Abstract
Objective
To assess the performance of convolutional neural networks (CNNs) for automated detection of keratoconus (KC) in standalone Scheimpflug-based dynamic corneal deformation videos.
Design
Retrospective cohort study.
Participants
We retrospectively analyzed datasets with records of 734 nonconsecutive, refractive surgery candidates, and patients with unilateral or bilateral KC.
Methods
We first developed a video preprocessing pipeline to translate dynamic corneal deformation videos into 3-dimensional pseudoimage representations and then trained a CNN to directly identify KC from pseudoimages. We calculated the model's KC probability score cut-off and evaluated the performance by subjective and objective accuracy metrics using 2 independent datasets.
Main Outcome Measures
Area under the receiver operating characteristics curve (AUC), accuracy, specificity, sensitivity, and KC probability score.
Results
The model accuracy on the test subset was 0.89 with AUC of 0.94. Based on the external validation dataset, the AUC and accuracy of the CNN model for detecting KC were 0.93 and 0.88, respectively.
Conclusions
Our deep learning-based approach was highly sensitive and specific in separating normal from keratoconic eyes using dynamic corneal deformation videos at levels that may prove useful in clinical practice.
Financial Disclosure(s)
Proprietary or commercial disclosure may be found in the Footnotes and Disclosures at the end of this article.
Keywords: Artificial intelligence, Deep learning, Convolutional neural network, Keratoconus, Scheimpflug-based dynamic corneal deformation videos
Keratoconus (KC) is a progressive ectatic disease characterized by corneal steepening and thinning due to deficits of structural integrity inducing irregular astigmatism and myopia with deterioration of visual acuity and vision-specific quality of life.1,2 Early diagnosis of KC is of foremost importance to halt disease progression.3 Corneal topography or tomography systems allow assessment of alteration of corneal shape and assist in KC classification and monitoring.4 However, these instruments cannot measure the mechanical stability of the cornea, which is thought to be the primary abnormality that triggers stromal thinning and steepening.5 Therefore, the early diagnosis of KC by analyzing corneal biomechanical properties has become a research hotspot aiming to measure the in vivo biomechanical properties of the cornea before the onset of topographic or tomographic changes.5,6 The Corvis ST (Oculus Optikgeräte GmbH) was introduced as a noncontact tonometer that records the ocular deformation response induced by a constantly defined air puff using an ultrahigh-speed Scheimpflug camera. The captured images are analyzed by the Corvis ST software to produce an estimate of intraocular pressure and several dynamic corneal response (DCR) parameters that provide a detailed assessment of corneal biomechanical properties. These parameters can be used to separate normal (N) from ectatic corneas.6, 7, 8, 9 A video function uses these images to release a slow-motion video of the corneal deformation reaction in response to the air pulse (4330 frames/second.). This highly precise video permits a viewable biomechanical analysis of the cornea.10
In this era of artificial intelligence, deep learning has greatly improved the diagnostic accuracy of some ocular diseases.11,12 Recently, deep learning has achieved high diagnostic performance in dry eye disease identification from ocular surface videos.13
In our study, we assessed the performance of deep convolutional neural networks (CNNs) for the automated detection of KC in Corvis ST videos as a complementary diagnostic tool to corneal topography and tomography data.
Patients and Methods
This study was conducted following ethical standards in the Declaration of Helsinki and its later amendments and was approved by the institutional review board of Federal University of São Paulo-UNIFESP/EPM (as the coordinator center) and the Hospital de Olhos-CRO, Guarulhos (as the affiliate center), and the Salouti Eye Center in Iran. Corresponding data use agreements were signed among contributing parties to use the data. We retrospectively analyzed datasets with records of 734 nonconsecutive, refractive surgery candidates, and patients with unilateral or bilateral KC, in these 2 centers located on 2 different continents which ensures subjects' ethnic variability in testing our approach capabilities to separate N from keratoconic corneas. Once required, each subject provided informed written consent to participate in the study before using their data, and data were de-identified in Brazil and Iran before any further processing. A total of 232 Subjects (131 N and 101 KC) were enrolled from the Hospital de Olhos-CRO, Guarulhos (Brazil-Dataset 1), and 502 Subjects (259 N and 243 KC) were enrolled from the Salouti Eye Center (Iran-Dataset 2).
All participants had a complete ophthalmic examination including Scheimpflug-based corneal tomography using the Pentacam HR and corneal biomechanical assessment using the Corvis ST (Oculus Opikgeräte GmbH).
The inclusion criterion for the KC population was the presence of clear signs of bilateral KC in corneal maps (derived from the Pentacam) such as Index of surface variance (deviation of the corneal radius with respect to the mean value) > 30, KC index (the ratio between the mean radius of curvature values in the upper and lower corneal segments) > 1.07, and minimum radius of curvature (the index that corresponds to the point of maximum anterior curvature) < 7.5. The topographical KC classification had to be at least stage 1 (topographical KC ≥ 1). The inclusion criteria for the N participants were the presence of Corvis ST examination and a Pentacam’s Belin-Ambrósio enhanced ectasia index total deviation < 1.6 standard deviation from normative values. Exclusion criteria were any previous ocular surgery or disease such as corneal collagen crosslinking or intracorneal ring segment implantation and myopia > 10.00 diopters. Moreover, to confirm the diagnosis of KC or N, all cases from each clinic were blindly reviewed by a third-party anterior segment expert (S.T.) to confirm inclusion criteria.
Measurements in each center were performed by the same experienced examiners, captured by automatic release to ensure user independence, and only examinations with good quality scores were included after a second manual, frame-by-frame analysis, made by an independent masked examiner to ensure the quality of each acquisition. For each participant, only 1 eye was randomly included in the analysis to avoid bias due to the relationship between bilateral eyes in the subsequent analysis.
DCR Parameters
The Corvis ST measures the corneal response to an applied predefined air impulse using an ultrahigh-speed Scheimpflug camera that captures 4330 frames per second for only 33 ms and eventually generates a video with 140 frames. Dynamic corneal response and related corneal thickness (pachymetric) parameters are calculated by analysis of the full process of the corneal dynamic response recorded. The stiffness parameter at the first applanation (SP A1)6 and 2 established indices including the Corvis biomechanical index (CBI)7 and the tomographic and biomechanical index (TBI)9 were used for comparative analysis. The biomechanically corrected intraocular pressure (bIOP) was used for correlations.
The stiffness parameter at the first applanation is defined as the resultant pressure divided by the deflection amplitude at the first applanation (A1). The resultant pressure is defined as the adjusted air pressure at A1 minus bIOP. The adjusted air pressure represents the load of the air pressure (calculated by converting the spatial and temporal velocity profiles of the air puff to pressure) impinging on the cornea at A1.6 Corvis biomechanical index includes several DCR parameters in addition to Ambrósio’s Relational Thickness to the horizontal profile.7 This combined index is calculated by logistic regression analysis where a final beta is transformed into a logistic sigmoid function.7 The TBI is generated by combining DCR and tomographic parameters using the leave-one-out crossvalidation technique implemented by a random forest classifier.9 The DCR and selected tomographic parameters were exported and linked to the related video and participants' demographic data. Each video was tagged by a serial number and the specific class (N or KC).
Corvis ST Video Frames Preprocessing
A total of 734 high-quality slow-motion video clips from 734 eyes showing the corneal deformation as a result of a constant air pulse were exported from the ST Corvis machines in both study centers. Recorded videos were sampled in audio video interleaved format at a resolution of 576 × 214 at 13 frames per second yielding a video length of 10 seconds. Each video is a synchronized release of the cross-sectional corneal image covering the horizontal 8.8 mm of the cornea. The video frames show the sequence of corneal shape change from convex to concave passing through the A1 state. When the air puff reaches its maximum the cornea is at its highest concavity. When the air puff is switched off the cornea returns to its original shape passing through the second applanation state. When the cornea finally returns to its natural convex shape, the video frame capturing ends. During the recording process, a blue light-emitting diode (LED) light (470 nm wavelength, ultraviolet free) illuminates the area, and the dispersed light from the cornea is recorded.7
Preprocessing Steps
The following fully automated video frame processing steps were performed using the OpenCV library (version 4.5.4, http://opencv.org).
-
1-
The Corvis ST videos display other features not related to the cornea as light is also dispersed from other anterior segment structures. This results in various background random shadows in addition to the printed timecode in ms and the Oculus Opikgeräte GmbH logo. Supplementary videos N1 and KC1 represent 2 examples of raw Corvis ST videos. For consistency, the same sampled videos will be used to show successive preprocessing steps afterward. All these background features are regarded as noise in our preprocessing and were removed in the first preprocessing step by extracting the cornea foreground image; this step produced abstract corneal sectional frames over a black smooth background (See supplementary video N2 and KC2).
-
2-
Replacement of corneal stromal image heterogeneity (produced by light dispersion by corneal stroma) by smooth white mask and binarizing each frame (See supplementary video N3 and KC3).
-
3-
All frames are registered, guided by the position of the peripheral segments of the corneal mask in each frame. Therefore, overlapping the successive corneal masks can convey information about pure deflection of the cornea, being insensitive to shifts or rotation of the entire eyeball caused by the air puff.
-
4-
This is followed by the process of skeletonization of the foreground corneal white mask. Skeletonization is the process of inducing morphological thinning that successively erodes away pixels from the boundary (while preserving the connectivity of the endpoints of the corneal periphery) until no more thinning is possible.14 This process effectively produces a compact representation of the corneal shape that preserves many of the topological characteristics of the deformation process. For simplicity, this curve will be referred to as the "corneal skeleton" afterward (See supplementary videos N4 and KC4). Figure S1 depicts all these preprocessing steps.
-
5-
The vertical (y-axis) shift in the position of each pixel along the corneal skeleton with reference to the position of the corresponding pixel located along a horizontal reference plane (corneal skeleton or horizontal line) is calculated for each frame (simple subtraction). This process was repeated in pixel-wise steps for each pixel along the corneal skeleton for 225 pixels on each side of the corneal apex position for each frame. This step yielded a 2-dimensional (2D) numerical (integers) array for each video. Each array has 140 horizontal lines (rows) and 450 vertical lines (columns). Each row describes vertical positional differences (along the y-axis) of each pixel along the corneal skeleton in a single frame compared with a reference horizontal plane (reference corneal skeleton or horizontal line). Each column is related to a specific pixel position along the x-axis of the corneal skeleton (225 pixels nasal and 225 pixels temporal to the position of the pixel at the corneal apex in the first frame).
-
6-
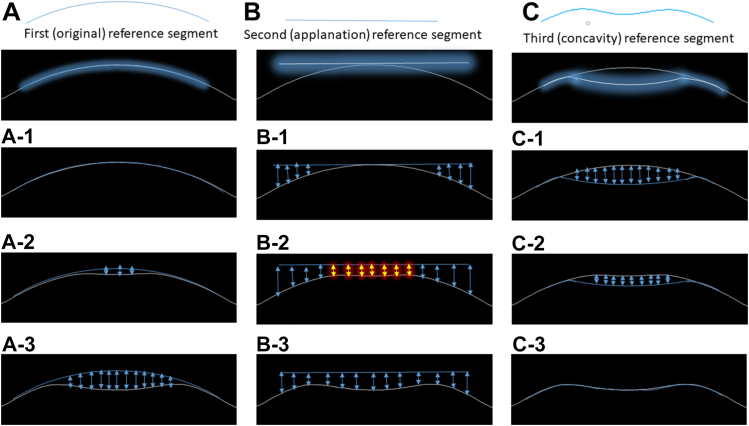
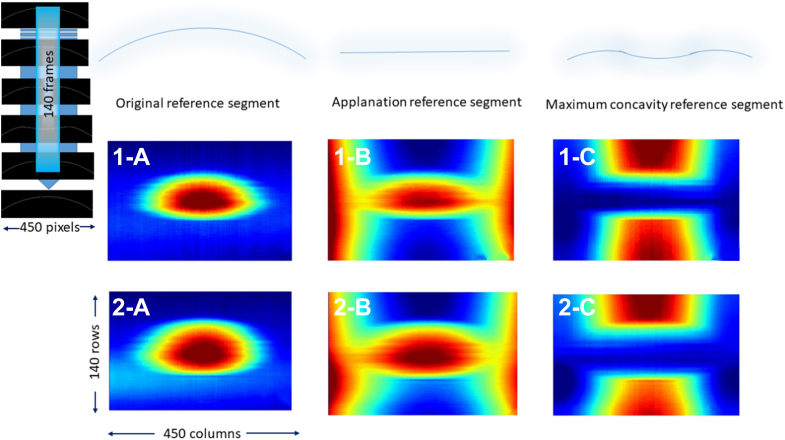
We developed 3 different types of 2D arrays for each video by comparing the position of each pixel in the corneal skeleton in each frame to a reference corneal skeleton or a horizontal line. Each 2D array type highlights a specific corneal “landmark” position and relatively penalizes the other positions according to the shape of the reference segment (plane). The first reference segment is a curve overlapping the original corneal unaltered shape (original 2D array). The second reference segment is a horizontal tangent to the position of the corneal apex at the original unaltered shape (applanation 2D array). The third reference segment is an arc overlapping the position of the cornea at maximum concavity shape (concavity 2D array). Figure 2 shows the process of calculation of each 2D array type. Figure 3 shows the 3 types of arrays extracted from supplementary video N4 and KC4 as heatmaps to facilitate visualization of 2D array numeric elements.
-
7-
The 3 different types of 2D arrays related to the same video were concatenated to get a 3-dimensional (3D) array (3-channel pseudoimage). Figure S4 depicts examples of the 3D arrays generated by concatenating 3 2D arrays for visualization (sourced from corresponding arrays shown in Figure 3). For simplicity, these 3D arrays will be referred to as "pseudoimages" afterward.
-
8-
Each pseudoimage was labeled according to the class of the corresponding source video. Pseudoimages corresponding to Dataset 2 were randomly split into training/validation (70%) and testing (30%) subsets. Pseudoimages corresponding to Dataset 1 were used for external validation of the model performance (external validation subset).
Figure 2.
Flow chart of the method of calculation of distance between each pixel on the corneal skeleton relative to the corresponding pixel in the reference segment. This process is repeated for each extracted corneal skeleton to get 140 numerical rows representing 140 video frames. A–C, represent 3 reference segments used and (1, 2, 3) represent samples of the corneal skeleton at its original position, applanation, and maximum concavity consecutively. The arrows represent the distance calculated between each reference segment and the corneal skeleton at the sampled corneal position (in pixels). The highlighted arrows in B-2 are expected to represent similar values during applanation.
Figure 3.
The 3 types of extracted 2-dimensional numerical arrays are represented as heatmaps to facilitate visualization. 1-a, 1-b, and 1-c represent the original reference segment map, the applanation reference segment map, and the maximum concavity reference segment map consecutively. These arrays were extracted from supplementary video N4. 2-a, 2-b, and 2-c represent the original reference segment map, the applanation reference segment map, and the maximum concavity reference segment map consecutively. These arrays were extracted from supplementary video KC4. Hot colors represent larger values (distance in pixels), while cool colors represent smaller values.
Classification and Validation
Dataset Final Preparation
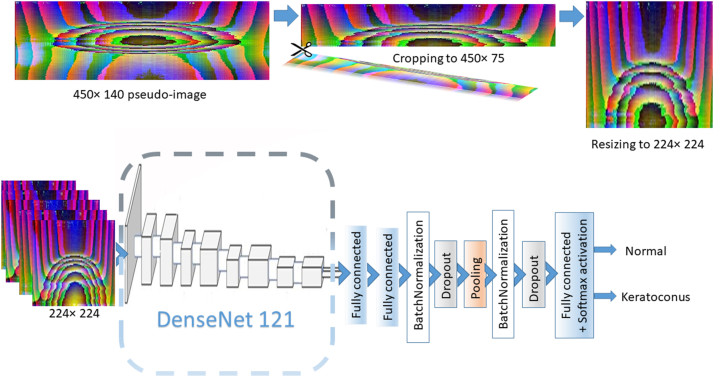
Although each of the component 2D of these pseudoimages represents a summary of corneal deformation throughout the whole deformation process, we opted to use a slice of these pseudoimages representing changes in the first 75 frames only (450 pixels width × 75 pixels height). The first 75 frames describe apex displacement from the undeformed state well beyond the A1 and are closely related to the SP A1 parameter. The SP A1 index is a strong indicator of the corneal biomechanical properties and has been proven to be useful in screening for KC with high sensitivity and specificity and is not influenced by scleral properties.6 Each slice was normalized and resized to 224 × 224 pseudoimage. This slicing process was implemented in training/validation, testing, and external validation subsets.
Deep CNN Architecture
To solve this binary classification problem, we trained a DenseNet121-based CNN architecture from scratch.15 The model was adapted to be compatible with input pseudoimages size of 224 × 224 and the output (classifying) layer was truncated and replaced by a light custom-designed model on top with Softmax activation to provide the likelihood of 2 classes of KC and N. The model's architecture is shown in Figure 5.
Figure 5.
Schematic diagram showing the structure of the employed custom network created on top of DenseNet121 model after truncation of the last classifying layer: First and second fully connected layers (1024 and 512 nodes respectively); Batch Normalization layer; Dropout (0.5), GlobalAveragePooling2D; Batch Normalization; Dropout (0.5); final classifying fully connected layer (2 nodes with Softmax activation). Pseudoimages extracted from videos were cropped to represent the early 75 video frames and then resized to 224 × 224 before being fed into the model.
Data Augmentation and Model Training
To minimize the risk of overfitting, all training/validation pseudoimage samples were augmented by in-place (on-the-fly) traditional data augmentation during training time with care to avoid corrupting the spatial and temporal relationships between array elements.16 This included minimal width shift, shear, and zoom. The model was trained for 600 epochs to assure improved generalizability (an epoch is an iteration over the entire input data provided). Categorical crossentropy was used as the loss function.17 The optimization was performed using stochastic gradient descent optimizer.18
Models Testing
After training, the model performance was assessed on the test subset. We used several objective metrics including accuracy, recall, precision, specificity, F1 score, confusion matrix, receiver operating characteristic curve (ROC), and area under the curve (AUC) to evaluate model.19 Model classification probability score for the KC class (where 0.0 represent the maximum probability of the N class and 1.0 represent the maximum probability for the KC class) was proposed as a possible index for KC detection after defining the optimal cut-off value using the Youden index. This index will be referred to as the "KC probability score" afterward. To get the finalized model the whole Dataset 2 pseudoimage slices (all available training and testing subsets) were used to repeat model training using the same parameters aiming to boost model performance on external validation (Dataset 1) by the added value of training on all available pseudoimage slices in Dataset 2.
Model Performance Benchmarking
To obtain benchmark performance metrics that allow comparison of the network performance in KC detection, we used SP A1 parameter, CBI, and TBI indices individually from Dataset 2 for training 3 Naive Bayes Classifiers (NBCs) to solve this binary classification problem.20 The corresponding parameters in Dataset 1 were used for validation of the classification performance of NBCs. Similarly, the performance of the finalized trained CNN model on pseudoimage slices in Dataset 1 was assessed and compared to the NBCs performance using confusion matrix, ROC curve, AUC, and detection error tradeoff curve.19 The detection error tradeoff curve allows for easier visual assessment of the overall performance of classification algorithms compared with ROC curves, without the need to magnify the top left corner of the ROC plots when models have high-AUCs. Also, it facilitates operating point analysis for extraction of the optimum point at which the false-negative error rate will improve. A pairwise comparison of the ROC curve of our algorithm with the ROC curves of the SP A1 parameter, CBI, and TBI indices was performed using the method described by DeLong et al.21
Correlations
To investigate the association between parameters, a pairwise correlation of age, central corneal thickness (CCT), bIOP, SP A1, CBI, TBI, and KC probability scores were evaluated using the Spearman rank correlation coefficient.
Class Activation Maps
Class activation maps (CAMs) highlight the regions that were more important for the CNN to identify a particular class, enabling a visual inspection of the model decision basis.22 We implemented a global average pooling (shape 7 × 7) before the final Softmax activation to produce the desired CAMs by projecting back the weights of the output layer and multiplying each feature map spatial average (calculated by global average pooling) with the corresponding weight to obtain the CAMs. Although the adopted preprocessing approach generated concatenated 3D numeric arrays, these pseudoimages can still convey average information about the most important frame sequence and area of the horizontal corneal section that guided the model decision. According to our preprocessing, pseudoimage slicing, and resizing, as every CAM has 7 × 7 component squares, each component square represents 64.3 (450/7) pixels of the registered corneal skeleton frames in the horizontal meridian (spatial dimension) and 10.7 (75/7) consecutive video frames vertically (temporal dimension).
Statistical Analysis
All statistical analyses were performed using Scipy (Scientific Computing tools for Python, version 1.8.1)23 and scikit-learn libraries of Python (version 1.1.1.).24 Scikit-learn is a Python module for machine learning built on top of Scipy. Normally distributed data were presented as the mean ± standard deviation. The probability distribution of each parameter was checked using the Kolmogorov–Smirnov 1-sample test for goodness of fit.
Python programming language (version 3.9.13) was used for developing models. Keras open-source software library (version 2.7.0) was used as an interface to the TensorFlow library (version 2.7.0).
Sample size calculation was done using NumPy (Numerical Python) which is the core scientific computing library in Python. We implemented NumPy t test for 2 independent samples. A sample size of ≥ 223 subjects was necessary for each group (effect size = 0.25, alpha error = 0.05, power = 0.75).
The independent samples t test was used to analyze parameters with normal distribution, while the Wilcoxon rank-sum test was used for nonparametric data to determine whether the data were significantly different between groups. For all analyses, P ≤ 0.05 was considered statistically significant. The NBC was selected as a widely used machine learning algorithm to address this binary classification problem for SP A1, CBI, and TBI as this model is simple yet effective in dealing with different types of input data. The simplicity of NBC can effectively minimize the risk of overfitting on relatively small datasets (as is the case in our external validation subset). In addition, as NBC is quite simple, it typically converges quicker than discriminative models like logistic regression.20
Area under the curve was calculated to validate the probability score estimate using a Python-based fast implementation of DeLong's algorithm and was also used for computing the statistical significance of comparing 2 AUCs.21 The optimum cutoff point for the probability index was calculated using Youden’s J statistic. J lies within a range from −1 to +1. A diagnostic test can be considered to yield reasonable results for positive values of J. Higher values indicate better performance of the diagnostic test. The pairwise correlation was calculated using the Spearman rank correlation coefficient. Deep-learning computations were performed on a single graphics processing unit.
Results
A total of 734 eyes (358 right eyes and 376 left eyes) of 734 patients with KC and normal participants were included. Table 1 shows the baseline characteristics of the study participants. Considering the KC population in Datasets 1 and 2, there was a statistically significant difference between mean keratometry, maximal keratometry, CCT, Belin-Ambrósio enhanced ectasia total deviation index, and SP A1 values. This difference ensures the presence of sufficient variability in the external validation subset (Dataset 1) for a better assessment of model robustness in extracting characteristic features from unseen data.
Table 1.
Demographic Characteristics of the Study Groups
| Parameter | Dataset 1 |
Dataset 2 |
P∗ | P† | ||||
|---|---|---|---|---|---|---|---|---|
| Normal | Keratoconus | P | Normal | Keratoconus | P | |||
| N | 101 | 131 | N.A. | 259 | 243 | N.A. | ||
| Race, n | N.A. | N.A | ||||||
| White | 58 | 64 | 240 | 225 | ||||
| Mixed | 22 | 29 | - | - | ||||
| African | 6 | 18 | - | 2 | ||||
| Asian | - | 2 | 2 | 4 | ||||
| Unreported | 15 | 18 | 17 | 12 | ||||
| Eye (right/left) | 45/56 | 59/72 | 0.941 | 123/136 | 131/112 | 0.151 | 0.616 | 0.102 |
| Gender (male/female) | 56/45 | 52/79 | 0.017 | 102/157 | 124/119 | 0.009 | 0.006 | 0.036 |
| Age (years) | 0.096 | 0.075 | 0.102 | 0.090 | ||||
| Median | 28 | 27 | 29 | 28 | ||||
| IQR | 8.0 | 9.5 | 9.0 | 8.0 | ||||
| Range | 19, 49 | 18, 52 | 22, 52 | 19, 50 | ||||
| Km (D) | <0.001 | <0.001 | 0.063 | <0.001 | ||||
| Median | 43.2 | 48.5 | 43.6 | 49.6 | ||||
| IQR | 2.08 | 3.44 | 1.84 | 5.35 | ||||
| Range | 41.2, 47.2 | 44.2, 55.5 | 40.7, 47.7 | 45.9, 54.2 | ||||
| Kmax(D) | <0.001 | <0.001 | 0.082 | 0.035 | ||||
| Median | 44.8 | 51.7 | 44.6 | 50.6 | ||||
| IQR | 3.06 | 5.80 | 2.98 | 6.70 | ||||
| Range | 40.8, 47.2 | 46.2, 58.4 | 40.2, 46.8 | 54.3, 45.6 | ||||
| IOP (mmHg) | <0.001 | 0.002 | <0.001 | 0.025 | ||||
| Median | 17 | 14 | 14 | 15 | ||||
| IQR | 3.50 | 4.50 | 3.25 | 4.42 | ||||
| Range | 13, 22 | 10, 19 | 11, 22 | 9, 23 | ||||
| CCT (μm) | <0.001 | <0.001 | <0.001 | <0.001 | ||||
| Median | 518 | 453 | 529 | 465 | ||||
| IQR | 37.00 | 46.75 | 32.25 | 58.32 | ||||
| Range | 464, 602 | 378, 531 | 457, 593 | 390, 494 | ||||
| BAD-D | <0.001 | <0.001 | 0.014 | <0.001 | ||||
| Median | 0.65 | 7.80 | 0.55 | 6.18 | ||||
| IQR | 0.58 | 3.02 | 0.42 | 3.55 | ||||
| Range | −0.24, 1.45 | 1.78, 13.08 | −0.11, 1.51 | 1.66, 12.33 | ||||
| SP A1 | <0.001 | <0.001 | 0.003 | < 0.001 | ||||
| Median | 111.00 | 72.77 | 113.09 | 63.57 | ||||
| IQR | 20.15 | 22.17 | 19.28 | 27.12 | ||||
| Range | 91.14, 140.76 | 25.16, 112.36 | 87.33, 152.76 | 28.14, 107.33 | ||||
| CBI | <0.001 | <0.001 | 0.16 | 0.582 | ||||
| Mean ± SD | 0.05 ± 0.03 | 0.95 ± 0.03 | 0.09 ± 0.07 | 0.92 ± 0.06 | ||||
| Range | 0.00, 0.51 | 0.42, 1.00 | 0.00, 0.62 | 0.44, 1.0 | ||||
| TBI | <0.001 | <0.001 | 0.454 | 0.622 | ||||
| Mean ± SD | 0.09 ± 0.07 | 0.99 ± 0.00 | 0.07 ± 0.05 | 0.99 ± 0.01 | ||||
| Range | 0.00, 0.35 | 0.96, 1.0 | 0.00, 0.45 | 0.85, 1.0 | ||||
BAD-D = Belin-Ambrósio enhanced ectasia total deviation index; CBI = Corvis biomechanical index; CCT = central corneal thickness; D = diopter; IOP = intraocular pressure; IQR = interquartile range after removing outliers; Km = mean keratometry; Kmax = maximal keratometry; N = number of subjects; N.A. = not applicable; SD = standard deviation; SP A1 = stiffness parameter at first applanation; TBI = tomography and biomechanical index.
The bold type signifies P ≤ 0.05.
Between Dataset 1 normal and Dataset 2 normal subgroup.
Between Dataset 1 keratoconus and Dataset 2 keratoconus subgroup.
The mean time needed for individual video preprocessing (including cleaning) to develop each type of 2D array was 44.32 ± 1.6, 44.80 ± 1.3, and 44.59 ± 2.1 seconds for the first (original), second (applanation), and third (concavity) arrays respectively. This time can be reduced by preprocessing the first 75 frames used in the studied pseudoimage slice to 23.61 ± 0.9 and 23.31 ± 0.2 seconds for the first and second array, respectively. As the calculation of the third array mandates the identification of the first frame featuring the cornea at maximum concavity, this whole video preprocessing time can be reduced by preprocessing the first 95 frames to 29.95 ± 1.2. Inference time per pseudoimage slice for the adopted model was 47.2 ± 3.3 ms in our single graphics processing unit.
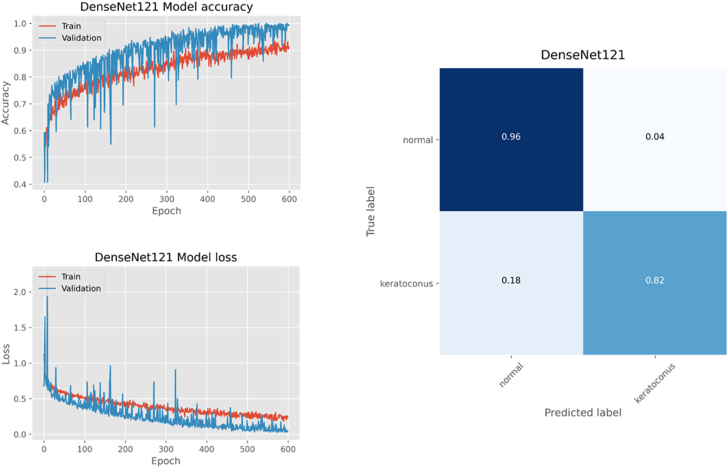
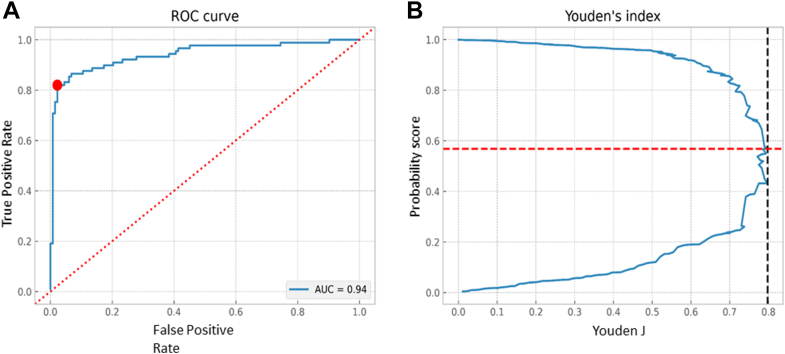
The training progress and confusion matrix for model performance on the test subset is shown in Figure 6. The model accuracy on the test subset was 0.89. The ROC curve analysis showed an AUC of 0.94 and the Youden index was highest with 0.86 at the threshold of 0.585 which yielded the best separation between N (< 0.585) from KC (> 0.585) eyes (Figure 7). At this threshold, the sensitivity, specificity, precision, and F1 scores were 0.96, 0.82, 0.84, and 0.90 for the N group, respectively, and 0.82, 0.96, 0.95, and 0.88 for the KC group, respectively.
Figure 6.
Epoch accuracy/loss during model training/validation (Dataset 2 training/validation subset with data augmentation). The confusion matrix shows the performance of the trained model on the test subset (Dataset 2 test subset).
Figure 7.
A, Receiver operating characteristic (ROC) curve for binary (keratoconus vs. normal) classification task by the trained model on the test subset (Dataset 2 test subset) with an area under the curve (AUC) of 0.942. The ROC curve is marked by a red dot at the site closest to the perfect classification point. B, Plot showing probability score at the cut-off point and Youden Index.
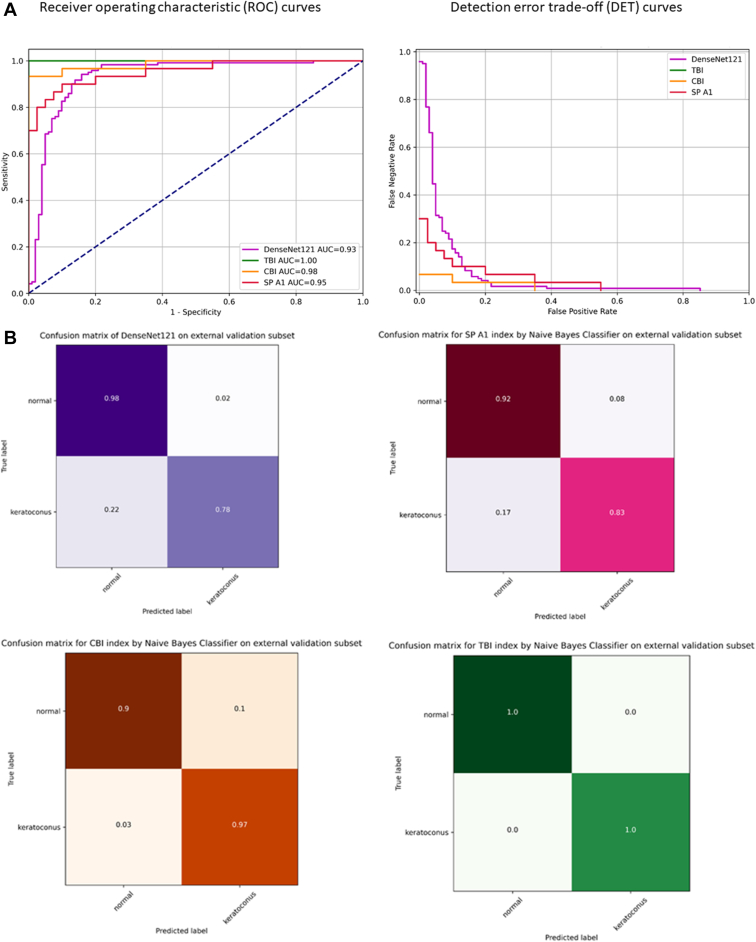
Tables 2 and 3 show the predictive accuracy and pairwise AUC comparisons of SP A1, CBI, and TBI NBC(s) and the adopted model using the external validation subset (Dataset 1), performed using Delong's method. Figure 8A, B show the ROC and detection error tradeoff curves and confusion matrices of the NBC(s) compared to model performance.
Table 2.
External Validation (Dataset 1) Classification Accuracy, Precision, Recall (sensitivity), F1 Score, and Receiver Operating Characteristic Analysis with the AUC for the SP A1 Parameter, CBI, and TBI Using Naive Bayes Classifier and for the Processed Pseudoimage Slices Using the Adopted DenseNet 121 Model
| Parameter | Class | Recall | Specificity | Precision | F1 Score | Accuracy | Mean AUC ± SE |
|---|---|---|---|---|---|---|---|
| DenseNet121 | Normal | 0.98 | 0.78 | 0.82 | 0.89 | 0.88 | 0.934 ± 0.026 |
| Keratoconus | 0.78 | 0.98 | 0.98 | 0.87 | |||
| SP A1 | Normal | 0.92 | 0.83 | 0.84 | 0.88 | 0.88 | 0.954 ± 0.020 |
| Keratoconus | 0.83 | 0.92 | 0.91 | 0.87 | |||
| CBI | Normal | 0.90 | 0.97 | 0.97 | 0.93 | 0.94 | 0.982 ± 0.010 |
| Keratoconus | 0.97 | 0.90 | 0.91 | 0.94 | |||
| TBI | Normal | 1.0 | 1.0 | 1.0 | 1.0 | 1.00 | 1.000 ± 0.000 |
| Keratoconus | 1.0 | 1.0 | 1.0 | 1.0 |
AUC = area under the receiver operating characteristic curve; CBI = Corvis biomechanical index; SE = standard error; SP A1 = stiffness parameter at first applanation; TBI = tomography and biomechanical index.
Table 3.
External Validation (Dataset 1) Receiver Operating Characteristic Curves and AUC Analysis for the SP A1 Parameter, CBI, and TBI Using Naive Bayes Classifier and for the Processed Pseudoimages Using DenseNet 121
| Parameter (Pairwise Comparison) | Mean Δ AUC ± SE | 95% CI | Z Statistic | P Value |
|---|---|---|---|---|
| SP A1 and DenseNet121 | 0.020 ± 0.024 | 0.0432, 0.0832 | 0.83 | 0.198 |
| CBI and DenseNet121 | 0.048 ± 0.021 | 0.0020, 0.0980 | 2.28 | 0.013 |
| TBI and DenseNet121 | 0.066 ± 0.026 | 0.0211, 0.1108 | 2.53 | 0.006 |
| SP A1 and CBI | 0.028 ± 0.013 | 0.0130, 0.0690 | 2.15 | 0.016 |
| SP A1 and TBI | 0.046 ± 0.020 | 0.0115, 0.0804 | 2.30 | 0.009 |
| CBI and TBI | 0.018 ± 0.010 | 0.0007, 0.0352 | 2.00 | 0.021 |
Δ AUC = difference between the area under the curve; CBI = Corvis biomechanical index; CI = confidence interval; ROC = receiver operating characteristic; SE = standard error; SP A1 = stiffness parameter at first applanation; TBI = tomography and biomechanical index.
The bold type signifies P ≤ 0.05.
Figure 8.
A, B, Receiver operating characteristic (ROC) curves, detection error trade-off curves, and confusion matrices for binary (keratoconus vs. normal) classification task of the external validation subset (Dataset 1) by 3 Naive Bayes classifiers, trained on Dataset 2 stiffness parameter at first applanation (SP A1), Corvis biomechanical index (CBI), and the tomographic and biomechanical index (TBI), compared to the performance of the trained adopted DenseNet121 based model on the cropped resized external validation dataset (Dataset 1) pseudoimages.
Youden index was highest with 0.810 at the threshold of 0.521 with a cut-off value of 0.521 to discriminate N (< 0.521) from KC (> 0.521) which yielded the best separation between N and KC cases in the external validation subset. The TBI had the highest predictive accuracy. The performance of the model was comparable to SP A1 NBC with no statistically significant difference in AUC comparison.
Figure S9 shows the Spearman rank correlation analysis of SP A1, CBI, TBI, and KC probability score with age, CCT, and bIOP. The KC probability score had a significant correlation with CCT, SP A1, CBI, and TBI in the KC group. In the N group, the correlation of the KC probability score with CCT and TBI was not significant. The linear relationship of the KC probability score with TBI and CCT in KC cases compared to a nonlinear relationship in N cases may reflect the bidirectional noise inherent to the probability score of the N group. In addition, the relation between corneal thickness and the biomechanical properties of the cornea is not essentially linear in the N group, especially considering the wider range of CCT (464–602 μm) in the N population in Dataset 1. The TBI was the only index that was independent of the bIOP in both the N and KC groups.
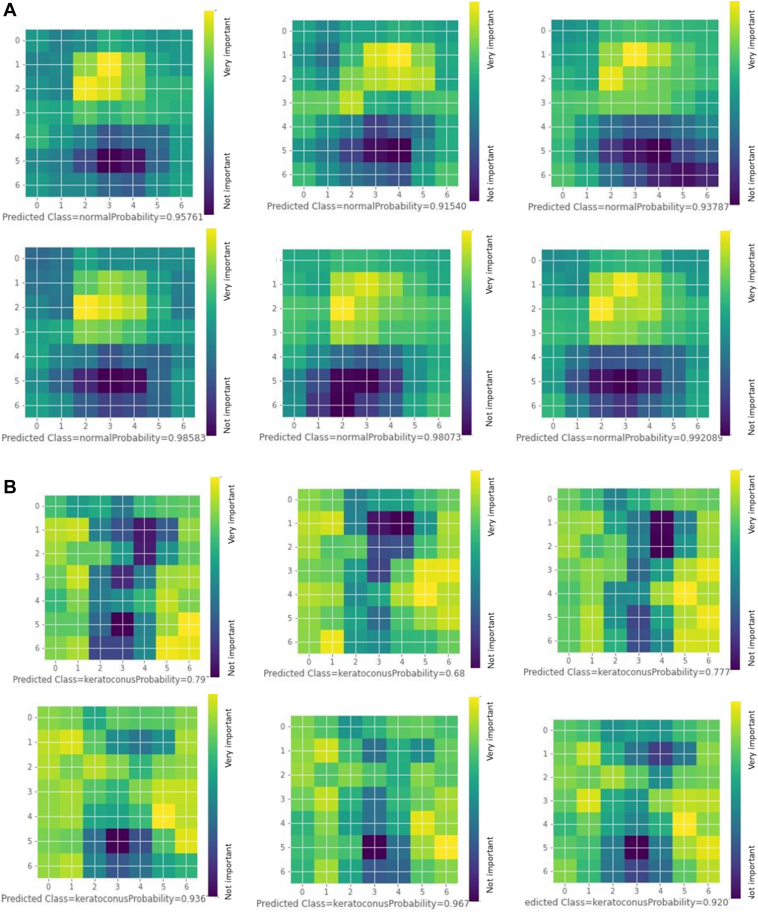
In Figure 10A, we present 6 examples of CAMs generated by the adopted model on test subset samples that were correctly classified as N. The CAMs appear to carry plausible spatial clinical inference and overall close similarity. Horizontally, the most important inferential regions were limited to the central and paracentral cornea (the 3 central squares roughly correspond to the central 3.5 mm of the scanned cornea according to preprocessing). Vertically, the highlighted areas represent the earlier frames at the time of A1 (second–fourth squares roughly correspond to the 11th–43rd frames) with lesser weights assigned to frames afterward. Accordingly, we may infer that the most important parameter that guided the model for high N class probability was the regular occurrence of the A1 in an expected morphological and chronological pattern. Figure 10B represents 6 examples of CAMs generated by the adopted model on test subset samples that were correctly classified as KC. The model appears to use more peripheral corneal regions compared to those used for classifying the N group (outside the 3 central squares), with the corneal apex persistently appearing as a cold spot, especially in latter frames. Vertically, there was variable activation along most frames without clear relevance of certain frame sequences for classification in contrast to the N group.
Figure 10.
A, Selected examples of normal group class activation maps (CAMs) showing the areas of the cornea and frame sequence that are most important for the model classification decision. All were correctly predicted with prediction probability entitled. B, Selected examples of keratoconus group CAMs showing the areas of the cornea and frame sequence that are most important for the model classification decision. All were correctly predicted with prediction probability entitled.
Discussion
Noncontact ultrahigh-speed corneal imaging with a Scheimpflug camera has allowed a new perspective for the analysis of the corneal deformation process using advanced image processing techniques10 that can capture changes in tangent modulus values of the cornea reported to occur in softer ectatic corneas.25, 26 These changes can be identified by emerging deep learning approaches to discriminate between N and KC eyes. To the best of our knowledge, this is the first study that has used this technique for the detection of KC in Corvis ST corneal deformation videos. In this study, we attempted to use deep learning to design a complementary KC probability index using only Corvis ST videos without using corneal topography or tomography data for the detection of KC.
Our preprocessing pipeline could achieve dimensionality reduction of video data to 2D space which would offer a great chance for implementing various machine learning algorithms to analyze and extract more numeric features that can be used as summarized novel biomechanical indices for KC diagnosis. Another study is already in progress to evaluate this possibility.
The use of the Belin-Ambrósio enhanced ectasia total deviation index as an inclusion criterion revealed the potentially similar robust sensitivity and specificity profiles of this metric and the TBI as previously demonstrated by other studies.9,27, 28, 29
Following the training and testing of the adopted model on Dataset 2, its diagnostic capability to discriminate between N and KC eyes was assessed on the external validation subset (Dataset 1) to avoid overfitting and to reassess the optimum cut-off value of the KC probability score in a different dataset. The model showed high sensitivity and specificity with an AUC of 0.93 in the external validation subset, suggesting a reasonable generalization to unseen videos and confirming the diagnostic capability of our model.
In the external validation dataset, NBCs based on CBI and TBI outperformed our CNN. This could be explained by the fact that several DCR parameters besides the pachymetric parameter Ambrósio’s Relational Thickness, contribute to CBI calculation, and additional tomographic parameters are involved for TBI derivation. Meanwhile, our model is based on only numeric monitoring of the corneal dynamic response extracted from a limited number of frames (75 frames per slice).7, 8, 9,27, 28, 29, 30 Nevertheless, SP A1 was the parameter with the closest matching NBC performance to the adopted network. This was not surprising as the development of the SP A1 involves the calculation of the time and position of the A1.6 These parameters are expected to be well represented in the calculated 3D array slices used in model training and were featured in the model CAMs.
The significant correlation of the KC probability score with the SP A1, CBI, and TBI in the KC group is interesting as these parameters are not directly calculated from deformation response imaging as in our approach but involve the use of numerical simulations and machine learning algorithms.6,7,9 So, this association highlights the validity of our simpler but consistent approach and supports the possible correlation of this score with the severity of KC. It also suggests possible validity to diagnose patients with subclinical KC (sub-tomographic ectasia) similar to other DCR parameters, which is a subject of another follow-up progressing study.31,32 The comparative analysis of the CCT with the bIOP of the N group showed no significant correlation, demonstrating the established ability of the bIOP correction algorithm to compensate for corneal thickness as an important confounding factor.33
In our approach, we opted to use pseudoimage slices rather than the whole 3D array to allow comparison with the SP A1. Stiffness parameter at the first applanation is considered the single DCR parameter of the highest sensitivity and specificity to diagnose KC compared with any other DCR parameter value6 (in contrast to CBI and TBI, which are summary indices derived based on several other parameters). This parameter is calculated in relation to the time and position of the A1, which should be featured in the selected part of the 3D array.7,9,28,32 In addition, limiting the number of frames was required in preprocessing to shorten model inference time, making our approach more practical for clinical application. Operating a high-performance graphics processing unit or a multithreaded central processing unit may further reduce this time to allow near real-time video classification during recording by integrating this model in future Corvis ST machines.
Our preprocessing technique encompassed many basic steps needed in video classification architecture. It allowed data conversion (video to frames), enhancing/smoothing, noise removal, image segmentation, and feature extraction, paving the way for feature matching and feature classification by the adopted CNN model. This problem-solver approach was designed specifically for Corvis ST videos, exploiting the consistency of the video time steps and the temporal/spatial profile of the air puff.6 Herber et al31 used linear discriminant analysis and random forest algorithms to develop classification and staging models for KC using DCR and corneal thickness-related (pachymetric) parameters calculated by Corvis ST. Our approach cannot be directly compared with this study as we have utilized raw Corvis ST videos without using established DCR parameters, corneal topography, or tomography data.
Recently, Tan et al33 used corneal contour data points extracted from each Corvis ST video frame to calculate the CCT, the time to the A1, corneal radius of curvature at the highest concavity, and distance between the initial position of the corneal apex and the nadir at the highest concavity. Then, the calculated parameters were used to train a feed-forward neural network for distinguishing N and KC eyes. Compared to our studied population, they used different inclusion criteria and included coethnic (Chinese) participants of a proportionally lesser sample size that included some bilateral eyes. Additionally, other than age, no other characteristics of the participants were provided, making the generalization of their findings challenging. Our approach differs technically from this study. We used a CNN model to exploit the extracted 3D images directly without extracting any single parameter that may bias the analysis while Tan et al extracted 4 parameters and then used simple neural network models to identify KC.
In this study, CAMs were useful to provide insight into discriminative pseudoimage regions used by the model. It was observed that the model consistently used clinically meaningful corneal regions and video frames related to the time and position of the A1 for identifying the N class. However, there was not a clear activation pattern that can highlight the frame sequence used for KC classification. This pattern loss may represent a by-product of data augmentation techniques used during model training, resulting in model confusion and lower sensitivity for the KC group which can be improved by avoiding this technique in case of availability of larger datasets.11,13
Our study had several limitations too. We did not assess the diagnostic potential of using different slices or the whole of 3D arrays or the component 2D arrays. The preprocessing ignored the possible use of available corneal thickness data in the imaged horizontal profile. However, we primarily intended to propose a purely biomechanical method to detect KC. Adding extra tomographic parameters may improve detection scores and accuracy but goes beyond the scope and goals of this work.
In addition, we did not analyze corneal vibrations imaged during the deformation process. The characteristic changes in corneal vibration were analyzed in other studies using dedicated image processing methods to develop characteristic parameters for early detection of KC.34
Another limitation is using traditional image data augmentation techniques that may introduce biases due to difficulty in identifying safe, label-preserving transformations in this pseudoimage domain that may have constituted a source of model confusion, especially considering the rigorous arrangement of the 3D array elements. This may be improved by adhering to data-specific augmentation at a distortion magnitude well below label-changing levels.35 However, the rationale for using traditional data augmentation on these finely calculated arrays with precise spatial and temporal relations is that the component 2D arrays monitor the corneal biomechanical response from different geometric perspectives so that features embedded in 1 array are not strictly registered with the others. So, biases introduced by augmentation in 1 2D array may not be relevant for the others, resulting in overall maintained robust representations in the mother 3D array. That was an important motive to use the concatenated 3D array instead of analyzing a single 2D array type as a grey-scale pseudoimage.
Also, we decided to exclude subclinical KC cases from the dataset to create the KC probability score. However, the distinction between N and KC eyes can be accomplished by standard topography or tomography and a population with very asymmetric ectasia with normal topography (subclinical KC) would have been the best population to test the capabilities of the KC probability score.7, 8, 9 Unfortunately, patients with very asymmetric ectasia are rarely found in the analyzed records, and building repositories of scarce datasets is further restricted by patient privacy concerns.36,37 In this respect, the developed index needs further refinement and validation to be a clinically useful diagnostic tool in the industry. We hope that the model performance can be improved using a larger dataset that is populated with cases of subclinical KC. Additionally, to overcome issues of imbalanced datasets and patient privacy, generative artificial intelligence can hopefully provide generative video models such as variational autoencoders, generative adversarial networks, diffusion models, flow-based models, and transformer-based models. Each model has its own strengths and limitations.38 These models may be used to synthesize new videos similar to a modest-sized training dataset of subclinical KC Corvis ST videos to help overcome class scarcity and allow the widespread availability of such data. Finally, the Corvis ST analyzes deformation videos of only 1 corneal section. Because of this limitation, it may be unable to detect potentially significant asymmetric biomechanical abnormalities exhibited across the nonimaged corneal sections of patients with KC.
Conclusions
Our study introduces the KC probability score as a novel add-on DCR index developed by advanced image processing with the implementation of deep learning for KC diagnosis from Corvis ST videos. This index has shown high sensitivity and specificity on both the test subset and external validation dataset from another continent. These findings suggest the potential use of the preprocessing pipeline for machine learning algorithms and propose the integration of the model in future multi-modal models to augment the current KC diagnostic armamentarium. Future work using additional independent datasets is required to validate the KC probability score as an add-on parameter to other DCR indices within other population characteristics, especially in the subclinical KC domain.
Manuscript no. XOPS-D-23-00037.
Footnotes
Supplemental material available atwww.aaojournal.org.
Disclosure:
All authors have completed and submitted the ICMJE disclosures form.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
HUMAN SUBJECTS: Human subjects were not included in this study. This study was conducted following ethical standards in the Declaration of Helsinki and its later amendments and was approved by the institutional review board of Federal University of São Paulo-UNIFESP/EPM (as the coordinator center) and the Hospital de Olhos-CRO, Guarulhos (as the affiliate center), and the Salouti Eye Center in Iran. Once required, each subject provided informed written consent to participate in the study before using their data, and data were de-identified in Brazil and Iran before any further processing. No animals subjects were included in this study.
Author Contributions:
Conception and design: Abdelmotaal, Yousefi
Analysis and interpretation: Abdelmotaal, Yousefi
Data collection: Abdelmotaal, Hazarbassanov, Salouti, Nowruzzadeh, Taneri
Obtained funding: Yousefi
Overall responsibility: Abdelmotaal, Hazarbassanov, Taneri, Al-Timemy, Lavric, Yousefi
Contributor Information
Hazem Abdelmotaal, Email: hazem@aun.edu.eg.
Rossen Mihaylov Hazarbassanov, Email: hazarbassanov@gmail.com.
Siamak Yousefi, Email: siamak.yousefi@uthsc.edu.
Supplementary Data
Raw video (keratoconus) before preprocessing.
KC1 video after cleaning background noise.
KC2 video with a white corneal mask.
KC3 video with the corneal skeleton.
Raw video (normal cornea) before preprocessing.
N1 video after cleaning background noise.
N2 video with a white corneal mask.
N3 video with the corneal skeleton.
References
- 1.Wagner H., Barr J.T., Zadnik K. Collaborative longitudinal evaluation of keratoconus (CLEK) study: methods and findings to date. Cont Lens Anterior Eye. 2007;30:223–232. doi: 10.1016/j.clae.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labiris G., Giarmoukakis A., Sideroudi H., et al. Impact of keratoconus, cross-linking, and cross-linking combined with photorefractive keratectomy on self-reported quality of life. Cornea. 2012;31:734–739. doi: 10.1097/ICO.0b013e31823cbe85. [DOI] [PubMed] [Google Scholar]
- 3.Rabinowitz Y.S. Keratoconus. Surv Ophthalmol. 1998;42:297–319. doi: 10.1016/s0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 4.Ambrosio R., Jr., Belin M.W. Imaging of the cornea: topography vs tomography. J Refract Surg. 2010;26:847–849. doi: 10.3928/1081597X-20101006-01. [DOI] [PubMed] [Google Scholar]
- 5.Roberts C.J., Dupps W.J., Jr. Biomechanics of corneal ectasia and biomechanical treatments. J Cataract Refract Surg. 2014;40:991–998. doi: 10.1016/j.jcrs.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts C.J., Mahmoud A.M., Bons J.P., et al. Introduction of two novel stiffness parameters and Interpretation of air puff-induced biomechanical deformation parameters with a dynamic Scheimpflug analyzer. J Refract Surg. 2017;33:266–273. doi: 10.3928/1081597X-20161221-03. [DOI] [PubMed] [Google Scholar]
- 7.Vinciguerra R., Ambrósio R., Jr., Elsheikh A., et al. Detection of keratoconus with a new biomechanical index. J Refract Surg. 2016;32:803–810. doi: 10.3928/1081597X-20160629-01. [DOI] [PubMed] [Google Scholar]
- 8.Vinciguerra R., Ambrósio R., Jr., Roberts C.J., et al. Biomechanical characterization of subclinical keratoconus without topographic or tomographic abnormalities. J Refract Surg. 2017;33:399–407. doi: 10.3928/1081597X-20170213-01. [DOI] [PubMed] [Google Scholar]
- 9.Ambrósio R., Jr., Lopes B.T., Faria-Correia F., et al. Integration of scheimpflug-based corneal tomography and biomechanical assessments for enhancing ectasia detection. J Refract Surg. 2017;33:434–443. doi: 10.3928/1081597X-20170426-02. [DOI] [PubMed] [Google Scholar]
- 10.Ali N.Q., Patel D.V., McGhee C.N.J. Biomechanical responses of healthy and keratoconic corneas measured using a noncontact Scheimpflug-based tonometer. Invest Ophthalmol Vis Sci. 2014;55:3651–3659. doi: 10.1167/iovs.13-13715. [DOI] [PubMed] [Google Scholar]
- 11.Abdelmotaal H., Mostafa M.M., Mostafa A.N.R., et al. Classification of color-coded Scheimpflug camera corneal tomography images using deep learning. Transl Vis Sci Technol. 2020;9:30. doi: 10.1167/tvst.9.13.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Timemy A.H., Mosa Z.M., Alyasseri Z., et al. A hybrid deep learning construct for detecting keratoconus from corneal maps. Transl Vis Sci Technol. 2021;10:16. doi: 10.1167/tvst.10.14.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdelmotaal H., Hazarbasanov R., Taneri S., et al. Detecting dry eye from ocular surface videos based on deep learning. Ocul Surf. 2023;28:90–98. doi: 10.1016/j.jtos.2023.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Chen W., Sui L., Xu Z., Lang Y. 2012 International Conference on Systems and Informatics (ICSAI2012), Yantai, China. 2012. Improved Zhang-Suen thinning algorithm in binary line drawing applications; pp. 1947–1950. [DOI] [Google Scholar]
- 15.Xu X., Lin J., Tao Y., Wang X. 2018 7th International Conference on Digital Home (ICDH), Guilin, China. 2018. An Improved DenseNet Method Based on Transfer Learning for Fundus Medical Images; pp. 137–140. [DOI] [Google Scholar]
- 16.Araújo T., Aresta G., Mendonça L., et al. Data augmentation for improving proliferative diabetic retinopathy detection in eye fundus images. IEEE Access. 2020;8:182462–182474. [Google Scholar]
- 17.Goodfellow I., Bengio Y., Courville A. MIT Press; Cambridge, MA: 2016. Deep Learning. [Google Scholar]
- 18.Ruder S. An overview of gradient descent optimization algorithms. arXiv. 2016 doi: 10.48550/arXiv.1609.04747. [DOI] [Google Scholar]
- 19.Powers D.M.W. Evaluation: from precision, recall, and F-measure to ROC, informedness, markedness & correlation. J Mach Learn Technol. 2011;2:37–63. [Google Scholar]
- 20.Webb G.I. In: Encyclopedia of Machine Learning. Sammut C., Webb G.I., editors. Springer; Boston, MA: 2011. Naïve Bayes; pp. 713–714. [Google Scholar]
- 21.Xu Fast implementation of DeLong’s algorithm for comparing the areas under correlated receiver operating characteristic curves. IEEE Signal Process Lett. 2014;21(11):1389–1393. [Google Scholar]
- 22.Zhou B., Khosla A., Lapedriza A., et al. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV. 2016. Learning Deep Features for Discriminative Localization; pp. 2921–2929. [DOI] [Google Scholar]
- 23.Oliphant T.E. Python for scientific computing. Comput Sci Eng. 2007;9:10–20. [Google Scholar]
- 24.Pedregosa F., Varoquaux G., Gramfort A., et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 25.Anderson K., El-Sheikh A., Newson T. Application of structural analysis to the mechanical behaviour of the cornea. J R Soc Interface. 2004;1:3–15. doi: 10.1098/rsif.2004.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira-Mendes J., Lopes B.T., Faria-Correia F., et al. Enhanced ectasia detection using corneal tomography and biomechanics. Am J Ophthalmol. 2019;197:7–16. doi: 10.1016/j.ajo.2018.08.054. [DOI] [PubMed] [Google Scholar]
- 27.Kataria P., Padmanabhan P., Gopalakrishnan A., et al. Accuracy of Scheimpflug-derived corneal biomechanical and tomographic indices for detecting subclinical and mild keratectasia in a South Asian population. J Cataract Refract Surg. 2019;45:328–336. doi: 10.1016/j.jcrs.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y., Guo L.L., Tian L., et al. Comparative analysis of the morphological and biomechanical properties of normal cornea and keratoconus at different stages. Int Ophthalmol. 2021;41:3699–3711. doi: 10.1007/s10792-021-01929-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herber R., Ramm L., Spoerl E., et al. Assessment of corneal biomechanical parameters in healthy and keratoconic eyes using dynamic bidirectional applanation device and dynamic Scheimpflug analyzer. J Cataract Refract Surg. 2019;45:778–788. doi: 10.1016/j.jcrs.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 30.Koh S., Inoue R., Ambrósio R., Jr., et al. Correlation between corneal biomechanical indices and the severity of keratoconus. Cornea. 2020;39:215–221. doi: 10.1097/ICO.0000000000002129. [DOI] [PubMed] [Google Scholar]
- 31.Herber R., Pillunat L.E., Raiskup F. Development of a classification system based on corneal biomechanical properties using artificial intelligence predicting keratoconus severity. Eye Vis (Lond) 2021;8:21. doi: 10.1186/s40662-021-00244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vinciguerra R., Elsheikh A., Roberts C.J., et al. Influence of pachymetry and intraocular pressure on dynamic corneal response parameters in healthy patients. J Refract Surg. 2016;32:550–561. doi: 10.3928/1081597X-20160524-01. [DOI] [PubMed] [Google Scholar]
- 33.Tan Z., Chen X., Li K., et al. Artificial intelligence-based diagnostic model for detecting keratoconus using videos of corneal force deformation. Transl Vis Sci Technol. 2022;11:32. doi: 10.1167/tvst.11.9.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koprowski R., Wilczyński S. Corneal vibrations during intraocular pressure measurement with an air-puff method. J Healthc Eng. 2018;2018 doi: 10.1155/2018/5705749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shorten C., Khoshgoftaar T.M. A survey on image data augmentation for deep learning. J Big Data. 2019;6:60. doi: 10.1186/s40537-021-00492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao L., Zhang L., Liu C., Wu S. Handling imbalanced medical image data: a deep-learning-based one-class classification approach. Artif Intell Med. 2020;108 doi: 10.1016/j.artmed.2020.101935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price W.N., Cohen I.G. Privacy in the age of medical big data. Nat Med. 2019;25:37–43. doi: 10.1038/s41591-018-0272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhagwatkar R., Bachu S., Fitter K., Kulkarni A., Chiddarwar S. 2020 International Conference on Power, Instrumentation, Control and Computing (PICC), Thrissur, India. 2020. A Review of Video Generation Approaches; pp. 1–5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw video (keratoconus) before preprocessing.
KC1 video after cleaning background noise.
KC2 video with a white corneal mask.
KC3 video with the corneal skeleton.
Raw video (normal cornea) before preprocessing.
N1 video after cleaning background noise.
N2 video with a white corneal mask.
N3 video with the corneal skeleton.