Abstract
Background:
Neoadjuvant chemotherapy (NAC) is the standard of care in muscle-invasive bladder cancer (MIBC). However, treatment is intense, and the overall benefit is small, necessitating effective biomarkers to identify patients who will benefit most.
Objective:
To characterize cell-free DNA (cfDNA) methylation in patients receiving NAC in SWOG S1314, a prospective cooperative group trial, and to correlate the methylation signatures with pathologic response at radical cystectomy.
Design, setting, and participants:
SWOG S1314 is a prospective cooperative group trial for patients with MIBC (cT2-T4aN0M0, ≥5 mm of viable tumor), with a primary objective of evaluating the coexpression extrapolation (COXEN) gene expression signature as a predictor of NAC response, defined as achieving pT0N0 or ≤pT1N0 at radical cystectomy. For the current exploratory analysis, blood samples were collected prospectively from 72 patients in S1314 before and during NAC, and plasma cfDNA methylation was measured using the Infinium MethylationEPIC BeadChip array.
Intervention:
No additional interventions besides plasma collection.
Outcome measurements and statistical analysis:
Differential methylation between pathologic responders (≤pT1N0) and nonresponders was analyzed, and a classifier predictive of treatment response was generated using the Random Forest machine learning algorithm.
Results and limitations:
Using prechemotherapy plasma cfDNA, we developed a methylation-based response score (mR-score) predictive of pathologic response. Plasma samples collected after the first cycle of NAC yielded mR-scores with similar predictive ability. Furthermore, we used cfDNA methylation data to calculate the circulating bladder DNA fraction, which had a modest but independent predictive ability for treatment response. In a model combining mR-score and circulating bladder DNA fraction, we correctly predicted pathologic response in 79% of patients based on their plasma collected at baseline and after one cycle of chemotherapy. Limitations of this study included a limited sample size and relatively low circulating bladder DNA levels.
Conclusions:
Our study provides the proof of concept that cfDNA methylation can be used to generate classifiers of NAC response in bladder cancer patients.
Patient summary:
In this exploratory analysis of S1314, we demonstrated that cell-free DNA methylation can be profiled to generate biomarker signatures associated with neoadjuvant chemotherapy response. With validation in additional cohorts, this minimally invasive approach may be used to predict chemotherapy response in locally advanced bladder cancer and perhaps also in metastatic disease.
Keywords: Cell-free DNA, Methylation, Muscle-invasive bladder cancer, Predictive biomarker, Machine learning, Neoadjuvant chemotherapy
1. Introduction
For patients with muscle-invasive bladder cancer (MIBC), cisplatin-based neoadjuvant chemotherapy (NAC) has been the standard of care [1]. However, this treatment is toxic and poorly tolerated in a significant number of MIBC patients. Furthermore, the pathologic response rate is approximately 40%, suggesting that many patients may not benefit from NAC [2]. In recent years, significant efforts have been directed at elucidating MIBC molecular subtypes and testing their association with chemotherapy response. Known factors associated with cisplatin chemotherapy response include the p53-like subtype [3,4], DNA damage repair insufficiency [5,6], and mutations in ERCC2, ERBB2, and FGFR3 [7–10]. Though promising, these biomarker candidates are still undergoing clinical validation and have yet to be widely adopted in the clinic. The parent trial of this study, SWOG S1314 trial, was designed to evaluate the coexpression extrapolation (COXEN) score, a tumor tissue gene-expression model aimed at predicting NAC response in MIBC patients [11]. COXEN was found to have a statistically significant association with downstaging when applied to a pooled group including both treatment arms (gemcitabine and cisplatin [GC] and dose-dense methotrexate, vinblastine, adriamycin, and cisplatin [ddMVAC]). However, the primary analysis did not confirm a statistically significant correlation between treatment-specific COXEN scores and NAC response, underscoring the continuing unmet need for a biomarker to guide the use of NAC in MIBC patients.
An analysis of cell-free DNA (cfDNA) can impact cancer treatment decisions in many ways [12], including the monitoring of disease relapse [13,14], detection of actionable somatic mutations [15], and screening for cancer [16]. In MIBC, the presence of tumor-specific mutations in cfDNA has been associated with a high risk of recurrence and response to adjuvant therapy [17]. In addition to mutations, cfDNA also contains DNA methylation information associated with important epigenetic gene regulation and cellular function in normal and malignant tissues [18]. Methylation is tissue specific and can be used to interrogate cellular components in tumor tissue as well as cfDNA [19]. Recently, cfDNA methylation profiling has demonstrated its potential in early cancer detection [20,21], as well as the molecular subtyping of cancer [22].
Plasma cfDNA methylome represents a broad spectrum of cellular states in both host and tumor cells, and may therefore correlate with NAC response. To test this, we collected patient plasma from MIBC patients in S1314 at two time points: before the initiation of NAC and after one cycle of NAC. We characterized cfDNA methylation in these samples using the MethylationEPIC microarray, and we examined the correlation between the cfDNA methylome before and during chemotherapy and pathologic response to NAC at the time of radical cystectomy.
2. Patients and methods
2.1. Clinical cohort
SWOG S1314 was a phase 2 study with 1:1 randomization between GC and ddMVAC chemotherapy conducted by SWOG and other member groups of the National Clinical Trials Network [11]. The purpose of the trial was to evaluate whether either the prespecified GC or the ddMVAC COXEN score dichotomies were associated with a favorable response to NAC at radical cystectomy. Each patient had his/her pathology responses annotated as a complete pathologic response (pT0N0; complete response [CR]), downstaging response (≤pT1N0; partial response [PR]), or stable or progressive disease (residual muscle-invasive disease; no response). For our analysis, we define the responders (Rs) as all the patients with complete or downstaging responses (≤pT1N0) and the nonresponders (NRs) as the patients who had persistent pT2 or worse disease.
2.2. Sample collection and processing
Patient samples were collected with informed consent in accordance with a protocol amendment to S1314 that was reviewed and approved by CTEP Central institutional review board (IRB) as well as by each treating institution’s IRB. The collection of the healthy donor blood was approved by the IRB of the University of Southern California (IRB no. HS-11–00054). Cell-free DNA was extracted and subjected to the Infinium MethylationEPIC BeadChip array (Illumina, San Diego, CA, USA).
2.3. Data analysis and machine learning
All analyses were performed in R version 4.0.2 (R Foundation for Statistical Computing, Vienna, Austria). The methods for methylation array data processing can be found in the Supplementary material and Supplementary Figure 1.
After identification of differentially methylated loci (DMLs) between Rs and NRs, we utilized a random forest (RF) model to generate a classifier (methylation-based response score [mR-score]) with a modified leave-one-out cross-validation procedure (Supplementary material and Supplementary Fig. 2). As a sensitivity analysis, we also created a classifier using Elastic Net. For the on-treatment mR-score, we used all 72 pretreatment cfDNA methylation data to train an RF model using the same model-training parameters but without the leave-one-out procedure. The 57 on-treatment cfDNA methylation data were then inputted into the trained model to obtain the on-treatment mR-score for each patient.
The relative contributions of different cell types to cfDNA was calculated using non-negative least squares linear regression based on tissue-characteristic probes and their methylation values in our samples, as described by Moss et al [19].
Please see the Supplementary material for detailed methods for the data analysis.
3. Results
3.1. Sample collection and patient characteristics
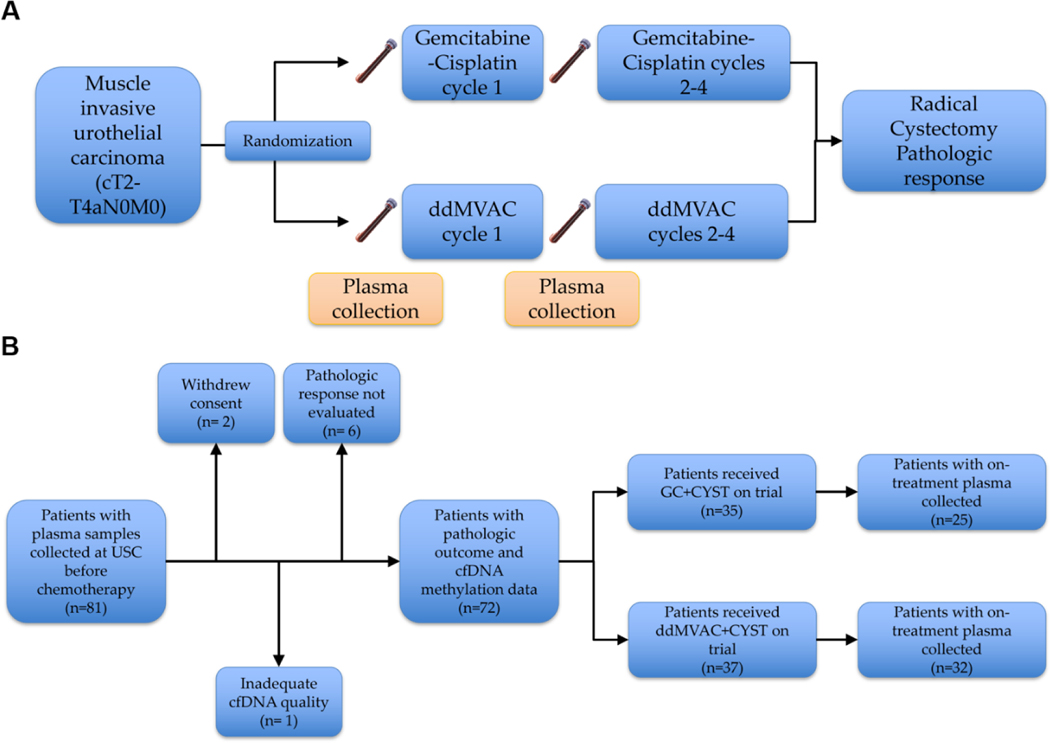
In S1314, 81 patients consented and submitted pre-NAC blood samples. Of those, 73 patients were evaluable in this analysis, having received cisplatin-based NAC on trial followed by cystectomy to determine their pathologic response on trial (Fig. 1 and Supplementary Table 1). One sample had poor cfDNA quality on the array and was removed from the analysis. The clinicopathologic characteristics were summarized in Table 1. None of the characteristics (sex, age, clinical stage, performance status, and cfDNA concentration) were significantly associated with pathologic response. The overall patient characteristics in patients analyzed in the study are similar to the S1314 primary COXEN analysis cohort (Supplementary Table 2).
Fig. 1 –
Sample collection and patient characteristics. (A) Study and sample collection schema. (B) Consort diagram.
cfDNA = cell-free DNA; CYST = cystectomy; ddMVAC = dose-dense methotrexate, vinblastine, adriamycin, and cisplatin; GC = gemcitabine and cisplatin; USC = University of Southern California.
Table 1 –
Patient characteristics for pretreatment cfDNA analysis
| Total patients (N = 72) | Responder (R) (N = 37) | Nonresponder (NR) (N = 35) | p value |
|---|---|---|---|
|
| |||
| Sex (male/female) | 32/5 | 30/5 | >0.9 |
| Age, median (first to third quartile) | 62.7 (55.7–68.9) | 65.9 (62.0–69.4) | 0.2 |
| Clinical stage (T2/T3 or T4a) | 34/3 | 29/6 | 0.3 |
| ECOG performance status (0/1) | 28/9 | 26/9 | >0.9 |
| Treatment arms (ddMVAC + CYST/GC + CYST) | 22/15 | 15/20 | 0.2 |
| cfDNA concentration (ng/7.5 ml), median (first to third quartile) | 85.3 (62.5–147.9) | 113.4 (75.0–141.0) | 0.3 |
cfDNA = cell-free DNA; CYST = cystectomy; ddMVAC = dose-dense methotrexate, vinblastine, adriamycin, and cisplatin; ECOG = Eastern Cooperative Oncology Group; GC = gemcitabine and cisplatin.
3.2. Use of mR-score as a predictive biomarker for NAC response
As the MethylationEPIC array has been used in the past predominantly for tumor tissue analysis, we conducted a lead-in analysis validating the feasibility of using this platform for cfDNA analysis and selected 20 ng cfDNA as the loading amount to be used across all samples (Supplementary material and Supplementary Fig. 3). Next, we used the MethylationEPIC array to characterize patient samples. When comparing methylation between Rs (including CR and PR) and NRs, 23 799 DMLs were identified with a p value cutoff of 0.05. However, none of the individual DMLs were significant after a multiple test correction using the Benjamini-Hochberg procedure, indicating that no single DML had a strong predictive value to distinguish between Rs and NRs.
As individual DMLs exhibited a modest association with treatment response, we applied machine learning algorithms to create a composite biomarker from a large number of DMLs. As DMLs less methylated in NRs were more frequently located on CpG islands and clustered into differentially methylated regions (DMRs) (Supplementary Fig. 3), we selected DMLs less methylated in NRs within the DMRs as the starting point for our machine learning. Using the top 500 less methylated in NR (lmNR) DMLs, we found that the t-distributed stochastic neighbor embedding (tSNE) plot clustered Rs away from NRs (Fig. 2A).
Fig. 2 –
The mR-score as a predictive biomarker for NAC response. (A) A tSNE plot clustered responders (Rs) and nonresponders (NRs) based on the top 500 lmNR loci. Healthy donors were clustered with R. (B) The mR-scores in different response groups. There was no statistically significant difference among the groups. (C) The mR-scores in patients receiving different NAC regimens. (D) Scatter diagram and correlation between mR-scores and the Elastic Net scores. Spearman correlation coefficient R = 0.77 (p = 2.2e-16).
CR = patients with pathologic T0 response; DDMVAC + CYST = dose-dense methotrexate, vinblastine, adriamycin, and cisplatin followed by cystectomy; GC + CYST = gemcitabine-cisplatin followed by cystectomy; lmNR = less methylated in NR; mR-score = methylation-based response score; NAC = neoadjuvant chemotherapy; NR = patients without a pathologic response; PR = patients with downstaged partial response; tSNE = t-distributed stochastic neighbor embedding.
We subsequently utilized a resampling procedure to calculate an mR-score for each sample. This mR-score is trained by comparing the methylation signature between Rs and NRs, and is designed to predict the probability of NRs. Therefore, a high mR-score is associated with an NR, whereas a low mR-score is associated with an R. Specifically, for each patient, we first created ten randomly selected 62-sample training sets not containing the patient of interest. The DMLs were selected within the training set and used to train an RF model. Each trained model provided a prediction score, and the mR-score for each patient was determined as the median of the predicted score by the ten trained models. Using the RF model, we achieved a biomarker with a receiver operating characteristic (ROC) area under the curve (AUC) of 0.636 (95% confidence interval [CI] 0.498–0.773). We also observed a progressive correlation, wherein the median mR-scores were lowest in CR, higher in PR, and highest in NR (Fig. 2B). When used to predict CR, the mR-score had a similar AUC of 0.656 (95% CI 0.509–0.803).
As patients were randomized to receive one of two cisplatin-containing regimens in S1314, we investigated the predictive ability of an mR-score across the treatment arms. Although the limited patient number precluded any meaningful statistical analysis, we observed similar predictive abilities to both chemotherapy regimens (Fig. 2C).
We also used Elastic Net [23], another well-known machine learning algorithm, to build a classifier and obtained similar results (AUC 0.639, 95% CI 0.503–0.774). A strong correlation was observed between the mR-scores and the Elastic Net–based prediction scores (Spearman correlation coefficient 0.77, p = 2.2e-16; Fig. 2D).
3.3. On-treatment mR-scores correlated with response
We investigated whether the RF model trained on pretreatment samples could be used to analyze and assign mR-scores to plasma cfDNA samples collected on C2D1, after receiving the first cycle of NAC. This was not meant as an independent validation cohort, but rather to determine whether the models developed using the baseline cfDNA methylation could be equally effective for predicting response using samples collected later in the treatment course. Of the 72 patients in our sample cohort, 57 had plasma collected after the first cycle of chemotherapy. Again, none of the baseline clinical and laboratory characteristics of these 57 patients were significantly associated with pathologic response (Supplementary Table 3).
The cfDNA methylation data from the 57 on-treatment samples were analyzed and assigned mR-scores using the RF model that had been trained on the 72 pretreatment samples. The performance of the on-treatment mR-score was slightly better, with an AUC of 0.720 (95% CI 0.582–0.857).
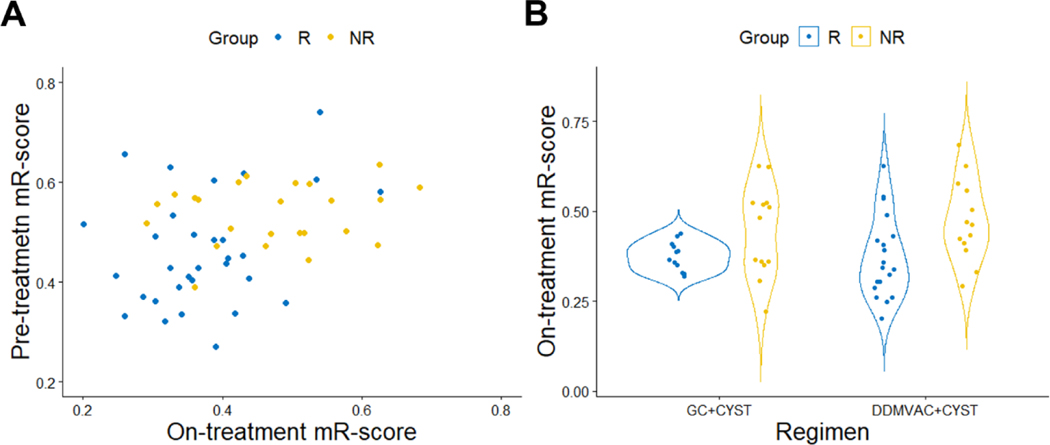
There was also a significant correlation between the pretreatment and on-treatment mR-scores (Spearman correlation coefficient 0.39, p = 0.003; Fig. 3A). Similarly, we observed similar predictive abilities to both chemotherapy regimens using the on-treatment mR-score (Fig. 3B).
Fig. 3 –
On-treatment mR-scores correlated with response. (A) Weak correlation between on-treatment and pretreatment mR-scores. Spearman correlation coefficient R = 0.39 (p = 0.003). (B) On-treatment mR-scores in patients receiving different NAC regimens.
DDMVAC + CYST = dose-dense methotrexate, vinblastine, adriamycin, and cisplatin followed by cystectomy; GC + CYST = gemcitabine-cisplatin followed by cystectomy; mR-score = methylation-based response score; NAC = neoadjuvant chemotherapy; NR = nonresponder; R = responder.
3.4. A combined risk-stratification model to predict NAC response using circulating bladder DNA fraction and mR-score
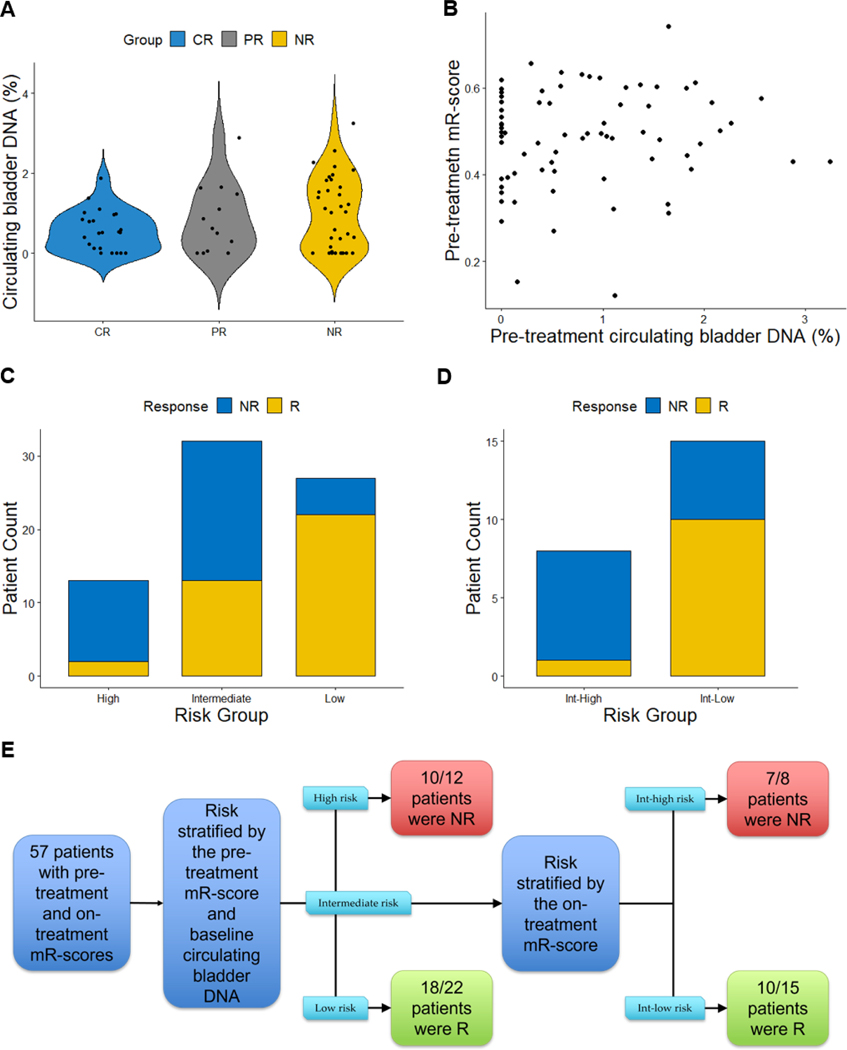
It has repeatedly been demonstrated that elevated circulating tumor DNA (ctDNA) correlates with poor prognosis in many cancer types [13]. Circulating tumor DNA levels are usually estimated using deep sequencing, wherein the allele frequency of detected somatic mutations is used to represent the proportion of ctDNA present in the total cfDNA (the rest of the cfDNA is derived from normal host tissues) [24]. However, no cfDNA mutational profiles were available for the purpose of calculating this estimate. As an alternative, novel methods have recently been reported for estimating the relative prevalence of tissue origin of cfDNA based on tissue-specific methylation patterns [19]. We hypothesized that using this approach, the fraction of circulating bladder DNA may be calculated and may serve as a surrogate for bladder ctDNA. When comparing the circulating bladder DNA percentages in samples from CR, PR, and NR, progressively higher circulating bladder DNA percentages were observed (Fig. 4A), and healthy donor samples did not have a meaningful level of circulating bladder DNA. As a biomarker for NAC response, the circulating bladder DNA yielded an AUC of 0.600 (95% CI 0.463–736). When used to predict CR, circulating bladder DNA yielded a similar AUC of 0.616 (95% CI 0.488–0.745). Of the Rs, 84% (31/37) had circulating bladder DNA <1.11% as compared with 51% (18/35) of NRs, suggesting that low circulating bladder DNA had high sensitivity for identifying NAC Rs.
Fig. 4 –
A proposed risk-stratification model to predict NAC response using circulating bladder DNA fraction and mR-score. (A) Circulating bladder DNA (%) in patients in different response groups. There was no statistically significant difference among the groups. (B) Correlation between mR-scores and circulating bladder DNA (%) obtained before chemotherapy. Spearman correlation coefficient R = 0.073 (p = 0.5). (C) Using mR-score and circulating bladder DNA fraction to risk-stratify patients before chemotherapy. Tentative cutoffs were selected at the Youden’s index for demonstration purpose only, and further biomarker development is needed before the final cutoffs are selected. Patients with high mR-scores or high circulating bladder DNA were given 1 point each. The high-risk group was defined as patients with a high pretreatment mR-score (>0.496) and high circulating bladder DNA fraction (>1.11%), the low-risk group was defined as patients with a low pretreatment mR-score and low circulating bladder DNA fraction, and the intermediate-risk group was defined as the patients not in the high- or low-risk groups. (D) Intermediate-risk group patients were further risk stratified into intermediate-high (Int-high) and intermediate-low (Int-low) groups using their on-treatment mR-scores. The Int-high risk group was defined as the intermediate-risk group patients with a high on-treatment mR-score (>0.433); the Int-low risk group was defined as the intermediate-risk group patients with a low on-treatment mR-score. (E) Proposed approach that may utilize cfDNA methylation for patient risk stratification. Out of 57 patients, 45 were correctly classified in our cohort using this approach; model performance will require further validation in independent cohorts.
cfDNA = cell-free DNA; CR = patients with pathologic T0 response; mR-score = methylation-based response score; NAC = neoadjuvant chemotherapy; NR = patients without pathologic response; PR = patients with downstaged partial response; R = responders including patients with pathologic T0 response and downstaged partial response.
We observed that there was no linear correlation between circulating bladder DNA fractions and pretreatment mR-scores (Spearman correlation coefficient 0.073; Fig. 4B). Given that these two candidate biomarkers were independent of one another, we proposed and tested a risk-stratification model that combined the pretreatment circulating bladder DNA fraction and mR-score, followed by the on-treatment mR-score to stratify the intermediate-risk patients. As a proof-of-concept, we chose Youden’s index as the cutoff for these three measures, and patients were assigned to risk groups based on their test values relative to these cutoffs (Fig. 4C–E). In this cohort, the proposed model had an overall predictive accuracy of 79% (45/57 patients with pretreatment and on-treatment cfDNA were classified correctly).
4. Discussion
In patients with MIBC, cisplatin-based NAC offers a modest clinical benefit but is attended by treatment toxicity. To date, no clinically validated predictive biomarker has widely been adopted to predict NAC response. Consequently, many patients are exposed to the toxicities of NAC with little or no benefit while delaying potentially curative surgery. Hence, there is a continuing critical unmet need for biomarkers that identify patients most likely to benefit [25]. Here, we report an exploratory analysis of the SWOG S1314 trial using cfDNA methylation as a biomarker to predict response to NAC in MIBC patients. Seeking a streamlined assay for analysis of large-sample cohorts, we used the MethylationEPIC array to analyze cfDNA methylation from the 72-patient cohort with available plasma cfDNA in S1314. To our knowledge, this is the first report using the MethylationEPIC array to profile cfDNA methylation, with a majority of the MethylationEPIC array probes measured at high quality in all our patients. Compared with previous reports using bisulfite sequencing [26] or methylation immunoprecipitation sequencing [21], our approach offers the benefits of time efficiency, cost effectiveness, and a mature data analysis workflow, allowing for a high-throughput analysis of clinical samples with high reproducibility [27].
As there was no single methylation site with significant predictive value for the NAC response, we utilized Random Forest machine learning to combine multiple DMLs into one biomarker, the methylation-based resistance score (mR-score, which predicts NAC response with an ROC AUC of 63.6%). Although CR is a well-recognized endpoint prognostic of overall survival [28,29], here we chose to analyze CR and PR together as a classifier for NAC Rs versus NRs because studies demonstrated that a downstaging PR to NAC also confers survival benefits [30–32]. Perhaps more importantly, as NAC is the standard of care for cisplatin-eligible patients supported by level I evidence, we believe that it is more important to distinguish those who definitely will not derive any response or benefit (NRs) from all other patients (PR + CR). Encouragingly, plasma cfDNA collected after the first cycle of chemotherapy yielded a similarly predictive mR-score as the one calculated from prechemotherapy plasma, reflecting a robust and reproducible ability to predict NAC response versus nonresponse both before and during chemotherapy treatment. Furthermore, we analyzed the estimated pretreatment fraction of circulating bladder DNA as a surrogate for ctDNA and showed that it is an independent biomarker with high sensitivity for the identification of chemotherapy Rs. When using circulating bladder DNA and mR-scores together in a combined risk-stratification model, these accurately predicted response to NAC in 79% of the patients in the S1314 cohort. For patients predicted not to respond to NAC, upfront surgery or clinical trials may be offered in lieu of toxic and ineffective chemotherapy. In the current proof-of-concept model using the 57-patient cohort, this approach would avert treatment with ineffective NAC in ten patients at baseline and in another seven patients after the first cycle, at a cost of withholding beneficial NAC from three patients.
The cfDNA methylation-based biomarker strategies offer several advantages: First, DNA methylation is unique as an analyte in that it is slow to degrade but also functional, reflecting dynamic shifts in transcription patterns. Second, cfDNA provides methylation patterns and transcriptional regulation from tumor and normal cells throughout body, potentially reflecting not only tumor biology, but also host-tumor (eg, immune cell) interactions. The comprehensive information obtained in the cfDNA methylome may be complementary to tumor-specific information obtained in the standard pathology analysis and tumor tissue profiling. Third, cfDNA sampling is minimally invasive, enabling serial sample collection throughout treatment. In our study, we demonstrated that the mR-score obtained after the first cycle of chemotherapy treatment can be used to supplement the information obtained before chemotherapy to further risk-stratify patients receiving NAC. This on-treatment mR-score may prove to be especially helpful in patients not tolerating NAC and considering proceeding straight to surgery.
There are limitations associated with this study cohort. As an exploratory proof-of-principle cohort of limited size, its findings will require validation in additional large prospective cohorts. In addition, MIBC patients undergoing surgical resection have relatively limited disease burden compared with patients with metastatic disease, and therefore more limited tumor-specific methylation data are obtainable from the plasma cfDNA. Compared with reports of ctDNA fraction as high as 80% in patients with metastatic bladder cancer [33], we observed a relatively low circulating bladder DNA fraction of 0–3%. Furthermore, our study may be confounded by the transurethral resection of bladder tumor (TURBT)-bonus effect, wherein the diagnostic TURBT itself may have eradicated the tumor, especially in patients with cT2 disease (88% of patients in our study). While the TURBT-bonus effect may be reflected in the amount of ctDNA, our models were trained not on ctDNA methylation features but on all cfDNA methylation, of which bladder-derived DNA comprised a small portion (0–3%). Consequently, the mR-score was not driven by residual disease, as evidenced by its lack of correlation with circulating bladder DNA (Fig. 4B). Rather, the mR-score reflected collective host-tumor methylation profiles and therefore was less susceptible to the potential effect of TURBT-bonus on baseline ctDNA amounts. Indeed, it is likely that the majority of the DMLs we observed were associated with host factors including the immune system (peripheral blood leukocytes). Ultimately, this may prove to be a strength of the cfDNA-based approach, because the leukocyte contribution to cfDNA methylation profiles may help predict the response to immunotherapy, a newly established standard of care in metastatic urothelial carcinoma [34]. Indeed, this total plasma methylome approach can be developed further and validated in conjunction with other predictive biomarkers in the setting of metastatic disease, where it would potentially reflect both tumor and host methylation states associated with response to chemotherapy or immunotherapy.
5. Conclusions
We report a new approach using cfDNA methylation and machine learning to generate an mR-score—the probability of nonresponse to NAC in patients with MIBC. In a proposed risk-stratification model, we combine the mR-score with methylation-based quantitation of circulating bladder DNA to correctly predict NAC response in nearly 80% of analyzable patients in S1314, a prospective multicenter cooperative group trial.
Supplementary Material
Financial disclosure:
Amir Goldkorn certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Yi-Tsung Lu is an employee of, owns stock in, and has received travel and accommodation expenses from Seagen. Gareth Morrison is an employee of Private Health Management. Joshua J. Meeks participated in advisory boards for Ferring, AstraZeneca, BMS, Janssen, Foundation Medicine, Merck, and UroGen. Seth P. Lerner has funding for clinical trials from Endo, FKD, JBL (SWOG), Genentech (SWOG), QED, UroGen, Vaxiion, and Viventia; consulting fees from Aura Bioscience, C2i Genomics, FerGene, Genentech, Merck, Pfizer/EMD Serono, Stimit, UroGen, Vaxiion, and Verity; patent for TCGA classifier; and honoraria from Annenberg, Clinical Care Options, Grand Rounds Urology, Ology, and UroToday. Thomas W. Flaig reports grants and personal fees from Novartis, Bavarian Nordic, Dendreon, GTx, Janssen Oncology, Medivation, Sanofi, Pfizer, Bristol-Myers Squibb, Roche/Genentech, Exelixis, Aragon Pharmaceuticals, Sotio, Tokai Pharmaceuticals, AstraZeneca/MedImmune, Lilly, Astellas Pharma, Agensys, Seattle Genetics, La Roche-Posay, Merck, and Aurora Oncology, outside the submitted work. Ian M. Thompson Jr reports grants from NCI during the conduct of the study. David McConkey reports grants and personal fees from Bioclin, personal fees from Janssen and H3 Biomedicine, and grants from AstraZeneca, outside the submitted work. Amir Goldkorn has a patent for US Patent # 8,551,425 B2, 2013 issued and licensed to Corestone Biosciences, Circulogix. No disclosures were reported by the other authors.
Funding/Support and role of the sponsor:
Yi-Tsung Lu is supported by the Tower Cancer Research Foundation 2018 Career Development Award and the Conquer Cancer Foundation of the American Society of Clinical Oncology/Nebraska Oncology Society 2019 Young Investigator Award. Suhn K. Rhie is supported by the National Institute of Health (K01CA229995, R21CA260082, and R21HG011686) and the USC Norris Comprehensive Cancer Center (NCCC) Genomic and Epigenomic Regulation Grant. Kimberly D. Siegmund is supported by the USC NCCC data science core (P30CA014089). Amir Goldkorn is supported by the National Cancer Institute (R01CA257610, R01CA172436, and P30CA014089), as well as by the Kure It Foundation, the Coull Foundation, and the Hope Foundation for Cancer Research. This work was also supported by National Institute of Health, National Cancer Institute, and National Clinical Trials Network grants U10CA180888 and U10CA180819. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Data sharing: The cfDNA EPIC microarray data were deposited in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE193208.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Meeks JJ, Bellmunt J, Bochner BH, et al. A systematic review of neoadjuvant and adjuvant chemotherapy for muscle-invasive bladder cancer. Eur Urol 2012;62:523–33. [DOI] [PubMed] [Google Scholar]
- [2].Advanced Bladder Cancer Meta-analysis Collaboration. Neoadjuvant chemotherapy in invasive bladder cancer: update of a systematic review and meta-analysis of individual patient data advanced bladder cancer (ABC) meta-analysis collaboration. Eur Urol 2005;48:202–5; discussion 205–6. [DOI] [PubMed] [Google Scholar]
- [3].Choi W, Porten S, Kim S, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014;25:152–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Seiler R, Ashab HAD, Erho N, et al. Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur Urol 2017;72:544–54. [DOI] [PubMed] [Google Scholar]
- [5].Plimack ER, Dunbrack RL, Brennan TA, et al. Defects in DNA repair genes predict response to neoadjuvant cisplatin-based chemotherapy in muscle-invasive bladder cancer. Eur Urol 2015;68:959–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liu D, Plimack ER, Hoffman-Censits J, et al. Clinical validation of chemotherapy response biomarker ERCC2 in muscle-invasive urothelial bladder carcinoma. JAMA Oncol 2016;2:1094–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Groenendijk FH, de Jong J, Fransen van de Putte EE, et al. ERBB2 mutations characterize a subgroup of muscle-invasive bladder cancers with excellent response to neoadjuvant chemotherapy. Eur Urol 2016;69:384–8. [DOI] [PubMed] [Google Scholar]
- [8].Yang Z, Zhang R, Ge Y, et al. Somatic FGFR3 mutations distinguish a subgroup of muscle-invasive bladder cancers with response to neoadjuvant chemotherapy. EBioMedicine 2018;35:198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gil-Jimenez A, van Dorp J, Contreras-Sanz A, et al. Assessment of predictive genomic biomarkers for response to cisplatin-based neoadjuvant chemotherapy in bladder cancer. Eur Urol 2023;83:313–7. [DOI] [PubMed] [Google Scholar]
- [10].Van Allen EM, Mouw KW, Kim P, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov 2014;4:1140–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Flaig TW, Tangen CM, Daneshmand S, et al. A randomized phase II study of coexpression extrapolation (COXEN) with neoadjuvant chemotherapy for bladder cancer (SWOG S1314; NCT02177695). Clin Cancer Res 2021;27:2435–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med 2018;379:1754–65. [DOI] [PubMed] [Google Scholar]
- [13].Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov 2017;7:1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Patel KM, van der Vos KE, Smith CG, et al. Association of plasma and urinary mutant DNA with clinical outcomes in muscle invasive bladder cancer. Sci Rep 2017;7:5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zill OA, Banks KC, Fairclough SR, et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin Cancer Res 2018;24:3528–38. [DOI] [PubMed] [Google Scholar]
- [16].Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, Consortium C. Sensitive and specific multicancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol 2020;31:745–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Powles T, Assaf ZJ, Davarpanah N, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature 2021;595:432–7. [DOI] [PubMed] [Google Scholar]
- [18].Greenberg MVC, Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol 2019;20:590–607. [DOI] [PubMed] [Google Scholar]
- [19].Moss J, Magenheim J, Neiman D, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun 2018;9:5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li W, Li Q, Kang S, et al. CancerDetector: ultrasensitive and non-invasive cancer detection at the resolution of individual reads using cell-free DNA methylation sequencing data. Nucleic Acids Res 2018;46:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018;563:579–83. [DOI] [PubMed] [Google Scholar]
- [22].Beltran H, Romanel A, Conteduca V, et al. Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J Clin Invest 2020;130:1653–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Das J, Gayvert KM, Bunea F, Wegkamp MH, Yu H. ENCAPP: elastic-net-based prognosis prediction and biomarker discovery for human cancers. BMC Genomics 2015;16:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wyatt AW, Annala M, Aggarwal R, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst 2017;109:djx118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Freiha F, Reese J, Torti FM. A randomized trial of radical cystectomy versus radical cystectomy plus cisplatin, vinblastine and methotrexate chemotherapy for muscle invasive bladder cancer. J Urol 1996;155:495–9; discussion 499–500. [PubMed] [Google Scholar]
- [26].Wu A, Cremaschi P, Wetterskog D, et al. Genome-wide plasma DNA methylation features of metastatic prostate cancer. J Clin Invest 2020;130:1991–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 2016;17:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Grossman HB, Natale RB, Tangen CM, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med 2003;349:859–66. [DOI] [PubMed] [Google Scholar]
- [29].Petrelli F, Coinu A, Cabiddu M, Ghilardi M, Vavassori I, Barni S. Correlation of pathologic complete response with survival after neoadjuvant chemotherapy in bladder cancer treated with cystectomy: a meta-analysis. Eur Urol 2014;65:350–7. [DOI] [PubMed] [Google Scholar]
- [30].Rosenblatt R, Sherif A, Rintala E, et al. Pathologic downstaging is a surrogate marker for efficacy and increased survival following neoadjuvant chemotherapy and radical cystectomy for muscle-invasive urothelial bladder cancer. Eur Urol 2012;61:1229–38. [DOI] [PubMed] [Google Scholar]
- [31].Martini A, Jia R, Ferket BS, et al. Tumor downstaging as an intermediate endpoint to assess the activity of neoadjuvant systemic therapy in patients with muscle-invasive bladder cancer. Cancer 2019;125:3155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cajipe M, Wang H, Elshabrawy A, et al. Pathological downstaging following radical cystectomy for muscle-invasive bladder cancer: survival outcomes in the setting of neoadjuvant chemotherapy versus transurethral resection only. Urol Oncol 2020;38:231–9. [DOI] [PubMed] [Google Scholar]
- [33].Vandekerkhove G, Todenhofer T, Annala M, et al. Circulating tumor DNA reveals clinically actionable somatic genome of metastatic bladder cancer. Clin Cancer Res 2017;23:6487–97. [DOI] [PubMed] [Google Scholar]
- [34].Ghasemzadeh A, Bivalacqua TJ, Hahn NM, Drake CG. New strategies in bladder cancer: a second coming for immunotherapy. Clin Cancer Res 2016;22:793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.