Abstract
The Pseudomonas aeruginosa Mus clinical isolate produces OXA-18, a pI 5.5 class D extended-spectrum β-lactamase totally inhibited by clavulanic acid (L. N. Philippon, T. Naas, A.-T. Bouthors, V. Barakett, and P. Nordmann, Antimicrob. Agents Chemother. 41:2188–2195, 1997). A second β-lactamase was cloned, and the recombinant Escherichia coli clone pPL10 expressed a pI 7.4 β-lactamase which conferred high levels of amoxicillin and ticarcillin resistance and which was partially inhibited by clavulanic acid. The 2.5-kb insert from pPL10 was sequenced, and a 266-amino-acid protein (OXA-20) was deduced; this protein has low amino acid identity with most of the class D β-lactamases except OXA-2, OXA-15, and OXA-3 (75% amino acid identity with each). OXA-20 is a restricted-spectrum oxacillinase and is unusually inhibited by clavulanic acid. OXA-20 is a peculiar β-lactamase because its translation initiates with a TTG (leucine) codon, which is rarely used as a translational origin in bacteria. Exploration of the genetic environment of oxa20 revealed the presence of the following integron features: (i) a second antibiotic resistance gene, aacA4; (ii) an intI1 gene; and (iii) two 59-base elements, each associated with either oxa20 or aacA4. This integron is peculiar because it lacks the 3′ conserved region, and therefore is not a sul1-associated integron like most of them, and because its 3′ end is located within tnpR, a gene involved in the transposition of Tn5393, a gram-negative transposon. P. aeruginosa Mus produces two novel and unrelated oxacillinases, OXA-18 and OXA-20, both of which are inhibited by clavulanic acid.
Analysis of the known β-lactamase sequences permits their division into four classes, designated A to D, based on their amino acid contents (1). Plasmid-mediated β-lactamases are observed in Pseudomonas aeruginosa isolates in fewer than 2% of samples, according to a study conducted at the Royal London Hospital in the United Kingdom in 1991 (29). TEM-1 and TEM-2 have been observed in this species (32), where they confer resistance to amino-penicillins, carboxy-penicillins, and ureido-penicillins. PSE (Pseudomonas-specific enzyme)-type β-lactamases (with the exception of PSE-2, which is, in fact, an oxacillinase), also called carbenicillin-hydrolyzing enzymes, are found primarily in this bacterial species but have also been identified in members of the family Enterobacteriaceae. PSE-1 or CARB-2 is the most frequent plasmid-mediated β-lactamase found in P. aeruginosa (38).
The OXA-type (oxacillin-hydrolyzing) enzymes are frequently observed in P. aeruginosa. They usually confer resistance to amoxicillin and cephalothin and possess high-level hydrolytic activity against cloxacillin, oxacillin, and methicillin. Their activities are usually poorly or not inhibited by clavulanic acid (3). All oxacillin-hydrolyzing β-lactamases belong to Ambler class D (1) and thus possess an active serine site like class A and C β-lactamases do (22). Ambler class D includes OXA-1 to OXA-18, as well as PSE-2 (OXA-10). While some oxacillinases are variants differing by only single amino acid changes [for example, OXA-1 and OXA-4; OXA-10 (PSE-2), OXA-11, and OXA-14] and others demonstrate a significant degree of amino acid identity (for example, OXA-1, OXA-7, OXA-5, and OXA-10; OXA-2 and OXA-3), most of them have only low levels of amino acid identity to each other (20 to 30%) (44, 45).
Most of the oxacillinase genes identified to date, except the gene for LCR-1, oxa12, and oxa18, are located on the variable region of integrons (39, 41). Integrons determine a site-specific recombination system capable of capturing and subsequently expressing genes that are contained in gene cassettes (for a review, see reference 41). Integrons contain a recombination site, attI, into which the captured genes are integrated and carry in the 5′ conserved region an integrase gene, intI (see Fig. 6A) (41). This enzyme mediates both the insertion and the excision of the resistance genes. Three distinct classes of elements which include all three of the features (intI, attI, and promoter) that define integrons have been described so far (41). Class 1 includes the majority of the integrons found in clinical isolates to date, class 2 includes the transposon Tn7 and relatives, and class 3 contains one single integron thus far (2, 41). Members of each integron class have nearly identical integrases, while integrases vary significantly between the classes. Each gene cassette includes a gene associated with a recombination site known as a 59-base element (59-BE) located downstream of the gene. 59-BEs differ substantially in length from 57 to 141 bp, but they are all bounded by an inverse core site (RYYYAAC) at the left-hand side closest to the 3′ end of the gene coding region and a core site (GTTRRRY) at the right-hand side (47). Once inserted into an integron, a small part of the 59-BE (the conserved motif TTRRRY at the 5′ end) is found at the start of the linearized cassette and the remainder (ending with a conserved G at the 3′ end) is downstream of the gene (5, 16, 41). These two motifs seem to be necessary for the recombination of the resistance genes (30, 41). This mechanism explains how plasmids may accumulate such a diversity of resistance genes.
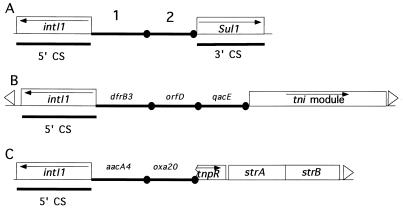
FIG. 6.
Locations of gene cassettes within class 1 integrons. The cassettes are represented by solid lines, and their 59-BEs are represented by closed circles. (A) General structure of naturally occurring class 1 integrons. The intI1 gene, which codes for the integrase responsible for the cassette movement, is contained in the 5′ conserved sequence (5′ CS). The 3′ CS found downstream of the integrated cassettes includes the sulfonamide resistance gene sul1. Inserted genes are indicated by 1 and 2. (B) General structure of a Tn402 type of integron. It is a class 1 integron lacking the 3′ CS. Four transposition genes (tni) present are indicated downstream of the cassettes (41). (C) Schematic structure of the oxa20-associated integron found in P. aeruginosa Mus.
A clinical isolate, P. aeruginosa Mus, showed resistance both to extended-spectrum cephalosporins and to aztreonam. Isoelectric focusing revealed that this strain produced three β-lactamases with pIs of 5.5, 7.4, and 8.2. While the pI 8.2 enzyme likely corresponded to a chromosomal cephalosporinase, the pI 5.5 enzyme, named OXA-18 (39), had a broad substrate profile, hydrolyzing amoxicillin, ticarcillin, cephalothin, ceftazidime, cefotaxime, and aztreonam but neither imipenem nor cephamycins. OXA-18 is a peculiar class D β-lactamase because it confers high resistance to expanded-spectrum cephalosporins and is totally inhibited by clavulanic acid (39).
Here, we describe the third β-lactamase of the same P. aeruginosa isolate, a novel chromosomally mediated restricted-spectrum oxacillinase. We analyzed the gene coding for this enzyme by cloning and sequencing it and by comparing the gene sequence with that of other class D β-lactamases. We determined the enzymatic properties of the enzyme and attempted to characterize its genetic determinant. This class D β-lactamase has moderate hydrolysis activity against oxacillin and higher activity against early cephalosporins. Its activity is partially inhibited by clavulanic acid, and its gene is located on a non-sul1 type of integron. In addition, we further characterized the genetic environment of this integron.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this work are listed in Table 1. P. aeruginosa Mus was isolated in 1995, at the Hôpital Saint-Antoine, Paris, France, from a biliar drain of a hospitalized patient from Sicily, Italy. The clinical case was described previously (39). This strain was also resistant to amikacin, chloramphenicol, gentamicin, kanamycin, netilmicin, streptomycin, tobramycin, and sulfonamides.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or referencea |
|---|---|---|
| Strains | ||
| E. coli JM1090 | endA1 hsdR17 gyrA96 Δ(lac proA) recAB1 relA supE44 thi F′ (lacIqlacZΔM15 proAB+ traD36) | 52 |
| In vitro-obtained ciprofloxacin-resistant E. coli JM109 | Ciprofloxacin resistant | 39 |
| E. coli NCTC 50192 | 154-, 66-, 48-, and 7-kb reference plasmids | 11 |
| P. aeruginosa Mus | OXA-18 and the studied β-lactamase | 39 |
| P. aeruginosa PU21 | ilv leu Strr Rifr | 20 |
| In vitro-obtained ciprofloxacin-resistant P. aeruginosa PU21 | Ciprofloxacin resistant | 39 |
| A. hydrophila 76-14 | Wild-type phenotype | IPSC |
| In vitro-obtained ciprofloxacin-resistant A. hydrophila 76-14 | Ciprofloxacin resistant | 39 |
| Plasmids | ||
| pBK-CMV phagemid | Neomycinr/kanamycinr | Stratagene |
| pBR322 | Recombinant plasmid containing 560-bp SspI-PstI internal fragment of blaTEM-1 | 48 |
| pHUC37 | Recombinant plasmid containing 435-bp PstI-NotI internal fragment of blaSHV-3 | 34 |
| pPZ1 | Recombinant plasmid containing 1.1-kb SnaBI internal fragment of blaPER-1 | 35 |
| pPL1 | Recombinant plasmid containing 2.6-kb genomic Sau3A fragment containing blaOXA-18 | 39 |
| pPL10 | Recombinant plasmid containing 2.5-kb genomic Sau3A fragment containing blaOXA-20 | This report |
| pPL11 | Recombinant plasmid containing 12-kb genomic Sau3A fragment containing blaOXA-20 | This report |
IPSC, Institut Pasteur strain collection.
Antimicrobial agents and MIC determinations.
The antimicrobial agents used in this study were obtained from standard laboratory powders and were used immediately after their solubilization. The agents and their sources were as follows: amoxicillin, clavulanic acid, cloxacillin, and ticarcillin, Smith-Kline-French-Beecham (Nanterre, France); aztreonam and cefepime, Bristol-Myers Squibb (Paris La Défense, France); ceftazidime, Glaxo (Paris, France); cefamandole, cephalothin, and moxalactam, Eli Lilly (Saint-Cloud, France); piperacillin and tazobactam, Lederle (Oullins, France); sulbactam, Pfizer (Orsay, France); cefotaxime and cefpirome, Hoechst-Roussel (Paris, France); cefoxitin and imipenem, Merck Sharp & Dohme-Chibret (Paris, France).
MICs were determined by an agar dilution technique on Mueller-Hinton agar (Diagnostics Pasteur) with an inoculum of 104 CFU. All plates were incubated at 37°C for 18 h. MICs of β-lactams were determined alone or in combination with a fixed concentration of clavulanic acid (2 μg/ml).
Hybridization.
Dot blots were performed with the ECL nonradioactive kit (Amersham, Les Ulis, France) as described by the manufacturer. The probes (Table 1) consisted of the 1.1-kb SnaBI fragment from recombinant plasmid pPZ1 for blaPER-1, the 450-bp PstI-NotI fragment from recombinant plasmid pHUC37 for blaSHV-3, the 560-bp SspI-PstI fragment from plasmid pBR322 for blaTEM-1, and the 450-bp PstI-NotI fragment from recombinant plasmid pPL1 for blaOXA-18.
Plasmid content and mating-out assays.
DNA of P. aeruginosa Mus plasmid was prepared by four different methods as described by Danel et al. (10), Hansen and Olsen (18), Kado and Liu (23), and Takahashi and Nagano (50). Plasmid DNA was analyzed by electrophoresis on an 0.8% agarose gel (BRL Life Technologies, Eragny, France) containing 0.25 μg of ethidium bromide (Pharmacia Biotech, Orsay, France) per ml. Plasmid DNAs of standard sizes were extracted from Escherichia coli NCTC 50192. The extracted material was subjected to electroporation into E. coli JM109. Recombinant bacteria were plated on amoxicillin-containing (100 μg/ml) Trypticase soy agar plates.
Direct transfer of resistance genes into in vitro-obtained ciprofloxacin-resistant P. aeruginosa PU21, E. coli JM109, or Aeromonas hydrophila 76.14 was attempted by liquid and solid mating-out assays at 30 and 37°C. Transconjugant selection was performed on Trypticase soy agar plates (Diagnostics Pasteur) containing ciprofloxacin (3 μg/ml) and either amoxicillin (100 μg/ml) or ticarcillin (150 μg/ml).
Cloning experiments and analysis of recombinant plasmids.
Genomic DNA of P. aeruginosa Mus was extracted as described before (35). Fragments from genomic DNA that was partially Sau3AI digested (Pharmacia Biotech) were ligated into the BamHI (Pharmacia Biotech) site of phagemid pBK-CMV (Stratagene, La Jolla, Calif.) as previously described (35).
Recombinant plasmid DNAs were prepared by using Qiagen columns (Coger, Paris, France). Plasmid mapping was performed after double-restriction analysis (43). Fragment sizes were estimated by comparison to the 1-kb DNA ladder molecular weight standard (BRL Life Technologies).
Isoelectric focusing.
Cultures were grown overnight at 37°C in 20 ml of Trypticase soy broth with 100 μg of amoxicillin per ml. Bacterial suspensions were disrupted by sonication (two times, for 30 s at 20 Hz each time; Vibra Cell 300 phospholyser; Bioblock, Illkirch, France) and centrifuged (30 min, 10,000 × g, 4°C). The supernatant containing the crude enzyme extracts was subjected to analytical isoelectric focusing on a pH 3.5 to 9.5 ampholine polyacrylamide gel (Ampholine PAG plate; Pharmacia Biotech) for 90 min at a constant voltage of 1,500 V (50 mA, 30 W). The focused β-lactamases were detected by overlaying the gel with 1 mM nitrocefin (Glaxo, Paris, France) in 100 mM phosphate buffer (pH 7.0). The pI values were determined and compared to those of known β-lactamases, i.e., 5.4 for TEM-1, 5.6 for TEM-2, 7.0 for SHV-3, and 8.2 for SHV-5 (3).
β-Lactamase purification.
A one-liter culture of E. coli JM109(pPL10) was grown overnight. The bacteria were harvested for 10 min at 6,000 × g, and the pellet (4 g) was resuspended in 15 ml of 50 mM BisTris (pH 7.1) {[bis(2-hydroxyethyl) imino]tris(hydroxymethyl)methane} at 4°C. The bacterial cells were disrupted by ultrasonic treatment as described above. Residual cells and debris were removed by centrifugation (48,000 × g for 30 min at 4°C). Nucleic acids were precipitated by the addition of 0.2 M spermine (7%, vol/vol) and centrifugation at 100,000 × g for 60 min at 4°C. The supernatant was dialyzed overnight at 4°C against 2 liters of 50 mM BisTris (pH 7.1) and was loaded onto a column (2.5 cm [diameter] by 5 cm) of Q Sepharose Fast Flow (Pharmacia Co. Ltd., Uppsala, Sweden) equilibrated in the dialysis buffer. The β-lactamase, which did not bind, was eluted in the unadsorbed fraction and was loaded onto a Superose 12 gel filtration column (Pharmacia) equilibrated in 20 mM Tris buffer (pH 7.6) containing 0.5 mM dithiothreitol and 0.06% sodium azide. Fractions containing activity, which was detected with the chromogenic cephalosporin nitrocefin (36), were obtained after 40 min at a flow rate of 0.3 ml/min. The preparation thus obtained was concentrated and stored at −20°C after addition of an equal volume of glycerol. Purity was assessed by electrophoresis on a sodium dodecyl sulfate (SDS)-12% polyacrylamide gel (24) stained with Coomassie blue R-250 (Sigma Chemicals, St. Louis, Mo.). The enzyme concentration was estimated with a densitometer (Densylab; Bioprobe) with a standard bovine serum albumin scale analyzed under the same conditions used as a reference.
Kinetic measurements.
All kinetic measurements were performed at 30°C in 100 mM sodium phosphate (pH 7.0). The initial rates of hydrolysis were determined spectrophotometrically with a Uvikon 940 spectrophotometer. The following wavelengths and absorption coefficients were used: for benzylpenicillin, 232 nm, Λɛ = 1,100 M−1 cm−1; for ampicillin and ticarcillin, 235 nm, Λɛ = 1,050 M−1 cm−1; for cephalothin, 262 nm, Λɛ = 7,960 M−1 cm−1; for cephaloridine, 255 nm, Λɛ = 9,360 M−1 cm−1; for aztreonam, 318 nm, Λɛ = 640 M−1 cm−1; for cefoxitin, 265 nm, Λɛ = 7,380 M−1 cm−1; for ceftazidime, 260 nm, Λɛ = 8,660 M−1 cm−1; for oxacillin and cloxacillin, 263 nm, Λɛ = 1,000 M−1 cm−1. Kinetic parameters were determined by recording the initial rates at different substrate concentrations and by analyzing the results with the regression analysis program LEONARA written by Cornish-Bowden (7). The kcat and Km values were estimated by using a nonlinear least-squares regression method with dynamic weights (7). The 50% inhibitory concentration (IC50) was determined as the clavulanate concentration that reduced the hydrolysis rate of 100 μM benzylpenicillin by 50% under conditions in which the enzyme was preincubated with various concentrations of inhibitor for 5 min at 30°C before addition of the substrate.
Determination of relative molecular mass.
The relative molecular mass of plasmid pPL10 β-lactamase was estimated by SDS-polyacrylamide gel electrophoresis analysis. Crude extracts and marker proteins were boiled for 10 min in a 1% SDS–3% mercaptoethanol solution and then subjected to electrophoresis on a 12% polyacrylamide gel (200 V, 4 h, at room temperature). Renaturation of β-lactamase activity after denaturing electrophoresis was performed as described previously (31).
DNA sequencing and protein analysis.
Both strands of the 2.5-kb cloned DNA fragment from pPL10 were sequenced while part of pPL11 was sequenced only on one strand, both with an Applied Biosystems sequencer (ABI 311). The nucleotide sequence and the deduced protein sequence were analyzed by using the Genetics Computer Group (GCG) software package (Biotechnology Center, University of Wisconsin—Madison, Madison). Multiple sequence alignment of deduced peptide sequences was carried out with the GCG program Pileup, which uses a simplification of the progressive alignment method of Feng and Doolittle (13). Among the known class D β-lactamases, 13 were compared to OXA-20: OXA-1 and OXA-7 from E. coli (37, 45); OXA-2 and OXA-3 from Salmonella typhimurium (9, 44); OXA-5, OXA-10, OXA-11, OXA-14, OXA-15, OXA-18, and LCR-1 from P. aeruginosa (8, 11, 12, 15, 19, 39); OXA-9 from Klebsiella pneumoniae (51); and OXA-12 (ASB-1) from Aeromonas sobria (40). A dendrogram was derived from the multiple-sequence alignment by a parsimony method using the phylogeny package PAUP (Phylogenetic Analysis Using Parsimony) version 3.0 (49).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the GenBank nucleotide database under accession no. AF024602.
RESULTS
Preliminary hybridizations and cloning of the restricted-spectrum β-lactamase gene.
Preliminary hybridization experiments indicated that P. aeruginosa Mus did not harbor any known class A β-lactamase resistance gene (39). blaTEM-1, blaSHV-3, and blaPER-1 probes failed to provide positive hybridization signals.
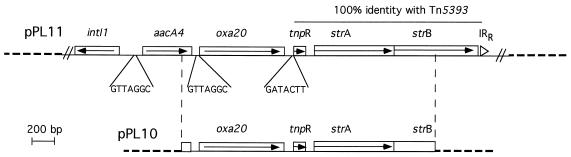
Total DNA from P. aeruginosa Mus was partially digested with restriction endonuclease Sau3AI and ligated to BamHI-digested plasmid pBK-CMV. The ligation product was transformed into E. coli JM109 by electroporation. Recombinant strains were selected onto Trypticase soy agar plates containing either amoxicillin (50 μg/ml) or kanamycin (30 μg/ml). Several recombinant colonies expressing a high level of amoxicillin, cephalothin, and ticarcillin resistance that was inhibited by clavulanic acid were obtained. The recombinant plasmids expressing the restricted-spectrum β-lactamase resistance phenotype were extracted and analyzed. The insert sizes ranged from 2.5 to 20 kb. A detailed restriction map was generated for the plasmid containing a 2.5-kb insert (pPL10) (Fig. 1). To determine whether the novel β-lactamase was related to oxa18, hybridization experiments were performed. No cross hybridization was observed. A larger plasmid, pPL11, was used to sequence the genetic environment of oxa20 (Fig. 1).
FIG. 1.
Schematic map of the recombinant plasmids pPL10 and pPL11, which code for OXA-20, the pI 7.4 restricted-spectrum β-lactamase from the P. aeruginosa Mus clinical strain. The thin solid line represents the cloned inserts from P. aeruginosa Mus, while the dotted lines indicate the vector pBK-CMV. The open boxes represent the genes, and the arrows indicate their translational orientations. The recombinational hot spot sequences or composite core sites (GTTRRRY) are also indicated. The designation of the gene names is referenced in the text. IRR, right inverted repeat.
Plasmid DNA and transfer of resistance.
No plasmid was detected in P. aeruginosa Mus. Direct mating-out experiments failed to transfer the β-lactam resistance marker into P. aeruginosa PU21, E. coli JM109, or A. hydrophila 76-14. Moreover, attempts to transform P. aeruginosa total DNA into E. coli JM109 by electroporation failed to give any β-lactam-resistant E. coli. The oxa20 gene thus seems to be chromosomally located.
Antibiotic susceptibility.
The MICs of β-lactams revealed high levels of resistance of P. aeruginosa Mus to amino-, carboxy-, and ureido-penicillins and to restricted- and extended-spectrum cephalosporins (Table 2). MICs of β-lactams for E. coli JM109 harboring recombinant plasmid pPL10 demonstrated resistance mainly to penicillins. MICs of aztreonam, ceftazidime, moxalactam, cefoxitin, and imipenem for E. coli JM109(pPL10) were unchanged relative to those for E. coli JM109 alone. Resistance to aztreonam, ceftazidime, and cefepime was therefore due to OXA-18 and/or the presumed cephalosporinase in the P. aeruginosa Mus clinical isolate. All β-lactam MICs, except those of moxalactam and imipenem, were decreased in the presence of clavulanic acid (2 μg/ml).
TABLE 2.
MICs of β-lactams for P. aeruginosa Mus, E. coli JM109 harboring recombinant plasmid pPL10, and reference strain E. coli JM109
| Antibiotic(s) | MIC (μg/ml) for:
|
||
|---|---|---|---|
| P. aeruginosa Musa | E. coli JM109(pPL10)b | E. coli JM109 | |
| Amoxicillin | >512 | 512 | 2 |
| Amoxicillin-Clac | >512 | 32 | 2 |
| Ticarcillin | 256 | 512 | 2 |
| Ticarcillin-Cla | 64 | 32 | 1 |
| Piperacillin | 64 | 4 | 1 |
| Piperacillin-Cla | 32 | 1 | 0.5 |
| Cephalothin | >1,024 | 32 | 4 |
| Cephalothin-Cla | >512 | 8 | 2 |
| Cefamandole | >512 | 8 | 1 |
| Cefamandole-Cla | >512 | 2 | 1 |
| Cefoxitin | >1,024 | 16 | 8 |
| Cefoxitin-Cla | >512 | 8 | 8 |
| Ceftazidime | 128 | 0.5 | 0.25 |
| Ceftazidime-Cla | 8 | 0.5 | 0.25 |
| Cefotaxime | 128 | 0.06 | 0.06 |
| Cefotaxime-Cla | 8 | 0.06 | 0.06 |
| Cefepime | 16 | 0.12 | 0.06 |
| Cefepime-Cla | 4 | 0.03 | 0.06 |
| Cefpirome | 32 | 0.5 | 0.12 |
| Cefpirome-Cla | 8 | 0.06 | 0.06 |
| Moxalactam | 64 | 0.25 | 0.25 |
| Moxalactam-Cla | 64 | 0.12 | 0.12 |
| Aztreonam | 256 | 0.25 | 0.12 |
| Aztreonam-Cla | 16 | 0.12 | 0.06 |
| Imipenem | 8 | 0.25 | 0.06 |
| Imipenem-Cla | 8 | 0.25 | 0.06 |
P. aeruginosa Mus produces the restricted-spectrum β-lactamase OXA-20 along with an extended spectrum β-lactamase OXA-18 and an undetermined cephalosporinase.
E. coli JM109 harboring recombinant plasmid pPL10 produced the studied β-lactamase.
Cla, clavulanic acid at a fixed concentration of 2 μg/ml.
Properties of the OXA-20 β-lactamase.
Analytical isoelectric focusing revealed that P. aeruginosa Mus had three distinct β-lactamase activities of pI 5.5, 7.4, and 8.2. E. coli JM109 harboring the recombinant plasmid pPL10 had only one β-lactamase activity, of pI 7.4 (data not shown). The relative molecular mass of the cloned mature β-lactamase from E. coli JM109 harboring pPL10 was estimated to be 29 kDa (data not shown).
Determination of the kinetic parameters by using a purified OXA-20 preparation showed that this β-lactamase had good activity against penicillin G, ampicillin, and cephalothin, an early cephalosporin (kcat/Km values of 2.4 to 5.9 μM−1 · s−1) (Table 3). The hydrolytic activity of OXA-20 against oxacillin and cephaloridine was significant but lower than that against the three previous drugs (kcat/Km of 0.3 μM−1 · s−1) (Table 3). Cloxacillin, ticarcillin, and aztreonam were characterized by the lowest hydrolysis activities (kcat/Km values lower than 0.09 μM−1 · s−1) (Table 3). Hydrolysis of expanded-spectrum cephalosporins was not measurable. The IC50 of clavulanic acid was 2.2 μM.
TABLE 3.
Steady-state kinetic parameters of OXA-20 β-lactamasea
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (μM−1 · s−1) |
|---|---|---|---|
| Oxacillin | 116 ± 5 | 329 ± 34 | 0.35 ± 0.020 |
| Cloxacillin | 70 ± 7 | 788 ± 114 | 0.088 ± 0.005 |
| Penicillin Gb | 26 ± 0.90 | 4.4 ± 1.30 | 5.9 ± 1.70 |
| Ampicillin | 80 ± 1.40 | 33 ± 2.20 | 2.4 ± 0.13 |
| Ticarcillin | 4 ± 0.45 | 147 ± 34 | 0.03 ± 0.004 |
| Cephalothin | 13 ± 0.05 | 5 ± 0.20 | 2.6 ± 0.08 |
| Cephaloridine | 20 ± 0.90 | 69 ± 5.00 | 0.3 ± 0.009 |
| Aztreonam | 6 ± 0.50 | 69 ± 12.00 | 0.09 ± 0.009 |
| Ceftazidime | NDc | ND | ND |
| Cefoxitin | ND | ND | ND |
Values are means ± standard errors.
The IC50 for penicillin inhibition by clavulanate was 2.2 ± 0.25 μM.
ND, not determinable (the initial rate was lower than 0.001 μM−1 · s−1).
Sequence analysis of the OXA-20 β-lactamase gene.
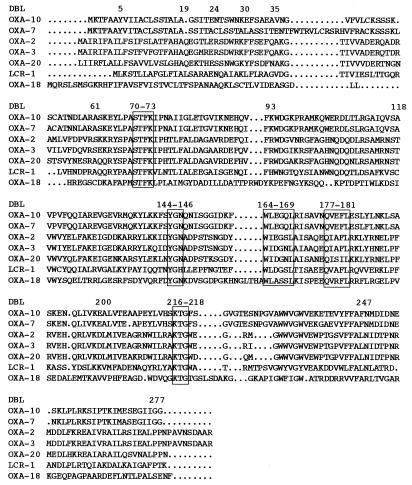
Both strands of the 2.5-kb cloned DNA fragment were sequenced entirely. Analysis of this insert for coding regions revealed a sufficiently large open reading frame (ORF) of 798 bp encoding a 266-amino-acid preprotein approximately 30.6 kDa in size. The DNA sequence of this gene, along with flanking sequences, is shown in Fig. 2. Within the corresponding protein, a serine-threonine-phenylalanine-lysine (S-T-F-K) tetrad was found at positions 70 to 73 (Fig. 3); it included the conserved serine and lysine amino acid residues characteristic of β-lactamases possessing a serine active site (22) or penicillin-binding proteins (21). Four structural elements characteristic of class D β-lactamases were found: Y-G-N at positions 144 to 146, W-L-E-G-S-L at positions 164 to 169, Q-X-X-I-L at positions 177 to 181, and K-T-G at positions 216 to 218 (Fig. 3).
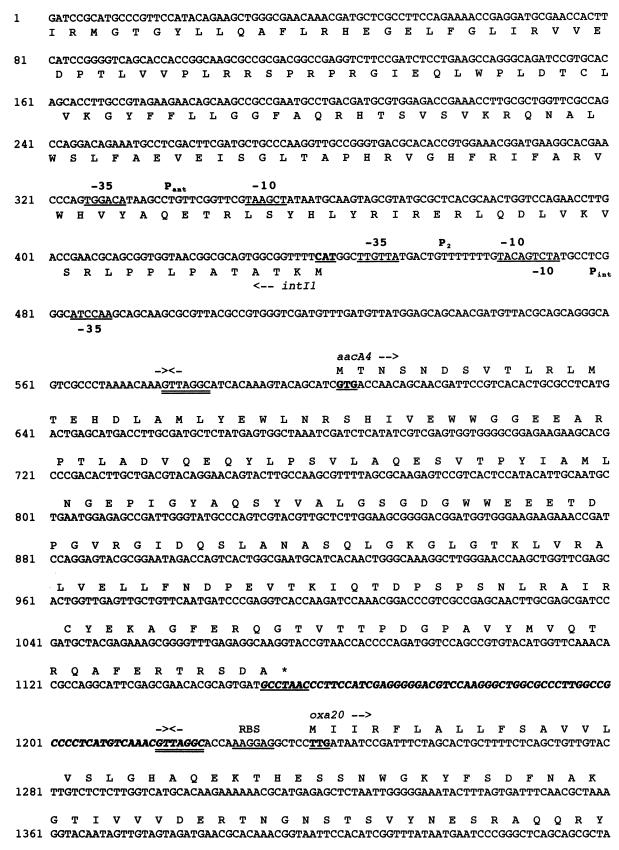
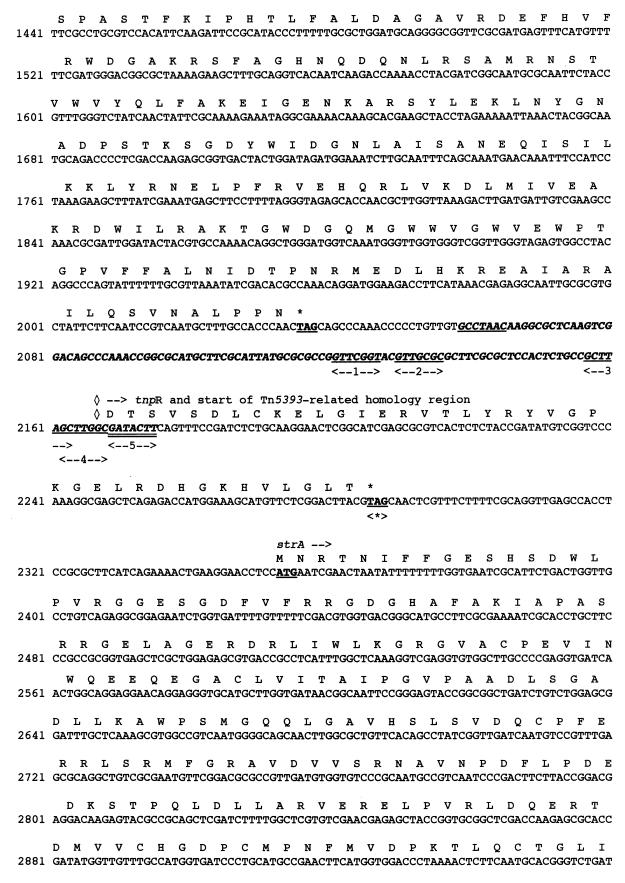
FIG. 2.
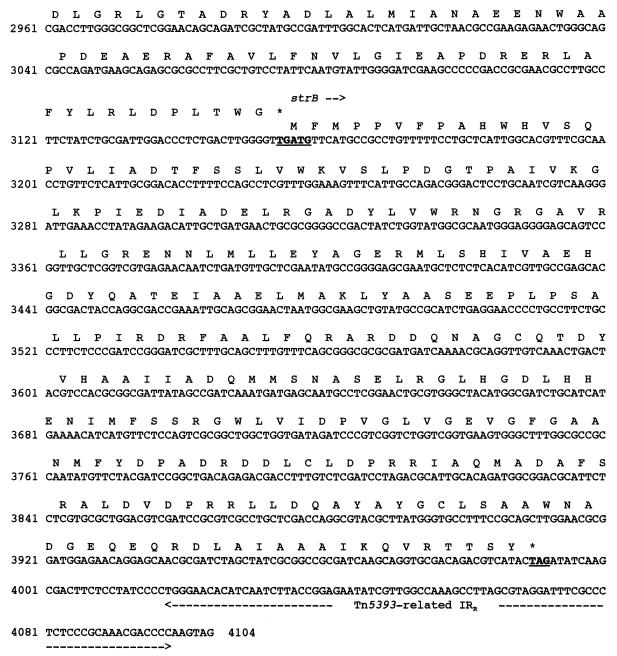
Nucleotide sequence of a 4,104-bp fragment of pPL11 containing the β-lactamase coding region. The deduced amino acid sequence is designated in single-letter code below the nucleotide sequence. The recombinational hot spot sequences or composite core sites (GTTRRRY) are double underlined. Facing arrows indicate putative integrase cleavage sites within the composite core sites. The five putative 3′ ends of the oxa20 59-BE are indicated by single underlining associated with a number between two arrows. The 59-BEs are in boldface italic type. The start and stop codons of the various genes are boldfaced and underlined. Gene names, followed by an arrow indicating their translational orientation, are indicated below their initiation codons. The positions and designations of the ORFs are as follows: intI1, at positions 441 to 1, codes for the integrase of the integron; aacA4, at positions 659 to 1156, codes for AAC(6′)-Ib; oxa20, at positions 1238 to 2038, codes for OXA-20, the restricted-spectrum class D β-lactamase; tnpR, at positions 2169 to 2288, codes for the 3′ end of the resolvase of Tn5393; strA, at positions 2353 to 3156, codes for the streptomycin resistance gene A; and strB, at positions 3156 to 3992, codes for the streptomycin resistance gene B. The −35 and −10 sequences of the Pant, P2, and Pint promoters are underlined. The Tn5393-related right inverted repeat (IRR) is indicated by broken underlining. RBS, ribosome binding site of oxa20; <∗>, IS1133 insertion site in Tn5393 (4); ◊, identity breakpoint with Tn5393-related sequence.
FIG. 3.
Alignment of the amino acid sequence of OXA-20 with those of six class D β-lactamases representing each branch of the phylogenetic tree displayed in Fig. 4. Dots indicate gaps within the alignment. Standard numbering for class D enzymes is indicated by the label DBL (44). The boxes indicate conserved regions within the OXA family of β-lactamases. The enzymes included in the alignment are detailed in Materials and Methods.
The overall GC content of this gene was 45%, which does not lie within the expected range (60.1 to 69.5%) for P. aeruginosa genes (except for pilin genes) (42). The GC content value is more typical of Enterobacteriaceae. The translation stop codon (TAG), found at positions 2036 to 2038, corresponded to the one most commonly used in P. aeruginosa and enterobacterial genes. The initiation codon of that enzyme is, however, peculiar since it starts with TTG (coding for leucine), which is only rarely used as an initiation codon in bacteria (28).
Homology with other β-lactamases.
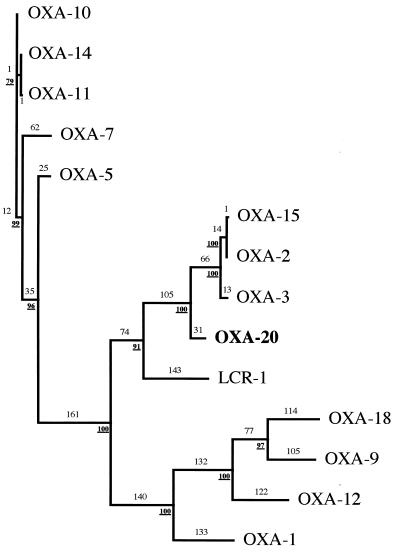
The peptide sequence deduced from the OXA-20 β-lactamase structural gene has less than 20% amino acid identity with the sequences of OXA-5, OXA-7, OXA-9, OXA-10 (PSE-2), OXA-11, and OXA-12. OXA-2, OXA-3, and OXA-15 were the three oxacillin-hydrolyzing β-lactamases with the highest levels identity (75% identity for each), while LCR-1 has 36% amino acid identity. OXA-18, which is present in the same bacterial strain (39), has only 16% amino acid identity. The enzyme is a novel class D β-lactamase and thus was named OXA-20. A dendrogram was constructed to relate OXA-20 to 13 other class D β-lactamases (Fig. 4). OXA-20 was most highly related to OXA-2, OXA-15, OXA-3, and, to a lesser extent, LCR-1.
FIG. 4.
Dendrogram obtained for 14 class D β-lactamases according to parsimony (48). Branch lengths (indicated in lightface numbers) are to scale and proportional to the number of amino acid changes. The values at branching points (numbers in boldface type) refer to the number of times a particular node was found in 100 bootstrap replications. The distance along the vertical axis has no significance.
Analysis of the genetic environment of oxa20.
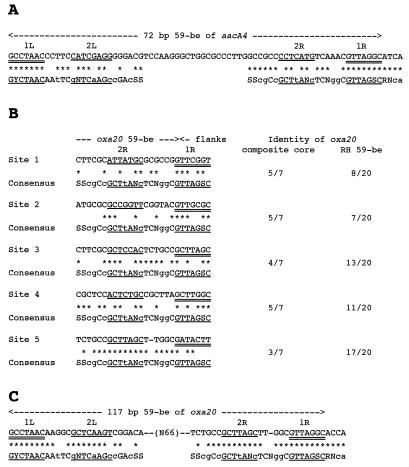
Sequence analysis of the 2.5-kb segment described above strongly suggested that oxa20 is a gene cassette located on an integron. Exploration of the genetic environment of oxa20 revealed the presence of the following integron features: (i) a second antibiotic resistance gene, aacA4 [coding for AAC(6′)-Ib]; (ii) an intI gene (coding for an integrase) associated with an attI recombinant site; and (iii) two 59-BEs, each associated with an antibiotic resistance gene (Fig. 1 and 2). oxa20 is associated with gene cassette-specific sequences such as a core site GTTRRRY and an inverse core site RYYYAAC (Fig. 2). The oxa20 gene cassette starts at position 1217 with the 5′ sequence TTAGGC, while the cassette could end at five potential G’s (Fig. 2 and 5B). Because the sequence immediately following the oxa20 cassette is not identifiable as a cassette, it is difficult to precisely locate the end of the cassette unless the consensus sequence is used (47). There are five potential GTTRRRY sequences (Fig. 2 and 5, sites 1 to 5). However, the sequence that best conforms to the RH 59-BE consensus is site 5 (GATACTT). The cassette seems to end at the G at base 2169 and the 59-BE is 117 bp long. The nucleotide sequence upstream of oxa20 revealed an ORF which was identical to the 6′-N-aminoglycoside acetyltransferase gene, aacA4, which is found primarily in P. aeruginosa (14). The nucleotide sequence of this gene along with the deduced amino acid sequence are shown in Fig. 2. The product of aacA4, AAC(6′)-Ib, is a 185-amino-acid protein that confers resistance to tobramycin and amikacin and presents features corresponding to a gene cassette with perfect recombination hot spots and a 72-bp 59-BE.
FIG. 5.
Structure of 59-BEs. The inverse core and core sequences are double underlined. L1, L2, R1, and R2 are four regions found to be highly conserved within class 1 59-BEs (47). (A) Sequence comparison of the 72-bp aacA4 59-BE present in the circular form of the aacA4 gene cassette with a consensus sequence (47). (B) Comparison of the 3′ ends of five putative oxa20 59-BE sequences and the consensus sequence. Identity values are given as the number of shared nucleotides per total number of nucleotides. The composite core site represents the 7 nucleotides of the sequence GTTRRRY, while the RH 59-BE represents the 3′ end of oxa20 59-BE (until the conserved G of the putative composite core site). (C) Comparison of the sequence of the 117-bp oxa20 59-BE present in the circular form of the oxa20 gene cassette with a consensus sequence (47). Consensus bases in uppercase letters are present in two-thirds or more of the 59-BEs and bases in lowercase letters are present in half or more of the 59-BEs. S, C or G.
This oxa20-containing integron is peculiar since it lacks the 3′ conserved region (Fig. 6) and therefore is not a classical sul1-associated integron like most integrons are. Analysis of the sequence of the integrase gene revealed 97.3% sequence identity with the integrase intI1 of class 1 integrons such as In1 (17). At 216 bp upstream of the first recombination hot spot, GTTAGGC, promoter −35 (TGGACA) and −10 (TAAGCT) sequences were found. The spacing between the two sequences was 17 bp. This promoter, Pant (Fig. 2), which is located within the integrase gene and which is responsible for the expression of the genes located in the integron (6), was shown to be a weak one (6). A potential secondary promoter, P2, was found, but this promoter is likely to be inactive since the spacing between the −35 (TTGTTA) and the −10 (TACAGT) sequences corresponded to only 14 bp (6). P2 has the potential to become a strong promoter by insertion of 3 bp into the spacer region (6). Besides P2, no obvious secondary promoter sequence was found.
Analysis of the genetic environment of the oxa20-containing integron.
The integron containing oxa20 seems to be located on a class II gram-negative transposon related to Tn5393 (4). Indeed, downstream of the oxa20 gene cassette, 117 bp of the 3′ end of tnpR, the resolvase gene of transposon Tn5393 was found. Two streptomycin resistance genes, strA and strB, were identified on the basis of their DNA sequence homology. In Tn5393 of Erwinia amylovora, strA is separated from tnpR by a 1.2-kb insertion sequence designated IS1133. Since this insertion sequence is absent in the transposon found in P. aeruginosa Mus (Fig. 2), we suggest that this transposon be called Tn5393-related.
DISCUSSION
The starting point of this work was the observation that a P. aeruginosa clinical isolate presented an extended-spectrum resistance phenotype with a marked synergistic effect between clavulanic acid and ceftazidime or aztreonam. In the course of cloning the gene of the extended-spectrum β-lactamase, a second enzyme, with a pI of 7.4, was identified. Analysis of the nucleotide sequence of oxa20 demonstrated that the deduced protein sequence had homology to the sequences of Ambler class D β-lactamases (1) (class 2d by the Bush et al. classification [3]). Several interesting features emerged from the analysis of the nucleotide and the deduced amino acid sequences. (i) Analysis of the GC content (45%) and codon usage suggested that oxa20 very likely has an enterobacterial origin. In contrast, oxa18, the other oxacillinase gene present in the same strain, had a GC content of 61%, which is typical of a P. aeruginosa origin (39). (ii) oxa20 lacks an efficient translational initiation codon. The codon found, TTG, is considered in E. coli to be at least eight times less efficient in initiation than the ATG (methionine) codon (28). (iii) A potential promoter which fits E. coli or P. aeruginosa promoter consensus sequences (42) was not identified 5′ to the coding sequence. (iv) Protein sequence alignment with sequences of other class D β-lactamases showed that OXA-20 has the highest level of sequence identity with OXA-2 (75%), OXA-3 (75%), and OXA-15 (75%) (9, 12, 44). OXA-20 is therefore not a simple point-mutant derivative from any known oxacillinase nor is it related to OXA-18.
OXA-20 confers high-level resistance to amoxicillin, ticarcillin, and piperacillin but not to cefotaxime, ceftazidime, or aztreonam. Resistance to penicillin is partially reversed by clavulanic acid. Such inhibition was observed previously for other oxacillin-hydrolyzing enzymes such as OXA-12 (40), a restricted-spectrum oxacillinase from A. sobria, and the extended-spectrum OXA-18 enzyme from the same P. aeruginosa isolate (39). These results correlate with the hydrolytic properties of OXA-20, which is a typical class D β-lactamase that hydrolyzes oxacillin and cloxacillin faster than benzylpenicillin, like OXA-2 and OXA-3 (25, 26).
In the clinical strain P. aeruginosa Mus, no plasmid was found; therefore, it is likely that OXA-20 is chromosomally mediated. Most of the oxacillin- and carbenicillin-hydrolyzing β-lactamase genes isolated from P. aeruginosa have been described as part of transposons (27, 33, 52), and all of the oxacillinase genes identified so far, except oxa12, oxa18, and the gene for LCR-1, are located on the variable region of class 1 integrons (41). The structure of class 1 integrons, the most abundant and naturally occurring integrons, can be described as follows (Fig. 6A): the resistance genes are surrounded by two conserved regions called 5′CS, encoding the integrase, and a conserved region, 3′CS, coding for sulfonamide resistance. The 5′CS region was found on cloned fragments from the clinical isolate P. aeruginosa Mus, but the 3′CS region was not. The inserted genes are flanked at their 5′ end by the core motif GTTRRRY and at their 3′ end by an inverse core motif, RYYYAAC, which is part of the 59-BE (5). These sequences are present around oxa20 (Fig. 1 and 2). Therefore, oxa20 is a gene cassette located on an integron, like most oxacillinase genes. The 59-BEs constitute a loosely related family of imperfect inverted repeats which differ from each other in their sequence and length (6). However, the two outermost stretches of sequence conform to consensus sequences (47). The aacA4 gene cassette has a 72-bp 59-BE that conforms well to the consensus sequence (Fig. 5A) (47). This gene, which codes for amikacin and tobramycin resistance, is often cassette associated and frequently found on integrons (41). For oxa20, a 117-bp 59-BE which conforms well to the consensus sequence is found (Fig. 5C) (47). The composite core site 3′ of oxa20, GATACTT, is different from the consensus core site, GTTRRRY. A GTTRRRY-to-GATRRRY mutation leads to a 98% drop in integration activity (47). Therefore, this oxa20 composite core site, GATACTT, may represent a rather inefficient recombination site. In addition, this site is located within the beginning of the tnpR homology region and could thus be an explanation for why the 3′CS and part of the Tn5393-related transposon were not found. An IntI1-mediated deletion or cointegration event caused by recombination between the oxa20 59-BE and a secondary site in the Tn5393-related transposon could lead to the structure found. Indeed, a cointegration of this type could lead to incorporation of a plasmid containing the integron into the chromosome, if the Tn5393-related transposon were already there.
Tn5393 was first identified in E. amylovora, Erwinia herbicola, and Pseudomonas syringae pr. papulans and then in many other gram-negative bacteria. The reason for the spread of this transposon can be attributed to the presence of strA and strB, two genes responsible for streptomycin resistance. The finding that bacteria from plants have the streptomycin resistance genes found in bacteria from human and veterinary isolates extends the importance of this resistance determinant. One major difference between the Tn5393 found in E. amylovora and that in P. aeruginosa Mus is still observed. Indeed, the Tn5393-related transposon from P. aeruginosa Mus lacks an insertion sequence, IS1133, which is located between strA and tnpR in a regular Tn5393 transposon (4) and is likely unable to transpose since tnpR is interrupted by the integron.
Several factors may influence the level of expression of a particular antibiotic resistance gene located on an integron. The most obvious are transcription and translation initiation signals (the efficiency of which can readily be altered by mutation) or the copy number (plasmid or chromosomal origin). oxa20 is most likely chromosomally encoded, and therefore its copy number is low. The translational initiation of oxa20 corresponds to a leucine, which is the least efficient initiation codon in E. coli (28). The ribosomal binding site of oxa20 is, however, close to consensus (28). Expression of resistance genes in the inserted cassettes of integrons is dependent on the transcription signals and on the position of the inserted cassette within the insert region. The main promoter, Pant, (−35 [TGGACA] and −10 [TAAGCT]), which is responsible for the expression of the genes in the insert region (Fig. 2), is 20-fold less active than the −35 (TTGACA) and −10 (TAAACT) promoter found in integrons such as In4 and in In6 (6). The weak promoter, Pant, can be found alone in naturally occurring integrons, but it can also be found together with a secondary downstream promoter named P2 (Fig. 2), which may compensate for the low activity of the weak Pant. In the OXA-20 integron, P2 is likely inactive because the −35 and the −10 sequences are separated by only 14 bp. The three G’s usually inserted between the preexisting −35 and −10 boxes in some naturally occurring integrons are missing here (6). The level of antibiotic resistance by any particular gene cassette was shown to be the highest when the gene was in the cassette located closest to Pant and was reduced when the cassette was situated downstream of one or more cassettes (6). oxa20 is the second gene, and therefore a lower level of expression relative to that of aacA4 is expected. Overall, it seems that the expression of oxa20 is likely to be rather poor based on transcription and translation initiation signals and on position within the insert unit.
P. aeruginosa Mus produces two novel and unrelated oxacillinases, OXA-18 and OXA-20, both of which are inhibited by clavulanic acid. This work gives further insight on the genomic plasticity of P. aeruginosa and provides another example of the biological variability of integrons and of antibiotic resistance genes.
ACKNOWLEDGMENTS
We thank V. Jarlier, in whose laboratory part of this work was performed, L. N. Philippon for valuable advice, and J. C. Petit, in whose laboratory the P. aeruginosa Mus strain was originally isolated.
This work was financed by a grant from the Faculté de Médecine, Paris-Sud, Le Kremlin-Bicêtre, France, and by the Institut National de la Santé et de la Recherche Médicale (grant CRI950601).
REFERENCES
- 1.Ambler R P. The structure of β-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiou C S, Jones A L. Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other gram-negative bacteria. J Bacteriol. 1993;175:732–740. doi: 10.1128/jb.175.3.732-740.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collis C M, Hall R M. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol. 1992;174:1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collis C M, Hall R M. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–162. doi: 10.1128/aac.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornish-Bowden A. Fundamentals of enzyme kinetics. Seattle, Wash: Portland Press, Inc.; 1995. Graphs of the Michaelis-Menten equation; pp. 30–37. [Google Scholar]
- 8.Couture F, Lachapelle J, Levesque R C. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol Microbiol. 1992;6:1693–1705. doi: 10.1111/j.1365-2958.1992.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 9.Dale J W, Godwin D, Mossakowska D, Stephenson P, Wall W. Sequence of the OXA-2 β-lactamase: comparison with other penicillin-reactive enzymes. FEBS Lett. 1985;191:39–44. doi: 10.1016/0014-5793(85)80989-3. [DOI] [PubMed] [Google Scholar]
- 10.Danel F, Hall L M C, Gur D, Akalin H E, Livermore D M. Transferable production of PER-1 β-lactamase in Pseudomonas aeruginosa. J Antimicrob Chemother. 1995;35:281–294. doi: 10.1093/jac/35.2.281. [DOI] [PubMed] [Google Scholar]
- 11.Danel F, Hall L M C, Gur D, Livermore D M. OXA-14, another extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1881–1884. doi: 10.1128/aac.39.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danel F, Hall L M C, Gur D, Livermore D M. OXA-15, an extended-spectrum variant of OXA-2 β-lactamase isolated from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:785–790. doi: 10.1128/aac.41.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng D F, Doolittle R F. Progressive alignment and phylogenetic tree construction of protein sequences. Methods Enzymol. 1990;183:375–387. doi: 10.1016/0076-6879(90)83025-5. [DOI] [PubMed] [Google Scholar]
- 14.Galimand M, Lambert T, Gerbaud G, Courvalin P. Characterization of the aac(6′)-Ib gene encoding an amynoglycoside 6′-N-acetyltransferase in Pseudomonas aeruginosa BM2656. Antimicrob Agents Chemother. 1993;37:1456–1462. doi: 10.1128/aac.37.7.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall L M C, Livermore D M, Gur D, Akova M, Akalin H E. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:1637–1644. doi: 10.1128/aac.37.8.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-bp element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 17.Hall R M, Vockler C. The region of the IncN plasmid R46 coding for resistance to β-lactam antibiotics, streptomycin, spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res. 1987;15:7491–7501. doi: 10.1093/nar/15.18.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen J B, Olsen R H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978;135:227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huovinen P, Huovinen S, Jacoby G A. Sequence of PSE-2 β-lactamase. Antimicrob Agents Chemother. 1988;32:134–136. doi: 10.1128/aac.32.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacoby G A. Properties of R plasmids determining gentamicin resistance by acetylation in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1974;6:239–252. doi: 10.1128/aac.6.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joris B, Ghuysen J M, Dive G, Renard A, Dideberg O, Charlier P, Frère J M, Kelly J A, Boyington J C, Moews P C, Knox J R. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem J. 1988;250:313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joris B, Ledent P, Dideberg O, Fonzé E, Lamotte-Brasseur J, Kelly J A, Ghuysen J M, Frère J M. Comparison of the sequences of class A β-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991;35:2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Ledent P, Raquet X, Joris B, Van Beeumen J, Frère J M. A comparative study of class-D β-lactamases. Biochem J. 1993;292:555–562. doi: 10.1042/bj2920555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ledent P, Frère J M. Substrate induced inactivation of the OXA-2 beta-lactamase. Biochem J. 1993;295:871–878. doi: 10.1042/bj2950871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levesque R C, Jacoby G A. Molecular structure and interrelationships of multiresistance β-lactamase transposons. Plasmid. 1988;19:21–29. doi: 10.1016/0147-619x(88)90059-5. [DOI] [PubMed] [Google Scholar]
- 28.Lewin B. Genes V. Oxford, United Kingdom: Oxford University Press; 1994. [Google Scholar]
- 29.Liu P Y F, Gur D, Hall L M C, Livermore D M. Survey of the prevalence of β-lactamases amongst 1000 Gram-negative bacilli isolated consecutively at the Royal London Hospital. J Antimicrob Chemother. 1992;30:429–447. doi: 10.1093/jac/30.4.429. [DOI] [PubMed] [Google Scholar]
- 30.Martinez E, De La Cruz F. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 1990;9:1275–1281. doi: 10.1002/j.1460-2075.1990.tb08236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massida O, Rossolini G M, Satta G. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-β-lactamases. J Bacteriol. 1991;173:4611–4617. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthew M. Plasmid-mediated β-lactamases of gram-negative bacteria: properties and distribution. J Antimicrob Chemother. 1979;5:349–358. doi: 10.1093/jac/5.4.349. [DOI] [PubMed] [Google Scholar]
- 33.Mossakowska D, Ali N A, Dale J W. Oxacillin-hydrolysing β-lactamases. A comparative analysis at nucleotide and amino-acid sequence levels. Eur J Biochem. 1989;180:309–318. doi: 10.1111/j.1432-1033.1989.tb14649.x. [DOI] [PubMed] [Google Scholar]
- 34.Nicolas M H, Jarlier V, Honore N, Philippon A, Cole S T. Molecular characterization of the gene encoding SHV-3 β-lactamase responsible for transferable cefotaxime resistance in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1989;33:2096–2100. doi: 10.1128/aac.33.12.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nordmann P, Naas T. Sequence analysis of PER-1 extended-spectrum β-lactamase from Pseudomonas aeruginosa and comparison with class A β-lactamases. Antimicrob Agents Chemother. 1994;38:104–114. doi: 10.1128/aac.38.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Callaghan C H, Morris A, Kirby S M, Shingler A M. Novel method for the detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouellette M, Bissonnette L, Roy P H. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 β-lactamase gene. Proc Natl Acad Sci USA. 1987;84:7378–7382. doi: 10.1073/pnas.84.21.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philippon A, Thabaut A, Nevot P. Pseudomonas aeruginosa et β-lactamines. In: Courvalin P, Goldstein F, Philippon A, Sirot J, editors. L’Antibiogramme. Paris, France: MPC-Vidéom; 1985. pp. 103–110. [Google Scholar]
- 39.Philippon L N, Naas T, Bouthors A-T, Barakett V, Nordmann P. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2188–2195. doi: 10.1128/aac.41.10.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasmussen B A, Keeney D, Yang Y, Bush K. Cloning and expression of a cloxacillin-hydrolyzing enzyme and cephalosporinase from Aeromonas sobria AER 14M in Escherichia coli: requirement for an E. coli chromosomal mutation for efficient expression of the class D enzyme. Antimicrob Agents Chemother. 1994;38:2078–2085. doi: 10.1128/aac.38.9.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Recchia G D, Hall R M. Gene cassettes: a new class of mobile elements. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 42.Ronald S, Farinha M A, Allan B J, Kropinski A M. Cloning and physical mapping of transcriptional regulatory (sigma) factors from Pseudomonas aeruginosa. In: Galli E, Silver S, Witholt B, editors. Pseudomonas: molecular biology and biotechnology. Washington, D.C: American Society for Microbiology; 1992. pp. 249–257. [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sanschagrin F, Couture F, Levesque R C. Primary structure of OXA-3 and phylogeny of oxacillin-hydrolyzing class D β-lactamases. Antimicrob Agents Chemother. 1995;39:887–893. doi: 10.1128/aac.39.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scoulica E, Aransay A, Tselentis Y. Molecular characterization of the OXA-7 β-lactamase gene. Antimicrob Agents Chemother. 1995;39:1379–1382. doi: 10.1128/aac.39.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stokes H W, Hall R M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 47.Stokes H W, O’Gorman D B, Recchia G D, Parsekhian M, Hall R M. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 48.Sutcliffe J G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci USA. 1978;75:3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swofford D L. PAUP (version 3.0): phylogenetic analysis using parsimony. Champaign, Ill: Illinois Natural History Survey; 1989. [Google Scholar]
- 50.Takahashi S, Nagano Y. Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J Clin Microbiol. 1984;20:608–613. doi: 10.1128/jcm.20.4.608-613.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tolmasky M E, Crosa J H. Genetic organization of antibiotic resistance genes (aac(6′)-Ib, aadA, and oxa-9) in the multiresistance transposon Tn1331. Plasmid. 1993;29:31–40. doi: 10.1006/plas.1993.1004. [DOI] [PubMed] [Google Scholar]
- 52.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]