Abstract
The cytosine analog 2′-deoxy-3′-thiacytidine (3TC) has been shown to be an effective treatment for chronic hepatitis B virus (HBV) infection. However, several liver transplant patients who were undergoing treatment with 3TC for HBV infection experienced a breakthrough of virus while on 3TC. The predominant virus found in these patients’ sera contained either a valine or isoleucine for the methionine in the highly conserved YMDD nucleotide binding site in the HBV polymerase. To determine the biological relevance of the Met-to-Val substitution, we mutated a plasmid that contained a cDNA copy of the HBV pregenomic RNA such that when virus replication occurred during transient transfection of HepG2 cells, an M539V polymerase variant was produced. We found that in transiently transfected cells, this variant was approximately 330-fold less sensitive to the antiviral effects of 3TC and produced 7-fold less viral DNA than the wild type.
It has been established that 2′-deoxy-3′-thiacytidine (3TC), a cytosine analog, is effective for inhibiting hepatitis B virus (HBV) replication in chronically infected individuals (2, 5, 8). However, in three separate clinical trials, several immunosuppressed patients who were undergoing 3TC therapy for chronic HBV infection experienced a reoccurrence of viremia during their treatment (1, 9, 11). When the virus from these patients’ sera was genotyped, it was determined that they contained a mutation in the polymerase gene that caused either a valine or an isoleucine substitution for the methionine located in the highly conserved YMDD nucleotide binding domain of the HBV polymerase. Moreover, when the pretreatment and posttreatment sera from one of these patients were placed on primary hepatocytes that were treated with various concentrations of 3TC, more 3TC was required to inhibit the production of HBV DNA in the cells mixed with the posttreatment serum than with the pretreatment serum (1).
To determine if the Met-to-Val substitution in the YMDD motif of the polymerase was alone sufficient to alter the 3TC sensitivity of HBV, we wanted to create a plasmid that when transfected into human hepatoblastoma cells would produce the M539V polymerase variant of HBV. This plasmid then could be used to study the biological significance of the M539V substitution in HBV.
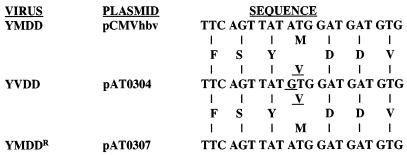
To create a plasmid capable of producing the M539V polymerase variant of HBV upon transfection into HepG2 cells, we used site-directed mutagenesis (Quickchange mutagenesis system; Stratagene) to mutate a cDNA copy of the pregenomic RNA (pgRNA) of HBV that was contained within plasmid pCMVhbv (3). Primers AV112 (5′-GCTTTCAGTTATGTGGATGATGTGG-3′) and AV113 (5′-CCACATCATCCACATAACTGAAAGD-3′) were used to change the first base in codon 539 of the polymerase gene from adenine to guanine, which should result in a methionine-to-valine substitution at this position (Fig. 1). The mutation in plasmid pAT0304 was verified by sequence analysis of approximately 400 bases of both strands of DNA in the immediate area of codon 539 of the polymerase gene (data not shown).
FIG. 1.
Nucleotide sequence of the YMDD nucleotide binding domain of the polymerase gene from plasmids pCMVhbv, pAT0304, and pAT0307. The nucleotide sequence of the HBV polymerase nucleotide binding domain was determined by sequence analysis of approximately 400 bp from both strands of DNA from plasmids pCMVhbv, pAT0304, and pAT0307 (fmol DNA Cycle Sequencing System; Promega). Single-letter abbreviations are used to denote the predicted amino acid sequence. Underlined nucleotide and amino acid sequences are those that differ from the wild-type sequence.
The 3TC sensitivity of the virus was determined by transfecting 5 × 105 HepG2 cells with 4 μg of either pCMVhbv or pAT0304 and pZeoSV-Lacz (Invitrogen, San Diego, Calif.) by using a liposomal system as described by the manufacturer (LipofectACE; GIBCO BRL/Life Technologies, Gaithersburg, Md.). Twenty-four hours after transfection, media containing various concentrations of 3TC were added to the cells. Four days after transfection, we collected the media and cells and isolated both extracellular and intracellular core-associated viral DNA as previously described (6).
We found that cells transfected with either plasmid produced virus whose replication could be inhibited by 3TC in a dose-responsive manner, as measured by the inhibition of the production of intracellular core-associated viral DNA (Fig. 2). The amount of viral DNA was normalized to transfection efficiency by β-galactosidase activity which was encoded by the lacZ gene on pZeoSV-Lacz. The normalized 50% effective concentration (EC50) of 3TC for wild-type virus (pCMVhbv) was 0.03 ± 0.02 μM (EC90, 0.3 ± 0.2 μM), which is comparable to what has been published previously (6). The normalized EC50 of 3TC for the YVDD variant (pAT0304) was approximately 10 μM (EC90, >10 μM). Therefore, the A-to-G mutation which should cause a methionine-to-valine substitution in the HBV polymerase reduced the potency of 3TC by approximately 330-fold. Similar results were obtained when we determined the potency of 3TC against the wild type and the M539V variant as measured by inhibition of the production of extracellular core-associated DNA (data not shown).
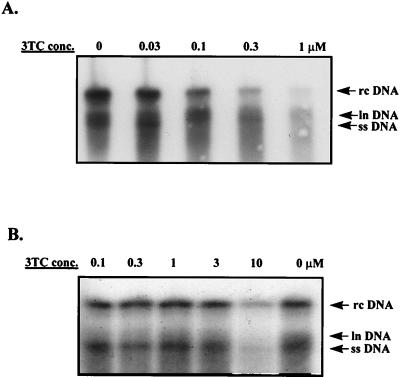
FIG. 2.
Sensitivity of the M539V variant to 3TC. HepG2 cells were transfected with pCMVhbv or pAT0304 and pZeoSV-Lacz. Twenty-four hours after transfection, the cells were treated with various concentrations of 3TC for 4 days. Intracellular viral core particles were isolated as previously described (6). Briefly, viral DNA was released from the core particles by proteinase K treatment followed by phenol extraction and ethanol precipitation. It was then separated by size by electrophoresis, transferred to a nylon filter, and detected with a radioactively labeled HBV-specific probe. The autoradiography exposure time was 16 h. (A) Core-associated HBV DNA isolated from HepG2 cells transfected with pCMVhbv and treated with various concentrations of 3TC. (B) Core-associated HBV DNA isolated from HepG2 cells transfected with pAT0304 and treated with various concentrations of 3TC. rc, relaxed circular; ln, linear; ss, single stranded. Note that there was a 3.5-fold difference in the amount of HBV DNA in the 0 drug lane in panels A and B after normalization to transfection efficiency.
While determining the 3TC sensitivity of the two viruses, we consistently observed that the cells which had been transfected with pAT0304 produced less viral DNA than cells transfected with an equal amount of pCMVhbv (Fig. 2A and B, compare the 0 drug lanes). When normalized to transfection efficiency, this difference was approximately 7-fold (range, 3- to 10-fold). This difference in the amounts of HBV DNA produced between the wild type and the M539V variant also was seen in the amount of extracellular core-associated viral DNA produced (data not shown).
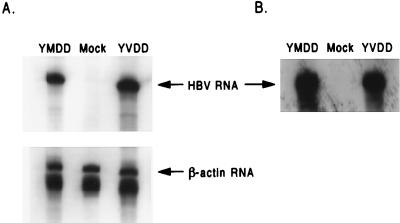
To determine if the difference in the amount of HBV DNA produced by the wild type and the M539V variant was due to a difference in the ability of pCMVhbv and pAT0304 to transcribe HBV RNA from the cytomegalovirus immediate-early (CMV IE) promoter located in these two plasmids, we isolated cytoplasmic RNA from HepG2 cells transfected with either pCMVhbv or pAT0304 and quantified the amount of viral RNA in the cytoplasm of the transfected cells by RNase protection assay with probes for HBV RNA (527 bases) and β-actin RNA (249 bases) as previously described (7). We found that after normalization to transfection efficiency by β-galactosidase levels and loading differences by β-actin RNA levels, the cytoplasmic fraction of HepG2 cells transfected with either pCMVhbv or pAT0304 contained equivalent amounts of HBV RNA (Fig. 3A). Therefore, the difference between the production of HBV DNA from the wild type and the M539V variant was not due to a difference in the ability of plasmid pCMVhbv or pAT0304 to produce viral RNA during the transient transfection of HepG2 cells.
FIG. 3.
RNase protection assays of cytoplasmic and core-associated viral RNA. (A) HepG2 cells were transfected with either pCMVhbv (YMDD), pAT0304 (YVDD) or pGEM3z (Mock), and pZeoSV-Lacz. Three days after transfection, RNA was isolated from the cytoplasmic fraction of the cell lysates as directed by the manufacturer (RNeasy kit; Qiagen). HBV RNA and β-actin RNA were detected and quantified by RNase protection assay (RPA-II kit; Ambion) by using riboprobes for HBV RNA (527 bases) and β-actin RNA (249 bases). The autoradiography exposure time was 16 h. (B) HepG2 cells were transfected with either pCMVhbv (YMDD), pAT0304 (YVDD), or pGEM3z (Mock). Three days after transfection, core-associated viral RNA was isolated from the intracellular immature core particles. Viral RNA was detected and quantified as described above. The autoradiography exposure time was 96 h.
One of the many roles that have been ascribed to the HBV polymerase in virus replication is to assist in the packaging of pgRNA into the immature core particle (3). To determine if the methionine-to-valine substitution in the polymerase of the 3TC-resistant variant affected the formation of immature core particles containing viral RNA, we isolated immature core particles from the cytoplasms of HepG2 cells which had been transfected with either pCMVhbv or pAT0304 and quantified the amount of core-associated viral RNA by RNase protection assay as described previously (7). We found that 5 × 105 HepG2 cells transfected with either pCMVhbv or pAT0304 contained similar amounts of core-associated HBV RNA in their cytoplasms (Fig. 3B). Therefore, the M539V polymerase variant was able to package pgRNA as effectively as the wild-type virus.
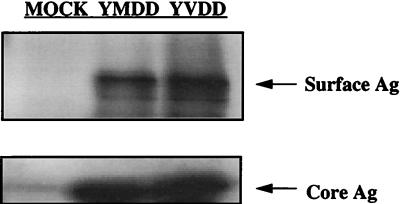
It is known that the four open reading frames of the HBV genome overlap; therefore, it is possible that a mutation in one of the open reading frames may also cause a mutation in one of the others. For example, the mutation in the polymerase gene responsible for the Met-to-Val substitution at codon 539 also causes an Ile-to-Met substitution at amino acid 195 of the HBV surface antigen (sAg). To determine if the M539V polymerase or the I195M sAg variations had any effect on the production of sAg in the transfected cells, we transfected HepG2 cells with pCMVhbv and pAT0304, as well as pZeoSV-Lacz, as described above. Three days after transfection, we lysed the cells and determined the level of β-galactosidase activity in the cell lysates. After correction for transfection efficiency, equivalent amounts of cell lysate were analyzed for the quantity of sAg by Western blot analysis with polyclonal antibodies specific for sAg (a kind gift from Fox Chase Cancer Center). After correction for loading variation by the quantification of HBV core Ag (cAg) with a polyclonal antibody specific for cAg (BioDesign International, Kennebunk, Maine), we found that the wild type and the M539V polymerase variant of HBV produced equivalent amounts of sAg (Fig. 4). Therefore, the I195M sAg substitution in the 3TC-resistant variant did not affect the levels of sAg in the transiently transfected cell.
FIG. 4.
Western blotting of sAg and cAg from HepG2 cells transfected with either pCMVhbv or pAT0304. HepG2 cells were transfected with either pCMVhbv (YMDD), pAT0304 (YVDD) or pGEM3z (Mock), and pZeoSV-Lacz. Three days after transfection, the cells were lysed. The cell lysates were assayed for β-galactosidase activity (Beta-Galactosidase Enzyme Assay System; Promega, Madison, Wis.), and the levels of sAg and cAg were determined by Western blot analysis with polyclonal antibodies specific for each of these proteins (1:2,000 dilution). Alkaline phosphatase-conjugated rabbit and goat secondary antibodies (1:1,000 dilution) were used for sAg and cAg, respectively. Detection was done with the ECL enhanced chemiluminescence kit (Amersham). The molecular masses of the sAg and cAg were approximately 27 and 21 kDa, respectively.
Since it was not possible to sequence the entire cDNA copy of the HBV genome in pAT0304 to determine if any other mutations may exist in this plasmid that may have affected 3TC sensitivity, we used site-directed mutagenesis to repair the mutation in the first position of codon 539 of the polymerase gene (G to A). This was done as described above with the mutagenic primers AV126 (5′-GCTTTCAGTTATATGGATGATGTGG-3′) and AV127 (5′-CCACATCATCCATATAACTGAAAGC-3′). We verified that the repair of the original mutation had occurred by sequencing approximately 400 bases of both strands of DNA surrounding codon 539 of the polymerase gene in the plasmid that was isolated after mutagenesis. This plasmid, pAT0307, had a sequence identical to that of pCMVhbv for those bases that were sequenced (data not shown).
We would predict that this plasmid when transfected into HepG2 cells should produce a virus that was wild type for 3TC sensitivity. When tested, the virus which resulted from the transfection of HepG2 cells with pAT0307 was as sensitive to the antiviral effects of 3TC as the wild-type virus produced from pCMVhbv (i.e., EC50, 0.04 ± 0.03 μM [pAT0307] versus 0.03 ± 0.02 μM [pCMVhbv]; EC90, 0.3 ± 0.2 μM [pAT0307] versus 0.3 ± 0.2 μM [pCMVhbv]). In addition, the repair of this mutation restored the level of production of intracellular core-associated HBV DNA in transfected HepG2 cells to that of the wild type (Fig. 5). Similar results were seen when extracellular core-associated HBV DNA was quantified (data not shown).
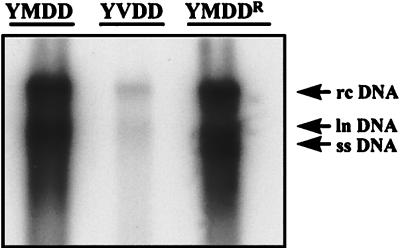
FIG. 5.
Reversion of the reduced viral DNA synthesis phenotype associated with the YVDD variant. HepG2 cells were transfected with 4 μg of pCMVhbv (YMDD), pAT0304 (YVDD), or pAT0307 (YMDDR). Three days after transfection, immature core particles were isolated as previously described (6). Viral DNA was released from the core particles by proteinase K treatment followed by phenol extraction and ethanol precipitation. It was then separated by size by electrophoresis, transferred to a nylon filter, and detected with a radioactively labeled HBV-specific probe. The autoradiography exposure time was 16 h. Note that there was a 10-fold difference in the amount of viral DNA produced by pCMVhbv and pAT0307 and pAT0304 after normalization to transfection efficiency. rc, relaxed circular; ln, linear; ss, single stranded.
These studies confirm that the Met-to-Val substitution in the YMDD nucleotide binding site of the HBV polymerase is responsible for and sufficient to decrease the sensitivity of HBV to the antiviral effects of 3TC. In the transient transfection system described above, this change was approximately 330-fold. A similar mutation in the HIV reverse transcriptase gene has been shown to exert a similar effect on the sensitivity of that virus to 3TC as well as several other cytosine analogs (4, 10, 12).
In addition, in HepG2 cells transiently transfected with pAT0304, the cytoplasmic fraction of the cell lysates contained approximately sevenfold less core-associated viral DNA than cells that had been transiently transfected with pCMVhbv. Since cells transfected with pCMVhbv and pAT0304 had similar levels of cytoplasmic and core-associated viral RNA, the difference in the level of HBV DNA expression probably is due to the Met-to-Val substitution having a detrimental effect on the ability of the polymerase to synthesize viral DNA from pgRNA. However, we do not know the in vivo biological significance of this observation, because the titer of the YVDD variant in the patient’s sera after virus breakthrough typically is equivalent to the titer of the wild-type virus previous to 3TC therapy (3, 5, 8).
Finally, the determination that the M539V substitution in the HBV polymerase is responsible for the creation of a 3TC-resistant variant of HBV provides a valuable tool for HBV drug discovery. However, since the transient transfection system described above must be performed with 60-mm-diameter plates to get enough core-associated HBV DNA to quantify, it is not amenable for large-scale screening of other nucleoside analogs for their potency against the M539V polymerase variant. We recently have described a stably transfected, inducible cell line that when grown under the proper conditions produces the M539V polymerase variant of HBV and have described how it can be used to screen compounds for activity against the 3TC-resistant variant (7).
Acknowledgments
We thank Raymond Schinazi for the gift of 3TC, Christoph Seeger for valuable scientific discussion and assistance, and Michael Otto for critically reviewing the manuscript.
REFERENCES
- 1.Bartholomew M M, Jansen R W, Jeffers L J, Reddy K R, Johnson L C, Bunzenddahl H, Condreay L D, Tzakis A G, Schiff E R, Brown N A. Hepatitis-B-virus resistance to lamivudine given for recurrent infection after orthotopic liver transplantation. Lancet. 1997;349:20–22. doi: 10.1016/S0140-6736(96)02266-0. [DOI] [PubMed] [Google Scholar]
- 2.Deinstag J L, Perrillo R P, Schiff E F, Bartholomew M, Vicary C, Rubin M. A preliminary trial of 3TC for chronic hepatitis B infection. N Engl J Med. 1995;333:1657–1661. doi: 10.1056/NEJM199512213332501. [DOI] [PubMed] [Google Scholar]
- 3.Fallows D A, Goff S P. Mutations in the ɛ sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J Virol. 1995;69:3067–3073. doi: 10.1128/jvi.69.5.3067-3073.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Q, Gu Z, Hiscott J, Dionne G, Wainberg M A. Generation of drug-resistant variants of human immunodeficiency virus type 1 by in vitro passage in increasing concentrations of 2′,3′-dideoxycytidine and 2′,3′-dideoxythiacytidine. Antimicrob Agents Chemother. 1993;37:130–133. doi: 10.1128/aac.37.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grellier L, Mutimer D, Ahmed M, Brown D, Burroughs A K, Tolles K, McMaster P, Beranek P, Kennedy F, Kibbler H, McPhillips P, Elias E, Dusheiko G. 3TC prophylaxis against reinfection in liver transplantation for hepatitis B cirrhosis. Lancet. 1996;348:1212–1215. doi: 10.1016/s0140-6736(96)04444-3. [DOI] [PubMed] [Google Scholar]
- 6.Ladner S K, Otto M J, Barker C S, Zaifert K, Wang G-H, Guo J-T, Seeger C, King R W. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ladner S K, Miller T J, Otto M J, King R W. The M539V polymerase variation responsible for 3TC-resistance also confers cross-resistance to other nucleoside analogs. Antiviral Chem Chemother. 1998;9:65–72. [PubMed] [Google Scholar]
- 8.Lai C-H, Ching C-K, Tung A K-M, Li E, Young J, Hill A, Wong B C-Y, Dent J, Wu P-C. 3TC is effective in suppressing hepatitis B virus DNA in Chinese hepatitis B surface antigen carriers: a placebo-controlled trial. Hepatology. 1997;25:241–244. doi: 10.1002/hep.510250144. [DOI] [PubMed] [Google Scholar]
- 9.Ling R, Mutimer D, Ahmed M, Boxall E, Elias E, Dusheiko M, Harrison T J. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with 3TC. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 10.Schinazi R F, Lloyd R M, Jr, Ngyen M-H, Cannon D L, McMillan A, Ilksoy N, Chu C K, Liotta D C, Bazmi H Z, Mellors J W. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother. 1993;37:875–881. doi: 10.1128/aac.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tipples G A, Ma M M, Fischer K P, Bain V G, Kneteman N M, Tyrrell K L J. Mutation in HBV RNA-dependent DNA polymerase confers resistance to 3TC in vitro. Hepatology. 1996;24:714–717. doi: 10.1002/hep.510240340. [DOI] [PubMed] [Google Scholar]
- 12.Tisdale M, Kemp S D, Parry N M, Ladner B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5633–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]