Abstract
The high mobility group protein HMGB1 is a highly abundant chromosomal protein known to interact preferentially with DNA that is branched, bent or otherwise structurally altered. Biologically the protein is thought to facilitate promoter attachment by transcription factors. Recently, however, HMGB1 has been shown to have biological roles beyond that of an architectural DNA-binding protein. Here we investigate the binding interactions of recombinant HMGB1 proteins with two branched RNA’s E. coli 5S rRNA and the group I intron ribozyme from Azoarcus pre tRNAIle. Using competitive electrophoretic mobility and circular dichroism binding assays, we show that HMGB proteins bind both substrates with high affinity. We also report that a recombinant rat HMGB protein, rHMGB1b, inhibits RNA cleavage by the ribozyme. These results raise the possibility that HMGB proteins possess structure dependent RNA binding activity and can modulate RNA processing as well as transcription.
Keywords: High Mobility Group Proteins, four-way junction DNA, ribozyme, DNA-binding, RNA-binding
High mobility group (HMG) proteins are among the most abundant non-histone chromosomal proteins, present at levels ca. 106 per nucleus. HMGB1 is a canonical HMG protein that binds strongly to cruciform or bent DNA and has been implicated in a variety of nuclear functions such as transcription, DNA repair, recombination and chromatin fiber assembly [1, 2]. Due to its high copy number in mammalian cells, HMGB1 has been considered a nuclear housekeeping product. However, HMGB1 has been shown to be a key component of alternate cellular pathways that mediate neuronal differentiation, stem cell recruitment and innate immunity; current research focuses on defining the role of HMGB1 as a pro-inflammatory cytokine [3].
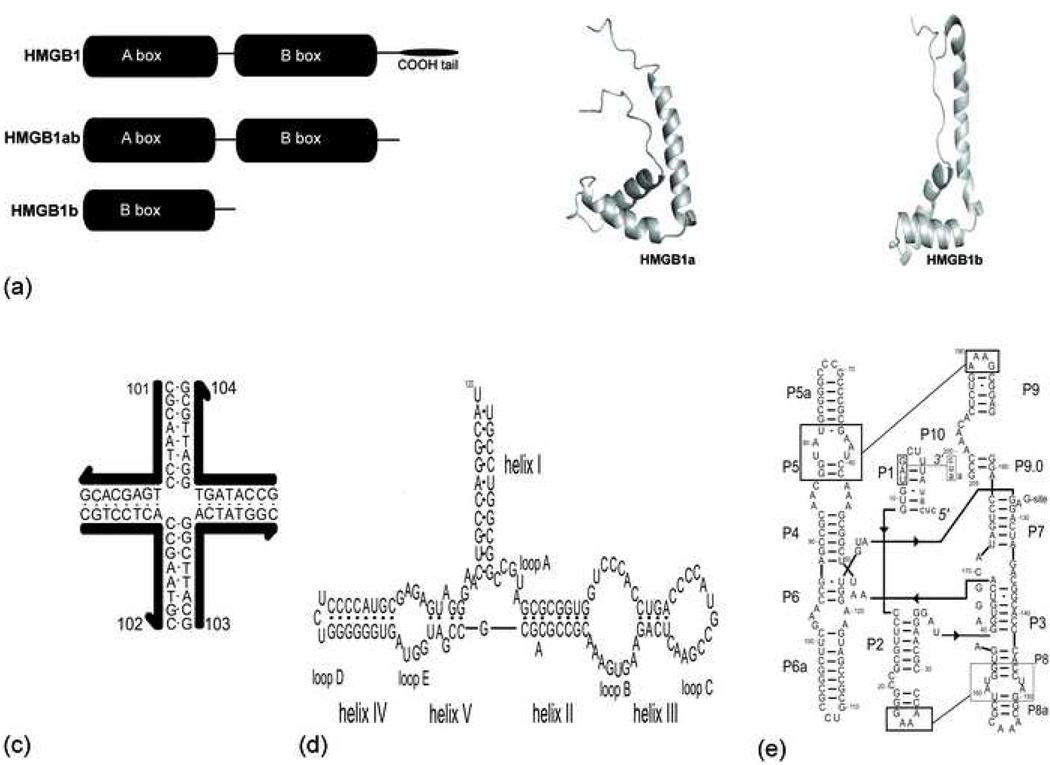
HMGB1 has a tripartite structure, consisting of two HMG box subunits (A and B) together with an acidic C-terminus (Figure 1a). Each box subunit possesses a characteristic global fold of three α-helices arranged in an L-shaped structure [2]. Each HMG box is composed of a conserved sequence of ~80 amino acids that are rich in basic, aromatic and proline residues typically present in DNA-binding proteins [1]. Individual boxes of HMGB1 bind weakly to duplex DNA but strongly to bent and distorted DNA structures such as four-way junctions (4WJs), super-coiled and cisplatin modified DNA [4, 5]. We have shown previously that recombinant HMG proteins containing tandem HMG boxes, arranged in either an AB or BA alignment, bind 4WJ DNA with higher affinity than individual HMG boxes [6].
Figure 1 (a-d):
(a) Schematic representation and NMR structure of recombinant HMG box proteins. HMGB1a and HMGB1b tructures correspond to PDB coordinate files 1aab and 1hme. (b) Schematic of the four-way junction DNA, J1. (c) Secondary structure of E. coli 5S rRNA. (d) Schematic of the group I pre-tRNAIle ribozyme, figure is modified from the model presented by Cech [27].
While HMGB1 is a well-established DNA binding protein, its role in alternate cellular pathways is not fully characterized, raising the possibility that HMG proteins may interact with a wider variety of nucleic acid substrates. For example the canonical HMG protein, HMGD, binds strongly to double-stranded regions of two HIV-1 regulatory RNA structures: the transactivation response region (TAR) and the Rev binding protein element (RBE) [7]. Other DNA-binding proteins have been reported to interact with RNA as well. The tumor suppressor p53 has also been found to bind RNA in a sequence-nonspecific manner that promotes RNA-RNA annealing [8].
To explore the role of RNA binding in the potential functions of HMGB1, we have investigated the non-specific binding interactions of recombinant HMGB1 proteins with unfractionated E. coli tRNA. We found no evidence for interaction in the case of tRNA, so we turned next to longer branched RNA species, including 5S, 16S and 23S E. coli rRNA’s. Experiments using labeled junction DNA as a probe revealed competition with each of these RNA’s. Figure 2 shows the data for 5S rRNA. Electrophoretic mobility shift (EMSA) and circular dichroism (CD) binding assays indicate that HMGB proteins, HMGB1b and HMGB1ab, bind 5S rRNA with high affinity. To detect functional consequences of HMG interactions with branched RNA, we tested the effect of HMG binding on the RNA splicing activity of the group I intron ribozyme from Azoarcus pre-tRNAIle (L-3). Binding of rHMGB1b to the ribozyme significantly reduces its RNA cleavage activity. Taken together, our data suggest that HMGB1 interacts with branched RNA species and thus may play a role in RNA processing.
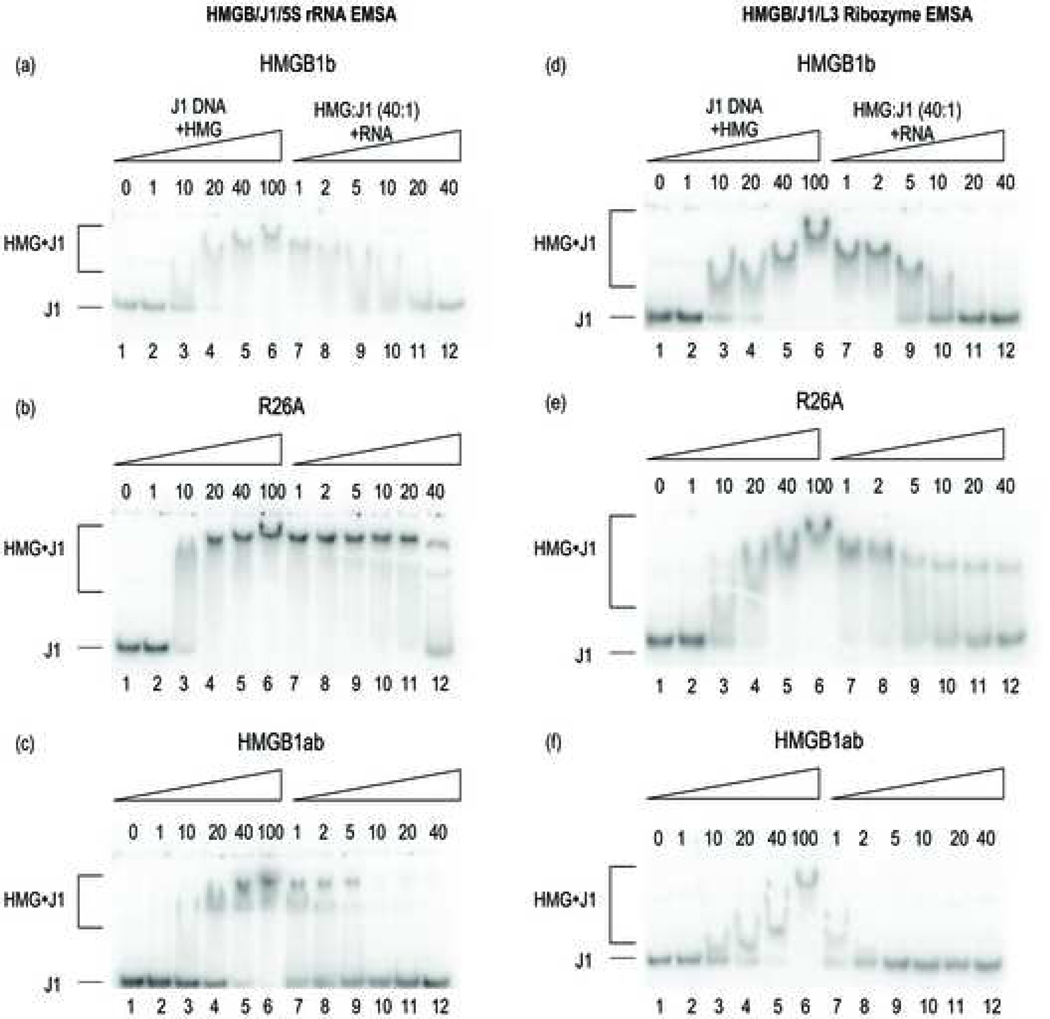
Figure 2 (a-f):
(a-c) Competitive EMSAs of HMG:J1 complex(es) in the presence of increasing 5S rRNA competitor RNA. Lanes 2–6 indicate the molar excess of protein concentration to J1. Lanes 7–12 indicate the molar excess of competitor RNA to J1. (d-f) Competitive EMSAs of HMGB:J1 complex(es) in the presence of increasing L-3 ribozyme competitor RNA.
Materials and methods
HMGB1 protein constructs
HMGB1b and HMGB1b/R26A (R26A), were expressed from the HMGB1b/pHB1 clone that was kindly provided by S.J. Lippard (Department of Chemistry, Massachusetts Institute of Technology). The alanine replacement mutation of R26A was introduced using the Kunkel method [9]. The di-domain protein, HMGB1ab, was cloned and inserted into the pGEX-4T-3 vector as described previously [6]. Recombinant HMG proteins were expressed, purified and characterized by methods described previously [6]. H1 and core histone proteins were kindly provided by Greg Bowman (T.C. Jenkins Department of Biophysics, Johns Hopkins University).
4WJ formation
The 4WJ, J1, was the DNA substrate employed in competitive EMSAs (Figure 1b) [10]. Oligonucleotide 101 was radiolabeled at its 5’ terminus with [γ−32P]ATP using T4 polynucleotide kinase. The radiolabeled strand (101) was purified with a Bio-Spin 6 column (BioRad). J1 was annealed by mixing the radiolabeled strand (101) with a 5-fold excess of the unlabelled oligonucleotides in 50 mM Tris-HCl (pH 7.5)/10 mM MgCl2. The mixture was lyophilized and suspended in annealing buffer: 50 mM Tris-HCl (pH 7.5) and 10 mM MgCl2, incubated for 2 min at 90o C and cooled to room temperature.
5S rRNA E. coli
5S rRNA (120-nt) was purchased from Sigma-Aldrich. 5S rRNA stock samples were diluted in 10 mM TrisHCl (pH 7.5), 0.1 M NaCl and 1mM MgCl2 before use.
(L-3) Azoarcus Ribozyme
The (L-3) Azoarcus ribozyme was prepared as previously described [11]. The ribozyme was engineered by deleting the first two nucleotides of the intron and the last two guanosines (G205 and G206) at the intron’s 3′-end. The nucleotides U4, U5 and U6 were mutated to G4, G5 and C6 respectively. The ribozyme transcripts were gel purified and resuspended in 10 mM Tris-HCl (pH 7.5) and 0.01 mM EDTA before use.
Electrophoretic mobility shift assays (EMSAs)
Radiolabeled J1 (12.5 nM) was incubated with HMG proteins in the absence and in the presence of competitor RNA. Each reaction mixture was incubated in binding buffer: 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM MgCl2 and 10% (w/v) glycerol in a final volume of 20 μl on ice for 30 min prior to loading onto 15% polyacrylamide gels in 0.5X TAE [20 mM Trisma (pH 8.0), 8 mM acetate, 0.1 mM MgCl2) at 4 ˚C for ~3.5 h. Gels were dried and analyzed using a Storm PhosphorImager.
Circular dichroism measurements
Circular dichroism (CD) spectra were recorded using an AVIV 202 spectrometer (Aviv Associates, Lakewood, NJ). RNA-HMG binding interactions were determined by recording the change in the CD spectrum of each RNA. Each RNA (1.0 μM) was incubated on ice for 30 min with each HMG protein (10 μM) in binding buffer: 20 mM HEPES, 30 mM NH4Cl, 0.2 M KCl, 2 mM DTT, 0.5 mM MgCl2 and 10% glycerol at 4°C. CD spectra were measured in a 0.1 cm path-length quartz cuvette and collected from 305 – 200 nm in 0.5 nm increments at 4°C.
Ribozyme cleavage assays
Details of ribozyme cleavage assays presented in Supplementary data [11, 12].
Results
Competitive Electrophoretic Mobility Assays
Competitive gel shift assays were conducted to detect binding of HMG proteins to the cognate DNA substrate, J1, in the presence each RNA. Three proteins were investigated: HMGB1b, R26A and HMGB1ab. R26A is an HMGB1b alanine replacement mutant that binds J1 with 2–3 fold higher affinity than the natvie protein [13]. Hence, the resulting complex provides a more stable species to evaluate the effect(s) of each RNA on HMG-J1 binding affinity. We have shown previously, using fluorescence binding assays and analytical ultracentrifugation, that HMGB1b binds J1 with a binding stoichiometry of 4:1 [14]. A 4:1 complex consists of four protein monomers bound to each branch point of the junction. Typical 4:1 complexes migrate as a single band without discrete intermediate binding species.
First labeled J1 was titrated with increasing amounts of each protein in the absence of competitor to demonstrate that each protein forms a stable complex (Figure 2, lanes 3–6). In each case, HMG proteins bound J1 to form a single major complex that did not reveal intermediate binding species. The single domain proteins, HMGB1b and R26A, have been shown to bind J1 to form 4:1 protein-DNA complexes [14]. We have not experimentally determined the binding stoichiometry of the HMGB1ab:J1 complex but the lack of discrete intermediate binding species indicates that resulting complex is stable. After establishing that each protein binds J1, complexes formed in the presence of 40-fold excess HMG-to-J1 (Figure 2, lane 5) were used to investigate the effect of competitor species. Each complex was titrated with increasing amounts of unlabeled RNA competitor (Figure 2, lanes 7–12). Although accurate binding constants were not determined, the concentrations at which 50% loss of J1 binding could be detected.
Figure 2a illustrates the behavior of the HMGB1b:J1 complex in the presence of 5S rRNA. The complex remains intact until the concentration of 5S rRNA reaches a 5-fold molar excess over J1 (lane 9). HMGB1b:J1 completely dissociates in the presence of 10-fold excess of 5S rRNA (lane 10). On the other hand, the R26A:J1 complex does not dissociate until 5S rRNA reached a 40-fold molar excess (Figure 2b, lane 12). The enhanced affinity of R26A for J1 is consistent with our previously reported results [13]. Next, we investigated the binding of J1 with the tandem HMG box protein, HMGB1ab. In this case we find that HMGB1ab displays the weakest affinity for J1 in the presence of 5S rRNA. The HMGB1ab:J1 complex is roughly half-dissociated in the presence of equamolar 5S rRNA (Figure 2c, lane 7) and completely dissociated in the presence of 10-fold excess of 5S rRNA (lane 10).
We also used competitive EMSAs to monitor the effect of a different RNA species, the L-3 ribozyme, on HMG:J1 binding affinity. Figure 2d displays the binding affinity profile of the HMGB1b:J1 complex in the presence of L-3 ribozyme. The complex remains tightly associated until the concentration of L-3 ribozyme reaches 5-fold molar excess over J1 (lane 9). At this point approximately 50% of the HMGB1b:J1 complex is dissociated. The complex dissociates completely in the presence of 20-fold excess of L-3 ribozyme (lane 11). In an analogous experiment, approximately 35% of the R26A:J1 complex dissociates in the presence of a 5-fold excess of competitor (lane 9, Figure 2e). Despite the initial reduction in J1 binding affinity, the complex remains intact in the presence of increasing levels of L-3 ribozyme (lanes 10–12) demonstrating higher affinity of R26A for J1. Finally, we investigated effect of the ribozyme on the HMGB1ab:J1 complex. The addition of ribozyme caused a drastic reduction in the affinity of HMGB1ab for J1 (Figure 2f). At equamolar levels of ribozyme and J1, approximately 50% of the complex dissociates and the J1 complex dissociates completely in the presence of 2-fold excess of the ribozyme (lanes 7–8).
Circular Dichroism Spectroscopy
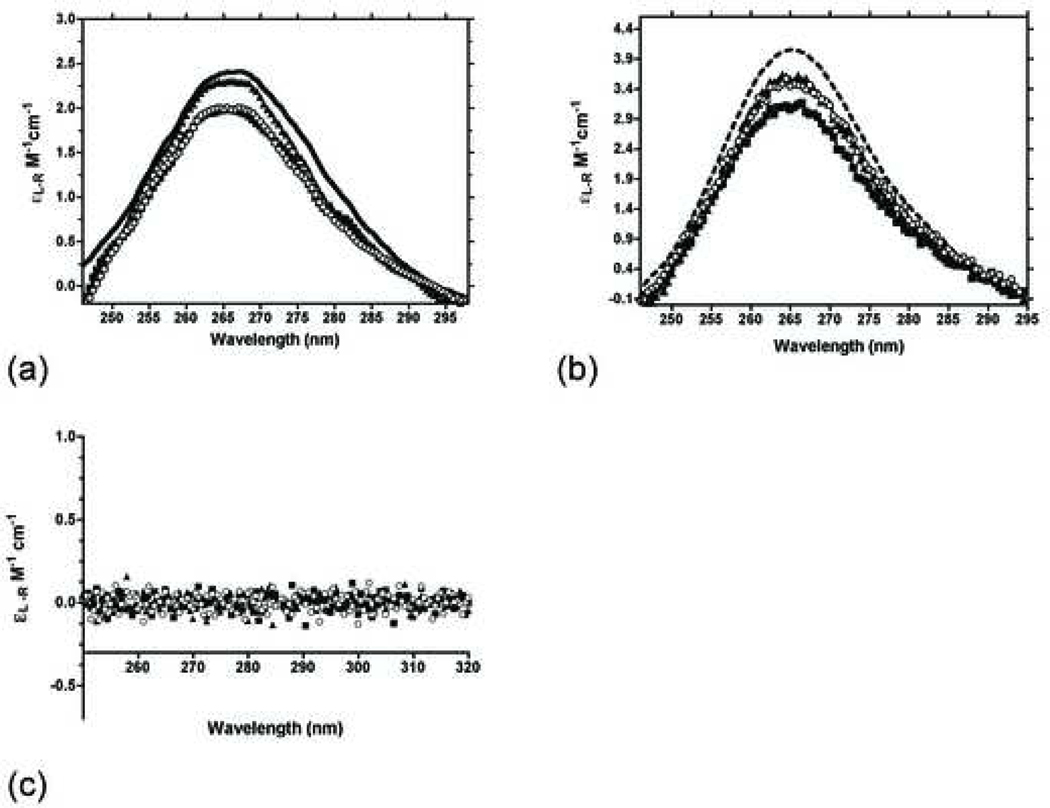
Next we established that HMG proteins interact directly with each RNA substrate in the absence of DNA. We monitored the circular dichroism spectra of 5S rRNA and L-3 ribozyme in the presence of HMG proteins to determine if protein binding produces a spectral change in the RNA. The secondary structure(s) of both RNA’s contain substantial helical structure with ellipticity maxima between 265 and 268 nm (Figure 3).
Figure 3 (a-c):
(a) CD spectra of the 1μM 5S rRNA in the absence and presence of 10 μM of each HMG protein at 4°C. (b) CD spectra of 1μM L-3 ribozyme in the absence and presence of 10 μM of each HMG protein at 4°C. (c) CD spectra of 10 μM of each HMG protein in the absence of RNA.
Binding interactions between HMG proteins to the RNA’s were investigated by recording the change in CD maxima between 260–270 nm. The spectrum of 5S rRNA undergoes a moderate change in the presence of the proteins. The 5S rRNA signal is reduced by approximately 20% in the presence of HMGB1b (Figure 3a). The tandem box protein, HMB1ab, generated a nearly identical change in the 5S rRNA signal. The binding interaction between R26A and 5S rRNA did not trigger an appreciable change in 5S rRNA secondary structure. It is unclear why the R26A mutant displays a reduced binding interaction with 5S rRNA.
The CD spectra of the L-3 ribozyme were altered significantly following incubation with each protein (Figure 3b). In each case HMG binding destabilizes the structure of the ribozyme. As a control, CD spectra of each protein were measured from 250 – 320 nm in the absence of RNA to ensure that the signal recorded was due solely to RNA (Figure 3c).
Ribozyme Activity Assay
Our data thus far point to tight binding between HMG proteins and 5S rRNA and the L-3 ribozyme. Does binding affect the ribozyme activity? Cleavage reactions were initiated by adding a 9-nt long radiolabeled oligonucleotide substrate to the pre-folded ribozyme. We monitored ribozyme activity following co-incubation with limiting and excess HMGB1b. The ribozyme activity was significantly reduced at protein concentrations > 1 nM (Figure 4a). This reduction in ribozyme activity indicates that protein binding perturbs the catalytically active conformation of the ribozyme.
Figure 4 (a,b):
Cleavage inhibition of L-3 ribozyme by HMGB1b. 9-nt radiolabed substrate was co-incubated with the ribozyme and increasing amounts of proteins as described in Material and methods. Cleavage product formation (fP) plotted vs. protein concentration. (b) Bar graph representation of fP inhibition in the presence of DNA binding proteins (HGMB1b, H1 and histone proteins). fP plotted vs. the net ionic charge of each protein.
Does this reduction in ribozyme activity reflect a nonspecific effect between a positively charged protein and the RNA substrate? To address this issue, we monitored the ribozyme activity in the presence of a series of positively charged proteins, including histone proteins [H1 and core histone mixture (H2A/H2B, H3 & H4)] and a ribosomal protein S17. Like HMG proteins, histone proteins are rich in basic amino acids with helix-loop-helix motifs [15]. S17 is a small β-barrel rRNA protein that binds the 16S rRNA of 30S ribosomal small subunit [16]. L-3 ribozyme was co-incubated with (0.001 – 1000 nM) of each protein and ribozyme activity was determined as described in Materials and methods. The mean residual ribozyme activity (fP) at each protein concentration is plotted vs. the net charge of each protein (Figure 4b). Of the proteins investigated, HMGB1b generated the largest reduction in ribozyme activity. The order of inhibition of the remaining proteins was: core histones < S17 < H1. The data did not show any correlation between net protein charge and the inhibitory effect, implying that the inhibition of ribozyme activity in presence of HMGB1b may reflect a specific interaction between the protein and the ribozyme.
Discussion
We and others have shown that the di-domain protein, HMGB1ab, has a higher binding affinity for junction DNA than single domain proteins [6, 17]. However, in the presence of RNA, the HMGB1ab:J1 complex proves to be less stable than complexes formed by single-domain HMG proteins. The source of instability of the HMGB1ab:J1 complex may be related to the detailed disposition of HMG box subunits near the junction branch point. Using a combination of NMR and footprinting assays Webb and Thomas have shown that the A box of HMGB1ab localizes at the junction branch point, while the B box favors the arms of the junction [18]. If the branch point itself is in a dynamic state, the HMGB1ab:J1 complex may dissociate more readily in the presence of a competitor species. This could account for the complete dissociation of the HMGB1ab:J1 complex in the presence of 2-fold excess ribozyme (Figure 2f).
Both HMGB1b and R26A have a higher relative binding affinity for J1 than the di-domain protein. HMGB1b and R26A presumably have more specific/stronger interactions with the arms of the junction and less of an interaction with the branch point. The HMGB1b:J1 complex is more stable at lower competitor concentrations [RNA < 10-fold excess] but each complex dissociates completely at competitor concentrations of ≥ 20-fold excess. Interestingly, the R26A mutant forms the most stable complex of the proteins investigated. The R26A:J1 complex remains intact throughout each RNA competitor titration. We presume that the increase in binding affinity of this mutant reflects more favorable packing interactions between the concave binding surface of R26A and the DNA backbone of J1.
Direct binding between each RNA and individual HMG proteins were evaluated by monitoring the change in the CD spectra of 5S rRNA and L-3 ribozyme. The CD band of the RNA’s centered near 265 nm measures both helical and tertiary structure [11]. The CD binding assays follow a similar trend to the EMSA data. The di-domain protein, HMGB1ab interacts most strongly with RNA. Both single domain proteins bind L-3 ribozyme to generate a similar change in ribozyme secondary structure but R26A does not shown appreciable binding with 5S rRNA. We assume that the decrease in the CD signal of each RNA represents a loss of secondary or tertiary structure.
As shown in Figure 4, HMGB1b significantly reduced ribozyme activity. Since HMGB1b has a Tm of 46°C the fact that the protein reduces ribozyme activity following a lengthy incubation (15 min.) at 50°C, provides additional evidence for a strong interaction between HMGB1b and the ribozyme. There are a number of possible explanations for inhibition by HMGB1b; one of the most obvious would be electrostatic interactions between (any) basic DNA-binding protein and the highly electronegative ribozyme. It has been shown previously that the cationic antibiotic neomycin B inhibits self-splicing group I introns and the hammerhead ribozyme [19, 20]. In this case the inhibition is attributed to competitive electrostatic interactions between the positively charged ammonium groups of the antibiotic and metal ions near the ribozyme catalytic core [19].
To address the issue of electrostatic inhibition and determine if other classical DNA-binding proteins display a similar effect on the ribozyme we tested the effect of histones on ribozyme activity. The average ribozyme inhibition values of each protein were plotted against the net charge of each protein (Figure 4b). HMGB1b displays the greatest reduction of L-3 ribozyme activity despite having the lowest net charge. H1 has a higher affinity for 4WJ DNA than HMGB1 [21]. Histones and H1 also have a greater chromatin residence time than HMGB1 [22]. The fact that histone proteins have a higher binding affinity for DNA may make them less promiscuous than HMGB proteins.
HMGB1 is a canonical DNA-binding protein that binds preferentially to a variety of distorted DNA structures including 4WJs, bulged/bent DNA and superhelical DNA. These characteristics are thought to implicate HMGB1 in the process of chromatin remodeling [1]. While both HMGB1 and histones are highly abundant, HMGB1 is delocalized within the nucleus with a low chromatin residence time [22]. Thus the protein can translocate within the nucleus [23]. HMGB1 is also released from the nucleus either passively in cells undergoing necrotic cell death or actively in certain cell types (i.e. monocytes and macrophages) [24, 25]. The presence of HMGB1 in the cytoplasm and extra-cellular space triggers a number of responses that are currently under investigation [3]. Whether the protein acts within or outside the nucleus, HMGB1 can access and interact with a variety of alternative nucleic acid substrates, including certain RNA’s as we show here.
In summary our data suggest that HMGB proteins may be associated with RNA related cellular pathways as well as with DNA. HMGB1 might then serve as a “general” nucleic acid chaperone capable of influencing RNA processing as well as regulating transcription levels. More detailed studies will be required to determine the role of HMGB1-rRNA interactions in light of recent data showing that a number of transcription factors bind RNA species as well [26].
Supplementary Material
Footnotes
HMGB interactions with RNA.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- [1].Bustin M, Reeves R, High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function, Prog Nucleic Acid Res Mol Biol 54 (1996) 35–100. [DOI] [PubMed] [Google Scholar]
- [2].Thomas JO, Travers AA, HMG1 and HMG2 and related architectural DNA-binding proteins, Trends Biochem Sci 26 (2001) 167–174. [DOI] [PubMed] [Google Scholar]
- [3].Lotze MT, Tracey KJ, High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal, Nat Rev Immunol 5 (2005) 331–342. [DOI] [PubMed] [Google Scholar]
- [4].Stros M, Launholt D, Grasser KD, The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins, Cell. Mol. Life Sci. 64 (2007) 2590–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pil P, Lippard SJ, Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin., Science 256 (1992) 234–237. [DOI] [PubMed] [Google Scholar]
- [6].Taudte S, Xin H, Bell AJ, Kallenbach NR, Interaction between HMG boxes., Protein Eng. 14 (2001) 1015–1023. [DOI] [PubMed] [Google Scholar]
- [7].Arimondo PB, Gelus N, Hamy F, Payet D, Travers A, Bailly C, The chromosomal protein HMG-D binds to the TAR and RBE RNA of HIV-1., FEBS Letters 485 (2000) 47–52. [DOI] [PubMed] [Google Scholar]
- [8].Oberosler P, Hloch P, Ramsperger U, Stahl H, p53-catalyzed annealing of complementary single-stranded nucleic acids, EMBO J 12 (1993) 2389–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kunkel TA, Bebenek K, McClary J, Efficient site-directed mutagenesis using uracil-containing DNA, Methods Enzymol 204 (1991) 125–139. [DOI] [PubMed] [Google Scholar]
- [10].Kallenbach NR, Ma RI, Seeman NC, An immobile nucleic acid junction constructed from oligonucleotides, Nature 305 (1983) 829–831. [Google Scholar]
- [11].Chauhan S, Caliskan G, Briber RM, Perez-Salas U, Rangan P, Thirumalai D, Woodson SA, RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme, J Mol Biol 353 (2005) 1199–1209. [DOI] [PubMed] [Google Scholar]
- [12].Zaug AJ, Davila-Aponte JA, Cech TR, Catalysis of RNA cleavage by a ribozyme derived from the group I intron of Anabaena pre-tRNA(Leu), Biochemistry 33 (1994) 14935–14947. [DOI] [PubMed] [Google Scholar]
- [13].Taudte S, Xin H, Kallenbach NR, Alanine mutagenesis of high-mobility-group-protein-1 box B (HMG1-B), Biochem J 347 Pt 3 (2000) 807–814. [PMC free article] [PubMed] [Google Scholar]
- [14].Xin H, Taudte S, Kallenbach NR, Limbach MP, Zitomer RS, DNA binding by single HMG box model proteins, Nucleic Acids Res 28 (2000) 4044–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Baxevanis AD, Landsman D, Histone and histone fold sequences and structures: a database, Nucleic Acids Res 25 (1997) 272–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Golden BL, Hoffman DW, Ramakrishnan V, White SW, Ribosomal protein S17: characterization of the three-dimensional structure by 1H and 15N NMR, Biochemistry 32 (1993) 12812–12820. [DOI] [PubMed] [Google Scholar]
- [17].Grasser KD, Teo S-H, Lee K-B, Broadhurst RW, Rees C, Hardman CH, Thomas JO, DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1). Eur. J. Biochem. 253 (1988) 787–795. [DOI] [PubMed] [Google Scholar]
- [18].Webb M, Thomas JO, Structure-specific binding of the two tandem HMG boxes of HMG1 to four-way junction DNA is mediated by the A domain., J. Mol. Bio. 294 (1999) 373–387. [DOI] [PubMed] [Google Scholar]
- [19].Hoch I, Berens C, Wesfhof E, Schroeder R, Antibiotic inhibition of RNA catalysis: neomycin B binds to the catalytic core of the td group I intron displacing essential metal ions., J. Mol. Biol. 282 (1998) 557–569. [DOI] [PubMed] [Google Scholar]
- [20].Clouet-d’Orval B, Stage TK, Uhlenbeck OC, Neomycin inhibition of the hammerhead ribozyme involves ionic interactions., Biochemstry 34 (1995) 11186–11190. [DOI] [PubMed] [Google Scholar]
- [21].Hill DA, Reeves R, Competition between HMG-I(Y), HMG-1 and histone H1 on four-way junction DNA, Nucleic Acids Res 25 (1997) 3523–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T, Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins., Mol. Cell. Biol. 24 (2004) 6393–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Agresti A, Scaffidi P, Riva A, Caiolfa VR, Bianchi ME, GR and HMGB1 interact only within chromatin and influence each other’s residence time., Mol. Cell. 18 (2005) 109–121. [DOI] [PubMed] [Google Scholar]
- [24].Wang H, HMG-1 as a late mediator of endotoxin lethality in mice., Science 285 (1999) 248–251. [DOI] [PubMed] [Google Scholar]
- [25].Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, Agresti A, Bianchi ME, Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion, Embo J 22 (2003) 5551–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cassiday LA, Maher LJ 3rd, Having it both ways: transcription factors that bind DNA and RNA, Nucleic Acids Res 30 (2002) 4118–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tanner MA, Cech TR, Activity and thermostability of the small self-splicing group 1 intron in the pre-tRNAIle of the purple bacterium Azoarcus., RNA 2 (1996) 74–83. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.