Key Points
Question
Can clinicians without coding expertise or access to well-labeled private data sets use self-training and automated machine learning to create high-performing diabetic retinopathy classification models?
Findings
In this quality improvement study, models designed without coding, private data sets, or extensive labeling demonstrated high performance comparable to bespoke and US Food and Drug Administration–approved diabetic retinopathy classification models.
Meaning
These findings suggest that the approach described here can advance democratization of artificial intelligence through clinician-driven model development.
This quality improvement study evaluates clinician-driven, self-trained use of automated machine learning and public data sets in designing high-performing diabetic retinopathy classification models.
Abstract
Importance
Democratizing artificial intelligence (AI) enables model development by clinicians with a lack of coding expertise, powerful computing resources, and large, well-labeled data sets.
Objective
To determine whether resource-constrained clinicians can use self-training via automated machine learning (ML) and public data sets to design high-performing diabetic retinopathy classification models.
Design, Setting, and Participants
This diagnostic quality improvement study was conducted from January 1, 2021, to December 31, 2021. A self-training method without coding was used on 2 public data sets with retinal images from patients in France (Messidor-2 [n = 1748]) and the UK and US (EyePACS [n = 58 689]) and externally validated on 1 data set with retinal images from patients of a private Egyptian medical retina clinic (Egypt [n = 210]). An AI model was trained to classify referable diabetic retinopathy as an exemplar use case. Messidor-2 images were assigned adjudicated labels available on Kaggle; 4 images were deemed ungradable and excluded, leaving 1744 images. A total of 300 images randomly selected from the EyePACS data set were independently relabeled by 3 blinded retina specialists using the International Classification of Diabetic Retinopathy protocol for diabetic retinopathy grade and diabetic macular edema presence; 19 images were deemed ungradable, leaving 281 images. Data analysis was performed from February 1 to February 28, 2021.
Exposures
Using public data sets, a teacher model was trained with labeled images using supervised learning. Next, the resulting predictions, termed pseudolabels, were used on an unlabeled public data set. Finally, a student model was trained with the existing labeled images and the additional pseudolabeled images.
Main Outcomes and Measures
The analyzed metrics for the models included the area under the receiver operating characteristic curve (AUROC), accuracy, sensitivity, specificity, and F1 score. The Fisher exact test was performed, and 2-tailed P values were calculated for failure case analysis.
Results
For the internal validation data sets, AUROC values for performance ranged from 0.886 to 0.939 for the teacher model and from 0.916 to 0.951 for the student model. For external validation of automated ML model performance, AUROC values and accuracy were 0.964 and 93.3% for the teacher model, 0.950 and 96.7% for the student model, and 0.890 and 94.3% for the manually coded bespoke model, respectively.
Conclusions and Relevance
These findings suggest that self-training using automated ML is an effective method to increase both model performance and generalizability while decreasing the need for costly expert labeling. This approach advances the democratization of AI by enabling clinicians without coding expertise or access to large, well-labeled private data sets to develop their own AI models.
Introduction
Deep learning (DL) algorithms are promising tools for medical image interpretation, especially within imaging-heavy specialties such as radiology, dermatology, and ophthalmology.1 However, not many models make it into production.2,3 Clinician-driven machine learning (ML) may have a higher likelihood of implementation because frontline clinicians are presumably best suited to select relevant use cases and design ML for patient-relevant end points such as visual acuity, treatment burden, or functional outcomes. Despite recent advances, supervised learning, the most used form of DL, relies on large amounts of labeled data.4,5 The scarcity of expert clinician time may be a substantial challenge when performing medical image labeling.6,7,8 Given that ML model performance is reliant on high-quality and reproducible ground-truth labels, this typically entails time-consuming and costly labeling efforts for health care ML.9,10,11 Accordingly, clinician-driven DL projects may be limited by the need for (1) coding expertise, (2) powerful computing resources, and (3) time and cost to generate high-quality labels.12
A lack of technical and coding expertise can be a barrier to clinician-driven ML. Our group has demonstrated a potential solution: automated ML. This framework enables ML model building without coding by largely automating the ML pipeline, including data set management, neural architecture search, and hyperparameter tuning.13 A number of automated, publicly available ML platforms enable domain experts, including health care professionals, without coding expertise to train their own high-performing DL models.14,15,16,17,18,19 Our aforementioned work demonstrates high performance and the ability to reproduce models demonstrating signals such as sex prediction from retinal fundus photographs.20,21 In this article, we build on that work by using automated ML and constraining the availability of other necessary resources, including private data sets and costly expert labels, to determine if underresourced clinicians can design well-performing models.
Semisupervised learning (SSL) is a potential solution for label scarcity because it makes use of unlabeled data, which are vastly more abundant.22 A type of SSL termed self-training entails initially training a “teacher model” with a labeled data set using supervised learning, then using the resulting teacher model to generate predictions, termed pseudolabels, on another unlabeled data set. Next, a “student model” is trained using an expanded data set consisting of the initially labeled data set combined with the additional data set and its pseudolabels (Figure 1). In the medical imaging context, this process typically involves using (1) a smaller data set labeled by clinician experts and (2) a larger unlabeled data set, such as a clinic’s entire corpus of unlabeled images and/or a public data set.23,24,25 This process may be repeated (ie, designating the student model as a teacher model to generate new pseudolabels and subsequently training a new student model iteration) to learn both generalized task representations and increase algorithm performance.22,26,27
Figure 1. Self-Training.
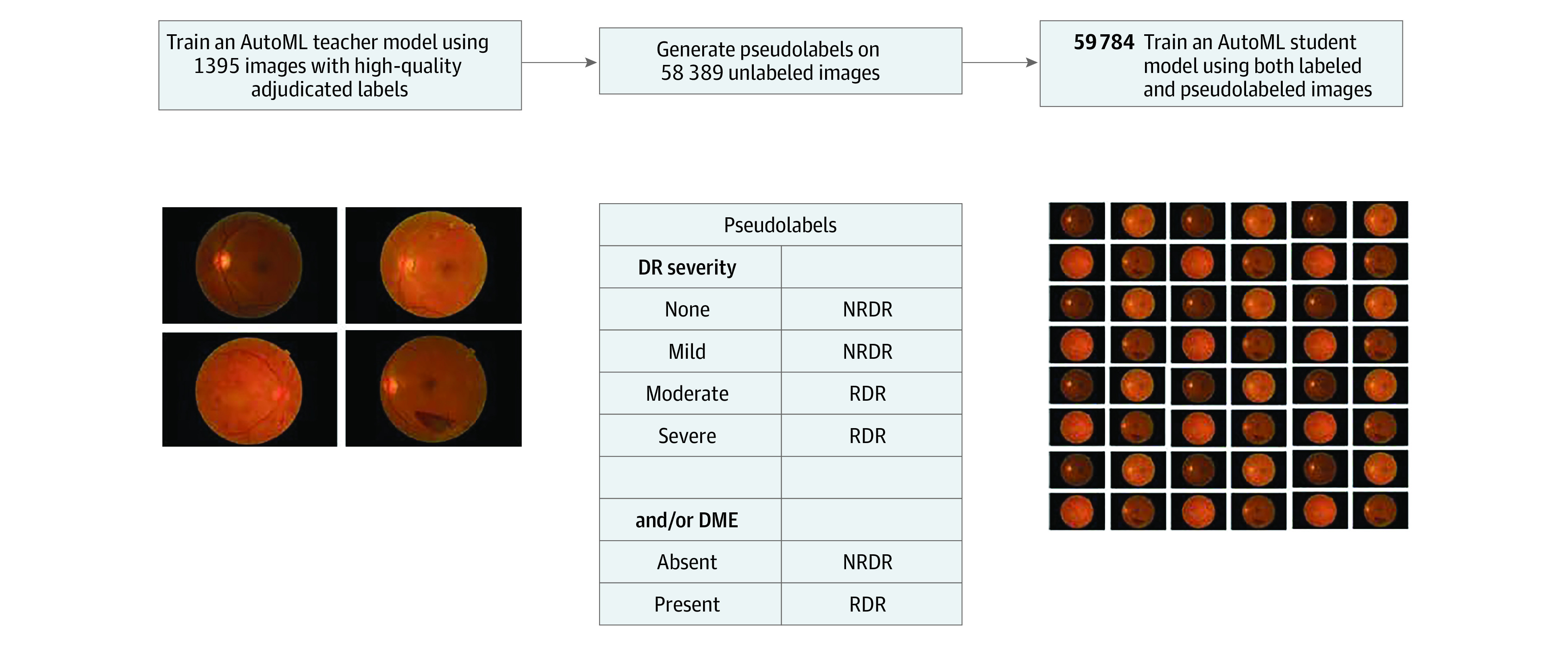
Illustration of the self-training approach, which involves 3 steps: (1) a teacher model is trained on labeled data, (2) the teacher model is used to generate pseudolabels on unlabeled data, and (3) a student model is trained on labeled and pseudolabeled images. AutoML indicates automated machine learning; DME, diabetic macular edema; DR, diabetic retinopathy; NRDR, nonreferable diabetic retinopathy; RDR, referable diabetic retinopathy.
We illustrate this framework in diabetic retinopathy (DR), a leading cause of visual impairment in up to one-third of patients with diabetes.28,29 According to projections, diabetes will affect 439 million adults by 2030, and 2.4 million eyes per day would require retinal examination worldwide.30,31 To address this public health problem, several artificial intelligence (AI) models have been approved by the US Food and Drug Administration (FDA) for DR classification and referral.32,33
In this study, we leveraged automated ML and SSL using public DR data sets to enable clinician-driven ML. We aimed to facilitate model design by clinicians with little coding expertise, computing resources, or large, well-labeled data sets. Specifically, we trained a representative model to classify referable DR (RDR), demonstrating performance similar to FDA-approved algorithms.32,33
Methods
This quality improvement study was approved by Moorfields Eye Hospital. Informed consent was waived because deidentified data were used. The study followed the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) reporting guideline.
The study was conducted from January 1, 2021, to December 31, 2021. We used 2 public data sets with retinal images from patients in France (Messidor-2) and the UK and US (EyePACS) and 1 data set with retinal images from patients of an Egyptian medical retina clinic (Egypt). The Kafrelsheikh University Institutional Review Board approved the Egypt study, and participants gave a combination of written and oral informed consent and received no compensation or incentives.
Public Data Sets
Public data sets were used both to (1) enable replication of our work and (2) demonstrate that the task may be achieved without institution-specific data (Figure 2).34 Two 45° retinal color fundus photograph (CFP) data sets were selected: EyePACS (n = 58 689 JPG images) and Messidor-2 (n = 1748 PNG images).35,36 Gold-standard public labels from Google (Kaggle), as adjudicated through an iterative process involving multiple rounds were applied to the Messidor-2 images9 to train the teacher model. Four images were adjudicated as ungradable and were excluded, leaving 1744 images. Messidor-2 CFPs were obtained using Topcon TRC NW6 nonmydriatic cameras (Topcon Corporation) with a 45° field of view. Patient inclusion criteria and demographics for these data sets are published with the source data sets. Diabetic retinopathy grades were assigned per the International Classification of Diabetic Retinopathy (ICDR) protocol for both data sets,37 and diabetic macular edema was defined by hard exudates within 1 disc diameter of the fovea. To replicate the most common referral triage task encountered in screening programs and performed by FDA and CE–approved DL models, labels were binarized to RDR, comprising grades of moderate, severe, proliferative, and/or the presence of diabetic macular edema, and to nonreferable DR (NRDR), which represents the absence of RDR.7,8,33,38,39,40,41
Figure 2. Model Training and Validation Data Flow.
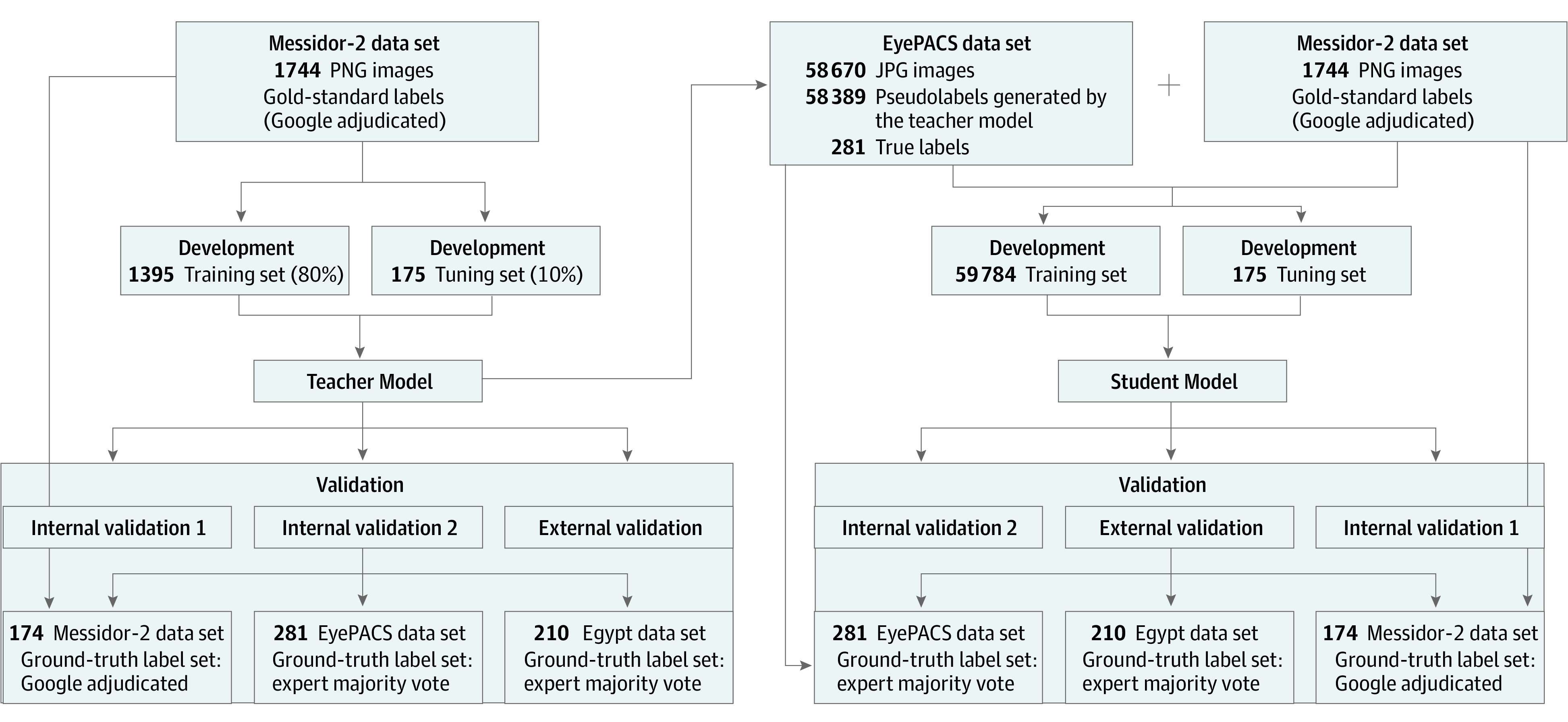
Data splits and flows for self-training and validation of student and teacher models. Once trained, the teacher model is used to apply pseudolabels to a new data set (EyePACS). Subsequently, the resulting labels and images are combined with the teacher model training data set to train a student model. Both models are internally and externally validated on adjudicated or arbitrated validation data sets.
To ensure high label quality for the EyePACS internal validation subset, 300 images were randomly selected from the EyePACS data set using the rand() function in Excel, version 16 (Microsoft Corp). These images were independently relabeled by 3 blinded retina specialists (M.B.G., J.H., and H.K.) using the ICDR protocol for diabetic retinopathy grade and diabetic macular edema presence. The images were recategorized to RDR and NRDR as described, and the majority grade was assigned for cases of disagreement. Nineteen images were ungradable, defined as either (1) field of view not encompassing the entire nerve and temporal vascular arcades or (2) without sufficient image quality to exclude microaneurysm-sized lesions. These images were excluded, leaving 281 images. Data set details are presented in Table 1.
Table 1. Data Set Characteristics.
| Characteristic | Teacher model (development set: teacher) | Student model (development set: student) | Teacher and student models (validation set) | ||||
|---|---|---|---|---|---|---|---|
| Training set (n = 1395) | Tuning set (n = 175) | Training set (n =59 784) | Tuning set (n = 175) | Internal validation 1 (n = 174) | Internal validation 2 (n = 281) | External validation (n = 210) | |
| Image source data set | Messidor-2 | Messidor-2 and EyePACS | Messidor-2 | EyePACS | Egypt | ||
| Label source | Google (Kaggle public) | Google (Kaggle public) and teacher model | Google (Kaggle public) | Expert | Expert | ||
| Labeling approach | Adjudication | Adjudication and self-training | Adjudication | Majority vote | Majority vote | ||
| Camera, with 45° field of view | Topcon TRC NW6 | Various cameras | Topcon TRC NW6 | Different cameras | Topcon DRI OCT Triton | ||
| Clinical environment | Public ophthalmology department | Public ophthalmology department and primary care | Public ophthalmology department | Primary care | Private ophthalmology practice | ||
External Validation Data Set
The external validation data set comprised 210 CFPs of 106 patients with diabetes who attended a private medical retina clinic in Tanta, Egypt. A Topcon DRI OCT Triton machine, version 10.11 (Topcon Corporation) was used to acquire 55° fundus photographs. All data were obtained retrospectively via convenience sampling through medical record review of clinical visits and were subsequently anonymized by 1 author (M.M.). The data set was labeled by 3 retina specialists (M.B.G., J.H., and H.K.) using an identical approach to the EyePACS validation.
Model Training
The DL models were trained using AutoML Vision on the Google Cloud Platform (GCP). As described in our prior work, this platform provides a graphical user interface for data upload, labeling, and model training (all without coding).12,13,34 Automated ML entails data set management, neural architecture search, and automated hyperparameter tuning. Images were uploaded to GCP buckets, and labels were uploaded to the GCP via CSV files containing labels and data set splits. External validations were performed via command-line interface batch prediction requests. Patient-level splits were maintained for the EyePACS data set; however, no patient-level data were provided for the Messidor-2 data set, for which we were unable to ensure that patient-level splits were maintained. Each cloud compute hour represents 8 parallel Nvidia Tesla V100 GPU-connected machines (Nvidia). All automated ML model training was specified to use maximum allowable cloud compute hours (800) with early stopping enabled, which automatically stopped training when no further improvement was noted. There were no local computer requirements for use of cloud-based platforms.
Teacher Model
The Messidor-2 data set with gold-standard adjudicated labels applied was randomly split to train, tune, and validation sets (80%, 10%, and 10%, respectively). Automated ML was used as described to train a DL model, which is henceforth referred to as the teacher model.
Student Model
Following teacher model training, the model was deployed on the GCP, and batch prediction was performed via the GCP software developer kit command-line interface to run inference on the EyePACS training data set (n = 58 389). This generated model predictions of RDR and NRDR for EyePACS training images. The resulting predictions were assigned as pseudolabels to the EyePACS training data set (n = 58 389), which was combined with the teacher model training data set (n = 1395). Subsequently, a student model was trained via automated ML using the combined training set (n = 59 784) with the teacher tune set (n = 175) (Figure 2).
Bespoke Model
To compare our code-free automated ML model approach with a traditional DL model designed via coding, we developed a bespoke-coded RDR student model using identical images, data splits, and label sets to the automated ML approach. Models were built in TensorFlow, version 1.15 (TensorFlow), with Python, version 3.7 (Python Software Foundation) and the sklearn library (Scikit-learn).42,43 We first compared the performance of 2 commonly used model architectures, InceptionV3 and ResNet50, in a general hyperparameter configuration. We selected InceptionV3 as the backbone of the bespoke model due to its superior performance. We then searched for optimized performance by grid-searching the hyperparameters, including the learning rate, optimizer momentum, and batch size. We implemented data augmentation on the training and tuning sets, including random rotation and flipping as well as color jitter to avoid model overfitting and to increase generalization. An InceptionV3 model pretrained on ImageNet weights, with a learning rate of 0.1, SGD momentum of 0.0, and batch size of 32, was trained for 20 epochs. All model layers were set as trainable.
Statistical Analysis
Performance metrics are reported at the Youden threshold. Confusion matrices were used to calculate accuracy, sensitivity, specificity, and F1 scores. Batch prediction results displayed softmax outputs for predictions, which were used to generate receiver operating characteristic curves and calculate the area under the receiver operating characteristic curve (AUROC) using sklearn inbuilt functions. The Fisher exact test was performed using a GraphPad online calculator.44 Two-tailed P values were calculated for failure case analysis. Data analysis was performed from February 1 to February 28, 2021.
Results
Internal Validation
The student model demonstrated improved AUROC and overall performance metrics compared with the teacher model (Figure 3). For the Messidor-2 validation data set, the AUROC values, accuracy, and F1 scores were 0.951, 93.7%, and 86.7% for the student model compared with 0.939, 92.0%, and 84.1% for the teacher model, respectively. Sensitivity and specificity were 84.8% and 96.9% for the student model and 80.4% and 96.1% for the teacher model, respectively. Full performance metrics are listed in Table 2.
Figure 3. Algorithm for Receiver Operating Characteristic Curves.
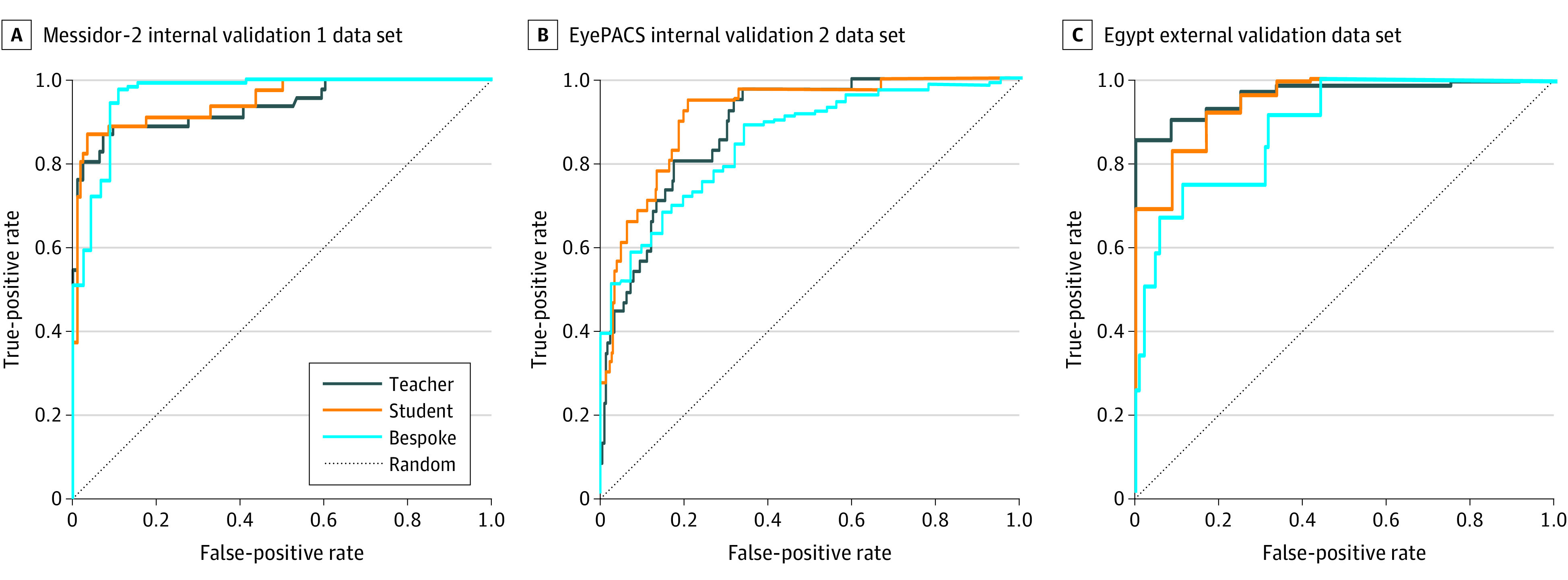
A, Receiver operating characteristic curves for teacher, student, and bespoke manually coded models on the Messidor-2 internal data set. B, Respective model performance on the EyePACS internal data set. C, Respective model performance on the Egypt external data set. Ground-truth labels were adjudicated (Messidor-2) or arbitrated (EyePACS and Egypt).
Table 2. Algorithm Performance Metrics.
| Model | AUROC | Accuracy, % | F1 score, % | Sensitivity, % | Specificity, % | PPV, % | NPV, % |
|---|---|---|---|---|---|---|---|
| Teacher | |||||||
| Messidor-2 internal validation 1 | 0.939 | 92.0 | 84.1 | 80.4 | 96.1 | 88.1 | 93.2 |
| EyePACS internal validation 2 | 0.886 | 84.3 | 51.1 | 56.1 | 89.2 | 46.9 | 92.2 |
| Egypt external validation | 0.964 | 93.3 | 96.4 | 94.4 | 75.0 | 98.4 | 45.0 |
| Student | |||||||
| Messidor-2 internal validation 1 | 0.951 | 93.7 | 87.6 | 84.8 | 96.9 | 90.7 | 94.7 |
| EyePACS internal validation 2 | 0.916 | 74.4 | 52.0 | 95.1 | 70.8 | 35.8 | 98.8 |
| Egypt external validation | 0.950 | 96.7 | 98.3 | 100 | 41.7 | 96.6 | 100 |
| Bespoke | |||||||
| Messidor-2 internal validation 1 | 0.985 | 96.5 | 94.8 | 96.5 | 96.9 | 89.1 | 96.1 |
| EyePACS internal validation 2 | 0.853 | 74.4 | 83.2 | 74.2 | 75.6 | 94.7 | 33.3 |
| Egypt external validation | 0.890 | 94.3 | 50.0 | 50.0 | 97.0 | 50.0 | 97.0 |
Abbreviations: AUROC, area under the receiver operating characteristic curve; NPV, negative predictive value; PPV, positive predictive value.
The EyePACS validation data set AUROC was improved for the student model compared with the teacher model. The AUROC values, accuracy, and F1 scores were 0.916, 74.4%, and 52.0% for the student model compared with 0.886, 84.3%, and 51.1% for the teacher model, respectively. Compared with Messidor-2 internal validation, there were increased false-positive predictions in both the student and teacher models (eTable in Supplement 1) and, thus, lower positive predictive values and F1 scores. The decreased number of false-negative predictions in the student model (n = 2) compared with the teacher model (n = 18) represents 16 patients who may have otherwise suffered vision-threatening consequences from undertreatment.
External Validation
Student model performance on the Egypt external validation data set demonstrated similarly high performance compared with the teacher model. The AUROC values, accuracy, and F1 scores were 0.950, 96.7%, and 98.3% for the student model and 0.964, 93.3%, and 96.4% for the teacher model, respectively (Table 2). Sensitivity and specificity were 100% and 41.7% for the student model and 94.4% and 75.0% for the teacher model, respectively. A markedly lower specificity in this relatively unbalanced data set containing greater than 90% RDR suggests that tuning the threshold for this care setting may be necessary for balanced model performance with respect to desired false-negative and false-positive rates.
Failure Case Analysis
We performed a post hoc failure analysis to better characterize cases in which models generated incorrect (false-positive or false-negative) predictions. Because we had access to 3 independent grades for each image in the EyePACS and Egypt validation data sets, we binarized the validation images with or without grader agreement with respect to RDR vs NRDR (Figure 1). In the Egypt validation data set, incorrect model predictions were significantly less likely to have grader agreement compared with correct (true-positive or true-negative) predictions (20.0% vs 92.7%; P < .001). Similarly, in the EyePACS validation data set, incorrect predictions demonstrated significantly less grader agreement compared with correct predictions (60.8% vs 87.4%; P < .001). Together, these findings suggest that disagreed-upon images may have inherent ground-truthing difficulty and demonstrate correspondingly increased classification difficulty for humans and models alike.
Bespoke DL Model
The AUROC of the bespoke model on the EyePACS validation data set was slightly worse compared with the automated ML student model. The AUROC values, accuracy, and F1 scores were 0.853, 74.4%, and 83.2% for the bespoke model compared with 0.916, 74.4%, and 52.0% for the automated ML model, respectively (Table 2). However, the performance of the bespoke model on the Messidor-2 validation data set was improved compared with the automated ML student model. The AUROC values, accuracy, and F1 scores were 0.985, 96.5%, and 94.8% for the bespoke model compared with 0.951, 93.7%, and 87.6% for the automated ML model, respectively. With regard to the Egypt external validation data set, the AUROC of the bespoke model was worse compared with the automated ML student model. The AUROC values, accuracy, and F1 scores were 0.890, 94.3%, and 50.0% for the bespoke model compared with 0.950, 96.7%, and 98.3% for the automated ML model, respectively.
Discussion
This study presents a self-training automated ML approach with public data sets for DR classification of retinal CFPs. This framework simultaneously addresses multiple barriers to the democratization of clinician-driven ML. Our findings suggest that leveraging small data sets with high-quality labels on large, unlabeled public data sets improves model performance. The resultant automated ML student model demonstrated improved AUROC values on internal validation (from 0.939 to 0.951 and 0.886 to 0.916) on the Messidor-2 and EyePACS data sets, respectively, and a similarly high AUROC on the external validation data set. Validation performance improved on the EyePACS data set with images from a variety of imaging hardware, suggesting enhanced generalizability, via incorporating the larger pseudolabeled data set.45 Compared with our bespoke model, the automated ML models demonstrated improved AUROC in both the EyePACS and external validation data sets (0.853 and 0.890 vs 0.916 and 0.950, respectively), suggesting that automated ML obviates the need for manual coding. Together, these findings suggest that clinicians without coding expertise or access to private and well-labeled data sets can design their own ML models. As ML tools are poised to become commonplace in ophthalmology and medicine, clinicians are best suited to direct AI design for solving clinically relevant problems. Using our framework, motivated clinicians can familiarize themselves with ML and derive value for their patients.
A post hoc analysis on the EyePACS and Egypt validation data sets determined that images with grader disagreement were more likely to receive incorrect model predictions (60.8% vs 87.4%; P < .001 and 20.0% vs 92.7%; P < .001, respectively). This finding suggests that edge cases with inherent ground- truth uncertainty are more likely to have incorrect predictions. Grader variability is a well-known issue with DR grading, which is a complex process that requires human identification of subtle retinal microvascular abnormalities.9 Despite gaining insights on model outputs through this analysis, model explainability is inherently more opaque with automated ML. Nevertheless, knowledge of model architectures and hyperparameters in bespoke models does not inherently provide explainability on a per-image basis. Our group is investigating automated ML explainability including saliency techniques such as integrated gradients and XRAI region-based attribution maps.46,47,48
Although our student models demonstrated high AUROC values, pooled metrics may obscure the possibility of inaccurate predictions on important subgroups.49 For example, the model may underperform in patients with proliferative DR and tractional retinal detachment. Prior studies have demonstrated that commercial DR algorithms have low sensitivity to detect these cases in clinical scenarios50 despite high sensitivity on publicly available data sets.51 This is termed hidden stratification and may be especially meaningful if worse performance occurs in severe disease.51,52 An analysis of hidden stratification is beyond our scope given the limited granularity of public data sets. However, we believe this topic represents a promising future direction.
Limitations
Our study has several limitations. Because patient-level data were not provided for the Messidor-2 data set, we were unable to ensure patient-level splits. To mitigate the potential for falsely increased performance, we performed an additional validation with a subset of the EyePACS data set regraded and arbitrated by 3 retina specialists. The Egypt data set consists of CFPs from a different camera, field of view, and higher RDR prevalence compared with our internal validation. On this data set, the student model had 11 fewer false-negative predictions than the teacher model, which in combination with a 100% negative predictive value is amenable to a screening use case. The data shift was presumably larger for the student model because it was mostly trained on EyePACS images comprising the least RDR prevalence. The Egypt data set acts as a “worst-case” stress test of data distribution shift and model generalization and is not used to compare student and teacher performance. The bespoke model may have overfit, as its performance was worse on this data set. A limitation of the automated ML platform is its inability to compare or provide uncertainty estimates between models; thus, we were unable to report CIs of performance variations.
Self-training has the potential for misclassification of unlabeled data by the teacher model.53 Although this may aggregate incorrect pseudolabels over successive rounds of self-training, we limited our approach to 1 round and demonstrated improved performance. A possible solution is to filter training data based on teacher model softmax outputs, in which predictions below a threshold are excluded from student training.10,26,54 However, exclusively using the most confident predictions may decrease generalizability, which would be detrimental in clinical use, as high variability occurs from image (eg, quality, device) and patient-related (eg, race and ethnicity, concurrent disease) factors.55
Although there are free tiers, cloud-based automated ML incurs scaled costs, and self-training on large data sets may become expensive. While many automated ML platforms, including the GCP, are compliant with Health Insurance Portability and Accountability Act and International Organization for Standardization requirements, ethics approvals are necessary before using these platforms on identifiable data sets. Before clinical use, any algorithm used as a medical device must undergo requisite pivotal trials to demonstrate safety and clinical outcomes, and it would benefit from multidisciplinary teams including computer scientists, statisticians, and clinicians.
Conclusions
In this quality improvement study, we leveraged a self-training, code-free automated ML approach to address barriers for democratization of clinician-driven ML, including the need for coding expertise, scarcity of data, and cost of high-quality labeling. Using public DR data sets to classify DR referral, we elucidated that self-training is an effective method to increase model performance and generalizability while decreasing the need for expensive expert labeling. To address patient-relevant clinical end points, medical ML models may be best designed by use case experts such as clinicians. The improved performance of our code-free automated ML models compared with our manually designed bespoke model suggests that automated ML may allow clinicians and researchers without coding expertise to achieve similar results as computer scientists. As the tools for ML continue to be made more readily available, our SSL approach has the potential to address the disparity of needing expensive clinical labeling, potentially enabling clinicians without coding expertise or access to large, well-labeled private data sets to develop their own models.
eTable. Model Confusion Matrices
Data Sharing Statement
References
- 1.Ting DSW, Pasquale LR, Peng L, et al. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol. 2019;103(2):167-175. doi: 10.1136/bjophthalmol-2018-313173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aristidou A, Jena R, Topol EJ. Bridging the chasm between AI and clinical implementation. Lancet. 2022;399(10325):620. doi: 10.1016/S0140-6736(22)00235-5 [DOI] [PubMed] [Google Scholar]
- 3.Gartner identifies the top strategic technology trends for 2021. News release. Gartner. Accessed November 27, 2022. https://www.gartner.com/en/newsroom/press-releases/2020-10-19-gartner-identifies-the-top-strategic-technology-trends-for-2021
- 4.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436-444. doi: 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 5.Mahajan D, Girshick R, Ramanathan V, et al. Exploring the limits of weakly supervised pretraining. arXiv. Preprint posted online May 2, 2018. doi: 10.1007/978-3-030-01216-8_12 [DOI]
- 6.Korot E, Wood E, Weiner A, Sim DA, Trese M. A renaissance of teleophthalmology through artificial intelligence. Eye (Lond). 2019;33(6):861-863. doi: 10.1038/s41433-018-0324-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellemo V, Lim ZW, Lim G, et al. Artificial intelligence using deep learning to screen for referable and vision-threatening diabetic retinopathy in Africa: a clinical validation study. Lancet Digit Health. 2019;1(1):e35-e44. doi: 10.1016/S2589-7500(19)30004-4 [DOI] [PubMed] [Google Scholar]
- 8.Heydon P, Egan C, Bolter L, et al. Prospective evaluation of an artificial intelligence-enabled algorithm for automated diabetic retinopathy screening of 30 000 patients. Br J Ophthalmol. 2021;105(5):723-728. doi: 10.1136/bjophthalmol-2020-316594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krause J, Gulshan V, Rahimy E, et al. Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology. 2018;125(8):1264-1272. doi: 10.1016/j.ophtha.2018.01.034 [DOI] [PubMed] [Google Scholar]
- 10.Xie Q, Luong MT, Hovy E, Le QV. Self-training with noisy student improves ImageNet classification. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). IEEE; 2020:10684-10695. doi: 10.1109/CVPR42600.2020.01070 [DOI] [Google Scholar]
- 11.Nguyen Q, Valizadegan H, Hauskrecht M. Learning classification models with soft-label information. J Am Med Inform Assoc. 2014;21(3):501-508. doi: 10.1136/amiajnl-2013-001964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faes L, Wagner SK, Fu DJ, et al. Automated deep learning design for medical image classification by health-care professionals with no coding experience: a feasibility study. Lancet Digit Health. 2019;1(5):e232-e242. doi: 10.1016/S2589-7500(19)30108-6 [DOI] [PubMed] [Google Scholar]
- 13.Korot E, Guan Z, Ferraz D, et al. Code-free deep learning for multi-modality medical image classification. Nat Mach Intell. 2021;3(4):288-298. doi: 10.1038/s42256-021-00305-2 [DOI] [Google Scholar]
- 14.Yang J, Zhang C, Wang E, Chen Y, Yu W. Utility of a public-available artificial intelligence in diagnosis of polypoidal choroidal vasculopathy. Graefes Arch Clin Exp Ophthalmol. 2020;258(1):17-21. doi: 10.1007/s00417-019-04493-x [DOI] [PubMed] [Google Scholar]
- 15.Antaki F, Kahwati G, Sebag J, et al. Predictive modeling of proliferative vitreoretinopathy using automated machine learning by ophthalmologists without coding experience. Sci Rep. 2020;10(1):19528. doi: 10.1038/s41598-020-76665-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JH, Kim YT, Lee JB, Jeong SN. A performance comparison between automated deep learning and dental professionals in classification of dental implant systems from dental imaging: a multi-center study. Diagnostics (Basel). 2020;10(11):910. doi: 10.3390/diagnostics10110910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Touma S, Antaki F, Duval R. Development of a code-free machine learning model for the classification of cataract surgery phases. Sci Rep. 2022;12(1):2398. doi: 10.1038/s41598-022-06127-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan KW, Wong CH, Ip HF, et al. Evaluation of the performance of traditional machine learning algorithms, convolutional neural network and AutoML Vision in ultrasound breast lesions classification: a comparative study. Quant Imaging Med Surg. 2021;11(4):1381-1393. doi: 10.21037/qims-20-922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Byrne C, Abbas A, Korot E, Keane PA. Automated deep learning in ophthalmology: AI that can build AI. Curr Opin Ophthalmol. 2021;32(5):406-412. doi: 10.1097/ICU.0000000000000779 [DOI] [PubMed] [Google Scholar]
- 20.Poplin R, Varadarajan AV, Blumer K, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng. 2018;2(3):158-164. doi: 10.1038/s41551-018-0195-0 [DOI] [PubMed] [Google Scholar]
- 21.Korot E, Pontikos N, Liu X, et al. Predicting sex from retinal fundus photographs using automated deep learning. Sci Rep. 2021;11(1):10286. doi: 10.1038/s41598-021-89743-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mey A, Loog M. A soft-labeled self-training approach. In: 2016 23rd International Conference on Pattern Recognition (ICPR). IEEE; 2016:2604-2609. [Google Scholar]
- 23.Azizi S, Culp L, Freyberg J, et al. Robust and data-efficient generalization of self-supervised machine learning for diagnostic imaging. Nat Biomed Eng. 2023;7(6):756-779. doi: 10.1038/s41551-023-01049-7 [DOI] [PubMed] [Google Scholar]
- 24.Azizi S, Mustafa B, Ryan F, et al. Big self-supervised models advance medical image classification. arXiv. Preprint posted online January 13, 2021. doi: 10.1109/ICCV48922.2021.00346 [DOI]
- 25.Sowrirajan H, Yang J, Ng AY, Rajpurkar P. MoCo-CXR: MoCo pretraining improves representation and transferability of chest X-ray models. arXiv. Preprint posted online October 11, 2020. doi: 10.48550/arXiv.2010.05352 [DOI]
- 26.Triguero I, García S, Herrera F. Self-labeled techniques for semi-supervised learning: taxonomy, software and empirical study. Knowl Inf Syst. 2015;42:245-284. doi: 10.1007/s10115-013-0706-y [DOI] [Google Scholar]
- 27.Chen T, Kornblith S, Norouzi M, Hinton G. A simple framework for contrastive learning of visual representations. arXiv. Preprint posted online February 13, 2020.
- 28.Lee R, Wong TY, Sabanayagam C. Epidemiology of diabetic retinopathy, diabetic macular edema and related vision loss. Eye Vis (Lond). 2015;2:17. doi: 10.1186/s40662-015-0026-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Saaddine JB, Chou CF, et al. Prevalence of diabetic retinopathy in the United States, 2005-2008. JAMA. 2010;304(6):649-656. doi: 10.1001/jama.2010.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeTore J, Rizzolo D. Telemedicine and diabetic retinopathy. JAAPA. 2018;31(9):1-5. doi: 10.1097/01.JAA.0000544310.25116.97 [DOI] [PubMed] [Google Scholar]
- 31.Sim DA, Mitry D, Alexander P, et al. The evolution of teleophthalmology programs in the United Kingdom: beyond diabetic retinopathy screening. J Diabetes Sci Technol. 2016;10(2):308-317. doi: 10.1177/1932296816629983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ipp E, Liljenquist D, Bode B, et al. ; EyeArt Study Group . Pivotal evaluation of an artificial intelligence system for autonomous detection of referrable and vision-threatening diabetic retinopathy. JAMA Netw Open. 2021;4(11):e2134254. doi: 10.1001/jamanetworkopen.2021.34254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abràmoff MD, Lavin PT, Birch M, Shah N, Folk JC. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digit Med. 2018;1:39. doi: 10.1038/s41746-018-0040-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan SM, Liu X, Nath S, et al. A global review of publicly available datasets for ophthalmological imaging: barriers to access, usability, and generalisability. Lancet Digit Health. 2021;3(1):e51-e66. doi: 10.1016/S2589-7500(20)30240-5 [DOI] [PubMed] [Google Scholar]
- 35.Decencière E, Zhang X, Cazuguel G, et al. Feedback on a publicly distributed image database: the Messidor database. Image Analysis & Stereology. 2014;33(3):231. doi: 10.5566/ias.1155 [DOI] [Google Scholar]
- 36.Dugas E, Jorge J, Cukierski W. Diabetic retinopathy detection. Kaggle. 2015. Accessed February 14, 2020. https://www.kaggle.com/c/diabetic-retinopathy-detection/data
- 37.Wilkinson CP, Ferris FL III, Klein RE, et al. ; Global Diabetic Retinopathy Project Group . Proposed international clinical diabetic retinopathy and diabetic macular edema disease severity scales. Ophthalmology. 2003;110(9):1677-1682. doi: 10.1016/S0161-6420(03)00475-5 [DOI] [PubMed] [Google Scholar]
- 38.Scanlon PH. The English National Screening Programme for diabetic retinopathy 2003-2016. Acta Diabetol. 2017;54(6):515-525. doi: 10.1007/s00592-017-0974-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tufail A, Rudisill C, Egan C, et al. Automated diabetic retinopathy image assessment software: diagnostic accuracy and cost-effectiveness compared with human graders. Ophthalmology. 2017;124(3):343-351. doi: 10.1016/j.ophtha.2016.11.014 [DOI] [PubMed] [Google Scholar]
- 40.Ting DSW, Cheung CYL, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318(22):2211-2223. doi: 10.1001/jama.2017.18152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ipp E, Shah VN, Bode BW, Sadda SR. Diabetic retinopathy (DR) screening performance of general ophthalmologists, retina specialists, and artificial intelligence (AI): analysis from a pivotal multicenter prospective clinical trial. Diabetes. 2019;68(suppl 1):599-P. doi: 10.2337/db19-599-P [DOI] [Google Scholar]
- 42.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12(85):2825-2830. [Google Scholar]
- 43.Abadi M, Agarwal A, Barham P, et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. arXiv. Preprint posted online March 14, 2016.
- 44.GraphPad . Fisher’s exact test online calculator. Accessed February 2, 2021. https://www.graphpad.com/quickcalcs/contingency1/
- 45.Rogers TW, Gonzalez-Bueno J, Garcia Franco R, et al. Evaluation of an AI system for the detection of diabetic retinopathy from images captured with a handheld portable fundus camera: the MAILOR AI study. Eye (Lond). 2021;35(2):632-638. doi: 10.1038/s41433-020-0927-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beqiri S, Rustemi E, Kelly M, Struyven R, Korot E, Keane PA. Qualitative comparison of AutoML explainability tools with bespoke saliency methods. Invest Ophthalmol Vis Sci. 2022;63(7):200-F0047. [Google Scholar]
- 47.Sundararajan M, Taly A, Yan Q. Axiomatic attribution for deep networks. arXiv. Preprint posted online March 4, 2017. doi: 10.48550/arXiv.1703.01365 [DOI]
- 48.Kapishnikov A, Bolukbasi T, Viégas F, Terry M. XRAI: better attributions through regions. arXiv. Preprint posted online June 6, 2019.
- 49.Goel K, Gu A, Li Y, Ré C. Model patching: closing the subgroup performance gap with data augmentation. arXiv. Preprint posted online August 15, 2020.
- 50.van der Heijden AA, Abramoff MD, Verbraak F, van Hecke MV, Liem A, Nijpels G. Validation of automated screening for referable diabetic retinopathy with the IDx-DR device in the Hoorn Diabetes Care System. Acta Ophthalmol. 2018;96(1):63-68. doi: 10.1111/aos.13613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abràmoff MD, Lou Y, Erginay A, et al. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Invest Ophthalmol Vis Sci. 2016;57(13):5200-5206. doi: 10.1167/iovs.16-19964 [DOI] [PubMed] [Google Scholar]
- 52.Oakden-Rayner L, Dunnmon J, Carneiro G, Ré C. Hidden stratification causes clinically meaningful failures in machine learning for medical imaging. Proc ACM Conf Health Inference Learn (2020). 2020;2020:151-159. doi: 10.1145/3368555.3384468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li M, Zhou ZH. SETRED: self-training with editing. In: Ho TB, Cheung D, Liu H, eds. Advances in Knowledge Discovery and Data Mining. Springer ; 2005:611-621. doi: 10.1007/11430919_71 [DOI] [Google Scholar]
- 54.Fazakis N, Kanas VG, Aridas CK, Karlos S, Kotsiantis S. Combination of active learning and semi-supervised learning under a self-training scheme. Entropy (Basel). 2019;21(10):988. doi: 10.3390/e21100988 [DOI] [Google Scholar]
- 55.Yip MYT, Lim G, Lim ZW, et al. Technical and imaging factors influencing performance of deep learning systems for diabetic retinopathy. NPJ Digit Med. 2020;3:40. doi: 10.1038/s41746-020-0247-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable. Model Confusion Matrices
Data Sharing Statement


