Abstract
Aims
To investigate the associations of GCKR and ADIPOQ variants with the risk of gestational diabetes mellitus (GDM) in Chinese women.
Methods
GCKR rs1260326, ADIPOQ rs266729, and rs1501299 were selected and genotyped in 519 GDM patients and 498 controls. Candidate SNPs were genotyped using multiplex polymerase chain reaction (PCR) combined with next-generation sequencing methods, and the association of these SNPs with GDM was analyzed.
Results
We found that GCKR rs1260326 was significantly associated with an increased risk of GDM in the allele model, the codominant model (CC vs. TT), the dominant model, the recessive model, and the genotypic model distributions (p = 0.0029, p = 0.0022, p = 0.0402, p = 0.0038, and p = 0.0028, respectively). The rs1260326 polymorphism was shown to be associated with 1 h-OGTT level and gravidity in GDM patients (CC vs. TT: p = 0.0475 and p = 0.0220, respectively). Diastolic blood pressure (DBP) was significantly higher in the GDM patients with the rs266729 GG genotype compared to those with the CC or CG genotype (p = 0.0444 and p = 0.0339, respectively). The DBP of the GDM patients with the rs1501299 GT genotype was lower than that of those with the GG genotype (p = 0.0197). There was a weak linkage disequilibrium value between the GCKR and ADIPOQ SNPs.
Conclusions
The genes GCKR and ADIPOQ may be involved in the pathophysiology of GDM.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00592-023-02165-1.
Keywords: Gestational diabetes mellitus, GCKR, ADIPOQ, SNPs
Introduction
Gestational diabetes mellitus (GDM) is defined as glucose intolerance with onset or first recognition during pregnancy, which is a common complication in pregnant women. GDM brings negative effects for pregnant women, fetuses, neonates, and children in the growth and development period, such as hypertensive disease in pregnancy, placental abruption, abortion, fetal macrosomia, fetal malformations, neonatal asphyxia, and cognitive dysfunction in children in the growth and development period. The risks of type 2 diabetes mellitus (T2DM) and cardiovascular disease in pregnant women and their offspring are increased [1]. GDM is caused by a number of factors, the most important of which are insulin resistance and pancreatic islet β cell dysfunction. The incidence of GDM in pregnant women with a family history of diabetes was significantly higher, which revealed that genes played an important role in the pathogenesis of GDM. In humans, single nucleotide polymorphisms (SNPs) are the most prevalent kind of genetic variation. They relate to single nucleotide modifications at certain genomic locations. SNPs can be utilized to predict GDM risk. A large amount of previous evidence has shown that some genetic variations were associated with GDM [2–4].
The GCKR gene is located on chromosome 2p23.3 and contains 19 exons. The encoded glucokinase regulatory protein (GKRP) regulates glycolysis by inhibiting the enzyme activity of the glycolytic enzyme glucokinase (GCK) at low glucose concentrations [5]. GCK is responsible for glucose phosphorylation in the glycolysis pathway. Therefore, it is crucial for preserving blood glucose homeostasis [6]. The overexpression of GCKR in the liver causes an increase in GCK activity, which lowers glucose levels while raising triglyceride levels. The ADIPOQ gene is located on chromosome 3q27 and contains 3 exons and 2 introns. The encoded adiponectin increases the phosphorylation of AMP-activated protein kinase (AMPK) and p38 mitogen-activated protein kinase (p38 MAPK), as well as the peroxisome proliferator-activated receptor (PPAR) ligand activity after binding with its receptors, which plays a role in the down-regulation of key gluconeogenesis enzymes, the promotion of fatty acid oxidation, and the increase of glucose uptake [7, 8].
GDM is the prophase of T2DM to some extent, and they have similar pathophysiological changes and genetic characteristics. At present, the association studies of SNPs and GDM genetic susceptibility are mainly based on the association studies of T2DM genetic susceptibility. Tracing the genetic origin of GDM may help clarify the pathogenesis of the disease. Therefore, our study explored the correlations between GCKR and ADIPOQ gene polymorphisms and GDM in Chinese women so as to provide a new basis and direction for the clinical treatment of GDM in the future.
Materials and methods
Study subjects
The subjects were continuously recruited from the same center (Department of Obstetrics and Gynecology, the Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang Province, China) from December 2016 to December 2018. There were 1157 pregnant women in the research, including 560 pregnant women with GDM and 597 pregnant women with a normal oral glucose tolerance test (OGTT). The diagnosis of GDM was based on a 75-g OGTT at 24–28 weeks’ gestation, according to the 2015 International Association of Diabetes and Pregnancy Study Groups (IADPSG) criteria [9]. The diagnosis of GDM was made when one of the following plasma glucose values in the OGTT was met or exceeded: fasting plasma glucose 92 mg/dL (5.1 mmol/L), 1-h plasma glucose 180 mg/dL (10.0 mmol/L), and 2-h plasma glucose 153 mg/dL (8.5 mmol/L) [9]. The exclusion criteria were pre-gestational diabetes mellitus (n = 20), multiple pregnancies (n = 17), hypertension (n = 21), ethnic minorities (n = 14), liver and renal disfunction (n = 16), complicated with systemic metabolic diseases such as thyroid dysfunction (n = 15), systemic lupus erythematosus (n = 6), rheumatoid disease (n = 5), and other diseases that may cause abnormal blood glucose during pregnancy (n = 7). During the genotyping process, cases that could not be completely genotyped were excluded (n = 19). After exclusion, 1017 pregnant women (519 GDM patients and 498 controls) were recruited (Supplementary Figure S1).
All participants agreed with the ethics of the study and signed informed consent, which was approved by the ethics committee of the Second Affiliated Hospital of Harbin Medical University.
Selection of SNPs
Recent genome-wide association studies (GWAS) identified GCKR and ADIPOQ as being associated with T2DM and the metabolic syndrome [10–12]. Blood glucose control and lipid balance were significantly influenced by GCKR and ADIPOQ gene polymorphisms. However, the influence of these genetic variants on GDM in the general population is unclear and controversial [13, 14]. Based on the results of T2DM GWAS [15], minor allele frequency (MAF) > 0.15 in the Chinese population, and tracking the relevant literature [16, 17], we finally selected three candidate SNPs (GCKR rs1260326, ADIPOQ rs266729, and rs1501299) that might be associated with GDM. GCKR rs1260326 (T > C) is a missense polymorphism that causes a Leu to Pro substitution (P446L). rs266729 (C > G) is located in the promoter region of the ADIPOQ gene, and rs1501299 (G > T) is located in intron 2 of the ADIPOQ gene.
Extraction and genotyping of DNA
Each subject had a peripheral venous blood sample (4–5 ml) collected into a 2% EDTA-Na2 anticoagulant tube, which was then stored at -80 °C until DNA extraction. TIANamp Genomic DNA Kit from Tiangen Biotech, China, was used to extract genomic DNA from peripheral blood samples. Genotyping of the selected GCKR rs1260326, ADIPOQ rs266729, and rs1501299 was tested using multiplex PCR combined with next-generation sequencing methods by the Shanghai Bio Wing Applied Biotechnology Company (http://www.biowing.com.cn) [18]. Primer3 online software (version 0.4.0, http://frodo.wi.mit.edu/) was used to amplify primer sequences. The primers used for amplification are as follows: for rs1260326 forward 5′-CTATAGTGGAGCAGGTGAAAGAG-3′ and reverse 5′-TCATATTCAAAGAAAAGCAGTGGC-3′; for rs266729 forward 5′-GTTTTGGATGTCTTGTTGAAGTTG-3′ and reverse 5′-CTAGAAAGTTTAGGCTTGAAGTGG-3′; for rs1501299 forward 5′-GTTATAGAGGCACCATCTACACTC-3′ and reverse 5′-GAGATCCAGGTAAGAATGTTTCTG-3′. TIANgel Midi Purification Kit (Tiangen Biotech, China) was utilized for purifying the PCR products after PCR amplification was performed. The purified PCR products were performed by Illumina HiSeq XTen platform with paired-end sequencing (2 × 150 bp) according to the manufacturer’s instructions. The Burrows-Wheeler Aligner (BWA, v0.7.12) was used to align the sequences to the human reference genome, and Samtools (v0.1.19) was used for SNP calling and genotyping [19]. Some samples were randomly selected for blind DNA replication for quality control in genotyping.
Statistics
The genotypic distributions of each SNP in the GDM patients and controls were tested for departure from Hardy–Weinberg equilibrium (HWE) using an exact test. Kolmogorov–Smirnov test was used to analyze the distribution of continuous patient demographic and clinical characteristic data. Normally distributed continuous data were compared using analysis of variance (ANOVA), and non-normally distributed continuous data were compared using rank sum test. Categorical, normally distributed continuous, and non-normally distributed data are shown as number (n) and percentage (%), mean ± standard deviation (SD), and median and interquartile range (IQR), respectively. The 95% confidence interval (CI) and odds ratio (OR) were calculated through multiple logistic regression analysis to evaluate the potential association between GCKR and ADIPOQ gene polymorphisms and GDM. Adjusted ORs were computed with adjustment for confounding factors that included maternal age, gestational age, BMI before pregnancy, BMI at enrollment, systolic blood pressure (SBP), diastolic blood pressure (DBP), birth weight, urea, prothrombin time (PT), activated partial thromboplastin time (APTT), and gravidity, then calculated using 10,000 permutations for each model to correct the multiple test. The statistical analyses were performed using IBM SPSS 24.0 Statistics and R 4.0.0. The SHEsis software was used to assess linkage disequilibrium between pair of SNPs [20].
Results
Quality control and SNP genotype
All of the tested SNPs were in agreement with the Hardy–Weinberg Equilibrium (HWE) in the GDM patients and controls of this study (p > 0.05), as shown in Supplementary Table S1. In addition, quality control was set up for the genotypes of several samples. The genotype calling rate in 115 quality control samples was 98.70%, which fully improved the reliability of the follow-up research results.
Clinical characteristics of the study population
The clinical characteristics of the GDM patients and controls were shown in Table 1. The maternal age, gestational age, BMI before pregnancy, BMI at enrollment, SBP, DBP, birth weight, urea, PT, APTT, and gravidity of the GDM patients and controls were significantly different (p < 0.05).
Table 1.
Clinical characteristics of the GDM patients and controls
| Variables | Controls | Patients | p |
|---|---|---|---|
| Numbers (n) | 498 | 519 | |
| Maternal age (year) | 29.00 (27.00–32.00) | 31.00 (28.00–34.00) | < .0001 |
| Age < 35 | 422 (84.7) | 392 (75.5) | 0.0002 |
| Age ≥ 35 | 76 (15.3) | 127 (24.5) | |
| Gestational age (week) | 39.00 (39.00–40.00) | 39.00 (38.00–40.00) | < .0001 |
| BMI before pregnancy (kg/m2) | 20.64 (19.00–22.66) | 22.43 (20.20–24.61) | < .0001 |
| Underweight, 18.5 | 91 (18.3) | 54 (11.0) | < .0001 |
| Normal weight, 18.5–23.9 | 339 (68.2) | 283 (57.9) | |
| Overweight, 24.0–27.9 | 54 (10.9) | 106 (21.7) | |
| Obese, ≥ 28 | 13 (2.6) | 46 (9.4) | |
| BMI at enrollment (kg/m2) | 26.95 (24.80–28.91) | 27.93 (25.56–30.41) | < .0001 |
| SBP (mmHg) | 116.00 (110.00–122.00) | 120.00 (113.00–126.00) | < .0001 |
| DBP (mmHg) | 78.00 (74.00–84.00) | 81.00 (75.00–87.00) | < .0001 |
| Birth weight (g) | 3350.00 (3000.00–3700.00) | 3450.00 (3150.00–3750.00) | 0.0055 |
| Urea (mmol/L) | 3.28 (2.71–3.89) | 3.42 (2.85–4.12) | 0.0031 |
| Creatinine (umol/L) | 48.00 (43.00–54.00) | 48.00 (42.00–56.00) | 0.8988 |
| PT (Sec) | 9.80 (9.50–10.10) | 9.90 (9.60–10.20) | 0.0216 |
| PTA (%) | 108.00 (101.00–116.00) | 107.00 (100.00–115.00) | 0.1272 |
| APTT (Sec) | 29.60 (28.20–31.10) | 29.10 (27.60–31.00) | 0.0087 |
| TT (Sec) | 13.00 (12.40–13.80) | 13.00 (12.50–13.70) | 0.5616 |
| Gravidity (n/%) | 1.00 (1.00–2.00) | 2.00 (1.00–2.00) | 0.0037 |
| 1 | 271 (54.4) | 236 (46.4) | 0.0149 |
| 2 | 137 (27.5) | 148 (29.1) | |
| ≥ 3 | 90 (18.1) | 125 (24.6) |
p < 0.05 was considered as statistically significant (bold)
BMI body mass index, SBP systolic blood pressure, DBP diastolic blood pressure, PT prothrombin time, PTA prothrombin activity, APTT activated partial thromboplastin time, TT thrombin time, p p value
Genotype and allele association analysis
The genotypes and alleles frequencies of GCKR rs1260326, ADIPOQ rs266729, and rs1501299 in the GDM patients and controls were further analyzed, as shown in Table 2. The frequency and distribution of rs1260326 genotypes and alleles were significantly different between the GDM patients and controls (p = 0.0208 and p = 0.0080, respectively). For rs1260326, after adjusting for maternal age, gestational age, BMI before pregnancy, BMI at enrollment, SBP, DBP, birth weight, urea, PT, APTT, and gravidity, the allele model, the codominant model (CC vs. TT), the dominant model, the recessive model, and the genotypic model distributions were different between the GDM patients and controls (p = 0.0029, p = 0.0022, p = 0.0402, p = 0.0038, and p = 0.0028, respectively). After 10,000 permutations, the results were still statistically significant (p < 0.05, all). The genotypes and alleles frequencies of rs266729 were not significantly different in the GDM patients compared with controls (p > 0.05, all), and there was no significant difference between the GDM patients and controls under any model (p > 0.05, all). The frequency and distribution of rs1501299 alleles were significantly different between the GDM patients group and the control group (p = 0.0320). An analysis of the study between the two groups showed a statistically significant difference under the codominant model (TT vs. GG) of the rs1501299 polymorphism (p = 0.0457). However, the significance disappeared after adjusting for confounding factors (p > 0.05).
Table 2.
Association of SNPs in GCKR and ADIPOQ with GDM risk
| SNP | Controls | Patients | pa | Model | pb | OR (95% CI) | pc | OR (95% CI) | pd | |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1260326 | ||||||||||
| TT | 164(33.1) | 142(28.1) | Allele | C vs. T | 0.0081 | 1.269(1.064–1.514) | 0.0029 | 1.381(1.117–1.709) | 0.0045 | |
| TC | 245(49.5) | 241(47.7) | Codominant | CC vs. TT | 0.0065 | 1.638(1.148–2.339) | 0.0022 | 1.984(1.279–3.077) | 0.0037 | |
| CC | 86(17.4) | 122(24.2) | 0.0208 | TC vs. TT | 0.3832 | 1.136(0.853–1.513) | 0.3180 | 1.192(0.844–1.684) | 0.3154 | |
| Dominant | CC + TC vs. TT | 0.0533 | 1.303(0.996–1.705) | 0.0402 | 1.406(1.015–1.948) | 0.0461 | ||||
| T | 573(57.9) | 525(52.0) | Recessive | CC vs. TC + TT | 0.0140 | 1.472(1.081–2.004) | 0.0038 | 1.735(1.195–2.519) | 0.0016 | |
| C | 417(42.1) | 485(48.0) | 0.0080 | Overdominant | TC vs. CC + TT | 0.8318 | 0.974(0.761–1.245) | 0.6463 | 0.933(0.693–1.255) | 0.6348 |
| Genotypic | CC vs. TC vs. TT | 0.0077 | 1.270(1.065–1.514) | 0.0028 | 1.385(1.119–1.714) | 0.0019 | ||||
| rs266729 | ||||||||||
| CC | 254(51.4) | 259(52.1) | Allele | G vs. C | 0.9735 | 0.997(0.819–1.213) | 0.5976 | 0.938(0.739–1.190) | 0.6073 | |
| CG | 206(41.7) | 201(40.4) | Codominant | GG vs. CC | 0.7975 | 1.067(0.649–1.754) | 0.5501 | 0.827(0.444–1.541) | 0.5544 | |
| GG | 34(6.88) | 37(7.44) | 0.8923 | CG vs. CC | 0.7400 | 0.957(0.738–1.241) | 0.7409 | 0.948(0.692–1.300) | 0.7482 | |
| Dominant | GG + CG vs. CC | 0.7257 | 1.045(0.817–1.336) | 0.9903 | 0.998(0.741–1.345) | 0.9627 | ||||
| C | 714(72.3) | 719(72.3) | Recessive | GG vs. CG + CC | 0.8503 | 1.048(0.646–1.698) | 0.6109 | 0.860(0.480–1.540) | 0.6268 | |
| G | 274(27.7) | 275(27.7) | 0.9735 | Overdominant | CG vs. GG + CC | 0.7968 | 1.033(0.806–1.325) | 0.8018 | 1.039(0.769–1.405) | 0.8241 |
| Genotypic | GG vs. CG vs. CC | 0.7189 | 1.037(0.851–1.264) | 0.8263 | 0.973(0.766–1.237) | 0.8103 | ||||
| rs1501299 | ||||||||||
| GG | 254(51.4) | 284(57.0) | Allele | T vs. G | 0.0324 | 0.804(0.658–0.982) | 0.0983 | 0.816(0.642–1.038) | 0.1124 | |
| GT | 198(40.1) | 186(37.3) | Codominant | TT vs. GG | 0.0457 | 0.596(0.359–0.990) | 0.1265 | 0.624(0.340–1.143) | 0.1350 | |
| TT | 42(8.50) | 28(5.62) | 0.0893 | GT vs. GG | 0.1930 | 0.840(0.646–1.092) | 0.2860 | 0.841(0.611–1.157) | 0.2883 | |
| Dominant | TT + GT vs. GG | 0.2355 | 0.861(0.673–1.102) | 0.3310 | 0.862(0.640–1.163) | 0.2889 | ||||
| G | 706(71.5) | 754(75.7) | Recessive | TT vs. GT + GG | 0.0576 | 0.619(0.377–1.016) | 0.1583 | 0.654(0.363–1.180) | 0.1518 | |
| T | 282(28.5) | 242(24.3) | 0.0320 | Overdominant | GT vs. TT + GG | 0.8256 | 0.972(0.757–1.249) | 0.8142 | 0.964(0.711–1.308) | 0.8317 |
| Genotypic | TT vs. GT vs. GG | 0.0834 | 0.839(0.688–1.023) | 0.1737 | 0.847(0.667–1.076) | 0.1695 | ||||
p, p value; p < 0.05 was considered as statistically significant (bold)
aComparison between GDM and controls
bCalculated using multivariate logistic regression analysis
cAdjusted by maternal age, gestational age, BMI before pregnancy, BMI at enrollment, SBP, DBP, birth weight, urea, PT, APTT, and gravidity
dAdjusted by maternal age, gestational age, BMI before pregnancy, BMI at enrollment, SBP, DBP, birth weight, urea, PT, APTT, and gravidity, then calculated using 10,000 permutations for each model to correct the multiple test
In ROC analysis, the AUC for clinical risk factors (maternal age, gestational age, BMI before pregnancy, BMI at enrollment, SBP, DBP, birth weight, urea, PT, APTT, and gravidity) was 0.729 (95% CI 0.694, 0.763) (Fig. 1). Addition of rs1260326 status (TC or CC) increased the predictive value of the model, giving an AUC of 0.735 (95% CI 0.701, 0.769).
Fig. 1.
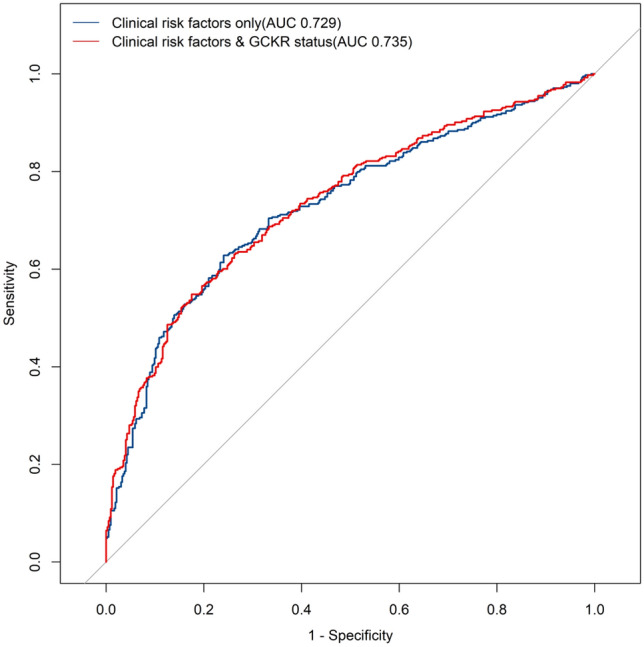
ROC curves for prediction of GDM. Clinical risk factors include maternal age, gestational age, BMI before pregnancy, BMI at enrollment, SBP, DBP, birth weight, urea, PT, APTT, and gravidity
We further investigated the association between GCKR rs1260326, ADIPOQ rs266729, and rs1501299 polymorphisms and clinical information in patients with GDM. Stratified analysis was performed to analyze the association between the genotypes of the three SNPs and maternal age, gestational age, BMI before pregnancy, BMI at enrollment, SBP, DBP, birth weight, urea, PT, APTT, gravidity, HbA1c, FBG, 1 h-OGTT, 2 h-OGTT. As shown in Tables 3, 4, and 5, a significant association was demonstrated between rs1260326 polymorphism and 1 h-OGTT level and gravidity in patients with GDM (CC vs. TT: p = 0.0475 and p = 0.0220, respectively). We observed that DBP was significantly higher in the GDM patients with the rs266729 GG genotype compared to those with the CC or CG genotype (p = 0.0444 and p = 0.0339, respectively). The DBP of the GDM patients with the rs1501299 GT genotype was lower than that of those with the GG genotype (p = 0.0197).
Table 3.
Analysis of GCKR rs1260326 genotype in GDM patients by clinical features
| Variables | TT | TC | CC | p | CC vs. TT | TC vs. TT | CC vs. TC |
|---|---|---|---|---|---|---|---|
| Maternal age (year) | 30.00 (28.00–35.00) | 31.00 (29.00–34.00) | 31.00 (28.00–35.00) | 0.8206 | 0.7879 | 0.9758 | 0.9005 |
| Gestational age (week) | 39.00 (38.00–40.00) | 39.00 (38.00–40.00) | 39.00 (38.00–40.00) | 0.5550 | 0.9562 | 0.5502 | 0.7863 |
| BMI before pregnancy (kg/m2) | 22.43 (20.20–24.22) | 22.31 (20.20–24.50) | 22.86 (20.06–25.34) | 0.7131 | 0.8789 | 0.9403 | 0.6986 |
| BMI at enrollment (kg/m2) | 28.28 (26.17–30.41) | 27.82 (25.44–30.22) | 28.28 (25.34–30.44) | 0.6514 | 0.8180 | 0.6201 | 0.9919 |
| SBP (mmHg) | 120.00 (114.00–126.00) | 120.00 (112.00–125.00) | 120.00 (113.50–125.50) | 0.5453 | 0.9997 | 0.6585 | 0.6125 |
| DBP (mmHg) | 81.00 (76.00–88.00) | 80.00 (75.00–85.50) | 80.50 (75.00–87.00) | 0.7112 | 0.8606 | 0.6866 | 0.9847 |
| Birth weight (g) | 3400.00 (3200.00–3700.00) | 3450.00 (3200.00–3800.00) | 3450.00 (3100.00–3750.00) | 0.5840 | 0.8904 | 0.5489 | 0.8941 |
| Urea (mmol/L) | 3.38 (2.78–4.01) | 3.43 (2.85–4.15) | 3.50 (3.00–4.11) | 0.7508 | 0.7530 | 0.8186 | 0.9797 |
| PT (s) | 9.90 (9.60–10.20) | 9.90 (9.60–10.30) | 9.80 (9.50–10.10) | 0.7881 | 0.8183 | 0.9948 | 0.8073 |
| APTT (s) | 28.60 (27.30–30.90) | 29.20 (27.70–31.10) | 29.20 (27.90–31.00) | 0.2568 | 0.3666 | 0.2819 | 0.9927 |
| Gravidity (n/%) | 1.00 (1.00–2.00) | 2.00 (1.00–3.00) | 2.00 (1.00–3.00) | 0.0181 | 0.0220 | 0.0561 | 0.7230 |
| HbA1c (%) | 5.60 (5.30–5.80) | 5.50 (5.30–5.80) | 5.60 (5.40–5.80) | 0.4588 | 0.9937 | 0.5740 | 0.5434 |
| FBG(mmol/L) | 5.27 (5.10–5.48) | 5.27 (5.03–5.50) | 5.21 (4.94–5.58) | 0.8189 | 0.9875 | 0.8898 | 0.8400 |
| 1 h-OGTT(mmol/L) | 8.59 (7.38–10.16) | 9.25 (7.95–10.40) | 9.52 (7.86–10.72) | 0.0384 | 0.0475 | 0.1213 | 0.6035 |
| 2 h-OGTT(mmol/L) | 8.15 (7.02–9.24) | 8.25 (7.10–9.17) | 8.55 (7.18–9.18) | 0.7188 | 0.7029 | 0.9734 | 0.8010 |
p < 0.05 was considered as statistically significant (bold)
BMI body mass index, SBP systolic blood pressure, DBP diastolic blood pressure, PT prothrombin time, APTT activated partial thromboplastin time, p p value
Table 4.
Analysis of ADIPOQ rs266729 genotype in GDM patients by clinical features
| Variables | CC | CG | GG | p | GG vs. CC | CG vs. CC | GG vs. CG |
|---|---|---|---|---|---|---|---|
| Maternal age (year) | 31.00 (28.00–34.00) | 30.00 (29.00–35.00) | 31.00 (29.00–35.00) | 0.5840 | 0.7485 | 0.6464 | 0.9681 |
| Gestational age (week) | 39.00 (38.00–40.00) | 39.00 (38.00–40.00) | 39.00 (39.00–40.00) | 0.6532 | 0.7265 | 0.9393 | 0.6304 |
| BMI before pregnancy (kg/m2) | 22.48 (20.26–24.78) | 22.36 (20.20–24.49) | 22.50 (20.00–23.88) | 0.8861 | 0.8870 | 0.9985 | 0.8804 |
| BMI at enrollment (kg/m2) | 27.92 (25.34–30.41) | 27.99 (26.03–30.44) | 28.52 (25.28–29.73) | 0.8257 | 1.0000 | 0.8131 | 0.9610 |
| SBP (mmHg) | 119.00 (113.00–125.00) | 120.00 (113.00–126.00) | 120.00 (112.00–130.00) | 0.4690 | 0.4605 | 0.8962 | 0.5705 |
| DBP (mmHg) | 80.86 ± 8.92 | 80.62 ± 9.19 | 84.11 ± 10.45 | 0.0983 | 0.0444 | 0.7813 | 0.0339 |
| Birth weight (g) | 3400.00 (3150.00–3750.00) | 3450.00 (3150.00–3750.00) | 3450.00 (3300.00–3800.00) | 0.6479 | 0.6447 | 0.9767 | 0.6654 |
| Urea (mmol/L) | 3.42 (2.84–4.20) | 3.49 (2.87–4.08) | 3.39 (2.85–3.87) | 0.9209 | 0.9860 | 0.9173 | 0.9931 |
| PT (s) | 9.90 (9.60–10.20) | 9.90 (9.60–10.20) | 10.00 (9.70–10.30) | 0.4185 | 0.4114 | 0.9978 | 0.4135 |
| APTT (s) | 29.10 (27.70–31.00) | 28.90 (27.40–30.90) | 29.90 (28.40–31.10) | 0.1999 | 0.3679 | 0.7042 | 0.1723 |
| Gravidity (n/%) | 2.00 (1.00–2.00) | 2.00 (1.00–3.00) | 2.00 (1.00–3.00) | 0.2659 | 0.6000 | 0.2871 | 0.9929 |
| HbA1c (%) | 5.60 (5.40–5.80) | 5.60 (5.30–5.80) | 5.75 (5.40–5.90) | 0.4720 | 0.4893 | 0.9840 | 0.4371 |
| FBG(mmol/L) | 5.22 (4.92–5.49) | 5.30 (5.10–5.57) | 5.23 (5.05–5.45) | 0.3219 | 0.9326 | 0.3723 | 0.5494 |
| 1 h-OGTT(mmol/L) | 9.42 (7.77–10.50) | 8.85 (7.73–10.13) | 9.45 (7.96–10.81) | 0.1531 | 0.9430 | 0.1845 | 0.3836 |
| 2 h-OGTT(mmol/L) | 8.20 (7.09–9.12) | 8.28 (7.31–9.12) | 8.91 (6.99–9.59) | 0.4705 | 0.5115 | 0.9269 | 0.4648 |
p < 0.05 was considered as statistically significant (bold)
BMI body mass index, SBP systolic blood pressure, DBP diastolic blood pressure, PT prothrombin time, APTT activated partial thromboplastin time, p p value
Table 5.
Analysis of ADIPOQ rs1501299 genotype in GDM patients by clinical features
| Variables | GG | GT | TT | p | TT vs. GG | GT vs. GG | TT vs. GT |
|---|---|---|---|---|---|---|---|
| Maternal age (year) | 31.00(29.00–35.00) | 30.00(28.00–34.00) | 30.00(28.00–33.00) | 0.5585 | 0.7791 | 0.6046 | 0.9778 |
| Gestational age (week) | 39.00(38.00–40.00) | 39.00(38.00–40.00) | 39.50(38.00–40.00) | 0.2691 | 0.3238 | 0.8523 | 0.2586 |
| BMI before pregnancy (kg/m2) | 22.43(20.20–24.77) | 22.49(20.13–24.27) | 22.04(19.28–24.61) | 0.8348 | 0.8218 | 0.9710 | 0.8925 |
| BMI at enrollment (kg/m2) | 27.92(25.65–30.44) | 27.97(25.53–29.86) | 27.85(24.61–29.40) | 0.6949 | 0.6643 | 0.9571 | 0.7830 |
| SBP (mmHg) | 120.00(113.00–126.00) | 120.00(112.00–126.00) | 119.00(110.00–125.00) | 0.4436 | 0.3871 | 0.9583 | 0.5458 |
| DBP (mmHg) | 81.84 ± 9.27 | 79.79 ± 9.34 | 80.59 ± 7.64 | 0.0641 | 0.5038 | 0.0197 | 0.6720 |
| Birth weight (g) | 3450.00(3150.00–3750.00) | 3425.00(3150.00–3775.00) | 3400.00(3250.00–3625.00) | 0.9025 | 0.8992 | 0.9973 | 0.9034 |
| Urea (mmol/L) | 3.41(2.78–4.12) | 3.45(2.93–4.20) | 3.49(3.02–3.68) | 0.6518 | 0.9987 | 0.6440 | 0.8776 |
| PT (s) | 9.90(9.60–10.20) | 9.90(9.60–10.20) | 9.90(9.50–10.30) | 0.8221 | 0.9699 | 0.8116 | 0.9987 |
| APTT (s) | 29.10(27.60–31.10) | 29.20(27.90–30.90) | 29.40(27.70–30.50) | 0.9584 | 0.9609 | 0.9983 | 0.9528 |
| Gravidity (n/%) | 2.00(1.00–3.00) | 2.00(1.00–2.00) | 1.00(1.00–2.00) | 0.5029 | 0.4859 | 0.9561 | 0.5405 |
| HbA1c (%) | 5.60(5.30–5.80) | 5.60(5.30–5.80) | 5.50(5.20–5.70) | 0.5227 | 0.5396 | 0.9671 | 0.5084 |
| FBG(mmol/L) | 5.21(4.90–5.49) | 5.29(5.11–5.51) | 5.31(5.16–5.60) | 0.0896 | 0.3042 | 0.1439 | 0.8533 |
| 1 h-OGTT(mmol/L) | 9.23(7.90–10.37) | 9.15(7.70–10.50) | 8.04(7.66–9.74) | 0.1753 | 0.1301 | 0.9825 | 0.2412 |
| 2 h-OGTT(mmol/L) | 8.25(7.09–9.20) | 8.52(7.30–9.27) | 8.49(6.70–9.08) | 0.5628 | 0.6370 | 0.8918 | 0.5694 |
p < 0.05 was considered as statistically significant (bold)
BMI body mass index, SBP systolic blood pressure, DBP diastolic blood pressure, PT prothrombin time, APTT activated partial thromboplastin time, p p value
Linkage disequilibrium among the three SNPs
The linkage disequilibrium among the three SNPs (GCKR rs1260326, ADIPOQ rs266729, and rs1501299) were examined. It was found that rs1260326 vs. rs266729 (D’ = 0.056, r2 = 0.001), rs1260326 vs. rs1501299 (D’ = 0.013, r2 = 0.000), and rs266729 vs. rs1501299 (D’ = 0.030, r2 = 0.001) were in weak linkage disequilibrium (Supplementary Figure S2).
Discussion
In the present study, we investigated the association between GCKR and ADIPOQ genetic variants and the risk of GDM in Chinese women. Through the analysis of the clinical data of GDM patients and controls, we found that the maternal age, gestational age, BMI before pregnancy, BMI at enrollment, SBP, DBP, birth weight, urea, PT, APTT, and gravidity of the GDM patients and controls were significantly different, especially the risk of GDM was higher in women with advanced maternal age (≥ 35 years old), which were consistent with the results of previous studies [21–23]. The rates of overweight, obesity, and higher gravidity (≥ 2) were significantly higher in the GDM patients compared with controls. The results also confirmed that GDM will increase the risk of hypertension in pregnancy and fetal macrosomia. Urea, PT, and APTT in patients with GDM were significantly different from those in controls. At present, studies have applied some biochemical indicators, such as APTT, to the prediction model of GDM [24].
To our knowledge, this is the first report that the GCKR rs1260326 polymorphism has been found to be associated with GDM. Our study showed that the C allele of rs1260326 increased the risk of GDM in Chinese women. The CC genotype of rs1260326 had a 1.984-fold increased risk of GDM in comparison to the TT genotype in the codominant model. GCKR rs1260326 is a missense polymorphism that causes leucine to proline substitution (P446L). With proline (encoded by the C allele of rs1260326) as opposed to leucine (encoded by the T allele of rs1260326) at position 446, GKRP responds more robustly to fructose-6-phosphate, resulting in more avid binding of glucokinase to GKRP, which leads to a decrease in glucokinase activity [25, 26]. Liu et al. [27] demonstrated that the C allele of rs1260326 was associated with greater insulin resistance. However, She et al. [28] reported the association between GCKR rs1260326 polymorphism and GDM in the Chinese Wuhan population, pointing out that there was no significant association between rs1260326 and GDM. There were few reports on the relationship between GCKR rs1260326 and GDM. Thus, the findings that GCKR rs1260326 may increase the risk of GDM need to be verified in the future.
Our results that the addition of genetic information to clinical risk factors modestly improved the prediction of GDM are consistent with several other researchers' findings [23, 29]. Clinical risk variables alone did not have as high a predictive value for GDM as those combined with the GCKR rs1260326 genotype. We identified genetic information associated with the risk of GDM, and the identified genetic polymorphisms could improve models’ predictive ability for GDM beyond classical risk factors and clinical markers. The findings may assist in early screening for and prevention of GDM and provide new insights into the mechanisms underlying GDM pathology.
A previous study showed that GCKR rs1260326 was significantly correlated with fasting blood glucose level [16]. Our subgroup analysis showed that GDM women with the rs1260326 CC genotype had a higher 1 h OGTT level compared to the TT genotype, which inspired us to speculate that there might be a timing effect on the association of GCKR SNPs with glycemic changes. Additional studies are warranted to validate the findings and clarify the underlying mechanism.
Our study found no evidence that the ADIPOQ rs266729 and rs1501299 were associated with GDM in Chinese women. Most studies on the correlation between ADIPOQ rs1501299 and GDM showed that there was no significant correlation between them [17, 30]. So far, only a study conducted by Shaat N et al. [31] suggested that the T allele of the ADIPOQ gene rs1501299 was associated with an increased risk of GDM in Scandinavia. rs1501299 is located in intron 2 of the ADIPOQ gene, which will be removed during mRNA post-transcriptional modification [32]. The correlation between ADIPOQ rs266729 and GDM was inconsistent in previous reports. In 2010, Liang Z et al. [33] found that the ADIPOQ gene rs266729 was related to GDM based on gene chip technology. In 2014, Beltcheva O et al. [30] reported that the G allele of the ADIPOQ gene rs266729 played a protective role in GDM to some extent. On the contrary, Pawlik A et al. [17] found that the G allele of the ADIPOQ gene rs266729 was associated with an increased risk of GDM in 2017. However, Silva et al. [34] believed that the ADIPOQ gene rs266729 had nothing to do with GDM, which was consistent with our research results. The reasons for these negative results remain unknown, but two possibilities should be considered. First, it may be because of genetic trait differences, as we know that genetic polymorphisms in human genes are distinct in different ethnicities, populations, and geographic regions. In addition, even though we might find a potential link between the disease-causing gene and the disease itself, GDM is a multi-factorial disease, and individual exposure to diverse environmental factors and genetic backgrounds may cause different results.
In addition, the subgroup analysis showed that, in the GDM patients, rs266729 and rs1501299 were associated with DBP, suggesting that the ADIPOQ gene polymorphisms genotypes may affect DBP in the GDM patients. An association between the T allele of rs1501299 and lower DBP has been reported in Amerindian subjects [35]. Significantly lower DBP in subjects with the mutated genotypes at rs1501299 were also reported in another study [36]. While the genetic background contributing specifically to the changes in DBP in GDM is still unknown, Studies have shown an inverse correlation between adiponectin and blood pressure [37, 38]. The exact role of these SNPs has yet to be determined, but it could be speculated that the ADIPOQ gene polymorphisms may influence adiponectin levels, which affect blood pressure by influencing endothelial function, regulating the renin-angiotensin system, and interacting with the sympathetic nervous system [39]. Therefore, it would be beneficial to conduct 24-h ambulatory blood pressure monitoring in further research and correlate the ADIPOQ gene polymorphisms with variations in blood pressure levels via adiponectin concentration.
So far as we know, this is the first study that analyzes the associations of the SNPs of GCKR rs1260326, ADIPOQ rs266729, and rs1501299 with GDM risk in the northeastern Han Chinese population and confirms that GCKR rs1260326 increased the risk of GDM. There were some deficiencies in this study. First of all, we did not detect the levels and activities of GCKR and ADIPOQ in the blood of the subjects, nor could we analyze the relationship between gene polymorphisms and their expression levels. Second, in the analysis of GDM genetic susceptibility, even after we adjusted for confounding variables such as maternal age, gestational age, BMI before pregnancy, BMI at enrollment, SBP, DBP, birth weight, urea, PT, APTT, and gravidity, these unmatched characteristics between groups may affect the results of the study. We also could not rule out the possibility that normal control pregnant women would develop GDM in subsequent pregnancies, which could result in grouping error and a reduction in the impact of genetic factors on the risk of GDM. Finally, our study population was limited to Han people in northeast China. People from different regions and ethnic backgrounds should be included, and replication of our findings in a broader population is warranted.
Conclusions
In summary, the CC genotype of rs1260326 increased the risk of GDM in Chinese women. The addition of rs1260326 to clinical risk factors modestly improved the prediction of GDM. GCKR SNPs may have a timing effect on glycemic changes in GDM patients. Different ADIPOQ rs266729 or rs1501299 genotypes carried by GDM women can have different effects on DBP. In addition, we identified weak linkage disequilibrium between GCKR and ADIPOQ SNPs.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all the participants in this study. This study was supported by the National Natural Science Foundation of China (No. 82071929).
Author contributions
MZ: data collection, data analysis, and drafting the manuscript. YL, YP and YW: data analysis and revision of the manuscript. YF, TJ and SX: data collection and data analysis. SL and WW: patient screening. LS and JT: experimental design and overall planning. All authors contributed to the article and approved of the final version for publication.
Funding
This study was supported by the National Natural Science Foundation of China (No. 82071929).
Data availability
SNP date is available in the Figshare database (https://figshare.com/), https://doi.org/10.6084/m9.figshare.23001491.v1.
Declarations
Conflict of interest
The authors declare that they have no competing interest.
Ethics approval and consent to participate
The study protocol was approved by the ethics committee of the Second Affiliated Hospital of Harbin Medical University (KY2019-118) and was performed in accordance with the principles of the Declaration of Helsinki.
Informed consent
Informed consent was obtained from all the study subjects before enrollment.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiawei Tian, Email: jwtian2004@163.com.
Litao Sun, Email: litaosun1971@aliyun.com.
References
- 1.Feng Y, Jiang CD, Chang AM, et al. Interactions among insulin resistance, inflammation factors, obesity-related gene polymorphisms, environmental risk factors, and diet in the development of gestational diabetes mellitus. J Matern Fetal Neonatal Med. 2019;32(2):339–347. doi: 10.1080/14767058.2018.1446207. [DOI] [PubMed] [Google Scholar]
- 2.Shaat N, Ekelund M, Lernmark A, et al. Association of the E23K polymorphism in the KCNJ11 gene with gestational diabetes mellitus. Diabetologia. 2005;48(12):2544–2551. doi: 10.1007/s00125-005-0035-0. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Zhang K, Li W, et al. Association of KCNQ1 gene polymorphism with gestational diabetes mellitus in a Chinese population. Diabetologia. 2009;52(11):2466–2468. doi: 10.1007/s00125-009-1500-y. [DOI] [PubMed] [Google Scholar]
- 4.Pappa KI, Gazouli M, Anastasiou E, et al. The Q192R polymorphism of the paraoxonase-1 (PON1) gene is associated with susceptibility to gestational diabetes mellitus in the Greek population. Gynecol Endocrinol. 2017;33(8):617–620. doi: 10.1080/09513590.2017.1302419. [DOI] [PubMed] [Google Scholar]
- 5.Matschinsky FM, Glaser B, Magnuson MA. Pancreatic beta-cell glucokinase: closing the gap between theoretical concepts and experimental realities. Diabetes. 1998;47(3):307–315. doi: 10.2337/diabetes.47.3.307. [DOI] [PubMed] [Google Scholar]
- 6.Van Schaftingen E. A protein from rat liver confers to glucokinase the property of being antagonistically regulated by fructose 6-phosphate and fructose 1-phosphate. Eur J Biochem. 1989;179(1):179–184. doi: 10.1111/j.1432-1033.1989.tb14538.x. [DOI] [PubMed] [Google Scholar]
- 7.Yamauchi T, Kamon J, Ito Y, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423(6941):762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- 8.Fang H, Judd RL. Adiponectin regulation and function. comprehensive. Physiology. 2018;8(3):1031–1063. doi: 10.1002/cphy.c170046. [DOI] [PubMed] [Google Scholar]
- 9.Gupta Y, Kalra B, Baruah MP, et al. Updated guidelines on screening for gestational diabetes. Int J Women's Health. 2015;7:539–550. doi: 10.2147/IJWH.S82046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simons N, Dekker JM, van Greevenbroek MM, et al. A common gene variant in glucokinase regulatory protein interacts with glucose metabolism on diabetic dyslipidemia: the combined CODAM and hoorn studies. Diabetes Care. 2016;39(10):1811–1817. doi: 10.2337/dc16-0153. [DOI] [PubMed] [Google Scholar]
- 11.Oh SW, Lee JE, Shin E, et al. Genome-wide association study of metabolic syndrome in Korean populations. PLoS ONE. 2020;15(1):0227357. doi: 10.1371/journal.pone.0227357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truong S, Tran NQ, Ma PT, et al. Association of ADIPOQ single-nucleotide polymorphisms with the two clinical phenotypes type 2 diabetes mellitus and metabolic syndrome in a Kinh Vietnamese population. Diabetes Metab Syndr Obes. 2022;15:307–319. doi: 10.2147/DMSO.S347830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai Y, Tang L, Li L, et al. The roles of ADIPOQ rs266729 and MTNR1B rs10830963 polymorphisms in patients with gestational diabetes mellitus: a meta-analysis. Gene. 2020;730:144302. doi: 10.1016/j.gene.2019.144302. [DOI] [PubMed] [Google Scholar]
- 14.Lin Z, Wang Y, Zhang B, Jin Z. Association of type 2 diabetes susceptible genes GCKR, SLC30A8, and FTO polymorphisms with gestational diabetes mellitus risk: a meta-analysis. Endocrine. 2018;62(1):34–45. doi: 10.1007/s12020-018-1651-z. [DOI] [PubMed] [Google Scholar]
- 15.Talmud PJ, Cooper JA, Morris RW, et al. Sixty-five common genetic variants and prediction of type 2 diabetes. Diabetes. 2015;64(5):1830–1840. doi: 10.2337/db14-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaxillaire M, Cavalcanti-Proença C, Dechaume A, et al. The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces type 2 diabetes risk in the DESIR prospective general French population. Diabetes. 2008;57(8):2253–2257. doi: 10.2337/db07-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pawlik A, Teler J, Maciejewska A, Sawczuk M, et al. Adiponectin and leptin gene polymorphisms in women with gestational diabetes mellitus. J Assist Reprod Genet. 2017;34(4):511–516. doi: 10.1007/s10815-016-0866-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen K, Zhou YX, Li K, et al. A novel three-round multiplex PCR for SNP genotyping with next generation sequencing. Anal Bioanal Chem. 2016;408(16):4371–4377. doi: 10.1007/s00216-016-9536-6. [DOI] [PubMed] [Google Scholar]
- 19.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 21.Aye IL, Powell TL, Jansson T. Review: adiponectin–the missing link between maternal adiposity, placental transport and fetal growth. Placenta. 2013;34(Suppl):S40–45. doi: 10.1016/j.placenta.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anghebem-Oliveira MI, Webber S, Alberton D, et al. The GCKR gene polymorphism rs780094 is a Risk factor for gestational diabetes in a Brazilian population. J Clin Lab Anal. 2017;31(2):e22035. doi: 10.1002/jcla.22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popova PV, Klyushina AA, Vasilyeva LB, et al. Association of common genetic risk variants with gestational diabetes mellitus and their role in GDM prediction. Front Endocrinol (Lausanne) 2021;12:628582. doi: 10.3389/fendo.2021.628582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Y, He A, Tang C, et al. Risk prediction models of gestational diabetes mellitus before 16 gestational weeks. BMC Pregnancy Childbirth. 2022;22(1):889. doi: 10.1186/s12884-022-05219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fagerberg L, Hallström BM, Oksvold P, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brouwers M, Jacobs C, Bast A, et al. Modulation of glucokinase regulatory protein: a double-edged sword. Trends Mol Med. 2015;21(10):583–594. doi: 10.1016/j.molmed.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Kuang A, Talbot O, et al. Metabolomic and genetic associations with insulin resistance in pregnancy. Diabetologia. 2020;63(9):1783–1795. doi: 10.1007/s00125-020-05198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.She L, Li W, Guo Y, et al. Association of glucokinase gene and glucokinase regulatory protein gene polymorphisms with gestational diabetes mellitus: a case-control study. Gene. 2022;824:146378. doi: 10.1016/j.gene.2022.146378. [DOI] [PubMed] [Google Scholar]
- 29.Krishnan M, Murphy R, Okesene-Gafa K, et al. The Pacific-specific CREBRF rs373863828 allele protects against gestational diabetes mellitus in Māori and Pacific women with obesity. Diabetologia. 2020;63(10):2169–2176. doi: 10.1007/s00125-020-05202-8. [DOI] [PubMed] [Google Scholar]
- 30.Beltcheva O, Boyadzhieva M, Angelova O, et al. The rs266729 single-nucleotide polymorphism in the adiponectin gene shows association with gestational diabetes. Arch Gynecol Obstet. 2014;289(4):743–748. doi: 10.1007/s00404-013-3029-z. [DOI] [PubMed] [Google Scholar]
- 31.Shaat N, Lernmark A, Karlsson E, et al. A variant in the transcription factor 7-like 2 (TCF7L2) gene is associated with an increased risk of gestational diabetes mellitus. Diabetologia. 2007;50(5):972–979. doi: 10.1007/s00125-007-0623-2. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Dai Q, Li W, et al. Association of vitamin D receptor gene polymorphisms with gestational diabetes mellitus-a case control study in Wuhan, China. BMC Pregnancy Childbirth. 2021;21(1):142. doi: 10.1186/s12884-021-03621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang Z, Dong M, Cheng Q, et al. Gestational diabetes mellitus screening based on the gene chip technique. Diabetes Res Clin Pract. 2010;89(2):167–173. doi: 10.1016/j.diabres.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Gueuvoghlanian-Silva BY, Torloni MR, Mattar R, et al. Profile of inflammatory mediators in gestational diabetes mellitus: phenotype and genotype. Am J Reprod Immunol (New York, NY:1989) 2012;67(3):241–250. doi: 10.1111/j.1600-0897.2011.01090.x. [DOI] [PubMed] [Google Scholar]
- 35.Arnaiz-Villena A, Fernández-Honrado M, Rey D, et al. Amerindians show association to obesity with adiponectin gene SNP45 and SNP276: population genetics of a food intake control and "thrifty" gene. Mol Biol Rep. 2013;40(2):1819–1826. doi: 10.1007/s11033-012-2236-1. [DOI] [PubMed] [Google Scholar]
- 36.Boumaiza I, Omezzine A, Rejeb J, et al. Association between eight adiponectin polymorphisms, obesity, and metabolic syndrome parameters in Tunisian volunteers. Metab Syndr Relat Disord. 2011;9(6):419–426. doi: 10.1089/met.2011.0035. [DOI] [PubMed] [Google Scholar]
- 37.Baden MY, Yamada Y, Takahi Y, et al. Association of adiponectin with blood pressure in healthy people. Clin Endocrinol (Oxf) 2013;78(2):226–231. doi: 10.1111/j.1365-2265.2012.04370.x. [DOI] [PubMed] [Google Scholar]
- 38.Sethna CB, Leonard MB, Gallagher PR, et al. Serum adiponectin levels and ambulatory blood pressure monitoring in pediatric renal transplant recipients. Transplantation. 2009;88(8):1030–1037. doi: 10.1097/TP.0b013e3181b9e1ec. [DOI] [PubMed] [Google Scholar]
- 39.Kaze AD, Musani SK, Bidulescu A, et al. Plasma adiponectin and blood pressure progression in African Americans: the Jackson heart study. Am J Hypertens. 2021;34(11):1163–1170. doi: 10.1093/ajh/hpab101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SNP date is available in the Figshare database (https://figshare.com/), https://doi.org/10.6084/m9.figshare.23001491.v1.


