Abstract
Background
Tumor antigenicity and efficiency of antigen presentation jointly influence tumor immunogenicity, which largely determines the effectiveness of immune checkpoint blockade (ICB). However, the role of altered antigen processing and presentation machinery (APM) in breast cancer (BRCA) has not been fully elucidated.
Methods
A series of bioinformatic analyses and machine learning strategies were performed to construct APM-related gene signatures to guide personalized treatment for BRCA patients. A single-sample gene set enrichment analysis (ssGSEA) algorithm and weighted gene co-expression network analysis (WGCNA) were combined to screen for BRCA-specific APM-related genes. The non-negative matrix factorization (NMF) algorithm was used to divide the cohort into different clusters and the fgsea algorithm was applied to investigate the altered signaling pathways. Random survival forest (RSF) and the least absolute shrinkage and selection operator (Lasso) Cox regression analysis were combined to construct an APM-related risk score (APMrs) signature to predict overall survival. Furthermore, a nomogram and decision tree were generated to improve predictive accuracy and risk stratification for individual patients. Based on Tumor Immune Dysfunction and Exclusion (TIDE) method, random forest (RF) and Lasso logistic regression model were combined to establish an APM-related immunotherapeutic response score (APMis). Finally, immune infiltration, immunomodulators, mutational patterns, and potentially applicable drugs were comprehensively analyzed in different APM-related risk groups. IHC staining was used to assess the expression of APM-related genes in clinical samples.
Results
In this study, APMrs and APMis showed favorable performances in risk stratification and therapeutic prediction for BRCA patients. APMrs exhibited more powerful prognostic capacity and accurate survival prediction compared to conventional clinicopathological features. APMrs was closely associated with distinct mutational patterns, immune cell infiltration and immunomodulators expression. Furthermore, the two APM-related gene signatures were independently validated in external cohorts with prognosis or immunotherapeutic responses. Potential applicable drugs and targets were mined in the APMrs-high group. APM-related genes were further validated in our in-house samples.
Conclusion
The APM-related gene signatures established in our study could improve the personalized assessment of survival risk and guide ICB decision-making for BRCA patients.
Keywords: Antigen processing and presentation machinery, Gene signatures, Breast cancer, Risk assessment, Gene mutation, Immunotherapy
1. Introduction
Breast cancer (BRCA) remains a significant threat to women's health and wellness, and its heterogeneity makes breast cancer classification therapy enter the era of precision treatment [1]. Although advances in early detection and treatment have reduced breast cancer mortality, almost all patients who develop the metastatic disease will die from it [2]. In addition, these patients often suffer from severe side effects of traditional treatments, including surgery, radiotherapy, chemotherapy and endocrine therapy. Considering the existing indicators are not sufficient to predict the patient's clinical outcomes and therapeutic responses, it is necessary to establish robust tools to further facilitate precision and individualized therapy.
Immunotherapy, represented by immune checkpoint blockade (ICB), has recently entered the cancer mainstream [3]. Recently, in the clinical trial Impassion 130, atezolizumab combined with nab-paclitaxel showed clinical benefit in patients with advanced programmed cell death ligand 1 (PD-L1) positive triple negative breast cancer (TNBC) [4]. With the success of immunotherapy, breast cancer, which was previously considered as "weak immunogenicity", also has entered the stage of immunotherapy [5]. Studies have shown that BRCA has extensive genomic alterations and a high tumor mutation burden (TMB) [6]. Subsequently, Mittendorf EA et al. pointed out that approximately 20 % of patients in TNBC, a subtype of breast cancer with an extremely poor prognosis, highly express PD-L1 [7]. Additionally, PD-L1 expression is also positively correlated with the density of tumor infiltrating lymphocytes (TILs) [8]. These findings provide a rationale for the assessment of immunotherapeutic approaches in BRCA patients. However, only a minority of patients respond to ICB, and there are currently no excellent biomarkers to identify which patients are more likely to respond to immunotherapy. Therefore, how to further improve the efficacy of immunotherapy is an urgent problem in clinical practice, although immunotherapy has significantly improved the prognosis for some patients with BRCA.
It is now well acknowledged that host recognition of cancer cells through the immune system forms an independent line of defense based on the ability to recognize and eliminate tumor cells, which is known as immunosurveillance [9,10]. CD8+ T cells reactive to tumor antigens are the primary mediators of anticancer immunity, and modulation of the CD8+ T cell response has become the main focus of cancer immunotherapies [11]. However, the tumor microenvironment is often immunosuppressive, leading to CD8+ T cell dysfunction and promoting tumor evasion of immune surveillance through various mechanisms [12]. Cancer rejection antigens are the targets of anticancer T cells [13]. Candidates for cancer rejection antigens include tumor-associated antigens (TAAs), viral antigens and tumor-specific antigens (TSAs), which are the targets of anticancer T cells such as CD8+ T cells [14]. To date, clinically effective CD8+ T cell responses appear to primarily recognize antigens derived from most TSAs, also known as neoantigens [15]. However, T cells cannot directly recognize tumor neoantigens. In humans, there are three main (and several minor) MHC class I molecules, also known as human leukocyte antigen (HLA) [16]. The primary function of MHC class I molecules is to present antigen peptides to CD8+ T cells for their immune surveillance activities [17]. This classical antigen presentation process, which is called the antigen processing and presentation machinery (APM), consists of four major steps: 1) antigen processing into short peptides; 2) peptide transport to the endoplasmic reticulum (ER); 3) assembly of the MHC Class I loading complex; 4) antigen presentation to CD8+ T cells [18]. To generate an effective antitumor response, tumor neoantigens must be directly presented by tumor cells, and must be taken up by professional antigen presenting cells (pAPCs), mainly dendritic cells (DCs), and finally presented to CD8+ T cells for recognition and killing [19]. Indeed, studies show that tumors have developed various mechanisms to limit MHC class I molecules presentation of antigens and evade immune recognition [18,20]. Moreover, aberrant expression or dysfunction of APM components affects the assembly of MHC class I peptide complexes and their eventual recognition by CD8+ T cells [21]. Recent researches reported that antigen presentation defects have already caused ICB-response failure [22,23].
In this study, we systematically measured antigen processing and presentation efficiency, and calculated the APM score using a single-sample gene set enrichment analysis (ssGSEA) algorithm based on the expression pattern of APM genes [24]. A set of BRCA-specific APM-related genes and two different APM-related clusters were identified. Then, we performed a series of bioinformatic and machine learning approaches to construct prognostic APM-related gene signatures and evaluate potential therapeutic responses. Differences in immune and mutational characterizations were further exhibited in different risk subgroups. Based on these comprehensive analyses, we hope our study could provide some new clues for improving therapeutic strategies and personalized management which could benefit the high-risk BRCA subsets.
2. Materials and methods
2.1. Data collection and preprocessing
Available transcriptome profiling data of BRCA samples and follow-up information were systematically searched in public databases. In this study, we included as many datasets as possible for data training and robust validation. Six microarray datasets with overall survival (OS) (GSE1456 [25], GSE20685 [26], GSE20711 [27], GSE42568 [28], GSE58812 [29] and METABRIC), five microarray datasets with recurrence-free survival (RFS) (GSE20711, GSE22219 [30], GSE45725 [31], GSE162228 [32] and METABRIC), and one RNA-seq dataset (TCGA-BRCA) were included in our study. In addition, the transcriptome profiling data and clinical outcomes of 25 stage IV melanoma patients who received immunotherapy were retrieved from GSE100797 [33].
RNA-seq data of TCGA-BRCA was obtained from The Cancer Genome Atlas (TCGA, https://portal.gdc. cancer.gov/) using R package “TCGAbiolinks”. All the raw CEL files and follow-up information of microarray datasets were obtained from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). Probe IDs were mapped to gene symbols according to the annotation file from each microarray platform. The maximal measurement was regarded as the final gene expression value when a gene symbol has multiple probes. All the microarray and RNA-seq data were normalized and log2 transformed following previously published literatures [34,35].
Somatic variants of TCGA-BRCA and METABRIC were called using MuTect2 and were sorted in the mutation annotation format (MAF) files. R package “maftools” was used to visualize the somatic mutation variants and frequency [36], and the function “somaticInteractions” was used to detect mutually exclusive or co-occurring set of genes, and “forestPlot” was used to compare the frequency of mutated genes between different risk groups.
2.2. Identification of BRCA-specific APM-related genes
The TCGA-BRCA dataset was used as the training cohort in this study. Single-sample gene set enrichment analysis (ssGSEA) and weighted gene co-expression network analysis (WGCNA) [37] were combined to screen for BRCA-specific APM-related genes. In detail, based on the APM gene signature reported by Wang et al. [22] and TCGA-BRCA RNA-seq data, the APM ssGSEA score was calculated for each BRCA sample with R package “GSVA” [24]. Subsequently, WGCNA algorithm was performed to construct a scale-free co-expression network, and a gene module which is mostly correlated with APM ssGSEA score was identified according to the correlation coefficients and corresponding significance. To verify the robustness of the BRCA-APM gene module, we performed Gene Ontology (GO) enrichment analysis to investigate the enriched signaling pathways of the BRCA-APM gene module using R package “clusterProfiler”. On the other hand, the non-negative matrix factorization (NMF) algorithm [38] was used to divide the TCGA-BRCA cohort into different clusters with an optimal factorization k value, and the fgsea algorithm was used to validate the alterations of immunity-related signaling pathways retrieved from the Molecular Signatures Database (MSigDB) between two BRCA-APM clusters [39].
2.3. Establishment of an APM-related risk score signature for survival prediction
Firstly, using R package “coxph”, Cox coefficients and p values were calculated for each BRCA-APM candidate gene in the TCGA-BRCA cohort, and the importance of candidate genes with p < 0.01 were further investigated using the random survival forest (RSF) algorithm [40]. A total of 1,000 decision trees were created in the RSF algorithm to ensure model stability. Secondly, candidate genes with relative importance > 0.4 were selected, and further submitted to the least absolute shrinkage and selection operator (Lasso) Cox regression analysis [41]. Lasso regularization adds a penalty parameter (λ) to the Cox model, and this action can lead to zero coefficients, which means some candidate genes are completely neglected for evaluation. Finally, an APM-related risk score (APMrs) signature for survival prediction was established based on the relative expression and Lasso Cox coefficient of individual genes as follows:
Furthermore, the prognostic value of the APMrs was validated in a series of independent cohorts including GSE1456, GSE20685, GSE20711, GSE42568, GSE58812, GSE22219, GSE45725, GSE162228 and METABRIC.
2.4. Construction of an APM-related signature for immunotherapy prediction
A computational method named Tumor Immune Dysfunction and Exclusion (TIDE) was used to estimate the response and resistance to ICB of the TCGA-BRCA samples [42]. Differentially expressed genes (DEGs) between responders and non-responders were identified with a threshold of adjusted p < 0.0001 based on reads count matrix and R package “DESeq2”. With a total of 1,000 decision trees, the random forest (RF) algorithm [43] was used to screen for the most important APM-related genes associated with the ICB response in the TCGA-BRCA cohort. A total of 23 candidate genes were overlapped in both mean decreased accuracy and Gini ranking methods, and then submitted to Lasso logistic regression model [44] to find out the most robust APM-related gene signature which is associated with ICB response.
An APM-related immunotherapeutic response score (APMis) signature was established to predict the immunotherapy response based on the relative expression and Lasso logistic coefficient of individual genes as follows:
An immunotherapy cohort (GSE100797) which contains 25 stage IV melanoma patients with clinical outcomes (immunotherapeutic response and progression-free survival) was used to validate the predictive capacity of APMis. Furthermore, the expression profiles of inhibitory and stimulatory immune checkpoints were investigated and compared in the APMis-low and -high groups of the validation cohort.
2.5. Single-cell RNA-sequencing (scRNA-seq) analysis
One scRNA-seq dataset named GSE176078 was obtained from the Gene Expression Omnibus (GEO), and the dataset GSE176078 includes scRNA-seq data (Chromium, 10X Genomics) of 26 primary BRCA tumors. The expression characteristics of specific genes in malignant, immune and stromal cells of BRCA were visualized using the Tumor Immune Single-Cell Hub (TISCH) database [45]. TISCH employed a global-scaling normalization method (“NormalizeData” function) in Seurat to scale the raw counts (UMI) in each cell to 10,000, and the expression of a gene in the cell was quantified as log2(TPM/10+1) to ensure relatively comparable.
2.6. Connectivity map (CMap) analysis
CMap is an open resource that applies transcriptome profiling data to probe the relationships between diseases and potential targets and to explore applicable drugs according to the dysregulation pattern of specific genes [10]. The differentially expressed genes (DEGs) between APMrs-low and -high groups were identified and submitted to CMap for analysis to explore potential targets and applicable drugs for high-risk BRCA patients. Top 30 compounds with the highest predictive scores and corresponding descriptions of mode-of-action (MoA) were displayed in a dot diagram. In addition, the representative compound-target pairs and the expression profile of these potential targets were shown in a Sankey plot.
2.7. Additional bioinformatic and statistical analyses
Three computational approaches including xCell, CIBERSORT and TIMER were applied to quantify the infiltration abundance of various immune cells based on the transcriptome profiling data of each sample [46]. The difference of cytolytic activity between APMrs-low and -high samples was compared with the expression profiles of three acknowledged markers GZMA, GZMB and PRF1 [47]. The differences of inhibitory immune checkpoints, stimulatory immune checkpoints, antigen presentation-related genes were also compared.
The Kaplan-Meier estimator was applied to depict survival curves, and the log-rank test was used to assess the survival difference between different subgroups, and the R package “survminer” was used to determine the best cut-off value with maximum statistics. Univariate and multivariate Cox regression analyses were stepwisely performed to evaluate the significance of each variable for prognosis. Independent risk factors for overall survival including age, stage and APMrs were included to construct an integrated survival decision tree to improve the risk stratification for BRCA patients using R package “rpart”, and its discriminative capacity was validated in the METABRIC cohort. To tailor an OS prediction model for individual patients, a nomogram was developed to predict the 1, 3, 5-year OS probability, and calibration curves were plotted to visualize the predictive accuracy of the nomogram. The receiver operating characteristic (ROC) analysis was used to evaluate the predictive accuracy of the APMis. The decision curve analysis (DCA) was performed to evaluate the 5-year overall survival net benefit for all the variables. Student's t-test was used to analyze differences between groups subject to a normal distribution. Two-sided p value or FDR q value < 0.05 was considered statistically significant. All analyses were performed in the SPSS Statistics 20.0, GraphPad Prism 8 and R 4.1.0 software.
2.8. Clinical samples and Immunohistochemistry (IHC)
We collected 30 breast cancer tissues from patients undergoing surgery at the First Affiliated Hospital of Nanjing Medical University, all of whom provided written consent. Our study was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (Ethics code 2022-SR-520).
For IHC staining, the tissues were paraffin-embedded, sliced into 5 μm sections, and deparaffinized and rehydrated. Antigen was retrieved in citrate buffer (pH 6.0) at 98 ℃ for 10 minutes. We used an IHC stain kit (KIT-9710; MXB Biotechnologies, China) and treated the continuous slices with endogenous enzymes blocking reagents and nonspecific blocking reagents before incubating them with antibodies against CD83 (1:500, Abcam) and NFKBIA (1:10000, Abcam) overnight. The next day, the slices were sequentially incubated with donkey anti-mouse/rabbit secondary antibodies and Streptomyces anti-biotin protein-peroxidase for 10 minutes. Chromogenic detection was performed using the DAB Detection Kit (DAB-2031, MXB Biotechnologies, China), and the sections were counterstained with hematoxylin.
We analyzed all slides using Aperio ImageScope software and assessed the density of positive cells per sample using Image Pro Plus. For positive cell counting, three representative, non-adjacent, non-overlapping fields of view were randomly selected and each core was magnified 400X. The density of positive cells was measured and averaged as cells/mm2. Finally, two independent pathologists examined the IHC staining.
3. Results
3.1. Identification of BRCA-specific APM-related genes
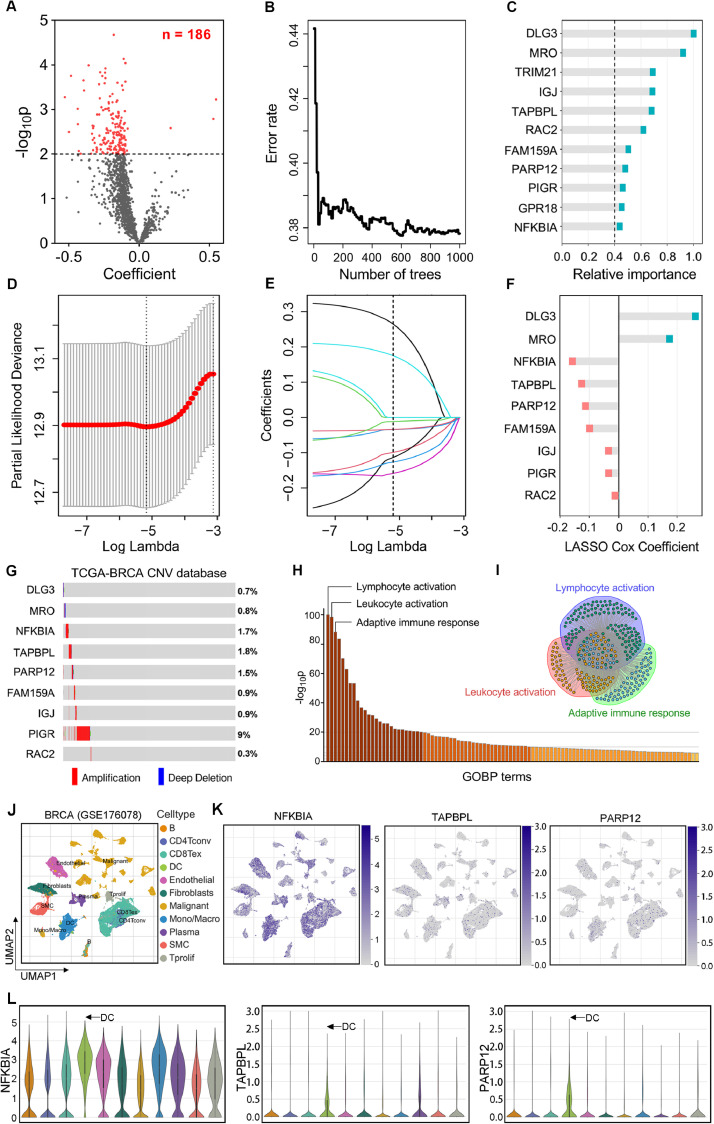
Based on a previously reported literature [22], we selected the following APM genes for quantification: B2M, CALR, CANX, ERAP1, ERAP2, HLA-A, HLA-B, HLA-C, PDIA3, PSMB5, PSMB6, PSMB7, PSMB8, PSMB9, PSMB10, TAP1, TAP2 and TAPBP. The ssGSEA approach was applied to quantify the APM score for each BRCA tumor tissue [24]. We observed that patients with lower APM exhibited worse OS in the TCGA-BRCA cohort (HR = 1.993, 95 % CI = 1.408 – 2.820, p < 0.001; Fig. S1), and the similar results were observed in progression-free survival (PFS) (HR = 1.736, 95 % CI = 1.218 – 2.475, p = 0.007) and cancer-specific survival (CSS) (HR = 2.188, 95 % CI = 1.373 – 3.488, p = 0.0061). Then, we performed WGCNA [37] on the transcriptome profiling data and APM ssGSEA scores to construct a scale-free co-expression network in the TCGA-BRCA cohort. A total of 36 gene modules were generated with a power of 7 as the optimal soft threshold (Fig. 1A & Fig. S2). The correlations between each gene module and the APM ssGSEA scores were analyzed by Pearson's coefficient, demonstrating the brown and darkgreen modules exhibited the highest correlation (r = 0.66, p = 2e-135 and r = 0.69, p = 4e-151) with APM (Fig. 1A). A total of 1665 genes involved in the two modules were considered as “APM-related module”, in which the module membership (MM) and gene significance (GS) revealed a highly positive correlation (r = 0.839, p < 0.001) (Fig. 1B). Subsequently, we submitted all the 1665 genes involved in the APM-related module to Gene Ontology for enrichment analysis. The results indicated that five most significant processes were annotated as leukocyte cell-cell adhesion, mononuclear cell differentiation, leukocyte cell-cell adhesion, regulation of T cell activation and T cell activation (Fig. 1C). NMF consensus clustering [38] was applied to classify the TCGA-BRCA cohort into two subgroups (cluster 1 (C1) and cluster 2 (C2), Fig. 1D) based on the expression matrix of the established 1665 APM related genes with the optimal NMF k value of 2 (Fig. S3). The boxplot showed that APM ssGSEA scores were significantly increased in C1 compared to C2 (p < 0.0001, Fig. 1E). To confirm the biological functions of these genes are related to APM in BRCA, we used all GOBP gene sets with a fgsea algorithm between the two distinct subgroups [39]. The top ten pathways enriched in the C1 with higher APM scores displayed dramatically higher activity of various immune processes, particularly T cell activation (Fig. 1F). Therefore, these results confirmed that the identified genes involved in APM-related modules can represent the characteristics of APM.
Fig. 1.
. A set of 1665 BRCA-specific APM-related genes were identified. (A) WGCNA was performed with transcriptome profiling data and APM ssGSEA scores in the TCGA-BRCA cohort to construct a scale-free co-expression network. A total of 36 non-grey modules were identified. The brown and darkgreen gene module exhibited the highest correlation with APM (r = 0.66, p = 2e-135 and r = 0.69, p = 4e-151) and were considered as “APM-related module”. (B) Scatter diagram depicted a highly positive correlation (r = 0.839, p < 0.001) between GS and MM in the “APM-related module”. (C) Gene Ontology for enrichment analysis was performed on 1665 genes involved in the “APM-related module”. Five most significant processes were displayed. (D) NMF consensus clustering was applied to divide the TCGA-BRCA cohort into two clusters based on the expression matrix of the established 1665 APM-related genes. (E) APM ssGSEA score was significantly elevated in cluster C1 compared to C2 (*** p < 0.001). (F) GSEA analysis based on fgsea algorithm demonstrated that C1 exhibited significantly higher activity of various immune processes compared with C2, particularly T cell activation. NES: normalized enrichment score; padj: adjusted p value.
3.2. Establishment of an APM-related gene signature for prognostic evaluation
Subsequently, the 1665 genes from the APM-related module were submitted to the univariate Cox proportional-hazards model. The volcano plot showed that 186 promising candidates were filtered out when the threshold of p value for Cox regression analysis was less than 0.01 (Fig. 2A). For further screening, the 186 genes were analyzed by the random survival forest (RSF) algorithm [40]. As the number of trees in the RSF model increases, the prediction error rate gets smaller, suggesting that the model becomes more robust as the forest structure becomes more complex (Fig. 2B). We selected genes with the criterion of relative importance greater than 0.4 as the optimal candidates. Ultimately, a total of 11 genes were identified and the important order for them is displayed in Fig. 2C. Subsequently, the most robust prognostic genes for OS were identified among the 11 candidate genes using the Lasso Cox regression algorithm [41]. 10-fold cross-validation was used to avoid overfitting, and an optimal λ value of 0.00566 was determined (Fig. 2D-E). A collection of 9 genes (DLG3, MRO, NFKBIA, TAPBPL, PARP12, FAM159A, IGJ, PIGR and RAC2) retained their respective Cox coefficients, and the distribution of corresponding coefficients for the gene signature is shown in Fig. 2F. Finally, the APMrs formula was developed to quantify the risk assessment of BRCA patients, with their individual gene expression value weighted by the LASSO Cox coefficients.
Fig. 2.
An APM-related risk score (APMrs) signature for OS was established. (A) The volcano plot showed that 186 promising candidates with p < 0.01 were identified among 1665 genes extracted from the “APM-related module”. (B) Random survival forest algorithm was applied to screen for the most important APM related genes correlated with OS. (C) Candidate genes with relative importance > 0.4 were selected and ranked. (D) 11 candidates entered into LASSO Cox regression model to identify the most robust genes correlated with OS. 10-fold Cross-validation was applied to overcome over-fitting effect. (E) An optimal λ value of 0.00566 was determined. (F) 9 genes were finally filtered with their individual Cox coefficients to establish a prognostic APM-related signature. (G) The oncoplot based on the cBioPortal database revealed the genomic alterations of the APMrs gene signature in TCGA-BRCA. (H) GO enrichment analysis was performed to explore the potential pathways underlying the established APMrs gene signature, and the three most important terms were labeled with “Lymphocyte activation”, “Leukocyte activation”, and “Adaptive immune response”. (I) Substantial genes were overlapped in the above-mentioned three biological processes. (J) The UMAP plot visualizes the distribution of 11 different cell categories in the BRCA TME. (K & L) The expression distribution of the three most important “protective” genes (NFKBIA, TAPBPL and PARP12) in the 11 cell clusters were shown in the UMAP and violin plots, and we observed that they are mainly enriched in the DC cells (arrow).
Subsequently, we used the cBioPortal database to reveal the genomic alterations of the APMrs gene signature in TCGA-BRCA. The oncoplot in Fig. 2G showed that somatic mutation rarely occurs in the gene signature, while copy number variation was frequently observed, especially in the PIGR gene. To explore the potential pathways underlying the established APMrs gene signature, DEGs between APMrs-low and -high samples were selected with a threshold p value of 0.0001, and GO enrichment analysis demonstrated that the three most important terms were labeled with “Lymphocyte activation”, “Leukocyte activation”, and “Adaptive immune response”, indicating the APMrs gene signature could reflect the dysfunction of immune response and induce the upstream regulation of leukocyte activation (Fig. 2H). Furthermore, we observed that substantial genes were overlapped in the above-mentioned three biological processes (Fig. 2I), which could partly interpret the critical role of the APMrs gene signature in the APM process and downstream immune cascade responses.
Based on the TISCH database, the UMAP plot was used to visualize the distribution of 11 different cell categories including B, CD4Tconv, CD8Tex, DC, endothelial, fibroblast, malignant, monocyte/macrophage, plasma, SMC and Tprolif cells in the BRCA TME (Fig. 2J). As shown in Fig. 2K,L, we assessed the expression patterns of the three most important “protective” genes named NFKBIA, TAPBPL and PARP12 in the 11 main cell clusters. We observed that all the three genes are enriched in the DC cells, which indicates that they are closely correlated with DCs activity and APM process.
3.3. Validation of APMrs in different independent BRCA cohorts
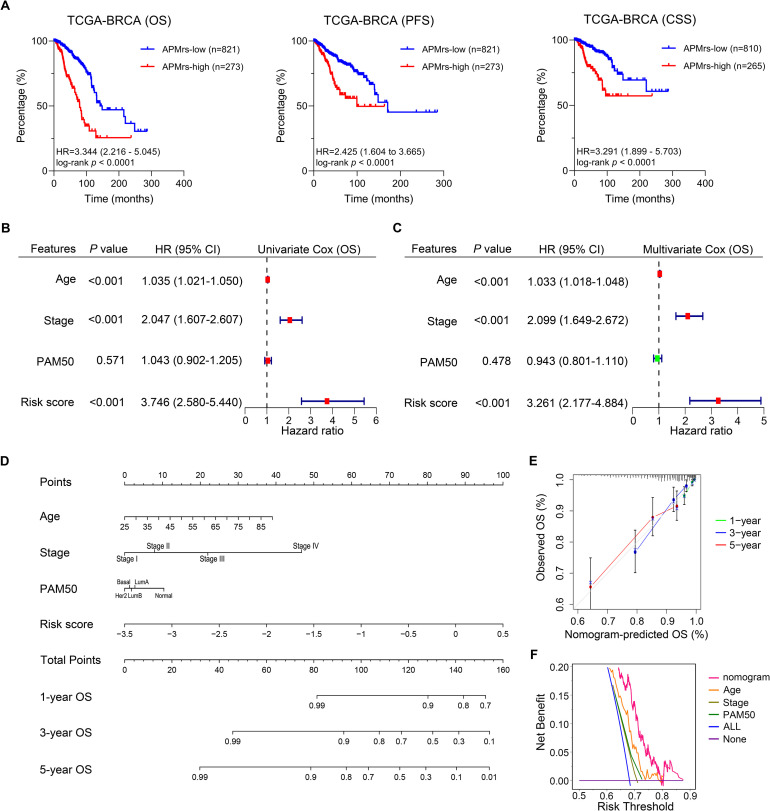
Next, we assessed the prognostic value of APMrs signature in BRCA patients. Kaplan-Meier analysis showed that patients with higher APMrs exhibited worse OS in the TCGA-BRCA cohort (HR = 3.344, 95 % CI = 2.216 – 5.045, p < 0.0001). Similar results were displayed in PFS (HR = 2.425, 95 % CI = 1.604 – 3.665, p < 0.0001) and CSS (HR = 3.291, 95 % CI = 1.899 – 5.703, p < 0.0001) (Fig. 3A). Furthermore, univariate and multivariate Cox regression analysis were performed and compare the prognostic capacity of APMrs and other traditional features including age, pathological stage and PAM50 subtypes (Basal-like, HER2 enriched, Luminal A, Luminal B and normal-like) in the pooled cohort. Three parameters (APMrs, age and stage) are independent risk factors for OS in TCGA-BRCA patients, and APMrs suggested the highest significance in univariate Cox regression analysis (HR = 3.746, 95 % CI = 2.580-5.440, p < 0.001; Fig. 3B) and multivariate Cox regression analysis (HR = 3.261, 95 % CI = 2.177-4.884, p < 0.001; Fig. 3C) among all the variables. To quantify risk assessment and predict 1-, 3- and 5-year OS probabilities for individual BRCA patients, a personalized scoring nomogram was constructed combining APMrs and other clinicopathological features (Fig. 3D). Calibration curves of 1-year (green line), 3-year (blue line) and 5-year (red line) OS prediction were close to the ideal performance (45-degree line), indicating the nomogram had a high OS accuracy prediction (Fig. 3E). In addition, the DCA analysis showed that the nomogram exhibited the highest 5-year overall survival net benefit when compared to other parameters including age, stage and PAM50 subtype (Fig. 3F).
Fig. 3.
The APM-related risk score (APMrs) signature could predict prognosis in TCGA-BRCA patients. (A) In the TCGA-BRCA cohort, Kaplan-Meier analysis demonstrated that patients with higher APMrs exhibited worse OS (HR = 3.344, 95 % CI = 2.216 – 5.045, p < 0.0001), worse PFS (HR = 2.425, 95 % CI = 1.604 – 3.665, p < 0.0001) and worse CSS (HR = 3.291, 95 % CI = 1.899 – 5.703, p < 0.0001). (B-C) Three parameters (APMrs, age and stage) are independent risk factors for overall survival in TCGA-BRCA patients, and APMrs exhibited the highest significance in univariate Cox regression analysis (HR = 3.746, 95 % CI = 2.580-5.440, p < 0.001) and multivariate Cox regression analysis (HR = 3.261, 95 % CI = 2.177-4.884, p < 0.001). (D) A nomogram was constructed to quantify risk assessment and predict OS probabilities for individual BRCA patients. (E) Calibration curves of survival prediction at different years (1-year: green line, 3-year: blue line, and 5-year: red line) were close to ideal performance (45-degree line). (F) DCA analysis showed that the nomogram exhibited the highest 5-year overall survival net benefit when compared to other parameters including age, stage and PAM50 subtype.
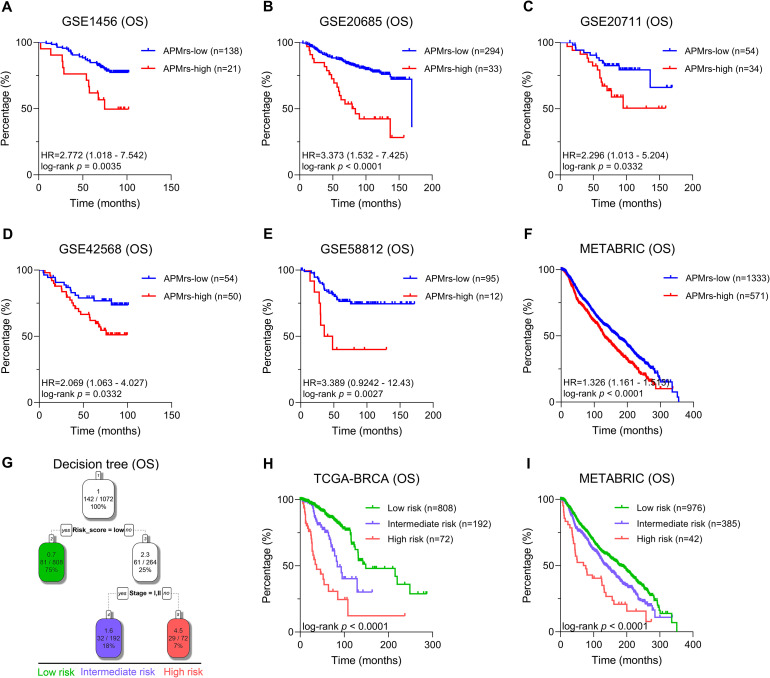
In addition, six datasets (GSE1456, GSE20685, GSE20711, GSE42568, GSE58812 and METABRIC) with overall survival information were used to validate the prognostic value of APMrs. Kaplan–Meier analysis demonstrated that BRCA patients with higher APMrs exhibited worse OS in each cohort (GSE1456: HR = 2.772, 95 % CI = 1.018–7.542, p = 0.0035; GSE20685: HR = 3.373, 95 % CI = 1.532–7.425, p < 0.0001; GSE20711: HR = 2.296, 95 % CI = 1.013–5.204, p = 0.0322; GSE42568: HR = 2.069, 95 % CI = 1.063–4.027, p = 0.0322; GSE58812: HR = 3.389, 95 % CI = 0.9242–12.43, p = 0.0027; METABRIC: HR = 1.326, 95 % CI = 1.161–1.513, p < 0.0001; Fig. 4A–F, respectively). Furthermore, we selected five datasets (GSE20711, GSE22219, GSE45725, GSE162228 and METABRIC) to validate the RFS prediction of APMrs. Fig. S4 showed similar results, with higher APMrs reflecting worse RFS (GSE20711: HR = 2.421, 95 % CI = 1.218–4.812, p = 0.0053; GSE22219: HR = 2.399, 95 % CI = 1.538–3.742, p = 0.0009; GSE45725: HR = 3.713, 95 % CI = 0.8807–15.66, p = 0.0039; GSE162228: HR = 8.955, 95 % CI = 1.029–77.90, p < 0.0001; METABRIC: HR = 1.248, 95 % CI = 1.066–1.461, p = 0.0038).
Fig. 4.
Validation and optimization of the APM-related risk score (APMrs) signature in BRCA patients. (A-F) Kaplan-Meier analysis demonstrated that patients with higher APMrs exhibited worse OS in the validation cohorts, such as GSE1456 (HR = 2.772, 95 % CI = 1.018–7.542, p = 0.0035), GSE20685 (HR = 3.373, 95 % CI = 1.532–7.425, p < 0.0001), GSE20711 (HR = 2.296, 95 % CI = 1.013–5.204, p = 0.0322), GSE42568 (HR = 2.069, 95 % CI = 1.063–4.027, p = 0.0322), GSE58812 (HR = 3.389, 95 % CI = 0.9242–12.43, p = 0.0027), and METABRIC (HR = 1.326, 95 % CI = 1.161–1.513, p < 0.0001). (G) An integrated survival decision tree was built to optimize the risk stratification for BRCA patients. (H-I) Significant differences of overall survival in TCGA-BRCA cohort (H, p < 0.0001) and in METABRIC cohort (I, p < 0.0001) were observed among different risk subgroups defined by the decision tree.
3.4. Construction and validation of a survival decision tree to improve risk stratification
Subsequently, we aimed to build an integrated prognostic model to improve risk stratification and risk assessment for BRCA patients. Considering APMrs, age and stage were independent risk factors for overall survival, we included the three parameters and built an integrated survival decision tree with the “rpart” R package based on the recursive partition analysis. As shown in the decision tree (Fig. 4G), three risk subgroups are defined based on two components including APMrs as the dominant branch along with pathological stage. Specifically, patients with low APMrs were defined as “low risk” group, whereas “intermediate risk” and “high risk” groups were defined as “High APMrs & stage I/II” and “High APMrs & stage III/IV”, respectively. Significant differences in OS were observed among the three risk subgroups (p < 0.001; Fig. 4H). The survival decision tree was further validated in the METABRIC cohort (p < 0.001; Fig. 4I), indicating its valuable discriminative capacity. Overall, the survival decision tree could greatly optimize risk stratification and survival prediction accuracy for BRCA patients.
3.5. Patterns of immune cell infiltration and immunomodulators expression in BRCA patients
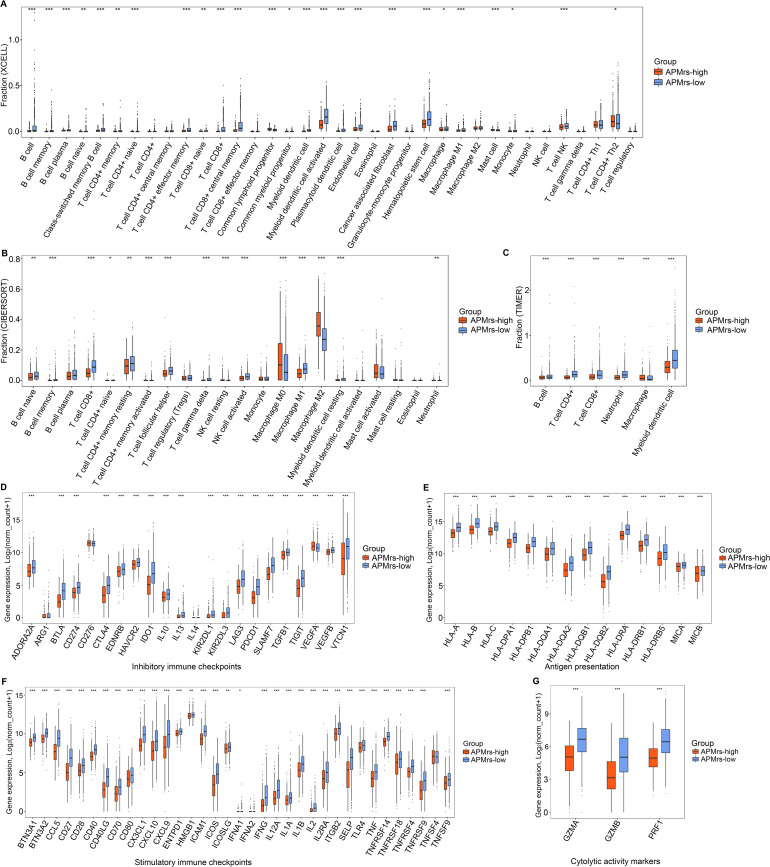
The presence of immune infiltration in tumors is a marker of favorable prognosis, especially in basal-like and HER2-positive breast cancer [48]. To further evaluate the infiltrating levels of various immune cells involved in BRCA, three algorithms including xCell, CIBERSORT and TIMER were performed [46]. Three stacked barplots represented by three different algorithms illustrated the distinct patterns of the relative abundance of 36, 22 and 6 immune cell types, and significantly distinct immune patterns were observed between APMrs-low and APMrs-high samples (Fig. 5A–C). Combining the three approaches, the APMrs-high group was closely associated with M0 Macrophages, M2 Macrophages and mast cells, corresponding to an immunosuppressive phenotype [49]. In contrast, the APMrs-low group showed increased infiltrations of CD8+ T cells, activated memory CD4+ T cells, follicular helper T cells, gamma delta T cells, activated myeloid dendritic cell, activated NK cells and M1 Macrophages, which were called the active-immune phenotype [50].
Fig. 5.
Patterns of immune cell infiltration and immunomodulators expression in BRCA patients. (A) Most of the 36 immune cell types were differentially distributed between the APMrs-high and -low group based on xCell algorithm. (B) Most of the 22 immune cell types were differentially distributed between the APMrs-high and -low group based on CIBERSORT algorithm. (C) All 6 immune cell types were differentially distributed between the APMrs-high and -low group based on TIMER algorithm. (D-E) Compared with APMrs-high group, representative immune regulatory molecules (including inhibitory immune checkpoints, stimulatory immune checkpoints, antigen presentation-related genes, and cytolytic activity markers) were significantly elevated in APMrs-low group. ***p < 0.001, **p < 0.01, *p < 0.05.
In addition, we compared the expression levels of four kinds of key immune regulatory molecules (including inhibitory immune checkpoints, stimulatory immune checkpoints, antigen presentation-related genes, and cytolytic activity markers) between two different APM risk groups. We observed that the APMrs-low group was marked by the significantly highest expression values of immunomodulatory molecules, particularly some classical immune checkpoints such as PDCD1, CD274, CTLA4, LAG3, TIGIT and TGFB1; while the APMrs-high group presented with the lowest expression levels (Fig. 5D–G). A study has identified that breast cancer with high expression of the antigen presentation-related genes such as HLA-A and HLA-B, has an increased immune T cell activation and favorable outcomes [20]. When CD8+ T cell activation or during clinical responses to anti-CTLA-4 and anti-PD-L1 immunotherapies, immune cytolytic activity (CYT) based on three key cytolytic effectors, granzyme A (GZMA), granzyme B (GZMB) and perforin (PRF1) are significantly upregulated [47]. These results demonstrated that the APMrs-low group may benefit more from immunotherapy.
3.6. Mutational landscapes in different risk groups of BRCA patients
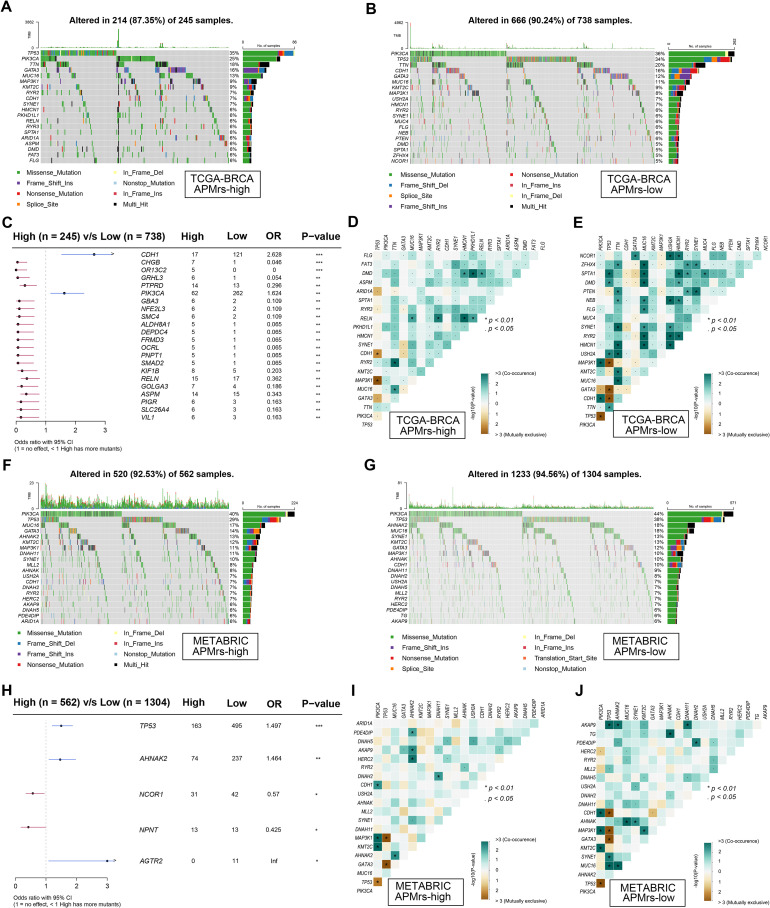
Considering immune infiltration is always strongly associated with mutation in solid tumors, we investigated the mutational landscape in different risk groups of BRCA. Oncoplots were respectively generated for the two different APM-related risk groups, and top 20 frequently mutated genes in each group were displayed (Fig. 6A,B). In detail, TP53, PIK3CA and TTN were the three genes with the highest mutation frequency in both groups. Significantly mutated genes were identified in the APMrs-low group compared to the APMrs-high group. As shown in a forest plot (Fig. 6C), CDH1 and PIK3CA are the two most frequently mutated genes in APMrs-low samples. In addition, the heatmaps illustrated the co-occurrence and mutually exclusive mutations of the top 20 frequently mutated genes in each cohort. More co-occurrence and mutually exclusive mutations could be observed in the APMrs-low group, which suggested that APMrs-low samples harbored more somatic mutations and higher immune infiltration in BRCA, thereby more likely to evoke immune responses and consequently have favorable prognosis (Fig. 6D,E). Similar results were observed in the METABRIC cohort. In detail, among the top 20 genes with mutation frequency in each group, PIK3CA and TP53 are two most frequently mutated genes (Fig. 6F-G). Compared to the APMrs-high group, TP53, AHNAK2 and AGTR2 are three most frequently mutated genes in APMrs-low group (Fig. 6H). Similarly, the APMrs-low group in METRBRIC displayed more events of co-occurrence and mutually exclusive mutations in comparison with the APMrs-high group (Fig. 6I,J). These evidences revealed distinct mutational landscapes in different APMrs groups.
Fig. 6.
Mutational landscape between different risk groups. In the TCGA-BRCA cohort, (A-B) Oncoplots were generated for different APM-related risk groups, and top 20 frequently mutated genes in each group were displayed. (C) Compared with APMrs-high group, CDH1 and PIK3CA are two most frequently mutated genes in APMrs-low group. (D-E) More co-occurrence mutations were observed in APMrs-low group when compared with APMrs-high group. In METABRIC cohort, (F-G) Oncoplots were generated for different APM-related risk groups, and top 20 frequently mutated genes in each group were displayed. (H) Compared with APMrs-high group, TP53, AHNAK2 and AGTR2 are three most frequently mutated genes in APMrs-low group. (I-J) More co-occurrence mutations were observed in APMrs-low group when compared with APMrs-high group. ***p < 0.001, **p < 0.01, *p < 0.05.
3.7. Establishment and validation of an APM-related signature for immunotherapy prediction
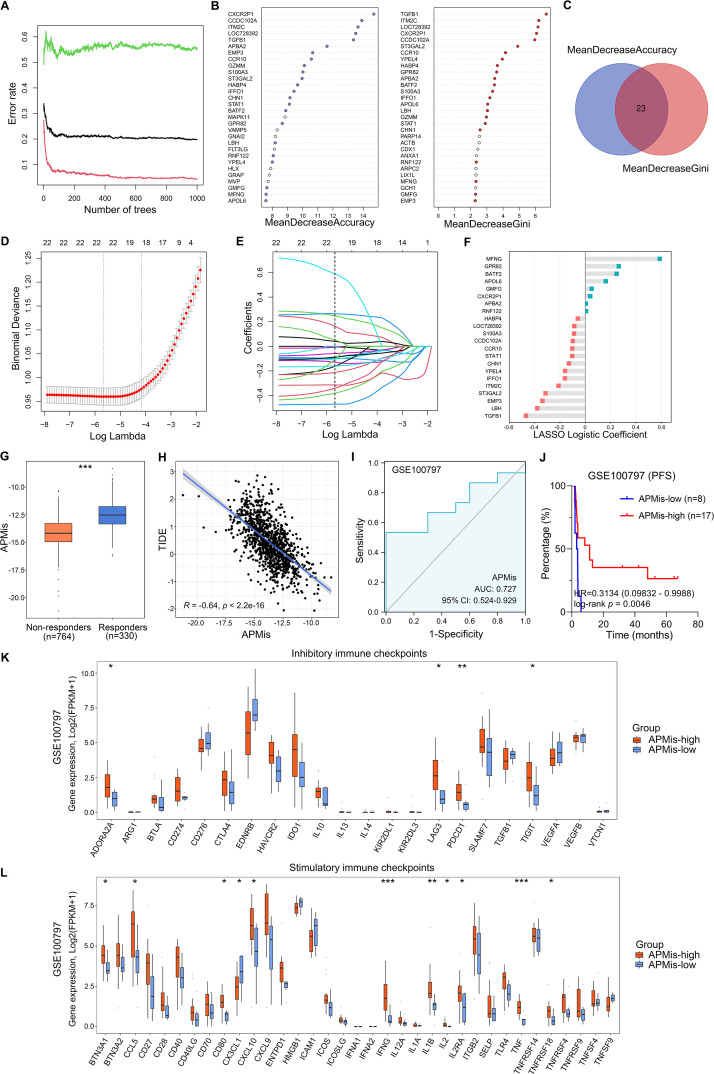
However, since the presence of immune infiltration does not imply immune function, we applied the TIDE algorithm and estimated potential ICB responses in all BRCA samples, and classified them into responders and non-responders [42]. Totally 348 DEGs between responders and non-responders were identified with a threshold of FDR q < 0.0001. Next, RF algorithm [43] was used to screen for the most significant genes associated with ICB response in the TCGA-BRCA cohort (Fig. 7A). The top 30 genes were ranked by importance according to the mean decreased accuracy and Gini ranking methods (Fig. 7B), and 23 overlapping genes were further selected (Fig. 7C). Subsequently, in the Lasso logistic regression analysis, 10-fold cross-validation was applied to overcome over-fitting effect (Fig. 7D), and an optimal λ value of 0.0035 was selected (Fig. 7E). Finally, a total of 22 genes (APBA2, APOL6, BATF2, CCDC102A, CCR10, CHN1, CXCR2P1, EMP3, GMFG, GPR82, HABP4, IFFO1, ITM2C, LBH, LOC728392, MFNG, RNF122, S100A3, ST3GAL2, STAT1, TGFB1, YPEL4) preserved their individual logistic coefficients (Fig. 7F).
Fig. 7.
Establishment and validation of an APM-related signature for immunotherapy prediction. (A) Random forest (RF) algorithm was used to screen for the most significant genes associated with ICB response in the TCGA-BRCA cohort. (B-C) Top 30 genes were ranked by importance according to the mean decreased accuracy and Gini ranking methods. And 23 genes showed overlap between the two ranking methods. (D-E) Lasso logistic regression analysis was further used to establish a robust signature to predict immunotherapeutic response. 10-fold cross-validation was applied to overcome over-fitting effect, and an optimal λ value of 0.0035 was selected. (F) 22 genes were finally screened with their individual logistic coefficients. (G) Compared with non-responders in the TCGA-BRCA cohort, the responders showed higher APMis. (H) TIDE scores were significantly negatively correlated with APMis in TCGA-BRCA samples (R = −0.64, p < 0.001). (I) APMis exhibited an AUC value of 0.727 to predict immunotherapeutic response in the training set GSE100797. (J) Kaplan–Meier analysis showed that patients with higher APMis exhibited better PFS in the GSE100797 (HR = 0.3134, 95 % CI = 0.09832–0.9988, p = 0.0046). (K-L) Compared with APMis-low group in the GSE100797, some representative immune checkpoints including LAG3, TIGIT, PDCD1, and IFNG were significantly elevated in APMis-high group. ***p < 0.001, **p < 0.01, *p < 0.05.
According to the established formula, the APM-related immunotherapeutic response score (APMis) was calculated for each sample based on the relative expression and Lasso logistic coefficient of individual genes. Fig. 7G showed that the responders in the TCGA-BRCA cohort exhibited higher APMis compared with non-responders. In addition, we correlated APMis with TIDE scores in all TCGA-BRCA patients and found a significant negative correlation (R = −0.64, p < 0.001, Fig. 7H). These results suggest that the lower the APMis, the higher the TIDE score, the greater the likelihood of immune dysfunction and immune escape, and the worse the immunotherapy effect. In the validation cohort GSE100797, APMis exhibited an AUC value of 0.727 (Fig. 7I) to predict immunotherapeutic response. Furthermore, the validation cohort is divided into APMis-low and -high groups, and the Kaplan–Meier analysis demonstrated that patients with higher APMis exhibited better progression-free survival (PFS) in the GSE100797 (HR = 0.3134, 95 % CI = 0.09832–0.9988, p = 0.0046; Fig. 7J). In addition, we compared the expression profiles of inhibitory and stimulatory immune checkpoints between the APMis-low and -high groups of the validation cohort. Compared with APMis-low samples, some representative immune checkpoints including LAG3, TIGIT, PDCD1, and IFNG were significantly elevated in APMis-high ones (Fig. 7K,L). In summary, APMis might serve as a promising biomarker for immunotherapy.
3.8. Screening of potential targets and applicable drugs for BRCA patients with high APMrs
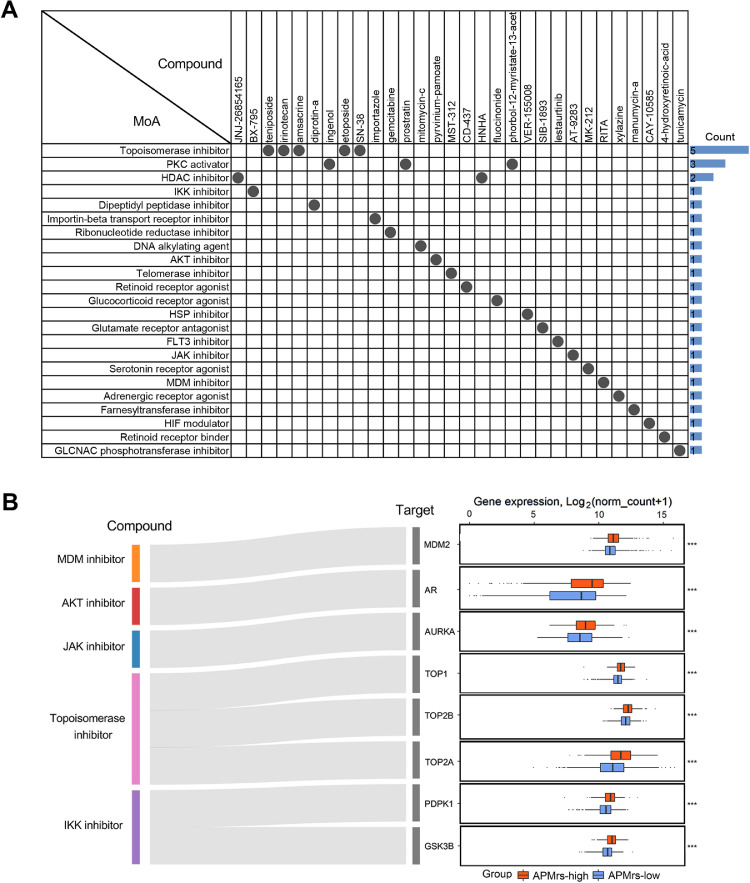
Recent polypharmacology studies demonstrated that compounds which are actionable toward more than one gene or molecular pathway should be valued [51]. A list of the top 150 dysregulated genes in APMrs-high groups was submitted to CMap to identify potential drugs applicable for APMrs-high patients. CMap mode-of-action (MoA) analysis revealed 23 mechanisms of action shared by 30 compounds with the highest prediction scores (Fig. 8A). In particular, five compounds, namely teniposide, irinotecan, amsacrine, etoposide and SN-38, shared the MoA of topoisomerase inhibitor. The Sankey diagram depicts the flow from representative compounds to potential targets and their expression profiles (Fig. 8B). All these target genes have specifically high expressions in APMrs-high BRCA samples, suggesting that targeting these genes with corresponding compounds might be a promising strategy for the personalized treatment of high-risk subsets.
Fig. 8.
Screening of potential targets and applicable drugs for BRCA patients with high APMrs. (A) CMap mode-of-action (MoA) analysis revealed 23 mechanisms of action shared by the top 30 compounds potentially applicable for APMrs-high patients. (B) The Sankey diagram showed the flow from representative compounds to potential targets and their expression profiles. ***p < 0.001.
3.9. Validation of APM-related genes in BRCA clinical samples
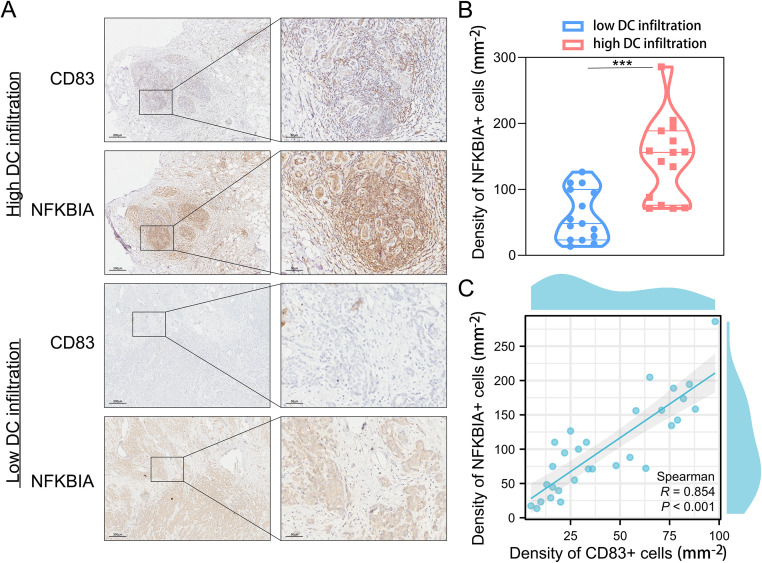
As shown in Fig. 2, we selected one of the most critical “protective” genes——NFKBIA in the APMrs for further clinical validation. Based on single-cell sequencing analysis, NFKBIA is predominantly enriched in DC cells (Fig. 2L), which is consistent with the correlation between APM-related genes and antigen presentation pathways. CD83, an acknowledged marker for DC cells, was selected to identify DC cells in the tissues. IHC staining was performed on 30 breast cancer tissues to determine the density of positive cells for NFKBIA and CD83 separately. According to the median density of CD83 positive cells, we divided clinical samples into high DC infiltration group and low DC infiltration group. The representative images of the expression of CD83 and NFKBIA in the two groups were displayed in Fig. 9A and co-localization of CD83 with NFKBIA was observed. Compared with low DC infiltration group, NFKBIA-positive cells expressed more in the high DC infiltration group (Fig. 9B). Spearman correlation analysis indicated a positive correlation between the density of CD83-positive cells and NFKBIA-positive cells (Fig. 9C). Therefore, the study provides clinical evidence to further establish the involvement of APMrs in the APM process regulated by DC cells.
Fig. 9.
The expression of CD83 and NFKBIA in BRCA clinical samples. (A) Representative images of CD83 staining and NFKBIA staining in the BRCA tissues, counterstained with DAPI (blue), in high DC infiltration group and low DC infiltration group. Scale bar, 200 μm (left panel); 50 μm (right panel). (B) The density of NFKBIA-positive cells is higher in high DC infiltration group than low DC infiltration group. ***p < 0.001. (C) The density of NFKBIA-positive cells is positively correlated with the density of CD83-positive cells (R=0.854, p < 0.001).
4. Discussion
Tumor immunogenicity, which is influenced by both tumor cell itself and the surrounding tumor microenvironment, acts as an important inherent feature of cancer and significantly affects the effectiveness of immunotherapy. Fundamental determinants of tumor immunogenicity include tumor antigenicity, and the efficiency of antigen processing and presentation [22]. Antigen presentation is accomplished through a series of intracellular events coordinated by multiple APM proteins [52]. Growing evidences have suggested that dysfunction of APM proteins were widely occurred in multiple cancer types, and the accompanied antigen presentation defects affect ICB effectiveness [42,53]. In the present study, we evaluated the tumor immunogenicity of breast cancer with a previously reported APM signature [22]. The APM signature and APM-induced risk score could predict prognosis of breast cancer patients, and significantly associated with distinct genomic alterations and immune infiltration. In addition, APMis, an immunotherapy response score established with multiple machine learning approaches serves as a favorable predictor for immunotherapy response.
The APM signature score was calculated based on the expression value of B2M, CALR, CANX, ERAP1, ERAP2, HLA-A, HLA-B, HLA-C, PDIA3, PSMB5, PSMB6, PSMB7, PSMB8, PSMB9, PSMB10, TAP1, TAP2 and TAPBP [22]. These APM proteins participate in the complex process of antigen presentation. In detail, tumor antigens are degraded by proteasomes and immunoproteasomes into short peptides, particularly PSMB8, PSMB9 and PSMB10. These peptides are then transferred to the ER with the help of the antigen-processing-associated transporter proteins (TAPs), which contain two non-covalently associated subunits TAP1 and TAP2. In the endoplasmic reticulum, MHC class I heavy chains and β2m assemble into the MHC Class I β2m complex under the synergistic action of the ER chaperones calpain, calreticulin, and the thiol oxidoreductase ERp57 (PDIA3). Trimmed by an ER-associated aminopeptidase (ERAP1) [54], these antigenic peptides are loaded on MHC class I β2m complexes, and then expressed on the cell surface by exocytosis via the Golgi apparatus, and subsequently recognized by CD8+ T cells. However, defects in the expression of MHC class I or proteins involved in the APM weaken the activity of antigen presentation, thereby promoting immune evasion. For example, reduction of MHC Class I expression was associated with reduced levels of CD8 T- cell infiltration in lung cancer [21].
In this study, we proposed a method to quantify APM based on the the transcriptome profiling data and specific genes involved in the APM process. Firstly, we identified a set of BRCA-specific APM-related genes which were closely associated with the differentiation, adhesion and activation of various immune cells within the tumor microenvironment. Based on the NMF algorithm, two distinct clusters (C1 and C2) were identified in the TCGA-BRCA cohort, and significantly different immunoreactivities were observed between the two clusters. Subsequently, a prognostic APM-related signature was established in the TCGA-BRCA cohort. Random survival forest algorithm and LASSO Cox regression models were combined to screen for the most robust candidate genes to establish the APM-related risk score (APMrs) signature for individual BRCA patients, and nine robust genes (DLG3, MRO, NFKBIA, TAPBPL, PARP12, FAM159A, IGJ, PIGR and RAC2) were finally identified. In multivariate Cox regression analysis, APMrs retained its prognostic capacity and accurate survival prediction after adjustment of clinicopathological confounders. Moreover, two integrated prognostic models (a scoring nomogram and a survival decision tree) were established to optimize survival risk stratification and prediction for BRCA patients. The discriminative capacity of the survival decision tree was further validated in the GEO dataset and METABRIC cohort. In addition, the landscapes of immune checkpoints, cytolytic activity signature and antigen presentation signature were further investigated in the two APM risk groups. We observed that immune infiltration (especially that of T cells) and most gene markers were significantly elevated in the APMrs-low group, suggesting differences of intrinsic tumor immunogenicity. Low APM risk score was also associated with more co-occurrence and mutually exclusive mutations, which reflects distinct mutational landscapes of driver genes and dysregulation of oncogenic pathways involved in tumorigenesis and progression. Taken together, these results suggested that APM-related risk score could serve as a reliable prognostic marker for BRCA patients.
Defective expression of MHC class I and APM proteins is common and has been reported to occur in 73 % to 90 % of patients, depending on tumor type [55]. Considering the important role of antigen presentation in antitumor immune responses, we hypothesized that defects in this pathway could result in poor response to ICB. To test this hypothesis, we constructed the immunotherapy score signature (APMis) with TIDE algorithm for BRCA patients who may benefit from ICB treatment. We found that patients with a higher APMis were more likely to respond to ICB in both breast cancer and melanoma patients who received immunotherapy, which suggested that APMis is a universal biomarker for immunotherapeutic prediction.
Furthermore, CMap database was employed to explore potential targets and applicable drugs for APMrs-high patients. Five topoisomerase inhibitors were highlighted with the highest prediction scores, and TOP1, TOP2A and TOP2B are common targets shared by the five compounds. Bai Y. et al have found that high expression of TOP1 and TOP2A were correlated to worse OS in epithelial ovarian carcinoma patients [56]. In non-small-cell lung cancer (NSCLC) patients, a relationship has been identified between topoisomerase isoforms and the clinicopathological features, such as grades, stages, chemotherapy and radiotherapy. Among the topoisomerase isoforms, TOP2A and TOP3A were associated with worse prognosis [57]. These evidences indicate that topoisomerase isoforms may also be potential targets in APMrs-high BRCA patients.
Although we comprehensively analyzed the immune landscape, mutational characterizations and treatment response in different APM subgroups, some limitations cannot be ignored in this study. First, although as many databases as possible were included in our study, the sampling bias could only be reduced but not completely eliminated. Second, our conclusions were induced from integrated analyses of clinical samples collected from public databases, and we only validated the expression of one gene in the APMrs model in in-house samples, therefore, in vivo and in vitro experiments are expected to verify the biological functions of more APM-related genes in BRCA tumorigenesis and progression. Third, since our study was based on many retrospective cohorts, prospective controlled studies with larger sample size are required for further validation.
In conclusion, our study indicated that APM-related genes could serve as useful markers in predicting clinical outcomes and therapeutic responses for breast cancer patients, if confirmed, has the potential to provide some new clues for improving therapeutic strategies which could benefit the high-risk BRCA subset patients.
Funding
This work was supported by grant from the National Natural Science Foundation of China (No. 81872365, No. 82203119), Natural Science Foundation of Jiangsu Province (No. BK20220733) and Key Research of Gusu School (GSKY20220105).
CRediT authorship contribution statement
Hong-yu Shen: Conceptualization, Formal analysis, Investigation, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. Jia-lin Xu: Conceptualization, Formal analysis, Investigation, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. Zhen Zhu: Conceptualization, Formal analysis, Investigation, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. Hai-ping Xu: Conceptualization, Formal analysis, Investigation, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. Ming-xing Liang: Conceptualization, Formal analysis, Investigation, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. Di Xu: Conceptualization, Formal analysis, Investigation, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. Wen-quan Chen: Conceptualization, Formal analysis, Investigation, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. Jin-hai Tang: Conceptualization, Formal analysis, Investigation, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. Zheng Fang: Conceptualization, Formal analysis, Investigation, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. Jian Zhang: Conceptualization, Formal analysis, Investigation, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2023.100942.
Contributor Information
Jin-hai Tang, Email: jhtang@njmu.edu.cn.
Zheng Fang, Email: fangzheng2021@njmu.edu.cn.
Jian Zhang, Email: dr_jianzhang@njmu.edu.cn.
Appendix. Supplementary materials
Data availability
All presented data and codes in this study are available from the corresponding author upon reasonable request.
References
- 1.Fitzmaurice C, Abate D, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study. JAMA Oncol. 2019;5:1749–1768. doi: 10.1001/jamaoncol.2019.2996. Global Burden of Disease Cancer C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emens LA. Breast cancer immunotherapy: facts and hopes. Clin. Cancer Res. 2018;24:511–520. doi: 10.1158/1078-0432.CCR-16-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paulson KG, Voillet V, McAfee MS, et al. Acquired cancer resistance to combination immunotherapy from transcriptional loss of class I HLA. Nat. Commun. 2018;9:3868. doi: 10.1038/s41467-018-06300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmid P, Adams S, Rugo HS, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 5.Stamm H, Oliveira-Ferrer L, Grossjohann EM, et al. Targeting the TIGIT-PVR immune checkpoint axis as novel therapeutic option in breast cancer. Oncoimmunology. 2019;8 doi: 10.1080/2162402X.2019.1674605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mittendorf EA, Philips AV, Meric-Bernstam F, et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol. Res. 2014;2:361–370. doi: 10.1158/2326-6066.CIR-13-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.AiErken N, Shi HJ, Zhou Y, et al. High PD-L1 expression is closely associated with tumor-infiltrating lymphocytes and leads to good clinical outcomes in chinese triple negative breast cancer patients. Int J Biol Sci. 2017;13:1172–1179. doi: 10.7150/ijbs.20868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Efremova M, Rieder D, Klepsch V, et al. Targeting immune checkpoints potentiates immunoediting and changes the dynamics of tumor evolution. Nat. Commun. 2018;9:32. doi: 10.1038/s41467-017-02424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J, Shi R, Zhang X, et al. Characterization of immune landscape in papillary thyroid cancer reveals distinct tumor immunogenicity and implications for immunotherapy. OncoImmunology. 2021;10 doi: 10.1080/2162402X.2021.1964189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beshnova D, Ye J, Onabolu O, et al. De novo prediction of cancer-associated T cell receptors for noninvasive cancer detection. Sci. Transl. Med. 2020;12:eaaz3738. doi: 10.1126/scitranslmed.aaz3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow MT, Ozga AJ, Servis RL, et al. Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity. 2019;50:1498. doi: 10.1016/j.immuni.2019.04.010. 512 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schietinger A, Philip M, Krisnawan VE, et al. Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity. 2016;45:389–401. doi: 10.1016/j.immuni.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng Q, Wu Y, Sui X, et al. POTN: a human leukocyte antigen-A2 immunogenic peptides screening model and its applications in tumor antigens prediction. Front. Immunol. 2020;11:02193. doi: 10.3389/fimmu.2020.02193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leisegang M, Engels B, Schreiber K, et al. Eradication of large solid tumors by gene therapy with a T-cell receptor targeting a single cancer-specific point mutation. Clin. Cancer Res. 2016;22:2734–2743. doi: 10.1158/1078-0432.CCR-15-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arora J, Pierini F, McLaren PJ, et al. HLA heterozygote advantage against HIV-1 Is driven by quantitative and qualitative differences in HLA allele-specific peptide presentation. Mol. Biol. Evol. 2020;37:639–650. doi: 10.1093/molbev/msz249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gornalusse GG, Hirata RK, Funk SE, et al. HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat. Biotechnol. 2017;35:765–772. doi: 10.1038/nbt.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leone P, Shin EC, Perosa F, et al. MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J. Natl. Cancer Inst. 2013;105:1172–1187. doi: 10.1093/jnci/djt184. [DOI] [PubMed] [Google Scholar]
- 19.Zamarin D, Postow MA. Immune checkpoint modulation: rational design of combination strategies. Pharmacol. Ther. 2015;150:23–32. doi: 10.1016/j.pharmthera.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Noblejas-Lopez MDM, Nieto-Jimenez C, Morcillo Garcia S, et al. Expression of MHC class I, HLA-A and HLA-B identifies immune-activated breast tumors with favorable outcome. Oncoimmunology. 2019;8 doi: 10.1080/2162402X.2019.1629780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JC, Davis C, Deshpande C, et al. Gene signature of antigen processing and presentation machinery predicts response to checkpoint blockade in non-small cell lung cancer (NSCLC) and melanoma. J. Immunother. Cancer. 2020;8 doi: 10.1136/jitc-2020-000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S, He Z, Wang X, et al. Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction. eLife. 2019;8 doi: 10.7554/eLife.49020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pawitan Y, Bjohle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao KJ, Chang KM, Hsu HC, et al. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer. 2011;11:143. doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dedeurwaerder S, Desmedt C, Calonne E, et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol. Med. 2011;3:726–741. doi: 10.1002/emmm.201100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke C, Madden SF, Doolan P, et al. Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis. 2013;34:2300–2308. doi: 10.1093/carcin/bgt208. [DOI] [PubMed] [Google Scholar]
- 29.Jezequel P, Loussouarn D, Guerin-Charbonnel C, et al. Gene-expression molecular subtyping of triple-negative breast cancer tumours: importance of immune response. Breast Cancer Res. 2015;17:43. doi: 10.1186/s13058-015-0550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buffa FM, Camps C, Winchester L, et al. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71:5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 31.Wang DY, Done SJ, Mc Cready DR, et al. Validation of the prognostic gene portfolio, clinicomolecular triad classification, using an independent prospective breast cancer cohort and external patient populations. Breast Cancer Res. 2014;16:R71. doi: 10.1186/bcr3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen YJ, Huang CS, Phan NN, et al. Molecular subtyping of breast cancer intrinsic taxonomy with oligonucleotide microarray and nanostring ncounter. Biosci. Rep. 2021;41 doi: 10.1042/BSR20211428. [DOI] [PMC free article] [PubMed] [Google Scholar]; BSR20211428.
- 33.Lauss M, Donia M, Harbst K, et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat. Commun. 2017;8:1738. doi: 10.1038/s41467-017-01460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi R, Bao X, Rogowski P, et al. Establishment and validation of an individualized cell cycle process-related gene signature to predict cancer-specific survival in patients with bladder cancer. Cancers (Basel) 2020;12 doi: 10.3390/cancers12051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi R, Bao X, Unger K, et al. Identification and validation of hypoxia-derived gene signatures to predict clinical outcomes and therapeutic responses in stage I lung adenocarcinoma patients. Theranostics. 2021;11:5061–5076. doi: 10.7150/thno.56202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung SY, Papp JC, Sobel EM, et al. Breast cancer risk and insulin resistance: post genome-wide gene-environment interaction study using a random survival forest. Cancer Res. 2019;79:2784–2794. doi: 10.1158/0008-5472.CAN-18-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J, Zhao T, Zhao D, et al. Development and validation of a hypoxia-related gene signature to predict overall survival in early-stage lung adenocarcinoma patients. Ther. Adv. Med. Oncol. 2020;12 doi: 10.1177/1758835920937904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bao M, Shi R, Zhang K, et al. Development of a membrane lipid metabolism-based signature to predict overall survival for personalized medicine in ccRCC patients. EPMA J. 2019;10:383–393. doi: 10.1007/s13167-019-00189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, Jiang L, Zhao R, et al. MiRNA-based model for predicting the TMB level in colon adenocarcinoma based on a LASSO logistic regression method. Medicine. 2021;100:e26068. doi: 10.1097/MD.0000000000026068. Baltimore. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun D, Wang J, Han Y, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49:D1420–D1D30. doi: 10.1093/nar/gkaa1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu ZY, Zhao M, Chen W, et al. Analysis of prognostic genes in the tumor microenvironment of lung adenocarcinoma. PeerJ. 2020;8:e9530. doi: 10.7717/peerj.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salgado R, Denkert C, Demaria S, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen YP, Wang YQ, Lv JW, et al. Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: implications for immunotherapy. Ann. Oncol. 2019;30:68–75. doi: 10.1093/annonc/mdy470. [DOI] [PubMed] [Google Scholar]
- 50.Sacher AG, St Paul M, Paige CJ, et al. Cytotoxic CD4(+) T cells in bladder cancer-a new license to kill. Cancer Cell. 2020;38:28–30. doi: 10.1016/j.ccell.2020.06.013. [DOI] [PubMed] [Google Scholar]
- 51.Medvedev A, Moeser M, Medvedeva L, et al. Evaluating biological activity of compounds by transcription factor activity profiling. Sci. Adv. 2018;4:eaar4666. doi: 10.1126/sciadv.aar4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cathro HP, Smolkin ME, Theodorescu D, et al. Relationship between HLA class I antigen processing machinery component expression and the clinicopathologic characteristics of bladder carcinomas. Cancer Immunol. Immunother. 2010;59:465–472. doi: 10.1007/s00262-009-0765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desbois M, Udyavar AR, Ryner L, et al. Integrated digital pathology and transcriptome analysis identifies molecular mediators of T-cell exclusion in ovarian cancer. Nat. Commun. 2020;11:5583. doi: 10.1038/s41467-020-19408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang SC, Momburg F, Bhutani N, et al. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a "molecular ruler" mechanism. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17107–17112. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai L, Michelakos T, Yamada T, et al. Defective HLA class I antigen processing machinery in cancer. Cancer Immunol. Immunother. 2018;67:999–1009. doi: 10.1007/s00262-018-2131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bai Y, Li LD, Li J, et al. Targeting of topoisomerases for prognosis and drug resistance in ovarian cancer. J. Ovarian Res. 2016;9:35. doi: 10.1186/s13048-016-0244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hou GX, Liu P, Yang J, et al. Mining expression and prognosis of topoisomerase isoforms in non-small-cell lung cancer by using Oncomine and Kaplan-Meier plotter. PLOS One. 2017;12 doi: 10.1371/journal.pone.0174515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All presented data and codes in this study are available from the corresponding author upon reasonable request.