Abstract
Background
The utilization of neoadjuvant therapy is progressively expanding in various clinical settings. However, the absence of a clinically validated biomarker to evaluate the treatment response remains a significant challenge in the field. Circulating tumor DNA (ctDNA) detection, a novel and emerging monitoring approach in the field of oncology, holds promise as a potential prognostic biomarker for patients with cancer. This meta‐analysis investigated the clinical significance of ctDNA detection as a predictive tool for cancer recurrence in patients receiving neoadjuvant treatment.
Methods
A comprehensive systematic literature search was conducted using public databases to identify relevant studies that investigated the association between ctDNA detection and cancer recurrence in patients receiving neoadjuvant treatment. Hazard ratios (HRs) and their corresponding 95% confidence intervals (95% CI) were calculated to assess the relationship between cancer recurrence and relevant factors. Cancer recurrence was considered the primary outcome.
Results
A total of 23 studies encompassing 1590 patients across eight different cancer types were included in the final analysis. Positive ctDNA detection was significantly associated with higher cancer recurrence, especially at post‐neoadjuvant treatment and post‐surgery time points. The risk values for the different cancer categories and geographic areas also differed significantly.
Conclusion
Our comprehensive meta‐analysis revealed a significant positive correlation between ctDNA detection and a higher risk of cancer recurrence in patients receiving neoadjuvant treatment. In addition, the risk of recurrence was influenced by variations in cancer type, timing of detection, and geographic region. These findings highlight the promising clinical applicability of ctDNA as a prognostic marker and monitoring approach for patients with cancer. However, the precise mechanism is unknown and more evidence is needed for further research.
Keywords: biomarker, cancer, circulating tumor DNA, meta‐analysis, neoadjuvant treatment
1. INTRODUCTION
Currently, cancer, which is one of the foremost causes of human mortality, exhibits a significantly high worldwide prevalence. The year 2020 witnessed a substantial escalation in the global burden of cancer, with over 19 million cases and approximately 9 million fatalities. 1 These estimations underscore the urgent need for continued research and interventions to combat this devastating disease. Despite advancements in surgical techniques, chemotherapy, radiotherapy, and emerging modalities, which have undoubtedly enhanced survival rates and quality of life among patients with cancer, the persistently high incidence of local recurrence and distant metastasis necessitates further investigation and innovative therapeutic strategies to address this ongoing challenge. Therefore, there is an urgent demand for the development of novel biomarkers that can facilitate the detection of cancer recurrence and monitoring of treatment response, preferably through noninvasive and patient‐friendly approaches, to address the unmet clinical needs in this critical area.
Circulating tumor DNA (ctDNA) holds immense promise as a valuable tool for cancer management and detection, assessing treatment response, and potentially enabling early cancer detection. Circulating cell‐free DNA (ccfDNA), a constituent naturally present in blood at typically low concentrations, exhibits elevated levels in various circumstances such as exercise, trauma, and cancer. 2 , 3 ctDNA carries specific mutations, which can be isolated and detected in a wide range of bodily fluids, constituting a subset of ccfDNA with potential diagnostic and prognostic implications. 4 It has shown great promise for detecting minimal residual disease (MRD) and predicting responses to specific therapies in clinical settings, while being minimally invasive and highly sensitive. 5
Neoadjuvant treatment has gained significant prominence in cancer treatment, which is mainly employed prior to surgery to effectively decrease the occurrence of metastasis and tumor volume. 6 Neoadjuvant treatment exhibits superior benefits across multiple dimensions for patients with cancer when compared to conventional adjuvant therapy. In the context of breast cancer research, noteworthy findings indicate that neoadjuvant treatment significantly contributes to tumor size reduction, increased rates of breast‐conserving surgery, and improved prognostic outcomes for patients with residual disease. 7 Furthermore, neoadjuvant chemotherapy has demonstrated the ability to enhance treatment compliance, potentially leading to improved patient outcomes, while the occurrence of postoperative complications and treatment‐related toxicities may restrict adherence to adjuvant treatment. 8 For instance, breast cancer mortality has emerged as the second most prominent cause of cancer‐related deaths in women. 9 Neoadjuvant treatment represents a valuable therapeutic approach for patients with breast cancer, enabling higher rates of breast conservation and direct assessment of treatment efficacy. 10
However, the existing literature on the correlation between ctDNA detection and cancer recurrence evaluation in patients undergoing neoadjuvant treatment is currently limited and subject to ongoing debate. In current clinical work, the assessment of pathological staging and microscopic residual disease score systems are the main approaches to estimate the risk of recurrence, 11 , 12 , 13 which are limited and difficult to perform. Therefore, there is an urgent clinical need for novel biomarkers to identify the responses to neoadjuvant treatment and recurrence. In this meta‐analysis, we evaluated the potential role of ctDNA detection in predicting cancer recurrence.
2. MATERIALS AND METHODS
Our meta‐analysis was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta‐Analyses guidelines. The International Prospective Register of Systematic Reviews registration number for this study is PROSPERO CRD 42023395312.
2.1. Data sources and literature searching
Five electronic databases, including PubMed, Cochrane Library, Embase, Web of Science, and JAMA, were symmetrically searched based on the MeSH words from the National Center of Biotechnology Information (NCBI). (((“Neoplasms”) AND “Circulating Tumor DNA”) AND “Neoadjuvant Treatment”) AND (“Recurrence” OR “Neoplasm Recurrence, Local”) were used as the search query. The patient, intervention, comparison, and outcome (PICO) framework was used, with no search restrictions. The search was performed independently by two authors, and any disagreements were resolved after discussion with a third author.
2.2. Inclusion and exclusion criteria
All of the authors formulated these criteria to identify eligible studies. The inclusion criteria were as follows: (I) publications on the recurrence of cancer patients treated with neoadjuvant treatment and ctDNA detection; (II) studies where participants were divided into two or more groups according to their ctDNA detection; (III) studies where participants underwent both neoadjuvant treatment and ctDNA detection; (IV) studies including sufficient and standard data; and (V) only English publications. The exclusion criteria were as follows: (I) duplicate publications and data; (II) literature with data from public databases; and (III) literature types such as reviews, case reports, meeting abstracts, and basic experimental research literature.
2.3. Data extraction and quality assessment
The screening of search results and data extraction were performed by two independent authors. Discrepancies were resolved through rigorous discussions. In cases of ongoing disputes, a third author was readily available for further resolution. All participants in the included studies underwent neoadjuvant treatment and ctDNA detection. The following information was collected in a predefined table, from each of the eligible studies, including author's name, publication year, country or area, the number of patients, study design, cancer type, neoadjuvant treatment program, ctDNA analysis method, ctDNA detection time points, ctDNA results, follow‐up duration, and recurrence outcomes. The authors were contacted in instances of data unavailability to retrieve the required information. The Newcastle–Ottawa scale (NOS) was used for the quality assessment of the included studies.
2.4. Outcomes and data analysis
The recurrence risk was the main outcome of our study. Recurrence details were collected from both the ctDNA‐positive and ctDNA‐negative participants. Review Manager 5.4 software for Mac was used to calculate the pooled hazard ratios (HRs) with corresponding 95% confidential intervals (CIs). To minimize the effects of heterogeneity, a random‐effects model was used to perform the dichotomous variance method. Statistical heterogeneity was assessed using the χ2 test and the I 2 test. Publication bias was estimated using a funnel plot, and sensitivity analysis was performed by removing literature from relatively low‐quality resources. p < 0.05 was considered statistically significant in this study. Moreover, to further explore the possible factors affecting this association, subgroup analyses including various cancer types, detection time points, and regions were performed.
3. RESULTS
3.1. Study identification
The initial systemic literature search included 341 articles, comprising 61 articles from PubMed, 22 from the Cochrane Library, 171 from Embase, 72 from the Web of Science, and 15 from JAMA. After removing duplicates and studies that did not meet the requirements, 23 articles were selected (Figure 1).
FIGURE 1.
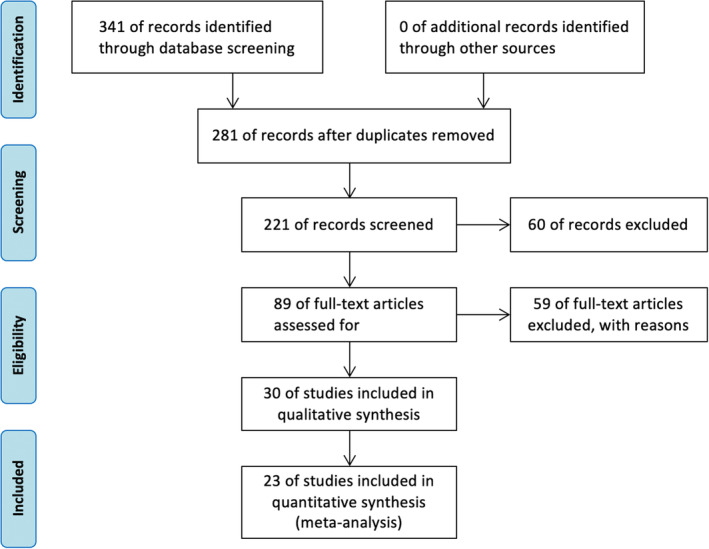
Flowchart of the literature search and selection.
3.2. Baseline characteristics of included research
The characteristics and details of the 23 included studies are summarized in Table 1. A total of 1590 patients, 8 cancer types (breast cancer, non‐small cell lung cancer, gastric cancer, colorectal cancer, renal cancer, esophageal cancer, melanoma, and bladder cancer), and 10 countries from 4 regions (America, Asia, Europe, and Australia) were included in these 23 studies, with publication year from 2016 to 2022. Among all the studies, 11 were prospective observational cohorts, 8 were retrospective cohorts, 2 were randomized controlled studies, and 2 were randomized controlled trials (RCTs). Of the studies included in the analysis, 11, 13, and 15 included pre‐neoadjuvant ctDNA detection (before neoadjuvant treatment), post‐neoadjuvant ctDNA detection (after neoadjuvant treatment but before surgery), and post‐surgery ctDNA detection (after surgery), respectively. The follow‐up duration was collected in 22 studies, ranging from 6.83 months to 61.2 months.
TABLE 1.
Baseline characteristics of included studies.
| Study | Year | Country | Follow‐up | Cancer type | NOS score |
|---|---|---|---|---|---|
| Alessandro Leal 14 | 2020 | America | Median: 42 months | Gastric cancer | 8 |
| Bernadett Szabados 15 | 2022 | England | Median: 25 months | Urothelial bladder cancer | 5 |
| Dengbo Ji 16 | 2020 | China | NA | Rectal cancer | 8 |
| Dongsheng Yue 17 | 2020 | China | Median: 6.83 months | Non‐small cell lung cancer | 8 |
| E. La Rocca 18 | 2020 | Italy | Median: 61.2 months | Breast cancer | 5 |
| E. Ortolan 19 | 2022 | Italy | Median: 36 months | Breast cancer | 7 |
| Emil Christensen 20 | 2022 | Denmark | Median: 96 months | Urothelial bladder cancer | 9 |
| Filipa Lynce 21 | 2019 | America | Median: 17 months | Breast cancer | 5 |
| Fre ´de ´ric Cailleux 22 | 2019 | America | Median: 36.36 months | Breast cancer | 6 |
| Georgina V Long 23 | 2022 | Australia | Median: 27 months | Melanoma | 7 |
| Haeyoung Kim 24 | 2022 | Korea | Median: 22 months | Breast cancer | 5 |
| Hiroyo Takahashi 25 | 2019 | Japan | Median: 23 months | Breast cancer | 6 |
| Jeanne Tie 26 | 2021 | Australia | Median: 50.5 months | Colorectal cancer | 6 |
| Jennifer Wo 27 | 2016 | America | Meidian: 23.1 months | Gastroesophageal cancer | 7 |
| Joana Vidal 28 | 2021 | Spain | Median: 38 months | Rectal cancer | 7 |
| Lisa S.M. Hofste 29 | 2020 | Netherlands | Median: 28 months | Rectal cancer | 9 |
| M.J.M. Magbanua 30 | 2021 | America | Median: 57.6 months | Breast cancer | 7 |
| Po‐Han Lin 31 | 2022 | America | Median: 61.2 months | Breast cancer | 8 |
| Shelize Khakoo 32 | 2020 | America | Median: 26.4 months | Rectal Cancer | 5 |
| Susan G. R. McDuff 33 | 2021 | America | Median: 20 months | Rectal cancer | 7 |
| V. F. Bonazzi 34 | 2020 | Australia | Median: 37 months | Esophageal cancer | 7 |
| Wenyang Liu 35 | 2020 | China | Median: 33.25 months | Rectal cancer | 8 |
| Yaqi Wang 36 | 2022 | China | Median: 21.5 months | Rectal cancer | 8 |
| Study | Number of patients | NAT treatment | Method of ctDNA analysis | ctDNA detection time points | Study type |
|---|---|---|---|---|---|
| Alessandro Leal 14 | 50 | Epirubicin, cisplatin, oxaliplatin, capecitabine | NGS, PCR | Post‐NAT | Randomized controlled study |
| Bernadett Szabados 15 | 36 | Cisplatin, atezolizumab | Multiplex PCR; NGS | Post‐surgery | Prospective study |
| Dengbo Ji 16 | 46 | Capecitabine alone, mFOLFOX6, CapeOx | NGS | Pre‐NAT | Retrospective study |
| 37 | Post‐NAT | ||||
| 32 | Post‐surgery | ||||
| Dongsheng Yue 17 | 22 | Nivolumab + platinum double chemotherapy 54%; docetaxel + cisplatin 18%; Pemetrexed + cisplatin 10%; nivolumab + ipilimumab 18% | ddPCR; UDS; LC‐MRD‐panel; NGS | Pre‐NAT | Retrospective study |
| 22 | Post‐NAT | ||||
| 22 | Post‐surgery | ||||
| E. La Rocca 18 | 25 | Anthracycline + taxane‐based (81.8%); Platinum‐based (12.1%) Anthracycline‐based (6.1%) | NGS; ddPCR | Post‐surgery | Retrospective study |
| E. Ortolan 19 | 26 |
Anthracycline/taxane (80.6%) Anthracycline/taxane + platins (12.9%) Other (6.5%) |
Primary‐tumor‐targeted gene sequencing; Patient‐ specific point mutations; ddPCR | Post‐NAT | Prospective study |
| 24 | Post‐surgery | ||||
| Emil Christensen 20 | 59 |
Gemcitabine + cisplatin (94%) Gemcitabine + carboplatin (1.5%) Gemcitabine (3%) Carboplatin+etoposide (1.5%) |
WES; UDS; NGS | Pre‐NAT | Prospective study |
| 63 | Post‐NAT | ||||
| 64 | Post‐surgery | ||||
| Filipa Lynce 21 | 30 | Taxane + anthracyclines; Taxanes only | NA | Pre‐NAT | Randomized controlled study |
| Fre ´de ´ric Cailleux 22 | 38 | NA | WES; Bespoke multiplex PCR; NGS | Pre‐NAT | Retrospective |
| 42 | Post‐NAT | ||||
| 40 | Post‐surgery | ||||
| Georgina V Long 23 | 35 | dabrafenib + trametinib | NA | Pre‐NAT | RCT |
| Haeyoung Kim 24 | 11 | Capecitabine | Gene sequencing | Pre‐NAT | Prospective study |
| Hiroyo Takahashi 25 | 54 | Paclitaxel | OS‐MSP | Post‐surgery | Prospective study |
| Jeanne Tie 26 | 54 | Oxaliplatin‐based | Safe‐SeqS assay; Tumor tissue mutation analysis; Plasma sample mutation analysis | Pre‐NAT | Prospective study |
| 49 | Post‐surgery | ||||
| Jennifer Wo 27 | 16 | FOLFIRINOX | Paired‐end sequencing; NGS | Post‐NAT | Prospective study |
| 14 | Post‐surgery | ||||
| Joana Vidal 28 | 52 |
Aflibercept+mFOLFOX6 (64%) mFOLFOX6 (36%) |
NGS | Pre‐NAT | RCT |
| 45 | Post‐NAT | ||||
| Lisa S.M. Hofste 29 | 16 | Carboplatin and paclitaxel + Gy radiation | UDS | Post‐surgery | Prospective study |
| M. J. M. Magbanua 30 | 75 | NA | UDS | Pre‐NAT | Retrospective study |
| 74 | Post‐NAT | ||||
| Po‐Han Lin 31 | 95 | Anthracycline; taxane; anti‐Her2 target therapy | NGS | Post‐surgery | Retrospective study |
| Shelize Khakoo 32 | 47 | NA | dd PCR | Post‐NAT | Prospective study |
| Susan G. R. McDuff 33 | 24 | Capecitabine or infusion fluorouracil | NGS; dd PCR | Post‐NAT | Retrospective study |
| 19 | Post‐surgery | ||||
| V. F. Bonazzi 34 | 36 | NA | Multiple tumor biopsies sequencing | Post‐surgery | Retrospective study |
| Wenyang Liu 35 | 42 | Capecitabine + oxaliplatin regimen | Multiplex PCR; LDS | Pre‐NAT | Prospective study |
| 35 | During‐NAT | ||||
| 60 | Post‐NAT | ||||
| Yaqi Wang 36 | 103 | Capecitabine + irinotecan | Panel sequencing | Post‐NAT | Prospective study |
| 103 | Post‐surgery |
Abbreviations: ddPCR, droplet digital polymerase chain reaction; LC‐MRD, lung cancer‐specific minimal residual disease; LDS, low‐depth sequencing; NA, not available; NAT, neoadjuvant treatment; NGS, next‐generation sequencing; OS‐MSP, one‐step methylation‐specific polymerase chain reaction; PCR, polymerase chain reaction; RCT, randomized clinical trial; Safe‐SeqS, safe‐Sequencing; UDS, ultradeep sequencing; WES, whole‐exome sequencing.
3.2.1. Positive‐ctDNA detection was positively correlated with higher recurrence possibility
In the current study, a comprehensive analysis was conducted using all available data to investigate the association between ctDNAs and recurrence. The results revealed that the positive ctDNA detection result was strongly associated with a higher recurrence rate (HR: 4.92, 95% CI: 3.00–8.09, p < 0.001, I 2 = 58%; Figure 2).
FIGURE 2.
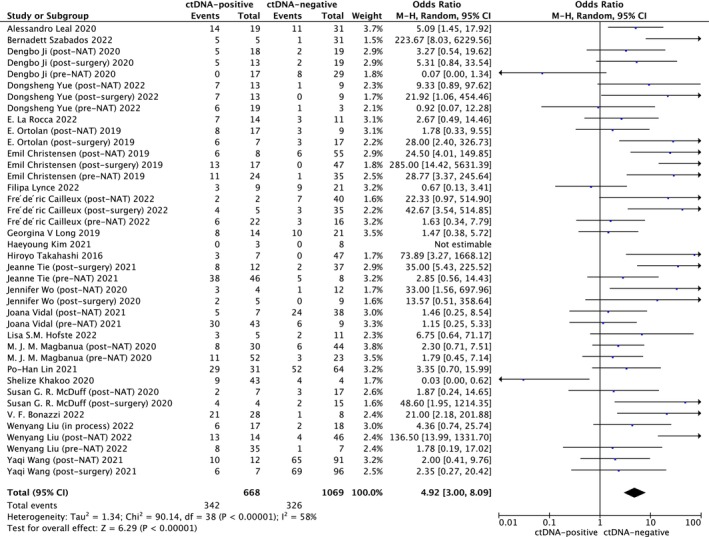
Forest plot showing the association between ctDNA detection and recurrence in cancer patients treated with neoadjuvant treatment. CI: confidential interval; Random: random‐effects model.
3.3. The risk of recurrence varied across different cancer types
Subgroup analyses were conducted to explore the association between ctDNA and recurrence across different cancer types, time points, and geographic regions, allowing for comprehensive evaluation of the data. (Table 2) In cancer category analysis, a statistical significance was observed in all the types including breast cancer (HR: 3.93, 95% CI: 1.76–8.78, p < 0.001, I 2 = 38%), rectal cancer (HR: 2.53, 95% CI: 1.05–6.10, p = 0.04, I 2 = 60%), and digestive tract cancer (HR: 3.51, 95% CI: 1.65–7.47, p = 0.001, I 2 = 57%). While all cancer types were positively related to the possibility of relapse, the HR values differed significantly. (Figure 3A).
TABLE 2.
Subgroup analysis details.
| Subgroup | (OR, 95% CI) | Number of studies | Number of patients | p | Heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|---|
| Tau2 | Chi2 | df | I 2 (%) | p | |||||
| Cancer type | |||||||||
| Breast cancer | 3.93 (1.76, 8.78) | 9 | 454 | <0.001 | 0.48 | 11.27 | 17 | 57 | 0.001 |
| Rectal cancer | 2.53 (1.05, 6.10) | 14 | 661 | 0.04 | 1.64 | 32.65 | 13 | 60 | 0.002 |
| Digestive tract cancer | 3.51 (1.65, 7.57) | 18 | 777 | 0.001 | 1.43 | 39.69 | 3 | 0 | 0.56 |
| Time points | |||||||||
| Pre‐NAT | 1.58 (0.80, 3.12) | 11 | 464 | 0.19 | 0.38 | 13.28 | 9 | 32 | 0.15 |
| Post‐NAT | 4.26 (1.81, 9.98) | 13 | 609 | <0.001 | 1.41 | 31.27 | 12 | 62 | 0.002 |
| Post‐surgery | 14.55 (6.87, 30.82) | 15 | 629 | <0.001 | 0.72 | 21.21 | 14 | 34 | 0.10 |
| Area | |||||||||
| America | 3.26 (1.46, 7.30) | 13 | 564 | 0.004 | 1.08 | 26.58 | 12 | 55 | 0.009 |
| Asia | 4.42 (1.70, 11.49) | 13 | 589 | 0.002 | 1.52 | 24.58 | 11 | 55 | 0.01 |
| Europe | 8.91 (2.89, 27.47) | 10 | 410 | <0.001 | 2.11 | 27.05 | 9 | 67 | 0.001 |
| Australia | 6.50 (1.40, 30.12) | 4 | 174 | 0.02 | 1.64 | 9.32 | 3 | 68 | 0.03 |
Abbreviations: CI, confidential interval; OR, odds ratio; NAT, neoadjuvant treatment.
FIGURE 3.
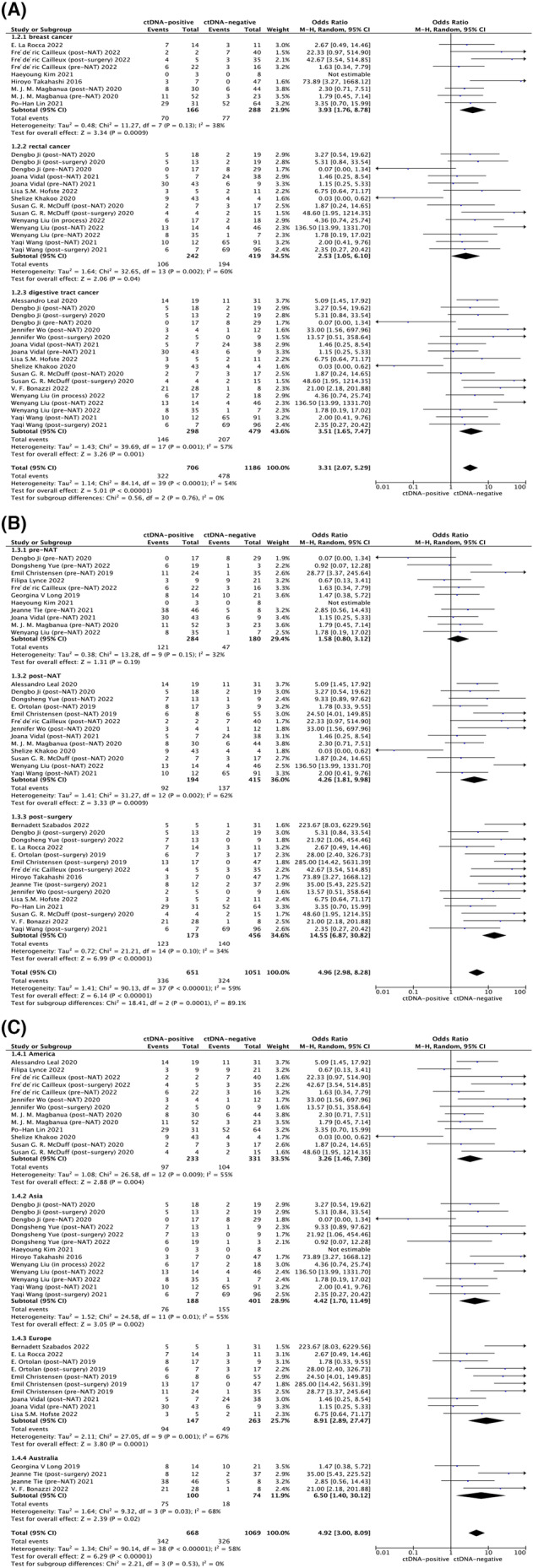
Subgroup analysis between ctDNA detection and recurrence in cancer patients treated with neoadjuvant treatment. (A) Subgroup analysis of cancer type. (B) Subgroup analysis of ctDNA detection time point. (C) Subgroup analysis of various area. CI, confidential interval; GI tract, gastrointestinal tract; NAT, neoadjuvant treatment; Random, random‐effects model.
3.4. Multi‐time points were related with recurrence rate
In the sub‐analysis between cancer recurrence and ctDNA detection time points presented in Figure 3B, a robust correlation was observed at the time points post‐neoadjuvant treatment (HR: 4.26, 95% CI: 1.81–9.98, p < 0.001, I 2 = 62%) and post‐surgery (HR: 14.55, 95% CI: 6.87–30.82, p < 0.001, I 2 = 34%), which indicated that a detectable ctDNA in patients who received the detection at post‐neoadjuvant treatment and post‐surgery time points was positively correlated with recurrence occurrence. However, no statistically significant differences were observed in pre‐neoadjuvant detection. (HR: 1.58, 95% CI: 0.80–3.12, p = 0.19, I 2 = 32%).
3.5. Relapse rate differed in various regions
Moreover, we conducted an analysis of diverse regions to explore potential variations in the association between ctDNA and cancer recurrence, which were also proved to be related with recurrence in America (HR: 3.26, 95% CI: 1.46–7.30, p = 0.004, I 2 = 55%), Asia (HR: 4.42, 95% CI: 1.70–11.49, p = 0.002, I 2 = 55%), Europe (HR: 8.91, 95% CI: 2.89–27.47, p < 0.001, I 2 = 67%), and Australia (HR: 6.50, 95% CI: 1.40–30.12, p = 0.02, I 2 = 68%) (Figure 3C).
3.6. Sensitivity analysis and publication bias
In the sensitivity analysis, the resulting trends remained the same as those in the above analysis after removing the five low‐quality comparisons, as shown in Figure S1. 15 , 18 , 21 , 24 , 32 (HR: 5.42, 95% CI: 3.34–8.75, p < 0.001, I 2 = 51%) The NOS scores of 23 included literatures ranged from 5 to 9. Funnel plots were chosen to estimate publication bias in this study and no obvious bias was observed. (Figure S2).
4. DISCUSSION
Our meta‐analysis indicated that a positive ctDNA result in patients with cancer who were treated with neoadjuvant therapy is strongly correlated with a higher incidence of recurrence. Additionally, we observed substantial variations in the risk values for recurrence based on the different cancer types, ctDNA detection time points, and the patients' geographic regions. The results revealed that the cancer categories of breast and digestive tract cancer and four regional classifications (America, Asia, Europe, and Australia) were positively related to recurrence. Significant associations were observed only at the post‐neoadjuvant treatment and post‐surgery time points, with no statistically significant relationships identified in the pre‐neoadjuvant treatment period. To the best of our knowledge, this meta‐analysis is the most comprehensive study exploring the relationship between ctDNA detection in patients treated with neoadjuvant therapy and cancer recurrence. We included the maximum number of studies and subgroup analyses including cancer category, detection time points, and regions. In addition, to enhance credibility and reduce heterogeneity, we ensured that all participants had undergone both ctDNA detection and neoadjuvant treatment, thereby strengthening the validity of our findings.
ctDNA is a novel monitoring approach that works in MRD detection, treatment response prediction, and resistance mechanism evaluation. 37 , 38 Currently, neoadjuvant treatment is considered a promising therapy for cancer before surgery. The neoadjuvant approach provides an ideal opportunity to discover novel biomarkers that can accurately predict the response to the specific treatment administered. 39 , 40 In addition, it has been demonstrated to improve prognosis and quality of life in several cancer types, including breast cancer, 41 locally advanced rectal cancer, 36 and non‐small cell lung cancer. 42 However, a valuable biomarker for assessing treatment efficiency is still needed. While several studies have explored ctDNA monitoring and its potential for recurrence prediction, 43 , 44 existing research specifically investigating its application in the context of patients receiving neoadjuvant treatment remains limited. Its predictive function for neoadjuvant treatment remains unknown and controversial. Shelize et al. found no difference in treatment response determined by RECIST between ctDNA‐positive and ctDNA‐negative patients at any time point. 45 Similarly, Dengbo ji et al. observed no correlation between circulating free DNA (cfDNA) and cancer recurrence prediction. 16 However, in Bernadett Szabados's research, ctDNA‐positive patients had a worse recurrence‐free survival than ctDNA‐negative patients. 15 In our research, we conducted this meta‐analysis to confirm that positive ctDNA detection is strongly associated with a higher cancer recurrence rate in patients receiving neoadjuvant treatment. ctDNA is associated with a higher tumor proliferation index and a more aggressive subtype. 46 Additionally, positive ctDNA detection indicated that tumors with high proliferation and increased cell turnover can release more tumor DNA fragments. Meanwhile, the detection of ctDNA in the bloodstream suggests the persistent presence of tumor cells or residual disease, thereby implying an elevated risk of cancer recurrence compared with cases where ctDNA is undetectable. This discovery indicates that ctDNA is a valuable biomarker for patients undergoing neoadjuvant treatment. Moreover, the status of ctDNA after neoadjuvant treatment can improve the performance of functional tumor volume as a predictor of metastatic recurrence. 22 Neoadjuvant treatment reduces tumor size and tumor burden, while the concentration of ctDNA is positively correlated with tumor burden. 47 Therefore, as neoadjuvant treatment leads to a decrease in tumor tissues, the concentration of ctDNA would also change correspondingly. Therefore, our study determined the key role of ctDNAs in monitoring the effects of neoadjuvant treatments.
The results also revealed that a positive ctDNA result was linked to various recurrence risks in diverse cancer types, which was reported for the first time. In patients with localized tumors, the ctDNA detection rate varies among the cancer types. ctDNAs were detected in 73% of patients with colorectal cancer, 57% of patients with gastroesophageal cancer, 48% of patients with pancreatic cancer, and 50% of patients with breast adenocarcinoma. 48 Based on our comprehensive analysis, ctDNA demonstrates potential as a highly valuable biomarker for breast cancer patients with neoadjuvant treatment, as evidenced by the significantly elevated odds ratio (OR: 3.93) and risk ratio (RR: 2.66). Notably, in the context of digestive tract cancer, the presence of detectable ctDNA was also correlated with cancer recurrence, but with different recurrence risks (OR: 3.51, RR: 1.87). Qiu et al. observed that among preoperative ctDNA‐positive patients, those with adenocarcinoma had a shorter recurrence‐free survival, whereas this finding was not evident in squamous cell carcinoma. 38 Therefore, ctDNA serves as a biomarker for assessing cancer prognosis and recurrence in patients with digestive tract malignancies; however, intrinsic mechanism research is still needed.
Current research on the efficacy of ctDNA detection as a biomarker at different time points remains controversial. Several studies have reported a meaningful relationship between ctDNA detection and recurrence prediction at pre‐neoadjuvant treatment, 14 , 20 post‐neoadjuvant treatment, 20 and post‐surgery time points, 17 , 20 whereas Bernadett Szabados reported no recurrence in ctDNA‐positive patients at pre‐neoadjuvant and post‐neoadjuvant time points. 15 After our analysis and investigations, the different risk values were observed at various time points. Interestingly, no statistical significance was observed when ctDNA detection was performed at the pre‐neoadjuvant treatment time point, while there was a strong association at the post‐neoadjuvant treatment and post‐surgery time points. Different stages of treatment and disease progression have an enhanced influence on the release and detection of ctDNA, consequently establishing a significant association. After neoadjuvant treatment or surgery, the tumor burden decreases but with the possibility of MRD. Thus, ctDNA analysis holds potential value in the neoadjuvant therapeutic context, as it allows for the identification of patients at a heightened risk of disease recurrence through the detection of MRD following neoadjuvant treatment. 49 In addition, we performed a subgroup analysis of different regions. The results showed a significant association between all four areas and different HR values. This phenomenon can be attributed to multiple factors, including biological heterogeneity, diverse cancer therapy protocols, variability in treatment responses, and the influence of environmental factors across different geographic areas.
However, this study has several limitations. First, despite having the highest number of included studies compared to similar studies, the sample size may still be insufficient to provide a comprehensive analysis. Additionally, the range of cancer types included in this study may not be sufficiently comprehensive to fully capture the heterogeneity of ctDNA‐associated recurrence. In addition, different neoadjuvant treatment regimens may have contributed to the heterogeneity.
5. CONCLUSION
This meta‐analysis showed that positive ctDNA detection results in patients with cancer who received neoadjuvant treatment is positively correlated with an increased likelihood of recurrence. Highlighting ctDNA's potential as a recurrence predictor in clinical settings, especially following neoadjuvant treatment and surgery. Furthermore, our findings underscore the variable risk profiles across various cancer types and geographic regions. However, further research and clinical trials are imperative to explore the concrete mechanisms and the precise association between ctDNA detection and recurrence in patients with cancer undergoing neoadjuvant treatment.
AUTHOR CONTRIBUTIONS
Jiaxin Zhou: Conceptualization (lead); data curation (lead); formal analysis (lead); investigation (lead); resources (lead); software (lead); writing – original draft (lead); writing – review and editing (lead). Haocong Mo: Formal analysis (equal); supervision (equal); validation (equal); writing – original draft (equal). Dahai Hu: Data curation (equal); formal analysis (equal); software (equal); writing – original draft (equal). Xiaoxu Zhao: Funding acquisition (equal); supervision (equal); validation (equal). Hong Zhou: Funding acquisition (equal); supervision (equal); validation (equal); writing – review and editing (equal). Jinghua Pan: Funding acquisition (equal); investigation (equal); methodology (equal); supervision (equal); writing – review and editing (equal).
FUNDING INFORMATION
This work was partially supported by the Guangdong Province Medical Science and Technology Research Fund Project (A2021056), and the Guangdong Basic and Applied Basic Research Foundation (2020A1515110639).
CONFLICT OF INTEREST STATEMENT
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting information
Figure S1 Forest plot of sensitivity analysis.
Figure S2 (A) Funnel plot for the analysis between ctDNA detection and recurrence in cancer patients treated with neoadjuvant treatment. (B) Funnel plot for the subgroup analysis of different cancer categories between ctDNA detection and recurrence in cancer patients treated with neoadjuvant treatment. (C) Funnel plot for the subgroup analysis in various ctDNA detection time points between ctDNA detection and recurrence in cancer patients treated with neoadjuvant treatment. (D) Funnel plot for the subgroup analysis of different regions between ctDNA detection and recurrence in cancer patients treated with neoadjuvant treatment.
Zhou J, Mo H, Hu D, Zhao X, Zhou H, Pan J. Association of ctDNA detection and recurrence assessment in patients with neoadjuvant treatment. Cancer Med. 2023;12:19794‐19806. doi: 10.1002/cam4.6544
Contributor Information
Xiaoxu Zhao, Email: zhaoxiaoxu@jnu.edu.cn.
Hong Zhou, Email: zhouhongjnu@foxmail.com.
Jinghua Pan, Email: panjh@jnu.edu.cn.
DATA AVAILABILITY STATEMENT
The data and materials (including Supplementary Material) used to support the findings in this research are available from the corresponding author upon request.
REFERENCES
- 1. Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, Bray F. Cancer statistics for the year 2020: An overview. Int J Cancer. Epub ahead of print. 2021; doi: 10.1002/ijc.33588 [DOI] [PubMed] [Google Scholar]
- 2. Kustanovich A, Schwartz R, Peretz T, Grinshpun A. Life and death of circulating cell‐free DNA. Cancer Biol Ther. 2019;20(8):1057‐1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wan JC, Massie C, Garcia‐Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223‐238. [DOI] [PubMed] [Google Scholar]
- 4. Jones J, Cain S, Pesic‐Smith J, et al. Circulating tumor DNA for malignant peripheral nerve sheath tumors in neurofibromatosis type 1. J Neurooncol. 2021;154(3):265‐274. [DOI] [PubMed] [Google Scholar]
- 5. Dawson S‐J, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199‐1209. [DOI] [PubMed] [Google Scholar]
- 6. Trimble E, Ungerleider R, Abrams J, et al. Neoadjuvant therapy in cancer treatment. Cancer. 1993;72(S11):3515‐3524. [DOI] [PubMed] [Google Scholar]
- 7. Takada M, Toi M. Neoadjuvant treatment for HER2‐positive breast cancer. Chin Clin Oncol. 2020;9(3):32. [DOI] [PubMed] [Google Scholar]
- 8. Ludmir EB, Palta M, Willett CG, Czito BG. Total neoadjuvant therapy for rectal cancer: an emerging option. Cancer. 2017;123(9):1497‐1506. [DOI] [PubMed] [Google Scholar]
- 9. Sun Y‐S, Zhao Z, Yang Z‐N, et al. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13(11):1387‐1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Selli C, Sims AH. Neoadjuvant therapy for breast cancer as a model for translational research. Breast Cancer (Auckl). 2019;13:1178223419829072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Langer R, Becker K. Tumor regression grading of gastrointestinal cancers after neoadjuvant therapy. Virchows Arch. 2018;472(2):175‐186. [DOI] [PubMed] [Google Scholar]
- 12. Smyth EC, Fassan M, Cunningham D, et al. Effect of pathologic tumor response and nodal status on survival in the Medical Research Council adjuvant gastric infusional chemotherapy trial. J Clin Oncol. 2016;34(23):2721‐2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Becker K, Mueller JD, Schulmacher C, et al. Histomorphology and grading of regression in gastric carcinoma treated with neoadjuvant chemotherapy. Cancer. 2003;98(7):1521‐1530. [DOI] [PubMed] [Google Scholar]
- 14. Leal A, van Grieken NCT, Palsgrove DN, et al. White blood cell and cell‐free DNA analyses for detection of residual disease in gastric cancer. Nat Commun. 2020;11(1):525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Szabados B, Kockx M, Assaf ZJ, et al. Final results of neoadjuvant Atezolizumab in cisplatin‐ineligible patients with muscle‐invasive urothelial cancer of the bladder. Eur Urol. 2022;82(2):212‐222. [DOI] [PubMed] [Google Scholar]
- 16. Ji D, Zhang D, Zhan T, et al. Tumor mutation burden in blood predicts benefit from neoadjuvant chemo/radiotherapy in locally advanced rectal cancer. Genomics. 2021;113(1 Pt 2):957‐966. [DOI] [PubMed] [Google Scholar]
- 17. Yue D, Liu W, Chen C, et al. Circulating tumor DNA predicts neoadjuvant immunotherapy efficacy and recurrence‐free survival in surgical non‐small cell lung cancer patients. Transl Lung Cancer Res. 2022;11(2):263‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. La Rocca E, De Santis MC, Silvestri M, et al. Early stage breast cancer follow‐up in real‐world clinical practice: the added value of cell free circulating tumor DNA. J Cancer Res Clin Oncol. 2022;148(6):1543‐1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ortolan E, Appierto V, Silvestri M, et al. Blood‐based genomics of triple‐negative breast cancer progression in patients treated with neoadjuvant chemotherapy. ESMO Open. 2021;6(2):100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Christensen E, Birkenkamp‐Demtröder K, Sethi H, et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra‐deep sequencing of plasma cell‐free DNA in patients with urothelial bladder carcinoma. J Clin Oncol. 2019;37(18):1547‐1557. [DOI] [PubMed] [Google Scholar]
- 21. Lynce F, Mainor C, Geng X, et al. Abstract PD9‐02: peripheral immune subsets and circulating tumor DNA (ctDNA) in patients (pts) with residual triple negative breast cancer (TNBC) treated with adjuvant immunotherapy and/or chemotherapy (chemo): the OXEL study. Cancer Res. 2022;82(4_Supplement):PD9‐02‐PD9‐02. [Google Scholar]
- 22. Cailleux F, Agostinetto E, Lambertini M, et al. Circulating tumor DNA after neoadjuvant chemotherapy in breast cancer is associated with disease relapse. JCO Precis Oncol. 2022;6:e2200148. [DOI] [PubMed] [Google Scholar]
- 23. Long GV, Saw RP, Lo S, et al. Neoadjuvant dabrafenib combined with trametinib for resectable, stage IIIB–C, BRAFV600 mutation‐positive melanoma (NeoCombi): a single‐arm, open‐label, single‐Centre, phase 2 trial. Lancet Oncol. 2019;20(7):961‐971. [DOI] [PubMed] [Google Scholar]
- 24. Kim H, Kim YJ, Park D, et al. Dynamics of circulating tumor DNA during postoperative radiotherapy in patients with residual triple‐negative breast cancer following neoadjuvant chemotherapy: a prospective observational study. Breast Cancer Res Treat. 2021;189(1):167‐175. [DOI] [PubMed] [Google Scholar]
- 25. Takahashi H, Kagara N, Tanei T, et al. Correlation of methylated circulating tumor DNA with response to neoadjuvant chemotherapy in breast cancer patients. Clin Breast Cancer. 2017;17(1):61‐69. e3. [DOI] [PubMed] [Google Scholar]
- 26. Tie J, Wang Y, Cohen J, et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: a prospective cohort study. PLoS Med. 2021;18(5):e1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wo JY, Clark JW, Eyler CE, et al. Results and molecular correlates from a pilot study of neoadjuvant induction FOLFIRINOX followed by chemoradiation and surgery for gastroesophageal adenocarcinomas neoadjuvant FOLFIRINOX and chemoradiation for GEA. Clin Cancer Res. 2021;27(23):6343‐6353. [DOI] [PubMed] [Google Scholar]
- 28. Vidal J, Casadevall D, Bellosillo B, et al. Clinical impact of presurgery circulating tumor DNA after total neoadjuvant treatment in locally advanced rectal cancer: a biomarker study from the GEMCAD 1402 TrialClinical impact of ctDNA in locally advanced rectal cancer. Clin Cancer Res. 2021;27(10):2890‐2898. [DOI] [PubMed] [Google Scholar]
- 29. Hofste LS, Geerlings MJ, von Rhein D, et al. Circulating tumor DNA‐based disease monitoring of patients with locally advanced esophageal cancer. Cancer. 2022;14(18):4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Magbanua MJM, Swigart LB, Wu H‐T, et al. Circulating tumor DNA in neoadjuvant‐treated breast cancer reflects response and survival. Ann Oncol. 2021;32(2):229‐239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin P‐H, Wang M‐Y, Lo C, et al. Circulating tumor DNA as a predictive marker of recurrence for patients with stage II‐III breast cancer treated with neoadjuvant therapy. Front Oncol. 2021;11:736769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khakoo S, Carter PD, Brown G, et al. MRI tumor regression grade and circulating tumor DNA as complementary tools to assess response and guide therapy adaptation in rectal cancer MRI tumor regression grade and ctDNA for rectal cancer. Clin Cancer Res. 2020;26(1):183‐192. [DOI] [PubMed] [Google Scholar]
- 33. McDuff SG, Hardiman KM, Ulintz PJ, et al. Circulating tumor DNA predicts pathologic and clinical outcomes following neoadjuvant chemoradiation and surgery for patients with locally advanced rectal cancer. JCO Precis Oncol. 2021;5:123‐132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bonazzi VF, Aoude LG, Brosda S, et al. ctDNA as a biomarker of progression in oesophageal adenocarcinoma. ESMO Open. 2022;7(3):100452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu W, Li Y, Tang Y, et al. Response prediction and risk stratification of patients with rectal cancer after neoadjuvant therapy through an analysis of circulating tumour DNA. EBioMedicine. 2022;78:103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y, Yang L, Bao H, et al. Utility of ctDNA in predicting response to neoadjuvant chemoradiotherapy and prognosis assessment in locally advanced rectal cancer: a prospective cohort study. PLoS Med. 2021;18(8):e1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pellini B, Chaudhuri AA. Circulating tumor DNA minimal residual disease detection of non‐small‐cell lung cancer treated with curative intent. J Clin Oncol. 2022;40(6):567‐575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qiu B, Guo W, Zhang F, et al. Dynamic recurrence risk and adjuvant chemotherapy benefit prediction by ctDNA in resected NSCLC. Nat Commun. 2021;12(1):6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Loibl S. Neoadjuvant treatment of breast cancer: maximizing pathologic complete response rates to improve prognosis. Curr Opin Obstet Gynecol. 2015;27(1):85‐91. [DOI] [PubMed] [Google Scholar]
- 40. Moding EJ, Nabet BY, Alizadeh AA, Diehn M. Detecting liquid remnants of solid tumors: circulating tumor DNA minimal residual disease. Cancer Discov. 2021;11(12):2968‐2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hassett MJ, Li H, Burstein HJ, Punglia RS. Neoadjuvant treatment strategies for HER2‐positive breast cancer: cost‐effectiveness and quality of life outcomes. Breast Cancer Res Treat. 2020;181(1):43‐51. [DOI] [PubMed] [Google Scholar]
- 42. Lee JM, Tsuboi M, Brunelli A. Surgical perspective on neoadjuvant immunotherapy in non‐small cell lung cancer. Ann Thorac Surg. 2022;114(4):1505‐1515. [DOI] [PubMed] [Google Scholar]
- 43. Wang R, Zhao A, Cao N, Li Z, Zhang G, Liu F. The value of circulation tumor DNA in predicting postoperative recurrence of colorectal cancer: a meta‐analysis. Int J Colorectal Dis. 2020;35(8):1463‐1475. [DOI] [PubMed] [Google Scholar]
- 44. Papakonstantinou A, Gonzalez NS, Pimentel I, et al. Prognostic value of ctDNA detection in patients with early breast cancer undergoing neoadjuvant therapy: a systematic review and meta‐analysis. Cancer Treat Rev. 2022;104:102362. [DOI] [PubMed] [Google Scholar]
- 45. Khakoo S, Carter PD, Brown G, et al. MRI tumor regression grade and circulating tumor DNA as complementary tools to assess response and guide therapy adaptation in rectal cancer. Clin Cancer Res. 2020;26(1):183‐192. [DOI] [PubMed] [Google Scholar]
- 46. Riva F, Bidard FC, Houy A, et al. Patient‐specific circulating tumor DNA detection during neoadjuvant chemotherapy in triple‐negative breast cancer. Clin Chem. 2017;63(3):691‐699. [DOI] [PubMed] [Google Scholar]
- 47. Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol. 2022;15(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early‐ and late‐stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC—challenges to implementing ctDNA‐based screening and MRD detection. Nat Rev Clin Oncol. 2018;15(9):577‐586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Forest plot of sensitivity analysis.
Figure S2 (A) Funnel plot for the analysis between ctDNA detection and recurrence in cancer patients treated with neoadjuvant treatment. (B) Funnel plot for the subgroup analysis of different cancer categories between ctDNA detection and recurrence in cancer patients treated with neoadjuvant treatment. (C) Funnel plot for the subgroup analysis in various ctDNA detection time points between ctDNA detection and recurrence in cancer patients treated with neoadjuvant treatment. (D) Funnel plot for the subgroup analysis of different regions between ctDNA detection and recurrence in cancer patients treated with neoadjuvant treatment.
Data Availability Statement
The data and materials (including Supplementary Material) used to support the findings in this research are available from the corresponding author upon request.


