Abstract
Post-translational modifications (PTMs) provide a rapid response to stimuli, finely tuning metabolism and gene expression and maintain homeostasis. Advances in mass spectrometry over the past two decades have significantly expanded the list of known PTMs in biology and as instrumentation continues to improve, this list will surely grow. While many PTMs have been studied in detail (e.g. phosphorylation, acetylation), the vast majority lack defined mechanisms for their regulation and impact on cell fate. In this review, we will highlight the field of PTM research as it currently stands, discussing the mechanisms that dictate site specificity, analytical methods for their detection and study, and the chemical tools that can be leveraged to define PTM regulation. In addition, we will highlight the approaches needed to discover and validate novel PTMs. Lastly, this review will provide a starting point for those interested in PTM biology, providing a comprehensive list of PTMs and what is known regarding their regulation and metabolic origins.
Keywords: acetylation/deacetylation, acylation, glycation, mass spectrometry, methylglyoxal, post-translational modification
Introduction
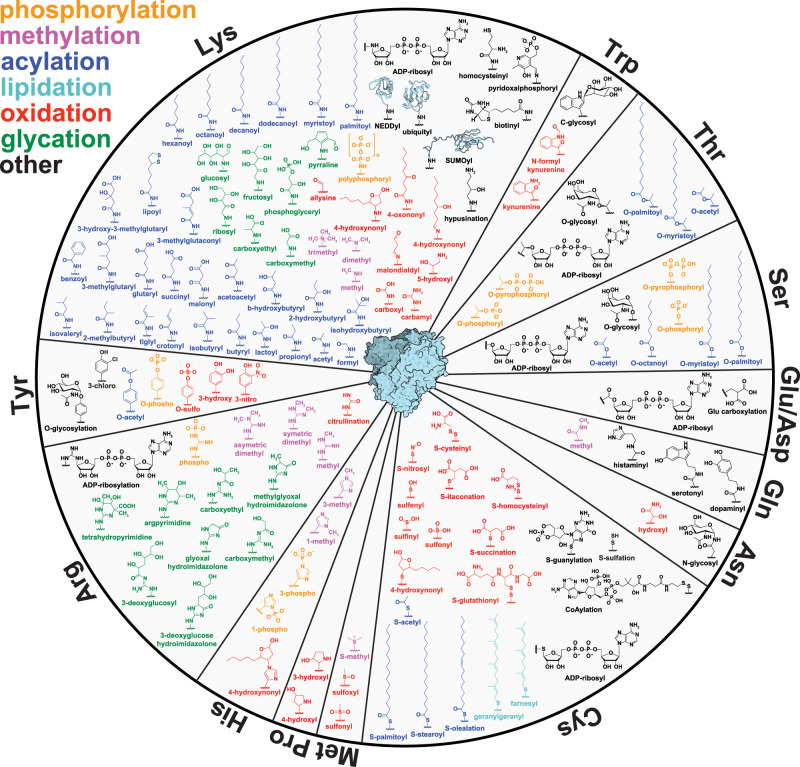
The ability of cells to rapidly detect and respond to stimuli is critical to maintaining homeostasis [1,2]. As protein synthesis is an energy demanding and slow process, altering protein function through the addition and removal of small molecules to target proteins provides a rapid response to restore homeostasis. This integration of post-translational modifications (PTMs) significantly expands the functional proteome, which estimations suggest to be >1 000 000 proteoforms from ∼20 000 genes [3]. PTMs are often derived from primary or secondary metabolic intermediates, serving as sensors for cellular nutrient status and often showing conservation across all domains of life [4–6]. The list of known PTMs is expansive, varying in target amino acid, chemical composition, and metabolic source (Figure 1 and Table 1) [6]. While some PTMs are well-characterized (e.g. Ser/Thr phosphorylation), many remain scantly investigated and their role in health and disease is largely unknown (e.g. His methylation).
Figure 1. A comprehensive view of the PTM landscape in the human proteome.
Table 1. PTMs found in the human proteome.
| PTM Class | Analyte | Variable Mass | Metabolic Precursor | Writer | Eraser | Reference |
|---|---|---|---|---|---|---|
| Methylation | methyl | Arg + 14.0157 | S-adenosylmethionine | Type I, II, III PRMTs | KDM4E, KDM5C | [148-153] |
| Gln + 14.0157 | S-adenosylmethionine | Fibrillarin | Unknown | [154] | ||
| Lys + 14.0157 | S-adenosylmethionine | KMTs | KDMs, JMJDs, LSD1 | [155] | ||
| Met + 14.0157 | S-adenosylmethionine | SETD3 | Unknown | [156] | ||
| asymmetricdimethyl | Arg + 28.0314 | S-adenosylmethionine | Type I PRMT | KDM3A1, KDM4A1, KDM6B1, KDM5C1 | [148-153] | |
| symmetricdimethyl | Arg + 28.0314 | S-adenosylmethionine | Type II, III PRMTs | JMJD61, KDM4E, KDM5C | [148-153] | |
| 1-methyl | His + 14.0157 | S-adenosylmethionine | METTL9 | Unknown | [157] | |
| 3-methyl | His + 14.0157 | S-adenosylmethionine | SETD3, METTL18, HPM1 | Unknown | [158] | |
| dimethyl | Lys + 28.0314 | S-adenosylmethionine | KMTs | KDMs, JMJDs, LSD1 | [155] | |
| trimethyl | Lys + 42.0471 | S-adenosylmethionine | KMTs | KDMs, JMJDs, LSD1 | [155] | |
| Glycation | 3-DG-H | Arg + 144.0422 | 3-deoxyglucosone | Non-enzymatic | Unknown | [159] |
| 3-DGArg | Arg + 162.0528 | 3-deoxyglucosone | Non-enzymatic | Unknown | [159] | |
| glyoxal hydroimidazolone | Arg + 39.9949 | Glyoxal | Non-enzymatic | Unknown | [160] | |
| methylglyoxal hydroimidazolone | Arg + 54.0106 | Methylglyoxal | Non-enzymatic | DJ-11 | [17, 20, 161] | |
| carboxymethyl | Arg + 58.0055 | Glyoxal | Non-enzymatic | Unknown | [160] | |
| Lys + 58.0055 | Glyoxal, 3-deoxyglucosone | Non-enzymatic | Unknown | [160] | ||
| carboxyethyl | Arg + 72.0212 | Methylglyoxal | Non-enzymatic | DJ-11 | [17, 20, 161] | |
| Lys + 72.0212 | Methylglyoxal | Non-enzymatic | DJ-11 | [16, 17, 20] | ||
| argpyrimidine | Arg + 84.0575 | Methylglyoxal | Non-enzymatic | Unknown | [161] | |
| tetrahydropyrimidine | Arg + 144.0422 | Methylglyoxal | Non-enzymatic | Unknown | [161, 162] | |
| pyrraline | Lys + 110.0368 | 3-deoxyglucosone | Non-enzymatic | Unknown | [159] | |
| ribosyl | Lys + 132.0423 | Ribose | Non-enzymatic | DJ-11, FN3K1 | [163] | |
| fructosyl | Lys + 162.0528 | Glucose | Non-enzymatic | FN3K1 | [164] | |
| glucosyl | Lys + 162.0528 | Fructose | Non-enzymatic | FN3K1 | 2 | |
| phosphoglyceryl | Lys + 167.9824 | 3-phosphoglycerate | Non-enzymatic | DJ-11, FN3K1 | [19] | |
| Phosphorylation | phosphorylation | Arg + 79.9663 | ATP | Unknown | Unknown | [165] |
| Ser + 79.9663 | ATP | Kinases | Phosphatases | [166, 167] | ||
| Thr + 79.9663 | ATP | Kinases | Phosphatases | [166, 167] | ||
| Tyr + 79.9663 | ATP | Kinases | Phosphatases | [166, 167] | ||
| 1-phosphoHis | His + 79.9663 | ATP | NME1/2 | PHPT1, LHPP, PP1/2A/2C, PGAM5 | [168, 169] | |
| 3-phosphoHis | His + 79.9663 | ATP | NME1/2 | PHPT1, LHPP, PP1/2A/2C, PGAM5 | [168, 169] | |
| pyrophosphorylation | Ser + 159.9327 | ATP; inositol pyrophosphate diphosphoinositol pentakisphosphate | Multi-step: Kinase 'priming' and subsequent non-enzymatic | Unknown | [170-172] | |
| Thr + 159.9327 | ATP; inositol pyrophosphate diphosphoinositol pentakisphosphate | Multi-step: Kinase 'priming' and subsequent non-enzymatic | Unknown | [170-172] | ||
| polyphosphorylation | Lys + (79.9663)n | Inorganic polyphosphate | Non-enzymatic | Endo/exopolyphosphatases | [173, 174] | |
| Oxidation | hydroxy | Asn + 15.9949 | 2-oxoglutarate | Asparaginyl hydroxylase | Unknown | [175] |
| citrullination | Arg + 0.9840 | H2O | Protein Arginine Deiminases (PADs) | Unknown | [176] | |
| S-sulfenyl | Cys + 15.9949 | Reactive oxygen/nitrogen species | Non-enzymatic | Reversible, Thioredoxins | [41] | |
| S-sulfinyl | Cys + 31.9898 | Reactive oxygen/nitrogen species | Non-enzymatic | Sulfiredoxins | [41] | |
| S-sulfonyl | Cys + 47.9847 | Reactive oxygen/nitrogen species | Non-enzymatic | Irreversible | [41] | |
| S-nitrosyl | Cys + 28.9901 | Reactive nitrogen species, S-Nitrosoglutathione | Non-enzymatic | Thioredoxin, GSH; Serves as an intermediate to disufide bond generation | [177, 178] | |
| S-succination | Cys + 116.0109 | Fumarate | Non-enzymatic | Unknown | [179] | |
| S-itaconation | Cys + 130.0267 | Itaconate | Non-enzymatic | Unknown | [89] | |
| S-cysteinyl | Cys + 119.0041 | Cysteine | Non-enzymatic | Reversible | [180, 181] | |
| S-homocysteinyl | Cys + 133.0198 | Homocysteine | Non-enzymatic | Reversible | [181] | |
| S-glutathionyl | Cys + 305.0681 | GSSG, GSNO, GSH | Glutathione S-transferases, Non-Enzymatic | Reversible, Thioredoxins, Glutaredoxins | [182] | |
| 5-hydroxylation | Lys + 15.9949 | 2-oxoglutarate | JMJD6, lysyl hydroxylases | Unknown | [183, 184] | |
| allysyl | Lys - 1.0316 | O2, H2O | Lysyl oxidases | Unknown | [185] | |
| carbamyl/homocitrullination | Lys + 43.0058 | OCN- | Non-enzymatic | Unknown | [186] | |
| carboxyl | Lys + 43.9898 | CO2 | Non-enzymatic | Reversible | [186] | |
| Glu + 43.9898 | Vitamin K, CO2, O2 | γ-glutamyl carboxylase | Unknown | [187, 188] | ||
| homocysteinyl | Lys + 174.0460 | Homocysteine thiolactone | Non-enzymatic | Unknown | [189] | |
| oxidation | Met + 15.9949 | Reactive oxygen species | Non-enzymatic, Methionine sulfoxide reductases | Methionine sulfoxide reductases | [190] | |
| dioxidation | Met + 31.9899 | Reactive oxygen species | Non-enzymatic | Unknown | [191] | |
| hydroxy | Pro + 15.9949 | 2-oxoglutarate | Prolyl hydroxylase | Unknown | [192] | |
| 4-HNE | Cys + 156.1151 | 4-hydroxy-2-nonenal | Non-enzymatic | Unknown | [193] | |
| His + 156.1151 | 4-hydroxy-2-nonenal | Non-enzymatic | Unknown | [193] | ||
| Lys + 140.1201 | 4-hydroxy-2-nonenal | Non-enzymatic | Unknown | [193] | ||
| Lys + 156.1151 | 4-hydroxy-2-nonenal | Non-enzymatic | Unknown | [193] | ||
| ketoamide | Lys + 154.0994 | 4-oxo-2-nonenal | Non-enzymatic | SIRT2 | [194-196] | |
| MDA | Lys + 54.0106 | Malondialdehyde | Non-enzymatic | Unknown | [197] | |
| Lys + 134.0368 | Malondialdehyde | Non-enzymatic | Unknown | [197] | ||
| N-formylkynurenine | Trp + 31.9899 | Reactive oxygen species | Non-enzymatic | Unknown | [198] | |
| kynurenine | Trp + 3.9949 | Reactive oxygen species | Non-enzymatic | Unknown | [198] | |
| 3-hydroxy | Tyr + 15.9949 | Reactive oxygen species | Non-enzymatic | Unknown | [199] | |
| O-sulfo | Tyr + 79.9568 | 3′-phosphoadenosine 5′-phosphosulfate | Tyrosyl-protein sulfotransferase | Unknown | [200] | |
| 3-nitroTyr | Tyr + 44.9851 | Reactive nitrogen species | Non-enzymatic | Unknown | [199] | |
| Acylation | S-acetyl | Cys + 42.0106 | Acetyl-CoA, Acetyl-GSH | Non-enzymatic | Unknown | [18] |
| S-palmitoyl | Cys + 238.2297 | Palmitoyl-CoA | DHHC domain proteins, Non-enzymatic | Acyl-protein thioesterases, Non-enzymatic hydrolysis | [102, 201] | |
| S-oleoyl | Cys + 266.2610 | Oleoyl-CoA | DHHC domain proteins, Non-enzymatic | Acyl-protein thioesterases, Non-enzymatic hydrolysis | [102, 202] | |
| S-stearoyl | Cys + 266.2610 | Stearoyl-CoA | DHHC domain proteins, Non-enzymatic | Acyl-protein thioesterases | [102, 201, 202] | |
| formyl | Lys + 27.9949 | Formylphosphate, 10-formyl-tetrahydrofolate2 | Non-enzymatic | HDAC1/2/3, SIRT3/6 | [63, 203, 204] | |
| acetyl | Lys + 42.0106 | Acetyl-CoA; Acetyl-GSH | KATs, Gcn5, p300/CBP, PCAF, NatA, Tip60, MOF; Non-Enzymatic | KDACs, SIRTs | [117, 205] | |
| propionyl | Lys + 56.0262 | Propionyl-CoA | p300/CBP, GNATs, MYSTs, HAT1, Non-Enzymatic | HDAC1/2/3, SIRT1/2/3 | [64, 206-208] | |
| crotonyl | Lys + 68.0262 | Crotonyl-CoA | p300/CBP, GNATs, MYSTs, Non-enzymatic | SIRT1/2/3, HDAC1/2/3 | [61, 206, 209, 210] | |
| butyryl | Lys + 70.0419 | Butyryl-CoA | Non-enzymatic, GNATs | SIRT1/2/3 | [64, 206, 207, 211] | |
| isobutyryl | Lys + 70.0419 | Isobutyryl-CoA | p300 | HDAC1/2/3, SIRT2 | [208, 209, 212] | |
| lactoyl | Lys + 72.0212 | lactyl-CoA2; lactoylGSH | p3002; Non-ezymatic | HDAC1/2/3 and SIRT2 | [16, 44, 95] | |
| tiglyl | Lys + 82.0419 | Tiglyl-CoA | Non-enzymatic | Unknown | [213] | |
| acetoacetyl | Lys + 84.0211 | Acetoacetyl-CoA | Non-enzymatic | Unknown | [213] | |
| 2-methylbutyryl | Lys + 84.0575 | 2-methylbutyryl-CoA | Unknown | Unknown | [213] | |
| isovaleryl2 | Lys + 84.0575 | isovaleryl-CoA | Unknown, Non-enzymatic | HDAC1/3 | [209, 214] | |
| malonyl | Lys + 86.0004 | Malonyl-CoA | KAT2A | SIRT5 | [141, 215] | |
| 2-hydroxybutyryl | Lys + 86.0368 | β-hydroxybutyryl-CoA | p300 | HDAC1/2/3, SIRT1/2 | [119, 145, 146, 216] | |
| β-hydroxyutyryl | Lys + 86.0368 | β-hydroxybutyryl-CoA | p300 | HDAC1/2/3, SIRT1/2 | [146] | |
| hexanoyl | Lys + 98.0732 | Hexanoyl-CoA | Unknown, Non-enzymatic | HDAC1/2/3/11, SIRT1/2/3/6 | [203, 217] | |
| succinyl | Lys + 100.0160 | Succinyl-CoA | CPT1a | SIRT5 | [211, 218-220] | |
| benzoyl | Lys + 104.0262 | Sodium benzoate | Lys acyltransferase2 | SIRT2 | [92] | |
| glutaryl | Lys + 114.0317 | Glutaryl-CoA | Unknown, Non-enzymatic | SIRT4/5 | [211, 221] | |
| 3-methylglutaconyl | Lys + 126.0317 | Methylglutaconyl-CoA | Unknown, Non-enzymatic | SIRT4 | [221] | |
| octanoyl | Lys + 126.1045 | Octanoyl-CoA | Unknown, Non-enzymatic | SIRT6, HDAC8/11 | [102, 197, 217, 222, 223] | |
| 3-methylglutaryl | Lys + 128.0473 | Methylglutayryl-CoA | Unknown, Non-enzymatic | SIRT4 | [211, 221] | |
| 3-hydroxy-3-methylglutaryl | Lys + 144.0423 | HMG-CoA | Unknown, Non-enzymatic | SIRT4 | [211, 221] | |
| decanoyl | Lys + 154.1358 | Decanoyl-CoA | Unknown, Non-enzymatic | SIRT6/7, HDAC1/2/3/11 | [208, 209, 217] | |
| dodecanoyl | Lys + 182.1671 | Dodecanoyl-CoA | Unknown, Non-enzymatic | HDAC8/11 | [217, 223] | |
| lipoyl | Lys + 188.0330 | Multi-step, involves octanoyl-acyl carrier protein | Multi-step: LIPT1, LIPT2, LIAS, glycine cleavage H protein. | HDAC11, SIRT2/4 | [197, 217, 224, 225] | |
| myristoyl | Lys + 210.1984 | Myristoyl-CoA | N-terminal Gly myristoyltransferases 1 and 2 | SIRT2/6, HDAC1/2/8/11 | [102, 217, 222] [209, 223, 226, 227] | |
| N6-(pyridoxalphosphate) | Lys + 229.0140 | Pyrixoxal 5'-phosphate | Non-enzymatic | Unknown | [228] | |
| palmitoyl | Lys + 238.2297 | Palmitoyl-CoA | Hedgehog acyltransferase | SIRT6, HDAC11 | [102, 217, 222] | |
| O-acetyl | Ser + 42.0106 | Acetyl-CoA | Unknown | Unknown | [229, 230] | |
| O-octanoyl | Ser + 126.1045 | Octanoyl-CoA | Ghrelin O-acyltransferase | Butyrylcholinesterase, Acyl-protein thioesterase 1, α2-macroglobulin, Notum | [231] | |
| O-myristoyl | Ser + 210.1984 | Myristate | Unknown | Unknown | [232] | |
| O-palmitoyl | Ser + 238.2297 | Palmitoyl-CoA | Membrane-bound O-acyl-transferases | Notum | [102] | |
| O-acetyl | Thr + 42.0106 | Acetyl-CoA | Unknown | Unknown | [229, 233] | |
| O-myristoyl | Thr + 201.1984 | Myristate | Unknown | Unknown | [232] | |
| O-palmitoyl | Thr + 238.2297 | Palmitoyl-CoA | Membrane-bound O-acyl-transferases | Notum | [102] | |
| O-acetyl | Tyr + 42.0106 | Acetyl-CoA | Unknown | Unknown | [229, 234] | |
| Lipidation | farnesylCys | Cys + 204.1878 | Farnesyl diphosphate | Farensyl transferase | Esterases2 | [102] |
| geranylgeranylCys | Cys + 272.2504 | Geranylgeranyl diphosphate | Geranylgeranyl transferase; Rab geranylgeranyl transferase | Esterases2 | [102] | |
| Other | N-glycosylation | Asn + Variable | N-acetylglucosamine | Oligosaccharyltransferases | PNGase/N-glycanase | [235] |
| O-glycosylation | Ser + Variable | N-acetylgalactosamine, N-acetylglucosamine, dolichol-P-mannose, fucose, glucose | O-GlcNAc transferase, O-glucosyltransferases | O-GlcNAcase | [235, 236] | |
| Thr + Variable | N-acetylgalactosamine, N-acetylglucosamine, dolichol-P-mannose, fucose, glucose | O-GlcNAc transferase, O-glucosyltransferases | O-GlcNAcase | [235, 236] | ||
| Tyr + Variable | N-acetylgalactosamine | O-GlcNAc transferase | Unknown | [197, 237] | ||
| S-guanyl | Cys + 343.0318 | 8-nitroguanosine 3',5'-cyclic monophosphate | Non-enzymatic | Unknown | [238] | |
| S-sulfhydryl | Cys + 31.9720 | Hydrogen sulfide | Non-enzymatic | Reversible | [239] | |
| CoAlation | Cys + 765.0995 | Coenzyme A | Non-enzymatic | Reversible | [240] | |
| histaminyl | Gln + 94.0531 | Histamine | Transglutaminase 2 | Unknown | [241] | |
| dopaminyl | Gln + 136.0524 | Dopamine | Transglutaminase 2 | Unknown | [242] | |
| serotonyl | Gln + 159.0685 | Serotonin | Transglutaminase 2 | Unknown | [243] | |
| biotinyl | Lys + 226.0776 | Biotin | Holocarboxylase synthetase | HDAC11, SIRT4 | [217, 244, 245] | |
| hypusination | Lys + 87.0684 | Spermidine | Multi-step process: deoxyhypusine synthase, deoxyhypusine hydroxylase | Unknown | [246, 247] | |
| ubiquityl | Lys + 114.042933 | Ubiquitin | Multi-step process involving E1, E2, and E3 ubiquitin ligases | Deubiquitination enzymes (Dubs) | [47, 48, 197, 248] | |
| NEDDyl | Lys + 114.042933 | NEDD | Multi-step process involving NAE, UBC12/UBE2F, and E3 ubiquitin ligases | COP9 signalosome, deneddylase 1 | [48, 249] | |
| SUMOyl | Lys + 114.042933 | SUMO | Multi-step process involving SAE1 or SAE2, Ubc9, and E3 SUMO ligases | Serine specific proteases (SENPs) | [48, 197, 250, 251] | |
| C-linked glycosylation | Trp + Variable | α-D-mannopyranose | C-Man glycosyltransferases | Unknown | [235, 236, 252, 253] | |
| 3-chloro | Tyr + 33.9610 | HOCl | Non-enzymatic | Unknown | [254] | |
| ADPRibosylation | Arg + 539.0466 | ADP-ribose | ADP-ribosyltransferases | Mono-ADP-ribosylhydrolases | [197] | |
| Cys + 539.0466 | ADP-ribose | ADP-ribosyltransferases | Mono-ADP-ribosylhydrolases | [197] | ||
| Glu + 539.0466 | ADP-ribose | ADP-ribosyltransferases | Mono-ADP-ribosylhydrolases | [197] | ||
| Lys + 539.0466 | ADP-ribose | ADP-ribosyltransferases | Mono-ADP-ribosylhydrolases | [197] | ||
| Ser + 539.0466 | ADP-ribose | ADP-ribosyltransferases | Mono-ADP-ribosylhydrolases | [197] | ||
| Thr + 539.0466 | ADP-ribose | ADP-ribosyltransferases | Mono-ADP-ribosylhydrolases | [197] | ||
| PolyADPRibosylation | Variable | ADP-ribose | Poly(ADP-ribosyl)polymerases | Poly-ADP-ribose Glycohydrolases | [197] |
Under debate;
Speculated;
Following tryptic digestion;
Most Lys acylations can be derived through non-enzymatic means; however, this does not exclude the possibility of a yet-to-be identified writer.
In the context of cell fate, there is no place where the regulation of PTMs is more critical than the nucleus [7]. Histone PTMs regulate chromatin structure, DNA accessibility, and serve as molecular handles for transcriptional machinery [8,9]. Over the past decade, advances in the sensitivity and specificity of mass spectrometry (MS) and DNA sequencing have led to a significant expansion in the number of functionally relevant histone PTMs [10]. To date, >100 types of PTMs have been identified on histones, many of which play a critical role in regulating transcriptional responses to endogenous and xenobiotic stimuli [11,12]. Given the consequences of a misplaced or the prolonged occupancy of a PTM, many are strategically placed onto target residues through ‘writers’ (e.g. kinases) and removed via ‘erasers’ (e.g. phosphatases) [13]. PTMs also serve as molecular handles for transcriptional machinery, providing a scaffold for ‘reader’ domain recognition (e.g. 14-3-3 domains) [13]. As many of these processes go awry in disease, significant effort has been placed on targeting the PTM landscape in chromatin, with numerous drugs currently on the market and/or making their way through clinical trials [14].
While many PTMs are tightly regulated through enzyme-catalyzed reactions, there is now mounting evidence that PTMs generated through non-enzymatic means may play a significant role in cell signaling, rather than serving as simple markers of toxicity [15–20]. A prime example of this is Lys acetylation, which is heavily enriched on glycolytic and TCA pathway enzymes, controlling metabolic flux and maintaining homeostasis [21]. Although Lys acyltransferases (e.g. p300) are established writers of these marks, the bulk of non-nuclear Lys acetylation is now postulated to arise from a non-enzymatic, S-to-N acyl-transfer from acetyl-CoA to an unmodified Lys residue [18]. This does classify these PTMs are unregulated, however, as Lys deacylases and sirtuins (SIRTs) remove modifications located at residues critical for function [22–25]. With >2 000 000 PTM sites now experimentally validated [26], one has to ask: Are all PTMs regulatory?
In this review, we will highlight the field of PTM research from the proverbial 30 000-foot view, solely focusing on modifications to amino acid sidechains and neglecting N- and C-terminal modifications. As an in-depth review could be written about nearly every PTM, our intent is not to take a deep dive into any single modification, but rather take a global view the field, the mechanisms that govern PTM site-specificity, and the technologies to investigate their role in health and disease. We will also close by discussing a growing notion that many PTMs simply serve as a metabolic reserve for nutrients and play a minimal role in the regulation of protein function. This review also aims to serve as a starting point and resource for those interested in studying PTMs using thorough analytical approaches.
PTMs are diverse and dynamically regulated
Although nearly every amino acid is prone to some degree of post-translational processing, the majority of reported PTMs reside on the nucleophilic side-chains of Lys, Arg, Cys, Ser, Thr, and Tyr [26] (Figure 1). As every protein contains amino acids with modifiable sidechains, it should not be surprising that nearly every protein has been reported, or predicted, to contain at least one PTM. Given the diversity in PTM composition and protein targets, a complete survey of the PTM landscape in biology is a seemingly insurmountable task; however, bioinformatic approaches have provided considerable insight into their prevalence [26,27]. dbPTM (https://awi.cuhk.edu.cn/dbPTM/) is an open access software that mines and curates existing PTM databases to provide functional and structural insights into >2 000 000 experimentally validated PTM sites [28]. This should be interpreted with some caution, however, as the prevalence of a select few PTMs (e.g. Ser/Thr phosphorylation, which accounts for >70% of experimentally validated PTM sites) are likely skewed due a strong focus on these pathways in biomedical research and ready access to tools/resources (i.e. phospho-‘specific' antibodies) [28]. While the importance of phosphorylation should not be understated, comprehensive studies evaluating its abundance have revealed a more modest estimation of ∼80 000 sites, with the nearly twice as many found to result from false-positive identifications [29].
Many PTMs are derived from primary metabolic intermediates and serve as sensors for nutrient status in the cell [6,7,30]. Acylations, for example, are a class of structurally diverse PTMs that exist on Lys, Ser, Thr, and Cys [31]. These PTMs range from a simple acetylation (C2H3O) to more complex modifications such as palmitoylation (C16H31O) (Figure 1) [6]. The bulk of acylations are derived from their parent acyl-CoA species and may be enzymatically or non-enzymatically placed onto proteins [30]. As a result, the abundance of protein acylation is largely reflective of local acyl-CoA concentrations [32,33]. In addition, acyl-GSH conjugates have also been identified as a major contributor to the acylLys pool, solely through a similar non-enzymatic S-to-N acyl transfer [16,18,34]; the contribution of acyl-CoA vs acyl-GSH towards the PTM pool, however, is unknown. Regardless of the route of addition, Lys acylations are dynamically regulated through their enzymatic removal via Lys deacylases and SIRTs, displaying half-lives ranging from minutes to >150 h [35]. These highly variable half-lives also differ among sites on the same protein, suggesting more active regulation of potentially regulatory sites, while allowing agnostic sites to maintain their occupancy [35]. Perhaps unsurprisingly, proteins involved with transcription/translation generally display shorter half-lives of acetylation sites, with many histones acetylation sites having half-lives ≤1 h [36]. Histone proteins have exceptionally long half-lives, with some primary cell lines displaying an average half-life on the order of weeks [37]. PTM status drastically impacts histone half-life, with Lys methylations (generally associated with heterochromatic, silenced regions of the genome) reducing histone turnover and acetylation (generally associated with euchromatic regions of the genome) increasing histone turnover [38]. While much work has been done in the field of acetylation, little is known about the relative half-lives of other Lys acylations and their impact on protein turnover.
Non-enzymatic PTMs are vital components to cell signaling and protein function, despite the long-held belief that they solely serve as markers of cell toxicity [39]. A prime example of this is Cys oxidation, which was originally used as an indication of oxidative stress and protein damage [40]. Cys oxidation is necessary for a myriad of biological processes, ranging from protein folding to transcriptional responses to stress [40]. Accumulation of these PTMs above the hormetic, or adaptive, threshold, however, is now appreciated as the critical determinant in establishing toxic responses [40]. Although a comprehensive investigation into Cys oxidation half-life has not been completed, estimations suggest sulfenylation, for example, is rapidly turned over, with half-lives <5 min [41]. Elegant studies have demonstrated that this quick turnover serves a regulatory role in metabolic flux, slowing output during periods of nutrient excess. GAPDH, a primary glycolytic enzyme, shows a near complete loss of activity <5 min after exposure to H2O2 due to the oxidation of an active site Cys and full restoration 30 min post-exposure [42]. This aligns with proposed role of Cys in biology, where up to 80% of Cys residues are predicted to be ‘functional’ [42,43].
Protein glycation has taken a similar path to Cys oxidation, long believed to be a measure of glucose toxicity and routinely used as a biomarker for glycemic control (i.e. HbA1c). This complex class of PTMs predominately reside on Arg and Lys, existing at concentrations comparable to other canonical, enzymatically regulated, protein modifications (e.g. asymmetric dimethylArg) [17]. Despite their non-enzymatic origins, protein glycation now has defined roles in the regulation of chromatin and metabolic flux [16,17,20]. Lys lactoylation, which is derived from the glycolytic by-products lactoylglutathione and lactate, are enriched on primary glycolytic enzymes, reducing metabolic output and slowing glycolytic flux [16,44]. These findings are critical in establishing a functional role for non-enzymatically generated PTMs, which should not classify a PTM as damaging or toxic to cell health but rather, important for maintaining homeostasis. Of course, like most other PTMs, the threshold dictating a homeostatic function from a toxic response (i.e. cell death) likely resides in its abundance [20,45]. Often, studies linking protein glycation to toxic responses rely on supraphysiological concentrations of glycating agents and thus careful consideration must be taken when employing model systems to explore the physiological role of non-enzymatic PTMs, such as glycation.
While small molecule modifications comprise the bulk of PTMs in biology, protein conjugates also have a critical role in cell biology. Lys ubiquitylation is an evolutionarily conserved PTM that regulates a multitude of biological processes, most notably protein degradation and turnover [46]. Ubiquitin is a 76 amino acid protein that is conjugated via an isopeptide bond with its C-terminal Gly carboxylate and a target Lys ε-amino group [47]. Akin to ubiquitylation, SUMOylation and NEDDylation are small ubiquitin-like protein modifications that are indispensable in eukaryotes [48]. These PTMs are enzymatically placed at precise Lys residues through a multi-step enzymatic process and, despite the structural and functional similarities in these three PTMs, the sites of modification vary widely; ubiquitylation is the most abundant (>60 000 sites), with SUMOylation (∼8000 sites) and NEDDylation (∼1000 sites) following [48]. Importantly, little overlap in target sites exists (∼100) between these three PTMs, suggesting functional independence despite the similarities in signaling and regulation [48]. What makes these PTMs particularly unique is their ability to undergo post-translational modifications themselves, through either or small molecule modifications (e.g. phosphorylation) or extensive homo- or hetero-poly-ubiquitin/NEDD/SUMO branching [48–50].
PTMs rarely exist in isolation as proteins are prone to undergo a wide array of diverse modifications simultaneously at numerous sites. Many PTMs trigger, or prime, proteins to undergo additional modification, which elicit alterations in function and/or localization [51,52]. This ‘PTM crosstalk’ is perhaps most well appreciated on histones where PTMs influence the binding of writers and erasers resulting in subsequent histone modifications [53]. For example, asymmetric dimethylation of Arg2 on histone H3 serves as a transcriptional repressive mark at promoters by preventing Lys methyltransferase 2A from methylating Lys4 of histone H3 [54,55]. Conversely, symmetric dimethylation of H3 Arg2 promotes trimethylation at Lys4 through binding of WDR5, a common component of coactivator complexes and euchromatic regions of the genome [54,55]. This is just one example of numerous instances of PTM crosstalk that have been elegantly defined in biology and certainly, this layering of PTMs adds additional layers of complexity in the study of PTMs [52,56].
Methods for the detection and study of PTMs
Antibody-based approaches
Antibody-based approaches remain a staple in PTM research, largely due to the ubiquity and relative ease of immunoassays. Pan-antibodies that broadly recognize a PTM, regardless of the target protein (e.g. anti-acetylLys) are useful tools to determine relative changes across the proteome in a set of biological samples. These antibodies are also routinely used for co-immunoprecipitations to investigate alterations on target proteins. It should be noted, however, that these approaches are constrained to the analysis of a single PTM, failing to provide information on its abundance or stoichiometry. In addition, poorly vetted and low-fidelity antibodies have polluted PTM research leading to false assertions as to the role of these modifications in biology and disease. Pertaining to histone PTMs, thorough investigations have revealed that >25% of all antibodies failed specificity tests [57,58]. Combinatorial PTMs also disrupt antibody specificity, whereby occupancy of nearby residues with PTMs can alter epitope recognition [57,58]. This is particularly relevant for histones, where the N-terminal tails are heavily modified with a diverse array of PTMs. While there are significant drawbacks to antibody-based approaches, they have no doubt transformed our ability to detect, enrich, and study PTMs on a broad scale. It is important moving forward, however, that antibodies are properly validated (i.e. knockout cell lines) and coupled to additional means of specificity (e.g. mass spectrometry, site-directed mutagenesis).
MS-based approaches
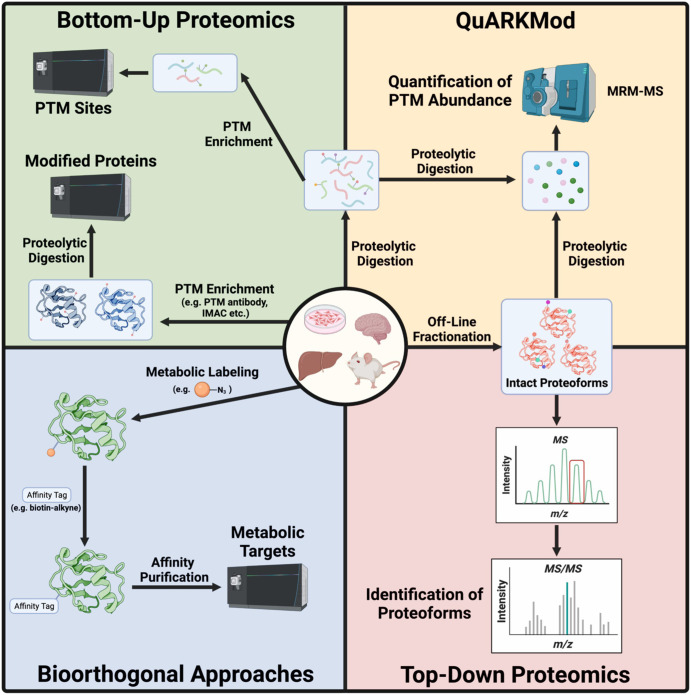
The past two decades have seen a rapid rise in both sensitivity and accuracy of mass spectrometers (MS) [59]. This has undoubtedly fueled the rapid expansion in the list of known PTMs in biology [28]. Although numerous MS-based approaches can be applied for PTM research, bottom-up proteomics remains the most prevalent and logical starting point. Here, proteins are proteolytically digested to peptides, separated via liquid-chromatography (LC), and analyzed using tandem mass spectrometry (MS/MS) for PTMs of interest. As PTMs are typically low in abundance compared with their unmodified counterparts, sample enrichment is almost always necessary to gain a full picture of the PTM profile [60]. By far the most common enrichment strategy is through immunoaffinity columns, using antibodies directed at either the protein or PTM of interest (Figure 2). In addition to antibody-based approaches, numerous off-line enrichment strategies can be employed: isoelectric focusing and SDS–PAGE can reduce proteome complexity, with the goal of isolating the protein(s) of interest [61–64]. Multi-dimensional chromatography approaches, such as high-pH [65,66], size exclusion [67], and strong cation exchange chromatography [68,69] have been widely utilized to increase resolution and dynamic range. When coupled with MS, these platforms can be used to generate high quality, accurate mass identification, quantification, and discovery of PTMs (Figure 2). This approach provides considerable flexibility in sample preparation, LC conditions, and ionization strategies; however, the most common proteomic workflow uses a tryptic digestion and reversed-phase LC coupled to an electrospray ionization MS. Trypsin, which cleaves after Lys and Arg, is a high-fidelity enzyme, yielding consistent peptides and reproducible MS data. There are significant caveats to this approach, however; tryptic digestion may not provide coverage across a desired site of modification, yielding peptides too large for MS detection (often <1500 m/z). A simple solution to this problem is to use an alternative protease (e.g. chymotrypsin, Glu-C), yielding peptides better suited for MS detection that span the site of interest . Conversely, proteins rich in Lys or Arg (e.g. histones) may yield tryptic peptides too short or polar for detection with standard reversed-phase LC conditions [70]. To address this, chemical derivatization strategies (e.g. propionylation) can be used to modify free Lys residues [70]. This propionylated Lys residue will result in a missed cleavage, leading to cleavage at the next available unmodified Arg (Figure 2). This strategy also increases the hydrophobicity of smaller peptides, resulting in increased retention on standard reversed-phase LC systems and improved MS detection [70]. A caveat to this approach, however, is the inability to differentiate endogenous vs. exogenous propionylation. To circumvent this problem, isotopically labeled propionic anhydride (e.g. d10) can be used during chemical derivitization [71].
Figure 2. Analytical approaches for the study of PTMs.
Created with BioRender.com.
As noted above, PTMs are often low in abundance and challenging to detect in crude proteome preparations using MS; thus, sample enrichment is typically required. This is most commonly achieved through immunoaffinity columns using proteolytically digested samples, selectively isolating only peptides containing the PTM of interest [72]. As unmodified peptides are lost during immunoprecipitation, reference proteome samples collected prior to enrichment are typically run in parallel using label-free quantitation approaches (i.e. spectral counting) [73]. Despite this experimental simplicity and adaptability to analyze large datasets, bias in sample preparation and data processing may hinder accuracy, precision, and reproducibility [73]. To circumvent this problem, numerous labeling approaches may be employed. Stable Isotope Labeling by Amino acids in Cell culture (SILAC) [74], chemical labeling using isobaric tags (i.e. iTRAQ) [75], and tandem mass tags [76] are routinely employed prior to immunoprecipitation. This provides a measure of relative quantitation in the form of heavy isotope to light isotope ratios. While these approaches provide relative quantification of proteins in a given dataset, stable isotope labeled protein or peptide standards can alternatively be spiked in prior to immunoprecipitation, yielding absolute quantification or a target protein/PTM [72].
Another popular strategy for the quantitation of PTMs in biological samples relies on multiple reaction monitoring (MRM)-MS [77]. Here, target sites and fragment ions are known and peptide masses are included in the MS parameters [77]. This results in increased sensitivity and provides improved sequence coverage to ensure that all sites are mapped [77]. From here, two strategies can be used for quantitation: (1) all modified peptide masses from a single site are summed and set as the denominator, with the target PTM set as the numerator, providing the relative abundance of a PTM at a given site [78]. With this approach, MS parameters must be optimized for each peptide as many PTMs are known to alter ionization efficiency thus biasing their abundance [78,79]; and (2) synthetic isotopically labeled peptides standards can be added for absolute quantification [77]. These approaches are routinely used for the analysis of histone PTMs due to the high degree of modification states and ease of histone isolations from complex biological samples, reducing interference from non-histone proteins. It should also be noted that these quantitation strategies are also amenable to standard shotgun proteomic approaches and are not unique to the MRM-MS platform.
The heterogeneity of peptides makes the discovery of PTMs a significant challenge, and while database algorithms have provided some hope (see below), these approaches are not overly amenable to most labs. To remove the complexity of peptides from the equation, QuARKMod, or the Quantitative Analysis of Arg (R) and Lys (K) Modifications (QuARKMod), relies on exhaustive proteolytic digestion, yielding single amino acids (Figure 2) [60]. Amino acids are then separated and detected using an MRM approach on a triple quadrupole MS. A major advantage to this approach is the increased sensitivity and, provided stable isotope standards are included, determination of PTM abundance, allowing for a comparison of all measured PTMs in a given sample [60]. The chromatographic separation of single amino acids also provides improved separation of isomeric species compared with that of peptides. This was demonstrated through the identification of lactoylLys, which is isomeric to the advanced glycation end-product, carboxyethylLys [16]. In addition, this method can be applied for PTM discovery using a precursor-ion scan for Lys or Arg fragments (84.1 m/z and 70.1 m/z, respectively) that remain consistent, regardless of the PTM [60]. While this approach provides an accurate measure of PTM abundance, this comes at the expense of site identification as all sequence information is lost following proteolytic digestion.
Undoubtedly the utility of bottom-up proteomic approaches has been instrumental in the discovery and quantitation of PTMs in biology. A significant shortcoming of these approaches, however, is that PTM stoichiometry on a single molecule is lost. Top-down proteomic approaches have aimed to address this, by ionizing intact proteins and analyzing subsequent fragment ions [80]. This yields complete sequence coverage, improves isoform resolution, and provides a comprehensive view of the PTM profile and interplay on a target protein (i.e. proteoforms) [81]. This was elegantly displayed with KRAS4b, one of four isoforms in the RAS family, which share high sequence homology (82–90%) [82]. KRAS4b undergoes post-translational processing of a conserved C-terminal CaaX motif, ultimately yielding carboxymethylation to the C-terminal Cys residue. This modification is essential for association with the plasma membrane and interaction with effector proteins. Typical bottom-up approaches yield identical peptides from each RAS isoform, making isoform-specific analysis an insurmountable task. Using top-down proteomics, 11 proteoforms of KRAS4b can be resolved, revealing a high degree of variability among cancer patients in this critical C-terminal processing [81]. Although the advantages to this approach are clear, there are significant hurdles preventing the mainstream application of top-down proteomics: cumbersome prefractionation of intact proteins, lower sensitivity compared with MRM approaches, limitations in protein size (<100 kDa), intact protein solubility (particularly with membrane proteins), the need for high-resolution mass analyzers, and complexity of data analysis [83,84].
Database algorithms for PTM identification
Due to the chemical heterogeneity of PTMs, MS and MS/MS spectra are often complex [62]. Database algorithms are thus a crucial component in the PTM workflow, mining spectra for PTMs of interest. Software such as X!Tandem [85] and Mascot [86] provide the user with the ability to search datasets for defined mass shifts on target residues of both known and unknown PTMs (Table 1). A major hinderance to these software packages is the limited number of variable modifications (i.e. PTMs) that can be searched in a single analysis, with each PTM resulting in an exponential rise in false-positive identifications. While data analysis software has become quite robust in recent years (<1% false discovery rate), it is still imperative to manually validate sites of interest, particularly for novel or poorly characterized PTMs. Separate software packages have also been developed to try and reduce the manual mining of MS spectra: PILOT_PTM can search an unrestricted number of PTMs on a template sequence (i.e. known site) [87]. Spectral Alignment-based Modified Peptide Identification, or SAMPEI, was developed based on the rationale that MS/MS spectra of protein are produced from both modified and unmodified peptides, and thus any statistics-based algorithm, such as X!Tandem, can be used for peptide sequence matches (PSMs) [88]. The remaining unmatched MS/MS spectra are used as target spectrum to identify peptide sequences with PTMs. Using a cellular model for inflammation, SAMPEI identified a mass shift corresponding to Cys +130.03 Da and Cys +146.02 Da, consistent with Cys itaconatylation and an oxidized product of this PTM, respectively [88]. These PTMs have known and appreciated roles in inflammatory signaling and have been identified on numerous metabolic enzymes [88,89]. This simple application was able to demonstrate the robustness of SAMPEI in the identification of unknown PTMs. However, since SAMPEI depends on statistical-based algorithms such as X!Tandem, the number of searched PTMs is limited.
Open search, which allows for a larger precursor mass tolerance resulting in the analysis of more spectra, is employed to explore the vast diversity of PTMs that are not accounted for in database search algorithms; however, achieving a comprehensive PTM search with practicality requires a sophisticated computational strategy. MSFragger, generates a theoretical fragment-ion index of peptides from in silico digestion of a protein database [90]. Mass binning and precursor mass ordering in the fragment-ion index enables rapid selection of candidate spectra matching experimental fragment ions, allowing all candidate spectra to be simultaneously scored against an experimental spectrum. The algorithm offers advantageous speed in open search compared with conventional database-search algorithms where theoretical spectra of in silico digested peptides are compared with experimental spectra. It is noteworthy that an inaccurate estimation of FDR occurs in a narrow-window search using a target-decoy strategy and this can be improved by accounting for all modifications under an open search strategy [90]. In addition, the localization of PTMs further reduces FDR. Some PTMs with identical or similar mass shifts on different amino acids cannot be distinguished without proper localization . PTMiner employs an interactive-leaning based algorithm for the accurate localization of both known and unknown PTMs in open search, which is instrumental in achieving <1% FDR [91].
The sequence alignment software, PTMap, has been immensely successfully in identifying novel PTMs, particulary on histones, including Lys succinylation, Lys crotonylation, Lys benzoylation, and Lys lactylation [61,92–95]. PTMap was the first unrestrictive search algorithm to use unmatched peaks in MS/MS spectra to eliminate false positives while simultaneously using matched peaks to select candidate peptides. To accomplish this, PTMap uses two steps to carry out the localization and putative mass of a PTM: first, iterative comparisons of the MS/MS spectra of experimental peptides to all theoretical MS/MS spectra of the candidate peptide isoforms with all possible PTMs localization. Second, PTMap looks for consecutive y- and b-ions or simultaneously modified y-and b-ions for confident identification of PTM site. While PTMap does not provide any insights into the molecular structure, high resolution mass accuracy does provide a necessary starting point by which hypotheses can be generated to formulate the composition of the unknown PTM.
Ensuring analytical rigor for increased reproducibility
MS-based proteomic studies have become commonplace in PTM research; however, appropriate analytical rigor must be maintained to ensure reproducibility. While many studies are focused on bulk PTM inventories using antibody-based pulldowns, it is important to manually validate and characterize sites of interest. This can be achieved using synthetic peptides to demonstrate co-elution and identical MS/MS fragmentations (i.e. y- and b-ions) [61,94]. This is particularly important when studying PTMs using low mass accuracy instrumentation (i.e. triple quads). For example, Lys acetylation and Lys trimethylation differ by only 0.0365 Da (Table 1) and are thus indistinguishable on low resolution instrumentation, yet have significant differences in their polarity and can be easily separated using RP-LC [60,77]. The importance of this synthetic peptide approach has been elegantly demonstrated with Lys 2-hydroxyisobutyrylation [96]. The mass shift resulting from this PTM (Lys +86.0368) can be derived from five possible isomers. Using synthetic peptides, co-elution with Lys 2-hydroxyisobutyrylation was demonstrated, showing this to be the dominant species in vivo [96]. Alternatively, stable isotope labeling can be used to verify PTMs and their metabolic origins. Here, a metabolic precursor (e.g. 13C6 glucose) is fed to cells and the incorporation of isotopic labels can be monitored on PTMs of interest (e.g. +2 Da for Lys acetylation) [97]. This added layer of confidence also provides the opportunity to monitor PTM half-life and protein turnover. These additional experimental measures increase analytical rigor are necessary to ensure reproducibility within the field.
Using chemical tools to study PTMs
Although immunoaffinity enrichments are commonplace in PTM research, chemical biology approaches are being adapted at an increasing rate and offer improved specificity and assay diversity. The advent of click chemistry-based probes has transformed our ability to study PTMs in cells with unparalleled specificity. By appending relatively inert bioorthogonal chemical tags (e.g. alkyne/azide) onto metabolic PTM precursors, it is possible to track the incorporation and regulation of these modifications over time, providing information on PTM half-life, protein turnover, and cellular fate (Figure 2). The first application of click chemistry for PTM research was applied for the study of mucin-type O-linked glycosylation [98]. Here, a bioorthogonal monosaccharide bearing an azide or alkyne functional group is fed to cells, resulting in the incorporation of tagged PTMs through the cells natural N-acetyl-a-galactosaminyltransferases [98,99]. Click chemistry is then employed using a variety of reporter tags to selectively visualize (e.g. alkynyl/azido fluorophores) or enrich (e.g. alkynyl/azido biotin) samples for modified proteins. As reliable, high-fidelity antibodies are often lacking in PTM research, these approaches provide unparalleled specificity, drastically reducing false-positive identifications and improper interpretation of results. As a result, this innovative approach has been expanded significantly, with ‘clickable’ metabolic analogs now available to study nearly every class of PTM [100–102].
The metabolic labeling of precursor molecules is the desired approach for the use of chemical biology probes, as they rely on the cells natural mechanism to deposit and regulate PTMs. This approach, however, is not possible for all PTMs as many are sterically restricted or too small (e.g. acetylation, oxidation) to allow for the incorporation of a chemical tag. Reactivity-based protein profiling has been instrumental in the study of numerous PTMs through an agnostic ‘addition by subtraction’ approach [103,104]. Using probes designed to react with unmodified amino acids (e.g. alkynyl-iodoacetamide and Cys), one can assert that a loss of signal stems from PTM occupancy [42,103]. This approach has been utilized extensively and probes have been developed to interrogate the Cys [103], Lys [104], Arg [105], Tyr [106], and Met [107] proteomes. The relative instability of some PTMs also makes sample processing a significant challenge that cannot be addressed with traditional immunoaffinity approaches. S-sulfinylation is a transient oxidation product of a Cys thiol that plays a major role in the redox state of the protein [108]. To overcome this challenge, dimedone-based reagents can be employed to selectively alkylate Cys S-sulfinylation, resulting in stabilization and chemical tagging of this transient PTM [109]. The dimedone warhead can be linked to an alkyne moiety whereby click chemistry can be employed to selectively enrich for S-sulfinylated proteins [109]. Using this approach, 193 protein targets of these labile PTMs have been identified, revealing Cys oxidation as a critical determinant in a host of biological processes, ranging from ER quality control to DNA repair [109].
Perhaps the original application of chemical biology for the study PTMs was the use of metal oxide affinity chromatography (i.e. IMAC) to selectively isolate phosphorylated proteins [110,111]. Here, proteins/peptides are loaded onto a metal oxide column (e.g. TiO2), selectively forming a complex with phosphorylated substrates. Modified samples are then eluted and can be monitored using either immunoblotting or LC–MS/MS [112]. This significantly reduces proteome complexity, removing unmodified species and selectively isolating phosphopeptides/proteins, resulting in increased phosphoproteomic depth and identification of lower stoichiometric sites. This approach has also been adapted for the study of phopshoglycerylLys modifications, which possess the necessary negatively charged phosphate moiety for IMAC, improving detection and quantitation of these low abundance PTMs species, which otherwise may have gone undiscovered [19].
What dictates specificity?
Traditionally, PTMs are viewed as tightly regulated and precisely placed through enzyme-controlled processes. PTM ‘writers’ often recognize a consensus sequence (i.e. a sequence motif) that marks a site for modification. For example, N-linked glycosylation occurs on Asn that reside in the Asn-X-Ser/Thr motif, where X is any amino acid except Pro [113]. This motif is recognized by an oligosaccharyltransferase (the ‘writer’), that transfers a precursor glycan to the target protein. While there are a few instances where N-linked glycosylation may occur outside of this motif (Asn-X-Cys), these PTMs are largely constrained to these sites [114]. Contrary to N-linked glycosylation, O-linked glycosylation occurs on Ser/Thr residues and lacks a consensus sequence, with the parameters that dictate site specificity still under debate [115]. This highlights perhaps the most important questions in the field of PTM research: What are the parameters that determine site-specificity? How are PTMs that lack a sequence motif regulated?
An argument could be made that the vast majority of PTMs are derived from non-enzymatic reactions (Table 1) and driven through site availability, local electrostatics, and mass action [18]. In support of this, proteomic inventories often display significant overlap in site-specificity between acylating species, despite lacking a clear sequence motif [72,116]. For example, Lys89 on enolase 1 (ENO1) is a surface exposed residue that is susceptible to acetylation [117], succinylation [116], lactylation [44], crotonylation [118], 2-hydroxyisobutyrylation [119], and benzoylation [120] (Figure 3). While the impact of modification at Lys89 has not been fully elucidated, other peripheral sites have shown significant alterations in stability depending on PTM status. Lactylation of Lys343 results in a significant reduction in stability, while Lys326 displays little to no impact [44]. Although a sequence motif does not exist for most Lys acylations, the propensity for a given site to undergo modification may be explained, in part, by the presence of Cys residues [34]. These Cys readily undergo transient modification (i.e. S-acylation), which is subsequently transferred through an S-to-N acyl transfer onto a nearby Lys residue [16,18,34]. From a biological perspective, these Cys–Lys pairs appear to be detrimental to health as long-lived species (e.g. primates) show less conservation compared with species with shorter lifespans (e.g. mice) [34]. Additional factors may be in play, but this reduced propensity to undergo non-enzymatic Lys acylation may play a larger role in metabolic flux and protein turnover than previously thought, dictating species longevity and/or aging-related disease.
Figure 3. Lys acylations are structurally diverse yet display significant redundancy in modification sites.
Proteomic inventories were mined to reveal a striking overlap in modification sites. While many of these sites are surface-exposed, some display a high propensity to modification despite being stearically hindered (e.g. Lys343 on ENO1 and Cys152 on GAPDH). PTM sites were curated from [44,92,116–120,141–147]. ENO1 (PDB: 2SPN) and GAPDH (PDB:3GPD).
This ‘mass action’ mode of PTM generation is perhaps best explained through the oxidation of Cys residues. Cys152 of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is heavily prone to modification, susceptible to sulfenylation [121], sulfinylation [122], nitrosylation [123], glutathionylation [124], and succination [125] (Figure 3). Cys152 resides in the active site of GAPDH and is critical for enzymatic activity [42]. This is notable as the PTMs that occupy this site are chemically diverse and are derived from vastly different metabolic sources, yet all serve to reduce GAPDH activity and glycolytic flux [42]. As Cys152 regulates metabolic flux, these oxidations are typically dynamically regulated, many of which display half-lives on the order of minutes [42]. Presumably, Cys152 remains easily accessible as a feedback mechanism to rapidly reduce glycolytic flux and the production of reactive intermediates to prevent further proteome oxidation and cellular damage.
Just as the site-specific addition of a PTM is critical to enzyme activity, efficient removal is also important to restore enzyme function. This action is performed by molecular ‘erasers’ which have been identified for many PTMs (Table 1). When eraser activity is compromised, PTMs accumulate and alter the metabolic landscape. For example, mice lacking the Lys deacylase, SIRT3, display increased Lys acetylation on long-chain specific acyl-CoA dehydrogenase (LCAD), leading to reduced hepatic fatty acid oxidation [126]. The important role of erasers is likely highlighted best when considering PTMs derived from non-enzymatic sources. Lys lactoylation, in addition to mitochondrial acylation, is predominately derived via non-enzymatic means [16,18]. Given the lack of an identified ‘writer’, these PTMs would seemingly modify any available Lys residue through simple mass action. Although this may be the case, there is often significant redundancy in site-specificity between acyl species (Figure 3) [72]. This implies that while the generation of a given PTM may occur stochastically, the sustained occupancy of any given site is regulated through its enzymatic removal. Thus, it is important to consider that while a sequence motif does not exist for all PTMs, mechanisms are likely in place to dictate site-specificity and sustain PTM occupancy as needed.
Separating regulatory PTMs from bystanders
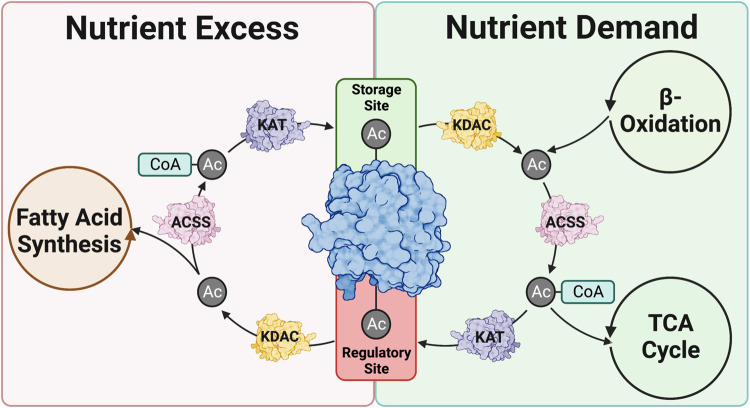
PTMs have historically been viewed as regulatory additions to proteins, providing a rapid response to regulate function [99,117]. In recent years, however, there is mounting evidence that many PTMs may simply be along for the ride, having no measurable impact of protein function [23]. Lys acetylation, for example, is a widely ubiquitous PTM that regulates a host of biological processes, ranging from metabolic flux to chromatin accessibility [6]. Current estimations indicate that 20–50% of all proteins are acetylated, yet many of these modifications do not impact protein function nor physiological outcome [127–129]. This has been elegantly demonstrated on histones, where certain Lys acetylation sites are now proposed to serve as a local store of acetate, readily available for rapid transfer to regulatory sites to alter the epigenomic landscape of the cell (Figure 4) [23]. This repurposing of acetate is an interesting hypothesis that likely holds true outside of the nucleus. KDACs and ACSS isoforms are expressed throughout the cell, making it possible that these reactions take place elsewhere, providing a rapid response to acetylate proteins when acetyl-CoA concentrations are limited. Indeed, as hig hlighted above, protein acylation levels are highly reflective of local acyl-CoA concentrations, so this may serve as an additional mechanism to provide acetyl groups on an ‘as needed' basis [32,33]. Furthermore, the focus on Lys acetylation may be too narrow in scope as KDACs and ACSSs are known to transfer other acyl substrates including propionate [130], crotonate [131], and butyrate [132]. Perhaps the most provocative hypothesis is that these PTMs are placed on proteins during periods of nutrient excess and removed when nutrients are limited, feeding cell metabolism, and maintaining homeostasis (Figure 4).
Figure 4. Hypothesized role for non-regulatory acylation sites on proteins.
Recent work has demonstrated the presence of non-regulatory acylation sites that can be taken back into acyl-CoA pools and redistributed to regulatory sites. We opine that this may serve as an additional metabolic feedback mechanism during periods of nutrient excess or demand. KDAC, lysine deacylase; ACSS, acyl-CoA synthetase short chain family member; KAT, Lys acytransferase. Created with BioRender.com.
Multiple factors dictate whether a PTM serves a functional and/or regulatory role. PTMs residing within the active site are often regulatory, altering substrate and/or cofactor binding. This is typically confirmed using recombinant protein, providing kinetic parameters of enzyme inhibition. Recombinant approaches, however, do not provide information on the role of PTMs in cellular trafficking, protein–protein binding, and/or half-life. These questions can be addressed using site-directed mutagenesis, altering the PTM site to either a ‘null’ amino acid, incapable of modification while maintaining necessary electrostatics (e.g. Lys to Arg) or to a PTM mimetic (e.g. Lys to Gln for acetylation) [133,134]. Although his can be informative, site-directed mutagenesis does not provide a direct measure of PTM function, but rather an absence of PTM occupancy. This shortcoming has been addressed through recent advances in genetic code expansion, which provides unparalleled investigations into PTM function on a site-by-site basis [135–137]. Here, unnatural amino acids are precisely incorporated into a protein of interest through a bioorthogonal aminoacyl-tRNA synthetase, which are evolved to recognize and incorporate an amino acid of interest into a target protein [136,137]. This approach has been used to identify lactoylation of Lys343, and not Lys326, as critical modifications mediating ENO1 stability [44]. Although a substantial library of aminoacyl-tRNA synthetases has been generated, this application has not been applied to all PTMs. Furthermore, the modified amino acid monomer must be synthesized and provided to cell culture media, which can prove challenging with more complex PTMs.
An alternative approach for the site-specific incorporation of PTMs into proteins relies on ultrafast split inteins [138,139]. Inteins are a family of protein domains that undergo self-excision as a post-translational modification. By splitting the intein into N- and C-terminal domains (IntN and IntC, respectively) it is possible to precisely incorporate a modification of interest into a target protein [139]. To accomplish this, a target protein is tagged with either IntN or IntC. Cells are then loaded with a synthetic polypeptide containing the opposing Int. When partner Ints interact, they undergo a rapid association and excision, resulting in chemical ligation of the peptide to the target protein [140]. A major limitation to this approach is the restriction to the N- or C-terminus of the target protein as polypeptides must be synthesized and remain cell-permeable [140]. While issues remain with both codon expansion and intein splicing, they have undoubtedly moved the field forward and provided critical information on the role of PTMs in cell metabolism and chromatin function.
Conclusions and outlook
PTMs are an integral part of biology, functioning as important signaling molecules to maintain homeostasis and adjust to metabolic and xenobiotic stressors. Consistent with this, there has long been a notion that most PTM sites are regulatory. Although many PTMs do serve this purpose, increasing evidence indicates that many PTMs may simply be along for the ride, providing no alterations in enzyme function. This idea has largely come to light with significant advances in chemical biology approaches. Notably, codon expansion allows for the precise incorporation of a PTM at a single site of investigation, rather than traditional ‘addition by subtraction’ approaches such as site-directed mutagenesis. Collectively, this has led to an intriguing hypothesis that PTMs may serve as a carbon sink, providing metabolic intermediates on demand through their enzymatic removal from protein substrates. As this field continues to experience a rapid expansion, it will be important for studies to maintain high analytical rigor, confirming observations with sound mechanistic studies. Advances in MS approaches have provided much of this added rigor, leading to increased reproducibility sound mechanistic insights into PTM biology.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Acknowledgements
This work was supported by National Institutes of Health Grants T32 GM008804 for N.K. and R35GM137910, R01DK133196 for J.J.G.
Abbreviations
- ACSS
acyl-CoA synthetase short chain family member
- ENO1
enolase 1
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- LC
liquid-chromatography
- MRM
multiple reaction monitoring
- MS
mass spectrometry
- PTMs
post-translational modifications
- QuARKMod
Quantitative Analysis of Arg (R) and Lys (K) Modifications
- SIRTs
sirtuins
Open Access
Open access for this article was enabled by the participation of University of Arizona in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with Individual.
CRediT Author Contribution
James J. Galligan: Conceptualization, Data curation, Supervision, Funding acquisition, Writing — review and editing. Naoya Kitamura: Conceptualization, Data curation, Writing — original draft.
References
- 1.Zhu, J. and Thompson, C.B. (2019) Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 20, 436–450 10.1038/s41580-019-0123-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metallo, C.M. and Vander Heiden, M.G. (2013) Understanding metabolic regulation and its influence on cell physiology. Mol. Cell 49, 388–398 10.1016/j.molcel.2013.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aebersold, R., Agar, J.N., Amster, I.J., Baker, M.S., Bertozzi, C.R., Boja, E.S.et al. (2018) How many human proteoforms are there? Nat. Chem. Biol. 14, 206–214 10.1038/nchembio.2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang, M., Xu, J.Y., Hu, H., Ye, B.C. and Tan, M. (2018) Systematic proteomic analysis of protein methylation in prokaryotes and eukaryotes revealed distinct substrate specificity. Proteomics 18, 1700300 10.1002/pmic.201700300 [DOI] [PubMed] [Google Scholar]
- 5.Hentchel, K.L. and Escalante-Semerena, J.C. (2015) Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol. Mol. Biol. Rev. 79, 321–346 10.1128/MMBR.00020-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jennings, E.Q., Fritz, K.S. and Galligan, J.J. (2022) Biochemical genesis of enzymatic and non-enzymatic post-translational modifications. Mol. Aspects Med. 86, 101053 10.1016/j.mam.2021.101053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diehl, K.L. and Muir, T.W. (2020) Chromatin as a key consumer in the metabolite economy. Nat. Chem. Biol. 16, 620–629 10.1038/s41589-020-0517-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenuwein, T. and Allis, C.D. (2001) Translating the histone code. Science 293, 1074–1080 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- 9.Kouzarides, T. (2007) Chromatin modifications and their function. Cell 128, 693–705 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 10.Huang, H., Sabari, B.R., Garcia, B.A., Allis, C.D. and Zhao, Y. (2014) Snapshot: histone modifications. Cell 159, 458–458.e451 10.1016/j.cell.2014.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Moreno, J.M., Fontecha-Barriuso, M., Martin-Sanchez, D., Sanchez-Nino, M.D., Ruiz-Ortega, M., Sanz, A.B.et al. (2020) The contribution of histone crotonylation to tissue health and disease: Focus on kidney health. Front. Pharmacol. 11, 393 10.3389/fphar.2020.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Susztak, K. (2014) Understanding the epigenetic syntax for the genetic alphabet in the kidney. J. Am. Soc. Nephrol. 25, 10–17 10.1681/ASN.2013050461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musselman, C.A., Lalonde, M.E., Cote, J. and Kutateladze, T.G. (2012) Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 10.1038/nsmb.2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng, Y., He, C., Wang, M., Ma, X., Mo, F., Yang, S.et al. (2019) Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal. Transduct. Target. Ther. 4, 62 10.1038/s41392-019-0095-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmel, R. and Fiedler, D. (2018) Features and regulation of non-enzymatic post-translational modifications. Nat. Chem. Biol. 14, 244–252 10.1038/nchembio.2575 [DOI] [PubMed] [Google Scholar]
- 16.Gaffney, D.O., Jennings, E.Q., Anderson, C.C., Marentette, J.O., Shi, T., Schou Oxvig, A.M.et al. (2020) Non-enzymatic lysine lactoylation of glycolytic enzymes. Cell Chem. Biol. 27, 206–213.e206 10.1016/j.chembiol.2019.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galligan, J.J., Wepy, J.A., Streeter, M.D., Kingsley, P.J., Mitchener, M.M., Wauchope, O.R.et al. (2018) Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks. Proc. Natl Acad. Sci. U.S.A. 115, 9228–9233 10.1073/pnas.1802901115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James, A.M., Hoogewijs, K., Logan, A., Hall, A.R., Ding, S., Fearnley, I.M.et al. (2017) Non-enzymatic N-acetylation of lysine residues by acetylCoA often occurs via a proximal S-acetylated thiol intermediate sensitive to glyoxalase II. Cell Rep. 18, 2105–2112 10.1016/j.celrep.2017.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moellering, R.E. and Cravatt, B.F. (2013) Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science 341, 549–553 10.1126/science.1238327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng, Q., Omans, N.D., Leicher, R., Osunsade, A., Agustinus, A.S., Finkin-Groner, E.et al. (2019) Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat. Commun. 10, 1289 10.1038/s41467-019-09192-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park, S.H., Ozden, O., Liu, G., Song, H.Y., Zhu, Y., Yan, Y.et al. (2016) SIRT2-Mediated deacetylation and tetramerization of pyruvate kinase directs glycolysis and tumor growth. Cancer Res. 76, 3802–3812 10.1158/0008-5472.CAN-15-2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jennings, E.Q., Ray, J.D., Zerio, C.J., Trujillo, M.N., McDonald, D.M., Chapman, E.et al. (2021) Sirtuin 2 regulates protein lactoylLys modifications. Chembiochem 22, 2102–2106 10.1002/cbic.202000883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendoza, M., Egervari, G., Sidoli, S., Donahue, G., Alexander, D.C., Sen, P.et al. (2022) Enzymatic transfer of acetate on histones from lysine reservoir sites to lysine activating sites. Sci. Adv. 8, eabj5688 10.1126/sciadv.abj5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinert, B.T., Moustafa, T., Iesmantavicius, V., Zechner, R. and Choudhary, C. (2015) Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO J. 34, 2620–2632 10.15252/embj.201591271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dittenhafer-Reed, K.E., Richards, A.L., Fan, J., Smallegan, M.J., Fotuhi Siahpirani, A., Kemmerer, Z.A.et al. (2015) SIRT3 mediates multi-tissue coupling for metabolic fuel switching. Cell Metab. 21, 637–646 10.1016/j.cmet.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramazi, S. and Zahiri, J. (2021) Posttranslational modifications in proteins: resources, tools and prediction methods. Database (Oxford) 2021, baab012 10.1093/database/baab012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khoury, G.A., Baliban, R.C. and Floudas, C.A. (2011) Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep. 1, 90 10.1038/srep00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Z., Li, S., Luo, M., Jhong, J.H., Li, W., Yao, L.et al. (2022) dbPTM in 2022: an updated database for exploring regulatory networks and functional associations of protein post-translational modifications. Nucleic Acids Res. 50, D471–D479 10.1093/nar/gkab1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalyuzhnyy, A., Eyers, P.A., Eyers, C.E., Bowler-Barnett, E., Martin, M.J., Sun, Z.et al. (2022) Profiling the human phosphoproteome to estimate the true extent of protein phosphorylation. J. Proteome Res. 21, 1510–1524 10.1021/acs.jproteome.2c00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trefely, S., Lovell, C.D., Snyder, N.W. and Wellen, K.E. (2020) Compartmentalised acyl-CoA metabolism and roles in chromatin regulation. Mol. Metab. 38, 100941 10.1016/j.molmet.2020.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee, S., Hao, Y.H. and Orth, K. (2007) A newly discovered post-translational modification–the acetylation of serine and threonine residues. Trends Biochem. Sci. 32, 210–216 10.1016/j.tibs.2007.03.007 [DOI] [PubMed] [Google Scholar]
- 32.Trefely, S., Huber, K., Liu, J., Noji, M., Stransky, S., Singh, J.et al. (2022) Quantitative subcellular acyl-CoA analysis reveals distinct nuclear metabolism and isoleucine-dependent histone propionylation. Mol. Cell 82, 447–462.e446 10.1016/j.molcel.2021.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simithy, J., Sidoli, S., Yuan, Z.F., Coradin, M., Bhanu, N.V., Marchione, D.M.et al. (2017) Characterization of histone acylations links chromatin modifications with metabolism. Nat. Commun. 8, 1141 10.1038/s41467-017-01384-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.James, A.M., Smith, A.C., Smith, C.L., Robinson, A.J. and Murphy, M.P. (2018) Proximal cysteines that enhance lysine N-acetylation of cytosolic proteins in mice are less conserved in longer-living species. Cell Rep. 24, 1445–1455 10.1016/j.celrep.2018.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kori, Y., Sidoli, S., Yuan, Z.F., Lund, P.J., Zhao, X. and Garcia, B.A. (2017) Proteome-wide acetylation dynamics in human cells. Sci. Rep. 7, 10296 10.1038/s41598-017-09918-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng, Y., Thomas, P.M. and Kelleher, N.L. (2013) Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nat. Commun. 4, 2203 10.1038/ncomms3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathieson, T., Franken, H., Kosinski, J., Kurzawa, N., Zinn, N., Sweetman, G.et al. (2018) Systematic analysis of protein turnover in primary cells. Nat. Commun. 9, 689 10.1038/s41467-018-03106-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zee, B.M., Levin, R.S., DiMaggio, P.A. and Garcia, B.A. (2010) Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin 3, 22 10.1186/1756-8935-3-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy, M.P., Bayir, H., Belousov, V., Chang, C.J., Davies, K.J.A., Davies, M.J.et al. (2022) Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat. Metab. 4, 651–662 10.1038/s42255-022-00591-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sies, H., Belousov, V.V., Chandel, N.S., Davies, M.J., Jones, D.P., Mann, G.E.et al. (2022) Defining roles of specific reactive oxygen species (ROS) in cell biology and physiology. Nat. Rev. Mol. Cell Biol. 23, 499–515 10.1038/s41580-022-00456-z [DOI] [PubMed] [Google Scholar]
- 41.Conte, L., and Carroll, M. and S, K. (2013) The redox biochemistry of protein sulfenylation and sulfinylation. J. Biol. Chem. 288, 26480–26488 10.1074/jbc.R113.467738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Reest, J., Lilla, S., Zheng, L., Zanivan, S. and Gottlieb, E. (2018) Proteome-wide analysis of cysteine oxidation reveals metabolic sensitivity to redox stress. Nat. Commun. 9, 1581 10.1038/s41467-018-04003-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Go, Y.M., Chandler, J.D. and Jones, D.P. (2015) The cysteine proteome. Free Radic. Biol. Med. 84, 227–245 10.1016/j.freeradbiomed.2015.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan, N., Wang, N., Yu, S., Zhang, H., Tang, S., Wang, D.et al. (2022) Cyclic immonium ion of lactyllysine reveals widespread lactylation in the human proteome. Nat. Methods 19, 854–864 10.1038/s41592-022-01523-1 [DOI] [PubMed] [Google Scholar]
- 45.Nokin, M.J., Durieux, F., Bellier, J., Peulen, O., Uchida, K., Spiegel, D.A.et al. (2017) Hormetic potential of methylglyoxal, a side-product of glycolysis, in switching tumours from growth to death. Sci. Rep. 7, 11722 10.1038/s41598-017-12119-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swatek, K.N. and Komander, D. (2016) Ubiquitin modifications. Cell Res. 26, 399–422 10.1038/cr.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim, W., Bennett, E.J., Huttlin, E.L., Guo, A., Li, J., Possemato, A.et al. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lobato-Gil, S., Heidelberger, J.B., Maghames, C., Bailly, A., Brunello, L., Rodriguez, M.S.et al. (2021) Proteome-wide identification of NEDD8 modification sites reveals distinct proteomes for canonical and atypical NEDDylation. Cell Rep. 34, 108635 10.1016/j.celrep.2020.108635 [DOI] [PubMed] [Google Scholar]
- 49.Perez Berrocal, D.A., Witting, K.F., Ovaa, H. and Mulder, M.P.C. (2019) Hybrid chains: a collaboration of ubiquitin and ubiquitin-like modifiers introducing cross-functionality to the ubiquitin code. Front. Chem. 7, 931 10.3389/fchem.2019.00931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koyano, F., Okatsu, K., Kosako, H., Tamura, Y., Go, E., Kimura, M.et al. (2014) Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 10.1038/nature13392 [DOI] [PubMed] [Google Scholar]
- 51.Leutert, M., Entwisle, S.W. and Villen, J. (2021) Decoding post-translational modification crosstalk With proteomics. Mol. Cell. Proteomics 20, 100129 10.1016/j.mcpro.2021.100129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hunter, T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 53.Lee, J.S., Smith, E. and Shilatifard, A. (2010) The language of histone crosstalk. Cell 142, 682–685 10.1016/j.cell.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kirmizis, A., Santos-Rosa, H., Penkett, C.J., Singer, M.A., Vermeulen, M., Mann, M.et al. (2007) Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449, 928–932 10.1038/nature06160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Migliori, V., Muller, J., Phalke, S., Low, D., Bezzi, M., Mok, W.C.et al. (2012) Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat. Struct. Mol. Biol. 19, 136–144 10.1038/nsmb.2209 [DOI] [PubMed] [Google Scholar]
- 56.Suganuma, T. and Workman, J.L. (2008) Crosstalk among histone modifications. Cell 135, 604–607 10.1016/j.cell.2008.10.036 [DOI] [PubMed] [Google Scholar]
- 57.Egelhofer, T.A., Minoda, A., Klugman, S., Lee, K., Kolasinska-Zwierz, P., Alekseyenko, A.A.et al. (2011) An assessment of histone-modification antibody quality. Nat. Struct. Mol. Biol. 18, 91–93 10.1038/nsmb.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuchs, S.M., Krajewski, K., Baker, R.W., Miller, V.L. and Strahl, B.D. (2011) Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr. Biol. 21, 53–58 10.1016/j.cub.2010.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yates, III, J.R. (2019) Recent technical advances in proteomics. F1000Res. 8, F1000 Faculty Rev-351 10.12688/f1000research.16987.1 [DOI] [Google Scholar]
- 60.Galligan, J.J., Kingsley, P.J., Wauchope, O.R., Mitchener, M.M., Camarillo, J.M., Wepy, J.A.et al. (2017) Quantitative analysis and discovery of lysine and arginine modifications. Anal. Chem. 89, 1299–1306 10.1021/acs.analchem.6b04105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan, M., Luo, H., Lee, S., Jin, F., Yang, J.S., Montellier, E.et al. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 10.1016/j.cell.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee, S., Tan, M., Dai, L., Kwon, O.K., Yang, J.S., Zhao, Y.et al. (2013) MS/MS of synthetic peptide is not sufficient to confirm new types of protein modifications. J. Proteome Res. 12, 1007–1013 10.1021/pr300667e [DOI] [PubMed] [Google Scholar]
- 63.Jiang, T., Zhou, X., Taghizadeh, K., Dong, M. and Dedon, P.C. (2007) N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc. Natl Acad. Sci. U.S.A. 104, 60–65 10.1073/pnas.0606775103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen, Y., Sprung, R., Tang, Y., Ball, H., Sangras, B., Kim, S.C.et al. (2007) Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6, 812–819 10.1074/mcp.M700021-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Batth, T.S., Francavilla, C. and Olsen, J.V. (2014) Off-line high-pH reversed-phase fractionation for in-depth phosphoproteomics. J. Proteome Res. 13, 6176–6186 10.1021/pr500893m [DOI] [PubMed] [Google Scholar]
- 66.Yang, F., Shen, Y., Camp, II, D.G. and Smith, R.D. (2012) High-pH reversed-phase chromatography with fraction concatenation for 2D proteomic analysis. Expert Rev. Proteomics 9, 129–134 10.1586/epr.12.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu, L., Li, X., Jiang, X., Zhou, H., Jiang, X., Kong, L.et al. (2007) Comprehensive peptidome analysis of mouse livers by size exclusion chromatography prefractionation and nanoLC-MS/MS identification. J. Proteome Res. 6, 801–808 10.1021/pr060469e [DOI] [PubMed] [Google Scholar]
- 68.Marino, F., Cristobal, A., Binai, N.A., Bache, N., Heck, A.J. and Mohammed, S. (2014) Characterization and usage of the EASY-spray technology as part of an online 2D SCX-RP ultra-high pressure system. Analyst 139, 6520–6528 10.1039/c4an01568a [DOI] [PubMed] [Google Scholar]
- 69.Wang, C., Ye, M., Wei, X., Bian, Y., Cheng, K. and Zou, H. (2016) A bead-based cleavage method for large-scale identification of protease substrates. Sci. Rep. 6, 22645 10.1038/srep22645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia, B.A., Mollah, S., Ueberheide, B.M., Busby, S.A., Muratore, T.L., Shabanowitz, J.et al. (2007) Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat. Protoc. 2, 933–938 10.1038/nprot.2007.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Plazas-Mayorca, M.D., Zee, B.M., Young, N.L., Fingerman, I.M., LeRoy, G., Briggs, S.D.et al. (2009) One-pot shotgun quantitative mass spectrometry characterization of histones. J. Proteome Res. 8, 5367–5374 10.1021/pr900777e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ali, H.R., Assiri, M.A., Harris, P.S., Michel, C.R., Yun, Y., Marentette, J.O.et al. (2019) Quantifying competition among mitochondrial protein acylation events induced by ethanol metabolism. J. Proteome Res. 18, 1513–1531 10.1021/acs.jproteome.8b00800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al Shweiki, M.R., Monchgesang, S., Majovsky, P., Thieme, D., Trutschel, D. and Hoehenwarter, W. (2017) Assessment of label-free quantification in discovery proteomics and impact of technological factors and natural variability of protein abundance. J. Proteome Res. 16, 1410–1424 10.1021/acs.jproteome.6b00645 [DOI] [PubMed] [Google Scholar]
- 74.Ong, S.E., Mittler, G. and Mann, M. (2004) Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods 1, 119–126 10.1038/nmeth715 [DOI] [PubMed] [Google Scholar]
- 75.Ross, P.L., Huang, Y.N., Marchese, J.N., Williamson, B., Parker, K., Hattan, S.et al. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 10.1074/mcp.M400129-MCP200 [DOI] [PubMed] [Google Scholar]
- 76.Thompson, A., Schafer, J., Kuhn, K., Kienle, S., Schwarz, J., Schmidt, G.et al. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 10.1021/ac0262560 [DOI] [PubMed] [Google Scholar]
- 77.Gao, J., Liao, R., Yu, Y., Zhai, H., Wang, Y., Sack, R.et al. (2014) Absolute quantification of histone PTM marks by MRM-based LC-MS/MS. Anal. Chem. 86, 9679–9686 10.1021/ac502333a [DOI] [PubMed] [Google Scholar]
- 78.Abshiru, N.A., Sikora, J.W., Camarillo, J.M., Morris, J.A., Compton, P.D., Lee, T.et al. (2020) Targeted detection and quantitation of histone modifications from 1,000 cells. PLoS ONE 15, e0240829 10.1371/journal.pone.0240829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sidoli, S., Kori, Y., Lopes, M., Yuan, Z.F., Kim, H.J., Kulej, K.et al. (2019) One minute analysis of 200 histone posttranslational modifications by direct injection mass spectrometry. Genome Res. 29, 978–987 10.1101/gr.247353.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Catherman, A.D., Skinner, O.S. and Kelleher, N.L. (2014) Top down proteomics: facts and perspectives. Biochem. Biophys. Res. Commun. 445, 683–693 10.1016/j.bbrc.2014.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ntai, I., Fornelli, L., DeHart, C.J., Hutton, J.E., Doubleday, P.F., LeDuc, R.D.et al. (2018) Precise characterization of KRAS4b proteoforms in human colorectal cells and tumors reveals mutation/modification cross-talk. Proc. Natl Acad. Sci. U.S.A. 115, 4140–4145 10.1073/pnas.1716122115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hobbs, G.A., Der, C.J. and Rossman, K.L. (2016) RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 129, 1287–1292 10.1242/jcs.182873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Savaryn, J.P., Catherman, A.D., Thomas, P.M., Abecassis, M.M. and Kelleher, N.L. (2013) The emergence of top-down proteomics in clinical research. Genome Med. 5, 53 10.1186/gm457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Melby, J.A., Roberts, D.S., Larson, E.J., Brown, K.A., Bayne, E.F., Jin, S.et al. (2021) Novel strategies to address the challenges in top-down proteomics. J. Am. Soc. Mass Spectrom. 32, 1278–1294 10.1021/jasms.1c00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Craig, R. and Beavis, R.C. (2003) A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 17, 2310–2316 10.1002/rcm.1198 [DOI] [PubMed] [Google Scholar]
- 86.Creasy, D.M. and Cottrell, J.S. (2002) Error tolerant searching of uninterpreted tandem mass spectrometry data. Proteomics 2, 1426–1434 [DOI] [PubMed] [Google Scholar]
- 87.Baliban, R.C., DiMaggio, P.A., Plazas-Mayorca, M.D., Young, N.L., Garcia, B.A. and Floudas, C.A. (2010) A novel approach for untargeted post-translational modification identification using integer linear optimization and tandem mass spectrometry. Mol. Cell. Proteomics 9, 764–779 10.1074/mcp.M900487-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cifani, P., Li, Z., Luo, D., Grivainis, M., Intlekofer, A.M., Fenyo, D.et al. (2021) Discovery of protein modifications using differential tandem mass spectrometry proteomics. J. Proteome Res. 20, 1835–1848 10.1021/acs.jproteome.0c00638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mills, E.L., Ryan, D.G., Prag, H.A., Dikovskaya, D., Menon, D., Zaslona, Z.et al. (2018) Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 10.1038/nature25986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kong, A.T., Leprevost, F.V., Avtonomov, D.M., Mellacheruvu, D. and Nesvizhskii, A.I. (2017) MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 14, 513–520 10.1038/nmeth.4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.An, Z., Zhai, L., Ying, W., Qian, X., Gong, F., Tan, M.et al. (2019) PTMiner: Localization and quality control of protein modifications detected in an open search and its application to comprehensive post-translational modification characterization in human proteome. Mol. Cell. Proteomics 18, 391–405 10.1074/mcp.RA118.000812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang, H., Zhang, D., Wang, Y., Perez-Neut, M., Han, Z., Zheng, Y.G.et al. (2018) Lysine benzoylation is a histone mark regulated by SIRT2. Nat. Commun. 9, 3374 10.1038/s41467-018-05567-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen, Y., Chen, W., Cobb, M.H. and Zhao, Y. (2009) PTMap–a sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. Proc. Natl Acad. Sci. U.S.A. 106, 761–766 10.1073/pnas.0811739106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang, Z., Tan, M., Xie, Z., Dai, L., Chen, Y. and Zhao, Y. (2011) Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 10.1038/nchembio.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang, D., Tang, Z., Huang, H., Zhou, G., Cui, C., Weng, Y.et al. (2019) Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 10.1038/s41586-019-1678-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dai, L., Peng, C., Montellier, E., Lu, Z., Chen, Y., Ishii, H.et al. (2014) Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 10, 365–370 10.1038/nchembio.1497 [DOI] [PubMed] [Google Scholar]
- 97.Lund, P.J., Kori, Y., Zhao, X., Sidoli, S., Yuan, Z.F. and Garcia, B.A. (2019) Isotopic labeling and quantitative proteomics of acetylation on histones and beyond. Methods Mol. Biol. 1977, 43–70 10.1007/978-1-4939-9232-4_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hang, H.C., Yu, C., Kato, D.L. and Bertozzi, C.R. (2003) A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc. Natl Acad. Sci. U.S.A. 100, 14846–14851 10.1073/pnas.2335201100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao, S., Xu, W., Jiang, W., Yu, W., Lin, Y., Zhang, T.et al. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 10.1126/science.1179689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Couvertier, S.M., Zhou, Y. and Weerapana, E. (2014) Chemical-proteomic strategies to investigate cysteine posttranslational modifications. Biochim. Biophys. Acta 1844, 2315–2330 10.1016/j.bbapap.2014.09.024 [DOI] [PubMed] [Google Scholar]
- 101.Parker, C.G. and Pratt, M.R. (2020) Click chemistry in proteomic investigations. Cell 180, 605–632 10.1016/j.cell.2020.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang, H., Zhang, X., Chen, X., Aramsangtienchai, P., Tong, Z. and Lin, H. (2018) Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev. 118, 919–988 10.1021/acs.chemrev.6b00750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weerapana, E., Wang, C., Simon, G.M., Richter, F., Khare, S., Dillon, M.B.et al. (2010) Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 10.1038/nature09472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hacker, S.M., Backus, K.M., Lazear, M.R., Forli, S., Correia, B.E. and Cravatt, B.F. (2017) Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 9, 1181–1190 10.1038/nchem.2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wanigasekara, M.S.K., Huang, X., Chakrabarty, J.K., Bugarin, A. and Chowdhury, S.M. (2018) Arginine-selective chemical labeling approach for identification and enrichment of reactive arginine residues in proteins. ACS Omega 3, 14229–14235 10.1021/acsomega.8b01729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun, F., Suttapitugsakul, S. and Wu, R. (2021) An Azo coupling-Based chemoproteomic approach to systematically profile the tyrosine reactivity in the human proteome. Anal. Chem. 93, 10334–10342 10.1021/acs.analchem.1c01935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin, S., Yang, X., Jia, S., Weeks, A.M., Hornsby, M., Lee, P.S.et al. (2017) Redox-based reagents for chemoselective methionine bioconjugation. Science 355, 597–602 10.1126/science.aal3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nelson, K.J., Klomsiri, C., Codreanu, S.G., Soito, L., Liebler, D.C., Rogers, L.C.et al. (2010) Use of dimedone-based chemical probes for sulfenic acid detection methods to visualize and identify labeled proteins. Methods Enzymol. 473, 95–115 10.1016/S0076-6879(10)73004-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leonard, S.E., Reddie, K.G. and Carroll, K.S. (2009) Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem. Biol. 4, 783–799 10.1021/cb900105q [DOI] [PubMed] [Google Scholar]
- 110.Thingholm, T.E. and Larsen, M.R. (2016) The Use of titanium dioxide for selective enrichment of phosphorylated peptides. Methods Mol. Biol. 1355, 135–146 10.1007/978-1-4939-3049-4_9 [DOI] [PubMed] [Google Scholar]
- 111.Graham, M.E., Thaysen-Andersen, M., Bache, N., Craft, G.E., Larsen, M.R., Packer, N.H.et al. (2011) A novel post-translational modification in nerve terminals: O-linked N-acetylglucosamine phosphorylation. J. Proteome Res. 10, 2725–2733 10.1021/pr1011153 [DOI] [PubMed] [Google Scholar]
- 112.Humphrey, S.J., Karayel, O., James, D.E. and Mann, M. (2018) High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat. Protoc. 13, 1897–1916 10.1038/s41596-018-0014-9 [DOI] [PubMed] [Google Scholar]
- 113.Kornfeld, R. and Kornfeld, S. (1985) Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54, 631–664 10.1146/annurev.bi.54.070185.003215 [DOI] [PubMed] [Google Scholar]
- 114.Valliere-Douglass, J.F., Eakin, C.M., Wallace, A., Ketchem, R.R., Wang, W., Treuheit, M.J.et al. (2010) Glutamine-linked and non-consensus asparagine-linked oligosaccharides present in human recombinant antibodies define novel protein glycosylation motifs. J. Biol. Chem. 285, 16012–16022 10.1074/jbc.M109.096412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Julenius, K., Molgaard, A., Gupta, R. and Brunak, S. (2005) Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15, 153–164 10.1093/glycob/cwh151 [DOI] [PubMed] [Google Scholar]
- 116.Weinert, B.T., Scholz, C., Wagner, S.A., Iesmantavicius, V., Su, D., Daniel, J.A.et al. (2013) Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851 10.1016/j.celrep.2013.07.024 [DOI] [PubMed] [Google Scholar]
- 117.Choudhary, C., Kumar, C., Gnad, F., Nielsen, M.L., Rehman, M., Walther, T.C.et al. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- 118.Xu, W., Wan, J., Zhan, J., Li, X., He, H., Shi, Z.et al. (2017) Global profiling of crotonylation on non-histone proteins. Cell Res. 27, 946–949 10.1038/cr.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang, H., Tang, S., Ji, M., Tang, Z., Shimada, M., Liu, X.et al. (2018) p300-mediated lysine 2-hydroxyisobutyrylation regulates glycolysis. Molecular cell. 70, 663–678.e666 10.1016/j.molcel.2018.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tan, D., Wei, W., Han, Z., Ren, X., Yan, C., Qi, S.et al. (2022) HBO1 catalyzes lysine benzoylation in mammalian cells. iScience 25, 105443 10.1016/j.isci.2022.105443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang, J., Gupta, V., Tallman, K.A., Porter, N.A., Carroll, K.S. and Liebler, D.C. (2015) Global, in situ, site-specific analysis of protein S-sulfenylation. Nat. Protoc. 10, 1022–1037 10.1038/nprot.2015.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reisz, J.A., Wither, M.J., Dzieciatkowska, M., Nemkov, T., Issaian, A., Yoshida, T.et al. (2016) Oxidative modifications of glyceraldehyde 3-phosphate dehydrogenase regulate metabolic reprogramming of stored red blood cells. Blood 128, e32–e42 10.1182/blood-2016-05-714816 [DOI] [PubMed] [Google Scholar]
- 123.Chakravarti, R., Aulak, K.S., Fox, P.L. and Stuehr, D.J. (2010) GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc. Natl Acad. Sci. U.S.A. 107, 18004–18009 10.1073/pnas.1008133107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Barinova, K.V., Serebryakova, M.V., Eldarov, M.A., Kulikova, A.A., Mitkevich, V.A., Muronetz, V.I.et al. (2020) S-glutathionylation of human glyceraldehyde-3-phosphate dehydrogenase and possible role of Cys152-Cys156 disulfide bridge in the active site of the protein. Biochim. Biophys. Acta. Gen. Subj. 1864, 129560 10.1016/j.bbagen.2020.129560 [DOI] [PubMed] [Google Scholar]
- 125.Kornberg, M.D., Bhargava, P., Kim, P.M., Putluri, V., Snowman, A.M., Putluri, N.et al. (2018) Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453 10.1126/science.aan4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hirschey, M.D., Shimazu, T., Goetzman, E., Jing, E., Schwer, B., Lombard, D.B.et al. (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 10.1038/nature08778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Davidson, M.T., Grimsrud, P.A., Lai, L., Draper, J.A., Fisher-Wellman, K.H., Narowski, T.M.et al. (2020) Extreme acetylation of the cardiac mitochondrial proteome does not promote heart failure. Circ. Res. 127, 1094–1108 10.1161/CIRCRESAHA.120.317293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gil, J., Ramirez-Torres, A., Chiappe, D., Luna-Penaloza, J., Fernandez-Reyes, F.C., Arcos-Encarnacion, B.et al. (2017) Lysine acetylation stoichiometry and proteomics analyses reveal pathways regulated by sirtuin 1 in human cells. J. Biol. Chem. 292, 18129–18144 10.1074/jbc.M117.784546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fisher-Wellman, K.H., Draper, J.A., Davidson, M.T., Williams, A.S., Narowski, T.M., Slentz, D.H.et al. (2019) Respiratory phenomics across multiple models of protein hyperacylation in cardiac mitochondria reveals a marginal impact on bioenergetics. Cell Rep. 26, 1557–1572.e1558 10.1016/j.celrep.2019.01.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Miller, K.D., Pniewski, K., Perry, C.E., Papp, S.B., Shaffer, J.D., Velasco-Silva, J.N.et al. (2021) Targeting ACSS2 with a transition-State mimetic inhibits triple-negative breast cancer growth. Cancer Res. 81, 1252–1264 10.1158/0008-5472.CAN-20-1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sabari, B.R., Tang, Z., Huang, H., Yong-Gonzalez, V., Molina, H., Kong, H.E.et al. (2015) Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 58, 203–215 10.1016/j.molcel.2015.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hao, F., Tian, M., Zhang, X., Jin, X., Jiang, Y., Sun, X.et al. (2021) Butyrate enhances CPT1A activity to promote fatty acid oxidation and iTreg differentiation. Proc. Natl Acad. Sci. U.S.A. 118, e2014681118 10.1073/pnas.2014681118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin, Y.H., Schmidt, W., Fritz, K.S., Jeong, M.Y., Cammarato, A., Foster, D.B.et al. (2020) Site-specific acetyl-mimetic modification of cardiac troponin I modulates myofilament relaxation and calcium sensitivity. J. Mol. Cell. Cardiol. 139, 135–147 10.1016/j.yjmcc.2020.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li, M., Luo, J., Brooks, C.L. and Gu, W. (2002) Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 277, 50607–50611 10.1074/jbc.C200578200 [DOI] [PubMed] [Google Scholar]
- 135.Wang, L., Xie, J. and Schultz, P.G. (2006) Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 35, 225–249 10.1146/annurev.biophys.35.101105.121507 [DOI] [PubMed] [Google Scholar]
- 136.Sun, Y., Chen, Y., Xu, Y., Zhang, Y., Lu, M., Li, M.et al. (2022) Genetic encoding of epsilon-N-L-lactyllysine for detecting delactylase activity in living cells. Chem. Commun. (Camb) 58, 8544–8547 10.1039/d2cc02643k [DOI] [PubMed] [Google Scholar]
- 137.Li, Y., Wang, S., Chen, Y., Li, M., Dong, X., Hang, H.C.et al. (2020) Site-specific chemical fatty-acylation for gain-of-function analysis of protein S-palmitoylation in live cells. Chem. Commun. (Camb) 56, 13880–13883 10.1039/d0cc06073a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Burton, A.J., Haugbro, M., Gates, L.A., Bagert, J.D., Allis, C.D. and Muir, T.W. (2020) In situ chromatin interactomics using a chemical bait and trap approach. Nat. Chem. 12, 520–527 10.1038/s41557-020-0474-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shah, N.H., Dann, G.P., Vila-Perello, M., Liu, Z. and Muir, T.W. (2012) Ultrafast protein splicing is common among cyanobacterial split inteins: implications for protein engineering. J. Am. Chem. Soc. 134, 11338–11341 10.1021/ja303226x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.David, Y., Vila-Perello, M., Verma, S. and Muir, T.W. (2015) Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins. Nat. Chem. 7, 394–402 10.1038/nchem.2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Peng, C., Lu, Z., Xie, Z., Cheng, Z., Chen, Y., Tan, M.et al. (2011) The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics 10, M111 012658 10.1074/mcp.M111.012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tan, M., Peng, C., Anderson, K.A., Chhoy, P., Xie, Z., Dai, L.et al. (2014) Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 19, 605–617 10.1016/j.cmet.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shaw, P.G., Chaerkady, R., Zhang, Z., Davidson, N.E. and Pandey, A. (2011) Monoclonal antibody cocktail as an enrichment tool for acetylome analysis. Anal. Chem. 83, 3623–3626 10.1021/ac1026176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Colak, G., Pougovkina, O., Dai, L., Tan, M., Te Brinke, H., Huang, H.et al. (2015) Proteomic and biochemical studies of lysine malonylation suggest its malonic aciduria-associated regulatory role in mitochondrial function and fatty acid oxidation. Mol. Cell. Proteomics 14, 3056–3071 10.1074/mcp.M115.048850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Huang, H., Luo, Z., Qi, S., Huang, J., Xu, P., Wang, X.et al. (2018) Landscape of the regulatory elements for lysine 2-hydroxyisobutyrylation pathway. Cell Res. 28, 111–125 10.1038/cr.2017.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang, H., Zhang, D., Weng, Y., Delaney, K., Tang, Z., Yan, C.et al. (2021) The regulatory enzymes and protein substrates for the lysine beta-hydroxybutyrylation pathway. Sci. Adv. 7, eabe2771 10.1126/sciadv.abe2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu, Q., Li, W., Wang, C., Fan, P., Cao, L., Wu, Z.et al. (2017) Ultradeep lysine crotonylome reveals the crotonylation enhancement on both histones and nonhistone proteins by SAHA treatment. J. Proteome Res. 16, 3664–3671 10.1021/acs.jproteome.7b00380 [DOI] [PubMed] [Google Scholar]
- 148.Fuhrmann, J. and Thompson, P.R. (2016) Protein arginine methylation and citrullination in epigenetic regulation. ACS Chem. Biol. 11, 654–668 10.1021/acschembio.5b00942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chang, B., Chen, Y., Zhao, Y. and Bruick, R.K. (2007) JMJD6 is a histone arginine demethylase. Science 318, 444–447 10.1126/science.1145801 [DOI] [PubMed] [Google Scholar]
- 150.Islam, M.S., McDonough, M.A., Chowdhury, R., Gault, J., Khan, A., Pires, E.et al. (2019) Biochemical and structural investigations clarify the substrate selectivity of the 2-oxoglutarate oxygenase JMJD6. J. Biol. Chem. 294, 11637–11652 10.1074/jbc.RA119.008693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Walport, L.J., Hopkinson, R.J., Chowdhury, R., Schiller, R., Ge, W., Kawamura, A.et al. (2016) Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 7, 11974 10.1038/ncomms11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jarrold, J. and Davies, C.C. (2019) PRMTs and arginine methylation: Cancer's best-Kept secret? Trends Mol. Med. 25, 993–1009 10.1016/j.molmed.2019.05.007 [DOI] [PubMed] [Google Scholar]
- 153.Uhlmann, T., Geoghegan, V.L., Thomas, B., Ridlova, G., Trudgian, D.C. and Acuto, O. (2012) A method for large-scale identification of protein arginine methylation. Mol. Cell. Proteomics 11, 1489–1499 10.1074/mcp.M112.020743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tessarz, P., Santos-Rosa, H., Robson, S.C., Sylvestersen, K.B., Nelson, C.J., Nielsen, M.L.et al. (2014) Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 505, 564–568 10.1038/nature12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Luo, M. (2018) Chemical and biochemical perspectives of protein lysine methylation. Chem. Rev. 118, 6656–6705 10.1021/acs.chemrev.8b00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dai, S., Holt, M.V., Horton, J.R., Woodcock, C.B., Patel, A., Zhang, X.et al. (2020) Characterization of SETD3 methyltransferase-mediated protein methionine methylation. J. Biol. Chem. 295, 10901–10910 10.1074/jbc.RA120.014072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Davydova, E., Shimazu, T., Schuhmacher, M.K., Jakobsson, M.E., Willemen, H., Liu, T.et al. (2021) The methyltransferase METTL9 mediates pervasive 1-methylhistidine modification in mammalian proteomes. Nat. Commun. 12, 891 10.1038/s41467-020-20670-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wilkinson, A.W., Diep, J., Dai, S., Liu, S., Ooi, Y.S., Song, D.et al. (2019) SETD3 is an actin histidine methyltransferase that prevents primary dystocia. Nature 565, 372–376 10.1038/s41586-018-0821-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Niwa, T. (1999) 3-Deoxyglucosone: metabolism, analysis, biological activity, and clinical implication. J. Chromatogr. B Biomed. Sci. Appl. 731, 23–36 10.1016/s0378-4347(99)00113-9 [DOI] [PubMed] [Google Scholar]
- 160.Ray, D.M., Jennings, E.Q., Maksimovic, I., Chai, X., Galligan, J.J., David, Y.et al. (2022) Chemical labeling and enrichment of histone glyoxal adducts. ACS Chem. Biol. 17, 756–761 10.1021/acschembio.1c00864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.McEwen, J.M., Fraser, S., Guir, A.L.S., Dave, J. and Scheck, R.A. (2021) Synergistic sequence contributions bias glycation outcomes. Nat. Commun. 12, 3316 10.1038/s41467-021-23625-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sjoblom, N.M., Kelsey, M.M.G. and Scheck, R.A. (2018) A systematic study of selective protein glycation. Angew. Chem. Int. Ed. Engl. 57, 16077–16082 10.1002/anie.201810037 [DOI] [PubMed] [Google Scholar]
- 163.Maksimovic, I., Zheng, Q., Trujillo, M.N., Galligan, J.J. and David, Y. (2020) An azidoribose probe to track ketoamine adducts in histone ribose glycation. J. Am. Chem. Soc. 142, 9999–10007 10.1021/jacs.0c01325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sanghvi, V.R., Leibold, J., Mina, M., Mohan, P., Berishaj, M., Li, Z.et al. (2019) The oncogenic action of NRF2 depends on de-glycation by fructosamine-3-kinase. Cell 178, 807–819.e821 10.1016/j.cell.2019.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wakim, B.T. and Aswad, G.D. (1994) Ca2+-calmodulin-dependent phosphorylation of arginine in histone 3 by a nuclear kinase from mouse leukemia cells. J. Biol. Chem. 269, 2722–2727 PMID: [PubMed] [Google Scholar]
- 166.Ubersax, J.A. and Ferrell, Jr, J.E. (2007) Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541 10.1038/nrm2203 [DOI] [PubMed] [Google Scholar]
- 167.Tarrant, M.K. and Cole, P.A. (2009) The chemical biology of protein phosphorylation. Annu. Rev. Biochem. 78, 797–825 10.1146/annurev.biochem.78.070907.103047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Fuhs, S.R., Meisenhelder, J., Aslanian, A., Ma, L., Zagorska, A., Stankova, M.et al. (2015) Monoclonal 1- and 3-phosphohistidine antibodies: new tools to study histidine phosphorylation. Cell 162, 198–210 10.1016/j.cell.2015.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hunter, T. (2022) A journey from phosphotyrosine to phosphohistidine and beyond. Mol. Cell 82, 2190–2200 10.1016/j.molcel.2022.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bhandari, R., Saiardi, A., Ahmadibeni, Y., Snowman, A.M., Resnick, A.C., Kristiansen, T.Z.et al. (2007) Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc. Natl Acad. Sci. U.S.A. 104, 15305–15310 10.1073/pnas.0707338104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chanduri, M., Rai, A., Malla, A.B., Wu, M., Fiedler, D., Mallik, R.et al. (2016) Inositol hexakisphosphate kinase 1 (IP6K1) activity is required for cytoplasmic dynein-driven transport. Biochem. J. 473, 3031–3047 10.1042/BCJ20160610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Saiardi, A. (2016) Protein pyrophosphorylation: moving forward. Biochem. J. 473, 3765–3768 10.1042/BCJ20160710C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Azevedo, C., Desfougeres, Y., Jiramongkol, Y., Partington, H., Trakansuebkul, S., Singh, J.et al. (2020) Development of a yeast model to study the contribution of vacuolar polyphosphate metabolism to lysine polyphosphorylation. J. Biol. Chem. 295, 1439–1451 10.1074/jbc.RA119.011680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Azevedo, C., Livermore, T. and Saiardi, A. (2015) Protein polyphosphorylation of lysine residues by inorganic polyphosphate. Mol. Cell 58, 71–82 10.1016/j.molcel.2015.02.010 [DOI] [PubMed] [Google Scholar]
- 175.Rodriguez, J., Haydinger, C.D., Peet, D.J., Nguyen, L.K. and von Kriegsheim, A. (2020) Asparagine hydroxylation is a reversible post-translational modification. Mol. Cell. Proteomics 19, 1777–1789 10.1074/mcp.RA120.002189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mondal, S. and Thompson, P.R. (2019) Protein arginine deiminases (PADs): biochemistry and chemical biology of protein citrullination. Acc. Chem. Res. 52, 818–832 10.1021/acs.accounts.9b00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wolhuter, K., Whitwell, H.J., Switzer, C.H., Burgoyne, J.R., Timms, J.F. and Eaton, P. (2018) Evidence against stable protein S-nitrosylation as a widespread mechanism of post-translational regulation. Mol. Cell 69, 438–450.e435 10.1016/j.molcel.2017.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gould, N.S., Evans, P., Martinez-Acedo, P., Marino, S.M., Gladyshev, V.N., Carroll, K.S.et al. (2015) Site-specific proteomic mapping identifies selectively modified regulatory cysteine residues in functionally distinct protein networks. Chem. Biol. 22, 965–975 10.1016/j.chembiol.2015.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mustafa, A.K., Gadalla, M.M., Sen, N., Kim, S., Mu, W., Gazi, S.K.et al. (2009) H2s signals through protein S-sulfhydration. Sci. Signal. 2, ra72 10.1126/scisignal.2000464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lin, H., Su, X. and He, B. (2012) Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem. Biol. 7, 947–960 10.1021/cb3001793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gadgil, H.S., Bondarenko, P.V., Pipes, G.D., Dillon, T.M., Banks, D., Abel, J.et al. (2006) Identification of cysteinylation of a free cysteine in the Fab region of a recombinant monoclonal IgG1 antibody using Lys-C limited proteolysis coupled with LC/MS analysis. Anal. Biochem. 355, 165–174 10.1016/j.ab.2006.05.037 [DOI] [PubMed] [Google Scholar]
- 182.Jakubowski, H. (2019) Homocysteine modification in protein structure/function and human disease. Physiol. Rev. 99, 555–604 10.1152/physrev.00003.2018 [DOI] [PubMed] [Google Scholar]
- 183.Grek, C.L., Zhang, J., Manevich, Y., Townsend, D.M. and Tew, K.D. (2013) Causes and consequences of cysteine S-glutathionylation. J. Biol. Chem. 288, 26497–26504 10.1074/jbc.R113.461368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Unoki, M., Masuda, A., Dohmae, N., Arita, K., Yoshimatsu, M., Iwai, Y.et al. (2013) Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6). J. Biol. Chem. 288, 6053–6062 10.1074/jbc.M112.433284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mantri, M., Loik, N.D., Hamed, R.B., Claridge, T.D., McCullagh, J.S. and Schofield, C.J. (2011) The 2-oxoglutarate-dependent oxygenase JMJD6 catalyses oxidation of lysine residues to give 5S-hydroxylysine residues. Chembiochem 12, 531–534 10.1002/cbic.201000641 [DOI] [PubMed] [Google Scholar]
- 186.Serra-Bardenys, G. and Peiro, S. (2022) Enzymatic lysine oxidation as a posttranslational modification. FEBS J. 289, 8020–8031 10.1111/febs.16205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.King, D.T., Zhu, S., Hardie, D.B., Serrano-Negron, J.E., Madden, Z., Kolappan, S.et al. (2022) Chemoproteomic identification of CO(2)-dependent lysine carboxylation in proteins. Nat. Chem. Biol. 18, 782–791 10.1038/s41589-022-01043-1 [DOI] [PubMed] [Google Scholar]
- 188.Ayombil, F. and Camire, R.M. (2020) Insights into vitamin K-dependent carboxylation: home field advantage. Haematologica 105, 1996–1998 10.3324/haematol.2020.253690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang, N., Li, B.Q., Gao, S., Ruan, J.S. and Cai, Y.D. (2012) Computational prediction and analysis of protein gamma-carboxylation sites based on a random forest method. Mol. Biosyst. 8, 2946–2955 10.1039/c2mb25185j [DOI] [PubMed] [Google Scholar]
- 190.Zhang, Q., Bai, B., Mei, X., Wan, C., Cao, H., Dan, L.et al. (2018) Elevated H3K79 homocysteinylation causes abnormal gene expression during neural development and subsequent neural tube defects. Nat. Commun. 9, 3436 10.1038/s41467-018-05451-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lim, J.C., You, Z., Kim, G. and Levine, R.L. (2011) Methionine sulfoxide reductase A is a stereospecific methionine oxidase. Proc. Natl Acad. Sci. U.S.A. 108, 10472–10477 10.1073/pnas.1101275108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Badgett, M.J., Boyes, B. and Orlando, R. (2017) The separation and quantitation of peptides with and without oxidation of methionine and deamidation of asparagine using hydrophilic interaction liquid chromatography with mass spectrometry (HILIC-MS). J. Am. Soc. Mass Spectrom. 28, 818–826 10.1007/s13361-016-1565-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Arsenault, P.R., Heaton-Johnson, K.J., Li, L.S., Song, D., Ferreira, V.S., Patel, N.et al. (2015) Identification of prolyl hydroxylation modifications in mammalian cell proteins. Proteomics 15, 1259–1267 10.1002/pmic.201400398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Barrera, G., Pizzimenti, S., Ciamporcero, E.S., Daga, M., Ullio, C., Arcaro, A.et al. (2015) Role of 4-hydroxynonenal-protein adducts in human diseases. Antioxid. Redox Signal. 22, 1681–1702 10.1089/ars.2014.6166 [DOI] [PubMed] [Google Scholar]
- 195.Zhu, X. and Sayre, L.M. (2007) Long-lived 4-oxo-2-enal-derived apparent lysine michael adducts are actually the isomeric 4-ketoamides. Chem. Res. Toxicol. 20, 165–170 10.1021/tx600295j [DOI] [PubMed] [Google Scholar]
- 196.Galligan, J.J., Rose, K.L., Beavers, W.N., Hill, S., Tallman, K.A., Tansey, W.P.et al. (2014) Stable histone adduction by 4-oxo-2-nonenal: a potential link between oxidative stress and epigenetics. J. Am. Chem. Soc. 136, 11864–11866 10.1021/ja503604t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cui, Y., Li, X., Lin, J., Hao, Q. and Li, X.D. (2017) Histone ketoamide adduction by 4-Oxo-2-nonenal is a reversible posttranslational modification regulated by Sirt2. ACS Chem. Biol. 12, 47–51 10.1021/acschembio.6b00713 [DOI] [PubMed] [Google Scholar]
- 198.Abo, M., Li, C. and Weerapana, E. (2018) Isotopically-labeled iodoacetamide-alkyne probes for quantitative cysteine-reactivity profiling. Mol. Pharm. 15, 743–749 10.1021/acs.molpharmaceut.7b00832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Taylor, S.W., Fahy, E., Murray, J., Capaldi, R.A. and Ghosh, S.S. (2003) Oxidative post-translational modification of tryptophan residues in cardiac mitochondrial proteins. J. Biol. Chem. 278, 19587–19590 10.1074/jbc.C300135200 [DOI] [PubMed] [Google Scholar]
- 200.Campolo, N., Issoglio, F.M., Estrin, D.A., Bartesaghi, S. and Radi, R. (2020) 3-Nitrotyrosine and related derivatives in proteins: precursors, radical intermediates and impact in function. Essays Biochem. 64, 111–133 10.1042/EBC20190052 [DOI] [PubMed] [Google Scholar]
- 201.Stewart, V. and Ronald, P.C. (2022) Sulfotyrosine residues: Interaction specificity determinants for extracellular protein-protein interactions. J. Biol. Chem. 298, 102232 10.1016/j.jbc.2022.102232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fujimoto, T., Stroud, E., Whatley, R.E., Prescott, S.M., Muszbek, L., Laposata, M.et al. (1993) P-selectin is acylated with palmitic acid and stearic acid at cysteine 766 through a thioester linkage. J. Biol. Chem. 268, 11394–11400 PMID: [PubMed] [Google Scholar]
- 203.Okubo, K., Hamasaki, N., Hara, K. and Kageura, M. (1991) Palmitoylation of cysteine 69 from the COOH-terminal of band 3 protein in the human erythrocyte membrane. acylation occurs in the middle of the consensus sequence of F–I-IICLAVL found in band 3 protein and G2 protein of Rift Valley fever virus. J. Biol. Chem. 266, 16420–16424 PMID: [PubMed] [Google Scholar]
- 204.Madsen, A.S., Andersen, C., Daoud, M., Anderson, K.A., Laursen, J.S., Chakladar, S.et al. (2016) Investigating the sensitivity of NAD+-dependent sirtuin deacylation activities to NADH. J. Biol. Chem. 291, 7128–7141 10.1074/jbc.M115.668699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wisniewski, J.R., Zougman, A. and Mann, M. (2008) Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. 36, 570–577 10.1093/nar/gkm1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Marmorstein, R. and Zhou, M.M. (2014) Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6, a018762 10.1101/cshperspect.a018762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sabari, B.R., Zhang, D., Allis, C.D. and Zhao, Y. (2017) Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 18, 90–101 10.1038/nrm.2016.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kulkarni, R.A., Worth, A.J., Zengeya, T.T., Shrimp, J.H., Garlick, J.M., Roberts, A.M.et al. (2017) Discovering targets of non-enzymatic acylation by thioester reactivity profiling. Cell Chem. Biol. 24, 231–242 10.1016/j.chembiol.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Moreno-Yruela, C., Zhang, D., Wei, W., Baek, M., Liu, W., Gao, J.et al. (2022) Class I histone deacetylases (HDAC1-3) are histone lysine delactylases. Sci. Adv. 8, eabi6696 10.1126/sciadv.abi6696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Moreno-Yruela, C., Galleano, I., Madsen, A.S. and Olsen, C.A. (2018) Histone deacetylase 11 Is an epsilon-N-myristoyllysine hydrolase. Cell Chem. Biol. 25, 849–856.e848 10.1016/j.chembiol.2018.04.007 [DOI] [PubMed] [Google Scholar]
- 211.Jiang, G., Li, C., Lu, M., Lu, K. and Li, H. (2021) Protein lysine crotonylation: past, present, perspective. Cell Death Dis. 12, 703 10.1038/s41419-021-03987-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wagner, G.R., Bhatt, D.P., O'Connell, T.M., Thompson, J.W., Dubois, L.G., Backos, D.S.et al. (2017) A class of reactive acyl-CoA species reveals the non-enzymatic origins of protein acylation. Cell Metab. 25, 823–837.e828 10.1016/j.cmet.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhu, Z., Han, Z., Halabelian, L., Yang, X., Ding, J., Zhang, N.et al. (2021) Identification of lysine isobutyrylation as a new histone modification mark. Nucleic Acids Res. 49, 177–189 10.1093/nar/gkaa1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Baldensperger, T., Sanzo, S.D., Ori, A. and Glomb, M.A. (2019) Quantitation of reactive acyl-CoA species mediated protein acylation by HPLC-MS/MS. Anal. Chem. 91, 12336–12343 10.1021/acs.analchem.9b02656 [DOI] [PubMed] [Google Scholar]
- 215.Montgomery, D.C., Sorum, A.W. and Meier, J.L. (2015) Defining the orphan functions of lysine acetyltransferases. ACS Chem. Biol. 10, 85–94 10.1021/cb500853p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhang, R., Bons, J., Scheidemantle, G., Liu, X., Bielska, O., Carrico, C.et al. (2023) Histone malonylation is regulated by SIRT5 and KAT2A. iScience 26, 106193 10.1016/j.isci.2023.106193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Xie, Z., Zhang, D., Chung, D., Tang, Z., Huang, H., Dai, L.et al. (2016) Metabolic regulation of gene expression by histone lysine beta-hydroxybutyrylation. Mol. Cell 62, 194–206 10.1016/j.molcel.2016.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kutil, Z., Novakova, Z., Meleshin, M., Mikesova, J., Schutkowski, M. and Barinka, C. (2018) Histone deacetylase 11 is a fatty-acid deacylase. ACS Chem. Biol. 13, 685–693 10.1021/acschembio.7b00942 [DOI] [PubMed] [Google Scholar]
- 219.Du, J., Zhou, Y., Su, X., Yu, J.J., Khan, S., Jiang, H.et al. (2011) Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 10.1126/science.1207861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rardin, M.J., He, W., Nishida, Y., Newman, J.C., Carrico, C., Danielson, S.R.et al. (2013) SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 18, 920–933 10.1016/j.cmet.2013.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kurmi, K., Hitosugi, S., Wiese, E.K., Boakye-Agyeman, F., Gonsalves, W.I., Lou, Z.et al. (2018) Carnitine palmitoyltransferase 1A has a lysine succinyltransferase activity. Cell Rep. 22, 1365–1373 10.1016/j.celrep.2018.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Anderson, K.A., Huynh, F.K., Fisher-Wellman, K., Stuart, J.D., Peterson, B.S., Douros, J.D.et al. (2017) SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab. 25, 838–855.e815 10.1016/j.cmet.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Jiang, H., Khan, S., Wang, Y., Charron, G., He, B., Sebastian, C.et al. (2013) SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 10.1038/nature12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Aramsangtienchai, P., Spiegelman, N.A., He, B., Miller, S.P., Dai, L., Zhao, Y.et al. (2016) HDAC8 catalyzes the hydrolysis of long chain fatty acyl lysine. ACS Chem. Biol. 11, 2685–2692 10.1021/acschembio.6b00396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Xie, Y., Chen, L., Wang, R., Wang, J., Li, J., Xu, W.et al. (2019) Chemical probes reveal Sirt2's new function as a robust ‘eraser’ of lysine lipoylation. J. Am. Chem. Soc. 141, 18428–18436 10.1021/jacs.9b06913 [DOI] [PubMed] [Google Scholar]
- 226.Cao, X., Zhu, L., Song, X., Hu, Z. and Cronan, J.E. (2018) Protein moonlighting elucidates the essential human pathway catalyzing lipoic acid assembly on its cognate enzymes. Proc. Natl Acad. Sci. U.S.A. 115, E7063–E7072 10.1073/pnas.1805862115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Jing, H., Zhang, X., Wisner, S.A., Chen, X., Spiegelman, N.A., Linder, M.E.et al. (2017) SIRT2 and lysine fatty acylation regulate the transforming activity of K-Ras4a. eLife 6, e32436 10.7554/eLife.32436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kosciuk, T., Price, I.R., Zhang, X., Zhu, C., Johnson, K.N., Zhang, S.et al. (2020) NMT1 and NMT2 are lysine myristoyltransferases regulating the ARF6 GTPase cycle. Nat. Commun. 11, 1067 10.1038/s41467-020-14893-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Huq, M.D., Tsai, N.P., Lin, Y.P., Higgins, L. and Wei, L.N. (2007) Vitamin B6 conjugation to nuclear corepressor RIP140 and its role in gene regulation. Nat. Chem. Biol. 3, 161–165 10.1038/nchembio861 [DOI] [PubMed] [Google Scholar]
- 230.Zhang, K., Chen, Y., Zhang, Z., Tao, S., Zhu, H. and Zhao, Y. (2010) Unrestrictive identification of non-phosphorylation PTMs in yeast kinases by MS and PTMap. Proteomics 10, 896–903 10.1002/pmic.200900510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Britton, L.M., Newhart, A., Bhanu, N.V., Sridharan, R., Gonzales-Cope, M., Plath, K.et al. (2013) Initial characterization of histone H3 serine 10 O-acetylation. Epigenetics 8, 1101–1113 10.4161/epi.26025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Davis, T.R., Pierce, M.R., Novak, S.X. and Hougland, J.L. (2021) Ghrelin octanoylation by ghrelin O-acyltransferase: protein acylation impacting metabolic and neuroendocrine signalling. Open Biol. 11, 210080 10.1098/rsob.210080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Alioua, A., Li, M., Wu, Y., Stefani, E. and Toro, L. (2011) Unconventional myristoylation of large-conductance Ca(2)(+)-activated K(+) channel (Slo1) via serine/threonine residues regulates channel surface expression. Proc. Natl Acad. Sci. U.S.A. 108, 10744–10749 10.1073/pnas.1008863108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Price, C.T. and Kwaik, Y.A. (2013) One bacterial effector with two distinct catalytic activities by different strains. EMBO Rep. 14, 753–754 10.1038/embor.2013.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Schlitzer, A. and Ginhoux, F. (2013) DNGR-ing the dendritic cell lineage. EMBO Rep. 14, 850–851 10.1038/embor.2013.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Schjoldager, K.T., Narimatsu, Y., Joshi, H.J. and Clausen, H. (2020) Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 21, 729–749 10.1038/s41580-020-00294-x [DOI] [PubMed] [Google Scholar]
- 237.Reily, C., Stewart, T.J., Renfrow, M.B. and Novak, J. (2019) Glycosylation in health and disease. Nat. Rev. Nephrol. 15, 346–366 10.1038/s41581-019-0129-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Halim, A., Brinkmalm, G., Ruetschi, U., Westman-Brinkmalm, A., Portelius, E., Zetterberg, H.et al. (2011) Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid beta-peptides in human cerebrospinal fluid. Proc. Natl Acad. Sci. U.S.A. 108, 11848–11853 10.1073/pnas.1102664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sawa, T., Zaki, M.H., Okamoto, T., Akuta, T., Tokutomi, Y., Kim-Mitsuyama, S.et al. (2007) Protein S-guanylation by the biological signal 8-nitroguanosine 3′,5′-cyclic monophosphate. Nat. Chem. Biol. 3, 727–735 10.1038/nchembio.2007.33 [DOI] [PubMed] [Google Scholar]
- 240.Tsuchiya, Y., Peak-Chew, S.Y., Newell, C., Miller-Aidoo, S., Mangal, S., Zhyvoloup, A.et al. (2017) Protein CoAlation: a redox-regulated protein modification by coenzyme A in mammalian cells. Biochem. J. 474, 2489–2508 10.1042/BCJ20170129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Vowinckel, J., Stahlberg, S., Paulmann, N., Bluemlein, K., Grohmann, M., Ralser, M.et al. (2012) Histaminylation of glutamine residues is a novel posttranslational modification implicated in G-protein signaling. FEBS Lett. 586, 3819–3824 10.1016/j.febslet.2012.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lepack, A.E., Werner, C.T., Stewart, A.F., Fulton, S.L., Zhong, P., Farrelly, L.A.et al. (2020) Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science 368, 197–201 10.1126/science.aaw8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Farrelly, L.A., Thompson, R.E., Zhao, S., Lepack, A.E., Lyu, Y., Bhanu, N.V.et al. (2019) Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 567, 535–539 10.1038/s41586-019-1024-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Healy, S., McDonald, M.K., Wu, X., Yue, W.W., Kochan, G., Oppermann, U.et al. (2010) Structural impact of human and Escherichia coli biotin carboxyl carrier proteins on biotin attachment. Biochemistry 49, 4687–4694 10.1021/bi901612y [DOI] [PubMed] [Google Scholar]
- 245.Mathias, R.A., Greco, T.M., Oberstein, A., Budayeva, H.G., Chakrabarti, R., Rowland, E.A.et al. (2014) Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell 159, 1615–1625 10.1016/j.cell.2014.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wator, E., Wilk, P., Biela, A., Rawski, M., Zak, K.M., Steinchen, W.et al. (2023) Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders. Nat. Commun. 14, 1698 10.1038/s41467-023-37305-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Park, M.H., Cooper, H.L. and Folk, J.E. (1981) Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl Acad. Sci. U.S.A. 78, 2869–2873 10.1073/pnas.78.5.2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Stringer, D.K. and Piper, R.C. (2011) Terminating protein ubiquitination: Hasta la vista, ubiquitin. Cell Cycle 10, 3067–3071 10.4161/cc.10.18.17191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Enchev, R.I., Schulman, B.A. and Peter, M. (2015) Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 16, 30–44 10.1038/nrm3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tammsalu, T., Matic, I., Jaffray, E.G.. Ibrahim, A.F.M., Tatham, M.H. and Hay, R.T. (2014) Proteome-wide identification of SUMO2 modification sites. Sci. Signal. 7, rs2 10.1126/scisignal.2005146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Hendriks, I.A., Lyon, D., Su, D., Skotte, N.H., Daniel, J.A., Jensen, L.J.et al. (2018) Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun. 9, 2456 10.1038/s41467-018-04957-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Niwa, Y., Suzuki, T., Dohmae, N. and Simizu, S. (2016) Identification of DPY19L3 as the C-mannosyltransferase of R-spondin1 in human cells. Mol. Biol. Cell 27, 744–756 10.1091/mbc.E15-06-0373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Bloch, J.S., John, A., Mao, R., Mukherjee, S., Boilevin, J., Irobalieva, R.N.et al. (2023) Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase. Nat. Chem. Biol. 19, 575–584 10.1038/s41589-022-01219-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Hazen, S.L., Crowley, J.R., Mueller, D.M. and Heinecke, J.W. (1997) Mass spectrometric quantification of 3-chlorotyrosine in human tissues with attomole sensitivity: a sensitive and specific marker for myeloperoxidase-catalyzed chlorination at sites of inflammation. Free Radic. Biol. Med. 23, 909–916 10.1016/s0891-5849(97)00084-1 [DOI] [PubMed] [Google Scholar]