Abstract
Pancreatic islets consist of multiple cell types that produce hormones required for glucose homeostasis, and islet dysfunction is a major factor in type 1 and type 2 diabetes. Numerous studies have assessed transcription across individual cell types using single-cell assays; however, there is no canonical reference of gene expression in islet cell types that is also easily accessible for researchers to query and use in bioinformatics pipelines. Here we present an integrated map of islet cell type–specific gene expression from 192,203 cells from single-cell RNA sequencing of 65 donors without diabetes, donors who were type 1 diabetes autoantibody positive, donors with type 1 diabetes, and donors with type 2 diabetes from the Human Pancreas Analysis Program. We identified 10 distinct cell types, annotated subpopulations of several cell types, and defined cell type–specific marker genes. We tested differential expression within each cell type across disease states and identified 1,701 genes with significant changes in expression, with most changes observed in β-cells from donors with type 1 diabetes. To facilitate user interaction, we provide several single-cell visualization and reference mapping tools, as well as the open-access analytical pipelines used to create this reference. The results will serve as a valuable resource to investigators studying islet biology.
Article Highlights
No canonical reference exists for cell type–specific gene expression from single-cell RNA sequencing of pancreatic islets.
We generated an integrated single-cell RNA sequencing map from healthy and diseased pancreatic islets and included user-friendly resources for visualizing and querying the data.
Most significant changes in gene expression exist in β-cells from donors with type 1 diabetes.
Our study provides a suite of tools and resources for the diabetes community to easily access cell type–specific gene expression analyses in pancreatic islets.
Introduction
The islets of Langerhans in the pancreas are clusters of endocrine cells including α-, β-, δ-, and γ-cell types, which each produce hormones that regulate blood glucose levels (1). Dysfunction of β-cells is one of the major pathologies of both type 1 and type 2 diabetes, which collectively affect >500 million individuals worldwide (2,3). Other cell types in the microenvironment around islets also contribute to the modulation of islet function and diabetes risk such as endothelial and immune cells (4,5). The regulation of gene activity establishes the identity of specific cell types as well as changes in response to environmental stimuli and disease states, and gene activity can be measured by sequence-based expression profiling (6). Understanding the gene expression profiles of islet cell types can therefore provide insight into their function and can also reveal how cells are altered in diabetes.
Single-cell technologies enable profiling the expression levels of genes in individual cells, which can then be used to define the gene regulatory profiles of specific cell types (7,8). Numerous studies have assayed gene expression in individual islet cells using single-cell techniques (9–13). These studies have defined gene expression profiles of endocrine and nonendocrine cell types in the pancreas, heterogeneous subpopulations of cells representing cellular states within cell types, and changes in disease states, including type 1 and type 2 diabetes. A caveat to these studies is that they have been performed using limited sample numbers, and in some cases, limited cell numbers, and there has been inconsistency in the results across studies, particularly when describing heterogeneity and changes in disease (14,15). In addition, the results of islet single-cell studies are not often made easily accessible to researchers, particularly those who are not experts in single-cell data analysis, to visualize and query the expression of a gene in each cell type or changes in cell type expression in disease.
The Human Pancreas Analysis Program (HPAP) was developed to comprehensively collect and profile pancreatic islet tissue from human donors to understand the pathogenesis of type 1 and type 2 diabetes (16,17). The data generated by HPAP for each donor include bulk and single-cell RNA sequencing (scRNA-seq) and Assay for Transposase-Accessible Chromatin (ATAC)-seq data as well as histology, genotyping, cellular phenotyping, and other data types. Data generated by HPAP are made freely available to researchers via a web portal PANC-DB (https://hpap.pmacs.upenn.edu/) where the raw sequence files from each individual donor can be directly downloaded (16). The rich set of raw islet donor data provided by this resource can then be used by researchers to create integrated resources that make HPAP accessible to the wider community studying islet biology and diabetes to develop testable hypotheses.
In this study, we created a reference map of gene expression in pancreatic islet cell types using scRNA-seq data from 65 donors available in HPAP. Using this reference map, we created several additional resources including 1) marker gene lists for every islet cell type and subpopulation, 2) normalized expression levels of genes in each islet cell type, and 3) changes in gene expression in type 1 diabetes, type 1 diabetes autoantibody positive (Aab+), and type 2 diabetes states in each islet cell type. We host these data in several interactive applications to enable researchers to visualize, query, and analyze this reference. Finally, we provide the open-source analytical pipelines used to create the reference map and annotations. These resources are available at www.isletgenomics.org.
Research Design and Methods
HPAP
Organ procurement and processing was performed by the HPAP, as previously described (16). In HPAP, isolated human islets were cultured for 4 days, on average, after isolation. On the day of harvest, islets were handpicked from culture and dissociated using 0.05% trypsin for 9 min prior to stopping the reaction with 100% FBS. The single cell suspension was passed through a 35-µm nylon cell strainer and resuspended in PBS + 10% FBS prior to scRNA-seq. scRNA-seq data from isolated and dissociated pancreatic islets were made publicly available by HPAP, and raw fastq files for experiments from 67 donors (10 donors with type 1 diabetes, 17 donors with type 2 diabetes, 29 with no diabetes [ND], and 9 with ND but Aab+) were downloaded from the PANC-DB data portal. Cell Ranger 6.0.1 (10× Genomics) software was used to perform alignment to the Genome Research Consortium human build 38 (GRCh38) reference genome and generate count matrices.
Preliminary Filtering
Barcodes were filtered for a minimum of 500 expressed genes per cell and <15% mitochondrial reads. Two samples (HPAP-027 and HPAP-093) were removed since the mean number of expressed genes per cell after this filtering step was markedly lower than for other samples (<1,000).
Ambient RNA Correction
Ambient RNA removal was performed to account for extracellular RNA contamination that may get trapped in a droplet during library generation. SoupX 1.6.1 (18) was used on raw feature barcode matrices for ambient RNA removal on the remaining 65 samples using the automated contamination fraction estimation method. Raw count values for each sample were corrected using the SoupX contamination estimates and the round-to-integer feature, ensuring resulting counts remain integers for use in downstream analyses (19).
Data Processing and Clustering
The SoupX-corrected count matrices were merged and log-normalized with a scale factor of 1,000. The variance stabilizing transformation method was used to find the 2,000 most variable features. Data were scaled, and principal component analysis was performed with 20 principal components using Seurat 4.2.0 (20). Harmony 0.1.1 (21) was used for batch correction using donor, 10× Genomics assay chemistry (10× 3′ v2 or 10× 3′ v3), and tissue source (Network for Pancreatic Organ Donors with Diabetes or University of Pennsylvania) as covariates. Uniform manifold approximation and projection and neighbors were calculated using the reduction from Harmony. Clustering was performed in Seurat 4.2.0 using the Leiden algorithm at a resolution of 0.5.
Postclustering Doublet Removal
Scrublet 0.2.3 (22) was used to identify doublets with the default parameters (expected doublet rate of 6%, minimum counts of two, minimum cells of three, minimum gene variability percentile of 85%, and 30 principal components). For each sample, RNA count matrices were extracted, saved in the MatrixMarket format, and input into Scrublet with default parameters. There were 4,382 barcodes flagged as doublets, and we removed these barcodes from the merged Seurat object and reperformed Harmony integration and clustering (resolution 0.3) with the remaining barcodes, as described above. We further curated a set of cell type–specific marker genes, and clusters that contained marker genes for two or more different cell types were further subclustered using the Leiden algorithm at resolutions of 0.15–0.25. Any subclusters expressing multiple cell type marker genes were presumed to be residual doublets, and we manually removed these subclusters, representing a total of 13,036 barcodes. We then reperformed Harmony integration and clustering using the final set of barcodes, as described above.
Cell Type Gene Expression Profiles
We aggregated reads from cells in each cell type and created “pseudo”-bulk counts from contamination-corrected RNA counts. We calculated transcripts per million (TPMs) for each donor in each cell type using GENCODE v38 GRCh38.p13 (23) gene size annotations. Differential gene expression analyses were performed using DESeq2 1.34.0 (24), comparing cell type profiles between samples from control ND donors and Aab+ donors with type 1 diabetes, and donors with type 1 or type 2 diabetes using a Wald test. Sex, scaled age, scaled BMI, 10× Genomics kit chemistry, and tissue procurement source were included in the model as covariates. Genes were only tested for a cell type if half of the samples per condition had at least five counts. Multiple test correction was performed using the Benjamini-Hochberg false detection rate (FDR) correction at a threshold of 0.10.
We performed preranked fast gene set enrichment analyses (25,26) for up- and downregulated genes in β-cells using Gene Ontology, Kyoto Encyclopedia of Genes and Genomes pathway, and Reactome terms. Ribosomal genes were excluded, and the remaining genes were ranked by the −log10 P values multiplied by the effect size. We restricted our analysis to gene sets with ≤500 genes and used the Monte Carlo approach in fast gene set enrichment analyses to identify significantly up- or downregulated pathways at FDR <0.10. For visualization, redundant pathway terms were removed.
We assessed whether differentially expressed genes in either type 1 or type 2 diabetes map to known risk loci by using publicly available type 1 or type 2 diabetes fine-mapped credible sets (27,28) and creating a 1 megabase (Mb) window flanking the lead variants. All genes within 1 Mb windows were annotated using GENCODE v38 (23) and intersected with significant genes from differential expression analyses for that disease.
Cell Type Proportion Analysis
We compared the proportion of β-cells and α-cells as a function of the total number of endocrine cells (α, β, δ, and γ+ε) between ND and each disease state using a Wilcoxon rank sum test. We excluded HPAP-019 from this analysis as this donor was enriched for β-cells prior to scRNA-seq experiments. To assess whether there was an association between β-cell proportion and age of onset and disease duration, we used a generalized linear model of log-scaled β-cell proportions as the response and disease duration and age of onset as predictors. For donors with a range listed for disease duration, the mean was used. Age of onset was calculated by subtracting disease duration from age at death.
Cell Type–Specific Marker Genes
We identified cluster-specific marker genes by comparing pseudo-bulk contamination-corrected RNA counts of gene expression in a given cluster with the remaining clusters across ND donors using a Wald test in DESeq2 1.34.0 (24). P values were corrected for multiple testing using Benjamini-Hochberg FDR at a threshold of 0.10. Genes were considered specific to a cluster if they had a minimum of five counts in 25% of samples from ND donors samples, logtwofold change threshold >1, and an adjusted P value <0.05. For cell types with multiple clusters, we used the same approach to compare pseudo-bulk profiles between clusters of just the same cell type across ND donors. Marker genes for each cell type or subtype were ranked by multiplying the effect size by the −log10 P value. The top 10 marker genes for cell types and subtypes are reported in Supplementary Tables 4 and 5, respectively, and full results are provided as Supplementary Data.
Data and Resource Availability
The raw sequence data are available on the PANC-DB website. Processed files and derived annotations generated by this study are available at isletgenomics.org. Custom code is available at https://github.com/Gaulton-Lab/HPAP-scRNA-seq.
Results
Reference Map of Single-Cell Expression in Islets
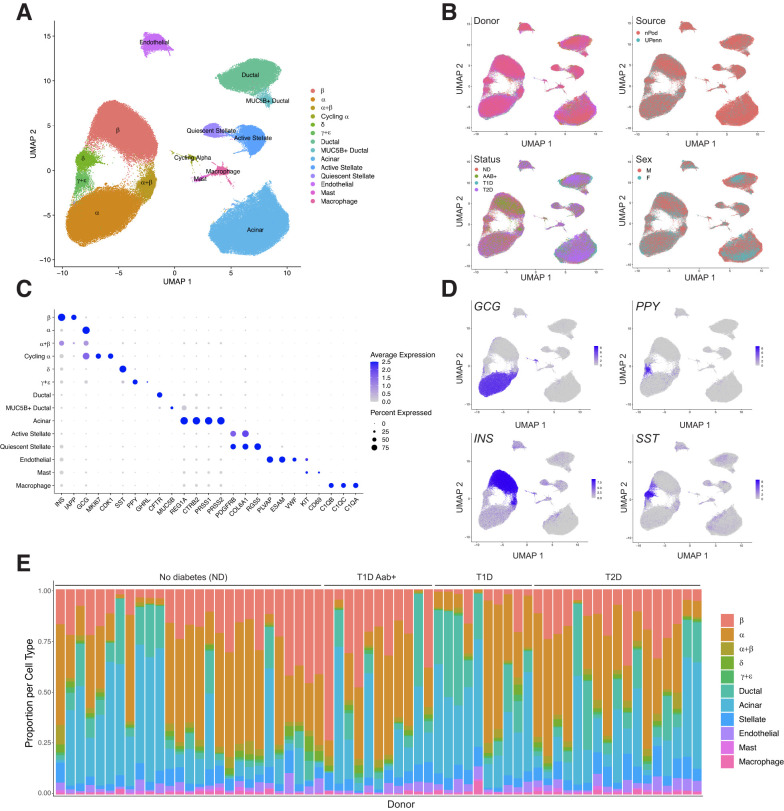
We downloaded scRNA-seq data from 67 donors in PANC-DB, and donor characteristics such as sex and age are listed in Supplementary Table 1. After prefiltering barcodes for each sample based on >500 expressed genes, we excluded two samples with lower average expressed genes per cell. With the remaining 65 samples, we performed processing and clustering using a custom pipeline. In brief, this pipeline consists of ambient RNA background correction, dimension reduction of log-normalized counts, batch correction, Leiden clustering, and postclustering doublet removal (see Research Design and Methods). The resulting map had 192,203 cells that mapped to 14 distinct clusters (Fig.1A). In the final map, on average, samples had 2,957 cells with 16,908 unique molecular identifiers and 2,724 expressed genes per cell. Clusters were broadly consistent across samples, and no clusters were preferentially represented by a small number of samples (Supplementary Fig. 1). We also observed little evidence for residual batch effects in the clusters driven by donor or other variables (Fig.1B and Supplementary Table 2).
Figure 1.
Map of gene expression in pancreatic islet cell types. A: Uniform manifold approximation and projection (UMAP) plot showing clustering of scRNA-seq profiles from 192,203 cells from HPAP donors in the PANC-DB website. Clusters are labeled based on cell type and subtype identity using known marker genes. B: Cells labeled based on variables such as donor, sex, disease status, and tissue source. C: Dot plot showing normalized expression level and percentage of expressing cells for selected marker genes in each cluster. D: Cells labeled with expression level of islet cell type hormones insulin (INS), glucagon (GCG), somatostatin (SST), and pancreatic polypeptide (PPY). E: Proportion of cells generated from each donor from each cell type, grouped by disease state. F, female; M, male; POD, Network for Pancreatic Organ Donors with Diabetes; T1D, type 1 diabetes; T2D, type 2 diabetes; UPenn, University of Pennsylvania.
We next annotated the identity of clusters using a curated set of well-established cell type and marker genes (Supplementary Table 3). This revealed 10 total cell types, including endocrine α- (GCG), β- (INS), δ- (SST), and γ- (PPY) cells, as well as nonendocrine acinar (REG1A), ductal (CFTR), endothelial (PLVAP), stellate (PDGFRA), macrophage (C1QA/B/C), and mast cells (KIT, CD69) (Fig.1C and D). We also observed evidence for ε-cells (GHRL) within the γ-cell cluster (Fig.1C), so we labeled this cluster as γ-+ε-cells. Although islet cell type identity can be annotated using a small number of marker genes, knowledge of a larger set of genes specifically expressed in each cell type can provide potential additional insight into what drives cell identity. We therefore identified genes in each cell type with highly specific expression relative to other cell types in the study (Supplementary Table 4) (see Research Design and Methods). In addition to canonical markers INS, IAPP, and MAFA, we also identified β-cell–specific genes previously implicated in β-cell function such as ADCYAP1 (29), WSCD1 (30), and HHATL (31), as well as those with no known β-cell function to our knowledge, such as SAMD11 and LRRTM3, which could be targeted in future studies.
We also identified several cell types with multiple distinct clusters (Fig.1A). In most cases, these clusters represent previously described cell subtypes or states; for example, we identified quiescent and activated states of stellate cells (32), a MUC5B+ subpopulation of ductal cells (32), and a subpopulation of “cycling” α-cells (33) (Fig.1A and C). We identified genes highly specific to each cell subtype when compared with other cells of that same cell type (Supplementary Table 5) (see Research Design and Methods). We also identified a cluster of 4,575 cells consisting of both α- and β-cells, which did not appear to be doublets based on the total reads per cell and expression patterns of insulin and glucagon (Supplementary Fig. 2). These cells may represent cellular states of α- and β-cells related to stress or signaling, as has been observed in other studies (15,33,34), although they more likely represent low-quality cells due to fewer total reads and expressed genes relative to other α- and β-cell clusters. Due to the ambiguity over what cell populations these cells exactly represent, we excluded the cluster from downstream analyses.
Given the sparsity of cellular profiles obtained from scRNA-seq, we next examined the expression of 16 genes in β-cells selected by a previous study (15) to determine the extent to which key β-cell genes are captured in β-cells from scRNA-seq of HPAP donors (Supplementary Fig. 3). At the individual cell level, detection of each gene was variable where highly expressed genes, such as INS, were observed in almost every cell (99.68% of β-cells), while other key genes with lower expression, such as PDX1 and GCK, were observed in only a minority of cells (34.1% and 12.1% of β-cells, respectively). By comparison, at the sample level, where reads from all cells in a sample are collapsed into pseudo-bulk profiles, we observed expression of almost every gene in all samples.
We next compared the proportions of each islet cell type across samples (Fig.1E). As the purity of the islet preparations in PANC-DB varies dramatically, we assessed the proportion of different endocrine cell types as a function of the total number of endocrine cells per sample. Among samples from donors without samples, there was substantial variability in the proportion of different islet cell types; for example, the proportion of β-cells in islets ranged from 0.19 to 0.71. When considering disease states, we observed decreased proportion of β-cells in islets from donors with type 1 diabetes compared with ND (average [avg.] ND = 0.40, avg. type 1 diabetes = 0.18; Wilcoxon P = 6.57 × 10−4), where β-cell proportion was not associated with age of onset or duration (P > 0.05). We observed a slight decrease in β-cells in donors with type 2 diabetes (avg. ND = 0.40, avg. type 2 diabetes = 0.376; Wilcoxon P = 0.36) and an increase in type 1 diabetes Aab+ (avg. ND = 0.40, avg. type 1 diabetes Aab+ = 0.45; Wilcoxon P = 0.324), although these estimates were not significant. While we also observed increased α-cell proportion in type 1 and type 2 diabetes (0.77 and 0.58, respectively; ND = 0.55), this is likely explained by the relative decrease in β-cells.
Changes in Islet Cell Type–Specific Gene Expression in Type 1 and Type 2 Diabetes
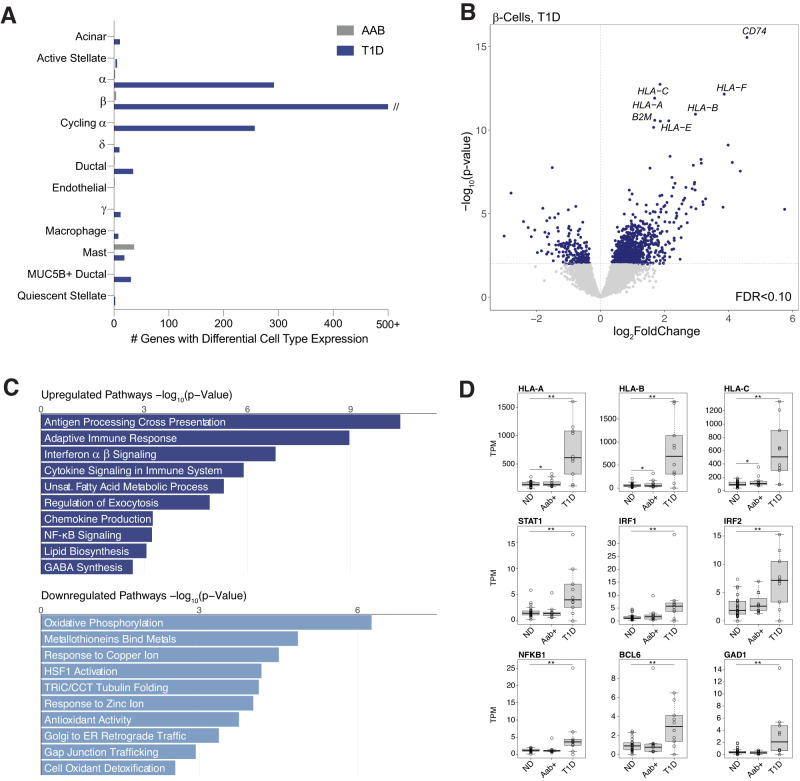
Identifying genes with changes in cell type activity in disease and predisease states can provide insight into disease pathogenesis. We therefore next determined changes in cell type gene expression in type 2 diabetes (n = 17), type 1 diabetes (n = 10), type 1 diabetes (Aab+) ND (n = 9) compared with ND control samples (n = 29). We tested for differential expression of genes in a cell type from pseudo-bulk profiles across samples accounting for sex, age, BMI, and technical covariates as well as ambient background RNA signal (see Research Design and Methods). In total, across all conditions, we identified 1,701 genes with significant changes in expression (FDR <0.10) in at least one cell type (Fig.2A).
Figure 2.
Changes in pancreatic islet cell type expression in type 1 diabetes. A: Number of genes in each cell type with significant changes (FDR <0.10) in expression in type 1 diabetes and type 1 diabetes Aab+ compared with ND donors using DESeq2. The x-axis is capped at 500 genes for legibility. B: Differential gene expression in β-cells in type 1 diabetes (T1D). Genes with upregulated expression are on the right side of the dashed line, and genes with downregulated expression are on the left side. The hard line represents the P value threshold corresponding to FDR <0.10. Genes with the most significant increase in expression are labeled. C: Gene sets enriched at FDR <0.10 among genes with upregulated and downregulated β-cell expression in type 1 diabetes. Gene sets are selected from the full set of significant results to highlight nonredundant biological terms and pathways. ER, endoplasmic reticulum; GABA, γ-aminobutyric acid; NF, nuclear factor; D: Normalized TPM β-cell expression level of HLA class I and selected cytokine response genes across ND, Aab+, and donors with 1 diabetes. **FDR <0.10, *Uncorrected P < 0.10 from DESeq2 analyses.
For type 1 diabetes, we identified 1,808 genes with significant changes in expression compared with ND donors (Supplementary Table 6). We observed the largest changes in β-cells (n = 1,305), although there were also significant changes in cycling α (n = 260), α (n = 35), acinar (n = 14), mast (n = 22), and other cell types. In β-cells, genes with the largest increases in expression included MHC class I (e.g., HLA-A, HLA-B) and MHC-related genes such as B2M and CD74 (Fig.2B). Genes upregulated in β-cells were broadly enriched (FDR <0.10) for antigen processing and presentation, interferon signaling, and immune response, among other processes, whereas downregulated genes were enriched for metal ion response, tubulin and gap junction activity, oxidative phosphorylation, and antioxidant activity (Fig.2C and Supplementary Table 7). In addition to MHC class I, we observed upregulation of cytokine response factors, such as IRF1/2, STAT1/4, and NFKB1, cell survival genes, such as BCL6, and the β-cell autoantigen GAD1 (Fig.2D). By comparison, few genes had significant changes in expression in individuals with type 1 diabetes Aab (Fig.2A). While MHC class I genes were nominally upregulated, we observed little change in cytokine response factors (Fig.2D). Among genes differentially expressed in type 1 diabetes, 98 map within 1 Mb of a type 1 diabetes risk locus, including INS, DLK1, and STAT4, highlighting candidate genes for underlying disease risk at these loci (Supplementary Table 8).
For type 2 diabetes, there were 84 genes with significant changes in any cell type compared with ND (Supplementary Table 9). Most of these genes were significant in β-cells (n = 79), although we also observed several significant genes in acinar, α-, and endothelial cells (Fig.3A). In β-cells, genes with the largest increase in expression in type 2 diabetes included TSHR, which is the receptor for thyroid stimulating hormone, SLC4A4, which is a bicarbonate cotransporter, and TNFRSF11B, which is a cytokine receptor for tumor necrosis factor family proteins (Fig.3B and D). Genes with upregulated expression in β-cells were broadly enriched (FDR <0.10) for RNA processing, hormone receptor activity, and cell growth and growth factor activity among other processes (Fig.3C and Supplementary Table 10). By comparison, downregulated genes were enriched for processes related to mitochondrial functions, such as oxidative phosphorylation, amino acid metabolism, and antioxidant activity. Several genes altered in type 2 diabetes, such as ASCL1, also map within 1 Mb of a known risk locus, suggesting candidate genes at these loci (Supplementary Table 8).
Figure 3.
Changes in pancreatic islet cell type expression in type 2 diabetes. A: Number of genes in each cell type with significant changes (FDR <0.10) in expression in type 2 diabetes (T2D) compared with ND donors using DESeq2. The x-axis is capped at 50 genes for legibility. B: Differential gene expression in β-cells in type 2 diabetes. Genes with upregulated expression are on the right side of the dashed line, and genes with downregulated expression are on the left side. The hard line represents the P value threshold corresponding to FDR <0.10. Genes with the most significant increase in expression are labeled. C: Gene sets enriched at FDR <0.10 among genes with upregulated and downregulated β-cell expression in type 2 diabetes. Gene sets are selected from the full set of significant results to highlight nonredundant biological terms and pathways. D: Normalized TPM β-cell expression level of selected differentially expressed genes across ND and donors with type 2 diabetes. **FDR <0.10, *Uncorrected P < 0.10 from DESeq2 analyses.
We next examined the overlap of genes with differential α- and β-cell expression in type 1 and type 2 diabetes with the results of multiple previous studies (9,35–38) (Supplementary Figure 4 and Supplementary Table 11). In type 1 diabetes, several studies assessed changes using a smaller set of HPAP donors, one of which used multiple differential analysis methods, and we observed moderate overlap with these results, including class I MHC genes and cytokine response factors. For type 2 diabetes, there was almost no overlap in differentially expressed genes in α- and β-cells with the results of previous bulk and single-cell studies included in the comparison (Supplementary Figure 4 and Supplementary Table 11). This is likely due to a multitude of factors, including differences in technology, analysis methods, covariates used, and tissue preparation, as well as limited sample and cell numbers for single-cell studies. However, several genes, such as ASCL1 and PPP1R1A (Fig.3D), which affect glucagon-like peptide 1 receptor–induced glucose-stimulated insulin secretion (39), had consistent changes across studies.
Reference Map and Resource Availability
The integrated map of gene expression in islet cell types generated by this study can be used to understand gene activity in physiological and disease states. In addition, this map can be used as part of bioinformatics pipelines, for example, to perform reference mapping of new scRNA-seq data sets. We therefore provide several resources and interactive applications to facilitate the wide use of this integrated map in a variety of downstream analyses. These resources are all available at http://www.isletgenomics.org.
First, we provide the islet cell type expression map in two interactive single cell browsers, CELLxGENE (40) and ShinyCell (41), which enable visualizing patterns across individual cells, for example, cell type identity, variables such as donor or library, technical factors such as number of features or percentage of mitochondrial reads, or the expression level of selected genes (Supplementary Fig. 5). In addition, we provide the expression map in Azimuth (20), which can be used for rapid on-the-fly reference mapping of new data sets (Supplementary Fig. 6).
Second, we provide annotations of activity in each islet cell type including marker genes, normalized gene expression levels, and changes in gene expression in type 1 diabetes, type 1 diabetes Aab+ and type 2 diabetes. We developed several interactive applications that enable users to select specific genes to view expression levels in each cell type as well as to visualize changes in cell type expression in different disease states (Supplementary Fig. 7).
Finally, the analytical pipelines used for data processing and clustering, defining expressed genes in each cell type, and defining differentially expressed genes are provided open access.
Discussion
Maps of gene expression levels in individual cell types within a heterogeneous tissue are valuable tools for hypothesis generation to understand cell type function and identity, gene activity, and changes in disease. In addition, these maps can be used for reference mapping of scRNA-seq data sets to facilitate annotation of cell identity and perform integrated analyses (20,42,43). While repositories such as PANC-DB provide access to a rich resource of raw sequence data and phenotypic information on human islet donors generated by HPAP (16), drawing insight from these data is a major challenge to researchers without single-cell data analysis expertise. Our study provides an integrated map of gene expression profiles in islet cell types and changes in disease derived from the scRNA-seq experiments in HPAP, which will help enable downstream analyses and hypothesis generation for many non–single-cell expert investigators.
There are several areas where the map can be further improved in future iterations. First, we were unable to separate a population of ε-cells, likely due both to the rarity of ε-cells and the sparsity of scRNA-seq profiles. By comparison, several studies profiling islets using different single-cell technology resolved small ε-cell populations (9,44). We also did not identify other rare cell types in the pancreas such as Schwann cells or lymphoid cell types. The samples profiled by HPAP are purified islets, and other cell types outside of the islet microenvironment are therefore underrepresented. Furthermore, the repertoire of discrete states that exist within each cell type, as well as any subtypes, for example, with distinct spatial localizations, remains to be resolved. Continued profiling of donors and cells from both purified islets and whole pancreas will help to define profiles for all pancreatic cell type and subtypes. Finally, even after accounting for ambient background RNA there is still residual expression of genes in off-target cell types, particularly for highly expressed genes in common cell types. As ambient RNA is a general feature of droplet-based assays (45), improvements in background correction for scRNA-seq are needed to estimate cell type–specific expression more accurately.
Genes with significant changes in cell type–specific expression provide insight into diabetes pathogenesis. In type 1 diabetes, we observed marked upregulation of processes related to MHC class I antigen presentation and cytokine signaling response. By comparison, we did not observe significant changes in type 1 diabetes Aab+ donors, which suggests modest effects in β-cells in these individuals in contrast to a recent report (35). As expected, there was significant reduction in β-cell numbers in type 1 diabetes; however, approximately half of the β-cells remained in donors with type 1 diabetes, which supports that there is persistent β-cell mass even in long-standing type 1 diabetes (46–48). In type 2 diabetes, we observed downregulation of mitochondrial function, which contributes to oxidative stress, impaired insulin secretion, and β-cell dysfunction (49–51), as well as changes in other processes implicated in β-cell function, such as amino acid metabolism (52), RNA processing (53), and cell growth (54). We also observed minimal reduction of β-cell numbers in type 2 diabetes, consistent with studies that have shown β-cell dysfunction but limited β-cell loss in type 2 diabetes (55).
The limited overlap with differential genes identified in previous studies, particularly for type 2 diabetes, is likely due to multiple factors. First, islets profiled in HPAP are isolated from cadaveric donors and then cultured, which will induce changes in cell type profiles and which will lead to differences compared with approaches such as pancreatectomy of living donors (37,38). Second, previous studies of single cells have had limited sample sizes and cell numbers (9). Larger sample sizes than those currently available even in HPAP are needed to determine the extent to which gene profiles change in type 2 diabetes, especially given heterogeneity among individuals in disease processes (56). Larger sample sizes will also enable finer grained partitioning of samples to understand gene regulatory differences between phenotypic subgroupings. For example, many type 1 diabetes Aab+ samples in HPAP are single GAD+ (35), yet there is wide diversity in the autoantibody profiles of individuals with different rates of progression (57,58). Finally, expanded profiling of islets in predisease, for example, from impaired glucose-tolerant donors, will help understand changes that occur during disease progression (59).
Another key difference across studies is the analysis method used for differential expression. Here we used sample pseudo-bulk profiles in differential analyses considering biological and technical properties of the samples and ambient RNA correction. Other recent studies performed differential analyses instead, using the profiles of individual cells (35,36), which can dramatically affect the results. For example, a previous analysis of 16 HPAP donors (35) identified 11,434 differential genes in type 1 diabetes in β-cells with an individual cell approach but only 53 differential genes with a pseudo-bulk approach. While pseudo-bulk approaches may be conservative in averaging intersample variation, individual cell approaches likely inflate results, particularly when not properly considering the nonindependence of cells from the same donor and variable cell numbers across donors (60). Other approaches to control for unwanted variation have also been applied to HPAP data, such as matching case subjects and control subjects on donor characteristics (36). As our integrated map contains data from all current HPAP donors, this map can be subset for matched analyses.
In summary, our map of islet cell type–specific expression and associated resources of cell type–specific gene activity in physiological and disease states provided by this study will be a valuable reference to the islet and diabetes research community.
This article contains supplementary material online at https://doi.org/10.2337/figshare.23925765.
Article Information
Funding. This work was supported by the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grant numbers DK-105554, DK-114650, and DK-120429 to K.J.G. This article used data acquired from the Human Pancreas Analysis Program (HPAP-RRID:SCR_016202) Database, a Human Islet Research Network (RRID:SCR_014393) consortium (UC4-DK-112217, U01-DK-123594, UC4-DK-112232, and U01-DK-123716).
Duality of Interest. K.J.G. holds stock in Neurocrine Biosciences and has done consulting for Genentech. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. R.M.E. performed all single-cell data analyses. R.M.E., R.L.M., H.M.M., P.B., and M.-L.O. developed analytical pipelines for single-cell data processing and analysis. R.M.E. and K.J.G. wrote the manuscript. P.K. developed and implemented tools for data visualization and interaction. K.J.G. conceived of the study and obtained funding. K.J.G. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Funding Statement
This work was supported by the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grant numbers DK-105554, DK-114650, and DK-120429 to K.J.G. This article used data acquired from the Human Pancreas Analysis Program (HPAP-RRID:SCR_016202) Database, a Human Islet Research Network (RRID:SCR_014393) consortium (UC4-DK-112217, U01-DK-123594, UC4-DK-112232, and U01-DK-123716).
References
- 1. Da Silva Xavier G. The cells of the islets of Langerhans. J Clin Med 2018;7:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mobasseri M, Shirmohammadi M, Amiri T, Vahed N, Hosseini Fard H, Ghojazadeh M.. Prevalence and incidence of type 1 diabetes in the world: a systematic review and meta-analysis. Health Promot Perspect 2020;10:98–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saeedi P, Petersohn I, Salpea P, et al. ; IDF Diabetes Atlas Committee . Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pract 2019;157:107843. [DOI] [PubMed] [Google Scholar]
- 4. Aamodt KI, Powers AC. Signals in the pancreatic islet microenvironment influence β-cell proliferation. Diabetes Obes Metab 2017;19(Suppl. 1):124–136 [DOI] [PMC free article] [PubMed]
- 5. Brissova M, Aamodt K, Brahmachary P, et al. Islet microenvironment, modulated by vascular endothelial growth factor-A signaling, promotes β cell regeneration. Cell Metab 2014;19:498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Z, Gerstein M, Snyder M.. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haque A, Engel J, Teichmann SA, Lönnberg T.. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med 2017;9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kanter I, Kalisky T.. Single cell transcriptomics: methods and applications. Front Oncol 2015;5:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Segerstolpe Å, Palasantza A, Eliasson P, et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab 2016;24:593–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baron M, Veres A, Wolock SL, et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst 2016;3:346–360.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Camunas-Soler J, Dai XQ, Hang Y, et al. Patch-seq links single-cell transcriptomes to human islet dysfunction in diabetes. Cell Metab 2020;31:1017–1031.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li J, Klughammer J, Farlik M, et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep 2016;17:178–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lawlor N, George J, Bolisetty M, et al. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res 2017;27:208–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang YJ, Kaestner KH.. Single-cell RNA-seq of the pancreatic islets--a promise not yet fulfilled? Cell Metab 2019;29:539–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mawla AM, Huising MO.. Navigating the depths and avoiding the shallows of pancreatic islet cell transcriptomes. Diabetes 2019;68:1380–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaestner KH, Powers AC, Naji A; HPAP Consortium . NIH initiative to improve understanding of the pancreas, islet, and autoimmunity in type 1 diabetes: the Human Pancreas Analysis Program (HPAP). Diabetes 2019;68:1394–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shapira SN, Naji A, Atkinson MA, Powers AC, Kaestner KH.. Understanding islet dysfunction in type 2 diabetes through multidimensional pancreatic phenotyping: the Human Pancreas Analysis Program. Cell Metab 2022;34:1906–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Young MD, Behjati S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience 2020;9:giaa151 [DOI] [PMC free article] [PubMed]
- 19. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–3587.e29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019;16:1289–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wolock SL, Lopez R, Klein AM.. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst 2019;8:281–291.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res 2012;22:1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A.. Fast gene set enrichment analysis. 20 June 2016. [preprint]. bioRxiv:060012
- 27. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 2018;50:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chiou J, Geusz RJ, Okino ML, et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 2021;594:398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamamoto K, Hashimoto H, Tomimoto S, et al. Overexpression of PACAP in transgenic mouse pancreatic beta-cells enhances insulin secretion and ameliorates streptozotocin-induced diabetes. Diabetes 2003;52:1155–1162 [DOI] [PubMed] [Google Scholar]
- 30. Taneera J, Fadista J, Ahlqvist E, et al. Identification of novel genes for glucose metabolism based upon expression pattern in human islets and effect on insulin secretion and glycemia. Hum Mol Genet 2015;24:1945–1955 [DOI] [PubMed] [Google Scholar]
- 31. Bacos K, Perfilyev A, Karagiannopoulos A, et al. Type 2 diabetes candidate genes, including PAX5, cause impaired insulin secretion in human pancreatic islets. J Clin Invest 2023;133:e163612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tosti L, Hang Y, Debnath O, et al. Single-nucleus and in situ RNA-sequencing reveal cell topographies in the human pancreas. Gastroenterology 2021;160:1330–1344.e11 [DOI] [PubMed] [Google Scholar]
- 33. Chiou J, Zeng C, Cheng Z, et al. Single-cell chromatin accessibility identifies pancreatic islet cell type- and state-specific regulatory programs of diabetes risk. Nat Genet 2021;53:455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xin Y, Dominguez Gutierrez G, Okamoto H, et al. Pseudotime ordering of single human β-cells reveals states of insulin production and unfolded protein response. Diabetes 2018;67:1783–1794 [DOI] [PubMed] [Google Scholar]
- 35. Fasolino M, Schwartz GW, Patil AR, et al. ; HPAP Consortium . Single-cell multi-omics analysis of human pancreatic islets reveals novel cellular states in type 1 diabetes. Nat Metab 2022;4:284–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bosi E, Marchetti P, Rutter GA, Eizirik DL.. Human alpha cell transcriptomic signatures of types 1 and 2 diabetes highlight disease-specific dysfunction pathways. iScience 2022;25:105056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Solimena M, Schulte AM, Marselli L, et al. Systems biology of the IMIDIA biobank from organ donors and pancreatectomised patients defines a novel transcriptomic signature of islets from individuals with type 2 diabetes. Diabetologia 2018;61:641–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wigger L, Barovic M, Brunner AD, et al. Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories towards type 2 diabetes. Nat Metab 2021;3:1017–1031 [DOI] [PubMed] [Google Scholar]
- 39. Cataldo LR, Vishnu N, Singh T, et al. The MafA-target gene PPP1R1A regulates GLP1R-mediated amplification of glucose-stimulated insulin secretion in β-cells. Metabolism 2021;118:154734. [DOI] [PubMed] [Google Scholar]
- 40. CZ CELLxGENE. Available from https://chanzuckerberg.github.io/cellxgene/
- 41. Ouyang JF, Kamaraj US, Cao EY, Rackham OJL.. ShinyCell: simple and sharable visualization of single-cell gene expression data. Bioinformatics 2021;37:3374–3376 [DOI] [PubMed] [Google Scholar]
- 42. Kang JB, Nathan A, Millard N, et al. Efficient and precise single-cell reference atlas mapping with Symphony. Nat Commun 2021;12:5890 [DOI] [PMC free article] [PubMed]
- 43. Lotfollahi M, Naghipourfar M, Luecken MD, et al. Mapping single-cell data to reference atlases by transfer learning. Nat Biotechnol 2022;40:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Muraro MJ, Dharmadhikari G, Grün D, et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst 2016;3:385–394.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang S, Corbett SE, Koga Y, et al. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol 2020;21:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oram RA, Sims EK, Evans-Molina C.. Beta cells in type 1 diabetes: mass and function; sleeping or dead? Diabetologia 2019;62:567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Campbell-Thompson M, Fu A, Kaddis JS, et al. Insulitis and β-cell mass in the natural history of type 1 diabetes. Diabetes 2016;65:719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Butler AE, Galasso R, Meier JJ, Basu R, Rizza RA, Butler PC.. Modestly increased beta cell apoptosis but no increased beta cell replication in recent-onset type 1 diabetic patients who died of diabetic ketoacidosis. Diabetologia 2007;50:2323–2331 [DOI] [PubMed] [Google Scholar]
- 49. Burgos-Morón E, Abad-Jiménez Z, Marañón AM, et al. Relationship between oxidative stress, er stress, and inflammation in type 2 diabetes: the battle continues. J Clin Med 2019;8:1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eguchi N, Vaziri ND, Dafoe DC, Ichii H.. The role of oxidative stress in pancreatic β cell dysfunction in diabetes. Int J Mol Sci 2021;22:1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fex M, Nicholas LM, Vishnu N, et al. The pathogenetic role of β-cell mitochondria in type 2 diabetes. J Endocrinol 2018;236:R145–R159 [DOI] [PubMed] [Google Scholar]
- 52. Newsholme P, Krause M.. Nutritional regulation of insulin secretion: implications for diabetes. Clin Biochem Rev 2012;33:35–47 [PMC free article] [PubMed] [Google Scholar]
- 53. Moss ND, Sussel L.. mRNA processing: an emerging frontier in the regulation of pancreatic β cell function. Front Genet 2020;11:983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dickson LM, Rhodes CJ.. Pancreatic β-cell growth and survival in the onset of type 2 diabetes: a role for protein kinase B in the Akt? Am J Physiol Endocrinol Metab 2004;287:E192–E198 [DOI] [PubMed] [Google Scholar]
- 55. Marselli L, Suleiman M, Masini M, et al. Are we overestimating the loss of beta cells in type 2 diabetes? Diabetologia 2014;57:362–365 [DOI] [PubMed] [Google Scholar]
- 56. Gloyn AL, Powers AC.. There is more than one way to reach type 2 diabetes. Nat Metab 2021;3:894–895 [DOI] [PubMed] [Google Scholar]
- 57. Jacobsen LM, Bocchino L, Evans-Molina C, et al. The risk of progression to type 1 diabetes is highly variable in individuals with multiple autoantibodies following screening. Diabetologia 2020;63:588–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yu L, Zhao Z, Steck AK.. T1D autoantibodies: room for improvement? Curr Opin Endocrinol Diabetes Obes 2017;24:285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang G, Chiou J, Zeng C, et al. Integration of single-cell multiomic measurements across disease states with genetics identifies mechanisms of beta cell dysfunction in type 2 diabetes. 2 Jan 2023 [preprint]. bioRxiv:2023;2022.12.31.522386
- 60. Zimmerman KD, Espeland MA, Langefeld CD.. A practical solution to pseudoreplication bias in single-cell studies. Nat Commun 2021;12:738. [DOI] [PMC free article] [PubMed] [Google Scholar]