Abstract
Genomic testing is crucial for the management of ovarian cancer. DNA from biopsies at diagnostic laparoscopies or interval debulking surgery after neoadjuvant chemotherapy, has a high failure rate. At relapse, biopsies may not be feasible. The aim of our study was to evaluate the feasibility and usefulness of measuring genomic instability score (GIS) on cell-free DNA (cfDNA) from ascites.
Patients enrolled in a prospective study (NCT03010124) consented to analysis of biological samples. CfDNA was extracted from 1 to 4 ml of double-centrifuged fresh ascites. Targeted Next-generation sequencing (NGS) including TP53 mutation (TP53m) was performed on cfDNA to confirm the presence of tumor cfDNA. Single Nucleotide Polymorphism Array estimating somatic copy number alterations (SCNA) was performed to calculate GIS for Homologous-Recombination deficiency (HRD).
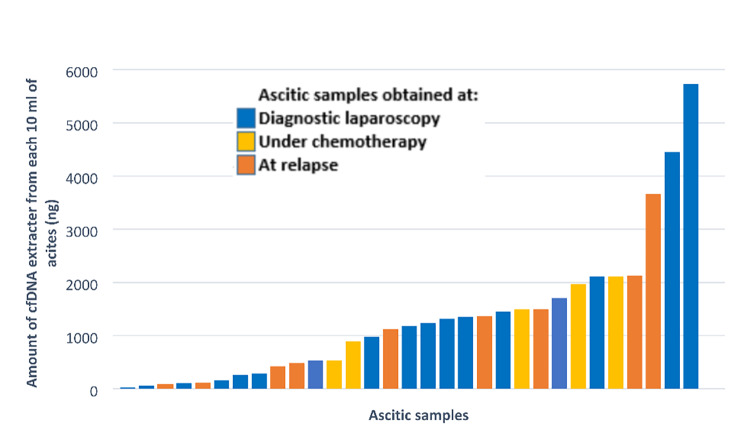
Twenty nine ascites were collected from 20 patients with suspected or confirmed OC. 93% (27/29) samples had detectable cfDNA (median 1120 ng [24-5732]) even when obtained during chemotherapy. A deleterious mutation was identified in 100%, with high allelic frequencies (median 60% [3.3–87%]), confirming that cfDNA was tumoral. SCNA analyses on 17 patients showed 11 high GIS, and 6 low GIS. 4 patients with confirmed BRCA mutation had a high GIS on ascites. When available from the same patient, SCNA profiles on ascites and tumor were superimposable.
Ascites is frequent at diagnosis and relapse and yields large amounts of tumoral cfDNA. SCNA analysis on ascitic cfDNA is feasible and can detect the same HRD scar as tumor testing. Ascites could provide an alternative to tumor sampling for HRD and BRCA testing.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40364-023-00533-1.
Keywords: Circulating Tumor DNA, Ovarian cancer, Homologous recombination deficiency, Genomic instability score, Ascites
Main text
To the editor,
Genomic testing is crucial for the management of ovarian cancer (OC). Approximately 25% of high-grade OC have germline or somatic BRCA1 or BRCA2 mutations [1]. 50% of high-grade OC are homologous recombination deficient (HRD). HRD is defined by the detection of a BRCA1 or BRCA2 mutation, or demonstration of high genomic instability, that can be measured by Genomic Instability Scores (GIS) [2]. These defects in homologous recombination increase tumor sensitivity to platinum chemotherapy and PARP inhibitors [3, 4]. 20% of patients with advanced OC, have non-contributive HRD testing on FFPE tumor based assays [5], often attributable to small biopsies obtained during upfront or interval debulking surgery. New radiologically guided biopsies are often not technically or safely feasible. In the last decade, the utility of liquid biopsies for cancer management has been demonstrated in several studies [6–8]. High-grade OC are characterized by almost universal somatic TP53 mutations (TP53m) which may be particularly suited to this approach [9].
In our study, we aimed to investigate the feasibility and clinical utility of tumor derived cfDNA from ascitic samples, as an alternative to tissue biopsy, to measure genomic instability as a surrogate for HRD.
Patients enrolled in a prospective study (NCT03010124) consented to analysis of biological samples. CfDNA was extracted from fresh ascites. Targeted Next-generation sequencing (NGS) including TP53m and SNParray for somatic copy number alterations (SCNA) analyses to calculate GIS were performed. Detailed material and methods are described in the supplementary material.
Overall, 29 ascites were collected from 20 patients with high-grade OC from March 2015 to January 2016. Ten ml of ascites were double centrifuged for cfDNA extraction. All samples (100%) had detectable cfDNA (median 1120 ng, range: 24-5732 ng) (Fig. 1).
Fig. 1.
High yield of tumoral cell-free DNA in ascites
Abbreviations: cfDNA: cell-free DNA
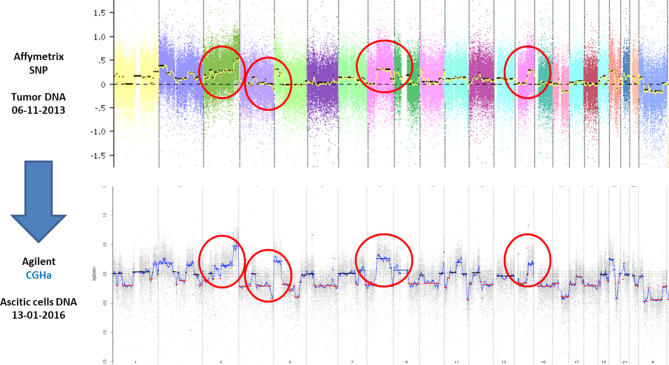
In 27/29 (93%) samples, a deleterious mutation was identified with high median allelic frequency: 60% (range 4–87%). Among 27 samples with confirmed cfDNA, a TP53m was detected in 25 (92%) samples confirming cfDNA is tumoral (Supplementary Table 2). We performed SCNA on cfDNA from ascites from patients with high-grade OC. Analysis were contributive and GIS was feasible on 17/20 (85%) patients. Samples were classified as high versus low genomic instability. 11/17 patients had a high GIS and were considered as homologous recombination deficient (HRD), and 6 patients a low GIS. All 5 patients with known germinal BRCA1/2 mutation included in this study had a high GIS score on ascites. When both were available from the same patient, the SCNA profiles derived from ctDNA in ascites and tumor sampling were superimposable (Fig. 2).
Fig. 2.
Comparison between SCNA profiles derived from tumor tissue and circulating tumor DNA
Abbreviations: CGHa: Comparative Genomic Hybridization Array, SCNA: Somatic Copy Number Alteration, SNP: Single Nucleotide Polymorphism Array
To determine the proportion of patients with sufficient ascites for ctDNA analysis, we performed a retrospective review of the operative reports from 100 consecutive patients who underwent a diagnostic and interval laparoscopy for high-grade OC. 97/100 patients had visible peritoneal fluid: volume of ascites ranged from 50 to 11,000 ml, with a median of 500 ml. After neoadjuvant chemotherapy, 40% of patients still had ascites with a volume ranging from 80 to 5000 ml, median of 150 ml. All these patients with ascites would have been candidates for genomic testing on cfDNA.
Ascites is frequent at diagnosis and relapse and yields large amounts of tumoral cfDNA. Our study demonstrates the feasibility of detecting somatic mutations in ascites of patients with high-grade OC. We provides a proof of concept that the measurement of genomic instability on cfDNA from ascites is feasible, detecting the same HRD scar as tumor testing. Our study has limitations. Our assay provides a rough measure of genomic instability and has not been validated against standard of care genomic instability tests. Although the cohort is small and the sampling heterogeneous, the exploratory results of our pilot study are very encouraging.
Very few data have been published on ctDNA in peritoneal fluid from patients with ovarian cancer. While standardization of assays remain to be addressed, this area of study may help improve the management of patients with OC in optimizing early detection, therapeutic choices and monitoring of drug resistance [6, 10].
When tumor biopsies are inaccessible or when the tumor tissue is insufficient, ascites could be an alternative to tumor sampling for HRD and BRCA testing. Our results should be validated in large prospective studies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank the patients and their families who have agreed that this clinical data could be collected and reported. We also thank all the caregivers from Gustave Roussy who treated those patients. We thank the biology team from Gustave Roussy who performed the genomic testing.
List of abbreviations
- cfDNA
Cell-free DNA
- GIS
Genomic instability score
- HRD
Homologous-Recombination deficiency
- NGS
Next-generation sequencing
- OC
Ovarian cancer
- SCNA
Somatic copy number alterations
- TP53m
TP53 mutation
Authors’ contributions
AL and ALF contributed to study conception andDesign. MK, REH, CS, SG, AM, FBD, JM and ECB contributed to data collection. MK, REH, RT, ER contributed to analysis and interpretation of results. MK and REH contribute to draft manuscript preparation. All authors reviewed the results and approved the final version of the manuscript.
Funding
The authors declare no funding support for this study.
Data Availability
All data generated or analysed during this study are included in this published article and its Additional information files.
Declarations
Ethics approval and consent to participate
The study was conducted in compliance with good clinical practice and had been authorized by Gustave Roussy Institutional Review Board. All patients included signed an informed consent form (OvBIOMark, NCT03010124).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests related to this study.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nielsen FC, van Overeem Hansen T, Sørensen CS. Hereditary breast and Ovarian cancer: new genes in confined pathways. Nat Rev Cancer. 2016;16(9):599–612. doi: 10.1038/nrc.2016.72. [DOI] [PubMed] [Google Scholar]
- 2.Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, Rendi MH, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and Peritoneal Carcinomas. Clin Cancer Res. 2014;20(3):764–75. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ledermann JA, Drew Y, Kristeleit RS. Homologous recombination deficiency and Ovarian cancer. Eur J Cancer Oxf Engl 1990. 2016;60:49–58. doi: 10.1016/j.ejca.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Ray-Coquard I, Pautier P, Pignata S, Pérol D, González-Martín A, Berger R, et al. Olaparib plus Bevacizumab as First-Line maintenance in Ovarian Cancer. N Engl J Med. 2019;381(25):2416–28. doi: 10.1056/NEJMoa1911361. [DOI] [PubMed] [Google Scholar]
- 6.Cheng X, Zhang L, Chen Y, Qing C. Circulating cell-free DNA and circulating tumor cells, the “liquid biopsies” in ovarian cancer. J Ovarian Res [Internet]. 2017 Nov 13 [cited 2020 Aug 19];10. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5683341/. [DOI] [PMC free article] [PubMed]
- 7.Asante DB, Calapre L, Ziman M, Meniawy TM, Gray ES. Liquid biopsy in Ovarian cancer using circulating Tumor DNA and cells: ready for prime time? Cancer Lett. 2020;468:59–71. doi: 10.1016/j.canlet.2019.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Kipps E, Tan DSP, Kaye SB. Meeting the challenge of Ascites in Ovarian cancer: new avenues for therapy and research. Nat Rev Cancer. 2013;13(4):273–82. doi: 10.1038/nrc3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng Y, Mei W, Ma K, Zeng C. Circulating Tumor DNA and minimal residual Disease (MRD) in solid tumors: current Horizons and Future perspectives. Front Oncol. 2021;11:763790. doi: 10.3389/fonc.2021.763790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its Additional information files.