Abstract
Yili River system hosts a diverse fauna of fishes and parasites. Gymnodiptychus dybowskii is a rare and endangered aboriginal cold-water fish inhabit in the Yili river system. Our research identified a new species Gyrodactylus gymnodiptychi n. sp. isolated from G. dybowskii in the Kunes River (Yili River, China). Morphological comparison revealed identifiable differences between the new species and other parasites, including Gyrodactylus aksuensis, and Gyrodactylus tokobaevi, which are two known parasites living in G. dybowskii inhabit in the Aksu River west of Frunze (Kyrgyzstan), as well as Gyrodactylus montanus living in Shizothorax intermedius inhabited in the Tadzhikistan or Uzbekistan. Especially, the dorsal bar of G. gymnodiptychi n. sp. was raised at both ends with a hollow, and its hamulus roots were curved inward. The BLASTN search of GenBank did not detect any other ITS1-5.8S-ITS2 rDNA sequences same as G. gymnodiptychi's. Using the Bayesian Information and Maximum Likelihood methods to analyze the ITS1-5.8S-ITS2 rDNA gene sequences, we constructed phylogenetic trees for G. gymnodiptychi n. sp. Accordingly, our morphological and molecular research indicated that G. gymnodiptychi n. sp. was not only a new species of parasites but also the first Gyrodactylus member identified in the Yili River in China.
Keywords: Gyrodactylus gymnodiptychi n. sp., Gymnodiptychus dybowskii, Monogenea, Phylogenetic analysis, China
Graphical abstract
Highlights
-
•
A new parasite species Gyrodactylus gymnodiptychi was identified in Yili River.
-
•
The fish Gymnodiptychus dybowskii was the host of Gyrodactylus gymnodiptychi.
-
•
Phylogenetic analysis indicated the new species a subgenus Limnonephrotus member.
1. Introduction
Yili River system is an international river flowing from the northeast Borohoro Mountains and southeast Halik Mountains to westward through the Yili Basin basin into Kazakhstan. The Borohoro and Halik Mountains are two branches of the Kazakhstan Tian Shan Mountains. Yili River flows 430 km long in China, extending to 3 major tributaries Tekes River, Kunes River, and Kashi River (Ren et al., 1998). Due to the growing freshwater fish farming industry, the cultivation and development of high-quality indigenous fish in the Yili River have become increasingly important for the economic development of the Yili River basin. As the industry shifts from natural ecological environments to intensive and large-scale aquaculture, the living conditions for fish have undergone significant changes. Yili River system hosts a diverse fauna with ten endemic species, including four Schizothoracinae fishes Gymnodiptychus dybowskii Kessler, 1987; Diptychus maculatus Steindachner, 1866; Schizothorax argentatus pseudaksaiensis Herzenstein, 1889; and Schizothorax argentatus Kessler, 1874; as well as six other fishes (Ren et al., 1998; Guo, 2012; Meng et al., 2018).
Monogeneans of the genus Gyrodactylus have been known for almost 190 years for their retention of fully grown daughters in utero until they themselves contain developing embryos (Bakke et al., 2007). Gyrodactylus was first described from bream (Abramis brama) by von Nordmann (1832). The growing invasion of Gyrodactylus into fish farms and wild fields has become a serious endemic disease leading to economic losses (Atkins, 1901; Embody, 1924; Guberlet et al., 1927; Williams, 1964; Johnsen and Jenser, 1991). It has the conservative morphology of particular structures and huge species diversity (Bakke et al., 2007). Soviet scholars have conducted extensive research on this subject including studies on fish parasites in Central Asian countries (Yamaguti, 1965; Gusev, 1985). Previous studies of parasites and their hosts inhabited in the upstream Yili River revealed that two fish subfamilies Schizothoracinae and Botiinae host nine species (eight genera) of parasites, including Dactylogyrus drjagini Bychowsky, 1936; Paradiplozoon schizothorazi Iksanov, 1965; Rhabdochona opienensis Hsü, 1933; Schyzocotyle acheilognathi Yamaguti, 1934; Allocreadium schizothoracis Pande, 1938; Acanthocephala sp.; Chilodonella cyprinid Moroff, 1902; Trichodina nobillis Chen, 1963; and Trichodina orientalis Chen et Hsieh in Anon.,1973. (Yao et al., 2013; Niu et al., 2017). Numerous parasites have been observed in fishes inhabited in the upper Yili River system (in China); however, they have not been scientifically studied.
Gymnodiptychus dybowskii belongs to the subfamily Schizothoracinae (Cyprinidae) and is a cold-water fish dwelling in alpine waters. Gymnodiptychus dybowskii inhabit in the Syrdarya River, the Balkhash Lake, and the Issyk-Kul Lake in Central Asia. In China, G. dybowskii inhabit in the Yili River system, the Junggar basin water system, and the Kaidu River of Xinjiang. Gymnodiptychus dybowskii is a rare and endangered aboriginal fish, and it represents an indigenous and ecologically essential species in Xinjiang aquatic ecosystems (Meng et al., 2018). In 2022, G. dybowskii was listed as a Class I key-protected aquatic wild animal by the People's Government of Xinjiang Uygur Autonomous Region. Up to now, studies have focused on the physiology and genetics of G. dybowski inhabit in the Yili River system (Niu et al., 2015; Guo et al., 2016); however, its monogenean parasites still remain to be studied. Only Gyrodactylus aksuensis and Gyrodactylus tokobaevi, the two known parasites infecting G. dybowskii, inhabit in the Aksu River west of Frunze (Kirghiz. S.S.R.) (Ergens and Karabekova, 1980).
Currently, identification and nomenclature of Gyrodactylus species are still based on morphological characteristics and host specificity. The morphological characterization is primarily based on the measurement of attachment structures (Bakke et al., 2002). Changes of morphological evolution of Gyrodactylus have been postulated to be associated with host shifting (Lindenstrøm et al., 2003; Shinn et al., 2004; Bakke et al., 2007). Changes of hamulus and marginal hook size may be associated with environmental changes, such as temperature (Kulemina and Skarlato, 1987; Dávidová et al., 2005). However, these environmental and host factors in association with changes of morphological characteristics still need to be clarified. Recent advancement of molecular technologies and combined uses of molecular and morphological data provide us a platform to further study Gyrodactylus taxa (Bueno-Silva and Boeger, 2014; Zahradníčková et al., 2016; García Vásquez et al., 2018).
In this study, we performed morphological characteristics of a novel Gyrodactylus organism isolated from the gill and fin of G. dybowskii inhabit in the Kunes River, upstream of Yili River system. We used molecular methods and GenBank to study the ITS1-5.8S-ITS2 rDNA gene sequences and construct phylogenetic trees for the organism to determine the new species G. gymnodiptychi n. sp.
2. Materials and methods
2.1. Fish and parasite sampling
Samples of G. dybowskii were captured using fyke nets from the Kunes River (82°34′49.61″ N; 84°44′30.41″ E) in the months of July 2018 and December 2019. After sampling, living fish were euthanized by severing the spinal cord posterior to the skull with a single cut. Gills and fins were then surgically isolated for gross examination and isolation of parasites within 24 h. Isolated parasites were preserved in 70% and 95% ethanol for morphometric and molecular analyses, respectively. The definitions of prevalence and mean intensity of infection were used following Bush et al. (1997). All animal procedures were approved by the Xinjiang Agricultural University Animal Care and Use Committee (No. 2019021).
2.2. Morphometric analysis
Twenty-eight intact individuals of G. gymnodiptychi n. sp. were isolated from fish bodies, and selected for morphological characterization. Nine individuals were fixed and stained in GAP to reveal features of hamulus, marginal hook, dorsal bar, and ventral bar. Nineteen individuals were fixed and stained with 4% PL to reveal features of hamulus, marginal hook, and male copulatory organ. Morphological features were microscopically analyzed using a Nikon ECLIPSE E200 imaging optical microscope. The length and width of the body, hamulus, ventral bar, dorsal bar and marginal hook were measured using an EZ-MET software (x86, 6.0.7543) (Shinn et al., 2004; Christison et al., 2005; García-Vásquez et al., 2007). Drawings of parasite morphological features were performed using a camera lucida.
2.3. Molecular analysis
Genomic DNA was extracted from parasites using the EasyPure® Genomix DNA kit (TransGen Biotech, Beijing, China). Fragments of the ITS1-5.8S-ITS2 ribosomal DNA (rDNA) amplified region were isolated by the PCR technique using the forward primer 5′-TTTCCGTAGGTGAACCT-3′ and the reverse primer 5′-TCCTCCGCTTAGTGATA-3′ (Cunningham, 1997). The amplification reaction was performed in a final volume of 50 μL, containing of 1 μL forward primer/reverse primer, 2 μL of DNA, and 25 μL of 2 × Super Master mix (Bio-Rad, CA, USA) and 21 μL of double distilled water. The samples were incubated in the following cycles: 1 cycle at 94 °C for 5 min, 30 cycles at 95 °C for 30 s, 65 °C for 30 s, and 72 °C for 75 s, and the final extension of 1 cycle at 72 °C for 10 min. PCR products were electrophoresed on agarose gels (1%), and DNA fragments were visualized by staining with GelStain (TransGen Biotech). DNA fragments were isolated and purified from agarose slices using an EasyPure® PCR purification kit (TransGen Biotech). Purified DNA fragments were constructed into a plasmid vector, using the pEASY®-T1 Cloning Kit (TransGen Biotech). Then, plasmid DNA was purified with a HiPure Plasmid MiniPrep Kit (TransGen Biotech), followed by sequencing the isolated ITS1-5.8S-ITS2 rDNA regions using the ABI Cycle Sequencer 3700 (Foster City, CA, USA).
2.4. Phylogenetic analysis
Phylogenetic status of Gyrodactylus gymnodiptychi n. sp. collected from the Kunes River was determined by comparing the ITS1-5.8S-ITS2 rDNA sequences of 46 Gyrodactylus species in the GenBank (Table 1). The sequences of Diplogyrodactylus martini Prikrylova, Matejusova, Musilova, Gelnar & Harris, 2009 and Gyrodactyloides bychowskii Albova, 1948 (family Gyrodactylidae) were used as two control outgroups. The sequences of the ITS1-5.8S-ITS2 rDNA genes in the GenBank were identified using the PhyloSuite 1.2.3, (Zhang et al., 2020), and DNA sequences were aligned using MAFFT v7 (Katoh and Standley, 2013). The aligned results were imported into Globcks 0.91b (Gerard and Jose, 2007) then the conserved sites were extracted using Gblocks 0.91b with the following parameter settings: minimum number of sequences for a conserved/flank position (25/25), maximum number of contiguous non-conserved positions (8), minimum length of a block (10), allowed gap positions (all). The model of molecular evolution were determined according to the corrected Bayesian Information (BI) and Maximum Likelihood (ML) using ModelFinder v1.6.8 (Kalyaanamoorthy et al., 2017). On the basis of the selected model, phylogenetic analyses were performed using two different algorithms: Bayesian Inference (BI) and Maximum Likelihood (ML). Both BI and ML analytical methods were used to determine phylogenetic relationship between parasites, followed by using the Bayesian information criterion (BIC) in ModelFinder v1.6.8 to identify optimal evolutionary model (Posada and Crandall, 1998). The BI analysis was performed by using the MrBayes 3.2 software with the parameter settings nst = 6 and rates = Invgamma (GTR+F+I+G4 model) (Ronquist and Huelsenbeck, 2003). Posterior probability of model parameters was evaluated with the Markov Chain Monte Carlo method (MCMC) running four chains, sampling every 100 generations, for 5,000,000 generations. After checking for convergence, the Burnin sample trees were discarded with the parameter setting at 0.25%. The remaining trees were calculated with the MrBayes 3.2.1. program to determine a 50% majority-rule consensus tree. On the other hand, ML analysis was reconstructed using IQ-TREE (Trifinopoulos et al., 2016) with 10,000 ultrafast bootstraps (Minh et al., 2013). The iTOL (https://itol.embl.de/) (Letunic and Bork, 2007) was used to visualise the phylogeny and architecture using files generated from PhyloSuite 1.2.3.
Table 1.
Sequence information of selected ITS1-5.8S-ITS2 rDNA of Gyrodactylus species used for phylogenetic analysis (* species sequenced in this study).
| Parasite | Host species | Locality | GenBank ID | Reference |
|---|---|---|---|---|
| G. ajimeNitta, 2021 | Niwaella delicata | Japan: Kyoto | LC545570 | Nitta (2021) |
| G. anguillae Ergens, 1960 | Anguilla australis | Australia: Victoria, Skipton | AB063294 | Hayward et al., 2012, unpublished |
| G. aphyae Malmberg, 1957 | Phoxinus phoxinus | Finland: River Merenoja, River Kovda system, White Sea basin | AF484528 | Ziętara et al. (2002) |
| G. arcuatus Bychowsky, 1933 | Gasterosteus aculeatus | Finland: Gulf of Bothnia, Baltic Sea | AF328865 | Ziętara et al. (2002) |
| G. brachymystacis Ergens, 1978 | Brachymystax lenok | China | GQ368237 | Gilmore et al. (2010) |
| G. branchialis Huyse, Malmberg & Volckaert, 2004 | Pomatoschistus marmoratus | France: Vaccares lagoon | DQ821770 | Huyse et al. (2006) |
| G. branchicus Malmberg, 1964 | Gasterosteus aculeatus | Russia: Kola Peninsula, White Sea | FJ435199 | Rokicka et al. (2009) |
| G. bubyri Osmanov, 1965 | Knipowitschia caucasica | Bulgaria: Atanasovsko Lake | KU355879 | Stoyanov et al. (2016) |
| G. bullatarudis Turnbull, 1956 | Poecilia reticulata | Trinidad and Tobago: Lopinot (Arouca) River | AY692024 | Cable et al. (2005) |
| G. cernuae Malmberg, 1957 | Gymnocephalus cernuus | Finland: River Oulujoki, Baltic Sea basin | AF484529 | Ziętara et al. (2002) |
| G. derjavini Mikailov, 1975 | Oncorhynchus mykiss | Iran: Caspian Sea basin | DQ323402 | Rokicka et al. (2007) |
| G. derjavinoides Malmberg, Collins, Cunningham & Jalali, 2007 | Salmo letnica | Macedonia: River Vardar system, Aegean Sea basin | EU304810 | Ziętara et al. (2010) |
| G. gymnodiptychi n. sp. | Gymnodiptychus dybowskii | China: Yili River | MH445968 | This study |
| G. gymnodiptychi n. sp. | Gymnodiptychus dybowskii | China: Yili River | MH445967 | This study |
| G. ginestrae Kvach, Ondracč;ková, Seifertová and Hulak, 2019 | Atherina boyeri | Ukraine: Black Sea, Gulf of Odessa | MK550602.2 | Kvach et al. (2019) |
| G. gracilihamatus Malmberg, 1964 | Gasterosteus aculeatus | Finland: Gulf of Bothnia, Baltic Sea | AF484532 | Ziętara et al. (2002) |
| G.gurleyi Price, 1937 | Goldfish Carassius auratus) | China | KC922453 | Li et al. (2014) |
| G. harengi Malmberg, 1957 | Clupea harengus | France: Ambleteuse | AJ309295 | Matejusová et al. (2003) |
| G. jiroveci Ergens and Bychowsky, 1967 | Barbatula barbatula | Czech Republic | AM502860 | Přikrylová et al. (2008) |
| G. jussii ZiÄ;TMtara & Lumme, 2003 | Phoxinus phoxinus | Finland: River Merenoja, River Kovda system, White Sea basin | AY061982 | Ziętara and Lumme (2003) |
| G. leptorhynchiCone et al., 2013 | Syngnathus leptorhynchus | Pacific coast of North America | JX110633 | Cone et al. (2013) |
| G. leucisci Zitnan, 1964 | Leuciscus leuciscus | Finland: River Oulujoki, Baltic Sea basin | AF484537 | Ziętara et al. (2002) |
| G. luciopercae Gussev, 1962 | Perca fluviatilis | Finland: Gulf of Bothnia, Baltic Sea | AF484541 | Ziętara et al. (2002) |
| G. macronychus Malmberg, 1957 | Phoxinus phoxinus | Finland: River Merenoja, River Kovda system, White Sea basin | AY061981 | Ziętara and Lumme (2003) |
| G. medakaNitta and Nagasawa, 2018 | Oryzias latipes | Japan: Tokushima | LC368477 | Nitta and Nagasawa (2018) |
| G. mongolicus Ergens and Dulmaa, 1970 | Oreoleuciscus potanini | Mongolia: Chono Kharaik river | OQ913868 | Lebedeva et al. (2023) |
| G. nemachili Bikhovski, 1936 | Oreoleuciscus potanini | Mongolia: Chono Kharaik river | OQ641772 | Lebedeva et al. (2023) |
| G. nipponensis Ogawa and Egusa, 1978 | Anguilla japonica | Japan: Shizuoka, Lake Hamana | AB063295 | Hayward et al. (2001) |
| G. notatae King et al., 2009 | Menidia menidia | Nova Scotia, Canada | FJ840489 | King et al. (2009) |
| G. orecchiaePaladini et al., 2009 | Sparus aurata | Adriatic Sea | FJ013097 | Paladini et al. (2009) |
| G. ostendicus Huyse and Malmberg, 2004 | Pomatoschistus marmoratus | France: Vaccares lagoon | DQ821768 | Huyse et al. (2006) |
| G. papernai Ergens and Bychowsky, 1967 | Salmo salar | Russia: River Vidlitsa, Lake Ladoga system, Baltic Sea Basin | EF446729 | Matejusová et al. (2001) |
| G. poeciliaeHarris and Cable (2000) | Poecilia caucana | Venezuela | AJ001844.2 | Harris and Cable (2000) |
| G. proterorhini Ergens, 1967 | Proterorhinus semilunaris | Bulgaria: Vidin, Danube | MK584285.2 | Kvach et al. (2019) |
| G. pseudonemacheili Ergens and Bychowsky, 1967 | Barbatula conilobus | Mongolia: Zavkhan river | OQ641764 | Lebedeva et al. (2023) |
| G. pterygialis Bychowsky and Polyansky, 1953 | Pollachius virens | Norway: Fjord near Bergen | AJ581657 | Matejusová et al. (2003) |
| G. pungitii Malmberg, 1964 | Pungitius pungitius | Finland: Lake Rytilampi, White Sea basin | AF484543 | Ziętara et al. (2002) |
| G. rarus Wegener, 1910 | Gasterosteus aculeatus | Finland: Gulf of Bothnia, Baltic Sea | FJ435196 | Rokicka et al. (2009) |
| G. rogatensis Harris, 1985 | Cottus gobio | Rogate (West Sussex, England) | AJ011411 | Cable et al. (1999) |
| G. rugiensis Glaser, 1974 | Pomatoschistus minutus | France: Vaccares lagoon | DQ821761 | Huyse et al. (2006) |
| G. salaris Malmberg, 1957 | Thymallus thymallus | Finland: River Oulankajoki, River Kovda system, White Sea basin | AF484544 | Ziętara et al. (2002) |
| G. salinae Paladini et al., 2011 | Aphanius fasciatus | hypersaline environment in Italy | JF950559 | Paladini et al. (2011) |
| G. tayshirensisLebedeva et al., 2023 | Barbatula conilobus | Mongolia: Zavkhan river | OQ641774 | Lebedeva et al. (2023) |
| G. truttae Gloser, 1974 | Salmo trutta | Poland: River Wisla system, Baltic Sea basin | EF464681 | Rokicka et al. (2007) |
| G. turnbulli Harris, 1986 | Poecilia reticulata | Poland: Gdansk aquarium | EF445942 | Lumme and Ziętara (2018) |
| G. zavkhanensisLebedeva et al., 2023 | Thymallus brevirostris | Mongolia: Zavkhan river | OQ641773 | Lebedeva et al. (2023) |
| Diplogyrodactylus martini Prikrylova, Matejusova, Musilova, Gelnar & Harris, 2009 | Polypterus senegalus | Senegal | AM943008 | Přikrylová et al. (2009) |
| Gyrodactyloides bychowskii Albova, 1948 | salmon | United Kingdom:Scotland | AJ249348 | Bruno et al. (2001) |
3. Results
3.1. Taxonomic summary
Gyrodactylus was collected from 69 G. dybowskii (fork length: 6.4–18.5 cm), with a prevalence of 91.3% (63 infected fish), and mean intensity was 7.29 (range, 1 to 22). All specimens represented a morphologically similar species but did not correspond to any other Gyrodactylus species already identified in the Kunes River.
Class Monogenoidea Bychowsky, 1937
Subclass Polyonchoinea Bychowsky, 1937
Order Gyrodactylidea Bychowsky, 1937
Family Gyrodactylidae Van Beneden and Hesse, 1863
Gyrodactylus gymnodiptychi n. sp.
Type host: Gymnodiptychus dybowskii Kessler, 1874.
Type locality: Kunes River, Xinjiang Uygur Autonomous Region, China (82°34′49.61″ N; 84°44′30.41″ E).
Site of infection: gills and fins.
Types material: The Holotypes XJLCC20191101 and the Paratypes XJLCC20191102-05 are deposited in the museum of Parasitology at the Xinjiang Agricultural University.
Genetic material: The ITS1-5.8S-ITS2 rDNA sequence was deposited in the GenBank (Accession numbers MH445967 and MH445968).
Etymology: The species was named by referring to the genus of host Gymnodiptychus dybowskii from which it parasitized.
Zoo bank: LSID urn:lsid:zoobank.org:pub:34E792F3-9ED8-45C3-A673-FC5C44577BAC.
3.2. Morphology
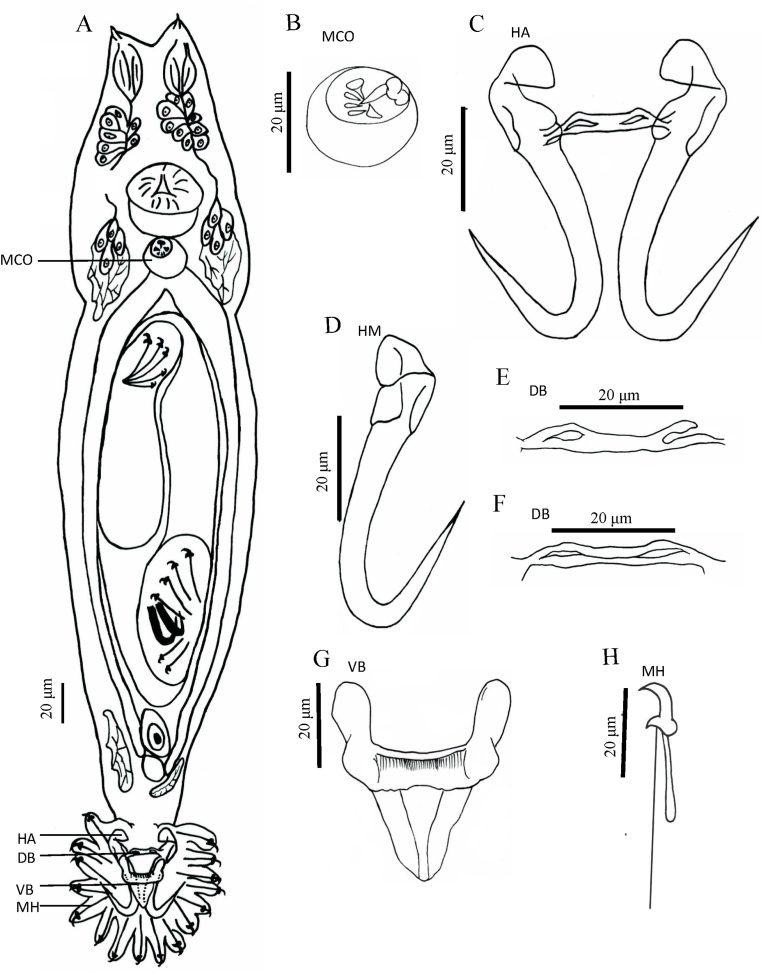
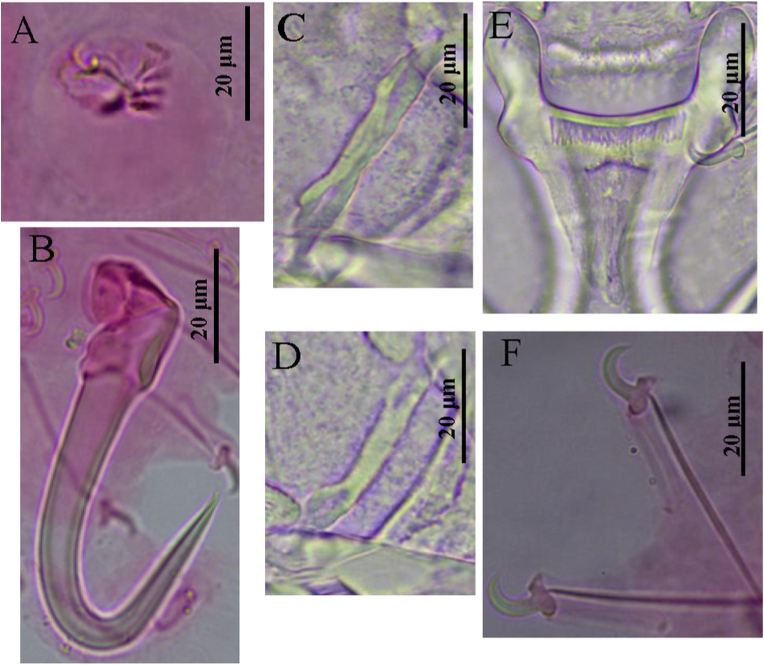
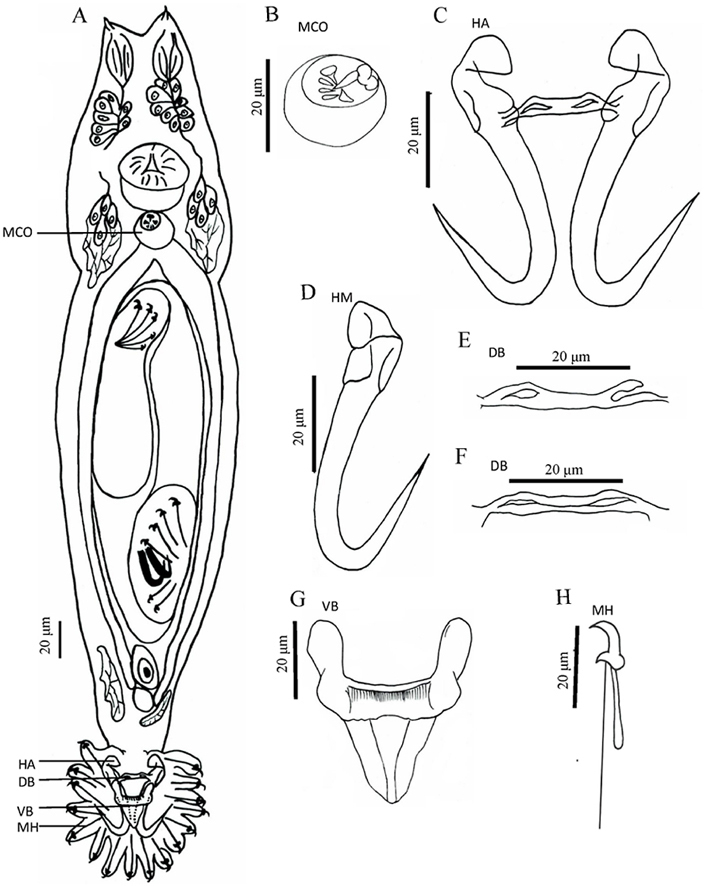
Based on 28 specimens. Body "gourd-like" shape, fusiform, a depression in the middle of body, total body length 368.0 (223.3–608.0) long, 80 (62.0–136.5) wide. Pharynx bulb 21.1 (13.8–29.7) long, 19.7 (13.6–28.2) wide (Fig. 1A and Table 2). The cecum was posterior to the anterior edge of the testes (Fig. 1A). MCO 12.3 (8.0–19.9) long, 8.5 (6.9–10.2) wide, armed with one central spine, two large spines and three small spines, posterior to pharyngeal bulb (Fig. 1, Fig. 2A). Hamuli 61.6 (57.5–73.7) long, shafts 52.3 (44.9–55.9) long, points 28.2 (18.8–39.0) long, slim; proximal shaft 8.4 (7.9–11.2) wide, curved. Aperture distance 19.1 (18.8–23.8) long, hamulus aperture angle 31.7° (26.9°–33.5°), hamulus root 22.4 (18.8–25.8) long, inward and curved (Fig. 1, Fig. 2B). Dorsal bar 29.2 (21.7–38.6) long, 2.2 (1.6–3.3) wide, the middle flat, straight, with a hollow at each end (Fig. 1, Fig. 2C & D). Ventral bar 40.7 (35.8–53.1) long, 8.0 (6.5–10.9) wide, ventral bar processes 12.1 (7.8–14.3) long, ovoid; ventral bar membrane 19.9 (15.9–22.3) long (Fig. 1, Fig. 2E). Marginal hook 38 (30.4–45.6) long, hook shaft 32.6 (24.1–37.9) long, rounded bottom; marginal hook sickle 8.7 (6.4–10.6) long, curved, tilted forward; sickle point 5.2 (3.6–5.7) wide, sickle distal 3.7 (2.7–5.0) wide. Marginal hook toe 2.23 (2.1–2.8) long, marginal hook aperture 7.1 (7.0–8.5) long, hook instep 1.0 (0.9–1.3) high, and filament loop 12.2 (12.0–16.1) long (Fig. 1, Fig. 2F).
Fig. 1.
Holotype of Gyrodactylus gymnodiptychi from the gills of Gymnodiptychus dybowskii. (A) Whole specimen (composite, ventral view), (B) Male copulatory organ (MCO), (C) Hamuli (HA), (D) Hamulus (HM), (E & F) Dorsal bar (DB), (G) Ventral bar (VB), and (H) Marginal hook (MH).
Table 2.
The comparison of Gyrodactylus gymnodiptychi with other morphologically similar species.
| Measurement |
N |
G. gymnodiptychi (n = 28) Present study |
G. tokobaevi (Ergens, 1980) |
G. montanus (Gussev, 1985) |
G.aksuensis (Ergens, 1980) |
|---|---|---|---|---|---|
| (length and width, μm) | Average (range) | Average (range) | Range | Average (range) | |
| Total body, length | 25 | 368.0 (223.3–608.0) | |||
| Total body, width | 28 | 80.0 (62.0–136.5) | |||
| Pharynx, length × width | 28 | 21.1 (13.8–29.7) × 19.7 (13.6–28.2) | |||
| Opisthaptor, length × width | 28 | 87.9 (62.0–110.7) × 87.3 (66.9–145.0) | |||
| Male copulatory organ, length × width | 13 | 12.3 (8.0–19.9) × 8.5 (6.9–10.2) | |||
| MCO spines | 6 | 1L, 5S | 1L, 8S | 1L, 6S | |
| Hamulus | |||||
| Total length | 28 | 61.6 (57.5–73.7) | 62–65 (65) | ||
| Aperture distance | 28 | 19.1 (18.8–23.8) | |||
| Point shaft width | 28 | 8.4 (7.9–11.2) | |||
| Point length | 28 | 28.2 (18.8–39.0) | 28–29 (29) | 21–33 | 13 (13–14) |
| Distal shaft width | 28 | 5.4 (4.2–6.9) | |||
| Shaft length HSL | 28 | 52.3 (44.9–55.9) | 42–44 (44) | 63–78 | 22 (22–23) |
| Inner curve length | 28 | 5.1 (3.63–6.61) | |||
| Aperture angle | 28 | 31.7° (26.9°–33.5°) | |||
| Point curve angle | 28 | 12.6° (8.2°–19.2°) | |||
| Inner aperture angle | 28 | 36.0° (33.0°–39.5°) | |||
| Root length | 28 | 22.4 (18.8–25.8) | 19–21 (21) | ||
| Ventral bar | |||||
| Length | 18 | 40.7 (35.8–53.05) | 29 (27–30) | 33–45 | 15 (15–16) |
| Width | 12 | 8.0 (6.5–10.9) | 7 (6–7) | 9–15 | 3 (3–4) |
| Process to mid–length | 1 | 4.6 (n = 1) | |||
| Mid–length | 1 | 8.5 (n = 1) | |||
| Process length | 9 | 12.1 (7.8–14.3) | |||
| Membrane length | 9 | 19.9 (15.9–22.3) | 17–20 (18) | ||
| Dorsal bar | |||||
| Length | 13 | 29.2 (21.7–38.6) | 22 (20–22) | 22–45 | 15 (15–16) |
| Width | 13 | 2.2 (1.6–3.3) | 3 (3–4) | 3–6 | 1 |
| Marginal hook | |||||
| Total length | 28 | 38 (30.4–45.6) | 29–31 | 40–53 | 21–22 |
| Shaft length | 28 | 32.6 (24.1–37.9) | |||
| Sickle length | 28 | 8.7 (6.5–10.6) | 6–7 | 7–8 | 5–5.5 |
| Sickle point width | 21 | 5.2 (3.6–5.7) | |||
| Toe length | 8 | 2.2 (2.1–2.8) | |||
| Sickle distal width | 24 | 3.7 (2.7–5.0) | |||
| Aperture | 7 | 7.1 (7.0–8.5) | |||
| Instep/arch height | 7 | 1.0 (0.9–1.3) | |||
| Filament loop | 20 | 12.2 (12.0–16.1) | |||
Fig. 2.
Light micrographs of the haptoral structures of Gyrodactylus gymnodiptychi from the gills of Gymnodiptychus dybowskii. (A) Male copulatory organ (MCO), (B) Hamulus (HA), (C & D) Dorsal bar (DB), (E) Ventral bar (VB), and (F) Marginal hook (MH).
3.3. Remarks
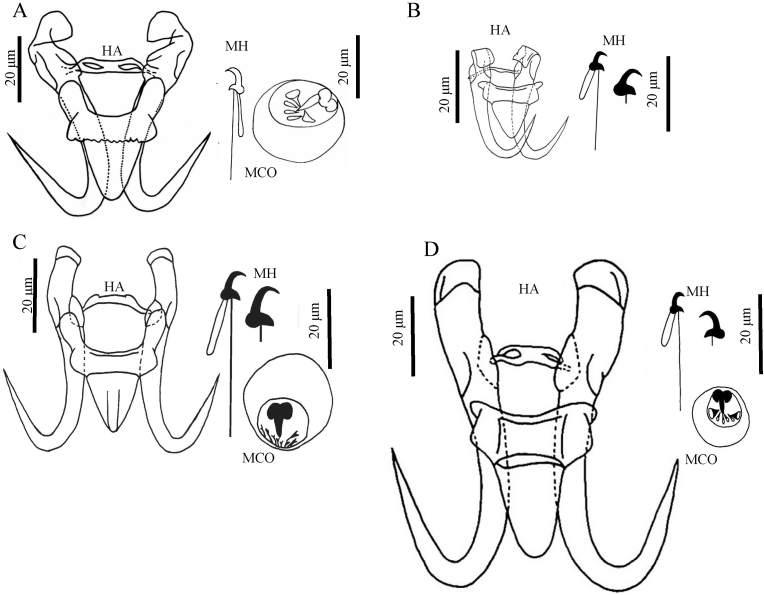
To understand the association of G. gymnodiptychi n. sp. with known members of Gyrodactylus, we compared the morphological features of G. gymnodiptychi n. sp. with Gyrodactylus aksuensis Ergens and Karabekova (1980); Gyrodactylus tokobaevi Ergens and Karabekova (1980); and Gyrodactylus montanus Bychowsky, 1957; Ergens and Karabekova (1980); Gusev, 1985). As depicted in Fig. 3, compared with G. aksuensis, the dorsal bar of G. gymnodiptychi n. sp. was raised at both ends with a hollow, but G. aksuensis was lanker and narrower than G. gymnodiptychi n. sp. (Fig. 3A and B). Gyrodactylus gymnodiptychi n. sp. exhibited similar ventral bar morphology to the G. tokobaevi (Fig. 3A and C). In both species, their ventral bar processes were prominent, but hamulus roots of G. gymnodiptychi n. sp. were curved inward. In addition, the dorsal bar of G. gymnodiptychi n. sp. had a straight center and a projection with a hollow at both ends, but the G. tokobaevi only had prominent ends without hollow. Additionally, the MCO of G. gymnodiptychi n. sp. had three spines fewer than G. tokobaevi (Fig. 3A and C). Gyrodactylus gymnodiptychi n. sp. exhibits similar dorsal bar morphology to the G. montanus. In both species, their dorsal bars had a hollow at both ends of the projection, but the hamulus root of G. gymnodiptychi n. sp. was curved inward and stouter than G. montanus. In addition, the ventral bar processes of G. gymnodiptychi n. sp. were more prominent than G. montanus (Fig. 3A and D). The results clearly revealed identifiable morphological differences between G. gymnodiptychi n. sp. and other Gyrodactylus members. In addition, G. gymnodiptychi n. sp. was the only one showing a hollow dorsal bar and curved hamulus root that were distinct from the other eleven species, carrying non-hollow dorsal bars and straight hamuli roots, of gyrodactylid isolated from the fish subfamily Schizothoracinae.
Fig. 3.
Morphological comparison of (A) Gyrodactylus gymnodiptychi, (B) Gyrodactylus aksuensis, (C) Gyrodactylus tokobaevi, and (D) Gyrodactylus montanus.
A - original drawing; B and C - from Ergens and Karabekova (1980); D - from Gusev (1985).
3.4. Molecular identification
The results determined that the two DNA sequences of 1202 bp (GenBank: MH445967) and 1199 bp (GenBank: MH445968) shared 99.46% identity, indicating that they were the same species. The results of a BLASTn search (Altschul et al., 1997) of the ITS1-5.8S-ITS2 fragment revealed no identical hits with entries in GenBank (Benson et al., 2007). Gyrodactylus gymnodiptychi n. sp. (GenBank: MH445967) appeared most closely related to Gyrodactylus tayshirensis (862/935, 92.19%, OQ641774) obtained from the Barbatula conilobus (Cypriniformes, Nemacheilidae, Barbatula) in Zavkhan river (Mongolia), Gyrodactylus jiroveci (834/906, 92.05%, AM502860) (Přikrylová et al., 2008) from Barbatula barbatula (Cypriniformes, Nemacheilidae, Barbatula) collected in Czech Republic, Gyrodactylus papernai (833/905, 92.04%, EF446729) (Ziętara et al., 2008) from Salmo salar (Salmoniformes, Salmonidae, Salmo) collected in Vidlitsa River, Lake Ladoga system (Russia), Gyrodactylus mongolicus (942/1045, 90.14%, GenBank: OQ913868) and Gyrodactylus nemachili (939/1044, 89.94%, OQ641772) obtained from the Oreoleuciscus potanini (Cypriniformes, Leuciscidae, Oreoleuciscus) in Chono Kharaik river (Mongolia), and Gyrodactylus zavkhanensis (933/1038, 89.88%, OQ641773) obtained from the Thymallus brevirostris (Salmoniformes, Salmonidae, Thymallus) in Zavkhan river (Mongolia). A BLASTn query of the 5.8S rDNA fragment detected 19 identical matches of species including Gyrodactylus mongolicus Ergens and Dulmaa, 1970 (OQ913866, OQ913868, OQ641769, OQ641768), Gyrodactylus cf. lagowskii (OQ672253), Gyrodactylus cf. konovalovi (OQ672250), Gyrodactylus cf. mantshuricusi (OQ672249, OQ672248), G. tayshirensis (OQ641774), Gyrodactylus zavkhanensis (OQ641773), Gyrodactylus nemachili Bikhovski, 1936 (OQ641772, OQ641771, OQ641770), Gyrodactylus pseudonemacheili Ergens and Bychowsky, 1967 (OQ641767, OQ641764, OQ641758, OQ641756), G. papernai Ergens and Bychowsky, 1967 (EF446729, AF484533).
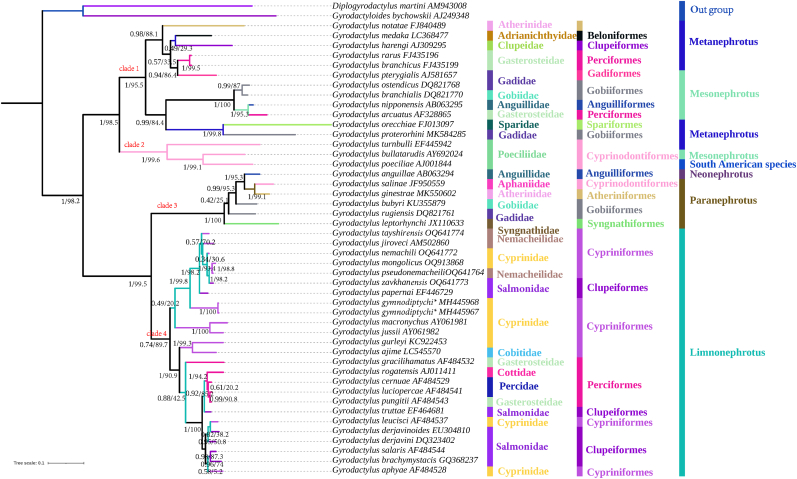
3.5. Phylogenetic analysis
We used BI and ML methods to topologically construct phylogenetic trees. As shown in Fig. 4, these two methods yielded two similar phylogenetic trees and only minor differences in statistical support values for some nodes. Both trees showed that all Gyrodactylus taxa were split into two major evolutionary lineages, and further divided into four clades. The first lineage consisted of two subgenera: clade 1 and clade 2. Clade 1 was composed of the subgenus Metaneprotus, which included six host families (Atherinidae, Adrianichthyidae, Clupeidae, Gasterosteidae, Sparidae, and Gadidae). Clade 2 consisted of the subgenus Mesonephrotus, which included four host families (Gadidae, Gobiidae, Sparidae, Gasterosteidae, and Poeciliidae). The second lineage consisted of two subgenera: clade 3 and clade 4. Clade 3 consisted of one species of Gyrodactylus in the subgenus Neonephrotus and five species of Gyrodactylus in the subgenus Paranephrotus. These species were found on hosts belonging to the families Anguillidae, Aphaniidae, Atherinidae, Gobiidae, Gadidae, and Syngnathidae. Additionally, G. gymnodiptychi n. sp. was discovered along with 23 species of Gyrodactylus in the subgenus Limnonephrotus, forming a sister group in clade 4. The nodes supporting this relationship were well supported in both the BI and ML trees. The hosts of Limnonephrotus species belonged to the families Nemacheilidae, Cyprinidae, Salmonidae, Cobitidae, Gasterosteidae, Cottidae, and Percidae. These phylogenetic trees commonly indicated that G. gymnodiptychi n. sp. was closely associated with several Gyrodactylus members isolated from fishes inhabited in the river of Russia, Mongolia and Czech Republic. A clade of G. gymnodiptychi n. sp. diverged first, then G. papernai, G. zavkhanensis, G. pseudonemacheili, G. mongolicus, G. nemachili, G. jiroveci and G. tayshirensis formed a clade.
Fig. 4.
Phylogenetic tree generated by the Bayesian Inference (BI) and Maximum Likelihood (ML) method based on ITS1-5.8S-ITS2 rDNA sequences of selected Gyrodactylus. Diplogyrodactylus martini and Gyrodactyloides bychowskii were used as the outgroup. The numbers at nodes indicate posterior probabilities and bootstrap branch support (%). Taxonomic identity were shown to the right: Family and Order of host fish, subgenus of Gyrodactylus. * Gyrodactylus gymnodiptychi in the phylogenetic tree.
4. Discussion
In this communication, we used morphological and molecular methods to identify a new species of parasite Gyrodactylus gymnodiptychi n. sp for the first time. The G. gymnodiptychi n. sp. was the only one showing a hollow dorsal bar and curved hamulus root that were distinct from the other eleven gyrodactylid members. Gyrodactylus gymnodiptychi n. sp. shared the same fish host G. dybowskii with two already known monogeneans G. tokobaevi and G. aksuensis.
The parasite fauna of fishes in theYili River is largely unknown. Although G. gymnodiptychi n. sp. shares the same host G. dybowskii with G. tokobaevi and G. aksuensis, our newly described species was isolated from G. dybowskii living in the Yili River, while G. tokobaevi and G. aksuensis inhabit in the unrelated Aksu River west of Frunze (Ergens and Karabekova, 1980). Eleven species of Gyrodactylus have been reported from subfamily Schizothoracinae, including G. hemivivinus Ergens and Daniyarov, 1976; G. kafirniganensis Ergens and Daniyarov, 1976; G. marjami Allamuratov and Gussev, 1969; G. montanus Bychowsky, 1957; G. narzikulovi Ergens and Dzhalilov, 1979; G. seravschani Osmanov, 1965; G. vicinus Bychowsky, 1957; G. aksuensis, 1980; G. tokobaevi; G. dzhalilovi Ergens and Ashurova, 1984; and G. editus Dzhalilov and Ashurova, 1980; Ergens and Karabekova (1980); Gusev, 1985). The studies revealed that in comparison with G. gymnodiptychi n. sp., G. narzikulovi and G. aksuensis, other 9 species showed relatively flat and straight hamuli roots (Ergens and Karabekova, 1980; Gusev, 1985). These species shared a feature of small ventral bar processes, except for G. gymnodiptychi n. sp. and G. tokobaevi, which had prominent ventral bar processes (Ergens and Karabekova, 1980; Gusev, 1985). In these species, our identified new G. gymnodiptychi n. sp. sharing the feature of a dorsal bar with a hollow at each end of the projection with G. montanus. Additionally, G. gymnodiptychi n. sp. was distinguishable from G. tokobaevi and G. montanus by its hollow at each end of dorsal bar and a curved hamulus root (Ergens and Karabekova, 1980; Gusev, 1985). The newly identified G. gymnodiptychi n. sp. was the only one carrying both a hollow at each end of dorsal bar and a curved hamulus root in contrast to other Schizothoracinae members carrying only one of the two features (Gusev, 1985).
The blast studies indicated that the ITS1-5.8S-ITS2 rDNAs of G. gymnodiptychi n. sp. isolated from the fish G. dybowskii were distinguishable from all other Gyrodactylus species listed in the GenBank. The constructed phylogenetic trees indicated the association of G. gymnodiptychi n. sp. with the subgenus Limnonephrotus. Two other subgenus members G. tayshirensis (OQ641774) and G. jiroveci (AM502860) shared high degrees of 92.19% and 92.05% homology of the ITS1-5.8S-ITS2 rDNA genes with G. gymnodiptychi n. sp., respectively; however, both of them are parasites of a G. dybowskii-unrelated genus host Barbatula identified in the Mongolia and Czech Republic (Přikrylová et al., 2008). Whether Barbatula members may host G. gymnodiptychi n. sp. remains to be determined.
In Xinjiang, eleven native members of the fish subfamily Schizothoracinae are Class I key-protected aquatic wild animals (Guo, 2012). Only the two members G. dybowskii and Schizthorax pseudaksaiensis are known hosts of monogeneans (Yao et al., 2013). The association of Schizothoracinae members and monogeneans is still largely unclear. Thus, it is important to advance our knowledge of monogeneans, such as Gyrodactylus members, which parasitize Schizothoracinae, in order to formulate effective methods for protecting Schizothoracinae members from monogenean-related diseases.
5. Conclusion
Newly investigated gyrodactylids from Gymnodiptychus dybowskii in Yili River are clearly distinguished from other members of the genus Gyrodactylus on the basis of morphological and genetic data. Gyrodactylus gymnodiptychi is proposed as a new species.
Funding
The present study was supported by the National Natural Science Foundation of China, China (Grants No. 31960737 and No. 31860738), the Natural Science Foundation of Xinjiang Uygur Autonomous Region, China (2021D01B56).
Availability of data and material
All data produced for this study are provided in the manuscript.
Authors contribution
Conception and Design, Sample Collection, Morphological analyses, Drew the morphological figures, Data analysis, and Manuscript Preparation: Wen-Run Zhang; Funding acquisition, Sample collection, Data curation, and Writing: Cui-Lan Hao; Sample collection: Kadirden Arken, Meng-Jie Rong, Sheng-Li Tian, Munira Kadir; Funding acquisition, Supervision: Cheng Yue. All authors read and approved the final version of the manuscript.
Data availability statement
The sequence data is uploaded to the NCBI GenBank and the raw sequences are available under the accession of MH445967 and MH445968.
Declaration of competing interest
The authors declared that they have no conflicts of interest to this work. We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Acknowledgments
We are grateful to Prof. HCR Wang for his critical scientific and editorial review of this manuscript.
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins C.G. The study of fish diseases. Trans. Am. Fish. Soc. 1901;30:82–89. [Google Scholar]
- Bakke T.A., Cable J., Harris P.D. The biology of gyrodactylid monogeneans: the "Russian-doll killers". Adv. Parasitol. 2007;64:161–376. doi: 10.1016/S0065-308X(06)64003-7. [DOI] [PubMed] [Google Scholar]
- Bakke T.A., Harris P.D., Cable J. Host specificity dynamics: observations on gyrodactylid monogeneans. Int. J. Parasitol. 2002;32:281–308. doi: 10.1016/s0020-7519(01)00331-9. [DOI] [PubMed] [Google Scholar]
- Benson D.A., Mark B., Lipman D.J., James O., Wheeler D.L. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno D.W., Collins C.M., Cunningham C.O., Mackenzie K. Gyrodactyloides bychowskii (Monogenea: Gyrodactylidae) from sea-caged Atlantic salmon Salmo salar in Scotland: occurrence and ribosomal RNA sequence analysis. Dis. Aquat. Org. 2001;45:191–196. doi: 10.3354/dao045191. [DOI] [PubMed] [Google Scholar]
- Bueno-Silva M., Boeger W.A. Neotropical Monogenoidea. 58. Three new species of Gyrodactylus (Gyrodactylidae) from Scleromystax spp. (Callichthyidae) and the proposal of COII gene as an additional fragment for barcoding gyrodactylids. Folia Parasitol. 2014;61:213–222. doi: 10.14411/fp.2014.028. [DOI] [PubMed] [Google Scholar]
- Bush A.O., Lafferty K.D., Lotz J.M., Shostak A.W. Parasitology meets ecology on its own terms: margolis et al. revisited. J. Parasitol. 1997;83:575–583. [PubMed] [Google Scholar]
- Cable J., Harris P.D., Tinsley R.C., Lazarus C.M. Phylogenetic analysis of Gyrodactylus spp. (Platyhelminthes: monogenea) using ribosomal DNA sequences. Can. J. Zool. 1999;77:1439–1449. [Google Scholar]
- Cable J., Oosterhout C.V., Barson N., Harris P.D. Gyrodactylus pictae n. sp. (Monogenea: Gyrodactylidae) from the Trinidadian swamp guppy Poecilia picta Regan, with a discussion on species of Gyrodactylus von Nordmann, 1832 and their poeciliid hosts. Syst. Parasitol. 2005;60:159–164. doi: 10.1007/s11230-004-6348-4. [DOI] [PubMed] [Google Scholar]
- Christison K.W., Shinn A.P., As J.G.V. Gyrodactylus thlapi n. sp. (monogenea) from Pseudocrenilabrus philander philander (weber) (cichlidae) in the okavango delta, Botswana. Syst. Parasitol. 2005;60:165–173. doi: 10.1007/s11230-004-6342-x. [DOI] [PubMed] [Google Scholar]
- Cone D.K., Appy R., Baggett L., King S., Gilmore S., Abbott C. A new gyrodactylid (Monogenea) parasitizing bay pipefish (Syngnathus leptorhynchus) from the Pacific Coast of North America. J. Parasitol. 2013;99:183–188. doi: 10.1645/GE-3224.1. [DOI] [PubMed] [Google Scholar]
- Cunningham C.O. Species variation within the internal transcribed spacer (ITS) region of Gyrodactylus (Monogenea:Gyrodactylidae) ribosomal RNA genes. J. Parasitol. 1997;83:215–219. [PubMed] [Google Scholar]
- Dávidová M., Jarkovsky J., Matějusová I., Gelnar M. Seasonal occurrence and metrical variability of Gyrodactylus rhodei itňan 1964 (Monogenea, Gyrodactylidae) Parasitol. Res. 2005;95:398–405. doi: 10.1007/s00436-005-1311-0. [DOI] [PubMed] [Google Scholar]
- Embody G.C. Notes on the control of Gyrodactylus on trout. Trans. Am. Fish. Soc. 1924;54:48–53. [Google Scholar]
- Ergens R., Karabekova D.U. Two new species of gyrodactylus (monogenea) from kirghizian Diptychus dybowskii (cypriniformes) Folia Parasitol. 1980;27:89–91. [Google Scholar]
- García-Vásquez A., Hansen H., Shinn A.P. A revised description of Gyrodactylus cichlidarum Paperna, 1968 (Gyrodactylidae) from the Nile tilapia, Oreochromis niloticus niloticus (Cichlidae), and its synonymy with G. niloticus Cone, Arthur et Bondad-Reantaso, 1995. Folia Parasitol. 2007;54:129–140. [PubMed] [Google Scholar]
- García Vásquez A., Pinacho Pinacho C.D., Martínez Ramírez E., Rubio Godoy M. Two new species of Gyrodactylus von Nordmann, 1832 from Profundulus oaxacae (Pisces: profundulidae) from Oaxaca, Mexico, studied by morphology and molecular analyses. Parasitol. Int. 2018;67:517–527. doi: 10.1016/j.parint.2018.03.003. [DOI] [PubMed] [Google Scholar]
- Gerard T., Jose C. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Gilmore S.R., Abbott C.L., Cone D.K. The placement of Gyrodactylus salmonis (Yin & Sproston) in the molecular phylogeny of studied members of the Gyrodactylus wageneri-group parasitizing salmonids. J. Fish. Dis. 2010;33:461–467. doi: 10.1111/j.1365-2761.2010.01154.x. [DOI] [PubMed] [Google Scholar]
- Guberlet J.E., Hansen W.A., Kavanagh J.A. vol. 2. University of Washington Publications in Fish eries; 1927. pp. 17–29. (Studies on the Control of Gyrodactylus). [Google Scholar]
- Guo Y. Xinjiang Science and Technology Press; Urumqi: 2012. The Fishes of Xinjiang. [Google Scholar]
- Guo Y., Hu W.G., Mo C., Wu F., Wang C.H., Ma D.C., He Y. An analysis of genetic diversity in Gymnodiptychus dybowskii based on mtDNA cytochrome b gene sequences. J. Dalian Ocean Univ. 2016;31:528–532. [Google Scholar]
- Gusev A.V. vol. 2. Leningrad Science Publishing Company; 1985. (Key to the Parasites of Freshwater Fish Fauna of the USSR). (in Russian) [Google Scholar]
- Harris P.D., Cable J. Gyrodactylus poeciliae n. sp. and G. milleri n. sp. (monogenea: Gyrodactylidae) from Poecilia caucana (steindachner) in Venezuela. Syst. Parasitol. 2000;47:79–85. doi: 10.1023/a:1006413804061. [DOI] [PubMed] [Google Scholar]
- Hayward C.J., Iwashita M., Ogawa K., Ernst I. Global spread of the eel parasite Gyrodactylus anguillae (monogenea) Biol. Invasions. 2001;3:417–424. [Google Scholar]
- Huyse T., Pampoulie C., Audenaert V., Volckaert F.A.M. First report of Gyrodactylus spp. (Platyhelminthes: monogenea) in the western Mediterranean Sea: molecular and morphological descriptions. J. Parasitol. 2006;92:682–690. doi: 10.1645/GE-690R.1. [DOI] [PubMed] [Google Scholar]
- Johnsen B.O., Jenser A.J. The Gyrodactylus story in Norway. Aquaculture. 1991;98:289–302. [Google Scholar]
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., Haeseler A.v., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S.D., Forest J.J., Cone D.K. Description of Gyrodactylus notatae n. sp. (monogenea: Gyrodactylidae) from menidia menidia (L.) (actinopterygii: Atherinidae) in nova scotia, Canada. Syst. Parasitol. 2009;74:23–27. doi: 10.1007/s11230-009-9185-7. [DOI] [PubMed] [Google Scholar]
- Kulemina I.V., Skarlato O. In: Investigations of Monogeneans in the USSR. Skarlato O.A., editor. AN SSSR; Leningrad: 1987. Size variability of the adhesive elements in some species of Gyrodactylus. [Google Scholar]
- Kvach Y., Ondračková M., Seifertová M., Hulak B. Gyrodactylus ginestrae n. sp. (monogenea: Gyrodactylidae), a parasite of the big-scale sand smelt, Atherina boyeri risso, 1810 (actinopterygii: Atherinidae) from the black sea. Parasitol. Res. 2019;118:3315–3325. doi: 10.1007/s00436-019-06483-8. [DOI] [PubMed] [Google Scholar]
- Lebedeva D., Ziętara M., Mendsaikhan B., Ermolenko A., Lumme J. Survivors from a pliocene climatic catastrophe: gyrodactylus (Platyhelminthes, monogenea) parasites of the relict fishes in the central asian internal drainage basin of Mongolia. Diversity. 2023;15:860. [Google Scholar]
- Letunic I., Bork P. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Li R.R., Li W.X., Wu X.D., Wang G.T. Identification of gyrodactylus species in goldfish (carassius auratus) through morphological study and the analysis of the rdna its sequence. Acta Hydrobiol. Sin. 2014;38:903–909. [Google Scholar]
- Lindenstrøm T., Collins C.M., Bresciani J., Cunningham C.O., Buchmann K. Characterization of a Gyrodactylus salaris variant: infection biology, morphology and molecular genetics. Parasitology. 2003;127:165–177. doi: 10.1017/s003118200300341x. [DOI] [PubMed] [Google Scholar]
- Lumme J., Ziętara M.S. Horizontal transmission of the ectoparasite Gyrodactylus arcuatus (Monogenea: Gyrodactylidae) to the next generation of the three-spined stickleback Gasterosteus aculeatus. Folia Parasitol. 2018;65:6. doi: 10.14411/fp.2018.006. [DOI] [PubMed] [Google Scholar]
- Matejusová I., Gelnar M., Verneau O., Cunningham C.O., Littlewood D. Molecular phylogenetic analysis of the genus Gyrodactylus (Platyhelminthes: monogenea) inferred from rDNA ITS region: subgenera versus species groups. Parasitology. 2003;127:603–611. doi: 10.1017/s0031182003004098. [DOI] [PubMed] [Google Scholar]
- Matejusová I., Gelnar M., McBeath A.J.A., Collins C.M., Cunningham C.O. Molecular markers for gyrodactylids (Gyrodactylidae: monogenea) from five fish families (Teleostei) Int. J. Parasitol. 2001;31:738–745. doi: 10.1016/s0020-7519(01)00176-x. [DOI] [PubMed] [Google Scholar]
- Meng W., Yang T., Liu Y., Halik M., Gao T. Comparative mitogenomic and phylogentic analyses of a schizothoracine fish, Gymnodiptychus dybowskii from two water systems in Xinjiang. Pakistan J. Zool. 2018;50:2119–2127. [Google Scholar]
- Minh B.Q., Nguyen M.A.T., Haeseler A.v. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta M. A new monogenean species, Gyrodactylus ajime n. sp. (Gyrodactylidae), parasitic on Niwaella delicata (Niwa), an endemic loach of Japan. Syst. Parasitol. 2021;98:307–319. doi: 10.1007/s11230-021-09979-z. [DOI] [PubMed] [Google Scholar]
- Nitta M., Nagasawa K. Gyrodactylus medaka n. sp. (Monogenea: Gyrodactylidae) parasitic on wild and laboratory-reared medaka Oryzias latipes (Beloniformes: Adrianichthyidae) in Japan. Parasitol. Int. 2018;67:651–658. doi: 10.1016/j.parint.2018.06.010. [DOI] [PubMed] [Google Scholar]
- Niu J.G., Zhang T., Liu H., Liu C.C., Zhang F., Cai L.G. Parasitic disease and its control in hatchlings of Gymnodiptychus dybowskii. Fish.Hebei. 2017;11:41–43. [Google Scholar]
- Niu Y.J., Ren D.Q., Chen S.A., Cai L.G., Niu J.G., Xie C.X. Growth characteristics of Gymnodiptychus dybowskii kessler in three tributaries of the Yili River in Xinjiang, China. J. Hydroecol. 2015;36:60–65. [Google Scholar]
- Paladini G., Cable J., Fioravanti M.L., Faria P.J., Di Cave D., Shinn A.P. Gyrodactylus orecchiae sp. n. (Monogenea: Gyrodactylidae) from farmed populations of gilthead seabream (Sparus aurata) in the Adriatic Sea. Folia Parasitol. 2009;56:21–28. doi: 10.14411/fp.2009.004. [DOI] [PubMed] [Google Scholar]
- Paladini G., Huyse T., Shinn A.P. Gyrodactylus salinae n. sp. (Platyhelminthes: monogenea) infecting the south European toothcarp Aphanius fasciatus (Valenciennes) (Teleostei, Cyprinodontidae) from a hypersaline environment in Italy. Parasites Vectors. 2011;4:100. doi: 10.1186/1756-3305-4-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Přikrylová I., Matějusová I., Jarkovský J., Gelnar M. Morphometric comparison of three members of the Gyrodactylus nemachili-like species group (Monogenea: Gyrodactylidae) on Barbatula barbatula L. in the Czech Republic, with a reinstatement of G. papernai Ergens & Bychowsky. Syst. Parasitol. 2008;69:33–44. doi: 10.1007/s11230-007-9106-6. 1967. [DOI] [PubMed] [Google Scholar]
- Přikrylová I., Matějusová I., Musilová N., Gelnar M., Harris P.D. A new gyrodactylid (Monogenea) genus on gray bichir, Polypterus senegalus (Polypteridae) from Senegal (West Africa) J. Parasitol. 2009;95:555–560. doi: 10.1645/GE-1652.1. [DOI] [PubMed] [Google Scholar]
- Ren M.L., Guo Y., Zhang Q.L., Li H., Adak, Cai L.G., Yong W.D., Ren B., Gao H. Heilongjiang Science and Technology Press; Heilongjiang: 1998. Fisheries Resources and Fishery of River Yili. [Google Scholar]
- Rokicka M., Lumme J., Ziętara M.S. Identification of Gyrodactylus ectoparasites in Polish salmonid farms by PCR-RFLP of the nuclear ITS segment of ribosomal DNA (Monogenea, Gyrodactylidae) Acta Parasitol. 2007;52:185–195. [Google Scholar]
- Rokicka M., Lumme J., Ziętara M.S. Two new Antarctic Gyrodactylus species (Monogenoidea): description and phylogenetic characterization. J. Parasitol. 2009;95:1112–1119. doi: 10.1645/GE-2002.1. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Shinn A.P., Hansen H., Olstad K., Bachmann L., Bakke T.A. The use of morphometric characters to discriminate specimens of laboratory-reared and wild populations of Gyrodactylus salaris and G. thymalli (Monogenea) Folia Parasitol. 2004;51:239–252. doi: 10.14411/fp.2004.029. [DOI] [PubMed] [Google Scholar]
- Stoyanov B., Huyse T., Pankov P., Georgiev B.B. Morphological and molecular identification of Gyrodactylus bubyri Osmanov, 1965 (monogenea: Gyrodactylidae) from caucasian dwarf goby, Knipowitschia caucasica (berg) (actinopterygii: gobionellidae) from a black sea lagoon. Parasitol. Res. 2016;115:1617–1625. doi: 10.1007/s00436-015-4899-8. [DOI] [PubMed] [Google Scholar]
- Trifinopoulos J., Nguyen L.T., Haeseler A.v., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams H.H. Some observations on the mass mortality of the freshwater fish Rutilus rutilus (L.) Parasitology. 1964;54:155–171. doi: 10.1017/s0031182000074448. [DOI] [PubMed] [Google Scholar]
- Yamaguti S. Interscience Publishers; New York, London: 1965. Systema Helminthum. Volume IV: Monogenea and Aspidocotylea. [Google Scholar]
- Yao W.J., Li W.X., Nie P. The helminth fauna of native Fisher from upstream Yili River. Acta Hydrobiol. Sin. 2013;37:168–171. [Google Scholar]
- Zahradníčková P., Barson M., Luus-Powell W.J., Přikrylová I. Species of Gyrodactylus von Nordmann, 1832 (Platyhelminthes: monogenea) from cichlids from Zambezi and Limpopo river basins in Zimbabwe and South Africa: evidence for unexplored species richness. Syst. Parasitol. 2016;93:679–700. doi: 10.1007/s11230-016-9652-x. [DOI] [PubMed] [Google Scholar]
- Zhang D., Gao F., Jakovli I., Zou H., Wang G.T. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- Ziętara M.S., Huyse T., Lumme J., Volckaert F.A. Deep divergence among subgenera of Gyrodactylus inferred from rDNA ITS region. Parasitology. 2002;124:39–52. doi: 10.1017/s0031182001008939. [DOI] [PubMed] [Google Scholar]
- Ziętara M.S., Kuusela J., Veselov A., Lumme J. Molecular faunistics of accidental infections of Gyrodactylus Nordmann, 1832 (Monogenea) parasitic on salmon Salmo salar L. and brown trout Salmo trutta L. in NW Russia. Syst. Parasitol. 2008;69:123–135. doi: 10.1007/s11230-007-9121-7. [DOI] [PubMed] [Google Scholar]
- Ziętara M.S., Lumme J. The crossroads of molecular, typological and biological species concepts: two new species of Gyrodactylus Nordmann, 1832 (Monogenea: Gyrodactylidae) Syst. Parasitol. 2003;55:39–52. doi: 10.1023/a:1023938415148. [DOI] [PubMed] [Google Scholar]
- Ziętara M.S., Rokicka M., Stojanovski S., Lumme J. Introgression of distant mitochondria into the genome of Gyrodactylus salaris: nuclear and mitochondrial markers are necessary to identify parasite strains. Acta Parasitol. 2010;55:20–28. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data produced for this study are provided in the manuscript.
The sequence data is uploaded to the NCBI GenBank and the raw sequences are available under the accession of MH445967 and MH445968.