquired with Zeiss Axio Imager M2 using a 40× NA 1.30 objective.
Microsome isolation
For studying the subcellular localization of the proteins, T-REx 293 cells were transfected with untagged pCDNA5/FRT/TO–DNAJB2 constructs (DNAJB2a p.*278Glyext*83 or a 1:1 mix of wild-type DNAJB2a and DNAJB2b). After ~20 h of tetracycline induction (500 ng/ml), cells were fractionated by ultracentrifugation essentially as described (42) and analysed by western blotting.
C2C12 myotubes
To create C2C12 cells with inducible expression of wild-type or p.*278Glyext*83 mutant DNAJB2a, C2C12 myoblasts were cotransfected with pSBtet-Hyg-DNAJB2 and pCMV(CAT)T7-SB100 (43) (a kind gift from Zsuzsanna Izsvák; Addgene plasmid #34879, RRID: Addgene_34 879). Polyclonal pools of stably transfected cells were selected in growth medium [pyruvate-free Dulbecco’s modified Eagle medium (DMEM) with 20% fetal calf serum, Glutamax and penicillin/streptomycin] with 200 μg/ml hygromycin B and maintained in growth medium with 50 μg/ml hygromycin B. The cells were grown on collagen-coated plates to confluency and differentiated to myotubes in differentiation medium (pyruvate-free DMEM with 2% heat-inactivated horse serum, l-glutamine, penicillin/streptomycin and 10% Opti-MEM I) for 4 days. One day before harvest, the cells were induced with doxycycline (2 μg/ml) to express DNAJB2 or left uninduced. The cells were fractionated with the ProteoExtract Subcellular Proteome Extraction kit according to the kit instructions and analysed by western blotting.
Protein turnover assays
For turnover studies, T-REx 293 cells were cotransfected with pCDNA5/FRT/TO–DNAJB2 construct(s) and pEGFP-V5. After 16 h of tetracycline induction (200 ng/ml), the medium was replaced with a chase medium containing 40 μg/ml cycloheximide (CHX). Immediately (t = 0) and at different time points (2, 4 and 6 h) of CHX treatment, cells were rinsed and frozen at −80°C. To block proteasomal or lysosomal protein turnover pathways, 10 μm MG132 or 20 mm NH4Cl + 100 μm leupeptin was included in the chase medium. DNAJB2 and GFP-V5 were analysed from the cell lysates by fluorescent western blotting using an Odyssey scanner. The amount of DNAJB2 was normalized to GFP-V5 (which does not turn over significantly during the chase period) to correct for transfection efficiency and was expressed relative to t = 0.
Sucrose density gradient centrifugation
To study DNAJB2 oligomerization, DNAJB2 constructs were expressed in T-REx 293 cells for 2 days and analysed by density gradient ultracentrifugation in 10–80% sucrose gradients as described previously for DNAJB6 (26).
Supplementary Material
Acknowledgements
We thank the Oxford Genomics Centre at the Wellcome Centre for Human Genetics (funded by Wellcome Trust grant reference 203141/Z/16/Z) for the generation and initial processing of the sequencing data, CSC—IT Center for Science Ltd for providing computational resources, and Merja Soininen and Anna Vainio for technical assistance.
Conflict of Interest statement. The authors declare no competing interests.
Contributor Information
Jaakko Sarparanta, Folkhälsan Research Center, Helsinki, Finland and Medicum, University of Helsinki, FI-00290 Helsinki, Finland.
Per Harald Jonson, Folkhälsan Research Center, Helsinki, Finland and Medicum, University of Helsinki, FI-00290 Helsinki, Finland.
Jens Reimann, Klinik und Poliklinik für Neurologie, Sektion Neuromuskuläre Erkrankungen, Universitätsklinikum Bonn, D-53127 Bonn, Germany.
Anna Vihola, Folkhälsan Research Center, Helsinki, Finland and Medicum, University of Helsinki, FI-00290 Helsinki, Finland; Neuromuscular Research Center, Tampere University Hospital and Fimlab Laboratories, FI-33520 Tampere, Finland.
Helena Luque, Folkhälsan Research Center, Helsinki, Finland and Medicum, University of Helsinki, FI-00290 Helsinki, Finland.
Sini Penttilä, Neuromuscular Research Center, Tampere University Hospital and Fimlab Laboratories, FI-33520 Tampere, Finland.
Mridul Johari, Folkhälsan Research Center, Helsinki, Finland and Medicum, University of Helsinki, FI-00290 Helsinki, Finland; Harry Perkins Institute of Medical Research, Centre for Medical Research, University of Western Australia, Nedlands WA, Australia.
Marco Savarese, Folkhälsan Research Center, Helsinki, Finland and Medicum, University of Helsinki, FI-00290 Helsinki, Finland.
Peter Hackman, Folkhälsan Research Center, Helsinki, Finland and Medicum, University of Helsinki, FI-00290 Helsinki, Finland.
Cornelia Kornblum, Klinik und Poliklinik für Neurologie, Sektion Neuromuskuläre Erkrankungen, Universitätsklinikum Bonn, D-53127 Bonn, Germany.
Bjarne Udd, Folkhälsan Research Center, Helsinki, Finland and Medicum, University of Helsinki, FI-00290 Helsinki, Finland; Neuromuscular Research Center, Tampere University Hospital and Fimlab Laboratories, FI-33520 Tampere, Finland.
Data availability
The data are available upon reasonable request. The DNAJB2 c.832 T > G p.(*278Glyext*83) variant has been submitted to LOVD by individual ID 00430268.
Funding
Folkhälsan Research Foundation; the Jane and Aatos Erkko Foundation; Medicinska Understödsföreningen Liv och Hälsa; the Sigrid Jusélius Foundation; Finska Läkaresällskapet.
References
- 1. Kampinga, H.H. and Craig, E.A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol., 11, 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kampinga, H.H., Andreasson, C., Barducci, A., Cheetham, M.E., Cyr, D., Emanuelsson, C., Genevaux, P., Gestwicki, J.E., Goloubinoff, P., Huerta-Cepas, J.et al. (2019) Function, evolution, and structure of J-domain proteins. Cell Stress Chaperones, 24, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheetham, M.E., Brion, J.P. and Anderton, B.H. (1992) Human homologues of the bacterial heat-shock protein DnaJ are preferentially expressed in neurons. Biochem. J., 284, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chapple, J.P. and Cheetham, M.E. (2003) The chaperone environment at the cytoplasmic face of the endoplasmic reticulum can modulate rhodopsin processing and inclusion formation. J. Biol. Chem., 278, 19087–19094. [DOI] [PubMed] [Google Scholar]
- 5. Westhoff, B., Chapple, J.P., van derSpuy, J., Höhfeld, J. and Cheetham, M.E. (2005) HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr. Biol., 15, 1058–1064. [DOI] [PubMed] [Google Scholar]
- 6. Claeys, K.G., Sozanska, M., Martin, J.J., Lacene, E., Vignaud, L., Stockholm, D., Laforêt, P., Eymard, B., Kichler, A., Scherman, D.et al. (2010) DNAJB2 expression in normal and diseased human and mouse skeletal muscle. Am. J. Pathol., 176, 2901–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blumen, S.C., Astord, S., Robin, V., Vignaud, L., Toumi, N., Cieslik, A., Achiron, A., Carasso, R.L., Gurevich, M., Braverman, I.et al. (2012) A rare recessive distal hereditary motor neuropathy with HSJ1 chaperone mutation. Ann. Neurol., 71, 509–519. [DOI] [PubMed] [Google Scholar]
- 8. Gess, B., Auer-Grumbach, M., Schirmacher, A., Strom, T., Zitzelsberger, M., Rudnik-Schöneborn, S., Röhr, D., Halfter, H., Young, P. and Senderek, J. (2014) HSJ1-related hereditary neuropathies: novel mutations and extended clinical spectrum. Neurology, 83, 1726–1732. [DOI] [PubMed] [Google Scholar]
- 9. Howarth, J.L., Kelly, S., Keasey, M.P., Glover, C., Lee, Y.B., Mitrophanous, K., Chapple, J.P., Gallo, J.M., Cheetham, M.E. and Uney, J.B. (2007) Hsp40 molecules that target to the ubiquitin-proteasome system decrease inclusion formation in models of polyglutamine disease. Mol. Ther., 15, 1100–1105. [DOI] [PubMed] [Google Scholar]
- 10. Sarparanta, J., Jonson, P.H., Kawan, S. and Udd, B. (2020) Neuromuscular diseases due to chaperone mutations: a review and some new results. Int. J. Mol. Sci., 21, 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen, H.J., Mitchell, J.C., Novoselov, S., Miller, J., Nishimura, A.L., Scotter, E.L., Vance, C.A., Cheetham, M.E. and Shaw, C.E. (2016) The heat shock response plays an important role in TDP-43 clearance: evidence for dysfunction in amyotrophic lateral sclerosis. Brain, 139, 1417–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schabhüttl, M., Wieland, T., Senderek, J., Baets, J., Timmerman, V., De Jonghe, P., Reilly, M.M., Stieglbauer, K., Laich, E., Windhager, R.et al. (2014) Whole-exome sequencing in patients with inherited neuropathies: outcome and challenges. J. Neurol., 261, 970–982. [DOI] [PubMed] [Google Scholar]
- 13. Gonzaga-Jauregui, C., Harel, T., Gambin, T., Kousi, M., Griffin, L.B., Francescatto, L., Ozes, B., Karaca, E., Jhangiani, S.N., Bainbridge, M.N.et al. (2015) Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Rep., 12, 1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teive, H., Kok, F., Raskin, S. and Arruda, W. (2015) Distal hereditary motor neuropathy with HSJ1 chaperone mutation, presenting with peripheral motor neuropathy, associated to Parkinsonism, and cerebellar ataxia. Case report. In XXI World Congress on Parkinson’s Disease and Related Disorders, Milan, Italy. Parkinsonism Related Disord, Vol. 22, e154. [Google Scholar]
- 15. Sanchez, E., Darvish, H., Mesias, R., Taghavi, S., Firouzabadi, S.G., Walker, R.H., Tafakhori, A. and Paisán-Ruiz, C. (2016) Identification of a large DNAJB2 deletion in a family with spinal muscular atrophy and parkinsonism. Hum. Mutat., 37, 1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu, X., Duan, X., Zhang, Y., Sun, A. and Fan, D. (2020) Molecular analysis and clinical diversity of distal hereditary motor neuropathy. Eur. J. Neurol., 27, 1319–1326. [DOI] [PubMed] [Google Scholar]
- 17. Frasquet, M., Rojas-García, R., Argente-Escrig, H., Vázquez-Costa, J.F., Muelas, N., Vílchez, J.J., Sivera, R., Millet, E., Barreiro, M., Díaz-Manera, J.et al. (2021) Distal hereditary motor neuropathies: mutation spectrum and genotype-phenotype correlation. Eur. J. Neurol., 28, 1334–1343. [DOI] [PubMed] [Google Scholar]
- 18. Sharifi, Z., Taheri, M., Fallah, M.S., Abiri, M., Golnabi, F., Bagherian, H., Zeinali, R., Farahzadi, H., Alborji, M., Tehrani, P.G.et al. (2021) Comprehensive mutation analysis and report of 12 novel mutations in a cohort of patients with spinal muscular atrophy in Iran. J. Mol. Neurosci., 71, 2281–2298. [DOI] [PubMed] [Google Scholar]
- 19. Yalcintepe, S., Gurkan, H., Dogan, I.G., Demir, S., Sag, S.O., Kabayegit, Z.M., Atli, E.I., Atli, E., Eker, D. and Temel, S.G. (2021) The importance of multiple gene analysis for diagnosis and differential diagnosis in Charcot Marie Tooth disease. Turk. Neurosurg., 31, 888–895. [DOI] [PubMed] [Google Scholar]
- 20. Liu, M., Xu, Y., Hong, D., Cong, L., Fan, Y. and Zhang, J. (2022) DNAJB2 c.184C>T mutation associated with distal hereditary motor neuropathy with rimmed vacuolar myopathy. Clin. Neuropathol., 41, 226–232. [DOI] [PubMed] [Google Scholar]
- 21. Saveri, P., Magri, S., Maderna, E., Balistreri, F., Lombardi, R., Ciano, C., Moda, F., Garavaglia, B., Reale, C., Lauria Pinter, G.et al. (2022) DNAJB2-related Charcot-Marie-Tooth disease type 2: pathomechanism insights and phenotypic spectrum widening. Eur. J. Neurol., 29, 2056–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thauvin-Robinet, C., Duplomb-Jego, L., Limoge, F., Picot, D., Masurel, A., Terriat, B., Champilou, C., Minot, D., St-Onge, J., Kuentz, P.et al. (2016) Homozygous FIBP nonsense variant responsible of syndromic overgrowth, with overgrowth, macrocephaly, retinal coloboma and learning disabilities. Clin. Genet., 89, e1–e4. [DOI] [PubMed] [Google Scholar]
- 23. Akawi, N., Ben-Salem, S., Lahti, L., Partanen, J., Ali, B.R. and Al-Gazali, L. (2016) A recessive syndrome of intellectual disability, moderate overgrowth, and renal dysplasia predisposing to Wilms tumor is caused by a mutation in FIBP gene. Am. J. Med. Genet. A, 170, 2111–2118. [DOI] [PubMed] [Google Scholar]
- 24. Cazurro-Gutiérrez, A., Marcé-Grau, A., Correa-Vela, M., Salazar, A., Vanegas, M.I., Macaya, A., Bayés, À. and Pérez-Dueñas, B. (2021) ε-Sarcoglycan: unraveling the myoclonus-dystonia gene. Mol. Neurobiol., 58, 3938–3952. [DOI] [PubMed] [Google Scholar]
- 25. Karamanos, T.K., Tugarinov, V. and Clore, G.M. (2019) Unraveling the structure and dynamics of the human DNAJB6b chaperone by NMR reveals insights into Hsp40-mediated proteostasis. Proc. Natl. Acad. Sci. U. S. A., 116, 21529–21538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sarparanta, J., Jonson, P.H., Golzio, C., Sandell, S., Luque, H., Screen, M., McDonald, K., Stajich, J.M., Mahjneh, I., Vihola, A.et al. (2012) Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat. Genet., 44(450–455), S451–S452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hageman, J., Rujano, M.A., vanWaarde, M.A., Kakkar, V., Dirks, R.P., Govorukhina, N., Oosterveld-Hut, H.M., Lubsen, N.H. and Kampinga, H.H. (2010) A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol. Cell, 37, 355–369. [DOI] [PubMed] [Google Scholar]
- 28. Echaniz-Laguna, A., Lornage, X., Lannes, B., Schneider, R., Bierry, G., Dondaine, N., Boland, A., Deleuze, J.F., Böhm, J., Thompson, J.et al. (2017) HSPB8 haploinsufficiency causes dominant adult-onset axial and distal myopathy. Acta Neuropathol., 134, 163–165. [DOI] [PubMed] [Google Scholar]
- 29. Al-Tahan, S., Weiss, L., Yu, H., Tang, S., Saporta, M., Vihola, A., Mozaffar, T., Udd, B. and Kimonis, V. (2019) New family with HSPB8-associated autosomal dominant rimmed vacuolar myopathy. Neurol. Genet., 5, e349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harms, M.B., Sommerville, R.B., Allred, P., Bell, S., Ma, D., Cooper, P., Lopate, G., Pestronk, A., Weihl, C.C. and Baloh, R.H. (2012) Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann. Neurol., 71, 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palmio, J., Jonson, P.H., Inoue, M., Sarparanta, J., Bengoechea, R., Savarese, M., Vihola, A., Jokela, M., Nakagawa, M., Noguchi, S.et al. (2020) Mutations in the J domain of DNAJB6 cause dominant distal myopathy. Neuromuscul. Disord., 30, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Inoue, M., Noguchi, S., Inoue, Y.U., Iida, A., Ogawa, M., Bengoechea, R., Pittman, S.K., Hayashi, S., Watanabe, K., Hosoi, Y.et al. (2023) Distinctive chaperonopathy in skeletal muscle associated with the dominant variant in DNAJB4. Acta Neuropathol., 145, 235–255. [DOI] [PubMed] [Google Scholar]
- 33. Bengoechea, R., Pittman, S.K., Tuck, E.P., True, H.L. and Weihl, C.C. (2015) Myofibrillar disruption and RNA binding protein aggregation in a mouse model of limb girdle muscular dystrophy 1D. Hum. Mol. Genet., 24, 6588–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bengoechea, R., Findlay, A.R., Bhadra, A.K., Shao, H., Stein, K.C., Pittman, S.K., Daw, J.A., Gestwicki, J.E., True, H.L. and Weihl, C.C. (2020) Inhibition of DNAJ-HSP70 interaction improves strength in muscular dystrophy. J. Clin. Invest., 130, 4470–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Evilä, A., Arumilli, M., Udd, B. and Hackman, P. (2016) Targeted next-generation sequencing assay for detection of mutations in primary myopathies. Neuromuscul. Disord., 26, 7–15. [DOI] [PubMed] [Google Scholar]
- 36. Kopanos, C., Tsiolkas, V., Kouris, A., Chapple, C.E., Albarca Aguilera, M., Meyer, R. and Massouras, A. (2019) VarSome: the human genomic variant search engine. Bioinformatics, 35, 1978–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen, S., Zhou, Y., Chen, Y. and Gu, J. (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 34, i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dobin, A., Davis, C.A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M. and Gingeras, T.R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B.et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hageman, J. and Kampinga, H.H. (2009) Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones, 14, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kowarz, E., Loscher, D. and Marschalek, R. (2015) Optimized sleeping beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J., 10, 647–653. [DOI] [PubMed] [Google Scholar]
- 42. Maruyama, K. and MacLennan, D.H. (1988) Mutation of aspartic acid-351, lysine-352, and lysine-515 alters the Ca2+ transport activity of the Ca2+-ATPase expressed in COS-1 cells. Proc. Natl. Acad. Sci. U. S. A., 85, 3314–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mátés, L., Chuah, M.K., Belay, E., Jerchow, B., Manoj, N., Acosta-Sanchez, A., Grzela, D.P., Schmitt, A., Becker, K., Matrai, J.et al. (2009) Molecular evolution of a novel hyperactive sleeping beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet., 41, 753–761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available upon reasonable request. The DNAJB2 c.832 T > G p.(*278Glyext*83) variant has been submitted to LOVD by individual ID 00430268.