Abstract
Acute kidney injury (AKI) presents a daunting challenge with limited therapeutic options. To explore the contribution of N6-methyladenosine (m6A) in AKI development, we have investigated m6A-modified mRNAs within renal tubular cells subjected to injuries induced by diverse stressors. Notably, while the overall level of m6A-modified RNA remains unaltered in renal tubular cells facing stress, a distinct phenomenon emerges—mRNAs bearing m6A methylation exhibit a pronounced tendency to accumulate within stress granules (SGs), structures induced in response to these challenges. Cumulation of m6A-modified mRNA in SGs is orchestrated by YTHDF1, a m6A ‘reader’ closely associated with SGs. Strikingly, AKI patients and various mouse AKI models showcase elevated levels of renal tubular YTHDF1. Depleting YTHDF1 within renal tubular cells leads to a marked reduction in m6A-modified mRNA accumulation within SGs, accompanied by an escalation in cell apoptosis under stress challenges. The significance of YTHDF1's protective role is further underscored by findings in AKI mouse models triggered by cisplatin or renal ischemia-reperfusion treatments. In particular, renal tubular-specific YTHDF1 knockout mice exhibit heightened AKI severity when contrasted with their wild-type counterparts. Mechanistic insights reveal that YTHDF1 fulfills a crucial function by safeguarding m6A-modified mRNAs that favor cell survival—exemplified by SHPK1—within SGs amid stress-challenged renal tubular cells. Our findings collectively shed light on the pivotal role of YTHDF1 in shielding renal tubules against AKI, through its adeptness in recruiting and preserving m6A-modified mRNAs within stress-induced SGs.
Keywords: Acute kidney injury, N6-methyladenosine, Stress granule, Cell survival, YTHDF1
Graphical abstract
Highlights
-
•
Renal tubular YTHDF1 is upregulated in AKI patients and diverse mouse AKI models.
-
•
Suppression of YTHDF1 within renal tubular cells augments apoptosis under stress.
-
•
Renal tubular-specific YTHDF1-KO intensifies AKI compared to their WT counterparts.
-
•
YTHDF1 shields tubular cells of AKI via safeguarding m6A-modified mRNAs within SGs.
1. Introduction
Acute kidney injury (AKI) is a serious clinical problem associated with high morbidity and mortality worldwide [1]. It has been widely reported that such a sudden decline in kidney function represented by AKI can be caused by renal ischemia-reperfusion (I/R), [2] sepsis [3] or nephrotoxicity such as cisplatin [4]. The molecular basis of AKI development and progression is still little known, despite extensive research, and there are no well-established treatments for AKI available [5].
Epigenetics has recently been a key modulating mechanism in AKI [5]. As the most enriched modifications in mRNAs, N6-methyladenosine (m6A) modulates various physiological and pathological developments, as metabolism, inflammation and apoptosis [[6], [7], [8]]. Shen et al. confirmed that m6A has a critical function of AKI due to cisplatin [9]. Through analyzing m6A RNA extracted from cisplatin-induced AKI mice, Li et al. also reported global increase of m6A levels after cisplatin treatment and that methylated mRNAs mainly regulated metabolism [10]. In line with this, Xing et al. identified m6A reader YTHDF1 as a key contributor for renal fibrosis [11]. Das et al. showed that mRNAs with higher fraction of m6A recovered for translation following stress relief, suggesting that m6A modification may facilitate mRNA recovery [12]. However, how does m6A-modification facilitate the mRNA recovery for protein translation once stress relief remains unclear.
AKI is generally accompanied by renal tubular cell proliferation and death under various stress conditions [13,14]. Many organelles in renal tubular cells, particularly endoplasmic reticulum (ER) and mitochondria, are engaged in this process [[15], [16], [17]]. Accumulating damaged mitochondria in renal tubular cells can result in cell apoptosis. Livingston et al. reported that clearance of damaged mitochondria protects kidney under ischemic condition [18]. In line with this, disruption of ER-mitochondrial coupling also results in tubular cell injury [19]. Xie et al. found that reticulon-1A disengaged mitochondrial hexokinase-1 from voltage-dependent anion channel-1 (VDAC1), leading to activation of apoptotic pathways in tubular epithelial cells [19]. Renal tubular cell apoptosis modulated with reactive oxygen species (ROS) and ER stress, [20] while reduction of ROS and ER stress by l-carnitine protects renal tubular cells [21]. For a similar reason, soluble epoxide hydrolase is identified as an important factor causing renal tubular mitochondrial dysfunction [22]. Like other cells types, renal tubular cells have developed various mechanisms to survive in these insults. One of mechanisms is to actively initiate integrated stress response to terminate global protein translation via phosphorylating eIF2α [23]. For instance, regulating ER stress can protect kidney cells [24]. Another mechanism for tubular cell survival is to selectively store the mRNAs of many cell survival genes in stress granules (SGs), a class of RNA granules can protect mRNAs from degradation [25]. These cell survival genes, including ribosomal protein S6 kinase and PKC, etc., can rapidly recover for protein translation following stress relief [26]. An elegant study by Wang et al. found that SGs formation in stressed proximal tubular cells is critical for cell survival [25]. Their study further demonstrated that G3BP1, a core protein of SG [27], is fundamental for keeping homeostasis under pressure stimuli. However, the mechanism underlying its protective function against AKI in stressful environments remain largely unknown.
In the present study, we substantiated mechanisms of m6A and SGs in regulating renal tubular function. We found that renal tubular cell YTHDF1 mitigates AKI through selectively recruiting m6A-methylated mRNAs, many of which are essential for cell proliferation and survival, into SGs and preserving them under various stress challenges. Through this mechanism, these m6A-modified mRNAs of cell proliferation and survival genes can evade degradation and maintain transcript abundance post stress. By generating renal tubular-specific YTHDF1-knockout mice, we further validated the protective role of renal tubular YTHDF1 against mouse AKI induced by cisplatin or I/R treatment.
2. Materials and Methods
2.1. Human specimens
Kidney tissues were obtained from AKI patients diagnosed in Jinling Hospital (Nanjing, China). Non-tubular injury para-cancerous kidney tissues of kidney carcinoma were used as the control. Medical School of Nanjing University authorized this research performed based on the Declaration of Helsinki and written consent of each patient.
2.2. Mouse models
8-week-old male C57BL/6J and proximal renal tubules specific YTHDF1 knockout (cKO) mice (GemPharmatech, Nanjing, China) used. YTHDF1Flox/Flox homozygous mice (C57BL/6J genetic background) were generated by CRISPR/Cas9 system, and mated with genetical background identical Ggt-cre mice [28] to obtain YTHDF1 cKO mice. Mouse genomic DNA was extracted using One Step Mouse Genotyping Kit (Vazyme, PD101-01). Genetic identification primers were presented in Supplementary Table S1. All animals fed with lab diet were kept in SPF environments with standard temperature and humidity. The Science and Technology Ethics Committee of Nanjing University (IACUC-2107007) authorized this research based on the ARRIVE and the National Research Council's guidelines. AKI resulted of cisplatin single intraperitoneal injection (20 mg/kg) into 8-week-old male mice. Control mice received saline only. The I/R surgery was proceed as stated earlier [25]. Mice were injected sodium pentobarbital intraperitoneally and maintained body temperature at 37 °C on a homeothermic station. The skin and muscle were incised to locate the renal artery, which was rapidly blocked with a non-invasive miniature arterial clip. The left kidney was clamped for 10–30 min and then released for blood circulation for 10–30 min. The parameters of renal function were measured by a bilateral I/R model, with 30 min ischemia on both sides, released the arterial clamps and sutured wounds, to reperfusion for 24 h. The control group was not treated for ischemia and the other operations were the same.
2.3. Cell culture and treatment
HK2 cells were cultured in DMEM/F12 (Gibco, 11330032) in 37 °C/5% CO2. Primary tubule cells were isolated and cultured from normal male C57BL/6J mice, male YTHDF1Flox/Flox homozygous mice (YTHDF1-WT) or YTHDF1-cKO mice according to a protocol described previously [29]. Primary renal tubular cells were cultured with additional 15 mM HEPES, 50 nM hydrocortisone, 1 × MEM nonessential amino acids, 5 μg/ml insulin, 5 μg/ml transferrin, and 50 nM selenium. Cells were exposed to cisplatin (Sigma-Aldrich, PHR1624; 20 μM, 12 h), H2O2 (Sigma-Aldrich, 88597; 10 mM, 4 h), 2-DG (Sigma-Aldrich, D8375; 50 mM, 1 h), 3-PO (Sigma-Aldrich, SML1343; 50 μM, 3 h), CCCP (Sigma-Aldrich, C2759; 10 μM, 1 h), NaAsO2 (Sigma-Aldrich, S7400; 0.5 mM, 1 h) or heat shock (42 °C, 1 h) to generate stress granules, respectively. In cell transfection assay, transfection reagents utilized from Invitrogen (13778150) and Promega (E2311). The siRNA sequences summarized in Supplementary Table S1.
2.4. Immunofluorescence staining
Cells were treated according to the prescribed conditions, fixed (Beyotime Biotechnology, P0099), permeabilized (Sigma-Aldrich, T9284) and blocked with 5% BSA. Cells were separately incubated with primary antibodies, secondary antibodies and DAPI. Finally, microscope coverslips containing cells were blocked with antifade reagent (Invitrogen, P36930) to avoid quenching of fluorescent dyes in fluorescence microscopy experiments. The procedure for patient and mouse kidney tissue was similar. 4 μm frozen sections of patients or mice kidney tissue were taken for immunofluorescence staining. Antibodies used in immunofluorescence staining: anti-AQP1 (Santa Cruz Biotechnology, sc-25287, 1:200); F4/80 (D2S9R) Rabbit mAb (Cell Signaling, 70076S, 1:100); anti-G3BP1 (13057-2-AP,1:500), anti-YTHDF1 (17479-1-AP, 1:500), anti-YTHDF2 (24744-1-AP, 1:100) and anti-YTHDF3 (25537-1-AP, 1:100) were from Proteintech; anti-G3BP1 (ab56574, 1:200) and anti-m6A (ab151230, 1:100) were from Abcam. Secondary antibodies were all from Invitrogen (A-21206, A-21203, 1:1000). All fluorescence pictures captured by LSM 880 (Zeiss) or TCS SP8 multiphoton system (Leica). Pictures were analyzed by ZEN Black (Zeiss), Leica Application Suite X (Leica) or ImageJ 1.44p.
2.5. Isolation of SGs
SGs were isolated as previously described [30]. Briefly, stress-treated HK2 cells containing stress granules were washed with pre-cooled PBS and the following manipulations were carried out on ice. HK2 cells containing 1 ml of pre-cooled lysate were stocked 30 times for 30 s in a Dounce homogenizer following 2,000g for 5 min centrifugation separating the nucleus (Nuc) from the cytoplasm (cyt). The soluble fraction (Sol) and SGs were detached by centrifugation at 18,000g for 20 min. These fractions were assayed for m6A RNA methylation quantification analysis, RT-qPCR, transcriptome sequencing and Western blot.
2.6. Western blot
Cells lysed with RIPA with 1 mM PMSF (Beyotime Biotechnology, ST506) and 1 × protease and phosphatase inhibitor (Beyotime Biotechnology, P1046) on ice for 40 min. For animal tissue samples, tissue was sheared into small pieces and added with lysis buffer with the same composition. The supernatant was obtained by centrifugation at 12,000g for 10 min at 4 °C and analyzed for protein quantification. Equal quantities of proteins were separated on gels (GenScript, M00654) and transferred to PVDF membranes (Merck, IPVH00010). After blocking, the membranes were incubated with primary and secondary antibodies conjugated to HRP and imaged by a chemiluminescence system. These antibodies were applied to WB: anti-Lamin B1 (12987-1-AP, 1:1000), anti-YTHDF1 (17479-1-AP, 1:1000), anti-YTHDF2 (24744-1-AP, 1:1000), anti-YTHDF3 (25537-1-AP, 1:1000) and SPHK1 (P10670-1-AP, 1:1000) were from Proteintech; anti-G3BP1 (Abcam, ab56574, 1:1000) and anti-GAPDH (sc-25778, Santa Cruz Biotechnology, 1:5000). Secondary antibodies were all from Jackson ImmunoResearch. Images were analyzed by ImageJ 1.44p.
2.7. m6A RNA methylation assay
The amount of m6A RNA was detected by EpiQuik kit (Epigentek, P-9005). 200ug of total RNA was bound using a highly effective RNA binding solution. RNA with m6A modification was captured with a detection m6A antibody. The absorbance value at 450 nm read with an enzyme calibrator was in proportion to the level of m6A methylation in RNA.
2.8. RNA-sequencing, MeRIP-sequencing and RIP-sequencing
We used TRIzol to isolate and purify total RNA. Poly(A) RNA (Invitrogen, 25–61005) was acquired, fragmented (NEBNext, E6150S) and cDNA was synthesized (Invitrogen, 1896649). For MeRIP or RIP sequencing, dynabeats were mixed with anti-m6A (Synaptic Systems, 202003, 1:100) or anti-YTHDF1 (Proteintech, 17479-1-AP, 1:100) in fragmented RNA, respectively. Finally, three biological replicates carried out double-end sequencing with illumine Novaseq 6000 in PE150 mode according to standard protocol. A gene-wide peek scan was performed to obtain information on the location of the peeks on the genome and the length of the peeks. Volcanoes were plotted for all genes in the differential expression analysis. KEGG enrichment was characterized on the website (http://www.kegg.jp/kegg) and presented as bubble plots by ggplot2. The intersection of the element lists was calculated using the Venn Diagrams website. The motif analysis was carried out using MEME2 (http://meme-suite.org) and HOMER (http://homer.ucsd.edu/homer/motif).
2.9. RT-qPCR
RNA was prepared by TRIzol or TRIzol LS (Invitrogen, 15596018, 10296010) and synthesized cDNA (Vazyme, R323-01). Levels of mRNA were analyzed by RT-PCR system (Vazyme, Q321-02) (Roche, Basel, Switzerland). Primers were presented in Supplementary Table S1.
2.10. Apoptosis assay
Cell apoptosis analysis was detected by Annexin V-Alexa Fluor647/PI staining (Yeasen Biotechnology, 40304ES60). HK2 or primary tubule cells were collected by digestion with EDTA-free trypsin, stained by Annexin V-Alexa Fluor647 and PI protecting from light, and assayed by flow cytometry (Thermo Fisher Scientific, A24858) within 1 h.
2.11. Renal function and histology
Blood was collected from mice to assess renal function. Blood creatinine (BioAssay Systems, DICT-500) and BUN (Jiancheng, C013-2-1) were tested. PAS, H&E and TUNEL (Beyotime Biotechnology, C1088, C1098) staining were performed with 4um thick slices of kidney tissue with a microscope (BX53, Olympus Corporation, Tokyo, Japan) taking pictures. The level of damaged tubules was calculated by renal tubular injuries (such as tubular atrophy and tubular dilatation).
2.12. m6A RNA immunoprecipitation qPCR
m6A RNA immunoprecipitation was performed using riboMeRIP m6A Transcriptome Profiling Kit (RiboBio, C11051-1). 100 μg total RNA was extracted, fragmented into fragments of about 200 nt with RNA fragmentation buffer, mixed with m6A anyibody and dynabeats. The eluted RNA was detected by qPCR and calculated by normalization input.
2.13. RNA immunoprecipitation qPCR
1 × 107 cells were lysed containing protease phosphatase inhibitor and RNase inhibitor for 30 min. The supernatant RNA was reacted with beads and antibodies and purified for qPCR assays calculating normalization input. DYKDDDDK-Tag (D6W5B) rabbit mAb (Cell Signaling, 14793S, 1:50) and anti-YTHDF1 (Proteintech, 17479-1-AP, 4 μg) antibodies were used.
2.14. mRNA smFISH
Cells were lined with microscope coverslips and treated according to the specified conditions. Fixed, permeabilized and incubated with prehybridization solution (LGC, SMF-WA1-60) with 30% formamide (Invitrogen, 17899). Cells were incubated with hybridization buffer (LGC, SMF-HB1-10) containing 0.5 μM of RNA smFISH probes overnight at 37 °C protected from light. The following steps were similar to those in immunofluorescence staining except that the buffer used was prepared with RNase-free buffer and RNase inhibitors were added. All probes were labeled with Cy5 dye at the N-terminal of sequences and displayed in Supplemental Table S2.
2.15. Statistical analyses
Biological repetitions varied across experiments and presented in the Materials and Methods or Figure legends. GraphPad Prism 9.0 was utilized for analysis of data and generation of figures. Normal distribution of data measured by the Shapiro-Wilk test. The statistical significance was confirmed by unpaired t-test, Mann-Whitney test, one-way ANOVA or two-way ANOVA. Results were presented as mean ± SD of each group and were recognized as statistically significant if the P-value was less than 0.05.
3. Results
3.1. Accumulation of m6A-modified mRNAs in SGs of renal tubular cells under stress condition
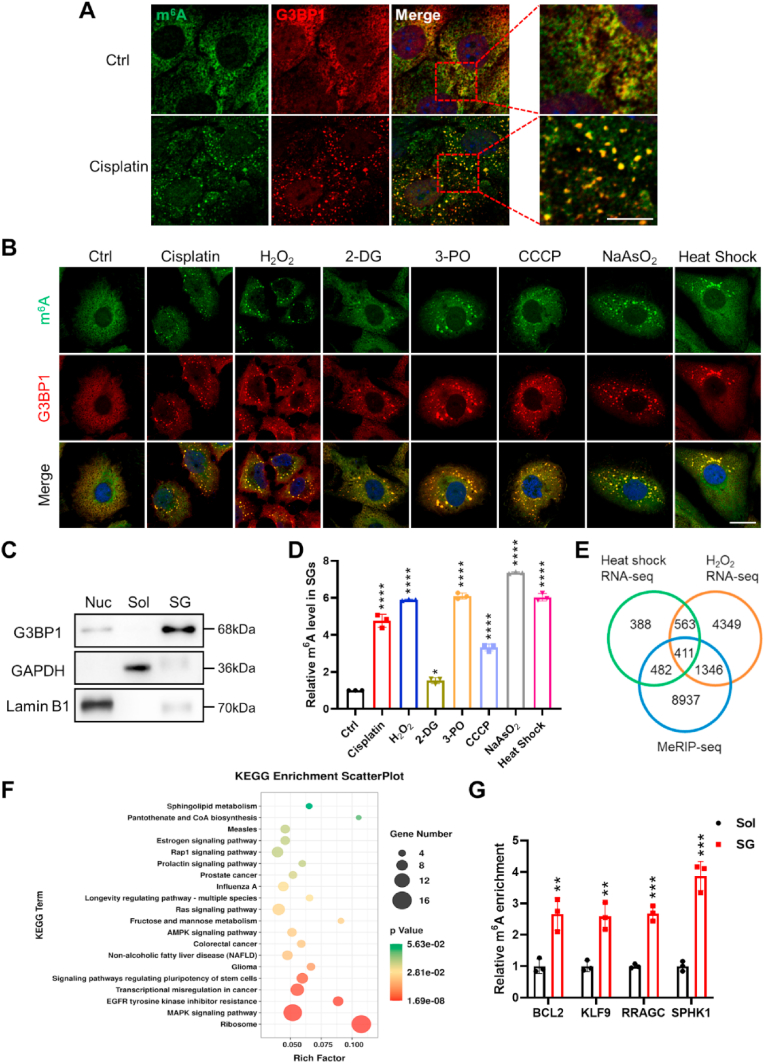
In a previous study, an increase in m6A-modified mRNAs within renal tubular cells was reported under conditions of acute kidney injury (AKI) induced by cisplatin treatment [31]. To delve into the role of m6A-modified mRNAs in modulating renal tubular cell function, we focused on mouse renal proximal tubular epithelial cells (PTECs), which were subjected to cisplatin treatment. As anticipated, the cisplatin treatment prompted the formation of numerous stress granules (SGs) that were marked by the G3BP1 antibody, a core protein of SGs within renal tubular cells (Fig. 1A, red). Remarkably, our findings revealed a prevailing co-localization of m6A-modified mRNAs (green) with G3BP1-labeled SGs in PTECs subsequent to cisplatin treatment (Fig. 1A). Strikingly, upon the removal of cisplatin from the PTECs, the aggregated SGs housing m6A-modified mRNAs exhibited a reduction in size, ultimately dispersing over time (Supplementary Fig. S1). Conversely, in the absence of cisplatin treatment, m6A-modified mRNAs were broadly disseminated across the cytoplasm of PTECs.
Fig. 1.
Accumulation of m6A-modified mRNAs in stress-induced SGs of renal tubular cells.
(A) Isolated mouse proximal tubular epithelial cells (PTECs) were subjected to treatment with or without cisplatin (20 μM, 12 h), followed by labeling with m6A antibody (green), G3BP1 antibody (red), and DAPI (blue), respectively. Scale bar, 5μm.
(B) HK2 cells were exposed to various stressors: cisplatin (20 μM, 12 h), H2O2 (10 mM, 4 h), 2-DG (50 mM, 1 h), 3-PO (50 mM, 3 h), CCCP (10 mM, 1 h), NaAsO2 (0.5 mM, 1 h) and heat shock (42 °C, 1 h). Scale bar, 20μm.
(C) SGs were isolated from H2O2-treated HK2 cells and subjected to Western blot analysis. Subcellular fractions included nucleus (Nuc), soluble cytosolic (Sol) and stress granule (SG) (three biological replicates).
(D) The enrichment of m6A-modified mRNAs was observed in SGs isolated from stressed HK2 cells by an m6A ELISA. Quantitative data were obtained from three biological replicates and analyzed using one-way ANOVA.
(E) Overlapping analysis of genes identified through RNA-sequencing and m6A-sequencing techniques.
(F) KEGG pathway analysis of the 411 genes mentioned in panel (E).
(G) Quantitative RT-qPCR was performed to assess m6A mRNA levels in Sol and SG fractions of H2O2-treated HK2 cells. Statistical analysis was conducted with three biological replicates using unpaired t-test.
For panels A and B, representative images were selected from a minimum of three biological replicates. All data are presented as mean ± SD. Statistical significance is denoted as *, P < 0.05, **, P < 0.01, ***, P < 0.001, and ****, P < 0.0001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To validate the selective accumulation of m6A-modified mRNAs in SGs of stress-challenged renal tubular cells, we exposed HK2 cells to various stress conditions, including cisplatin, [32] H2O2, [33] 2-deoxy-d-glucose (2-DG), [34] 3-(3-pyridinyl)-1-(4-pyridinyl)-2-propen-1-one (3-PO), [25] carbonyl cyanide m-chlorophenyl hydrazone (CCCP), [35] sodium arsenite (NaAsO2) [36] and heat shock [37]. Indeed, numerous SGs, labeled with G3BP1 (red), were formed in HK2 cells treated with cisplatin, H2O2, 2-DG, 3-PO, CCCP, NaAsO2 or heat shock (Fig. 1B). Conversely, in the absence of stress challenge (Ctrl), minimal SGs were observable. Similarly, under diverse stress conditions, m6A-modified mRNAs (highlighted in green) displayed a pronounced co-localization with G3BP1-tagged SGs in HK2 cells. In control HK2 cells, m6A-modified mRNAs were broadly dispersed throughout the cytoplasm. These findings collectively underscore the preferential accumulation of m6A-modified mRNAs within SGs in renal tubular cells subjected to diverse stressors. To substantiate this observation, we meticulously isolated the insoluble fraction (SG), the nuclear fraction (Nuc) and the soluble cytosolic fraction (Sol) from HK2 cells under distinct conditions (Fig. 1C). Subsequently, we evaluated the abundance of m6A-modified mRNAs specifically within the SG fraction. Consistent with the outcomes of the immunofluorescence labeling, the proportion of m6A-modified RNAs present within SGs demonstrated a marked escalation in response to stress challenges (Fig. 1D), while the overall cellular level of m6A-modified RNAs remained unaffected (Supplementary Fig. S2).
Subsequently, we delved into RNA sequencing analysis of RNAs extracted from SGs (Supplementary Fig. S3A) and applied MeRIP-sequencing analysis to HK2 cells. In alignment with preceding discoveries, [38,39] the typical m6A motifs were discernible within these m6A-modified mRNAs as evidenced by the MeRIP-sequencing data (Supplementary Fig. S3B). Notably, the coding sequences (CDS) and stop codons emerged as the primary regions of m6A enrichment (Supplementary Figs. S3C–D). Employing Venn analysis on genes obtained from HK2 cells under diverse stress scenarios, we identified a total of 411 m6A-modified genes congregating within SGs (Fig. 1E). These genes were predominantly associated with pathways involving ribosomes and MAPK signaling, as illuminated by KEGG pathway analysis (Fig. 1F). To confirm the enrichment of m6A-modified RNAs within SGs of stress-encountering HK2 cells, we performed RT-qPCR using specific m6A primers. We further examined the enrichment status of a selection of m6A-modified mRNAs—namely, BCL2, KLF9, RRAGC, and SPHK1—within the SGs and Sol fractions of HK2 cells challenged by H2O2 (Fig. 1G). Notably, the exposure to H2O2 significantly amplified the enrichment of these m6A-modified mRNAs within the SGs compared to the Sol fraction.
3.2. YTHDF1 protects renal tubular cells against AKI
Considering the accumulation of m6A-modified mRNAs within SGs during stress challenges in HK2 cells, we hypothesized that m6A readers might contribute to the rearrangement of mRNAs bearing m6A marks handling stressful conditions [40]. To explore this notion, we investigated the expression level and subcellular distribution of various m6A readers, namely YTHDF1-3, within HK2 cells experiencing stress challenges. The outcomes, as depicted in Fig. 2A, underscore that upon exposure to stressors such as cisplatin, H2O2 or CCCP, YTHDF1 and YTHDF3 were preferentially translocated to SGs, in contrast to YTHDF2. This trend of selective localization of YTHDF1 and YTHDF3 in SGs was further evident when HK2 cells were subjected to challenges involving 2-DG, 3-PO, NaAsO2 or heat shock (Fig. 2B–E). Given that YTHDF1 and YTHDF3 possess specific recognition capabilities for mRNAs bearing m6A modifications, their pronounced presence within SGs suggests a potential role in the accumulation of mRNAs possessing m6A marks within SGs amid stress-challenged conditions in HK2 cells.
Fig. 2.
Selective localization of m6A readers within stress-induced SGs in HK2 cells.
(A) Following exposure to stressors such as cisplatin (20 μM, 12 h), H2O2 (10 mM, 4 h) or CCCP (10 mM, 1 h), HK2 cells were subjected to immunolabeling with antibodies targeting G3BP1 (red) and m6A readers YTHDF1, YTHDF2 or YTHDF3 (green). Nuclei were counterstained with DAPI (blue).
(B) HK2 cells were stimulated with 2-DG (50 mM, 1 h). Immunolabeling was conducted using antibodies against YTHDF1/YTHDF3 (green) and G3BP1 (red), with nuclei staining by DAPI (blue).
(C) Cells were treated with 3-PO (50 mM, 3 h) followed by similar immunolabeling as mentioned earlier.
(D) HK2 cells were exposed to NaAsO2 (0.5 mM, 1 h), in which the subsequent immunolabeling utilized antibodies targeting YTHDF1/YTHDF3 (green) and G3BP1 (red). Nuclei were stained with DAPI (blue).
(E) Cells were treated with heat shock (42 °C, 1 h), where immunolabeling with YTHDF1/YTHDF3 (green) and G3BP1 (red) antibodies was performed, alongside DAPI staining for nuclei visualization.
Representative images in panels A to E were selected from a minimum of three independent biological replicates. Scale bars, 20 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
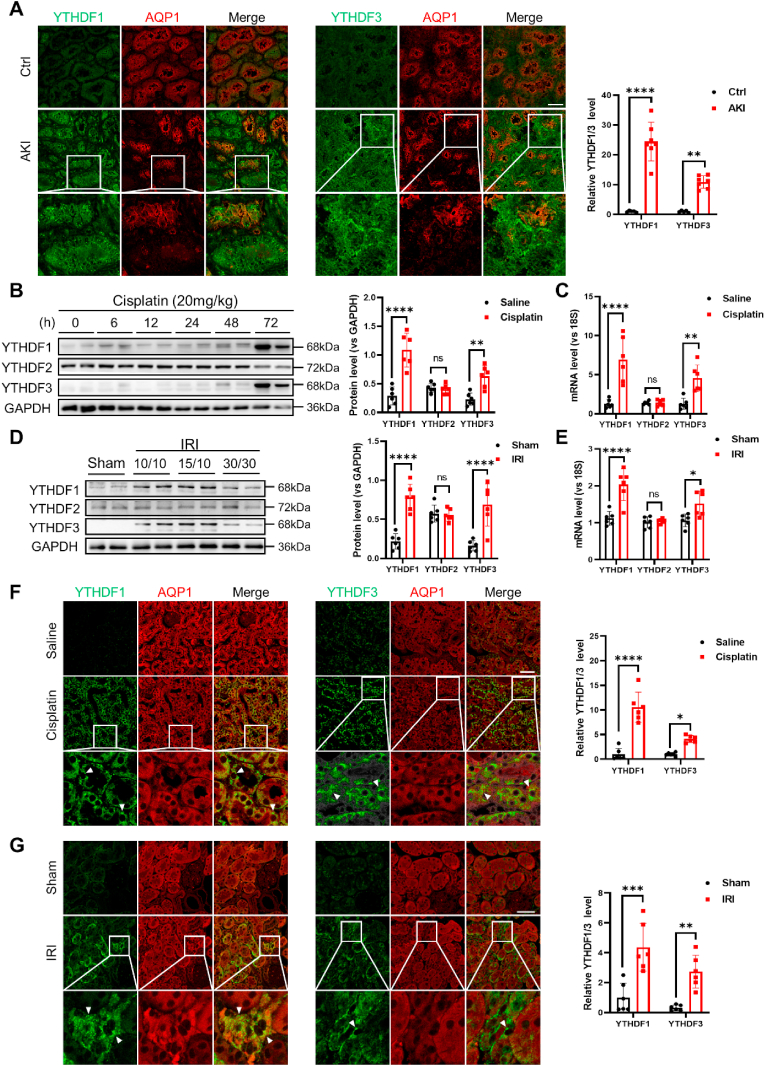
Our subsequent investigation involved the assessment of YTHDF1 and YTHDF3 levels in kidney tissue specimens from both human subjects and mouse models afflicted with acute kidney injury (AKI). In this endeavor, we procured kidney biopsies from 7 AKI patients and 5 control subjects at Jinling Hospital. For the identification of proximal tubule epithelial cells (PTECs), aquaporin 1 (AQP1) was selected as the designated marker [41]. As illustrated in Fig. 3A, the expression levels of YTHDF1 and YTHDF3 within PTECs of AKI patients demonstrated a noticeable elevation compared to those in control samples. Within murine models subjected to either cisplatin-induced nephrotoxicity or ischemia-reperfusion injury (IRI), it was observed that YTHDF1 and YTHDF3, but not YTHDF2, were considerably upregulated upon cisplatin treatment (Fig. 3B) or in response to ischemia-reperfusion (Fig. 3D). The increased levels of YTHDF1 and YTHDF3 within injured murine kidneys were also suggested to be attributed, at least in part, to transcriptional upregulation (Fig. 3C and E). Aligning with these findings, the immunofluorescence labeling of murine kidney tissues illustrated that mice subjected to cisplatin treatment (Fig. 3F) or IRI (Fig. 3G) exhibited prominently heightened YTHDF1 and YTHDF3 expression levels within PTECs, as compared to the control group.
Fig. 3.
Upregulation of renal tubular YTHDF1/YTHDF3 in AKI patients and experimental AKI mice.
(A) Immunofluorescence images display YTHDF1/YTHDF3 and AQP1 in kidney biopsies from 7 AKI patients and 5 controls. Scale bar, 50 μm
(B–C) In AKI mice treated with cisplatin, protein (B) and mRNA (C) levels of renal YTHDF1-3 were measured (6 biological replicates of mice subjected to cisplatin at specified times). The protein and mRNA levels of YTHDF1-3 were quantified in mice induced with cisplatin for 72 h
(D–E) Renal YTHDF1-3 protein (D) and mRNA (E) levels in mouse AKI models induced by ischemia-reperfusion injury (IRI) (6 biological replicates of mice undergoing IRI at prescribed intervals). The protein and mRNA levels of YTHDF1-3 were quantified in mice induced with ischemia for 10 min and reperfusion for 15 min.
(F) Immunofluorescence staining of YTHDF1/3 and AQP1 in the mouse cisplatin-induced AKI model (6 biological replicates of mice). Scale bar, 50 μm
(G) Immunofluorescence staining of YTHDF1/3 and AQP1 in the mouse IRI-induced AKI model (6 biological replicates of mice). Scale bar, 50 μm.
Data were subjected to analysis using two-way ANOVA and are presented as group means ± SD. Statistical significance is represented as *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. ns, no significance.
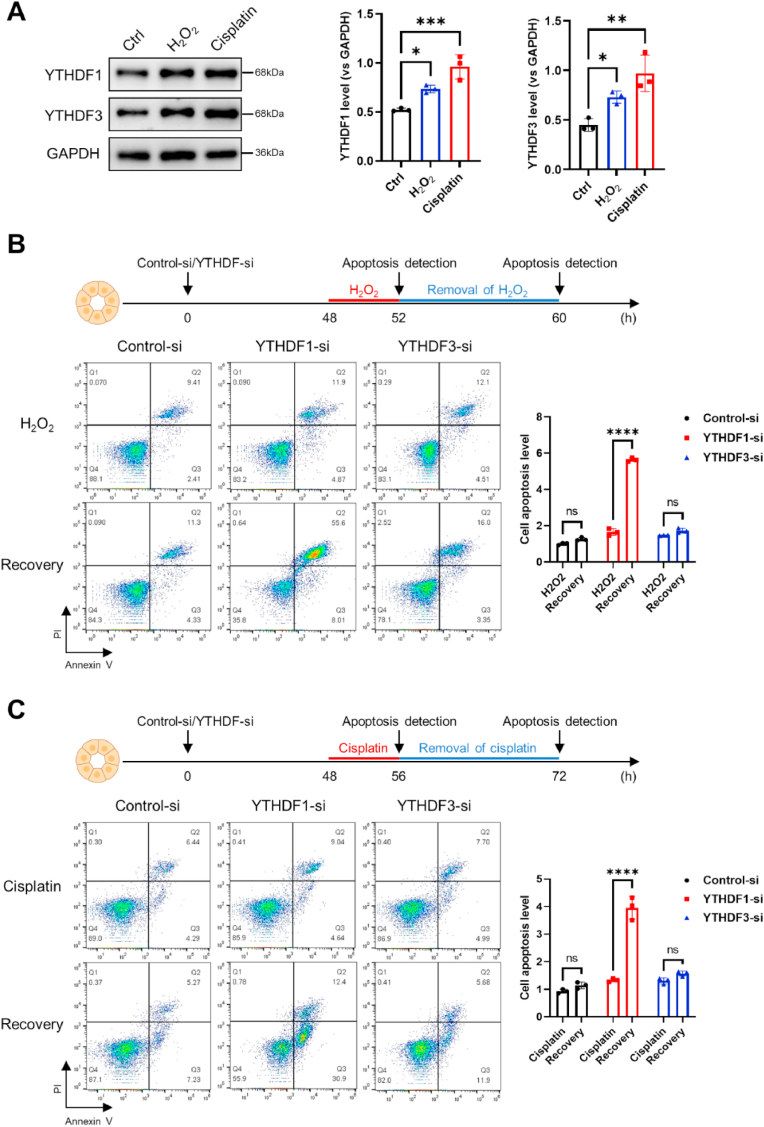
To elucidate the impact of YTHDF1 and YTHDF3 on the modulation of tubular cell function under stress conditions, we subjected HK2 cells to either H2O2 or cisplatin exposure, subsequently assessing the levels of YTHDF1 and YTHDF3. Remarkably, the protein levels of both YTHDF1 and YTHDF3 in HK2 cells demonstrated a surge upon treatment with H2O2 or cisplatin (Fig. 4A). Evidently, the augmentation of YTHDF1 and YTHDF3 within stress-challenged HK2 cells appeared to transpire at the transcriptional level (Supplementary Figs. S4A–B). Following this, we employed siRNA oligonucleotides to effectively downregulate YTHDF1 or YTHDF3 in HK2 cells prior to H2O2 (Fig. 4B) or cisplatin (Fig. 4C) exposure, subsequently assessing cell apoptosis through flow cytometry. Importantly, under normal culture conditions, the depletion of YTHDF1 or YTHDF3 had no discernible impact on cell apoptosis when compared to the control condition (Supplementary Fig. S4C). Interestingly, the outcome revealed that HK2 cell apoptosis induced by H2O2 (Fig. 4B) or cisplatin (Fig. 4C) treatment was significantly accentuated upon the knockdown of YTHDF1, but not YTHDF3, after the subsequent removal of H2O2 or cisplatin. This observation suggests a major protective role of YTHDF1 in stress-challenged HK2 cells. To validate the specific silencing of YTHDF1 and YTHDF3 in HK2 cells, we conducted Western blot and RT-qPCR analyses following transfection with YTHDF1 siRNA and YTHDF3 siRNA, respectively (Supplementary Fig. S5, A-B).
Fig. 4.
Defensive function of YTHDF1 against stress-induced renal tubular cell injury.
(A) YTHDF1 and YTHDF3 protein levels in HK2 cells treated with H2O2 or cisplatin were determined by Western blot analysis. Quantitative data from three biological replicates were subjected to one-way ANOVA for statistical analysis.
(B–C) Apoptosis evaluation of HK2 cells subjected to H2O2 (B, top) or cisplatin (C, top) treatment, as well as post-treatment removal of H2O2 (B, bottom) or cisplatin (C, bottom), was conducted using the Annexin/PI assay. Prior to the stress challenge, HK2 cells were transfected with control oligonucleotide (Control-si) or siRNA specific to YTHDF1 or YTHDF3, with assessments carried out 48 h after transfection. Quantitative data derived from three biological replicates were analyzed using a two-way ANOVA approach.
All data are represented as mean ± SD. Significance levels are indicated as *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. ns, no significance.
3.3. Renal tubular-specific YTHDF1 knockout mice display more severe AKI
To further corroborate the pivotal role of YTHDF1 in safeguarding PTECs against the acute injury instigated by cisplatin and ischemia-reperfusion treatments, we engineered mice with proximal tubular-specific YTHDF1 conditional knockout (cKO). Comprehensive genotyping, coupled with Western blot and immunofluorescence staining analyses, established the precise knockout of YTHDF1 within the renal proximal tubules of the cKO mice (Fig. 5A). Importantly, the GGT-Cre or floxed-only mice exhibited no discernible alterations pertaining to tubular structures or the expression of YTHDF1. The specific knockout of YTHDF1 within the proximal tubules notably exacerbated renal inflammation and dysfunction when subjected to cisplatin or ischemia-reperfusion challenges. As depicted in Fig. 5B, in stark contrast to the WT group, cKO mice displayed a prominent elevation in serum creatinine (Scr) and blood urea nitrogen (BUN) levels post cisplatin administration. Immunostaining with F4/80 antibody and TUNEL assays revealed that mice administered with cisplatin exhibited a substantially higher proportion of macrophage infiltration within the kidney tissues of cKO mice, accompanied by an augmented occurrence of renal cell apoptosis, as compared to WT mice (Fig. 5C–D). Consistent with these findings, Periodic Acid-Schiff (PAS) and Hematoxylin and Eosin (H&E) staining affirmed a heightened degree of tubular injury in cKO mice following cisplatin treatment when juxtaposed with WT mice (Fig. 5E). A similar phenomenon manifested in cKO mice subjected to ischemia-reperfusion, where elevated Scr and BUN levels were detected in cKO mice relative to WT mice following the ischemia-reperfusion (IRI) protocol (Fig. 5F). Moreover, the cKO mice subjected to IRI demonstrated a substantially greater extent of renal infiltration by inflammatory macrophages and a heightened occurrence of renal cell apoptosis (Fig. 5G–H). In accordance with these results, the assessment of kidney tissues through PAS and H&E staining corroborated the notion that the proximal tubular-specific YTHDF1 knockout exacerbated renal tubular injury induced by IRI in mice (Fig. 5I). Additionally, we examined the in vivo formation of SGs. Surprisingly, WT mice exhibited the formation of sizable SG aggregates, while cKO mice exhibited an absence of such sizable aggregates following both cisplatin-induced and IRI-induced acute injuries (Supplementary Fig. S6, A-B).
Fig. 5.
Exacerbation of mouse AKI induced by cisplatin or I/R in YTHDF1 cKO mice.
(A) On the left: Genotyping results. In the middle: Western blot analysis of YTHDF1 in renal tubules of both wild-type (WT) and conditional knockout (cKO) mice. On the right: Immunofluorescence staining of YTHDF1 in WT and cKO mice. A total of 6 biological replicates of mice were utilized for analysis. Scale bar, 50 μm
(B) Serum creatinine (Scr) and blood urea nitrogen (BUN) levels in mice with cisplatin-induced AKI are shown (6 biological replicates of mice).
(C) Immunofluorescence staining of F4/80 in mouse kidneys with cisplatin-induced AKI (6 biological replicates of mice). Scale bar, 100 μm
(D) Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) staining in mouse kidneys with cisplatin-induced AKI (6 biological replicates of mice). Scale bar, 100 μm
(E) Periodic acid-Schiff (PAS) and hematoxylin and eosin (H&E) staining in mouse kidneys with cisplatin-induced AKI (6 biological replicates of mice). Scale bar, 100 μm
(F) Scr and BUN levels in mice experiencing I/R-induced AKI (7 biological replicates of mice).
(G) Immunofluorescence staining of F4/80 in mouse kidneys with I/R-induced AKI (6 biological replicates of mice). Scale bar,100 μm
(H) TUNEL staining in mouse kidneys with I/R-induced AKI (6 biological replicates of mice). Scale bar, 100 μm
(I) PAS and H&E staining in mouse kidneys with I/R-induced AKI (6 biological replicates of mice). Scale bar, 100 μm.
Data were subjected to analysis using the Mann Whitney test and are presented as group means ± SD. Significance levels are indicated as *, P < 0.05, **, P < 0.01. ns, no significance.
3.4. YTHDF1 facilitates SG formation in renal tubular cells via recruiting m6A-modified mRNAs
Subsequently, we delved into the mechanisms underlying YTHDF1's protective role against acute kidney injury (AKI). Given that SGs serve as protective entities for tubular cells [25] and YTHDF1 selectively localizes to SGs under stress conditions (Fig. 2), an intriguing inquiry arose regarding the potential involvement of YTHDF1 in SG formation. To explore this aspect, we employed YTHDF1-siRNA to knock down YTHDF1 in HK2 cells prior to subjecting them to cisplatin challenge (Fig. 6A) or H2O2 exposure (Fig. 6B). Unexpectedly, the depletion of YTHDF1 significantly impeded the formation of SGs in stress-challenged HK2 cells, implying a crucial reliance on YTHDF1 for SG formation within these cells. This finding was further substantiated by the observation that isolated proximal tubular epithelial cells (PTECs) from YTHDF1 cKO mice exhibited notably fewer SGs when compared to WT mouse PTECs after being exposed to cisplatin or H2O2 (Fig. 6C–D). Notably, these findings are in line with a recent study that has demonstrated YTHDF1's facilitative role in the development of substantial SGs in U-2 OS cells [36]. Bolstering the notion of YTHDF1's protective effect on renal tubular cells against stress challenges, our assessment of cell apoptosis revealed that recovery from cisplatin (Fig. 6E) or H2O2 (Fig. 6F) treatment-induced apoptosis was markedly elevated in YTHDF1 cKO PTECs compared to WT PTECs.
Fig. 6.
Reduction of m6A-modified mRNA enrichments in SGs and augmentation of cellular apoptosis upon depletion of YTHDF1 in renal tubular cells.
(A–B) Following treatment of HK2 cells with cisplatin (A) or H2O2 (B), cells harboring YTHDF1 siRNA exhibited a transition from large SGs to smaller or absent ones, as observed in immunofluorescence images from dual staining of YTHDF1 and G3BP1. Scale bar, 20 μm
(C–D) PTECs isolated from WT and YTHDF1 cKO mice were treated with cisplatin (C) or H2O2 (D), and labeled with YTHDF1 antibody, G3BP1 antibody and DAPI, respectively. Scale bar, 20 μm
(E–F) Apoptosis of PTECs recovery from treatment with cisplatin (E) or H2O2 (F) assessed by Annexin/PI assay (3 biological replicates).
In panels A–D, the fluorescence intensities of SGs of 30–70 cells from three independent experiments were counted. The results were analyzed by unpaired t-test and presented as group means ± SD. **, P < 0.01, ****, P < 0.0001.
3.5. YTHDF1 preserves m6A-modified mRNAs in SGs under stress conditions
To substantiate whether YTHDF1 orchestrates the recruitment of crucial m6A-modified mRNAs, pivotal for cell survival, into SGs to safeguard them until the alleviation of stress, we embarked on an analysis of the m6A-modified mRNAs that potentially find sanctuary within SGs under stress conditions. The overlay of mRNAs bearing m6A modifications within SGs (411) as delineated in Fig. 1E, and mRNAs recognized by YTHDF1 (980) as identified through RIP-sequencing analysis, was represented in a Venn diagram (Fig. 7A). Intriguingly, this analysis revealed the existence of at least 17 m6A-modified mRNAs vital for cell survival, including SOCS3, BNC1, SPHK1, VGF, EVA1B, and ZFX, ensconced within SGs. To assess the contribution of YTHDF1 in facilitating the survival of these m6A-modified mRNAs under stress conditions, we adopted a YTHDF1-siRNA approach to deplete YTHDF1 in HK2 cells, which were subsequently treated with H2O2. Subsequently, SGs were isolated from these cells for RT-qPCR analysis (Fig. 7B, top). Additionally, we gauged the overall mRNA levels of these cells after the removal of H2O2 (Fig. 7B, bottom). The results from the RT-qPCR assay unequivocally demonstrated that in HK2 cells subjected to YTHDF1 depletion, the levels of the majority of these m6A-modified mRNAs were markedly diminished compared to those in control HK2 cells, whether within the confines of SGs or within the total mRNA pool. These findings lend robust support to the notion that YTHDF1 plays a pivotal role in the recruitment of m6A-modified mRNAs into SGs, safeguarding them from RNA degradation under stressful circumstances.
Fig. 7.
YTHDF1 safeguards m6A-modified mRNAs within SGs under stressful conditions.
(A) Venn analysis showcasing the intersection of m6A-modified mRNAs present in stress granules (SGs) and mRNAs bound by YTHDF1.
(B) Upper section: Transcriptional levels of genes as described in panel (A) within SGs induced by H2O2 treatment. Lower section: Transcriptional levels of genes from panel (A) within HK2 cells subjected to H2O2 stress and then reverted to normal medium for 8 h. Results were derived from 3 biological replicates and analyzed using an unpaired t-test.
(C) Left: MeRIP-qPCR quantification of m6A levels in SPHK1 mRNA in HK2 cells. Right: RIP-qPCR detection of SPHK1 mRNA association with YTHDF1. Data were gathered from 3 biological replicates and analyzed using a two-way ANOVA.
(D) Quantification of SPHK1 mRNA association with YTHDF1-WT or YTHDF1-Mut in HK2 cells. Flag-tagged YTHDF1-WT or YTHDF1-Mut was assessed via Western blot (left), and SPHK1 mRNA levels were quantified using RT-qPCR (right). The analysis was conducted across 3 biological replicates and evaluated with a two-way ANOVA.
(E) Single-molecule FISH (smFISH) images of SPHK1 mRNAs in HK2 cells treated with or without H2O2 (10 mM, 4 h). Notably, smFISH signals (magenta) colocalized with G3BP1 (red) and YTHDF1 (green) in transfected with control oligonucleotide (Control-si) group. These co-localized particles became smaller in transfected with YTHDF1 siRNA oligonucleotide (YTHDF1-si) group. Representative images were selected from a minimum of three biological replicates. Scale bars, 5 μm
(F) Protein levels of SPHK1 and YTHDF1 in YTHDF1-knockdown HK2 cells following H2O2 stress and subsequent return to normal medium for 8 h. Data were compiled from 3 biological replicates.
(G) Apoptosis of HK2 cells co-transfected with YTHDF1 siRNA and SPHK1 over expression plasmids under H2O2 conditions. The control group was transfected with the respective control siRNA and plasmid. A total of 3 biological replicates were utilized for the analysis, which was assessed using one-way ANOVA.
All outcomes were presented as mean ± SD. Significance is indicated as *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. ns, no significance. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In the subsequent phase of our investigation, we singled out m6A-modified SPHK1 (sphingosine kinase 1) mRNA as a focal point for in-depth analysis concerning its interaction with YTHDF1. Utilizing qPCR assays on immunoprecipitated m6A-modified RNAs, we discerned a pronounced presence of m6A methylation on SPHK1 mRNA (Fig. 7C, left). Through RNA immunoprecipitation using a YTHDF1 antibody, we validated the direct association between SPHK1 mRNA and YTHDF1 (Fig. 7C, right), and observed an augmentation of YTHDF1-associated SPHK1 mRNA (m6A-modified) levels following exposure to stress challenges. Subsequently, we introduced a Flag-YTHDF1 expression plasmid (YTHDF1-WT) alongside a Flag-YTHDF1 mutant plasmid (YTHDF1-Mut, K395A, Y397A) [42] (Supplementary Fig. S7) into HK2 cells (Fig. 7D, left). Employing RNA immunoprecipitation with a Flag antibody, we uncovered that YTHDF1-WT, but not YTHDF1-Mut, effectively immunoprecipitated SPHK1 mRNA (Fig. 7D, right), underscoring a specific interaction between YTHDF1 protein and m6A-modified SPHK1 mRNA.
Consonant with these findings, single-molecule FISH (smFISH) images of HK2 cells subjected to H2O2 treatment unveiled a concentration of m6A-modified SPHK1 mRNA (Fig. 7E, magenta) within G3BP1-labeled SGs (Fig. 7E, red) during stress challenge. Intriguingly, knockdown of YTHDF1 using YTHDF1 siRNA markedly impeded the stress-induced enrichment of SPHK1 mRNA within SGs (Fig. 7E, green). Given that intact m6A-modified SPHK1 mRNA can be utilized for protein synthesis upon relief from stress, we further gauged SPHK1 protein levels in HK2 cells, 8 h post-recovery from H2O2 exposure. As depicted in Fig. 7F, YTHDF1 knockdown notably diminished the H2O2-induced SPHK1 protein levels. Bolstering the significance of SPHK1 in terms of cell proliferation and survival, [43,44] apoptotic assays exhibited that the depletion of YTHDF1, leading to a decrease in SPHK1 levels, intensified H2O2-induced cell apoptosis in HK2 cells (Fig. 7G). Intriguingly, the amelioration of H2O2-induced cell apoptosis in YTHDF1-depleted HK2 cells was achieved through the substantial rescue of SPHK1 levels upon its overexpression. These findings collectively suggest that YTHDF1's protective role in HK2 cells may, at least in part, stems from its recruitment and preservation of m6A-modified SPHK1 mRNA.
4. Discussion
This study delved into the intricate mechanisms governing the interplay between stress granules (SGs) and m6A modifications, unraveling their impact on the regulation of renal tubular cell functionality. Our findings unveiled a pivotal role for the m6A reader YTHDF1 in orchestrating the synergistic influence of SGs and m6A modifications, ultimately affording protection to renal tubular cells amid diverse stress scenarios. YTHDF1 emerged as a key facilitator in SG formation and took on a vital responsibility in safeguarding renal tubules from acute injuries or stress-induced challenges. This protective function is executed through its adeptness in selectively recruiting and sheltering m6A-methylated mRNAs within SGs, thus promoting cellular resilience in the face of adversities.
SGs condensate consisted of RNA and proteins without membrane that dynamically assemble and disassemble in response to the appearance and release of stress stimuli, respectively. As mRNAs in SGs can be recovered for protein translation following stress relief, SGs may provide a protection for cells to survive the stress condition. Formation of SGs, generally accompanied with reduction and termination of protein translation has been considered as participating in humankind diseases [[45], [46], [47]]. It is well known that PTECs possess abundant mitochondria and are highly susceptible to metabolic stress such as oxidative stress or energy deprivation [48]. Recent study by Wang et al. showed that formation of SGs is a critical way for cells to adjust to pressure [25]. Mechanistically, termination of protein translation and formation of SGs under various stress conditions is usually initiated by the phosphorylation of eukaryotic initiation factor 2α (eIF2α) at Ser51 [25]. Previous studies in AKI reported that phosphorylation of eIF2α can be induced by mitochondria and ER stress [49]. Although the formation of SGs is related to cellular integrated stress response, the mechanisms that regulate SGs, including the size and type, under various stress conditions remain elusive. In addition to SG core protein, such as G3BP1, we also found that YTHDF1, as well as other m6A readers, are key components of SGs formed in HK2 cells under different stress conditions. Interestingly, depletion of YTHDF1 blocked the formation of large SGs in stress-challenged HK2 cells (Fig. 6), suggesting that YTHDF1 plays a key part in SG biogenesis in renal tubular cells. Given that YTHDF1 functions as a m6A reader, we postulate that continuous recruitment of m6A-methylated mRNAs into SGs by YTHDF1 is likely part of formation of SGs in stress-challenged renal tubular cells.
The m6A regulates RNA life cycle process, [50] although the mechanisms remain unknown, recent studies have demonstrated RNA modifications, especially m6A, are involved in the developmental mechanisms of kidney diseases [51]. Liu et al. reported that m6A RNA methylation was involved in sepsis-associated AKI [52]. Through comprehensive analysis of m6A methylome, Shen et al. showed an important role of that m6A in AKI with cisplatin [9]. Although the total level of m6A-modified mRNAs in renal tubular cells was not affected by stress challenges, m6A-modified mRNAs were preferentially relocated at SGs (Fig. 1). Our results suggest that m6A modification may protect mRNAs through selective storage in SGs. For example, SPHK1 mRNA can survive various stress conditions through m6A methylation and then being recruited and stored in SGs. When the stress is relieved, the m6A-modified SPHK1 mRNA in SGs will be rapidly recovered for SPHK1 protein production. Moreover, we found that selective cumulation mRNAs with m6A in SGs in stress-challenged renal tubular cells was mediated by YTHDF1. Depletion of renal tubular YTHDF1 markedly reduced the levels of panel of m6A-modified mRNAs, including SPHK1 mRNA, in HK2 cells recovered from stress challenge (Fig. 7). This finding of the function of YTHDF1 in facilitating cell proliferation and survival is consistent with an earlier report by Xing et al. who evidenced that YTHDF1 enhanced renal cell proliferation via upregulating Yes-associated protein (YAP) [11].
Sphingolipid metabolites play key roles in cellular proliferation and apoptosis. SPHK1 is a critical enzyme catalyzing the phosphorylation of sphingosine to sphingosine-1-phosphate (S1P), promoting cell proliferation and survival. Bajwa et al. found that SPHK2 deficiency attenuates kidney fibrosis [53]. In this study, we identified SPHK1 mRNA as one of m6A-modified mRNAs that were protected by YTHDF1 under stress condition. YTHDF1-dependent formation of SGs in stress-challenged renal tubular cells provide a dynamic platform for preserving m6A-modified SPHK1 mRNA. Given the significance of SPHK1 in promoting cell proliferation and survival, rapid recovery of m6A-modified SPHK1 mRNA after stress relief will enhance renal tubular wound healing and mitigate AKI. In conclusion, our results reveal that YTHDF1 mitigates acute renal tubule injury via safeguarding m6A-methylated mRNAs favoring cell proliferation and survival in SGs, and provide YTHDF1 as a therapeutic target for safeguarding against AKI.
Our study acknowledges three notable limitations warranting discussion. Firstly, the precise mechanism governing the upregulation and distribution of selective m6A readers within renal tubular cells during AKI conditions remains elusive. Secondly, the specific association of m6A reader YTHDF1 with stress-induced SGs within renal tubular cells presents an unanswered query. Is this association facilitated through a distinct interaction between YTHDF1 and the core SG protein G3BP1? Finally, we must acknowledge the potential variability in the impact of m6A modification on mRNA recruitment into SGs across different cell types. Notably, the work of Khong et al. has demonstrated limited effects of m6A modification on mRNA sequestration into SGs within wildtype and METTL3-deleted mES cells [54]. This research suggests a nuanced perspective, indicating that YTHDF proteins might play a minor role in RNA recruitment to SGs in mES cells challenged by arsenite. Clearly, further investigations are imperative to address these identified limitations comprehensively.
Authors' contributions
KZ, ZL and LL designed the research; WY, MZ, JL, SQ, FZ and ML performed experiments and data analysis; KZ and WY drafted the paper.
Funding
This work was supported by the National Key Research and Development Program of China [grant number 2018YFA0507100] and the National Natural Science Foundation of China [grant number 82170692].
Interest statement
None.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgements
Kidney samples were obtained from Renal Biobank of National Clinical Research Center of Kidney Diseases, Jiangsu Biobank of Clinical Resources, a part of The Open Project of Jiangsu Biobank of Clinical Resources (JSRB2021-03).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102921.
Contributor Information
Zhihong Liu, Email: liuzhihong@nju.edu.cn.
Ke Zen, Email: kzen@nju.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Sharfuddin A.A., Molitoris B.A. Pathophysiology of ischemic acute kidney injury. Nat. Rev. Nephrol. Apr 2011;7(4):189–200. doi: 10.1038/nrneph.2011.16. [DOI] [PubMed] [Google Scholar]
- 2.Raup-Konsavage W.M., Wang Y., Wang W.W., Feliers D., Ruan H., Reeves W.B. Neutrophil peptidyl arginine deiminase-4 has a pivotal role in ischemia/reperfusion-induced acute kidney injury. Kidney Int. Feb 2018;93(2):365–374. doi: 10.1016/j.kint.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z., Chen L., Liu H., et al. Gene signature for the prediction of the trajectories of sepsis-induced acute kidney injury. Crit. Care. Dec 21 2022;26(1):398. doi: 10.1186/s13054-022-04234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu S., Jia P., Fang Y., et al. Nuclear farnesoid X receptor attenuates acute kidney injury through fatty acid oxidation. Kidney Int. May 2022;101(5):987–1002. doi: 10.1016/j.kint.2022.01.029. [DOI] [PubMed] [Google Scholar]
- 5.Guo C., Dong G., Liang X., Dong Z. Epigenetic regulation in AKI and kidney repair: mechanisms and therapeutic implications. Nat. Rev. Nephrol. Apr 2019;15(4):220–239. doi: 10.1038/s41581-018-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. Oct 2019;20(10):608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 7.An Y., Duan H. The role of m6A RNA methylation in cancer metabolism. Mol. Cancer. Jan 12 2022;21(1):14. doi: 10.1186/s12943-022-01500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bechara R., Gaffen S.L. '(m(6))A' stands for 'autoimmunity': reading, writing, and erasing RNA modifications during inflammation. Trends Immunol. Dec 2021;42(12):1073–1076. doi: 10.1016/j.it.2021.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen J., Wang W., Shao X., et al. Integrated analysis of m6A methylome in cisplatin-induced acute kidney injury and berberine alleviation in mouse. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.584460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C.M., Li M., Zhao W.B., Ye Z.C., Peng H. Alteration of N6-methyladenosine RNA profiles in cisplatin-induced acute kidney injury in mice. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.654465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing J., He Y.C., Wang K.Y., Wan P.Z., Zhai X.Y. Involvement of YTHDF1 in renal fibrosis progression via up-regulating YAP. Faseb. J. Feb 2022;36(2) doi: 10.1096/fj.202100172RR. [DOI] [PubMed] [Google Scholar]
- 12.Das S., Santos L., Failla A.V., Ignatova Z. mRNAs sequestered in stress granules recover nearly completely for translation. RNA Biol. Jan 2022;19(1):877–884. doi: 10.1080/15476286.2022.2094137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price P.M., Safirstein R.L., Megyesi J. The cell cycle and acute kidney injury. Kidney Int. Sep 2009;76(6):604–613. doi: 10.1038/ki.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Q., Wang M., Gui Y., et al. Rheb1 protects against cisplatin-induced tubular cell death and acute kidney injury via maintaining mitochondrial homeostasis. Cell Death Dis. May 13 2020;11(5):364. doi: 10.1038/s41419-020-2539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrè S., Deng Y., Huen S.C., et al. Renal tubular cell spliced X-box binding protein 1 (Xbp1s) has a unique role in sepsis-induced acute kidney injury and inflammation. Kidney Int. Dec 2019;96(6):1359–1373. doi: 10.1016/j.kint.2019.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan Y., Xiao W., Lee K., et al. Inhibition of reticulon-1A-mediated endoplasmic reticulum stress in early AKI attenuates renal fibrosis development. J. Am. Soc. Nephrol. Jul 2017;28(7):2007–2021. doi: 10.1681/asn.2016091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su L., Zhang J., Gomez H., Kellum J.A., Peng Z. Mitochondria ROS and mitophagy in acute kidney injury. Autophagy. Feb 2023;19(2):401–414. doi: 10.1080/15548627.2022.2084862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livingston M.J., Wang J., Zhou J., et al. Clearance of damaged mitochondria via mitophagy is important to the protective effect of ischemic preconditioning in kidneys. Autophagy. Dec 2019;15(12):2142–2162. doi: 10.1080/15548627.2019.1615822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Y., E J., Cai H., et al. Reticulon-1A mediates diabetic kidney disease progression through endoplasmic reticulum-mitochondrial contacts in tubular epithelial cells. Kidney Int. Aug 2022;102(2):293–306. doi: 10.1016/j.kint.2022.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y., Yang D., Yang D., Jia R., Ding G. Role of reactive oxygen species-mediated endoplasmic reticulum stress in contrast-induced renal tubular cell apoptosis. Nephron Exp. Nephrol. 2014;128(1–2):30–36. doi: 10.1159/000366063. [DOI] [PubMed] [Google Scholar]
- 21.Lee Y.G., Chou H.C., Chen Y.T., et al. L-Carnitine reduces reactive oxygen species/endoplasmic reticulum stress and maintains mitochondrial function during autophagy-mediated cell apoptosis in perfluorooctanesulfonate-treated renal tubular cells. Sci. Rep. Mar 18 2022;12(1):4673. doi: 10.1038/s41598-022-08771-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang X.S., Xiang X.Y., Chen X.M., et al. Inhibition of soluble epoxide hydrolase attenuates renal tubular mitochondrial dysfunction and ER stress by restoring autophagic flux in diabetic nephropathy. Cell Death Dis. May 21 2020;11(5):385. doi: 10.1038/s41419-020-2594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokouchi M., Hiramatsu N., Hayakawa K., et al. Atypical, bidirectional regulation of cadmium-induced apoptosis via distinct signaling of unfolded protein response. Cell Death Differ. Aug 2007;14(8):1467–1474. doi: 10.1038/sj.cdd.4402154. [DOI] [PubMed] [Google Scholar]
- 24.Yan M., Shu S., Guo C., Tang C., Dong Z. Endoplasmic reticulum stress in ischemic and nephrotoxic acute kidney injury. Ann. Med. Aug 2018;50(5):381–390. doi: 10.1080/07853890.2018.1489142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S., Kwon S.H., Su Y., Dong Z. Stress granules are formed in renal proximal tubular cells during metabolic stress and ischemic injury for cell survival. Am. J. Physiol. Ren. Physiol. Jul 1 2019;317(1):F116–f123. doi: 10.1152/ajprenal.00139.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kedersha N., Ivanov P., Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. Oct 2013;38(10):494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang P., Mathieu C., Kolaitis R.M., et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell. Apr 16 2020;181(2):325–345.e28. doi: 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwano M., Plieth D., Danoff T.M., Xue C., Okada H., Neilson E.G. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J. Clin. Invest. Aug 2002;110(3):341–350. doi: 10.1172/jci15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L., Brooks C.R., Xiao S., et al. KIM-1-mediated phagocytosis reduces acute injury to the kidney. J. Clin. Invest. Apr 2015;125(4):1620–1636. doi: 10.1172/jci75417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. Jan 28 2016;164(3):487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J.N., Wang F., Ke J., et al. Inhibition of METTL3 attenuates renal injury and inflammation by alleviating TAB3 m6A modifications via IGF2BP2-dependent mechanisms. Sci. Transl. Med. Apr 13 2022;14(640) doi: 10.1126/scitranslmed.abk2709. [DOI] [PubMed] [Google Scholar]
- 32.Martin J.L., Terry S.J., Gale J.E., Dawson S.J. The ototoxic drug cisplatin localises to stress granules altering their dynamics and composition. J. Cell Sci. Jun 19 2023 doi: 10.1242/jcs.260590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emara M.M., Fujimura K., Sciaranghella D., Ivanova V., Ivanov P., Anderson P. Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. Jul 13 2012;423(4):763–769. doi: 10.1016/j.bbrc.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindström M., Chen L., Jiang S., et al. Lsm7 phase-separated condensates trigger stress granule formation. Nat. Commun. Jun 28 2022;13(1):3701. doi: 10.1038/s41467-022-31282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu F., Li X., Zhang P., et al. Melanoma differentiation-associated gene 5 is involved in the induction of stress granules and autophagy by protonophore CCCP. Biol. Chem. Jan 2016;397(1):67–74. doi: 10.1515/hsz-2015-0195. [DOI] [PubMed] [Google Scholar]
- 36.Fu Y., Zhuang X. m(6)A-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol. Sep 2020;16(9):955–963. doi: 10.1038/s41589-020-0524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markmiller S., Soltanieh S., Server K.L., et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. Jan 25 2018;172(3):590–604.e13. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. Apr 29 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 39.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. Jun 22 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ries R.J., Zaccara S., Klein P., et al. m(6)A enhances the phase separation potential of mRNA. Nature. Jul 2019;571(7765):424–428. doi: 10.1038/s41586-019-1374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kishore B.K., Krane C.M., Di Iulio D., Menon A.G., Cacini W. Expression of renal aquaporins 1, 2, and 3 in a rat model of cisplatin-induced polyuria. Kidney Int. Aug 2000;58(2):701–711. doi: 10.1046/j.1523-1755.2000.00216.x. [DOI] [PubMed] [Google Scholar]
- 42.Xu C., Liu K., Ahmed H., Loppnau P., Schapira M., Min J. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. Oct 9 2015;290(41):24902–24913. doi: 10.1074/jbc.M115.680389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ueda N. A rheostat of ceramide and sphingosine-1-phosphate as a determinant of oxidative stress-mediated kidney injury. Int. J. Mol. Sci. Apr 4 2022;23(7) doi: 10.3390/ijms23074010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okada T., Ding G., Sonoda H., et al. Involvement of N-terminal-extended form of sphingosine kinase 2 in serum-dependent regulation of cell proliferation and apoptosis. J. Biol. Chem. Oct 28 2005;280(43):36318–36325. doi: 10.1074/jbc.M504507200. [DOI] [PubMed] [Google Scholar]
- 45.Di Marco B., Dell'Albani P., D'Antoni S., et al. Fragile X mental retardation protein (FMRP) and metabotropic glutamate receptor subtype 5 (mGlu5) control stress granule formation in astrocytes. Neurobiol. Dis. Jul 2021;154 doi: 10.1016/j.nbd.2021.105338. [DOI] [PubMed] [Google Scholar]
- 46.Koppenol R., Conceição A., Afonso I.T., et al. The stress granule protein G3BP1 alleviates spinocerebellar ataxia-associated deficits. Brain. Jun 1 2023;146(6):2346–2363. doi: 10.1093/brain/awac473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y.J., Gendron T.F., Ebbert M.T.W., et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med. Aug 2018;24(8):1136–1142. doi: 10.1038/s41591-018-0071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiba T., Peasley K.D., Cargill K.R., et al. Sirtuin 5 regulates proximal tubule fatty acid oxidation to protect against AKI. J. Am. Soc. Nephrol. Dec 2019;30(12):2384–2398. doi: 10.1681/asn.2019020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y., Wei W., Fu J., Zhang T., Zhao J., Ma T. Forsythiaside A ameliorates sepsis-induced acute kidney injury via anti-inflammation and antiapoptotic effects by regulating endoplasmic reticulum stress. BMC Compl. Med. Ther. Feb 3 2023;23(1):35. doi: 10.1186/s12906-023-03855-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brocard M., Ruggieri A., Locker N. m6A RNA methylation, a new hallmark in virus-host interactions. J. Gen. Virol. Sep 2017;98(9):2207–2214. doi: 10.1099/jgv.0.000910. [DOI] [PubMed] [Google Scholar]
- 51.Luan J., Kopp J.B., Zhou H. N6-methyladenine RNA methylation epigenetic modification and kidney diseases. Kidney Int. Rep. Jan 2023;8(1):36–50. doi: 10.1016/j.ekir.2022.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu B., Ao S., Tan F., et al. Transcriptomic analysis and laboratory experiments reveal potential critical genes and regulatory mechanisms in sepsis-associated acute kidney injury. Ann. Transl. Med. Jul 2022;10(13):737. doi: 10.21037/atm-22-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bajwa A., Huang L., Kurmaeva E., et al. Sphingosine kinase 2 deficiency attenuates kidney fibrosis via IFN-γ. J. Am. Soc. Nephrol. Apr 2017;28(4):1145–1161. doi: 10.1681/asn.2016030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khong A., Matheny T., Huynh T.N., Babl V., Parker R. Limited effects of m(6)A modification on mRNA partitioning into stress granules. Nat. Commun. Jun 29 2022;13(1):3735. doi: 10.1038/s41467-022-31358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.