Abstract
Current strategies for delivery of proteins into cells face general challenges of endosomal entrapment and concomitant degradation of protein cargo. Efficient delivery directly to the cytosol overcomes this obstacle: we report here the use of biotin-streptavidin tethering to provide a modular approach to the generation of nanovectors capable of cytosolic delivery of biotinylated proteins. This strategy uses streptavidin to organize biotinylated protein and biotinylated oligo(glutamate) peptide into modular complexes that are then electrostatically self-assembled with a cationic guanidinium-functionalized polymer. The resulting polymer-protein nanocomposites demonstrate efficient cytosolic delivery of six biotinylated protein cargos of varying size, charge, and quaternary structure. Retention of protein function was established through efficient cell killing via delivery of the chemotherapeutic enzyme granzyme A. This platform represents a versatile and modular approach to intracellular delivery through non-covalent tethering of multiple components into a single delivery vector.
Keywords: protein delivery, polymer nanocarrier, supramolecular, biotin-streptavidin, cytosolic delivery
Graphical Abstract

INTRODUCTION
There are currently >130 FDA-approved protein therapeutics on the market1 but cell membrane impermeability has largely limited these approaches to extracellular targets.2,3 Intracellular protein delivery presents a transformative technology for biomedical applications and fundamental research.4–6 Proteins play critical roles in cell signal transduction7,8 and homeostasis,9,10 and many inherited diseases are caused by aberrant protein function.11,12
Most intracellular targets of protein delivery require access to the cytosol, including delivery to subcellular organelles and the nucleus.13 The vast majority of intracellular delivery strategies rely on endosomal uptake of modified proteins14 or nanovectors.15,16 Endosomal entrapment severely limits efficiency of these strategies,17 sequestering delivered cargo and preventing cytosolic access.18,19 Cargo entrapped in endosomes will generally be either exocytosed or degraded through the endo/lysosomal pathway,20 making endocytosis an ultimately challenging uptake route. Recent reports estimate that <10% of endocytosed cargo will escape into the cytosol,21 an outcome that can be improved by delivery vehicles engineered to trigger endosomal rupture and escape.22
Fusion of delivery vectors with the cell membrane provides a direct means of intracellular delivery, bypassing issues associated with endosomal entrapment.23,24 Liposomal vehicles can fuse with the cell membrane25,26 but face challenges related to opsonization and accelerated blood clearance.27,28 Polymer scaffolds are versatile platforms that provide flexibility of design through chemical diversity.29,30 Francis notably demonstrated intracellular protein delivery through site-selective conjugation of amphiphilic polymers to proteins.31 Other recent approaches have utilized cationic dendrimers32,33 or fluoropolymers34 to coordinate and deliver proteins to the cytosol,35,36 for diverse applications including cancer immunotherapy37 and therapeutic gene editing.38
Direct cytosolic protein delivery was previously achieved using guanidinium-functionalized gold nanoparticles39,40 and co-engineered proteins modified with terminal oligo(glutamate) “E-tags”. In this system, electrostatic guanidine-carboxylate interaction enabled self-assembly of supramolecular complexes. This strategy was recently adapted for guanidinium-functionalized poly(oxanorbornene) imide (PONI) polymers (PONI-Guan, Figure 1),41 providing highly efficient cytosolic delivery of several E-tagged proteins as demonstrated through nuclear co-localization with imaging flow cytometry, and enzymatic recombinase activity. Both strategies, however, require plasmid engineering to provide E-tagged proteins, generating a protein quite different from the native protein, and making the strategy cumbersome.
Figure 1.

a) Biotinylated components b-E20 and b-GFP complex (i) with streptavidin (STV). Addition of PONI-Guan polymer (ii) self-assembles into stable nanocomposites that deliver protein to the cytosol. b) Representative TEM micrograph of PONI-Guan/b-E20/b-GFP/STV nanocomposites (G/E 10). Scale bars = 20 nm. c) Confocal imaging (HEK-293T, G/E 10, 24 h incubation) reveals diffuse fluorescence throughout the cell. Scale bar = 10 μm.
Tetravalent biotin-streptavidin (STV) binding provides a modular high-affinity strategy for protein bioconjugation.42–45 A wide range of molecular and macromolecular systems can be biotinylated, making this a highly versatile platform for bioconjugation.45,46 We report here the use of biotin-streptavidin assemblies to provide cytosolic delivery of proteins into cells. In this system, biotinylated oligo(glutamate) (b-E20) and biotinylated proteins were first bound to STV (Figure 1). These conjugates were then self-assembled using PONI-Guan homopolymer to form discrete (200 – 350 nm on average) supramolecular polymer-protein nanocomposites. These complexes facilitated efficient cytosolic delivery, as demonstrated through microscopy and nuclear access of biotinylated green fluorescent protein (b-GFP) in mammalian cells. Cytosolic delivery of five additional fluorescent proteins of varying molecular weight, charge, and quaternary structure demonstrated versatility of this approach. Retention of enzymatic activity after delivery was validated by delivery of biotinylated granzyme A (b-GrA) with efficient cell killing. The modular nature of this platform provides a readily adaptable strategy for supramolecular conjugation and highly efficient cytosolic delivery of proteins.
RESULTS/DISCUSSION
Generation of biotin conjugates.
Green fluorescent protein (GFP, 27 kDa, pI 5.8) was used as a model protein cargo for its robust fluorescence and ability to passively diffuse into the nucleus to provide a clear readout of cytosolic access.47 b-GFP was prepared via amine-reactive crosslinking of wild-type GFP (wtGFP) via NHS ester coupling (Figure 2a and Figure S1). Analysis of conjugated GFP by electrospray ionization-mass spectrometry (ESI-MS)48 confirmed a distribution of labeling with ~6 biotins on average per b-GFP protein (Figure 2c–d). This result was validated using matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) (Figure S2). Binding between STV, b-GFP, and b-E20 was verified using a colorimetric HABA (4’-hydroxyazobenzene-2-carboxylic acid) displacement assay.49,50 Significantly, mono-biotinylated b-E20 and multi-biotinylated b-GFP displace comparable molar amounts of HABA, indicating b-GFP does not bind multiple STV proteins (Figure S3). Biotinylated E-tag (b-E20) was made likewise through coupling of commercially available E20 peptide with biotin NHS ester (Figure 2b). Details of these reactions are available in Supporting Information.
Figure 2.

Protein biotinylation through NHS ester coupling with a) lysine residues of wtGFP, and b) free terminal amine of E20 peptide. Detailed are available in Supporting Information. c) Representative ESI-MS spectra of wtGFP and d) b-GFP demonstrate mass differences. Average number of biotin-labeled residues estimated based on deconvoluted mass differences and relative intensity (see Supporting Information).
Additional proteins were biotinylated similarly through NHS ester coupling. α-chymotrypsin (25 kDa, pI 8.8), bovine serum albumin (BSA; 66 kDa, pI 4.7), apo-transferrin (77 kDa, pI 6.0), and DsRed (107 kDa, pI 11.0), were reacted with NHS-biotin at a 1:1 molar ratio to form b-α-chymotrypsin, b-BSA, b-apo-transferrin, and b-DsRed. The CRISPR-associated Cas9 protein (162 kDa, pI 8.0) and β-galactosidase (β-Gal, 465 kDa, pI 4.6) were conjugated at a 5:1 (NHS-biotin in excess) molar ratio to accommodate the larger size of these proteins. Cas9 notably features a nuclear localization signal (NLS) and was thus denoted as b-Cas9-NLS. Analysis of b-Cas9-NLS and b-β-gal by ESI-MS consistently resulted in poor ionization and low signal that could not be reliably deconvoluted (data not shown). For proteins prepared at a 1:1 molar ratio with NHS-biotin, ESI-MS analysis validated the presence of 1 biotin label per protein on average (Figure S4). Non-fluorescent proteins were subsequently labeled with Alexa Fluor 488 (AF488) to facilitate visualization in live cells.
Nanocomposite formation and characterization.
Complexes were first formed between STV, b-E20 peptide and b-GFP protein cargo through simultaneous mixture of the three components to avoid biased populations arising from cooperativity of STV-biotin binding.43,51 A 1:1:3 STV/b-E20/b-GFP molar ratio was chosen to maximize the population of STV complexed with three b-GFP proteins and a single b-E20 polypeptide (Figure S5). Significantly, assemblies generated by sequential component addition were delivery incompetent when complexed with PONI-Guan and largely remained attached at the cell surface (Figure S6).
PONI-Guan with 60 kDa molecular weight (synthetic details are available in Supporting Information) previously provided effective cytosolic delivery of proteins with engineered E-tags,41 and was used for self-assembly with the STV complexes. Nanocomposites were generated by simple mixture between aqueous PONI-Guan and STV/b-E20/b-GFP complexes, followed by incubation for 10 min at ambient temperature (Figure 3a). Formulations were based on charge ratio of total polymer guanidinium (G) groups to E-tag carboxylates (G/E ratio). A series of b-GFP nanocomposites were prepared through parametric variation of G/E ratio and characterized by DLS (Figures 3b and S7) and zeta potential (Figure 3c). The smallest complexes were formed at G/E 10, and all complexes featured a positive zeta potential (Figure 3c). TEM of the complexes indicated similar size and spherical morphology (Figure 3d). Complexation provided a degree of protection from degradation by protease, as compared with free protein (Figure S8), but less than with our previously reported platform.41 Nanocomposites were similarly generated with b-α-chymotrypsin, b-BSA, b-apo-transferrin, b-DsRed, b-Cas9-NLS, and b-β-gal at G/E 10 and analyzed by DLS. Complexes formed with an average size of 180–350 nm for all protein cargos evaluated except b-α-chymotrypsin, which exhibited an average size >800 nm (Figure S9).
Figure 3.

Characterization of supramolecular nanocomposites. a) Nanocomposite preparation through (i) STV conjugation with biotinylated components, followed by (ii) complexation with PONI-Guan homopolymer. b) Representative overlaid DLS spectra representing PONI-Guan/STV/b-E20/b-GFP (1:1:3 molar ratio, as described above) nanocomposites formulated at select G/E ratios. Size represented by number (size by intensity and volume are shown in Figure S7). c) Representative zeta potential measurements of PONI-Guan/STV/b-E20/b-GFP nanocomposites at select G/E ratios, with relevant controls. d) Representative TEM micrographs of PONI-Guan/STV/b-E20/b-GFP nanocomposites (G/E 10). Scale bar = 500 nm, inset scale bar = 20 nm.
Cytosolic protein delivery, and nuclear access of GFP.
Cytosolic delivery of b-GFP was demonstrated through incubation of PONI-Guan/STV/b-E20/b-GFP nanocomposites with HEK-293T (human embryonic kidney) cells in complete culture media (10% FBS) for 24 h at 37°C. Confocal microscopy revealed diffuse green fluorescence throughout the cytosol (Figure 4) indicative of cytosolic delivery. Diffuse nuclear fluorescence and co-localization with nuclear stain confirmed that GFP accessed the cytosol and diffused into the nucleus.52 Delivery at different time points confirmed that most delivery events occurred within 3–5 hours of initial incubation (Figure S10). All components were necessary to achieve cytosolic delivery (Figure S11). No significant toxicity was observed after incubation, and no obvious differences in cell morphology were observed (Figures S12 and S13). Efficient cytosolic delivery of b-GFP was also demonstrated in HeLa (human cervix adenocarcinoma) and CHO-K1 (hamster ovary epithelial-like) cells (Figure S14), demonstrating versatility of this delivery strategy.
Figure 4.

Representative confocal images of HEK-293T after 24 h incubation (10% FBS). Top row depicts b-GFP delivery at select G/E ratios (scale bar: 50 µm) with magnified image of G/E 10 (scale bar: 20 µm). Bottom row depicts cytosolic delivery of additional biotinylated proteins after 24 h incubation. Most delivery events occurred within 3–5 h. Nanocomposites were formulated at G/E 10. Scale bars: 50 µm. Diffuse fluorescence and nuclear access where appropriate signals cytosolic access.
To demonstrate generality of this delivery approach, a series of nanocomposites were generated using five different biotinylated proteins at G/E 10 and incubated with HEK-293T cells as above. After incubation, diffuse cytosolic fluorescence was observed with b-α-chymotrypsin, b-BSA, b-apo-transferrin, b-DsRed, and b-Cas9-NLS. The small size of b-α-chymotrypsin (25 kDa) allowed passive diffusion into the nucleus, further validating cytosolic access of delivered protein cargo. Delivery with larger proteins (>60 kDa) resulted in diffuse fluorescence with mostly dark nuclei, indicating cytosolic access without nuclear entry (Figure S15). Delivery of b-Cas9-NLS notably exhibited nuclear fluorescence due to NLS-driven nuclear import. Nanocomposites prepared with b-β-gal remained membrane-bound and did not facilitate cytosolic delivery (Figure S16), even with higher degrees of biotin labeling or formulation at different G/E ratios (data not shown). This is somewhat unsurprising given the extremely high molecular weight of β-galactosidase (465 kDa) and suggests an effective limitation of this delivery platform for very large cargos.
Quantification of GFP delivery by imaging flow cytometry.
Imaging flow cytometry provides a robust approach to quantifying delivery efficiency using large data sets.53 PONI-Guan/STV/b-E20/b-GFP formulations were prepared as described and incubated with HEK-293T cells for 24 h in serum-containing media. Flow imaging cytometry gating criteria was applied to the data as described in Supporting Information (Figures S17 and S18), and the cell population with co-localized nuclear fluorescence was quantified (Figure 5). Two positive controls were utilized: a commercial (Thermo Scientific) protein transfection kit (notably requiring serum-free delivery conditions) and engineered E-tagged GFP.41
Figure 5.
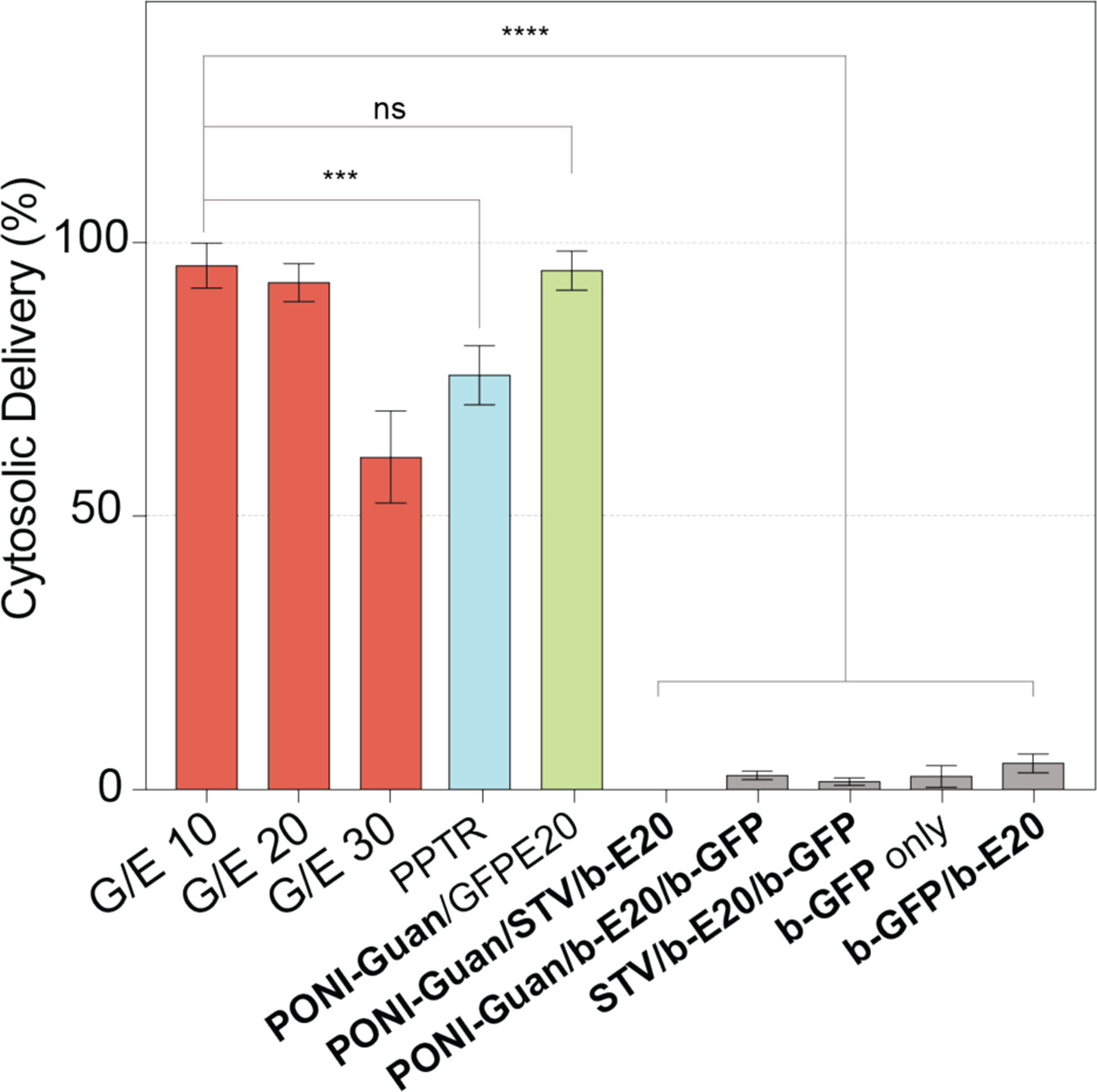
Imaging flow cytometry data presented as percentage of cell population, quantifying cytosolic/nuclear delivery of GFP. Nanocomposites were formulated as described above, at varying G/E ratios (red) or under various control conditions (gray). Delivery was compared with controls: b-GFP delivery (blue) using commercial Pierce Protein Transfection Reagent (PPTR, Thermo Scientific), incubated in serum-free media, and GFP-E20 (green) delivered by PONI-Guan at G/E 10.41 Data is average of 6 replicate wells. Error is standard deviation by population. Details of imaging flow cytometry experiments available in Supporting Information. Statistical analysis was performed through unpaired t-test; *** = p < 0.001; **** = p < 0.0001; ns is not significant.
A summary of delivery efficiencies using PONI-Guan/STV/b-E20/b-GFP nanocomposites in HEK-293T is shown in Figure 5. G/E ratios 10 and 20 demonstrated cytosolic delivery in >90% of cells. Delivery efficiency was notably comparable to PONI-Guan/GFP-E20 positive controls and outperformed the commercial transfection reagent.41 Delivery efficiency decreased as G/E ratio increased, suggesting a key role for surface charge density in cytosolic delivery.54,55 This trend is consistent with our previously reported results.41
Cholesterol provides membrane fluidity that is instrumental in facilitating cytosolic protein delivery through vehicle fusion with cell membranes in our prior nanoparticle39 and polymer41 protein delivery systems. Consistent with these reports, pretreatment of HEK-293T cells with cholesterol sequestration agents methyl-β-cyclodextrin (MβCD), α-cyclodextrin, and nystatin significantly decreased uptake of b-GFP.56,57 In contrast, delivery was minimally affected by pretreatment with inhibitors of clathrin-mediated endocytosis (chlorpromazine) 58, or macropinocytosis (imipramine) (Figure S19),59 supporting the bypassing of these pathways through a direct membrane-fusion-like process. Delivery was inhibited when incubated at 4˚C for 6h (Figure S20) due to contraction and decreased fluidity of the cell membrane.60,61,62,63
Delivery of therapeutic enzyme granzyme A.
Delivery of active enzymes into cells provides both a testbed for functional protein delivery as well as access to therapeutic applications.64 Granzyme A (GrA) is a serine protease that induces apoptosis and cell death,65,66,67 with selectivity observed for killing of malignant multidrug-resistant tumor cells.68 GrA homodimer (60 kDa) was purified and biotinylated using NHS-biotin to form b-GrA (Figure S21). b-GrA was complexed with STV, b-E20, and PONI-Guan polymer as described previously to form PONI-Guan/STV/b-E20/b-GrA nanocomposites, using an average of one E-tag per STV as above. Nanocomposites were formulated at G/E ratios ranging from 10 to 30, providing nanoparticles (Figure S22) consistent in size with those formed using b-GFP.
PONI-Guan/STV/b-E20/b-GrA nanoparticles were incubated with HEK-293T cells in serum-containing (10%) media. After incubation for 24 hours, cell killing was evaluated by Alamar Blue assay. Cell viability was significantly decreased at all G/E ratios for the PONI-Guan/STV/b-E20/b-GrA nanoparticles. (Figure 6). Likewise consistent with b-GFP delivery, pretreatment of cells with MβCD (7.5 mg/mL) prevented cell death, while pretreatment with an endocytosis inhibitor (CPM, 1.5 μg/mL) exhibited no significant difference from the treatment conditions. These results collectively demonstrate the ability of biotin-STV nanocomposites to deliver enzymes into the cell without compromising activity.
Figure 6.

Delivery of b-GrA causes decreased cell viability. Viability of HEK-293T cells following PONI-Guan/STV/b-E20/b-GrA treatment under varied conditions, as determined by Alamar Blue. G/E 10–30 (red) represents treatment with b-GrA nanocomposites at respective formulations. G/E 10+CPM (yellow) and G/E 10+MβCD (blue) show conditions that underwent pretreatment with small molecule inhibitor reagents. Controls are shown in gray. Data is average of 6 replicate wells. Error is standard deviation by population. Statistical analysis was performed through unpaired t-test; **** = p < 0.0001; ns is not significant. Details are available in Supporting Information.
CONCLUSIONS
In summary, biotin-streptavidin self-assembly provides a highly modular ‘plug-and-play’ approach to efficient intracellular delivery of proteins. The inherent simplicity of biotinylation facilitates access to effective E-tag delivery strategies that bypass endosomal pathways using membrane fusion-like processes. Cytosolic delivery and effective nuclear access of biotinylated GFP was quantified by imaging flow cytometry, demonstrating high efficiency compared with a commercial protein delivery reagent. Extension of this platform to accommodate five additional fluorescent protein cargos ranging in size, charge and structure demonstrated the versatility of this biotin-streptavidin tethering strategy for intracellular delivery. Delivery of cytotoxic protein granzyme A provided effective killing of cells, validating both retention of enzymatic activity and cytosolic access of delivered therapeutic protein. This modular platform immediately provides a robust approach to protein delivery ex vivo and in vitro, as well as a promising foundation for future in vivo therapeutic applications.
METHODS/EXPERIMENTAL
Materials.
All chemicals and solvents for syntheses were purchased from Fisher Scientific and Sigma-Aldrich, and used without further purification, unless otherwise stated. The chemicals were used as received. All reagents were purchased from Fisher Scientific and used as received for chemical synthesis. NIH-HEK-293T cells (ATCC CRL-3216) and CHO-K1 cells (ATCC CCL-61) were purchased from ATCC. Dulbecco’s Modified Eagle’s Medium (DMEM) (DMEM; ATCC 30–2002) and fetal bovine serum (Fisher Scientific, SH3007103) were used in cell culture. The yields of the compounds reported here refer to the yields of spectroscopically pure compounds after purification.
Nanocomposite preparation, characterization, and instrumentation.
PONI-Guan polymer/STV/b-E20/b-GFP nanocomposites were prepared in polypropylene microcentrifuge tubes (Fisher) by mixing in a specific order. STV (413 nM) was added first to a mixture of b-E20 (413 nM) and b-GFP (ranging from 413 nM to 1652 nM) by pipet and incubated at ambient temp. for ~10 minutes. After initial incubation period, PONI-Guan homopolymer (375 nM) was added to the nanocomposite mixture and allowed to incubate at ambient temperature for 10 minutes, escalating formation of final nanocomposites. Volume was corrected between samples using DPBS (HyClone). Samples were finally made up to volume with Dulbecco’s Modified Eagle Medium (DMEM, Gibco, 4500 mg/L glucose, HEPES-, L-Glutamine+, Sodium pyruvate+, phenol red+) supplemented with 10% (v/v) FBS (Gibco), and 1% antibiotics (Antibiotic-Antimycotic, Corning) and allowed to incubate a final 10 minutes. Size measurements were conducted in DPBS using a Malvern Zetasizer Nano-ZS. Alamar Blue (Invitrogen) viability studies were performed using a Molecular Devices Spectramax M2 with accompanying SoftMax Pro 7 software.
Electrospray ionization mass spectrometry.
ESI Mass spectrometry (Bruker MicroTOF-II) was used to confirm protein biotinylation. The pattern of peaks due to the multiple charges of the protein was deconvoluted using ESIprot software (online version: https://www.bioprocess.org/esiprot/esiprot_form.php). Mass of the protein was obtained at the given precision. Average degree of labeling was determined by comparing protein average mass differences (modified and unmodified) with the molecular weight of biotin. Biotinylated protein samples were prepared as previously described. Desalting was carried out through successive buffer exchange to MQ water using 10 kDa MWCO spin columns (Amicon). Samples were prepared for ESI by solvating the protein in a H2O:MeOH (1:1) solution with formic acid 0.1%.
Confocal microscopy.
Confocal microscopy imaging was performed using a Nikon A1 Spectral Detector Confocal. For all confocal studies, 1.5 × 105 cells were seeded in #1.5 circular confocal dishes (Mattek) for imaging 24 h prior. Confocal microscopy was used to assess delivery through fluorescence.
Supplementary Material
ACKNOWLEDGMENT
The authors would like to thank Dr. James Chambers and the Light Microscopy Core Facility at UMass Amherst as well as the Flow Cytometry Core Facility at UMass Amherst and Dr. Amy S. Burnside. The authors would like to especially acknowledge the UMass Amherst Mass Spectrometry Core Facility (RRID:SCR_019063) and Dr. Steven Eyles for his insight. pET26b-Granzyme A was a gift from Judy Lieberman (Addgene plasmid # 8823). pET21a-Streptavidin-Alive was a gift from Alice Ting (Addgene plasmid # 20860). DsRed2-pBAD was a gift from Michael Davidson (Addgene plasmid # 54608). pET-28b-Cas9-His was a gift from Alex Schier (Addgene plasmid # 47327).
Funding Sources
This research was supported by the NIH (EB022641 and DK DK121351).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Polymer synthesis and characterization; additional experimental details and related figures (PDF).
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Mullard A 2019 FDA Drug Approvals. Nat. Rev. Drug Discov 2020, 1, 79–84. [DOI] [PubMed] [Google Scholar]
- 2.Leader B; Baca QJ; Golan DE Protein Therapeutics: A Summary and Pharmacological Classification. Nat. Rev. Drug Discov 2008, 7, 21–39. [DOI] [PubMed] [Google Scholar]
- 3.Antignani A; Ho ECH; Bilotta MT; Qiu R; Sarnvosky R; FitzGerald DJ Targeting Receptors on Cancer Cells with Protein Toxins. Biomolecules 2020, 10, 1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scaletti F; Hardie J; Lee Y-W; Luther DC; Ray M; Rotello VM Protein Delivery into Cells Using Inorganic Nanoparticle–Protein Supramolecular Assemblies. Chem. Soc. Rev 2018, 47, 3421–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sánchez-Navarro M Advances in Peptide-Mediated Cytosolic Delivery of Proteins. Adv. Drug Deliv. Rev 2021, 171, 187–198. [DOI] [PubMed] [Google Scholar]
- 6.Ye Y; Yu J; Wen D; Kahkoska AR; Gu Z Polymeric Microneedles for Transdermal Protein Delivery. Adv. Drug Deliv. Rev 2018, 127, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer BJ; Yu J Protein Clusters in Phosphotyrosine Signal Transduction. J. Mol. Biol 2018, 430, 4547–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gopalan J; Wordeman L; Scott JD Kinase-Anchoring Proteins in Ciliary Signal Transduction. Biochem. J 2021, 478, 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De I; Dogra N; Singh S. The Mitochondrial Unfolded Protein Response: Role in Cellular Homeostasis and Disease. Current Molecular Medicine 2017, 17, 587–597. [DOI] [PubMed] [Google Scholar]
- 10.Ainslie A; Huiting W; Barazzuol L; Bergink S Genome Instability and Loss of Protein Homeostasis: Converging Paths to Neurodegeneration? Open Biol 2021, 11, 200296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kusum Y; Anurag Y; Priyanka V; Veda PP; Upendra ND Protein Misfolding Diseases and Therapeutic Approaches. Current Protein & Peptide Science 2019, 20, 1226–1245. [DOI] [PubMed] [Google Scholar]
- 12.Hegde RN; Subramanian A; Pothukuchi P; Parashuraman S; Luini A Rare ER Protein Misfolding-Mistrafficking Disorders: Therapeutic Developments. Tissue Cell 2017, 49, 175–185. [DOI] [PubMed] [Google Scholar]
- 13.Deprey K; Becker L; Kritzer J; Pluckthun A Trapped! A Critical Evaluation of Methods for Measuring Total Cellular Uptake versus Cytosolic Localization. Bioconjug. Chem 2019, 30, 1006–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Habault J; Poyet J-L Recent Advances in Cell Penetrating Peptide-Based Anticancer Therapies. Molecules 2019, 24, 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alamoudi K; Martins P; Croissant JG; Patil S; Omar H; Khashab NM Thermoresponsive Pegylated Bubble Liposome Nanovectors for Efficient SiRNA Delivery via Endosomal Escape. Nanomedicine 2017, 12, 1421–1433. [DOI] [PubMed] [Google Scholar]
- 16.He W; Xing X; Wang X; Wu D; Wu W; Guo J; Mitragotri S Nanocarrier‐Mediated Cytosolic Delivery of Biopharmaceuticals. Adv. Funct. Mater 2020, 30, 1910566. [Google Scholar]
- 17.Pei D; Buyanova M Overcoming Endosomal Entrapment in Drug Delivery. Bioconjug. Chem 2019, 30, 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brock DJ; Kustigian L; Jiang M; Graham K; Wang T-Y; Erazo-Oliveras A; Najjar K; Zhang J; Rye H; Pellois J-P Efficient Cell Delivery Mediated by Lipid-Specific Endosomal Escape of Supercharged Branched Peptides. Traffic 2018, 19, 421–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart MP; Lorenz A; Dahlman J; Sahay G Challenges in Carrier-Mediated Intracellular Delivery: Moving beyond Endosomal Barriers: Challenges in Carrier-Mediated Intracellular Delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol 2016, 8, 465–478. [DOI] [PubMed] [Google Scholar]
- 20.Luzio JP; Parkinson MDJ; Gray SR; Bright NA The Delivery of Endocytosed Cargo to Lysosomes. Biochem. Soc. Trans 2009, 37, 1019–1021. [DOI] [PubMed] [Google Scholar]
- 21.Smith SA; Selby LI; Johnston APR; Such GK The Endosomal Escape of Nanoparticles: Toward More Efficient Cellular Delivery. Bioconjug. Chem 2019, 30, 263–272. [DOI] [PubMed] [Google Scholar]
- 22.Du S; Liew SS; Li L; Yao SQ Bypassing Endocytosis: Direct Cytosolic Delivery of Proteins. J. Am. Chem. Soc 2018, 140, 15986–15996. [DOI] [PubMed] [Google Scholar]
- 23.Sun L; Gao Y; Wang Y; Wei Q; Shi J; Chen N; Li D; Fan C Guiding Protein Delivery into Live Cells Using DNA-Programmed Membrane Fusion. Chem. Sci 2018, 9, 5967–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ungermann C; Kümmel D Structure of Membrane Tethers and Their Role in Fusion. Traffic 2019, 20, 479–490. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann M; Hersch N; Gerlach S; Dreissen G; Springer R; Merkel R; Csiszár A; Hoffmann B Complex Size and Surface Charge Determine Nucleic Acid Transfer by Fusogenic Liposomes. Int. J. Mol. Sci 2020, 21, 2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Bahreman A, Daudey G, Bussmann J, Olsthoorn RCL, & Kros A Drug Delivery via Cell Membrane Fusion Using Lipopeptide Modified Liposomes. ACS Central Science, 2016, 2, 621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taira MC; Chiaramoni NS; Pecuch KM; Alonso-Romanowski S Stability of Liposomal Formulations in Physiological Conditions for Oral Drug Delivery. Drug Deliv 2004, 11, 123–128. [DOI] [PubMed] [Google Scholar]
- 28.Sercombe L; Veerati T; Moheimani F; Wu SY; Sood AK; Hua S Advances and Challenges of Liposome Assisted Drug Delivery. Front. Pharmacol 2015, 6, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lv J; Fan Q; Wang H; Cheng Y Polymers for Cytosolic Protein Delivery. Biomaterials 2019, 218, 119358. [DOI] [PubMed] [Google Scholar]
- 30.Kamaly N; Xiao Z; Valencia PM; Radovic-Moreno AF; Farokhzad OC Targeted Polymeric Therapeutic Nanoparticles: Design, Development and Clinical Translation. Chem. Soc. Rev 2012, 41, 2971–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sangsuwan R; Tachachartvanich P; Francis MB Cytosolic Delivery of Proteins Using Amphiphilic Polymers with 2-Pyridinecarboxaldehyde Groups for Site-Selective Attachment. J. Am. Chem. Soc 2019, 141, 2376–2383. [DOI] [PubMed] [Google Scholar]
- 32.Ren L; Lv J; Wang H; Cheng Y A Coordinative Dendrimer Achieves Excellent Efficiency in Cytosolic Protein and Peptide Delivery. Angew. Chem. Int. Ed Engl 2020, 59, 4711–4719. [DOI] [PubMed] [Google Scholar]
- 33.Kretzmann JA; Luther DC; Evans CW; Jeon T; Jerome W; Gopalakrishnan S; Lee Y-W; Norret M; Iyer KS; Rotello VM Regulation of Proteins to the Cytosol Using Delivery Systems with Engineered Polymer Architecture. J. Am. Chem. Soc 2021, 143, 4758–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z; Shen W; Ling J; Yan Y; Hu J; Cheng Y The Fluorination Effect of Fluoroamphiphiles in Cytosolic Protein Delivery. Nat. Commun 2018, 9, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Y Design of Polymers for Intracellular Protein and Peptide Delivery. Chin. J. Chem 2021, 39, 1443–1449. [Google Scholar]
- 36.Zhang S; Lv J; Gao P; Feng Q; Wang H; Cheng Y A pH-Responsive Phase-Transition Polymer with High Serum Stability in Cytosolic Protein Delivery. Nano Lett 2021, 21, 7855–7861. [DOI] [PubMed] [Google Scholar]
- 37.Xu J; Lv J; Zhuang Q; Yang Z; Cao Z; Xu L; Pei P; Wang C; Wu H; Dong Z; Chao Y; Wang C; Yang K; Peng R; Cheng Y; Liu Z A General Strategy Towards Personalized Nanovaccines Based on Fluoropolymers for Post-Surgical Cancer Immunotherapy. Nat. Nanotechnol 2020, 15, 1043–1052. [DOI] [PubMed] [Google Scholar]
- 38.Liu C; Wan T; Wang H; Zhang S; Ping Y; Cheng Y A Boronic Acid-Rich Dendrimer with Robust and Unprecedented Efficiency for Cytosolic Protein Delivery and CRISPR-Cas9 Gene Editing. Sci. Adv 2019, 5, eaaw8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mout R; Ray M; Tay T; Sasaki K; Yesilbag Tonga G; Rotello VM General Strategy for Direct Cytosolic Protein Delivery via Protein–Nanoparticle Co-Engineering. ACS Nano 2017, 11, 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y‐W; Mout R; Luther DC; Liu Y; Castellanos‐García L; Burnside AS; Ray M; Tonga GY; Hardie J; Nagaraj H; Das R; Phillips EL; Tay T; Vachet RW; Rotello VM In Vivo Editing of Macrophages through Systemic Delivery of CRISPR‐Cas9‐Ribonucleoprotein‐Nanoparticle Nanoassemblies. Adv. Therap 2019, 1900041. [DOI] [PMC free article] [PubMed]
- 41.Lee Y-W; Luther DC; Goswami R; Jeon T; Clark V; Elia J; Gopalakrishnan S; Rotello VM Direct Cytosolic Delivery of Proteins Through Coengineering of Proteins and Polymeric Delivery Vehicles. J. Am. Chem. Soc 2020, 142, 4349–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schendel LC; Sedlak SM; Gaub HE Switchable Reinforced Streptavidin. Nanoscale 2020, 12, 6803–6809. [DOI] [PubMed] [Google Scholar]
- 43.Waner MJ; Hiznay JM; Mustovich AT; Patton W; Ponyik C; Mascotti DP Streptavidin Cooperative Allosterism Upon Binding Biotin Observed by Differential Changes in Intrinsic Fluorescence. Biochem. Bio-phys. Rep 2019, 17, 127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reznik GO; Vajda S; Cantor CR; Sano T A Streptavidin Mutant Useful for Directed Immobilization on Solid Surfaces. Bioconjug. Chem 2001, 12, 1000–1004. [DOI] [PubMed] [Google Scholar]
- 45.Xu D, & Wegner SV Multifunctional Streptavidin–Biotin Conjugates with Precise Stoichiometries. Chem. Sci, 2020. 11, 4422–4429. [Google Scholar]
- 46.Udeshi ND; Pedram K; Svinkina T; Fereshetian S; Myers SA; Aygun O; Krug K; Clauser K; Ryan D; Ast T; Mootha VK; Ting AY; Carr SA Antibodies to Biotin Enable Large-Scale Detection of Biotinylation Sites on Proteins. Nat. Methods 2017, 14, 1167–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luther DC; Jeon T; Goswami R; Nagaraj H; Kim D; Lee Y-W; Rotello VM Protein Delivery: If Your GFP (or Other Small Protein) Is in the Cytosol, It Will Also Be in the Nucleus. Bioconjug. Chem 2021, 32, 891–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winkler R ESIprot: A Universal Tool for Charge State Determination and Molecular Weight Calculation of Proteins from Electrospray Ionization Mass Spectrometry Data. Rapid Commun Mass Spectrom 2010, 24, 285–294. [DOI] [PubMed] [Google Scholar]
- 49.Green NM Spectrophotometric Determination of Avidin and Biotin. Meth. In Enzymol 1970, 18, 418–424. [Google Scholar]
- 50.Sedlak SM; Bauer MS; Kluger C; Schendel LC; Milles LF; Pippig DA; Gaub HE Monodisperse Measurement of the Biotin-Streptavidin Interaction Strength in a Well-Defined Pulling Geometry. PLoS One 2017, 12, e0188722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu F; Zhang JZH; Mei Y The Origin of the Cooperativity in the Streptavidin-Biotin System: A Computational Investigation Through Molecular Dynamics Simulations. Sci. Rep 2016, 6, 27190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Timney BL; Raveh B; Mironska R; Trivedi JM; Kim SJ; Russel D; Wente SR; Sali A; Rout MP Simple Rules for Passive Diffusion Through the Nuclear Pore Complex. J. Cell Biol 2016, 215, 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rane AS; Rutkauskaite J; deMello A; Stavrakis S High-Throughput Multi-Parametric Imaging Flow Cytometry. Chem 2017, 3, 588–602. [Google Scholar]
- 54.Wexselblatt E; Esko JD; Tor Y On Guanidinium and Cellular Uptake. J. Org. Chem 2014, 79, 6766–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghosh PS; Kim C-K; Han G; Forbes NS; Rotello VM Efficient Gene Delivery Vectors by Tuning the Surface Charge Density of Amino Acid-Functionalized Gold Nanoparticles. ACS Nano 2008, 2, 2213–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahammad S; Parmryd I Cholesterol Depletion Using Methyl-β-Cyclodextrin. Methods Mol. Biol 2015, 1232, 91–102. [DOI] [PubMed] [Google Scholar]
- 57.Chen Y; Wang S; Lu X; Zhang H; Fu Y; Luo Y Cholesterol Sequestration by Nystatin Enhances the Uptake and Activity of Endostatin in Endothelium via Regulating Distinct Endocytic Pathways. Blood 2011, 117, 6392–6403. [DOI] [PubMed] [Google Scholar]
- 58.Sasso L; Purdie L; Grabowska A; Jones AT; Alexander C Time and Cell-Dependent Effects of Endocytosis Inhibitors on the Internalization of Biomolecule Markers and Nanomaterials: Endocytosis Inhibitors. J. Interdiscip. Nanomed 2018, 3, 67–81. [Google Scholar]
- 59.Lin H-P; Singla B; Ghoshal P; Faulkner JL; Cherian-Shaw M; O’Connor PM; She J-X; Belin de Chantemele EJ; Csányi G Identification of Novel Macropinocytosis Inhibitors Using a Rational Screen of Food and Drug Administration-Approved Drugs. Br. J. Pharmacol 2018, 175, 3640–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pan R; Xu W; Ding Y; Lu S; Chen P Uptake Mechanism and Direct Translocation of a New CPP for siRNA Delivery. Mol. Pharm 2016, 13, 1366–1374. [DOI] [PubMed] [Google Scholar]
- 61.Fu J; Yu C; Li L; Yao SQ Intracellular Delivery of Functional Proteins and Native Drugs by Cell-Penetrating Poly(disulfide)s. J. Am. Chem. Soc 2015, 137, 12153–12160. [DOI] [PubMed] [Google Scholar]
- 62.Takechi-Haraya Y; Aki K; Tohyama Y; Harano Y; Kawakami T; Saito H; Okamura E Glycosaminoglycan Binding and Non-Endocytic Membrane Translocation of Cell-Permeable Octaarginine Monitored by Real-Time-in-Cell NMR Spectroscopy. Pharmaceuticals 2017, 10, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walrant A; Matheron L; Cribier S; Chaignepain S; Jobin M-L; Sagan S; Alves ID Direct Translocation of Cell-Penetrating Peptides in Liposomes: A Combined Mass Spectrometry Quantification and Fluorescence Detection Study. Anal. Biochem 2013, 438, 1–10. [DOI] [PubMed] [Google Scholar]
- 64.Luan X; Wu Y; Shen Y-W; Zhang H; Zhou Y-D; Chen H-Z; Nagle DG; Zhang W-D Cytotoxic and Antitumor Peptides as Novel Chemotherapeutics. Nat. Prod. Rep 2021, 38, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Daalen KR; Reijneveld JF; Bovenschen N Modulation of Inflammation by Extracellular Granzyme A. Front. Immunol 2020, 11, 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lieberman J Granzyme A Activates Another Way to Die: Granzyme A-Mediated Cell Death. Immunol. Rev 2010, 235, 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mout R; Ray M; Tay T; Sasaki K; Yesilbag Tonga G; Rotello VM General Strategy for Direct Cytosolic Protein Delivery via Protein–Nanoparticle Co-Engineering. ACS Nano 2017, 11, 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grodzovski I; Lichtenstein M; Galski H; Lorberboum-Galski H IL-2-Granzyme A Chimeric Protein Overcomes Multidrug Resistance (MDR) Through a Caspase 3-Independent Apoptotic Pathway. Int. J. Cancer 2011, 128, 1966–1980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


