Summary
The host-microbiome associations occurring on the skin of vertebrates significantly influence hosts’ health. However, the factors mediating their interactions remain largely unknown. Herein, we used integrated technical and ecological frameworks to investigate the skin metabolites sustaining a beneficial symbiosis between tree frogs and bacteria. We characterize macrocyclic acylcarnitines as the major metabolites secreted by the frogs’ skin and trace their origin to an enzymatic unbalance of carnitine palmitoyltransferases. We found that these compounds colocalize with bacteria on the skin surface and are mostly represented by members of the Pseudomonas community. We showed that Pseudomonas sp. MPFS isolated from frogs’ skin can exploit acylcarnitines as its sole carbon and nitrogen source, and this metabolic capability is widespread in Pseudomonas. We summarize frogs’ multiple mechanisms to filter environmental bacteria and highlight that acylcarnitines likely evolved for another function but were co-opted to provide nutritional benefits to the symbionts.
Subject areas: Biological sciences, Microbiology, Microbial metabolism, Omics, Metabolomics
Graphical abstract
Highlights
-
•
Frogs’ dermosphere is a rich source of metabolites mediating symbiotic interactions
-
•
The skin of tree frogs produces and secretes macrocyclic acylcarnitines
-
•
Pseudomonas is the most abundant genus of the frogs’ skin bacterial community
-
•
Frogs’ Pseudomonas symbionts can use acylcarnitines as the sole nutrient source
Biological sciences; Microbiology; Microbial metabolism; Omics; Metabolomics
Introduction
Several microorganisms have established symbiotic interactions with diverse plants and animal groups like cnidarians, mollusks, insects, and vertebrates, including humans.1,2 In those cases where both partners benefit, the microorganisms can assist the host in nutrient assimilation,3 pathogen defense,4 and producing specialized metabolites used in social interactions.5 In return, the host can offer suitable conditions for their microbial symbiont, such as a humid environment and nutrients,1,6 and cooperate with them to inhibit microbial competitors.4
Among the organs exposed to the exterior world, the skin of vertebrates possesses specialized niches occupied by microorganisms interacting with each other and the host.1,6 Given the plethora of associations and diversity of chemical components, this organ can be viewed as an ecosystem7 exhibiting many parallelisms with the interface between plant roots and soil, the so-called rhizosphere.8,9 Like the rhizosphere, the skin surface endures variable external mechanical and physicochemical conditions. Likewise, it is a rich chemical environment with various molecule classes like mucins, lipids, peptides, and specialized metabolites derived from both the host and the microbiota.1,6 Also, similarly to plant metabolites in the rhizosphere, these substances may regulate the microbial community10 by suppressing (e.g., through inhibition factors)11,12 or stimulating (e.g., through nutrients) the growth of specific microbial consortia.1 However, unlike the soil, skin components acting as nutrient resources are limited and mostly unknown.1,6 Therefore, it is a harsh environment where microorganisms capable of catabolizing host-derived metabolites have strong ecological advantages.
In contrast to the skin of other vertebrates, amphibian skin secretes several metabolites produced endogenously or obtained through diet, which include alkaloids, biogenic amines, odorous volatiles, peptides, and steroids, the reason why it was described as a mine of active compounds.13,14 Still, although most of them have traditionally been studied as active agents against pathogens and predators, a new understanding of their biological functions and origin is currently emerging. For example, some of the compounds potentially mediating defensive functions15 and sexual interactions14 in the host are now known to be originated from symbiotic bacteria. In addition, these bacteria might produce metabolites like prodigiosin, sessilin, pyocyanin, and violacein that protect the host against the fungal pathogen Batrachochytrium dendrobatidis.16,17 On the host side, the skin can actively select and establish specific bacterial assemblages from rare environmental strains.18,19 This role was linked to amphibians’ antimicrobial peptides and characterized as a filtering mechanism that structures the microbial community.17,20 However, no study has assessed a more direct chemical benefit of the host to their symbionts, specifically, whether amphibians' skin metabolites can serve as a nutritional source for bacteria.
Here, we investigate the molecular determinants of the host that sustain an amphibian-bacteria symbiotic interaction. We used the tree frog Boana prasina and the bacterium Pseudomonas sp. MFPS14 isolated from its skin as a frog-symbiont system. Through a combination of omics technologies, we demonstrated that macrocyclic acylcarnitines are major metabolites endogenously produced by the frog’s skin glands, which are also present in the skin secretion of closely related species. Microscopy and mass spectrometry (MS) imaging revealed that these compounds are homogeneously distributed on the skin surface and colocalize with bacteria. Next, we examined the skin bacteria community structure of six frog species producing macrocyclic acylcarnitines and discovered that Pseudomonas is the most representative bacteria genus, suggesting that chemical and microbial profiles are closely linked. Through experiments with different carnitine derivatives, we show that Pseudomonas sp. MFPS catabolizes acylcarnitines and uses them for growth, while phylogenomic comparisons revealed that genes involved in such catabolism are widespread in Pseudomonas. Finally, using the rhizosphere as an ecological reference, we stress the significance of the dermosphere concept21 as a fundamental means for surveying the skin environment subjected to the influence of metabolites secreted by both the host and the microbial symbionts.
Results
Macrocyclic acylcarnitines are major components of frogs’ skin secretion
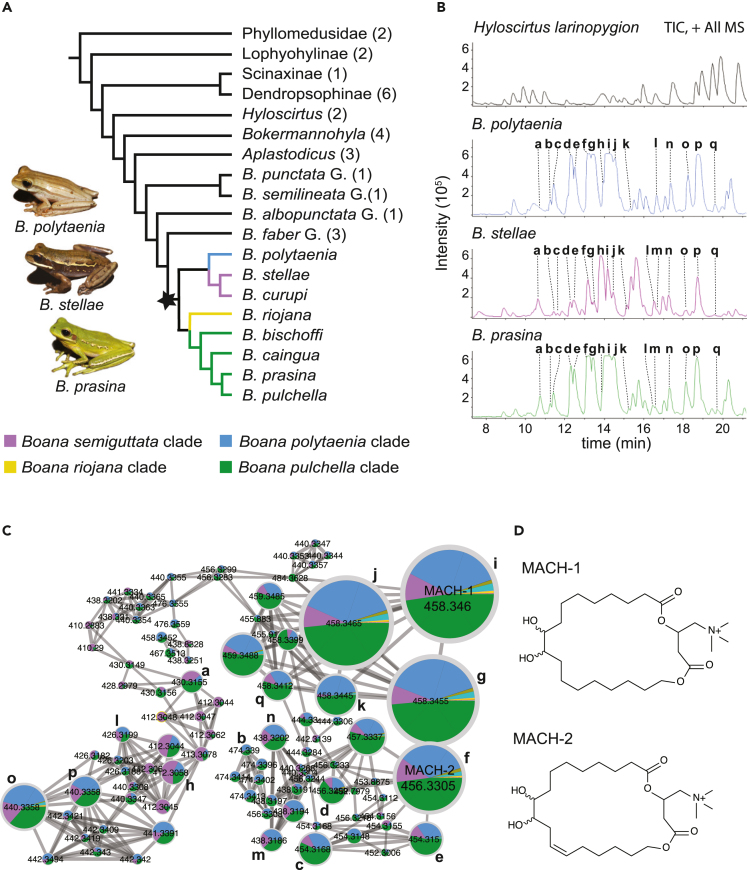
We examined the chemical profile of the water-soluble components present in the skin glandular secretions of the tree frog Boana prasina and 33 other tree frog species from 11 genera in Argentina, Colombia, and Brazil (Figure 1A; Table S1). The sampling was designed to understand the evolutionary context of the compounds found in our model species B. prasina, emphasizing the closely related species within the B. pulchella group. Our analyses revealed that the skin secretion of B. prasina and other species of this group has a similar chemical profile. They share the major chromatographic peaks, and their respective MS fragmentation patterns conform to derivatives of the same molecular family (Figures 1B and 1C). To elucidate the chemical nature of these metabolites, we extracted and purified two of the main compounds secreted by B. prasina and analyzed them by high-performance liquid chromatography (HPLC)-quadrupole time of flight (QTOF)- tandem mass spectrometry (MS/MS), MS/MS-based fragmentation, gas chromatography-mass spectrometry (GC-MS), and 1D-2D nuclear magnetic resonance (NMR) spectroscopy (see STAR Methods for details). These compounds had the molecular formulas C25H48NO6 and C25H46NO6 and were designated MACH-1 and MACH-2, respectively (Figure 1D). Spectroscopic analysis revealed them as macrocyclic acylcarnitines containing a C18 fatty acid chain substituted by two hydroxyl groups and one unsaturated bond at the C-12′ position in MACH-2 (Figures S1–S12; Tables S2 and S3). GC-MS analysis of derivatized compounds indicated that the position of the hydroxyl groups is at C′9 and C′10 of the fatty acid chain (Figures 1D and S13). Based on the molecular networking and the MS/MS fragmentation pattern analysis of MACH-1 and MACH-2, other compounds within this acylcarnitines’ molecular family were detected as part of the same homologous series (Figures 1C and S14–S18; Table S4). Although we observed compounds of this series in other species besides those within the B. pulchella group (Figure 1C), they only occurred as trace elements. The optical rotatory dispersion of MACH-1 was = −16 ± 4.
Figure 1.
Macrocyclic acylcarnitines are major components of the skin secretion of tree frogs within the Boana pulchella group
(A) A condensed phylogenetic relationship based on the analysis from Faivovich et al.22 of the South American tree frogs surveyed in this study. The star in the node indicates the root of the Boana pulchella group, whose representatives possess significant amounts of MACHs in their skin secretion. Blue, purple, yellow, and green branches correspond to the phylogenetic clades B. riojana, B. polytaenia, B. semiguttata, and B. pulchella, respectively. Images on the left depict representative specimens from the last three clades. Values in parentheses indicate the number of species examined.
(B) Total ion chromatograms in positive mode show the similar skin secretion profile of distinct clades within the B. pulchella group in contrast to the different profile of Hyloscirtus larinopygion.
(C) Based on LC-MS2 (ESI+) spectral and retention time similarities, molecular networking revealed that the primary metabolites of these tree frogs are MACH analogs (a–q in B), occurring only in trace amounts in a few other species. Colors in pie charts depict the different phylogenetics clades shown in A.
(D) Structures of MACH-1 and MACH-2 were identified from the skin secretion of Boana prasina by a combination of NMR, MS, and derivatization, followed by GC-MS. The putative structure of other MACH analogs is depicted in Figure S14.
Macrocyclic acylcarnitines are likely synthesized in frog’s skin
Long-chain acylcarnitines are essential intermediates for the β-oxidation of fatty acids in the mitochondrial matrix and energy production.23,24,25 For this reason, most studies have traditionally focused on organs and tissues with high energy demands, such as the skeletal muscle and the liver,25 yet, none has examined their significance in the skin. Therefore, we studied the skin transcripts and the lipid profile of male and female specimens and conducted comprehensive research on lipid metabolism-related genes to understand better the accumulation of acylcarnitines and the generation of macrocycle derivatives in this organ. As a result, we found that all components of an enzyme system for the production of MACH-1 and MACH-2 are expressed in the skin of Boana prasina (Table 1; Data S1; Figure 2). Notably, it involves enzymes typically active in other tissues like the lung and liver meditating activation and ω-hydroxylation of long-chain fatty acids.
Table 1.
Sequence identity of proteins involved in fatty acid metabolism and their expression levels in the skin of Boana prasina in comparison to proteins reported from different tissues in humans and Xenopus laevis
| Reaction/protein name | Humans |
Xenopus laevis |
Boana prasina |
|||
|---|---|---|---|---|---|---|
| Uniprot Acc. nº |
Identity % |
Uniprot Acc. nº |
Identity % |
Female tpm | Male tpm | |
| Fatty acid activation/ | ||||||
| acyl-CoA synthetase long-chain family member 1 | P33121 | 76.2 | Q7ZTJ3 | 85.5 | 0.6 | 2.5 |
| acyl-CoA synthetase long-chain family member 3 | O95573 | 74.4 | A0A1L8G4N3 | 88.4 | 0.2 | 0.4 |
| acyl-CoA synthetase long-chain family member 4 | O60488 | 82.2 | B3DLM0 | 89.5 | 0.2 | 0.6 |
| Acylcarnitine formation/ | ||||||
| carnitine palmitoyltransferase 1A | P50416 | 78.6 | A0A8J0V0P2 | 87.3 | 0.1 | 0.5 |
| carnitine palmitoyltransferase 1B | Q92523 | 64.9 | A0JMA3 | 82.4 | 2.9 | 5.6 |
| carnitine palmitoyltransferase 1C | Q8TCG5 | 59.6 | Q7ZWK5 | 88.1 | 0.3 | 0.7 |
| Fatty acid and carnitine recovering | ||||||
| carnitine palmitoyltransferase 2 | P23786 | 72.6 | Q7ZXE1 | 82.7 | 0.7 | 1.6 |
| Omega-hydroxylation | ||||||
| cytochrome P450 family 4 subfamily B member 1v1 | P13584 | 52.5 | A0A8J0V6C5 | 68.8 | 0.8 | 1.2 |
| cytochrome P450 family 4 subfamily B member 1v2 | P13584 | 52.9 | A0A8J0V6C5 | 73.6 | 5.6 | 4.3 |
| Acylcarnitine cyclization via lactonization | ||||||
| paraoxonase 2 | Q15165 | 56.9 | Q6IRR7 | 65.2 | 1.0 | 0.9 |
| Double bond oxygenation and epoxide opening | ||||||
| arachidonate 5-lipoxygenase | P09917 | 78.3 | A0A1L8FJT5 | 84.9 | 0.5 | 0.5 |
| epoxide hydrolase 2 | P34913 | 56.4 | Q6DCH2 | 73.0 | 10.0 | 13.0 |
Acc. nº, accession number; v1, variant 1; v2, variant 2; tpm, transcripts per million.
Figure 2.
Putative biosynthetic pathway for skin macrocyclic acylcarnitines
Enzymes in green were identified and annotated from the skin transcriptome of Boana prasina. Compounds in bold and with an asterisk were identified in the skin lipid profile and secretion analyses. Following acylcarnitine accumulation, the pathway likely involves four reaction steps (depicted in brackets).
The biosynthesis would begin with the activation of the oleate and linoleate, occurring at high concentrations in the frogs’ skin, by an acyl-coenzyme A (CoA) synthetase long chain (Tables 1 and S5; Figure 2). Afterward, these acyl-CoA thioesters would be conjugated with carnitine to form acylcarnitines by carnitine palmitoyltransferases (CPT1). Interestingly, we found that all three gene isoforms, CPT1A, CPT1B, and CPT1C, predominant in vertebrates’ liver, muscle, and brain, respectively,26,27 are expressed in the skin of B. prasina, albeit at different levels (Table 1; Data S1). The acylcarnitine esters would be initially accumulated in the skin gland because of the higher expression of CPT1 compared to CPT2 genes (4.1 and 4.7, in the female and male, respectively), which code for the enzyme responsible for the reconversion of acylcarnitines into the respective acyl-CoA esters and carnitine (Table 1; Data S1). Then, the synthesis would proceed through four reaction steps (Table 1; Data S1; Figure 2): (a) ω-hydroxylation mediated by cytochrome P450 family 4 subfamily B member 1, followed by (b) immediate cyclization, (c) epoxidation by a lipoxygenase such as arachidonate 5-lipoxygenase or another P450 monooxygenase, and (d) epoxide ring-opening and hydroxylation by an epoxide hydrolase such as the epoxide hydrolase 2. Given the limited information on macrocycles and diester of carnitines in vertebrates, we tentatively propose that the formation of the acylcarnitine macrocycle is mediated by the lactonizing activity of paraoxonase 2 or another unknown lipase.
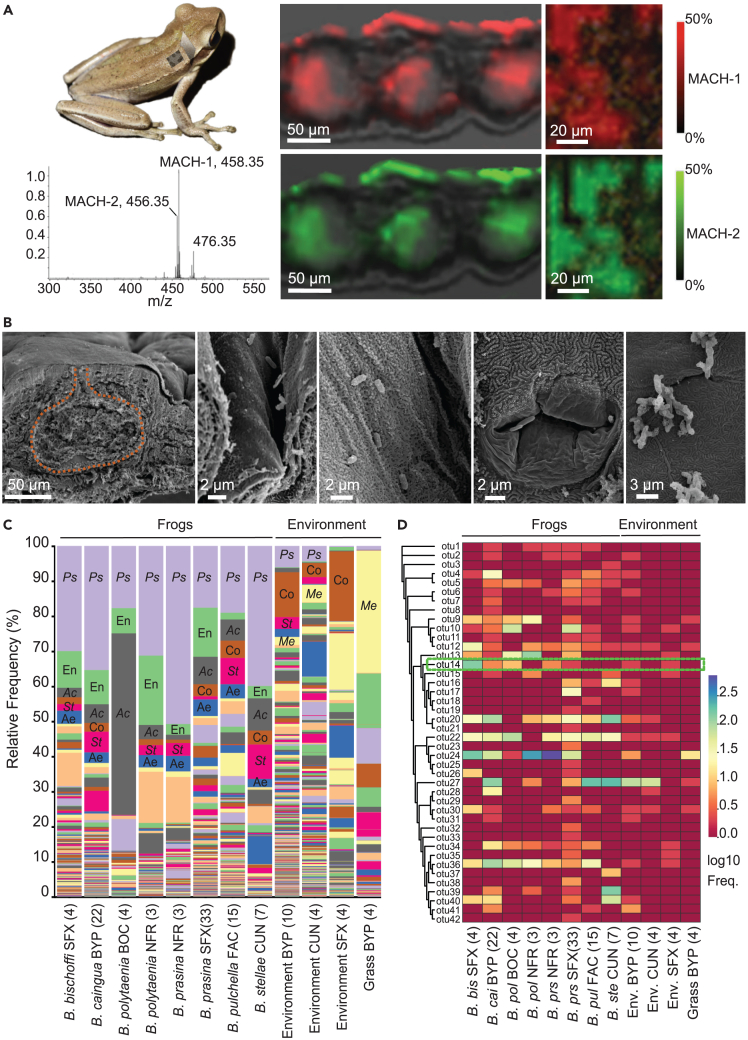
Macrocyclic acylcarnitines colocalize with bacterial cells on the frog skin
Analysis of transverse sections and the surface of dorsal skin by matrix-assisted laser ionization (MALDI) imaging (using MS1 and MS2 experiments) and by transmission and scanning electron micrographs (Figures 3A, 3B, S19, and S20) showed that MACH-1 and MACH-2 are found within serous glands and are distributed homogeneously over the skin surface (Figure 3A). Because this distribution corresponds to the serous gland granules, we can assume that these compounds accumulate in the granules within the glands and are spread on the skin surface after the glands release their content (Figures 3B and S19). We detected no bacteria within serous glands (Figures S19 and S20). In contrast, we found mixed bacterial communities of cocci and bacilli with and without flagella on the skin surface, mainly concentrated in skin folds and gland pores, where they colocalize with MACH-1 and MACH-2 (Figure 3B).
Figure 3.
Macrocyclic acylcarnitines and bacteria, mainly represented by Pseudomonas strains, colocalize in the frog skin
(A) Ions [M]+m/z 456.35 (MACH-2) and [M]+m/z 458.35 (MACH-1) detected by in situ MALDI IMS in the skin of Boana prasina within serous glands and at the surface as observed in transverse sections (Middle graph) and the dorsal surface (Right graph).
(B) Scanning electron micrographs showing secretion granules within serous glands (dotted lines on the Left graph) in transverse sections and bacteria distribution mainly in skin folds (Middle graphs) and around pore glands (Right graphs).
(C) Mean relative abundance of bacterial OTUs present in the skin of each population and environmental samples at the genus level. Pseudomonas (Ps) is predominant in frogs’ skin. Other relevant genera in frogs are Acinetobacter (Ac), Stenotrophomonas (St), and three unidentified genera of Enterobacteriaceae (En), Comamonadaceae (Co), and Aeromonadaceae (Ae), and Methylobacterium (Me) in environmental samples. Values in parentheses indicate the number of samples for each category. Acronyms of sites are listed in Table S1.
(D) Phylogenetic tree of OTUs sequences classified as Pseudomonas and heatmap showing the relative abundance of these OTUs in the skin of each population and environmental samples. The green dotted rectangle depicts the sequence associated with Pseudomonas sp. MPFS (OTU 14), a biologically significant symbiont isolated in our previous study14 and used in carnitine growth experiments (next section). Abbreviations, Freq., frequency.
Pseudomonas is prevalent in the bacterial skin community of its frog host
We systematically characterized the bacterial skin community of six species within the B. pulchella group from Brazil and Argentina using the V4 16S rRNA gene amplicon sequencing to explore a potential association between macrocyclic acylcarnitines and microorganisms. Alpha diversity was very similar between all populations except for two species from Argentina (B. pulchella [Fachinal, Misiones] and B. stellae [Cuñapiru, Misiones]), which showed higher and lower Faith phylogenetic diversity values, respectively (Figure S21). Eight genera at different abundance made up an average of 50% of the skin bacterial communities in all species of the B. pulchella group (Figure 3C). Notably, except for B. polytaenia from Serra da Bocaina (São Paulo, Brazil), Pseudomonas was the most representative genus in all frog populations, comprising 18%–50% of the abundance of the bacterial community. In contrast, the abundance of Pseudomonas in environmental samples was at a maximum of 8% (Figure 3C). Water samples and grass surfaces were dominated by an unidentified genus of the Comamonadacea family and Methylbacterium comprising 4%–20% and 35% of the bacterial community abundance, respectively.
Given the relevance of Pseudomonas sp. MPFS as a symbiont potentially mediating sexual interactions in B. prasina,14 we examined the relative abundance of different Pseudomonas at the operational taxonomic unit (OTU) level (Figure 3D). Overall, 42 OTUs were identified as Pseudomonas in frog skin and environmental samples. Of them, 15 OTUs, including that representing the frog symbiont Pseudomonas sp. MPFS (OTU 14) and two OTUs (OTUs 13 and 20) with identical sequences to Pseudomonas strains previously isolated from Boana prasina,14 were recovered in five or more frog populations from Argentina and Brazil with different relative abundances. However, none of them was found in all populations. Furthermore, despite one exception, these OTUs also occurred in environmental samples but at a much lower abundance. These findings suggest that environmental Pseudomonas can be found on the skin of tree frogs within the B. pulchella group, likely exploiting the host’s conditions for establishing and growing. Thus, we explored how macrocyclic acylcarnitines could influence the growth of the frog symbiont Pseudomonas sp. MPFS isolated from the skin of B. prasina.
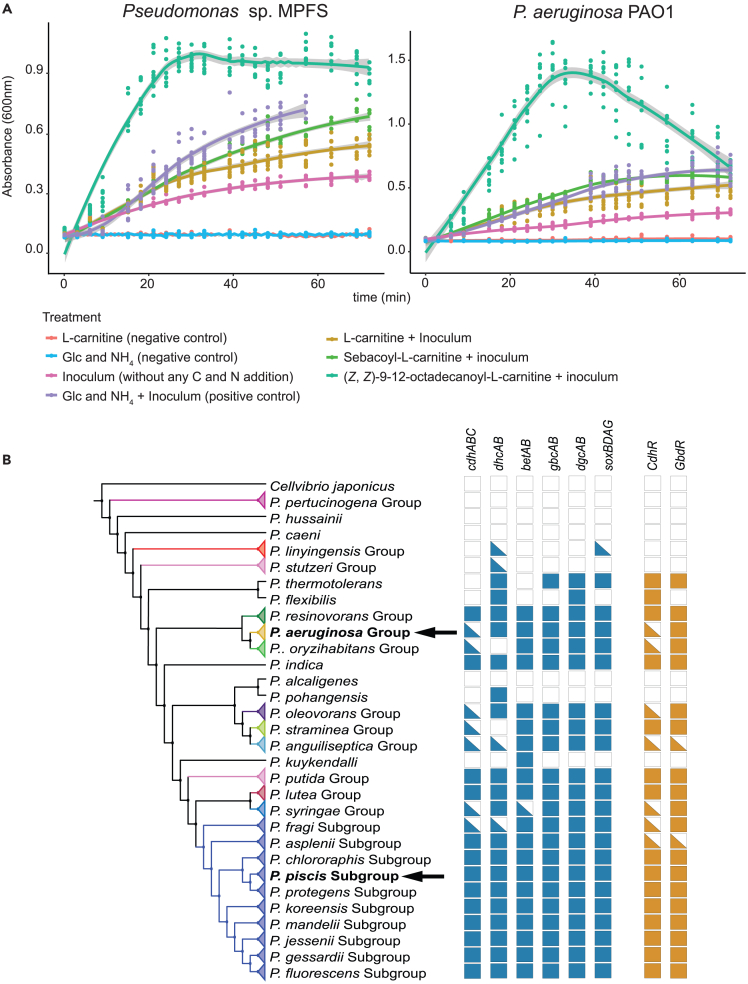
Pseudomonas symbionts can utilize acylcarnitine derivatives as nutrients
Given that Pseudomonas aeruginosa is an opportunistic pathogen capable of catabolizing acylcarnitines as sole carbon and nitrogen sources,28,29,30 we test whether Pseudomonas sp. MPFS can utilize acylcarnitines from B. prasina as an energy source. We measured the growth of this bacterium in three minimal media containing L-carnitine and two commercially available L-acylcarnitines with ten- and 18-carbon chain lengths as sole carbon and nitrogen source, using P. aeruginosa PAO1 as a positive control. We found that the growth of both strains was higher on media supplemented with L-carnitine, sebacoyl-L-carnitine, and (Z,Z)-9-12-octadecanoyl-L-carnitine compared to the control without any supplement (Figure 4A). Compared to a control medium supplemented with glucose and NH4 as carbon and nitrogen sources, the growth in the sebacoyl-L-carnitine medium was similar in P. aeruginosa PAO1 or slightly lower in Pseudomonas sp. MPFS (Figure 4B). However, both strains exhibited the maximum growth on (Z, Z)-9-12-octadecanoyl-L-carnitine. Notably, there was no stationary phase in the growth curve of P. aeruginosa PAO1, and a death phase immediately followed the log phase. Given the similarity between (Z,Z)-9-12-octadecanoyl-L-carnitine and MACH-2, in that the acyl chains of both compounds are derived from linoleate, we infer growth of Pseudomonas sp. MPFS is most likely similar on both compounds and the oleate derivative MACH-1.
Figure 4.
Carnitine and acylcarnitines serve as nutrients in Pseudomonas
(A) Growth curves of Pseudomonas sp. MPFS and P. aeruginosa (PAO1) show their capability to grow in media containing L-carnitine and L-acylcarnitines as the sole carbon and nitrogen source.
(B) Summary of the higher-level taxonomy of Pseudomonas (Left) with arrows depicting the position of the two strains used in (A). Genes involved in the catabolic pathway of carnitine (blue) or positive. Regulators (orange) of its synthetic pathway, previously described in P. aeruginosa (PAO1), are depicted as presence/absence in columns on the right. Filled boxes, triangles, and empty boxes show presence in all clade members, in some or none.
Next, we screened the genome of Pseudomonas sp. MPFS for genes involved in the catabolism of carnitines using the known gene inventory of P. aeruginosa PAO1 as a guide.29,31 This analysis confirmed that all genes and operons coding for enzymes involved in the metabolism and transcriptional regulation of carnitine to glycine and finally entering the central metabolism28 are also present in the genome of Pseudomonas sp. MPFS (Figure 4B; Data S2). Furthermore, we assessed the frequency of these genes within the Pseudomonas genus in a dataset of orthologous proteins from 141 strains, including representatives of all known genus groups and subgroups within the P. fluorescens group. As shown in Figure 4B, the enzymes participating in the carnitine-catabolism pathways are widespread within Pseudomonas, except in the ancestral groups P. pertucinogena, P. linyingensis, and P. stutzeri and a few isolated strains. These findings suggest that several Pseudomonas species possess key ecological advantages to occupy the skin of frogs enriched in these compounds by utilizing L-acyl-carnitines as a nutrient source for growth.
Discussion
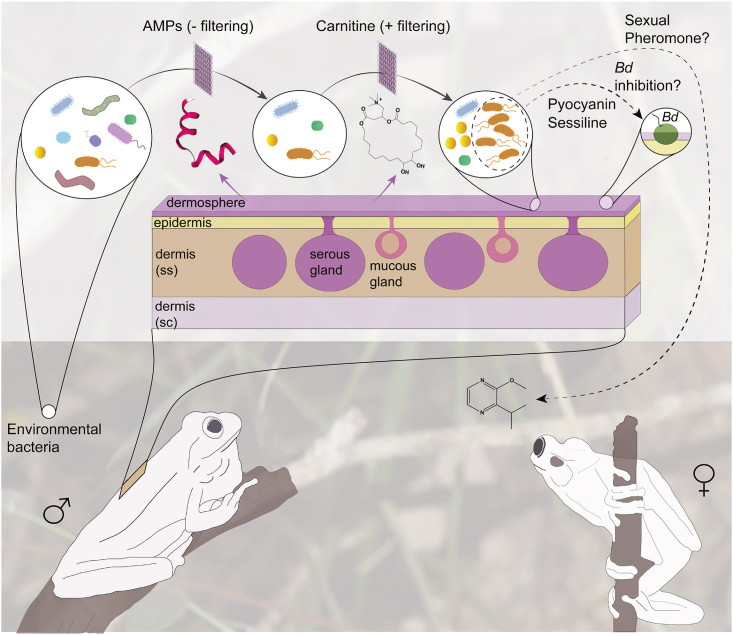
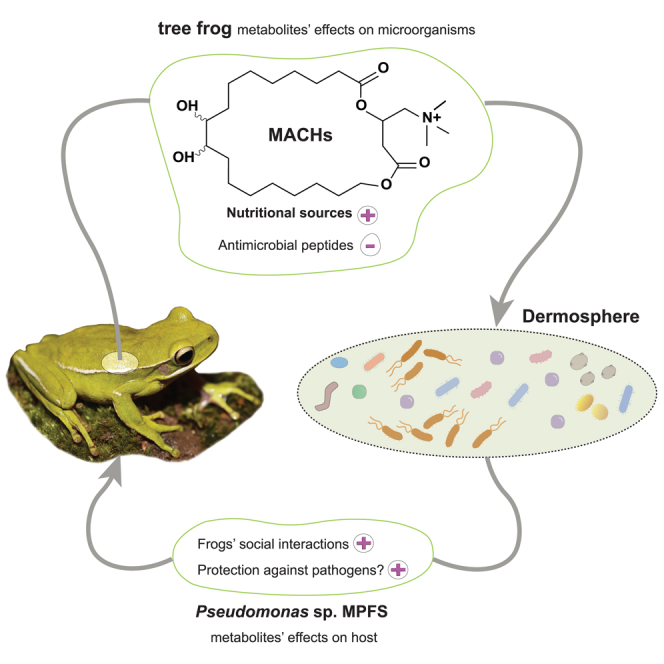
This study outlines the chemistry and molecular mechanisms underlying the interaction occurring on the skin of a diverse clade of tree frogs and their Pseudomonas symbionts. Based on our findings, we propose a chemical and ecological framework to investigate how symbiosis on amphibians’ skin is established and maintained over time (Figure 5). In this model, while most environmental bacteria can initially reach the skin surface of tree frogs, only those with specific adaptive resistance may overcome the high concentration of host antimicrobial peptides,17 which interacts with bacterial membranes through conserved motifs.32 At the same time, distinct bacterial lineages, such as members of the Pseudomonas community capable of utilizing macrocyclic acylcarnitines as nutritional metabolites, will own significant advantages over potential microbial competitors. Once Pseudomonas sp. MPFS and other Pseudomonas have settled on the skin, they can produce antifungal metabolites like pyocyanin and sessilin that would help the host fight the pathogen Batrachochytrium dendrobatidis.17 In addition, they can secrete odorous methoxypyrazines, likely mediating sexual interactions in the frog (Figure 5).14 Thus, this model would explain the low environmental abundance of Pseudomonas and its prevalence on the skin of tree frogs of the Boana pulchella group, as well as the mutual benefits for both partners.
Figure 5.
Simplified model for the chemical interaction between the host and Pseudomonas in the dermosphere of Boana prasina
Once on the skin, the pool of environmental bacteria may be filtered by negative (−) and positive (+) chemical factors secreted by the frog. Those that thrive in this environment may produce distinct metabolites mediating protection against pathogens and social behaviors. Abbreviations: AMPs, Antimicrobial peptides; Bd Batrachochytrium dendrobatidis; sc stratum compactum; ss stratum spongiosum.
We then used published information from diverse families worldwide to understand whether this model can be generalized to other amphibians. Collectively, data indicate that the skin is a unique niche that can be effectively occupied by only a few bacterial taxa, despite their low relative abundance in the environment.18,19,20,33 This effect is usually viewed as the consequence of host and phylogenetic factors, yet, the compounds and mechanisms involved remain poorly characterized.18,19,20 In this sense, recent efforts showed that antimicrobial skin peptides function as a host’s molecular filter for specific strains17,20,34,35 and likely exert their activity to conserved motifs.32 Given that most frogs have antimicrobial peptides and amphibians are the most common animal taxon in antimicrobial peptide databases,36 it is reasonable to assume that skin peptides could selectively inhibit microorganisms in many amphibians’ lineages. In contrast, biogenic amines13 (i.e., low-molecular-weight nitrogenated molecules similar to carnitine) secreted by several amphibians may serve as nutritional sources to the advantage of their symbiotic partners. As shown here, such compounds would particularly benefit species of Pseudomonas able to use them as carbon and energy sources.37 The significance of Pseudomonas in amphibians’ skin has been recognized in different families because of its predominance and the production of antifungal compounds like viscosin-like peptides38 and 2,4-diacetylphloroglucinol16 aiding the host in fighting chytrids. In addition, members of this community were recently shown to produce tetrodotoxin15 and methoxypyrazines,14 suggesting an active contribution in avoiding predators and sexual interactions, respectively. Thus, our proposed model suggests that amphibians’ metabolites of several lineages can actively regulate the establishment of beneficial symbionts through antimicrobial agents and nutrient provisions.
Although the skin of amphibians has been chemically examined over decades,13 recent studies have opened up exciting research perspectives.14,15,39 In this sense, our study represents the first characterization of a macrocyclic acylcarnitine in nature, together with a comprehensive report on the enzymes involved in the metabolism of acylcarnitines and long-chain fatty acids in amphibians’ skin. Specifically, the transcriptome analysis revealed the occurrence of enzymes that in mammals are predominantly expressed in extra-dermal tissues and also the first description of these enzymes in the skin of amphibians. For example, three members of the acyl-CoA synthetase long-chain family40 and the three isoforms of carnitine palmitoyltransferase 1, known to occur mainly in the liver (CPT1A), muscle (CPT1B), and brain (CPT1C),26 are expressed in the skin of B. prasina. Likewise, we found two distinct isoforms of the superfamily of cytochrome P450 enzymes within the family 4 subfamily B member 1 (CYP4B1) to be expressed in frogs’ skin, though in humans, it is predominantly expressed in lung and fat tissues.41 Such a diverse set of enzymes linked to long-chain fatty acid metabolism supports a very active role for frogs’ skin in regulating this class of compounds. In particular, the higher gene expression of CPT1 compared to CPT2 would explain the initial accumulation of acylcarnitines,42 which would become the substrate for subsequent reactions, including ⍵-hydroxylation and lactonization, lastly representing an alternative mechanism to regulate excess acyl groups.
The accumulation of acylcarnitines in humans is linked to incomplete fatty acid β-oxidation, insulin resistance, and oxidative stress, reflecting a dysregulated mitochondrial metabolism.43,44 However, the prevalence of acylcarnitines in the skin glands of frogs does not mark a metabolic disorder or a response to energetic demands. Instead, these compounds may contribute to cold acclimation, reduction of water loss, mitigation of ultraviolet A (UVA)-induced injury, and osmoprotectant as we observed these frogs to be active during freezing nights (near 0°C) and bask under direct sunlight during the day even at high altitudes. A combination of the marked amphipathic nature of acylcarnitines, the long aliphatic chain, and the charged quaternary ammonium moiety mainly supports these propositions. For example, cold acclimation in fishes is related to increased CPT1 activity and a net accretion of lipids45 and in frogs with the accumulation of plasma carnitine.46 Furthermore, depositing acylcarnitines on the skin surface would provide a waterproofing mechanism equivalent to the lipid mixture acting as a skin coating in other dry-adapted tree frogs.47 Lastly, it was established that L-carnitine could mitigate UVA-induced skin injury by suppressing oxidative stress and inflammatory responses.48 This protective role is supported by a recent study showing that 2-methylbutyrylcarnitine was upregulated in high-elevation frogs experimentally exposed to UV radiation.49
By characterizing amphibians’ macrocyclic acylcarnitines that likely evolved as an adaptation for unique physiological roles, which also currently modulate the establishment of beneficial symbionts, our findings add a new level of complexity for surveying coevolutionary interactions. The wide distribution of carnitine in nature25 and carnitine-catabolism pathways in Pseudomonas suggest they represent full-functioning mechanisms ready to be co-opted for symbiosis. Alternatively, but not mutually exclusive, the results indicate that carnitine’s catabolic genes have evolved in Pseudomonas because of their association with many multicellular organisms.17 Regardless of the process involved, we hypothesize that the coevolution between tree frogs and Pseudomonas relies on the active role of frogs’ metabolites favoring the maintenance of environmental-derived microorganisms that benefit the host in short- and midterm. This idea aligns with the holobiont concept, which postulates that some symbionts can increase their population size dramatically if their metabolites improve host fitness.3 Under this umbrella and in an analogy to the rhizosphere of plants,8 we recommend the term “dermosphere” proposed by Assis et al.21 to describe the skin region under the direct influence of chemicals released by the host and the associated microbiome. This term is more accurate than the mucosome50 because the noun “some” from Ancient Greek indicates “body”, denoting an enclosed compartment such as liposomes or peroxisomes, clearly not representing the open system of the skin surface. Many similarities can be recognized between the rhizosphere and the dermosphere. For instance, Hiltner proposed in his definition of rhizosphere more than 100 years ago that different plants’ root exudates foster the establishment of unique bacterial communities and that the composition of these communities would influence the host’s resistance to pathogens.8 Both effects are compatible with those reported here for the host’s specialized metabolites and the inhibition of the fungus B. dendrobatidis by various amphibians’ bacterial communities described elsewhere.51
In conclusion, our work provides broader functional and evolutionary pictures of amphibians’ metabolites while supporting an active role in shaping the skin microbiome. In our simplified model, the host secretes chemicals that indirectly, through inhibition of competitors, and positively, through nutrient provision, help the host’s symbionts, which, in turn, release chemicals that positively contribute to various aspects of the host’s biology. Additionally, our descriptions of lipid-related enzymes and macrocyclic acylcarnitines derivatives represent exciting research avenues for studying long-chain fatty acid metabolism in the skin and their potential link with unique physiological and protective functions in terrestrial vertebrates. This leads to the hypothesis that different host chemicals, such as acylcarnitines, primarily evolved as adaptations to other ecological roles and were subsequently co-opted to participate in symbiotic interactions. Our findings also allow for predicting a wide occurrence of similar mechanisms in the symbiosis between other vertebrate groups and their microbiomes, while the dermosphere framework may provide a more comprehensive view of the many metabolites’ roles in such interactions.
Limitations of the study
Our study focused on the interaction between a specific clade of tree frogs and their skin symbionts. Although we could demonstrate that a host-specific bacterium can exploit metabolites secreted by the skin of an amphibian’s species as its sole carbon and nitrogen source, our study has four major limitations. First, our study only relied on indirect evidence to extend our findings and models to other tree frogs and amphibians. Second, the biosynthetic proposal was based on enzymes identified from two transcriptomes. Third and fourth, our bacterial growth assays were conducted only on one bacterial strain as a representative of the bacterial symbiotic community of the host and using commercially available acylcarnitines rather than the macrocyclic acylcarnitines produced by the frogs. Future experiments should specifically examine MACH-1 and MACH-2, explore deeper the biosynthetic proposal, seek whether other symbionts also use amphibian skin’s metabolites as a nutritional source, investigate other metabolites that may benefit host-symbiont interaction, and include amphibians from distantly related clades.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Pseudomonas sp. MPFS | Brunetti et al.14,17 | N/A |
| Pseudomonas aeruginosa PAO1 | DSMZ (German Collection of Microorganisms and Cell Cultures) | DSM 22644 |
| Biological samples | ||
| Details of frog specimens is listed in Table S1 | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Ultra-pure water | Mili-Q, Millipore | N/A |
| Acetonitrile | J.T. Baker | ID No JT-9012-03 |
| TFA-Na+ | Sigma-Aldrich | ID No 132101 |
| Methanol d4 (99.8% purity) | Sigma-Aldrich | ID No 151947 |
| α-cyano-4-hydroxycinnamic acid matrix | Bruker | ID No 248-879-1 |
| Glucose | Sigma-Aldrich | ID No 1181302 |
| ClNH4 | Sigma-Aldrich | ID No 213330 |
| L-carnitine | Sigma-Aldrich | ID No 8.40092 |
| sebacoyl-L-carnitine | Sigma-Aldrich | ID No 16329 |
| (Z,Z)-9-12-octadecanoyl-L-carnitine | Sigma-Aldrich | ID No 76771 |
| Critical commercial assays | ||
| DNeasy blood and tissue extraction kit | Qiagen | ID No 69506 |
| Deposited data | ||
| GNPS metabolic data | This paper | MassIVE MSV0000924999 |
| Skin microbial community data | This paper | NCBI-SRA PRJNA Acc. No. 498895 |
| Pseudomonas sp. MPFS complete genome | Brunetti et al.17 | NCBI-Genbank Acc. No. NZ_CP066123.1 |
| Pseudomonas spp. OTU sequences | This paper | NCBI-Genbank Acc. No. OR335663–OR335704 |
| Raw transcriptome data | Brunetti et al.17 | NCBI-SRA PRJNA Acc. No. 675859 |
| Accession numbers for Pseudomonas spp. genomes | See Data S2 | See Data S2 |
| Experimental models: Organisms/strains | ||
| Pseudomonas sp. MPFS | Brunetti et al.14,17 | N/A |
| Pseudomonas aeruginosa PAO1 | DSMZ (German Collection of Microorganisms and Cell Cultures) | DSM 22644 |
| Oligonucleotides | ||
| Barcode primers of the V4 region of the bacterial 16S rRNA gene | https://earthmicrobiome.org/protocols-and-standards/16s/ | 515F (Parada) and 806R (Apprill) |
| Software and algorithms | ||
| DataAnalysis 4.0 | Bruker | https://www.bruker.com/protected/en/services/software-downloads/mass-spectrometry/software-solutions.html |
| MSConvert | MSConvert | https://proteowizard.sourceforge.io/index.html |
| MZmine2-2.53 | MZmine | http://mzmine.github.io/ |
| GNPS platform | GNPS | https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp |
| Cytoscape 3.8.0 | Cytoscape | https://cytoscape.org/index.html |
| TopSpin 3.5 | Bruker | https://www.bruker.com/en/products-and-solutions/mr/nmr-software/topspin.html |
| GCMS Solution | Shimadzu | https://www.shimadzu.com/an/products/gas-chromatograph-mass-spectrometry/gc-ms-software/gcmssolution/index.html |
| FlexImaging v.2.1 | Bruker | https://www.bruker.com/protected/en/services/software-downloads/mass-spectrometry/software-solutions.html |
| ITEM Software | ITEM Software | https://cfim.ku.dk/equipment/electron_microscopy/cm100/iTEM_Main_Brochure.pdf |
| QIIME2 v2020.11 | Qiime2 | https://qiime2.org/ |
| R-4.2.3 | R Core Team | https://www.R-project.org/ |
| eggnog-mapper v2 | Huerta-Cepas et al.52 | http://eggnog-mapper.embl.de/ |
| Kallisto | Bray et al.53 | https://github.com/pachterlab/kallisto |
| ProteinOrtho v4.26 | Lechner et al.54 | https://gitlab.com/paulklemm_PHD/proteinortho/ |
Resource availability
Lead contact
-
•
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Andrés E. Brunetti (aebrunetti@conicet.gov.ar, andresbrunetti@gmail.com).
Materials availability
-
•
This study did not generate new unique reagents.
Experimental model and study participant details
Frog specimens used
Twenty-one adult specimens of the South American treefrog Boana prasina were collected at two sites in São Paulo and Rio de Janeiro, Brazil (Table S1). In addition, adult specimens from 33 species representing 11 genera of the tree frog subfamilies Hylinae and Phyllomedusinae were collected in different localities from Brazil, Colombia, and Argentina (Table S1). Because of our particular interest in species related to B. prasina, specific efforts were made to collect representatives from the B. pulchella group, resulting in the collection of eight species (B. bischoffi, B. caingua, B. curupi, B. polyteania, B. prasina, B. pulchella, B. riojana, and B. stellae). These species represent four of the five recognized clades within this group. For all species, we included male and female specimens. Detailed information on species identity, location, and coordinates are listed in Table S1. In all cases, individuals were transported to the laboratory and kept in glass terraria at 22°C–26°C with a 12 h photoperiod, provided with dechlorinated tap water, access to refugia, and fed with small crickets. They were housed in these conditions for 2 to 4 days before sampling. After sampling, they were euthanized with a lethal dose of lidocaine 20% applied in the pelvic region. Vouchers are housed at “Célio F. B. Haddad Amphibians collections” at The State University of São Paulo, Rio Claro, Sao Paulo, Brazil; at the Museo de Herpetología Universidad de Antioquia (MHUA) in Medellín, Colombia; and at the Herpetological Collection of the Laboratorio de Genética Evolutiva, Instituto de Biología Subtropical (Unam-CONICET), Posadas, Misiones, Argentina.
Collection permits
All procedures involving frogs were carried out according to the regulations specified by Conselho Nacional de Controle de Experimentação Animal (CONCEA), Ministério da Ciência, Tecnologia e Inovação (MCTI) and were approved by the Ethics Committee on Animal Use (CEUA) of School of Pharmaceutical Sciences of Ribeirão Preto- (17.1.1074.60.0), Brazil. Collection permits were issued by Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) / SISBIO, Brasil (Permits 41508-8, 50071-1, 50071-2, 57098-1), Autoridad Nacional de Licencias Ambientales (ANLA), Colombia (Permits 1177-Oct 9th 2014 to the Universidad de Los Andes and 0524 May 27th 2014 to the Universidad de Antioquia), and Ministerio de Ecología y Recursos Naturales Renovables de Misiones (035/2017) and Instituto Misioneiro de Biodiversidad (IMiBio), Argentina (Permits 11/2021).
Method details
Acquisition of the skin glandular secretion
The skin glandular secretion of each frog was sampled from the dorsal skin region using the following protocol: the skin was cleaned and moistened with ultrapure water obtained from an Ultra Centrifugal Filter Unit (Milli-Q, Millipore). Subsequently, an electrical stimulus (1–2 V, pulse length 2–4 ms) was applied during 8–10 s using two platinum electrodes rubbed gently along the animal’s dorsum.55 Finally, the secretion was washed off with 20–30 mL of the sterilized water collected in falcon tubes (50 mL), frozen in liquid N2, lyophilized, and stored refrigerated at -20°C.
Skin metabolic profile: HPLC-DAD ESI MS and MS/MS
Analyses of liquid chromatography hyphenated to diode array detection and electrospray ionization tandem mass spectrometry (LC-DAD-MS/MS) were performed in an ultra-fast liquid chromatography (UFLC) instrument (Shimadzu, Japan). The UFLC instrument was coupled to a high-resolution mass spectrometer micrOTOF-QII (Bruker Daltonics, Billerica – USA). The dried glandular secretion was dissolved in water: acetonitrile (7:3) with 0.1% (v/v) trifluoroacetic acid (TFA) at a final concentration of 0.5 mg mL-1 and centrifuged. The supernatant was injected into a C18 - Luna Kinetex Phenomonex® (100 mm x 2.1 mm x 2.6 μm) column, with the temperature adjusted to 35°C. The mobile phase (flow 0.25 mL/min) consisted of water (A) and acetonitrile (B) in the following gradient: 0.0–35.0 min (15–80% B); 35.0–37.0 min (80–100% B); 37.0–42.0 min (100% B); 42.0–45.0 min (100–15% B); 45.0–48.0 min (15% B). qTOF acquisition parameters were as follows: capillary 3.5 kV, end plate offset 500 V, nebulizer 35 psi, dry gas (N2) with a flow of 8.0 L/min, and temperature of 300°C. Accurate masses were obtained using TFA-Na+ (sodiated trifluoroacetic acid) as the internal standard. Mass spectra were measured between m/z 90 to1200 in the positive ion mode. ESI-MS/MS fragments were generated by collision-induced dissociation using nitrogen as the collision gas (5–50 eV depending on the m/z of each precursor ion) and analyzed with the autoMSn mode using the Enhanced Resolution Mode for MS and UltraScan.
Processing and molecular networking of mass spectrometry data
Molecular networking56 workflows organize large data sets of tandem mass spectra based on the similarity between fragmentation patterns of different but related precursor ions. The LC-QTOF-MS/MS raw data were converted to mzXML format using the MSConvert software57 for molecular networking. First, isotope grouping, alignment, and data normalization were processed with MZmine2-2.5358 with the following parameters: an MS1 noise level of 1.0E103 and; an MS2 noise level of 1.0E102. Next, chromatograms were built with the ADAP chromatogram builder module with the following parameters: min highest intensity, 1.5E103; maximum m/z tolerance, 10 ppm. Next, they were deconvoluted using the ADAP (Wavelets) module59 with the following parameters: S/N threshold, 10; min feature height, 10; coefficient/area threshold, 110; peak duration range, 0.01–2.0 min; retention time wavelet range, 0.001–0.1 s. Next, the fragmentation spectra were paired to the deconvoluted peaks using 0.01 Da and 0.3 min windows, and the LC-MS features (isotopologues, adducts, and in-source fragments) were annotated using the peak grouping module with the following parameters: deisotope, true; m/z tolerance, ten ppm; retention time tolerance, 2 min; maximum charge, 3. Finally, the features were aligned with the Join Aligner module using the following parameters: tolerance, 5 ppm; weight for m/z, 75.0; retention time tolerance, 0.2 min; weight for retention time, 25.0.
The feature table and spectral data were exported and uploaded to the GNPS platform56 and analyzed with the feature-based molecular network workflow. Molecular features in the form of MS/MS spectra were putatively identified against matches to the MS2-based public spectral library based on the cosine score similarity (above 0.7) and setting six as the minimum number of shared peaks. The parameters and results will be available at the MassIVE repository from the GNPS platform.56 The results were subsequently visualized in Cytoscape 3.8.0.60 The molecular networking file contained the scan number, precursor m/z, and fragment m/z ions were colored at different taxonomic levels [species within the Boana pulchella group (i.e., B. pulchella clade, B. riojana clade, B. semiguttata clade, and B. polytaenia clade), group for other species within the genus Boana, genus for other species of Cophomantini, and subfamily in the remaining species].
Isolation and purification of acylcarnitines
The dried skin glandular secretion of ten individuals of Boana prasina was pooled (50 mg), resuspended in water (TFA 0.01%): methanol (4:6), and centrifuged. Afterward, the supernatant was fractionated by a chromatographic column containing Sephadex as the stationary phase (LH-20) using water (TFA 0.01%): and methanol (3:7) as the mobile phase. The purified fractions containing cyclic acylcarnitines were pooled, and the compounds were isolated using a C18 - Luna (Phenomonex® – 250 mm × 21.2 mm x 5 μm) preparative column equilibrated with 70% water (TFA 0.01%) (eluent A) and 30% methanol (eluent B). The elution profile was a gradient produced by a CBM-20A System controlling two LC-20AP HPLC pumps (Shimadzu, MD, USA) at a flow rate of 21 mL/min as follows: 0.0–45.0 min (30–70% B); 45.0–50.0 min (70−100% B); 50.0–55.0 min (100% B); 55.0–60.0 min (100−30% B); 60.0–65.0 (30% B). Eluents were monitored by UV absorbance at 220 nm. Fractions were collected using an automated fraction collector FRC-10A (Shimadzu, USA), concentrated, and vacuum-dried for subsequent analysis. Two main ions corresponding to MACH-1 (C25H48NO6; m/z 458) and MACH-2 (C25H46NO6; m/z 456) were detected by direct infusion into the ESI source by the syringe pump and analyzed by an ultrOTOF Q-II (Bruker Daltonics, Billerica – USA) mass spectrometer. The isolated cyclic acylcarnitines MACH-1 and MACH-2 were obtained as viscous, transparent oil yielding 0.6 mg (12%) and 0.4 mg (8%), respectively. They were next submitted to liquid chromatography-tandem mass spectrometry (LC-DAD-MS/MS) to assess their purity. The samples were lyophilized and stored at −20°C for further experiments (NMR, mass spectrometry, optical rotatory dispersion, and derivatization). Both purified compounds are colorless viscous oils.
Structural elucidation of MACH-1 and MACH-2
The purified samples of the cyclic acylcarnitines were dried under nitrogen gas, dissolved in methanol-d4 99.8% (Sigma-Aldrich), transferred to 3 mm micro-NMR tubes, and analyzed. The 1H NMR spectra were recorded at 25°C on a 14.1 Tesla Bruker Avance III spectrometer operating at a proton NMR frequency of 600.13 MHz (Bruker, Karlsruhe, Germany). Chemical shifts are given in δ units (ppm) relative to tetramethylsilane. All analyses were performed using TopSpin 3.5 (Bruker).
Several NMR experiments were performed for both cyclic acylcarnitines. Free induction decays (FIDs) were Fourier transformed, and the spectra were manually phased and baseline corrected. 1H -1H COSY experiments were performed using a spectral window of -0.33−9.88 ppm. 1H-13C direct coupling was studied by HMQC, using a spectral window of -0.33−9.88 ppm for the F2 dimension (1H) and -15−220 ppm for the F1 dimension (13C). Finally, 1H-13C HMBC data were obtained using a spectral width of -0.33−9.88 ppm for the F2 dimension (1H) and -20–210 ppm for the F1 dimension (13C).
Because of NMR’s limitations in assigning the positions of the two hydroxyl groups in the fatty acid chain, we derivatized and then analyzed the acylcarnitine m/z 458 with a Shimadzu (Kyoto, Japan) GC coupled to a Mass Spectrometer Detector (GC/MS QP2010 Plus). We first conducted mild alkaline hydrolysis for the derivatization process, for which the sample was dissolved in H2O: EtOH: KOH 2M (10:10:2) and heated at 40°C for 1 h. Next, it was acidified with HCl to a final pH = 5, and the free acids were extracted into ethyl acetate: diethyl ether (1:1) and derivatized (20 μl, BSTFA, 80°C, 1 h). The derivatized sample was then injected in the GC/MS (2 μl) equipped with a Restek Rtx® -5MS column (60 m×0.25 mm×0.25 μm) obtained from Restek (Bellefonte, PA, USA) using helium (purity of 99.9999%; White Martins, RS, Brazil) with a flow rate of 1.4 mL/min as the carrier gas. The oven temperature was programmed as follows: initial temperature of 70°C (5 min hold), 8°C/min to 330°C (60 min hold). The injector was operated in split mode (1:5) at a temperature of 280°C. A quadrupole mass spectrum detector was operated in electron impact mode at 70 eV and 250°C with a mass range of 35–700 u. The GC/MS spectra were analyzed using the GCMS Solution software.
High-resolution mass spectrometry analysis and 1D [13C (only for MACH01) and 1H] and 2D NMR experiments (HMBC, HMQC, and COSY) enable the unequivocal assignment of all the atoms in the 4-trimethylazaniumyl-butanoate moiety. In addition, these data allowed us to determine that the aliphatic region of MACH-1 and MACH-2 correspond to dehydroxylated C18 fatty acids likely derived from oleate and linoleate, respectively. Furthermore, NMR data allow determining that the two hydroxyl groups were in adjacent carbons because there were only two Carbon atoms with δ = 33 ppm. If, for instance, there were one or two carbon atoms between the two carbons bearing the -OH, we would expect two or four carbons, with NMR signals around δ 45 ppm and δ 33 ppm, respectively, which was not observed in the experiments. Finally, the precise location of the hydroxyls in the aliphatic chain was determined after fatty acid hydrolysis, followed by silylation and GCMS analysis, which allowed us to assign them to C9’ and C10' unambiguously.
Mass fragmentation patterns of MACH-1 and MACH-2
Purified fractions containing the cyclic acylcarnitines m/z 456 and m/z 458 from Boana prasina were submitted to high-resolution MS/MS analysis and MS/MS and MS3 analyses in the Ion Trap mass spectrometer AmaZon (Bruker Daltonics, Billerica – USA). Samples were introduced into the ESI source by a syringe pump and analyzed by a micrOTOFQ (Bruker Daltonics, Billerica – USA) mass spectrometer. Accurate mass analyses were performed using a solution of sodium trifluoroacetic acid [(TFA)n+Na]+ as the internal standard. The parameters were as follows: capillary voltage 3.5 kV, end plate offset of 500 V, the capillary temperature at 200°C; nitrogen as the sheath gas; drying gas flow at 4.0 L/min; drying gas temperature at 180°C; nebulizer gas pressure of 0.4 bar; positive ESI mode. Mass spectra were measured from m/z 50–600 in the positive ion mode. Fragmentation patterns of cyclic acylcarnitine were proposed considering the high-resolution MS/MS for molecular formulae determination (accurate molecular weights < 5ppm) and ions generated in the MS3 experiments (submitted as MassIVE dataset). Retention time (shift tolerance of ±0.05 min) was used as orthogonal information on peak annotation.
Optical rotatory dispersion analyses of MACH-1
The optical rotatory dispersion of the acylcarnitine MACH-1 was measured using a Jasco P2000 spectropolarimeter (Nikota, Japan) to determine whether the molecule is dextro- or levorotatory. After NMR analyses, the methanol-d4 was evaporated, and the sample was resuspended in methanol. Measures were taken at 25°C in triplicate using a 1 mL cuvette.
Analysis of skin lipids
After euthanasia, the dorsal skin of adult specimens of Boana prasina and B. bischoffi was removed, macerated under liquid nitrogen, and transferred to a 5 mL glass vial. The lipids were extracted using the protocol described by Bligh & Dyer (1959)61 as a guide. A 3.75 mL mixture of chloroform: methanol (2:1) was added to the macerated skin and submitted to ultrasound for 10 min and vortex for 2 min. Then, 1.25 mL of Mili-Q water was added to the homogenate and submitted to a second row of ultrasound and vortex. The homogenate was filtered through a Whatman 1 filter paper on a Buchner funnel with slight suction, the content was transferred to a test tube, left at room temperature for a few minutes, and centrifuged (3 min, 1000 rpm). The volume of the chloroform layer containing the lipid fraction was gently removed with a glass pipet, transferred to a 5 mL glass tube, evaporated to dryness under gaseous N2, and stored at 20°C.
The lipid fraction was dissolved in chloroform and separated in a thin-layer chromatography plate (silica gel 60 F254) using hexane: ethyl ether: acetic acid (80: 30: 2) as the mobile phase. Lipids were detected by iodine staining. Individual components were scraped from the plates and submitted to derivatization with alkylation reagents. Lipid derivatization was performed by dissolving the dry sample in 500 μL of BF3 in methanol, vortexed for 1 min, and maintaining it at 80°C for 10 min. Afterward, samples were immediately cooled on ice, followed by the addition of 500 μL of Mili-Q water and 1 mL of pentane, and then vortexed for 1 min. Next, the homogenate was centrifuged (3 min, 3000 rpm), the pentane phase containing the ester derivatives was collected gently, transferred to a 1.5 mL tube, evaporated to dryness under gaseous N2, and stored at -20°C.
The derivatized lipids were dissolved with 50–100 μL of hexane. Next, 2 μL were injected in the GC/MS Shimadzu QP2010 Plus (Kyoto, Japan) equipped with a ZB-Wax column (30 m × 0.25 mm×0.25 μm) obtained from Restek (Bellefonte, PA, USA). Helium (purity of 99.9999%; White Martins, RS, Brazil) with a 1.3 mL/min flow rate was used as the carrier gas. The oven temperature was programmed as follows: initial temperature of 50°C (3 min hold), 10°C/min to 200°C (5min hold), 5°C/min to a final temperature of 240°C (20 min hold). The injector was operated in splitless mode (1.5 min sampling time) at a temperature of 230°C. A quadrupole mass spectrum detector was operated in electron impact mode at 70 eV and 250°C with a mass range of 40–500 u. The spectra were analyzed using the GCMS Solution software.
Skin distribution of MACH-1 and MACH-2 by MALDI-IMS imaging
For observations of skin on transverse sections, dorsal skin samples from two male specimens of Boana prasina were removed, immediately embedded in carboxymethyl cellulose 2%, and frozen at -20°C. Then, the samples were sliced at 10 μm in a cryotome (CM1860 Cryostat, Leica Biosystem, Nussloch, Germany) at -20°C. Both transverse and whole skin sections were first mounted on indium tin oxide-coated glass slides for MALDI imaging (Bruker Daltonic, Bremen, Germany). Next, they were coated with Imageprep (Bruker Daltonic, Bremen, Germany) using sixty cycles with a solution of 7 mg mL−1 of an α-cyano-4-hydroxycinnamic acid matrix in 50% acetonitrile/water with 0.2% TFA. Each cycle was based on spraying, filling the chamber with nitrogen, and drying.
MALDI-Imaging analyses were performed using a MALDI-TOF/TOF (Bruker Daltonics, UltrafleXtreme, Bremen, Germany) system equipped with a smart beam-II laser system controlled by a FleXcontrol v.3.3 (Bruker Daltonics, Bremen, Germany), employing AutoXecute mode. IMS data were analyzed and normalized using FlexImaging v.2.1 (Bruker Daltonics). The settings of the instrument were as follows: positive ion reflector mode; ion source 1, 25.00 kV; ion source 2, 22.35 kV; lens, 7.75 kV; reflector, 26.60 kV; reflector 2, 13.30 kV; pulsed ion extraction, 100 ns; laser frequency, 100 Hz; and matrix suppression mass cutoff, m/z 500. The spectra were recorded across a mass range of m/z 100−600, accumulating 500 shots per spectrum. The images were collected from transverse sections and skin surface at a 20 μm spatial resolution in both x and y directions using MS positive ion mode. Quercetin and skin extracts of B. prasina were used as the external calibration of the mass spectrometer and to validate the detection of MACH-1 and MACH-2 by MALDI, respectively. The average mass deviation was below 10 ppm. MS/MS analysis directly from the skin was performed to confirm assignments of the two mains cyclic acylcarnitines in positive-ion reflector mode using LIFT fragmentation.
Transcriptome annotation, quantification, and biosynthetic proposal
The reference transcriptome assembled from one male and one female specimen in Brunetti et al.17 was annotated using the eggnog-mapper v2 available online52 with the following parameters: seed ortholog e-value 0.001, seed ortholog score 60, and query cover 20. The level of expressions of different genes was estimated from the cleaned reads of the male and the female using Kallisto.53 Based on the molecule structure, the annotation, and published information, we searched for enzymes associated with carnitine biosynthesis, the formation of the acylcarnitine ester ω-oxidation of fatty acids, epoxidation, and hydroxylation. The similarity of these proteins was compared with those annotated in humans and Xenopus laevis and retrieved from Uniprot. Transcriptome annotation and quantification were performed to determine whether the skin of Boana prasina possesses the enzymes for the biosynthesis of the cyclic acylcarnitines, to assess the relevance of these enzymes, and to propose a biosynthesis pathway for these compounds.
Electron microscopy analyses
Dorsal skin samples of Boana prasina were fixed with various concentrations of formaldehyde and glutaraldehyde and stored in a fixative solution upon electronic microscopy sample preparation. The skin was divided into fragments to perform scanning electron microscopy (SEM) and transmission electron microscopy (TEM).
For SEM, the skin was washed twice with TE buffer to remove the fixative. Dehydration was performed in a graded series of acetone (10, 30, 50, 70, and 90%) on ice, each step for 10 minutes, followed by two 10 minutes steps with 100% acetone at room temperature. Afterward, critical-point drying with liquid CO2 (CPD 30, Balzers, Liechtenstein) was done. The skin fragments were placed with conductive tapes on aluminum stubs and covered with a palladium–gold film by sputter coating (SCD 500, Bal-Tec, Liechtenstein). The skin was examined from two views, surface and transverse sections. Visualization was conducted with a field emission scanning electron microscope (Merlin, Zeiss, Germany) using the HESE2 Everhart Thornley SE detector, the in-lens SE detector, and an acceleration voltage of 5 kV.
For TEM, the fixed samples were washed for 40 minutes with 0.1 M EM-HEPES buffer (HEPES 0.1 M, 0.09 M sucrose, 10 mM CaCl2, 10 mM MgCl2, pH 6.9) and exposed to osmium tetroxide (1% in HEPES buffer) for 1 h at room temperature. After another washing step with HEPES buffer, samples were dehydrated with a graded series (10, 30, 50, 70, 90, and 2x 100%) of acetone, each step for 30 minutes on ice, respectively, at room temperature (100 % steps). In the 70% EtOH step, we added 2% uranyl acetate (UAc) and incubated overnight at 4°C. Samples were subsequently infiltrated with the low viscosity (LV) resin (1:1, 2:1, 2 × 100%), each step incubating for a minimum of 8 h. Samples were polymerized at 75°C for 15 h followed by a second polymerization step to mount the rotated flat-embedded sample block onto a resin pedestal to allow vertical sections of the skin. Ultrathin sections of approx. 50–70 nm was prepared with an Ultramicrotome (Reichelt Jung / Leica Microsystems, Germany) collected onto butvar-coated 300 mesh grids and counterstained with 4% aqueous uranyl acetate for 3 min and lead citrate for 15 seconds. Images were acquired with a transmission electron microscope TEM 910 and a Libra 120 Plus (both Zeiss, Germany) with an acceleration voltage of 80 kV and 120 kV, respectively, and at calibrated magnifications. Contrast and brightness were adjusted using the ITEM software.
Frogs’ sampling for microbial community analyses
Forty-five adult B. prasina specimens were collected at the margins of a small lake at São Francisco Xavier, São Paulo, Brazil (Table S1). Forty were released at the same collecting site after swabbing, and the remaining were kept as vouchers. In addition, adult specimens from five other species (B. bischoffi, B. caingua, B. polytaenia, B. pulchella, and B. stellae) of the B. pulchella group were collected at different sites in Brazil and Argentina. Each frog was handled with new gloves minimizing cross-contamination. Before swabbing, each specimen was rinsed with sterilized deionized water to eliminate transient bacteria. The dorsal skin was swabbed for all specimens within 12 h after collection using sterile MW113 swabs (Medical Wire & Equipment, Corsham, UK). In addition, environmental samples were collected by swabbing the water and grass at three sites. After sampling, each swab was immediately placed in sterile microcentrifuge vials and frozen in a -20°C freezer. Detailed information on the species, specimen numbers, location, coordinates, and voucher numbers for the frogs used in this section are listed in Table S1. Collection permits were issued by SISGEN (A1FC113, A9EC80A, A299B7D).
Bacterial community analysis
Genomic DNA was extracted from skin swabs and environmental and sterile water controls using the DNeasy blood and tissue extraction kit (Qiagen). The V4 region of the bacterial 16S rRNA gene was PCR-amplified using a dual-index approach with barcoded primers 515F and 806R.62 PCR reactions were performed in duplicates. Negative controls for the DNA extraction and PCR mix were included to check for contamination. PCR products of each sample were pooled, purified using a DNA Gel Extraction Kit (Norgen Biotek Corp, Torold, ON, Canada), and sequenced with paired-end 2x250 v2 chemistry on an Illumina MiSeq sequencer at TUCF genomics (Boston, MA, USA). The sequences of the amplicon libraries were deposited in the NCBI short read database (SI Appendix, Data S2 A, and BioProject ID obtained after submission).
The sequences were processed and analyzed in QIIME2 v2020.1163 on Ubuntu 20.4 LTS Linux. Briefly, the sequences were demultiplexed, paired-ends were merged, quality filtered, and denoised using Deblur workflow.64 The Operational Taxonomic Units (OTUs) in negative controls and less than one frog sample were filtered out. The remaining sequences were aligned, and the FastTree265 was used to build a phylogenetic tree of OTUs using QIIME 2 standard procedures. OTUs were classified using the q2-feature-classifier-sklearn against the Greengenes 13.8 database, with 99% similarity with the OTUs reference sequences (from the 515F/806R region of the sequences). Samples were rarefied to 1,500 reads. Descriptive analyses were conducted to assess bacterial richness and similarities between frog populations and the environment.
Because we were interested in the symbiosis between frogs from the Boana pulchella group and bacterial strains from the Pseudomonas genus, we extracted from the analysis those OTU sequences classified within this genus. These sequences were compared with the sequences reported in our previous work on the bacterial community analyses from different populations of B. prasina obtained from culture-dependent and culture-independent methods. In addition, a heatmap was constructed within Qiime2 to depict the relative abundance variation of those OTUs identified as Pseudomonas spp. in all samples.
Bacterial growth assays
Pseudomonas sp. MPFS isolated from the dorsal skin of Boana prasina and P. aeruginosa PAO1 were grown for 72 h at 30°C in Mueller-Hinton (MH) broth medium and inoculated at a final absorbance at 600 nm of ∼0.05 into M9 minimal medium supplemented with a specific carbon and nitrogen source.
Assays were conducted in 1 mL on 96-well polystyrene plates. For experiments, L-carnitine and two acylcarnitines (i.e., sebacoyl-L-carnitine and (Z,Z)-9-12-octadecanoyl-L-carnitine) purchased from Sigma-Aldrich were added to the M9 minimal medium66 at a final concentration of 20 mM without the addition of any other carbon and nitrogen source. Positive growth controls contained glucose and NH4Cl in the M9 medium. In contrast, negative controls either received the inoculum but lacked nitrogen and carbon sources or were supplemented with nitrogen and carbon sources but lacked the inoculum. Plates were incubated at 30°C, and OD600 was determined every 3 or 6 h for 72 h using a microplate spectrophotometer (Infinite M200 Pro, Tecan AG, Mannedorf, Switzerland). All tests were performed in triplicate according to the Clinical and Laboratory Standards Institute (2012). Growth curves were constructed for each treatment based on their data distribution using the locally weighted regression loess curves in R. A standard procedure was used to determine differences between treatments. Briefly, the maximum growth rate and 95% confidence interval were calculated from the log-linear part of growth curves from each AMP concentration and the control treatment without AMPs. When the confidence intervals for an AMP treatment and control did not overlap, they were judged to be significantly different.
Bacterial genome analyses
Since the description of its genome in 2000,67 Pseudomonas aeruginosa PAO1 has been the subject of many gene functional analyses, which yielded significant insights into the enzymes involved in the different metabolic processes of this bacterium. This information served as a reference for genomic analyses of host-symbiont systems in vertebrates, mainly because most studies have focused on P. aeruginosa as a human pathogen. Genes related to the catabolism of acylcarnitine as the sole source of carbon and nitrogen were obtained from published studies.29,31,68,69
Genes involved in the catabolism of acylcarnitines were identified in the genus Pseudomonas using the same data matrix from our previous work.17 This matrix was based on 159 strains, from which 141 corresponded to type strains (Data S2). In addition, gene orthology was assessed using ProteinOrtho v4.26,53 and the phylogenetic distribution of genes involved in the acylcarnitine catabolism was evaluated using the same phylogenetic hypothesis described previously.17
Acknowledgments
We are grateful to Ricardo Silva for his many helpful suggestions while conducting this work. We also thank Pedro P. Taucce, Camila Capel Godinho, José Godinho, Luiz Fernando Carmo, Carola Jovanovich, Helio Ricardo da Silva, Javier Torres, Diego Baldo, Juan Boeris, Andrea Caballero, Miriam C. Vera, Ariadne F. Sabbag, Carla M. Lopes, Delio Baêta, Fernando Vargas Salinas, and Mauricio Rivera Correa for their help during fieldwork. Izabel Cristina Casanova Turatti, Jose Carlos Tomaz, Valquiria A. Polisel Jabor, and Felipe Calil provided valuable assistance during mass spectrometry and bacterial growth experiments and Ina Schleicher helped us during EM sample preparation. We also thank Neelu Begum and Andrew H. Moeller for their critical comments and suggestions. This project was founded by São Paulo Research Foundation FAPESP grants #2013/50954-0, #2014/50265-3, 2020/02207-5, 2021/10639-5; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CNPq) (process #306623/2018-8); Agencia Nacional de Promoción Científica y Tecnológica grant number PICT 2020 #2939 and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) grant #PIP 2021-2023 to (AEB); FAPESP postdoctoral fellowships #2014/20915-6 and #2017/23725-1 (AEB); #2017/17648-4 (AB); #2012/21803-1 and #2014/01651-8 (AMCR); #2017/26162-8 (MLL); #2015/01001-6 (WGPM).
Author contributions
A.E.B., M.L.L., G.M.C., J.O., and N.P.L. designed research; A.E.B., M.L.L., A.B., B.B., C.B., M.M., F.C.N., J.N.M., and W.P.M. performed research; A.E.B., M.L.L., A.B., C.B., F.C.N., W.P.M., A.M.C.R., J.O., and N.P.L. analyzed data; A.E.B., M.L.L., and M.T.P. coordinated sampling and lab work; J.O. and N.P.L. supervised microbial and chemical analyses; A.E.B., M.L.L., A.B., F.C.N., G.M.C., and J.O. wrote the paper with contributions from all authors.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: October 4, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108109.
Contributor Information
Andrés E. Brunetti, Email: aebrunetti@conicet.gov.ar.
Jörg Overmann, Email: joerg.overmann@dsmz.de.
Norberto P. Lopes, Email: npelopes@fcfrp.usp.br.
Supplemental information
Data and code availability
-
•
Metabolomic data generated in this study are deposited at MassIVE: MSV0000924999
-
•
The sequences of the bacterial amplicon libraries and Pseudomonas spp. OTU sequences were deposited at NCBI-SRA PRJNA: 498895 and NCBI-GenBank: OR335663–OR335704, respectively.
-
•
The skin transcriptome of Boana prasina and the complete genome of Pseudomonas sp. MPFS, were available at NCBI-SRA PRJNA: 675859 and NCBI-GenBank: NZ_CP066123.1, respectively.
-
•
This study did not generate original codes. All data were analyzed following the protocols and instructions from the software, packages, and workflows available in the original publications or repositories.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Byrd A.L., Belkaid Y., Segre J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018;16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert S.F., Sapp J., Tauber A.I. A symbiotic view of life: we have never been individuals. Q. Rev. Biol. 2012;87:325–341. doi: 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg E., Zilber-Rosenberg I. Microbes drive evolution of animals and plants: the hologenome concept. mBio. 2016;7:e01395. doi: 10.1128/mBio.01395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLaren M.R., Callahan B.J. Pathogen resistance may be the principal evolutionary advantage provided by the microbiome. Philos. Trans. R. Soc. B. 2020;375:20190592. doi: 10.1098/rstb.2019.0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ezenwa V.O., Gerardo N.M., Inouye D.W., Medina M., Xavier J.B. Animal behavior and the microbiome. Science. 2012;338:198–199. doi: 10.1126/science.1227412. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y.E., Fischbach M.A., Belkaid Y. Skin microbiota-host interactions. Nature. 2018;553:427–436. doi: 10.1038/nature25177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tipton L., Darcy J.L., Hynson N.A. A developing symbiosis: enabling cross-talk between ecologists and microbiome scientists. Front. Microbiol. 2019;10:292. doi: 10.3389/fmicb.2019.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann A., Rothballer M., Schmid M. Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil. 2008;312:7–14. [Google Scholar]
- 9.Vieira S., Sikorski J., Dietz S., Herz K., Schrumpf M., Bruelheide H., Scheel D., Friedrich M.W., Overmann J. Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J. 2020;14:463–475. doi: 10.1038/s41396-019-0543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adair K.L., Douglas A.E. Making a microbiome: the many determinants of host-associated microbial community composition. Curr. Opin. Microbiol. 2017;35:23–29. doi: 10.1016/j.mib.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Mergaert P. Role of antimicrobial peptides in controlling symbiotic bacterial populations. Nat. Prod. Rep. 2018;35:336–356. doi: 10.1039/c7np00056a. [DOI] [PubMed] [Google Scholar]
- 12.Bosch T.C.G. Cnidarian-microbe interactions and the origin of innate immunity in metazoans. Annu. Rev. Microbiol. 2013;67:499–518. doi: 10.1146/annurev-micro-092412-155626. [DOI] [PubMed] [Google Scholar]
- 13.Erspamer V. In: Amphibian biology. Volume 1: The integument. Heatwole H., Barthalmus G., editors. Surrey Beatty & Sons; 1994. Bioactive secretions of the amphibian integument; pp. 178–350. [Google Scholar]
- 14.Brunetti A.E., Lyra M.L., Melo W.G.P., Andrade L.E., Palacios-Rodríguez P., Prado B.M., Haddad C.F.B., Pupo M.T., Lopes N.P. Symbiotic skin bacteria as a source for sex-specific scents in frogs. Proc. Natl. Acad. Sci. USA. 2019;116:2124–2129. doi: 10.1073/pnas.1806834116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaelli P.M., Theis K.R., Williams J.E., O’Connell L.A., Foster J.A., Eisthen H.L. The skin microbiome facilitates adaptive tetrodotoxin production in poisonous newts. Elife. 2020;9:e53898. doi: 10.7554/eLife.53898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodhams D.C., LaBumbard B.C., Barnhart K.L., Becker M.H., Bletz M.C., Escobar L.A., Flechas S.V., Forman M.E., Iannetta A.A., Joyce M.D., et al. Prodigiosin, violacein, and volatile organic compounds produced by widespread cutaneous bacteria of amphibians can inhibit two Batrachochytrium fungal pathogens. Microb. Ecol. 2018;75:1049–1062. doi: 10.1007/s00248-017-1095-7. [DOI] [PubMed] [Google Scholar]
- 17.Brunetti A.E., Bunk B., Lyra M.L., Fuzo C.A., Marani M.M., Spröer C., Haddad C.F.B., Lopes N.P., Overmann J. Molecular basis of a bacterial-amphibian symbiosis revealed by comparative genomics, modeling, and functional testing. ISME J. 2022;16:788–800. doi: 10.1038/s41396-021-01121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walke J.B., Becker M.H., Loftus S.C., House L.L., Cormier G., Jensen R.V., Belden L.K. Amphibian skin may select for rare environmental microbes. ISME J. 2014;8:2207–2217. doi: 10.1038/ismej.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenzie V.J., Bowers R.M., Fierer N., Knight R., Lauber C.L. Co-habiting amphibian species harbor unique skin bacterial communities in wild populations. ISME J. 2012;6:588–596. doi: 10.1038/ismej.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flechas S.V., Acosta-González A., Escobar L.A., Kueneman J.G., Sánchez-Quitian Z.A., Parra-Giraldo C.M., Rollins-Smith L.A., Reinert L.K., Vredenburg V.T., Amézquita A., Woodhams D.C. Microbiota and skin defense peptides may facilitate coexistence of two sympatric Andean frog species with a lethal pathogen. ISME J. 2019;13:361–373. doi: 10.1038/s41396-018-0284-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Assis A.B.d., Barreto C.C., Navas C.A. Skin microbiota in frogs from the Brazilian Atlantic Forest: Species, forest type, and potential against pathogens. PLoS One. 2017;12:e0179628. doi: 10.1371/journal.pone.0179628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faivovich J., et al. Phylogenetic relationships of the Boana pulchella group (Anura: Hylidae) Mol. Phylogenetics Evol. 2021;155:106981. doi: 10.1016/j.ympev.2020.106981. [DOI] [PubMed] [Google Scholar]
- 23.Rebouche C.J., Paulson D.J. Carnitine metabolism and function in humans. Annu. Rev. Nutr. 1986;6:41–66. doi: 10.1146/annurev.nu.06.070186.000353. [DOI] [PubMed] [Google Scholar]
- 24.Ramsay R.R., Zammit V.A. Carnitine acyltransferases and their influence on CoA pools in health and disease. Mol. Aspects Med. 2004;25:475–493. doi: 10.1016/j.mam.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Bremer J. Carnitine--metabolism and functions. Physiol. Rev. 1983;63:1420–1480. doi: 10.1152/physrev.1983.63.4.1420. [DOI] [PubMed] [Google Scholar]
- 26.Bonnefont J.-P., Djouadi F., Prip-Buus C., Gobin S., Munnich A., Bastin J. Carnitine palmitoyltransferases 1 and 2: biochemical, molecular and medical aspects. Mol. Aspects Med. 2004;25:495–520. doi: 10.1016/j.mam.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Lopes-Marques M., Delgado I.L.S., Ruivo R., Torres Y., Sainath S.B., Rocha E., Cunha I., Santos M.M., Castro L.F.C. The origin and diversity of Cpt1 genes in vertebrate species. PLoS One. 2015;10:e0138447. doi: 10.1371/journal.pone.0138447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meadows J.A., Wargo M.J. Carnitine in bacterial physiology and metabolism. Microbiology. 2015;161:1161–1174. doi: 10.1099/mic.0.000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meadows J.A., Wargo M.J. Transcriptional regulation of Carnitine catabolism in Pseudomonas aeruginosa by CdhR. mSphere. 2018;3:e00480-17. doi: 10.1128/mSphere.00480-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleber H.-P. Bacterial carnitine metabolism. FEMS Microbiol. Lett. 1997;147:1–9. doi: 10.1111/j.1574-6968.1997.tb10212.x. [DOI] [PubMed] [Google Scholar]
- 31.Wargo M.J., Hogan D.A. Identification of genes required for Pseudomonas aeruginosa carnitine catabolism. Microbiology. 2009;155:2411–2419. doi: 10.1099/mic.0.028787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunetti A.E., Fuzo C.A., Aguilar S., Rivera-Correa M., Marani M.M., Lopes N.P. The significance of hypervariability and conserved motifs in antimicrobial peptides from tree frogs. J. Nat. Prod. 2023;86:1761–1769. doi: 10.1021/acs.jnatprod.3c00224. [DOI] [PubMed] [Google Scholar]
- 33.Ellison S., Rovito S., Parra-Olea G., Vásquez-Almazán C., Flechas S.V., Bi K., Vredenburg V.T. The influence of habitat and phylogeny on the skin microbiome of amphibians in Guatemala and Mexico. Microb. Ecol. 2019;78:257–267. doi: 10.1007/s00248-018-1288-8. [DOI] [PubMed] [Google Scholar]
- 34.Loudon A.H., Kurtz A., Esposito E., Umile T.P., Minbiole K.P.C., Parfrey L.W., Sheafor B.A. Columbia spotted frogs (Rana luteiventris) have characteristic skin microbiota that may be shaped by cutaneous skin peptides and the environment. FEMS Microbiol. Ecol. 2020;96:fiaa168. doi: 10.1093/femsec/fiaa168. [DOI] [PubMed] [Google Scholar]
- 35.Woodhams D.C., Rollins-Smith L.A., Reinert L.K., Lam B.A., Harris R.N., Briggs C.J., Vredenburg V.T., Patel B.T., Caprioli R.M., Chaurand P., et al. Probiotics modulate a novel amphibian skin defense peptide that is antifungal and facilitates growth of antifungal bacteria. Microb. Ecol. 2020;79:192–202. doi: 10.1007/s00248-019-01385-9. [DOI] [PubMed] [Google Scholar]
- 36.Lazzaro B.P., Zasloff M., Rolff J. Antimicrobial peptides: Application informed by evolution. Science. 2020;368:eaau5480. doi: 10.1126/science.aau5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luengo J.M., Olivera E.R. Catabolism of biogenic amines in Pseudomonas species. Environ. Microbiol. 2020;22:1174–1192. doi: 10.1111/1462-2920.14912. [DOI] [PubMed] [Google Scholar]
- 38.Christian Martin H., Ibáñez R., Nothias L.-F., Boya P C.A., Reinert L.K., Rollins-Smith L.A., Dorrestein P.C., Gutiérrez M. Viscosin-like lipopeptides from frog skin bacteria inhibit Aspergillus fumigatus and Batrachochytrium dendrobatidis detected by imaging mass spectrometry and molecular networking. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-39583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taboada C., Brunetti A.E., Lyra M.L., Fitak R.R., Faigón Soverna A., Ron S.R., Lagorio M.G., Haddad C.F.B., Lopes N.P., Johnsen S., et al. Multiple origins of green coloration in frogs mediated by a novel biliverdin-binding serpin. Proc. Natl. Acad. Sci. USA. 2020;117:18574–18581. doi: 10.1073/pnas.2006771117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mashek D.G., Li L.O., Coleman R.A. Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. J. Lipid Res. 2006;47:2004–2010. doi: 10.1194/jlr.M600150-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Jarrar Y.B., Lee S.-J. Molecular functionality of cytochrome P450 4 (CYP4) genetic polymorphisms and their clinical implications. Int. J. Mol. Sci. 2019;20:4274. doi: 10.3390/ijms20174274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thumelin S., Esser V., Charvy D., Kolodziej M., Zammit V.A., McGarry D., Girard J., Pegorier J.P. Expression of liver carnitine palmitoyltransferase I and II genes during development in the rat. Biochem. J. 1994;300:583–587. doi: 10.1042/bj3000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dambrova M., Makrecka-Kuka M., Kuka J., Vilskersts R., Nordberg D., Attwood M.M., Smesny S., Sen Z.D., Guo A.C., Oler E., et al. Acylcarnitines: Nomenclature, biomarkers, therapeutic potential, drug targets, and clinical trials. Pharmacol. Rev. 2022;74:506–551. doi: 10.1124/pharmrev.121.000408. [DOI] [PubMed] [Google Scholar]
- 44.Aguer C., McCoin C.S., Knotts T.A., Thrush A.B., Ono-Moore K., McPherson R., Dent R., Hwang D.H., Adams S.H., Harper M.E. Acylcarnitines: potential implications for skeletal muscle insulin resistance. FASEB J. 2015;29:336–345. doi: 10.1096/fj.14-255901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodnick K.J., Sidell B.D. Cold acclimation increases carnitine palmitoyltransferase I activity in oxidative muscle of striped bass. Am. J. Physiol. 1994;266:R405–R412. doi: 10.1152/ajpregu.1994.266.2.R405. [DOI] [PubMed] [Google Scholar]
- 46.Voituron Y., Eugene M., Barré H. Survival and metabolic responses to freezing by the water frog (Rana ridibunda) J. Exp. Zool. Comp. Exp. Biol. 2003;299:118–126. doi: 10.1002/jez.a.10285. [DOI] [PubMed] [Google Scholar]
- 47.Lillywhite H.B. Water relations of tetrapod integument. J. Exp. Biol. 2006;209:202–226. doi: 10.1242/jeb.02007. [DOI] [PubMed] [Google Scholar]
- 48.Salama S.A., Arab H.H., Omar H.A., Gad H.S., Abd-Allah G.M., Maghrabi I.A., Al Robaian M.M. L-carnitine mitigates UVA-induced skin tissue injury in rats through downregulation of oxidative stress, p38/c-Fos signaling, and the proinflammatory cytokines. Chem. Biol. Interact. 2018;285:40–47. doi: 10.1016/j.cbi.2018.02.034. [DOI] [PubMed] [Google Scholar]
- 49.Fu T.-T., Sun Y.-B., Gao W., Long C.-B., Yang C.-H., Yang X.-W., Zhang Y., Lan X.-Q., Huang S., Jin J.-Q., et al. The highest-elevation frog provides insights into mechanisms and evolution of defenses against high UV radiation. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2212406119. e2212406119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woodhams D.C., Brandt H., Baumgartner S., Kielgast J., Küpfer E., Tobler U., Davis L.R., Schmidt B.R., Bel C., Hodel S., et al. Interacting symbionts and immunity in the amphibian skin mucosome predict disease risk and probiotic effectiveness. PLoS One. 2014;9:e96375. doi: 10.1371/journal.pone.0096375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Becker M.H., Walke J.B., Murrill L., Woodhams D.C., Reinert L.K., Rollins-Smith L.A., Burzynski E.A., Umile T.P., Minbiole K.P.C., Belden L.K. Phylogenetic distribution of symbiotic bacteria from Panamanian amphibians that inhibit growth of the lethal fungal pathogen Batrachochytrium dendrobatidis. Mol. Ecol. 2015;24:1628–1641. doi: 10.1111/mec.13135. [DOI] [PubMed] [Google Scholar]
- 52.Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen L.J., et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 54.Lechner M., Findeiß S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinf. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brunetti A.E., Hermida G.N., Iurman M.G., Faivovich J. Odorous secretions in anurans: Morphological and functional assessment of serous glands as a source of volatile compounds in the skin of the treefrog Hypsiboas pulchellus (Amphibia: Anura: Hylidae) J. Anat. 2016;228:430–442. doi: 10.1111/joa.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang M., Carver J.J., Phelan V.V., Sanchez L.M., Garg N., Peng Y., Nguyen D.D., Watrous J., Kapono C.A., Luzzatto-Knaan T., et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kessner D., Chambers M., Burke R., Agus D., Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pluskal T., Castillo S., Villar-Briones A., Orešič M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Myers O.D., Sumner S.J., Li S., Barnes S., Du X. Detailed investigation and comparison of the XCMS and MZmine 2 chromatogram construction and chromatographic peak detection methods for preprocessing mass spectrometry metabolomics data. Anal. Chem. 2017;89:8689–8695. doi: 10.1021/acs.analchem.7b01069. [DOI] [PubMed] [Google Scholar]
- 60.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 62.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amir A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Zech Xu Z., Kightley E.P., Thompson L.R., Hyde E.R., Gonzalez A., Knight R. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017;2:e00191-16. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price M.N., Dehal P.S., Arkin A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller J.H. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 67.Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 68.Bastard K., Smith A.A.T., Vergne-Vaxelaire C., Perret A., Zaparucha A., De Melo-Minardi R., Mariage A., Boutard M., Debard A., Lechaplais C., et al. Revealing the hidden functional diversity of an enzyme family. Nat. Chem. Biol. 2014;10:42–49. doi: 10.1038/nchembio.1387. [DOI] [PubMed] [Google Scholar]
- 69.Bazire P., Perchat N., Darii E., Lechaplais C., Salanoubat M., Perret A. Characterization of L-carnitine metabolism in Sinorhizobium meliloti. J. Bacteriol. 2019;201:e00772-18. doi: 10.1128/JB.00772-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Metabolomic data generated in this study are deposited at MassIVE: MSV0000924999
-
•
The sequences of the bacterial amplicon libraries and Pseudomonas spp. OTU sequences were deposited at NCBI-SRA PRJNA: 498895 and NCBI-GenBank: OR335663–OR335704, respectively.
-
•
The skin transcriptome of Boana prasina and the complete genome of Pseudomonas sp. MPFS, were available at NCBI-SRA PRJNA: 675859 and NCBI-GenBank: NZ_CP066123.1, respectively.
-
•
This study did not generate original codes. All data were analyzed following the protocols and instructions from the software, packages, and workflows available in the original publications or repositories.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.