Abstract
Immunological experiences lead to the development of specific T and B cell memory, which readies the host for a later pathogen rechallenge. Currently, immunological memory is best understood as a linear process whereby memory responses are generated by and directed against the same pathogen. However, numerous studies have identified memory cells that target pathogens in unexposed individuals. How “pre-existing memory” forms and impacts the outcome of infection remains unclear. In this review, we discuss differences in the composition of baseline T cell repertoire in mice and humans, factors that influence pre-existing immune states, and recent literature on their functional significance. We summarize current knowledge on the roles of pre-existing T cells in homeostasis and perturbation and their impacts on health and disease.
Introduction
The COVID-19 pandemic has now crossed the third-year mark since it began in late 2019. Rapid advances in vaccine and antiviral treatments have largely mitigated its widespread devastation, but SARS-CoV-2 infection remains a significant cause of death and disease burden with millions of people suffering from its long-term sequela (1–3). SARS-CoV-2 is not the only pathogen that poses a significant global threat. Major pandemics have plagued humankind throughout history (4). In recent decades, other viruses, such as HIV, Ebola virus, and Zika virus, have inflicted morbidity and mortality in many parts of the world. Climate change and human influences have contributed to spillover from animal reservoirs to humans and increased the opportunities for the evolution of novel variants. Global travel and trade have further blurred geographical boundaries and elevated the risk of another pandemic threat (5).
Public health officials, scientists, and national security experts are on the lookout for the next emerging pathogen. Fortunately, the immune system has also evolved to prepare for the unforeseeable and is armed for protection. Upon infection, innate immune cells sense pathogens through pattern recognition receptors and activate inflammatory processes to mediate the first line of defense (6). This is followed by adaptive immune cells, which are endowed with a diverse repertoire of receptors that specifically recognize and target a particular pathogen. Activated CD8+ T cells produce cytokines and mediate direct lysis of infected cells, whereas CD4+ T cells boost innate immune cells, support functional CD8+ T cells, and provide essential signals for B cell maturation and high-affinity Ab production (7–9). Protective Abs have multifaceted functions with direct pathogen neutralization capacity (10). Abs are also powerful arsenals that fix complements and engage innate effector cells to drive killing and opsonization (11). Together, these coordinated responses eliminate the threat, restore homeostasis, and establish a new baseline state of heightened vigilance.
Variations of this scenario play out repeatedly throughout the life of an individual in response to various forms of microbial challenges. A major unresolved question is how one’s response to a new pathogen is shaped by what came before in a long series of exposures. In turn, how does the current response impact the future state of health or disease? Immunological memory refers to the ability of the immune system to learn from past experiences. For the adaptive lymphocytes, a small subset of cells that have productively engaged the relevant microbes is maintained as long-lived memory cells. These cells retain the history of past encounters and are recalled in the event of a future rechallenge. T cell memory encompasses numerous distinct cellular states and is frequently subdivided based on cellular phenotype, function, and site of residence (Fig. 1). The advantage of memory T cells has been ascribed to their larger starting number, faster division rate, and accelerated effector responses (12–15). The ability to rapidly initiate a functional response underlies protective immunity from past infections and has been leveraged for disease prevention by vaccination. Immunological memory is primarily studied in the context of a previously encountered Ag. In this article, we expand on the classical framework of T cell memory to discuss memory responses to never-before-seen pathogens, referred to as pre-existing memory. We discuss the various ways that pre-existing memory could form, and we consider how they influence host responses to a novel pathogen. Because many key findings for T cell memory were initially discovered using animal models, we highlight relevant findings in mice for comparison. Finally, we reflect on open questions in the field and suggest future directions for leveraging pre-existing memory for protective immunity and beyond.
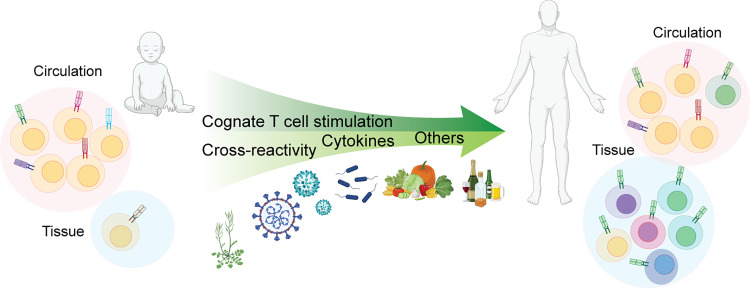
FIGURE 1.
The baseline T cell repertoire dynamically evolves over time.
The T cell repertoire is shaped by a combination of endogenous immune processes and external inputs from the environment. In young children, it consists mainly of diverse naive T cells with robust proliferative and differentiation potential. T cell stimulation from cognate Ags, cross-reactive epitopes, cytokine signals, and other changes in the cellular environment influence T cell differentiation and the establishment of immunological memory. Memory T cells exhibit rapid, specialized responses, express diverse trafficking receptors, and provide surveillance in circulation and tissue environments.
T cell memory in the absence of foreign Ags
T cells recognize specific peptide–histocompatibility complex (MHC) molecules through a heterodimeric TCR complex. During an infection, productive TCR engagement with pathogen-presenting dendritic cells activates naive T cells, leading to proliferation and effector differentiation. After pathogen clearance, a subset of previously activated cells acquires a memory program, preparing them for future defense against the same pathogen (16–18). This classical view that centers on foreign Ags as the primary driver of T cell memory differentiation has been challenged by the unexpected discovery that mice raised in gnotobiotic conditions, devoid of microbial exposures, still possess memory phenotype T cells (19, 20). It was also known that naive T cells transferred into various lymphopenic settings will spontaneously upregulate typical memory markers and undergo cell division (21, 22). Among cells that proliferated, a subset will continue to divide in the absence of microbes and food Ags (23, 24). In other studies, 10–30% of CD8+ T cells recognizing a model Ag or viral epitopes in unprimed mice exhibited a memory phenotype by CD44 expression. The abundance of these memory cells was not diminished in mice raised under germ-free conditions (25).
These key initial observations ushered in a period of intense investigation on how memory can be generated independent of environmental Ags. Memory T cells found in unprimed mice have been variously labeled as virtual memory (VM), endogenous memory, and homeostatic proliferating memory T cells depending on the experimental context (16, 26–28). Data from many laboratories have shown that the generation and maintenance of these cells require TCR tonic signaling in an MHC-dependent manner (24, 29–33). Taking advantage of CD5 expression to infer the strength of self-MHC interaction (34, 35), several groups have demonstrated that naive T cells with high CD5 levels give rise to more VM cells than those with low CD5 expression (33, 36, 37). Experimental manipulations that alter TCR signaling have been shown to further modify VM differentiation. For example, blocking costimulation by CD28 gene knockout and CTLA4-Ig treatment resulted in fewer VM CD4+ T cells (22, 33), whereas more CD8+ VM T cells were found in dedicator of cytokinesis 2–deficient mice that have an increase in TCR sensitivity to weak agonists (38).
Beyond TCR signaling, γ-chain cytokines are critical for VM differentiation. A crucial role for IL-15 in CD8+ VM induction was demonstrated by the absence of VM CD8+ T cells in IL-15−/−, IL-15Ra−/−, and CD122 (IL5Rb)−/− mice and the observation that memory cells can expand in unprimed animals after IL-15 stimulation (37, 39). CD8+ T cell proliferation and memory differentiation under lymphopenic conditions also require IL-7 (21, 32). By contrast, CD4+ T cells are less prone to differentiate without a known source of Ag in mice. For example, the CD4+ subset in unprimed mice contains fewer CD44highCD49dlow VM T cells compared with CD8+ T cells in the same animal (39). Direct tetramer analyses of foreign Ag–specific CD4+ T cells in unexposed adult mice also found the majority retained a naive phenotype (40). In general, homeostatic responses to cytokines appear to be subdued in CD4+ T cells. CD4+ T cells express lower levels of an IL-15R subunit (CD122) and monosialotetrahexosylganglioside, which are critical for optimal responses to IL-15 and IL-2 (32, 41). In addition, the activity of IL-7 on CD4+ T cells is blunted by reduced Ag presentation caused by IL-7–mediated downregulation of class II MHC molecules on IL-7Rα–expressing dendritic cells (42).
VM cells are functionally competent. Jameson and colleagues (43) studied OVA-specific VM cells using mice with a fixed TCR β-chain of OVA-specific OT-I that pairs with endogenously rearranged TCRα. This showed that VM CD8+ T cells produced less IFN-γ in response to OVA peptides in vitro but were comparably protective as true Ag-experienced memory cells against OVA-expressing L. monocytogenes infection (43). Protection is not strictly dependent on the recognition of cognate Ag. White et al. (37) showed a reduction in OVA-expressing L. monocytogenes bacteria load in mice that received VM-transgenic T cells recognizing an HSV epitope. Immune protection required IL-15 and was attributed to bystander killing of infected cells by IL-15–activated Ag-mismatched CD8+ T cells (37). Whether humans have an equivalent population of VM cells remains unclear. The study of VM in humans is complicated by divergent programs that regulate typical murine VM markers and clear limitations on direct manipulation of environmental exposures in people. Nevertheless, the discovery of homeostatic memory in mice is important because it raises broader questions on how endogenous basal signals are perceived by T cells and influence their differentiation and function.
Influences of the environment on T cell differentiation
Although memory phenotype T cells are present even in unprimed gnotobiotic mice, the living environment drives further maturation of T cells that are critical for protective and tolerogenic responses. The term “homeostasis” was coined by Walter Cannon in the early 1900s (44). It describes the coordinated responses that maintain internal stability and require dynamic integration between endogenously driven processes and external inputs. The natural environment contains an abundance of bacteria, fungi, and viruses that live in the soil and on plants and animals (45). An extensive network of adaptive and innate immune cells calibrates the immune responses to this complex microbial ecosystem, which drives early-life immune development, mediates tissue repair, and constrains inflammatory responses (46–49).
In addition to commensal organisms, the free-living environment also contains many pathogens. Latent viruses such as CMV cause lifelong infections globally (50, 51). Statistical estimates for the influenza virus showed that 3 to 11% of the population in the United States develop symptomatic infection each year (52). Since late 2019, more than six hundred million people worldwide have been infected and reinfected by SARS-CoV-2 and its many variants. For other major infectious diseases, the World Health Organization estimates that 38.4 million people live with HIV, 10.6 million people developed tuberculosis-related illnesses, and >247 million people became infected by malaria in 2021. The extent to which our immune system retains divergent inputs from the environment was addressed in a landmark twin study by Brodin et al. (53) Using covariance between monozygotic and dizygotic twins to estimate nonheritable influences, the authors showed that more than half of the 204 measured immune parameters were largely not genetically determined (53). Even for factors that showed strong concordance between young monozygotic twins, many diverged with age and discordance in CMV seropositivity. Thus, along with genetics, age, sex, and other host characteristics, environmental influences contribute to the heterogeneity of human immune responses (Fig. 1).
To address how the environment impacts T cell memory and responses, there is a growing effort to expose animal models to complex environments. Even without gnotobiotic containment, laboratory mice raised in hygienic specific-pathogen-free (SPF) conditions lack the breadth of exposures that a typical human would experience. Restricting microbial exposures is necessary to protect vulnerable genetically modified murine strains, but differences in Ag experiences also contribute to divergent data between many preclinical mice studies and human clinical trials (54–58). A seminal study by Masopust and colleagues (59) showed that mice raised in SPF conditions have T cells that largely mirror the naive state of a human newborn. By contrast, CD8+ T cells from mice trapped in the wild or obtained from the pet store have more differentiated T cells characterized by a granzyme B and KLRG1-expressing effector memory phenotype. Like adult humans, a greater population of T cells from free-living mice reside in nonlymphoid tissues (59, 60). These changes in T cell maturation and residency are transferable by cohousing SPF mice with pet-store mice, which confers resistance to infection by L. monocytogenes and other pathogens (59). Since the initial description of “dirty mice,” several other approaches have been developed to introduce diverse microbial exposures to laboratory mice. These include wild-mouse fecal transfers and “wildlings” created by transferring embryos into pseudopregnant wild mice (61, 62). Collectively, these models reveal key aspects of memory differentiation that are influenced by the environment. In doing so, they also improve the translational tools to better recapitulate human responses to vaccine and drug treatments (62, 63).
How do exposures to seemingly unrelated microbes in the living environment drive memory differentiation? The general concept that cells can adapt to prior inflammatory stimuli is not confined to T cells. Inflammation-driven epigenetic reprogramming and transcriptional changes underlie “trained memory,” which results in an altered baseline state in innate immune cells and epithelial tissues after recovery from an inflammatory insult (64). What is distinct for adaptive immune cells are their unique Ag recognition receptors. Because T cells express individualized TCRs, memory not only represents a variety of distinct cellular states but also modifies the composition of Ag-receptor repertoire. Cells expressing specific TCRs that can appropriately engage the relevant Ags are selectively expanded from a diverse precursor pool. Even when cytokines have a dominant role, as seen in VM in mice, memory differentiation is biased toward cells receiving stronger tonic signals from self-MHC complexes (33, 37).
Key studies from Selin, Welsh, and colleagues (65, 66) established the early conceptual framework for understanding how TCR recognition could bridge unrelated microbial experiences. By infecting mice sequentially with different viruses, they revealed that heterologous infections modified the hierarchy of responding T cells, and this involved activation of cross-reactive memory cells seeded by responses to an earlier pathogen (67–70). TCR cross-reactivity refers to the ability of a TCR to bind multiple distinct peptide-MHC (pMHC) complexes. The clonal selection theory proposed that each T cell recognizes a unique pMHC molecule. However, the number of distinct T cell clones, estimated to be on the order of 106, is far lower than the theoretical estimate of 109–1012 potential foreign Ags that can be presented by a given MHC molecule (71, 72). Several experimental approaches have been used to interrogate TCR cross-reactivity. These include synthetic peptide libraries (73, 74), yeast displays (75, 76), multiplexed tetramer staining (77, 78), Ag-specific lentiviral-mediated cell entry (79, 80), and reporter-based systems (81–83). Experimental estimates of cross-reactivity vary depending on screening platforms, epitope coverage, and the choice of TCRs, ranging from several thousand to more than one million peptides for a single receptor (73, 74, 76, 81). TCR cross-reactivity is enabled by the flexibility of CDR loops and receptor docking orientation (84–88). Further, TCRs focus on a small number of “hot spots” and are tolerant of variations in other peptide positions (89). Peptide binding to MHC molecules is also adaptable, with binding-induced register shifts and extensions from MHC binding grooves contributing to a greater range of peptide-loaded MHC complexes (90). Don Mason (72) argued in his seminal thesis that TCR cross-reactivity is necessary to overcome the practical constraints of having more potential foreign peptides than the space available to accommodate a similarly high number of T cells. From an evolutionary perspective, this expanded scope may have been critical for our survival by providing the host with greater protection against diverse pathogens and their variants.
The many drivers of pre-existing memory in human T cells
Unlike mouse models in which various endogenous and exogenous drivers of memory responses can be experimentally dissected, the sources that induce a particular naive T cell to acquire a memory program are more challenging to determine in humans. In probing the signals from past exposures in humans, we and others have identified pre-existing T cells to microbial Ags in unexposed individuals (Fig. 2). Using class II pMHC tetramers to identify virus-specific T cells, we found that uninfected blood donors could have a high proportion of CD4+ memory phenotype T cells that recognized HIV, CMV, and HSV with a negative Ab test for these infections (91). Albeit unlikely, unsuspected exposures are possible for these relatively common viruses. A more stringent test on T cell responses to the geographically restricted yellow fever virus (YFV) also showed memory phenotype in >50% of YFV-specific T cells from unexposed adults (92). Another study by Campion et al. (93) stimulated CD4+ T cells with overlapping peptides spanning HIV and Ebola proteome and similarly uncovered T cell responses within the memory phenotype sorted fraction from unexposed individuals. After the emergence of SARS-CoV-2, many laboratories have identified T cells that recognize SARS-CoV-2 in prepandemic blood samples (94–102). Several groups have also found pre-existing SARS-CoV-2–specific T cells in lymphoid and nonlymphoid tissues, generating excitement over the possibility that these T cells could mediate protective local immunity (103–105). Using adult and pediatric tonsils collected between 2015 and 2018, Niessl et al. (103) identified CD8+ T cell responses to SARS-CoV-2 peptides that were enriched in tonsils compared with matched blood. Responding CD8+ T cells included cytokine-producing tonsillar cells with increased expression for CXCR5 and tissue-resident memory T cell (TRM) markers, CD103 and CD69. Later studies on unexposed individuals have also identified cytokine responses to SARS-CoV-2 peptides by TRM cells in the bone marrow and bronchial alveolar lavage (104, 105).
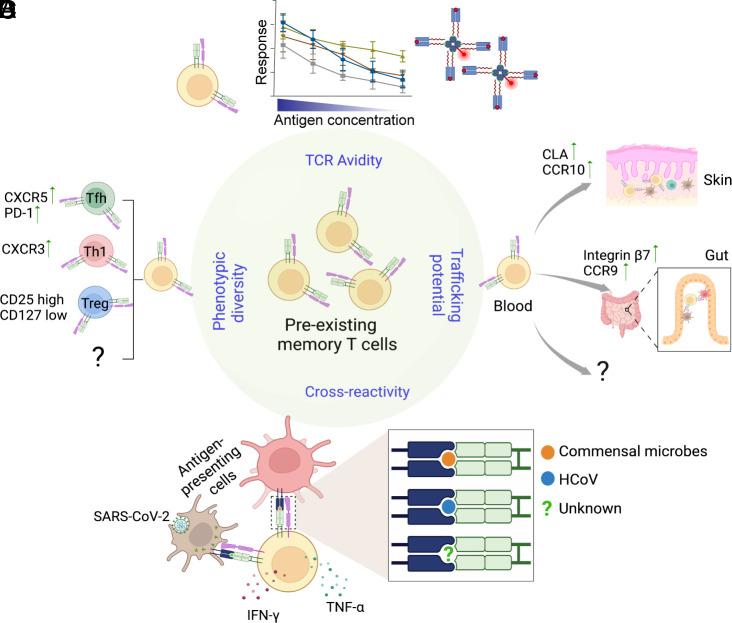
FIGURE 2.
The pre-existing memory repertoire is composed of a diverse array of T cells.
(A) Flexible TCR recognition enables T cell memory elicited by a particular Ag to recognize and respond to a range of other peptide–MHC complexes. For example, pre-existing memory to SARS-CoV-2 includes cross-reactive T cells that recognize Ags from HCoV, commensal microbes, and likely other yet-to-be-defined sources. (B) The pre-existing repertoire includes memory phenotype T cells that have already acquired disparate differentiation states. (C) Memory precursors express tissue-homing receptors. Different tissue tropism is likely imprinted by prior Ag encounters and reveals unique histories of antigenic experiences. (D) The memory repertoire established from past exposures consists of a diverse population of T cells with varying levels of TCR binding strength and responsiveness to cross-reactive Ags.
Pre-existing memory is typically associated with cross-reactive responses to related pathogens (Figs. 1, 2). For SARS-CoV-2, the widely circulating human coronavirus (HCoV) is the most obvious candidate because it shares high sequence homology in several genomic regions with SARS-CoV-2 (106). Cross-reactivity between SARS-CoV-2 and HCoV is supported by experimental evidence showing that several SARS-CoV-2 peptide-expanded T cell lines can indeed respond to analogous HCoV sequences (95, 107, 108). However, estimates of cross-reactivity to HCoV vary according to experimental systems and are infrequent in some donors (101). The existence of pre-existing memory T cells that recognize viruses without a close relative to simulate prior exposure, such as YFV, further suggests other avenues for acquiring an Ag-experienced state at baseline (92, 93). A broader consideration for the sources of cross-reactive Ags by Bartolo et al. (102) identified SARS-CoV-2–specific CD4+ T cells that recognized commensal bacteria-derived peptides. Using pMHC tetramers to probe Ag recognition of peripheral T cells collected before the pandemic, we found ∼20% of CD4+ T cells that recognized a spike peptide also costained with bacterial-peptide-loaded tetramers. This level of cross-recognition was comparable with HCoV-derived sequences. Cross-reactivity was further supported by responses to lysates made from the stool and cultured bacteria. Single-cell-derived SARS-CoV-2–specific T cell clones produced cytokines after in vitro stimulation by a skin microbe, Staphylococcus epidermidis, and gut bacteria, Prevotella copri and Bacteroides ovatus. Although it is unknown whether exposures to a particular organism had occurred in these donors, the commensal bacteria examined are common in the general population and are likely to colonize individuals in this study. Our ability to uncover cross-reactive responses after testing a handful of bacteria argues for an even greater likelihood that additional responses to bacteria, fungi, viruses, and other microbes could occur in the natural living environment (Fig. 2A). Consistent with this, Pothast et al. (109) generated CD4+ and CD8+ T cell clones from prepandemic samples and identified cross-reactive responses between SARS-CoV-2 and pp65 of CMV. The same cross-reactive αβTCR sequences were found in CD8+ T cells from multiple CMV+ donors, suggesting that shared exposures could leave a similar imprint on the basal immune repertoire across individuals.
In addition to TCR cross-reactivity, prior experiences can influence T cell baseline responses by modulating the cellular environment (Fig. 1). For example, early-life experiences regulate secretory IgA, which targets commensal bacteria to modulate the extent of systemic exposure (110). In IgA-deficient pediatric patients, the absence of fecal IgA was associated with systemic immune dysregulation that included higher levels of proinflammatory cytokines, altered CD8+ T cell phenotypes, and increased frequency of circulating T follicular helper (Tfh) cells (111). Recently, longitudinal multiomic profiling of influenza vaccine response discovered persistent immune changes characterized by an increase in GPR56+CD8+ T cells in male recoverees after mild COVID-19 (112). Instead of a classical TCR-driven response, GPR56+CD8+ T cells could be stimulated by IL-15 in vitro. In addition, their elevated basal frequency was associated with an early IFN-γ response, higher Ab titer, and an increase in plasmablast frequency after influenza vaccination. Thus, prior Ag experiences can shape T cell responses by modifying Ag availability, cytokine signals, and other relevant factors in the broader cellular environment. How extensive these changes may be, how long they last, and what determines the variability between individuals are major questions that remain unanswered. In the future, in-depth characterization of longitudinal human responses, in combination with mechanistic studies in animal models, may provide insights into how distinct immune pathways interconnect across time and space to shape the pre-existing immune landscape.
Human pre-existing responses to vaccines and infections
Are pre-existing memory T cells beneficial? After the emergence of SARS-CoV-2, many groups have examined the relationship between pre-existing T cells, Ab responses, and clinical outcomes. COVID-19 exhibits diverse clinical presentations, organ involvement, and symptom severity, ranging from asymptomatic infection to death (113–118). Using a closely monitored longitudinal cohort of health care workers recruited in March 2020, Swadling et al. (119) identified PCR and Ab-negative individuals who likely had an abortive SARS-CoV-2 infection. Resistance was attributed to cross-reactive T cell response against the highly conserved early transcribed replication-transcription complex, which was higher in seronegative health care workers works at baseline and further increased during the follow-up period (119). T cell responses to several conserved epitopes that included replication-transcription complex sequences were also elevated in another group of COVID-19–exposed PCR-negative individuals (120). Fine mapping of CD8+ T cell response by multimer staining showed a higher baseline for peptides with high sequence similarity to other HCoVs and a stronger response to these cross-reactive epitopes in patients with mild COVID-19 compared with those with severe disease (121). Cross-reactive CD8+ T cells were able to reduce intracellular SARS-CoV-2 copies in an airway epithelial cell line at low multiplicity of infection, suggesting a potential role in limiting viral spread during early stages of infection (109). With respect to CD4+ T cells, a higher reactivity to SARS-CoV-2 peptides at baseline has been linked to Tfh cell frequency and a more robust Ab response after vaccination and infection (122, 123). Because a substantial portion of the pre-existing response was directed toward an epitope in the HCoV homologous spike region (S-II), the decline in the overall S-II response with age was interpreted as a loss of protection from pre-existing memory in older individuals (122).
Although the earlier studies collectively support a protective effect from pre-existing memory, further studies into the nature of pre-existing cells have revealed heterogeneity within this population and the responses that they generate (Figs. 2, 3). In-depth analyses of pre-existing memory repertoire have uncovered cells with a broad range of Ag avidity, receptor diversity, and differentiation states (101, 102, 124). For example, single-cell RNA sequencing performed on SARS-CoV-2–reactive CD4+ T cells from unexposed donors uncovered Th1 and Tfh cell-related transcriptional profiles (101). CXCR3 and CXCR5 protein expression were also detected in a subset of SARS-CoV-2–specific T cells identified by direct ex vivo tetramer staining from prepandemic blood (Fig. 2B) (102). Furthermore, pre-existing memory cells likely have a range of tissue tropism based on divergent patterns of trafficking receptor expression (Fig. 2C). In unexposed individuals, distinct SARS-CoV-2–specific memory populations expressed gut homing receptors, Integrin β7 and CCR9, and skin trafficking receptors, cutaneous lymphocyte-associated Ag and CCR10 (102). With respect to T cell recognition, emerging data reveal a wide range of T cell binding strength and responsiveness to cross-reactive Ags within the pre-existing memory repertoire (Fig. 2D). This heterogeneity includes cells that exhibit reduced response to the current immunological threat (101, 107). Bacher et al. (101) uncovered memory phenotype CD4+ T cells in unexposed donors that recognized SARS-CoV-2 but required higher peptide concentration to respond. These pre-existing memory cells were largely nonreactive to HCoV and increased with age. Low-avidity responses were also enriched in patients with severe COVID-19 compared with mild disease, suggesting that weakly responsive memory precursors have limited protective effects (101). Fortunately, existing evidence suggests that a diverse T cell repertoire is capable of overcoming low-avidity pre-existing responses through dynamic reorganization of T cell hierarchy. Saggau et al. (124) used TCR sequencing to track CD4+ T cell responses to COVID-19 vaccination and found a replacement of pre-existing clonotypes with newly arising sequences in efficacious responses. Consistent with this, we had shown that high-frequency YFV-specific CD4+ precursors remained abundant during the first week after YFV vaccination but subsequently underwent limited expansion (92). Instead of further increasing the already expanded clones, the YFV vaccine preferentially boosted initially rare populations with a diverse TCR repertoire, including de novo responses and a subset of pre-existing memory precursors. Clonal reorganization was associated with preferential recruitment of high-avidity cells and resulted in improved viral Ag recognition by tetramer staining after vaccination. These data highlight the dynamic nature of the T cell repertoire, where a diverse range of T cells compete to engage with and respond to Ags during each encounter. As such, pre-existing memory may have broader implications beyond the immediate immune response. By actively competing with other T cells, they could shape the composition of the subsequent immune baseline and influence future responses to other immunologic challenges.
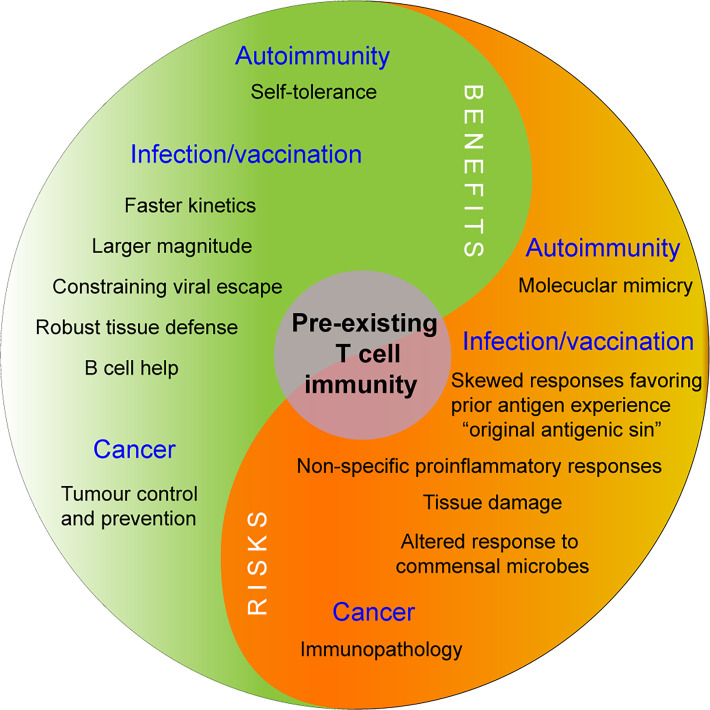
FIGURE 3.
Divergent influences of pre-existing T cell memory.
Pre-existing functional polarization may contribute to varied responses after stimulation. Having an existing reservoir of TRMs or the ability to rapidly mobilize tissue surveillance may facilitate local immunity but could also lead to immunopathology if inappropriately regulated. The impact of pre-existing memory T cells is likely context dependent and can have both positive and negative influences on immune health. Future strategies aimed at selectively modulating pre-existing memory may provide new opportunities to enhance protective immunity against cancer or infection and restore immune tolerance in autoimmune diseases.
Conclusion and open questions
Humans are exposed to various inflammatory and noninflammatory stimuli throughout our lifetime. The scientific community is now just beginning to untangle different aspects of these experiences and determine how they modify immune responses and health outcomes. The emergence of COVID-19 has highlighted how past exposures to similar and dissimilar stimuli are relevant factors to consider in human responses to pathogens, but the fundamental questions on this type of immunological memory extend beyond a single virus. Within each of us exist T cells that can recognize novel pathogens. This complex repertoire of T cells is influenced by our past immunological experiences. Some pre-existing T cells circulate, whereas others reside in tissues. A substantial portion of these cells are already Ag experienced and have specialized trafficking and differentiation programs. Layered experiences from various Ags and exposure conditions likely contribute to the diversity of pre-existing memory.
A major unaddressed question is how different pre-existing subsets individually and collectively contribute to a particular outcome. Which types of pre-existing responses are needed will likely also depend on the nature of the infection. Although memory cells have generally been viewed as a superior source of protective immunity, memory cells generated by past stimulus from a different source may or may not recall T cells that are best suited for the current immunological challenge. However, even for memory precursors with a lower avidity, the ability to quickly mobilize a sizable number of T cells could be critical at the very beginning of an infection or to control slower-replicating pathogens. As the immune responses evolve, the ability to recruit “best-fit” T cells to arm the immune system with the most responsive T cells may be needed for durable protective immunity. Having a diverse repertoire of T cells preserves the availability of alternative TCRs if currently expanded clones are unsuitable. For fast-evolving pathogens, a diverse T cell repertoire can serve to constrain the emergence of escape variants as mutations arise, thereby providing an additional layer of protection. Factors that alter the underlying clonal structure and cellular environment, such as advanced age, chronic infections, cancers, and autoimmune diseases, will likely influence T cell responses to pathogens. Decoding how pre-existing memory forms and functions is highly relevant for next-generation vaccine design. Advances in this area may enable approaches that elicit a particular type of pre-existing T cell population or use preconditioning exposures as novel strategies to tailor the immune responses toward a desired outcome in future studies.
Although we have focused on pre-existing T cell responses to pathogens, T cells also recognize Ags derived from other sources, such as commensal microbes, food Ags, and self-proteins. Given the breadth of T cell cross-reactivity and the longevity of some Ag-experienced T cells, we anticipate that there will be multidirectional cross-talks between homeostatic and antipathogen immunity. Indeed, molecular mimicry, whereby responses elicited by foreign Ags cause a misdirected response to similar-appearing self-antigens, has been suggested to drive many autoimmune diseases (125, 126). Infections can also alter the microbiome (127, 128), although the direct effects on commensal-reactive T cells are still unclear. Our data showing cross-reactivity between SARS-CoV-2 and skin and gut bacteria suggest that cross-talk via shared recognition is theoretically possible. Studies to date have largely focused on healthy individuals. Much remains unknown on how the baseline T cell repertoire is altered in the context of disease and how these changes, in turn, modify T cell homeostasis and responses to perturbation. In the future, understanding the roles of pre-existing memory in a disease setting may provide new opportunities for modulating the immune repertoire toward a tolerogenic or proinflammatory state in autoimmunity, cancer, and chronic infections.
Acknowledgments
The figures were created in part with BioRender.com.
Footnotes
This work was supported by the National Institutes of Health (R01AI134879 and R01AI66358 to L.F.S.) and the Department of Veterans Affairs (IMMA-020-15F and I01BX005422 to L.F.S.).
- HCoV
- human coronavirus
- pMHC
- peptide-MHC
- SPF
- specific-pathogen-free
- Tfh
- T follicular helper
- TRM
- tissue-resident memory T cell
- VM
- virtual memory
- YFV
- yellow fever virus
Disclosures
The authors have no financial conflicts of interest.
References
- 1. Davis, H. E., McCorkell L., Vogel J. M., Topol E. J.. 2023. Long COVID: major findings, mechanisms and recommendations. [Published erratum appears in 2023 Nat. Rev. Microbiol. 21: 408.] Nat. Rev. Microbiol. 21: 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flaxman, S., Whittaker C., Semenova E., Rashid T., Parks R. M., Blenkinsop A., Unwin H. J. T., Mishra S., Bhatt S., Gurdasani D., Ratmann O.. 2023. Assessment of COVID-19 as the underlying cause of death among children and young people aged 0 to 19 Years in the US. JAMA Netw. Open 6: e2253590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. El-Sadr, W. M., Vasan A., El-Mohandes A.. 2023. Facing the new Covid-19 reality. N. Engl. J. Med. 388: 385–387. [DOI] [PubMed] [Google Scholar]
- 4. Piret, J., Boivin G.. 2020. Pandemics throughout history. [Published erratum appears in 2022 Front. Microbiol. 13: 988058.] Front. Microbiol. 11: 631736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baker, R. E., Mahmud A. S., Miller I. F., Rajeev M., Rasambainarivo F., Rice B. L., Takahashi S., Tatem A. J., Wagner C. E., Wang L. F., et al. 2022. Infectious disease in an era of global change. Nat. Rev. Microbiol. 20: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diamond, M. S., Kanneganti T. D.. 2022. Innate immunity: the first line of defense against SARS-CoV-2. Nat. Immunol. 23: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laidlaw, B. J., Craft J. E., Kaech S. M.. 2016. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat. Rev. Immunol. 16: 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swain, S. L., McKinstry K. K., Strutt T. M.. 2012. Expanding roles for CD4(+) T cells in immunity to viruses. Nat. Rev. Immunol. 12: 136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crotty, S. 2019. T follicular helper cell biology: a decade of discovery and diseases. Immunity 50: 1132–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen, Y., Zhao X., Zhou H., Zhu H., Jiang S., Wang P.. 2023. Broadly neutralizing antibodies to SARS-CoV-2 and other human coronaviruses. Nat. Rev. Immunol. 23: 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang, A., Stacey H. D., D’Agostino M. R., Tugg Y., Marzok A., Miller M. S.. 2023. Beyond neutralization: Fc-dependent antibody effector functions in SARS-CoV-2 infection. Nat. Rev. Immunol. 23: 381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fraser, K. A., Schenkel J. M., Jameson S. C., Vezys V., Masopust D.. 2013. Preexisting high frequencies of memory CD8+ T cells favor rapid memory differentiation and preservation of proliferative potential upon boosting. Immunity 39: 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Veiga-Fernandes, H., Walter U., Bourgeois C., McLean A., Rocha B.. 2000. Response of naive and memory CD8+ T cells to antigen stimulation in vivo. Nat. Immunol. 1: 47–53. [DOI] [PubMed] [Google Scholar]
- 14. Rogers, P. R., Dubey C., Swain S. L.. 2000. Qualitative changes accompany memory T cell generation: faster, more effective responses at lower doses of antigen. J. Immunol. 164: 2338–2346. [DOI] [PubMed] [Google Scholar]
- 15. Pihlgren, M., Dubois P. M., Tomkowiak M., Sjogren T., Marvel J.. 1996. Resting memory CD8+ T cells are hyperreactive to antigenic challenge in vitro. J. Exp. Med. 184: 2141–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sprent, J., Surh C. D.. 2011. Normal T cell homeostasis: the conversion of naive cells into memory-phenotype cells. Nat. Immunol. 12: 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chakraborty, A. K., Weiss A.. 2014. Insights into the initiation of TCR signaling. Nat. Immunol. 15: 798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Akondy, R. S., Fitch M., Edupuganti S., Yang S., Kissick H. T., Li K. W., Youngblood B. A., Abdelsamed H. A., McGuire D. J., Cohen K. W., et al. 2017. Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552: 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dobber, R., Hertogh-Huijbregts A., Rozing J., Bottomly K., Nagelkerken L.. 1992. The involvement of the intestinal microflora in the expansion of CD4+ T cells with a naive phenotype in the periphery. Dev. Immunol. 2: 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang, T., Wei B., Velazquez P., Borneman J., Braun J.. 2005. Commensal microbiota alter the abundance and TCR responsiveness of splenic naive CD4+ T lymphocytes. Clin. Immunol. 117: 221–230. [DOI] [PubMed] [Google Scholar]
- 21. Schüler, T., Hammerling G. J., Arnold B.. 2004. Cutting edge: IL-7-dependent homeostatic proliferation of CD8+ T cells in neonatal mice allows the generation of long-lived natural memory T cells. J. Immunol. 172: 15–19. [DOI] [PubMed] [Google Scholar]
- 22. Min, B., McHugh R., Sempowski G. D., Mackall C., Foucras G., Paul W. E.. 2003. Neonates support lymphopenia-induced proliferation. Immunity 18: 131–140. [DOI] [PubMed] [Google Scholar]
- 23. Kieper, W. C., Troy A., Burghardt J. T., Ramsey C., Lee J. Y., Jiang H. Q., Dummer W., Shen H., Cebra J. J., Surh C. D.. 2005. Recent immune status determines the source of antigens that drive homeostatic T cell expansion. J. Immunol. 174: 3158–3163. [DOI] [PubMed] [Google Scholar]
- 24. Yi, J., Jung J., Hong S. W., Lee J. Y., Han D., Kim K. S., Sprent J., Surh C. D.. 2019. Unregulated antigen-presenting cell activation by T cells breaks self tolerance. Proc. Natl. Acad. Sci. USA 116: 1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haluszczak, C., Akue A. D., Hamilton S. E., Johnson L. D., Pujanauski L., Teodorovic L., Jameson S. C., Kedl R. M.. 2009. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J. Exp. Med. 206: 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jameson, S. C. 2021. The naming of memory T-cell subsets. Cold Spring Harb. Perspect. Biol. 12: a037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. White, J. T., Cross E. W., Kedl R. M.. 2017. Antigen-inexperienced memory CD8(+) T cells: where they come from and why we need them. Nat. Rev. Immunol. 17: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kawabe, T., Yi J., Sprent J.. 2021. Homeostasis of naive and memory T lymphocytes. Cold Spring Harb. Perspect. Biol. 13: a037879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ernst, B., Lee D. S., Chang J. M., Sprent J., Surh C. D.. 1999. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity 11: 173–181. [DOI] [PubMed] [Google Scholar]
- 30. Ge, Q., Rao V. P., Cho B. K., Eisen H. N., Chen J.. 2001. Dependence of lymphopenia-induced T cell proliferation on the abundance of peptide/MHC epitopes and strength of their interaction with T cell receptors. Proc. Natl. Acad. Sci. USA 98: 1728–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Viret, C., Wong F. S., C. A. Janeway, Jr. 1999. Designing and maintaining the mature TCR repertoire: the continuum of self-peptide:self-MHC complex recognition. Immunity 10: 559–568. [DOI] [PubMed] [Google Scholar]
- 32. Tan, J. T., Ernst B., Kieper W. C., LeRoy E., Sprent J., Surh C. D.. 2002. Interleukin (IL)-15 and IL-7 jointly regulate homeostatic proliferation of memory phenotype CD8+ cells but are not required for memory phenotype CD4+ cells. J. Exp. Med. 195: 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kawabe, T., Jankovic D., Kawabe S., Huang Y., Lee P. H., Yamane H., Zhu J., Sher A., Germain R. N., Paul W. E.. 2017. Memory-phenotype CD4+ T cells spontaneously generated under steady-state conditions exert innate Th1-like effector function. J. Immunol. 198(Suppl. 1): 150.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tarakhovsky, A., Kanner S. B., Hombach J., Ledbetter J. A., Muller W., Killeen N., Rajewsky K.. 1995. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science 269: 535–537. [DOI] [PubMed] [Google Scholar]
- 35. Mandl, J. N., Monteiro J. P., Vrisekoop N., Germain R. N.. 2013. T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity 38: 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Azzam, H. S., Grinberg A., Lui K., Shen H., Shores E. W., Love P. E.. 1998. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188: 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. White, J. T., Cross E. W., Burchill M. A., Danhorn T., McCarter M. D., Rosen H. R., O’Connor B., Kedl R. M.. 2016. Virtual memory T cells develop and mediate bystander protective immunity in an IL-15-dependent manner. Nat. Commun. 7: 11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mahajan, V. S., Demissie E., Alsufyani F., Kumari S., Yuen G. J., Viswanadham V., Huang A., Tran J. Q., Moon J. J., Irvine D. J., Pillai S.. 2020. DOCK2 sets the threshold for entry into the virtual memory CD8(+) T cell compartment by negatively regulating tonic TCR triggering. J. Immunol. 204: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sosinowski, T., White J. T., Cross E. W., Haluszczak C., Marrack P., Gapin L., Kedl R. M.. 2013. CD8alpha+ dendritic cell trans presentation of IL-15 to naive CD8+ T cells produces antigen-inexperienced T cells in the periphery with memory phenotype and function. J. Immunol. 190: 1936–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moon, J. J., Chu H. H., Pepper M., McSorley S. J., Jameson S. C., Kedl R. M., Jenkins M. K.. 2007. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cho, J. H., Kim H. O., Surh C. D., Sprent J.. 2010. T cell receptor-dependent regulation of lipid rafts controls naive CD8+ T cell homeostasis. Immunity 32: 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guimond, M., Veenstra R. G., Grindler D. J., Zhang H., Cui Y., Murphy R. D., Kim S. Y., Na R., Hennighausen L., Kurtulus S., et al. 2009. Interleukin 7 signaling in dendritic cells regulates the homeostatic proliferation and niche size of CD4+ T cells. Nat. Immunol. 10: 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee, J. Y., Hamilton S. E., Akue A. D., Hogquist K. A., Jameson S. C.. 2013. Virtual memory CD8 T cells display unique functional properties. Proc. Natl. Acad. Sci. USA 110: 13498–13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Starling, E. H. 1923. The wisdom of the body: the Harveian Oration, delivered before The Royal College of Physicians of London on St. Luke’s Day, 1923. BMJ 2: 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Banerjee, S., van der Heijden M. G. A.. 2023. Soil microbiomes and one health. Nat. Rev. Microbiol. 21: 6–20. [DOI] [PubMed] [Google Scholar]
- 46. Belkaid, Y., Harrison O. J.. 2017. Homeostatic immunity and the microbiota. Immunity 46: 562–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ivanov, I. I., Tuganbaev T., Skelly A. N., Honda K.. 2022. T cell responses to the microbiota. Annu. Rev. Immunol. 40: 559–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sakaguchi, S., Mikami N., Wing J. B., Tanaka A., Ichiyama K., Ohkura N.. 2020. Regulatory T cells and human disease. Annu. Rev. Immunol. 38: 541–566. [DOI] [PubMed] [Google Scholar]
- 49. Josefowicz, S. Z., Lu L. F., Rudensky A. Y.. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30: 531–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Staras, S. A., Dollard S. C., Radford K. W., Flanders W. D., Pass R. F., Cannon M. J.. 2006. Seroprevalence of cytomegalovirus infection in the United States, 1988-1994. Clin. Infect. Dis. 43: 1143–1151. [DOI] [PubMed] [Google Scholar]
- 51. Fowler, K., Mucha J., Neumann M., Lewandowski W., Kaczanowska M., Grys M., Schmidt E., Natenshon A., Talarico C., Buck P. O., Diaz-Decaro J.. 2022. A systematic literature review of the global seroprevalence of cytomegalovirus: possible implications for treatment, screening, and vaccine development. BMC Public Health 22: 1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tokars, J. I., Olsen S. J., Reed C.. 2018. Seasonal incidence of symptomatic influenza in the United States. Clin. Infect. Dis. 66: 1511–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brodin, P., Jojic V., Gao T., Bhattacharya S., Angel C. J., Furman D., Shen-Orr S., Dekker C. L., Swan G. E., Butte A. J., et al. 2015. Variation in the human immune system is largely driven by non-heritable influences. Cell 160: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Seok, J., Warren H. S., Cuenca A. G., Mindrinos M. N., Baker H. V., Xu W., Richards D. R., McDonald-Smith G. P., Gao H., Hennessy L., et al. ; Inflammation and Host Response to Injury, Large Scale Collaborative Research Program . 2013. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 110: 3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mak, I. W., Evaniew N., Ghert M.. 2014. Lost in translation: animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 6: 114–118. [PMC free article] [PubMed] [Google Scholar]
- 56. Shay, T., Jojic V., Zuk O., Rothamel K., Puyraimond-Zemmour D., Feng T., Wakamatsu E., Benoist C., Koller D., Regev A.; ImmGen Consortium . 2013. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc. Natl. Acad. Sci. USA 110: 2946–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Payne, K. J., Crooks G. M.. 2007. Immune-cell lineage commitment: translation from mice to humans. Immunity 26: 674–677. [DOI] [PubMed] [Google Scholar]
- 58. Medetgul-Ernar, K., Davis M. M.. 2022. Standing on the shoulders of mice. Immunity 55: 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Beura, L. K., Hamilton S. E., Bi K., Schenkel J. M., Odumade O. A., Casey K. A., Thompson E. A., Fraser K. A., Rosato P. C., Filali-Mouhim A., et al. 2016. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532: 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wijeyesinghe, S., Beura L. K., Pierson M. J., Stolley J. M., Adam O. A., Ruscher R., Steinert E. M., Rosato P. C., Vezys V., Masopust D.. 2021. Expansible residence decentralizes immune homeostasis. Nature 592: 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rosshart, S. P., Vassallo B. G., Angeletti D., Hutchinson D. S., Morgan A. P., Takeda K., Hickman H. D., McCulloch J. A., Badger J. H., Ajami N. J., et al. 2017. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171: 1015–1028.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rosshart, S. P., Herz J., Vassallo B. G., Hunter A., Wall M. K., Badger J. H., McCulloch J. A., Anastasakis D. G., Sarshad A. A., Leonardi I., et al. 2019. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365: eaaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fiege, J. K., Block K. E., Pierson M. J., Nanda H., Shepherd F. K., Mickelson C. K., Stolley J. M., Matchett W. E., Wijeyesinghe S., Meyerholz D. K., et al. 2021. Mice with diverse microbial exposure histories as a model for preclinical vaccine testing. Cell Host Microbe 29: 1815–1827.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Naik, S., Fuchs E.. 2022. Inflammatory memory and tissue adaptation in sickness and in health. Nature 607: 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Welsh, R. M., Selin L. K.. 2002. No one is naive: the significance of heterologous T-cell immunity. Nat. Rev. Immunol. 2: 417–426. [DOI] [PubMed] [Google Scholar]
- 66. Welsh, R. M., Che J. W., Brehm M. A., Selin L. K.. 2010. Heterologous immunity between viruses. Immunol. Rev. 235: 244–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Selin, L. K., Nahill S. R., Welsh R. M.. 1994. Cross-reactivities in memory cytotoxic T lymphocyte recognition of heterologous viruses. J. Exp. Med. 179: 1933–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen, H. D., Fraire A. E., Joris I., Brehm M. A., Welsh R. M., Selin L. K.. 2001. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat. Immunol. 2: 1067–1076. [DOI] [PubMed] [Google Scholar]
- 69. Chen, H. D., Fraire A. E., Joris I., Welsh R. M., Selin L. K.. 2003. Specific history of heterologous virus infections determines anti-viral immunity and immunopathology in the lung. Am. J. Pathol. 163: 1341–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Brehm, M. A., Pinto A. K., Daniels K. A., Schneck J. P., Welsh R. M., Selin L. K.. 2002. T cell immunodominance and maintenance of memory regulated by unexpectedly cross-reactive pathogens. Nat. Immunol. 3: 627–634. [DOI] [PubMed] [Google Scholar]
- 71. Jenkins, M. K., Chu H. H., McLachlan J. B., Moon J. J.. 2010. On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu. Rev. Immunol. 28: 275–294. [DOI] [PubMed] [Google Scholar]
- 72. Mason, D. 1998. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today 19: 395–404. [DOI] [PubMed] [Google Scholar]
- 73. Wooldridge, L., Ekeruche-Makinde J., van den Berg H. A., Skowera A., Miles J. J., Tan M. P., Dolton G., Clement M., Llewellyn-Lacey S., Price D. A., et al. 2012. A single autoimmune T cell receptor recognizes more than a million different peptides. J. Biol. Chem. 287: 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ishizuka, J., Grebe K., Shenderov E., Peters B., Chen Q., Peng Y., Wang L., Dong T., Pasquetto V., Oseroff C., et al. 2009. Quantitating T cell cross-reactivity for unrelated peptide antigens. J. Immunol. 183: 4337–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Adams, J. J., Narayanan S., Liu B., Birnbaum M. E., Kruse A. C., Bowerman N. A., Chen W., Levin A. M., Connolly J. M., Zhu C., et al. 2011. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity 35: 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Birnbaum, M. E., Mendoza J. L., Sethi D. K., Dong S., Glanville J., Dobbins J., Ozkan E., Davis M. M., Wucherpfennig K. W., Garcia K. C.. 2014. Deconstructing the peptide-MHC specificity of T cell recognition. Cell 157: 1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bentzen, A. K., Marquard A. M., Lyngaa R., Saini S. K., Ramskov S., Donia M., Such L., Furness A. J., McGranahan N., Rosenthal R., et al. 2016. Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat. Biotechnol. 34: 1037–1045. [DOI] [PubMed] [Google Scholar]
- 78. Zhang, S. Q., Ma K. Y., Schonnesen A. A., Zhang M., He C., Sun E., Williams C. M., Jia W., Jiang N.. 2018. High-throughput determination of the antigen specificities of T cell receptors in single cells. Nat. Biotechnol. 36: 1156–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dobson, C. S., Reich A. N., Gaglione S., Smith B. E., Kim E. J., Dong J., Ronsard L., Okonkwo V., Lingwood D., Dougan M., et al. 2022. Antigen identification and high-throughput interaction mapping by reprogramming viral entry. Nat. Methods 19: 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yu, B., Shi Q., Belk J. A., Yost K. E., Parker K. R., Li R., Liu B. B., Huang H., Lingwood D., Greenleaf W. J., et al. 2022. Engineered cell entry links receptor biology with single-cell genomics. Cell 185: 4904–4920.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kula, T., Dezfulian M. H., Wang C. I., Abdelfattah N. S., Hartman Z. C., Wucherpfennig K. W., Lyerly H. K., Elledge S. J.. 2019. T-scan: a genome-wide method for the systematic discovery of T cell epitopes. Cell 178: 1016–1028.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kisielow, J., Obermair F. J., Kopf M.. 2019. Deciphering CD4(+) T cell specificity using novel MHC-TCR chimeric receptors. [Published erratum appears in 2019 Nat. Immunol. 20: 663.] Nat. Immunol. 20: 652–662. [DOI] [PubMed] [Google Scholar]
- 83. Joglekar, A. V., Leonard M. T., Jeppson J. D., Swift M., Li G., Wong S., Peng S., Zaretsky J. M., Heath J. R., Ribas A., et al. 2019. T cell antigen discovery via signaling and antigen-presenting bifunctional receptors. Nat. Methods 16: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Reinherz, E. L., Tan K., Tang L., Kern P., Liu J., Xiong Y., Hussey R. E., Smolyar A., Hare B., Zhang R., et al. 1999. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science 286: 1913–1921. [DOI] [PubMed] [Google Scholar]
- 85. Reiser, J. B., Darnault C., Gregoire C., Mosser T., Mazza G., Kearney A., van der Merwe P. A., Fontecilla-Camps J. C., Housset D., Malissen B.. 2003. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat. Immunol. 4: 241–247. [DOI] [PubMed] [Google Scholar]
- 86. Garcia, K. C., Degano M., Pease L. R., Huang M., Peterson P. A., Teyton L., Wilson I. A.. 1998. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science 279: 1166–1172. [DOI] [PubMed] [Google Scholar]
- 87. Hennecke, J., Wiley D. C.. 2002. Structure of a complex of the human alpha/beta T cell receptor (TCR) HA1.7, influenza hemagglutinin peptide, and major histocompatibility complex class II molecule, HLA-DR4 (DRA*0101 and DRB1*0401): insight into TCR cross-restriction and alloreactivity. J. Exp. Med. 195: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sethi, D. K., Schubert D. A., Anders A. K., Heroux A., Bonsor D. A., Thomas C. P., Sundberg E. J., Pyrdol J., Wucherpfennig K. W.. 2011. A highly tilted binding mode by a self-reactive T cell receptor results in altered engagement of peptide and MHC. J. Exp. Med. 208: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Adams, J. J., Narayanan S., Birnbaum M. E., Sidhu S. S., Blevins S. J., Gee M. H., Sibener L. V., Baker B. M., Kranz D. M., Garcia K. C.. 2016. Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity. Nat. Immunol. 17: 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Riley, T. P., Hellman L. M., Gee M. H., Mendoza J. L., Alonso J. A., Foley K. C., Nishimura M. I., Vander Kooi C. W., Garcia K. C., Baker B. M.. 2018. T cell receptor cross-reactivity expanded by dramatic peptide-MHC adaptability. Nat. Chem. Biol. 14: 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Su, L. F., Kidd B. A., Han A., Kotzin J. J., Davis M. M.. 2013. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity 38: 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pan, Y. G., Aiamkitsumrit B., Bartolo L., Wang Y., Lavery C., Marc A., Holec P. V., Rappazzo C. G., Eilola T., Gimotty P. A., et al. 2021. Vaccination reshapes the virus-specific T cell repertoire in unexposed adults. Immunity 54: 1245–1256.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Campion, S. L., Brodie T. M., Fischer W., Korber B. T., Rossetti A., Goonetilleke N., McMichael A. J., Sallusto F.. 2014. Proteome-wide analysis of HIV-specific naive and memory CD4(+) T cells in unexposed blood donors. J. Exp. Med. 211: 1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Grifoni, A., Weiskopf D., Ramirez S. I., Mateus J., Dan J. M., Moderbacher C. R., Rawlings S. A., Sutherland A., Premkumar L., Jadi R. S., et al. 2020. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181: 1489–1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Mateus, J., Grifoni A., Tarke A., Sidney J., Ramirez S. I., Dan J. M., Burger Z. C., Rawlings S. A., et al. 2020. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 370: 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Le Bert, N., Tan A. T., Kunasegaran K., Tham C. Y. L., Hafezi M., Chia A., Chng M. H. Y., Lin M., Tan N., Linster M., et al. 2020. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584: 457–462. [DOI] [PubMed] [Google Scholar]
- 97. Peng, Y., Mentzer A. J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. 2020. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 21: 1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Braun, J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. 2020. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 587: 270–274. [DOI] [PubMed] [Google Scholar]
- 99. Nelde, A., Bilich T., Heitmann J. S., Maringer Y., Salih H. R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M., et al. 2021. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 22: 74–85. [DOI] [PubMed] [Google Scholar]
- 100. Weiskopf, D., Schmitz K. S., Raadsen M. P., Grifoni A., Okba N. M. A., Endeman H., van den Akker J. P. C., Molenkamp R., Koopmans M. P. G., van Gorp E. C. M., et al. 2020. Phenotype and kinetics of SARS-CoV-2–specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 5: eabd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bacher, P., Rosati E., Esser D., Martini G. R., Saggau C., Schiminsky E., Dargvainiene J., Schroder I., Wieters I., Khodamoradi Y., et al. 2020. Low-avidity CD4(+) T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity 53: 1258–1271.e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bartolo, L., Afroz S., Pan Y. G., Xu R., Williams L., Lin C. F., Tanes C., Bittinger K., Friedman E. S., Gimotty P. A., et al. 2022. SARS-CoV-2-specific T cells in unexposed adults display broad trafficking potential and cross-react with commensal antigens. Sci. Immunol. 7: eabn3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Niessl, J., Sekine T., Lange J., Konya V., Forkel M., Maric J., Rao A., Mazzurana L., Kokkinou E., Weigel W., et al. 2021. Identification of resident memory CD8(+) T cells with functional specificity for SARS-CoV-2 in unexposed oropharyngeal lymphoid tissue. Sci. Immunol. 6: eabk0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Diniz, M. O., Mitsi E., Swadling L., Rylance J., Johnson M., Goldblatt D., Ferreira D., Maini M. K.. 2022. Airway-resident T cells from unexposed individuals cross-recognize SARS-CoV-2. Nat. Immunol. 23: 1324–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Li, J., Reinke S., Shen Y., Schollmeyer L., Liu Y. C., Wang Z., Hardt S., Hipfl C., Hoffmann U., Frischbutter S., et al. 2022. A ubiquitous bone marrow reservoir of preexisting SARS-CoV-2-reactive memory CD4(+) T lymphocytes in unexposed individuals. Front. Immunol. 13: 1004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sette, A., Crotty S.. 2020. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. [Published erratum appears in 2020 Nat. Rev. Immunol. 20: 644.] Nat. Rev. Immunol. 20: 457–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Dykema, A. G., Zhang B., Woldemeskel B. A., Garliss C. C., Cheung L. S., Choudhury D., Zhang J., Aparicio L., Bom S., Rashid R., et al. 2021. Functional characterization of CD4+ T cell receptors crossreactive for SARS-CoV-2 and endemic coronaviruses. J. Clin. Invest. 131: e146922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Low, J. S., Vaqueirinho D., Mele F., Foglierini M., Jerak J., Perotti M., Jarrossay D., Jovic S., Perez L., Cacciatore R., et al. 2021. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science 372: 1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Pothast, C. R., Dijkland R. C., Thaler M., Hagedoorn R. S., Kester M. G. D., Wouters A. K., Hiemstra P. S., van Hemert M. J., Gras S., Falkenburg J. H. F., Heemskerk M. H. M.. 2022. SARS-CoV-2-specific CD4+ and CD8+ T cell responses can originate from cross-reactive CMV-specific T cells. eLife 11: e82050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Donald, K., Petersen C., Turvey S. E., Finlay B. B., Azad M. B.. 2022. Secretory IgA: linking microbes, maternal health, and infant health through human milk. Cell Host Microbe 30: 650–659. [DOI] [PubMed] [Google Scholar]
- 111. Conrey, P. E., Denu L., O’Boyle K. C., Rozich I., Green J., Maslanka J., Lubin J. B., Duranova T., Haltzman B. L., Gianchetti L., et al. 2023. IgA deficiency destabilizes homeostasis toward intestinal microbes and increases systemic immune dysregulation. Sci. Immunol. 8: eade2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sparks, R., Lau W. W., Liu C., Han K. L., Vrindten K. L., Sun G., Cox M., Andrews S. F., Bansal N., Failla L. E., et al. 2023. Influenza vaccination reveals sex dimorphic imprints of prior mild COVID-19. Nature 614: 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Pan, L., Mu M., Yang P., Sun Y., Wang R., Yan J., Li P., Hu B., Wang J., Hu C., et al. 2020. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am. J. Gastroenterol. 115: 766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Nobel, Y. R., Phipps M., Zucker J., Lebwohl B., Wang T. C., Sobieszczyk M. E., Freedberg D. E.. 2020. Gastrointestinal symptoms and coronavirus disease 2019: a case-control study from the United States. Gastroenterology 159: 373–375.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Baig, A. M., Sanders E. C.. 2020. Potential neuroinvasive pathways of SARS-CoV-2: deciphering the spectrum of neurological deficit seen in coronavirus disease-2019 (COVID-19). J. Med. Virol. 92: 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ramlah, R. 2021. Acute pancreatitis disease in the patient with COVID-19 infection: a systematic review. Ann. Hepatobiliary Pancreat. Surg. 25: S372. [Google Scholar]
- 117. Oran, D. P., Topol E. J.. 2020. Prevalence of asymptomatic SARS-CoV-2 infection. Ann. Intern. Med. 173: 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ludvigsson, J. F. 2020. Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr. 109: 1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Swadling, L., Diniz M. O., Schmidt N. M., Amin O. E., Chandran A., Shaw E., Pade C., Gibbons J. M., Le Bert N., Tan A. T., et al. 2022. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 601: 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kundu, R., Narean J. S., Wang L., Fenn J., Pillay T., Fernandez N. D., Conibear E., Koycheva A., Davies M., Tolosa-Wright M., et al. 2022. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat. Commun. 13: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Mallajosyula, V., Ganjavi C., Chakraborty S., McSween A. M., Pavlovitch-Bedzyk A. J., Wilhelmy J., Nau A., Manohar M., Nadeau K. C., Davis M. M.. 2021. CD8+ T cells specific for conserved coronavirus epitopes correlate with milder disease in patients with COVID-19. Sci. Immunol. 6: eabg5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Loyal, L., Braun J., Henze L., Kruse B., Dingeldey M., Reimer U., Kern F., Schwarz T., Mangold M., Unger C., et al. 2021. Cross-reactive CD4+ T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science 374: eabh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Mateus, J., Dan J. M., Zhang Z., Rydyznski Moderbacher C., Lammers M., Goodwin B., Sette A., Crotty S., Weiskopf D.. 2021. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science 374: eabj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Saggau, C., Martini G. R., Rosati E., Meise S., Messner B., Kamps A.-K., Bekel N., Gigla J., Rose R., Voss M., et al. 2022. The pre-exposure SARS-CoV-2-specific T cell repertoire determines the quality of the immune response to vaccination. Immunity 55: 1924–1939.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Albert, L. J., Inman R. D.. 1999. Molecular mimicry and autoimmunity. N. Engl. J. Med. 341: 2068–2074. [DOI] [PubMed] [Google Scholar]
- 126. Cusick, M. F., Libbey J. E., Fujinami R. S.. 2012. Molecular mimicry as a mechanism of autoimmune disease. Clin. Rev. Allergy Immunol. 42: 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Yeoh, Y. K., Zuo T., Lui G. C., Zhang F., Liu Q., Li A. Y., Chung A. C., Cheung C. P., Tso E. Y., Fung K. S., et al. 2021. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 70: 698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bernard-Raichon, L., Venzon M., Klein J., Axelrad J. E., Zhang C., Sullivan A. P., Hussey G. A., Casanovas-Massana A., Noval M. G., Valero-Jimenez A. M., et al. 2022. Gut microbiome dysbiosis in antibiotic-treated COVID-19 patients is associated with microbial translocation and bacteremia. Nat. Commun. 13: 5926. [DOI] [PMC free article] [PubMed] [Google Scholar]