Titcombe et al. report that the non-classical AP-1 nuclear factor BATF is essential for the growth arrest and yet continued survival of tolerant T cells that have entered an anergic state based on its ability to activate anergy genes and repress BIM-dependent cell death.
Abstract
T cells that encounter self-antigens after exiting the thymus avert autoimmunity through peripheral tolerance. Pathways for this include an unresponsive state known as anergy, clonal deletion, and T regulatory (Treg) cell induction. The transcription factor cues and kinetics that guide distinct peripheral tolerance outcomes remain unclear. Here, we found that anergic T cells are epigenetically primed for regulation by the non-classical AP-1 family member BATF. Tolerized BATF-deficient CD4+ T cells were resistant to anergy induction and instead underwent clonal deletion due to proapoptotic BIM (Bcl2l11) upregulation. During prolonged antigen exposure, BIM derepression resulted in fewer PD-1+ conventional T cells as well as loss of peripherally induced FOXP3+ Treg cells. Simultaneous Batf and Bcl2l11 knockdown meanwhile restored anergic T cell survival and Treg cell maintenance. The data identify the AP-1 nuclear factor BATF as a dominant driver of sustained T cell anergy and illustrate a mechanism for divergent peripheral tolerance fates.
Introduction
Self-reactivity is an expected and required feature of developing T lymphocytes. It ensures that mature T cell clones will recognize peptides presented on the host’s own major histocompatibility complex (MHC) molecules. After initial positive selection, however, T cell responsiveness to self-antigens is necessarily restrained to protect the host—first by central tolerance mechanisms in the thymus and secondarily by peripheral tolerance mechanisms as surviving clones enter the circulation (Hogquist et al., 2005; Mueller, 2010). Peripheral tolerance thus enables controlled T cell recognition of self-peptides not present in the thymus (Xing and Hogquist, 2012). Notably, these peripheral tolerance pathways prevent the development of autoimmune disease but may also impose a barrier to robust anticancer immunity (ElTanbouly and Noelle, 2021; Crespo et al., 2013).
Both T cell–intrinsic and –extrinsic peripheral tolerance pathways are in effect in the normal immune system. Upon TCR engagement by self-peptide–MHC ligand in the periphery, cell-intrinsic tolerance is maintained by either functional inactivation (anergy) or apoptotic cell death (deletion) (Mueller, 2010; ElTanbouly and Noelle, 2021). Anergy is characterized by a failure to sustain proliferation or produce autocrine growth factors in response to the continued presence of antigens (Schwartz, 2003). This state is induced when T cells repeatedly interact with peptide–MHC ligands in the absence of an infection or adjuvant (Kearney et al., 1994; Pape et al., 1998). Anergic CD4+ T cells upregulate the surface markers NRP1, CD73, and FR4, along with co-inhibitory receptors such as PD-1 and CTLA4 (Martinez et al., 2012; Kalekar et al., 2016; Tuncel et al., 2019). Although no mechanism has yet been shown, the cellular processes that govern anergy and deletion appear to be related as tolerized cells that escape peripheral deletion typically display features of anergy (Barron et al., 2008).
With certain TCR and cytokine inputs, peripheral CD4+ T cells may also differentiate into suppressive FOXP3+ T regulatory (Treg) cells and thereby enforce tolerance in a cell-extrinsic manner (Wing and Sakaguchi, 2010; Ohkura et al., 2013). Experiments have shown that a subset of anergic CD4+ T cells can also be induced to join the FOXP3+ Treg cell lineage with comparable protective capability (Kalekar et al., 2016; Kuczma et al., 2021). This raises the possibility of further mechanistic overlap in these various peripheral tolerance pathways. The potential for anergic T cells with their autoreactive TCRs to differentiate into FOXP3+ Tregs additionally offers a strong rationale for their persistent survival in the peripheral immune system (Kalekar and Mueller, 2017). Nonetheless, the factors that dictate a T cell’s trajectory along one pathway versus another are not well understood.
At a molecular level, anergy develops from TCR engagement in the absence of strong co-stimulatory signaling (e.g., CD28 ligation, IL-2R engagement, and mTOR activation) (Fathman and Lineberry, 2007; Chappert and Schwartz, 2010). Whereas complete T cell activation induces the combined activity of NFAT and activator protein 1 (AP-1) family transcription factors (TFs) at target genes, anergy induction and maintenance are thought to be devoid of AP-1 involvement (Macián, 2005; Fathman and Lineberry, 2007; Nurieva et al., 2011; Pereira et al., 2017). Decoupling NFAT activity from AP-1 is evidently sufficient to promote T cell anergy (Macián et al., 2000; Martinez et al., 2015). However, whether isolated NFAT transcriptional activity at anergy signature genes is the basis for functional unresponsiveness in vivo is not clear. AP-1 is a heterodimeric complex typically composed of FOS and JUN family subunits (Karin et al., 1997), and activation of these classical AP-1 complexes is deficient in anergic T cells (Mondino et al., 1996; Macián et al., 2002). In the decades since T cell anergy was first described (Jenkins and Schwartz, 1987), however, several additional proteins have been found to act within an AP-1 complex and bind to nearly identical DNA consensus sequence motifs (Garces de Los Fayos Alonso et al., 2018). One such AP-1 family member is the basic leucine zipper TF, ATF-like (BATF), which dimerizes with JUN and notably lacks the transactivation domain that is seen across most of the family (Murphy et al., 2013). The development and differentiation of several hematopoietic lineages are known to depend on BATF (Murphy et al., 2013; Wang et al., 2012), including nonlymphoid tissue FOXP3+ Treg cells (Delacher et al., 2020). In CD4+ helper T cells, BATF heterodimers are known to cooperatively bind with IFN regulatory factor 4 (IRF4) at unique AP-1–IRF composite elements (AICEs) (Glasmacher et al., 2012; Li et al., 2012), perhaps facilitating IRF4 and NFAT interactions at the DNA (Rengarajan et al., 2002). BATF protein undergoes clear upregulation upon TCR signaling but has been conflictingly shown to both promote and dampen effector T cell responses (Kurachi et al., 2014; Quigley et al., 2010).
In this study, we took an unbiased molecular approach to identify key TFs that underlie T cell anergy induction and maintenance. Chromatin accessibility and DNA-binding footprint analyses revealed that BATF serves as the dominant AP-1 nuclear factor in anergic T cells. Genetic manipulation of Batf further demonstrated its role as a key anergy-promoting transactivating factor and suggested that it also acts directly upstream of the Bcl2l11 (BIM) promoter as a repressor. Finally, upregulation of BIM expression in BATF-deficient cells led to the reduced long-term survival of both conventional and regulatory T cells experiencing chronic TCR stimulation. Identification of this BATF–BIM axis offers a direct mechanistic link between distinct peripheral tolerance pathways.
Results
Evidence for an AP-1 gene regulatory program operating in T cell anergy
To investigate TFs important to T cell anergy development and maintenance, previously established marker sets were used to physically sort polyclonal CD4+ T cell subsets from the secondary lymphoid organs (SLOs) of Foxp3(GFP)-reporter C57BL/6 (B6) mice (Kalekar et al., 2016). Anergic CD44high FOXP3− CD73high FR4high Nrp1+ T cells were compared with T effector/memory (Teff; CD44high FOXP3− CD73low FR4low), Treg (FOXP3+ CD25+), and naïve (CD44low) T cell populations. Divided aliquots from the same samples were processed in parallel to generate corresponding assays for transposase-accessible chromatin sequencing (ATAC-seq) and mRNA expression (RNA-seq) datasets. In total, we identified 9,365 differentially accessible open chromatin regions (peaks; ANOVA, false discovery rate [FDR] < 1e−2) and 4,349 differentially expressed genes (ANOVA, FDR < 5e−2) across the sorted sample subsets (Table S1). Principal component analysis (PCA) demonstrated highly similar T cell subpopulation relationships across both the ATAC-seq and RNA-seq datasets (Fig. 1 A), supporting the hypothesis that subset-specific gene expression (and thereby differentiation) is strongly correlated to chromatin accessibility. Anergic T cells clustered more closely to Treg cells and were the most distinct from naïve cells, fitting with the notion that both anergic and Treg cells are subject to chronic TCR engagement (Rudensky et al., 2006).
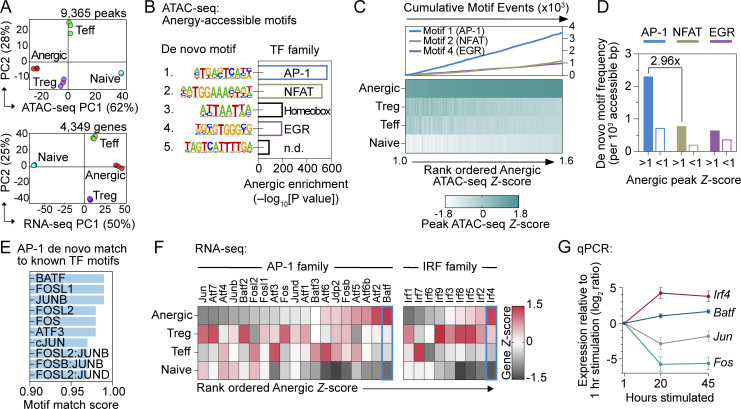
Figure 1.
Anergic T cell chromatin is poised for an AP-1–driven transcriptional program. (A) PCA of chromatin accessibility (top; n = 9,365 differentially regulated ATAC-seq peaks) and mRNA expression (bottom; n = 4,349 differentially regulated genes) of physically sorted CD4+ T cell subsets from pooled spleen and lymph nodes (SLOs). CD44high Foxp3(GFP)− CD73high FR4high Nrp1+ Anergic (red), CD44high Foxp3(GFP)− CD73low FR4low Teff (green), Foxp3(GFP)+ CD25+ Treg (purple), and CD44low Foxp3(GFP)− naïve (blue). Percentages indicate the amount of variance explained within each of the first two PC dimensions. (B) De novo TF motifs were identified within the anergic ATAC-seq signature peaks (anergic Z-score >1; 3,320 peaks) in comparison to all other differentially accessible peaks (anergic Z-score <1; 6,045 peaks). n.d. = not determined. (C) Heatmap of sample group ATAC-seq relative DNA accessibility for 3,320 peaks with anergic Z-score >1 (bottom) and enumerated de novo TF motif (as indicated in panel) occurrences within the corresponding peak regions (top). (D) Anergy-associated de novo TF motif frequencies within binned ATAC-seq peaks, comparing anergic ATAC-seq signature peaks (anergic Z-score >1; 3,320 peaks) to all other differentially accessible peaks (anergic Z-score <1; 6,045 peaks). (E) Anergy-associated AP-1 de novo TF motif similarity to known TF consensus binding sequences. (F) Heatmap of relative RNA-seq expression for AP-1 (left) and IRF (right) family TFs. (G) Relative mRNA expression measured by qPCR following in vitro stimulation of naïve CD4+ T cells with anti-CD3 and anti-CD28 mAbs. Data are mean ± SD (n = 3 unique cell samples at each time point, two independent experiments). Source data for A–F: n = 3 samples for each group except n = 2 for naïve ATAC-seq; five pooled mice, one experiment.
To determine the primary drivers of the anergy-specific gene regulatory program, the differentially regulated ATAC-seq dataset was scanned to discover de novo enriched DNA sequence motifs for comparison with known TF consensus binding sequences. In line with previous reports on anergy-associated TFs (Macián et al., 2002; Safford et al., 2005), we observed clear enrichment for motifs corresponding to the NFAT family (de novo rank #2) and EGR family (de novo rank #4) TFs within the 3,320 most strongly anergic peaks (anergic Z-score >1; Fig. 1 B). Surprisingly, an AP-1 family motif (de novo rank #1; P value = 1e−247) was the predominant TF signature identified within these anergy-associated peaks (Fig. 1 B), occurring nearly three times more often than the NFAT family motif (Fig. 1, C and D). Thus, ATAC-seq analysis of steady-state polyclonal CD4+ T cells reveals that an AP-1 family TF may play a previously unappreciated role in the T cell anergy gene regulatory program.
BATF as the candidate AP-1 mediator of T cell anergy
Dimeric AP-1 TF complexes bind to typified DNA sequence elements that differ slightly based on the exact TF subunit composition (Garces de Los Fayos Alonso et al., 2018). Due to the similarity of these consensus binding sequences (core motif TGAG/CTCA), it was not immediately possible to predict which family member might drive this unexpected AP-1 signature in anergic cells. We therefore incorporated transcriptional information from the same sorted samples and additional genomics analyses to identify the AP-1 TF occupant of these sites. In accordance with the protein’s strong motif match score (Fig. 1 E), RNA-seq comparison of the AP-1 gene family pointed to Batf as the top candidate with high mRNA upregulation in anergic cells (Fig. 1 F). High expression of Irf4, which interacts with BATF to bind at composite DNA elements (i.e., AICE), further supported the notion of a BATF-driven gene regulatory program in anergic cells. In contrast, the anergic subset showed relatively poor expression of the classical AP-1 subunit genes Fos and Jun (Fig. 1 F). We hypothesized that the divergence of Batf from classical AP-1 gene behavior in anergic cells could reflect the use of a distinct AP-1 complex in the context of chronic TCR engagement. Supporting this notion, Batf and Irf4 expressions arose later and were more stably maintained than classical AP-1 genes after priming (Fig. 1 G). Collectively, these findings were consistent with past descriptions of anergy-lacking classical AP-1 activation and pointed instead toward the non-classical AP-1 protein BATF as the relevant signature gene-regulating TF.
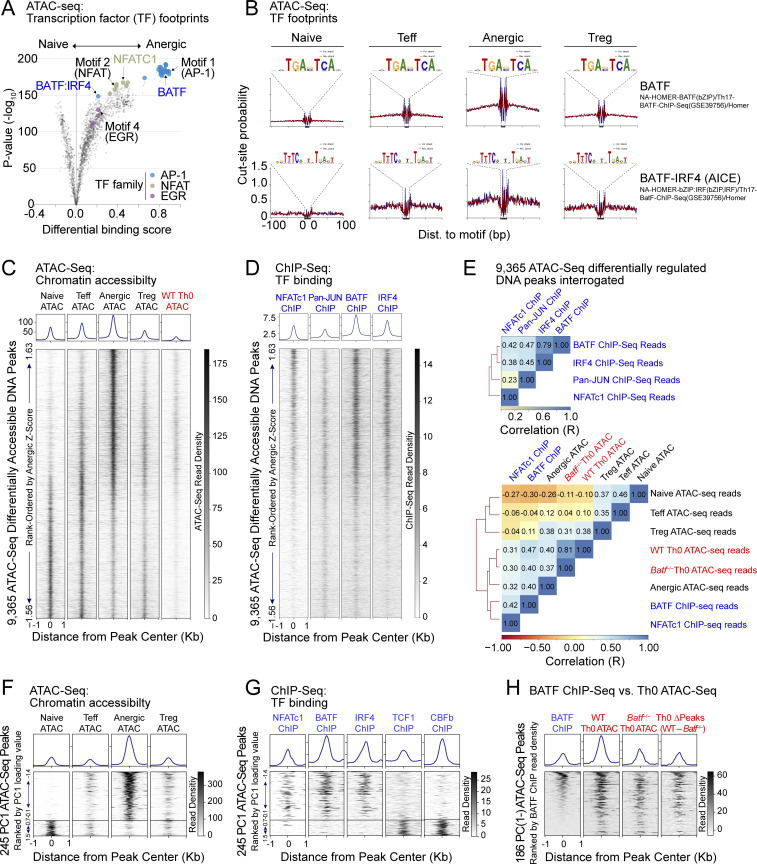
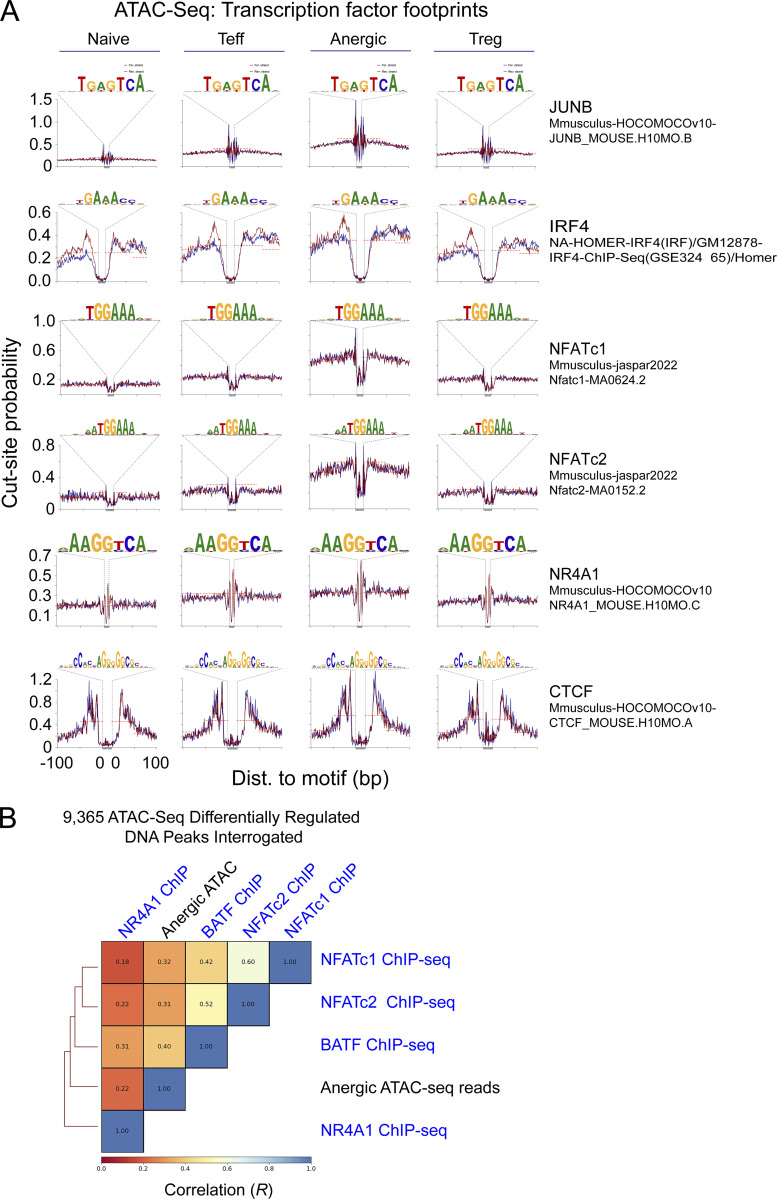
To assess for evidence of BATF binding at anergy-accessible DNA sequence motifs, all 38,509 called peaks were interrogated for discrete areas of relative ATAC-seq read signal-depletion (i.e., a TF footprint) at known nuclear factor consensus binding motifs. This analysis again suggested that AP-1 was the prevailing TF signature in anergic cells; additionally, it provided evidence that BATF binding sites were indeed occupied in the steady state (Fig. 2 A). High-resolution DNA read mapping confirmed TF footprints for BATF-IRF4 (AICE) and to a lesser degree BATF that were enhanced in the anergic group (Fig. 2 B). As expected, anergic T cells also demonstrated clear NFATc1, NFATc2, and IRF4 footprints (in addition to positive control CTCF chromatin binding) (Fig. S1 A). In contrast, most consensus sequence motifs for JUNB as well as the NR4A1 nuclear factor appeared vacant.
Figure 2.
BATF and IRF4 co-localize at BATF-IRF4 (AICE) motifs in anergic T cells. (A) Volcano plot summary of TF footprint enrichments determined by TOBIAS (Bentsen et al., 2020) from pairwise comparison of CD44low Foxp3(GFP)− naïve and CD44high Foxp3(GFP)− CD73high FR4high Nrp1+ anergic samples across 38,509 consensus ATAC-seq peaks. Colors denote footprints corresponding to anergy-associated TF families (as indicated), including the de novo motifs identified in Fig. 1. (B) Visualized footprints for BATF (top) and the BATF-IRF4 AICE (bottom) based on the ATACseqQC algorithm. Plots display Tn5 transposase cut-site probability within ±100 bp of all consensus binding motifs identified on chromosome 1 for both forward (blue) and reverse (red) de-duplicated and shifted ATAC-seq reads in each sample group (naïve, Teff, anergic, and Treg). (C and D) ATAC-seq read densities (C) and ChIP-seq read densities (D) within ±1 kb of the peak center for 9,365 differentially-accessible chromatin regions, displayed as a stack according to anergic Z-score. Histograms above represent the average read density across the same intervals for each sample group. ChIP-seq information was derived from public datasets for CD8+ T cell NFATc1 (Klein-Hessling et al., 2017), CD4+ T cell Pan-JUN (Li et al., 2012), CD4+ T cell BATF, and IRF4 (Iwata et al., 2017). 72 h in vitro stimulated (anti-CD3/CD28 mAb) WT Th0 ATAC-seq reads were mapped from publicly available ATAC-seq SRA files (Karwacz et al., 2017). (E) Pairwise Spearman correlation (R) and hierarchical clustering of ChIP-seq reads (top) and similar analysis of ATAC-seq reads as compared to BATF or NFATc1 ChIP-seq (bottom), when interrogated at 9,365 differentially accessible ATAC-seq peak intervals as shown in C and D. Batf−/− Th0 ATAC-seq reads were previously reported (Karwacz et al., 2017). (F) A total of 245 unique ATAC-seq peaks assigned to PC cluster groups PC(1−) and PC(1+) after TSNE plotting were ranked according to lowest PC1 loading value, and then were displayed as a stack with read densities in each sample group as indicated (lower panels). Histograms sum all reads across all selected peak intervals (upper panels). (G) Published ChIP-seq datasets for NFATc1, BATF, IRF4, TCF1, and CBFb were interrogated across the identical PC(1−) and PC(1+) ATAC-seq peak coordinate intervals as shown in F, with ChIP-seq read densities again displayed as stacks. (H) Published BATF ChIP-seq reads at 186 unique PC(1−) ATAC-seq peaks were ranked by highest BATF read density and then compared to published WT and Batf−/− Th0 ATAC-seq reads following 72 h of in vitro stimulation with anti-CD3/CD28 mAbs. One additional stack (Th0 ΔPeaks) represents the numerical difference between the WT and Batf−/− ATAC-seq read counts at each DNA coordinate. Source ATAC-seq data for all panels as in Fig. 1.
Figure S1.
ATAC-seq chromatin accessibility peaks are bound by BATF, IRF4, and NFAT proteins in anergic CD4+ T cells. (A) Using the ATACseqQC algorithm, chromosome 1 ATAC-seq reads were interrogated for the presence of JUNB, IRF4, NFATc1, NFATc2, NR4A1, and CTCF consensus DNA-binding motifs. Tn5 transposase cut-site probability across these sequences was then determined for both forward (blue) and reverse (red) de-duplicated and shifted DNA reads in each sample group (naïve, Teff, anergic, and Treg) as indicated. Cut-site protection (“footprint”) when present is indicated by the dashed line. (B) Pairwise Spearman correlation (R) and hierarchical clustering of anergic ATAC-seq reads and select ChIP-seq reads interrogated at 9,365 differentially accessible ATAC-seq peak intervals. Source ATAC-seq data for all panels as in Fig. 1.
We next used chromatin immunoprecipitation and sequencing (ChIP-seq) datasets to validate our footprint analyses and visualize known TF binding within anergy-accessible chromatin. Alignment of our 9,365 differentially accessible ATAC-seq peak reads with reported ChIP-seq reads confirmed that regions of unique chromatin accessibility in anergic cells directly correlated with NFATc1 binding (R = 0.32; Fig. 2, C–E). NFATc2 also demonstrated a similar positive correlation with the anergic group ATAC-seq reads (R = 0.31; Fig. S1 B). BATF was found to bind DNA within many of the same peaks as NFATc1 (R = 0.42), and this binding was again biased to anergy-associated peaks (R = 0.40, Fig. 2, C–E). IRF4 binding closely matched that seen for BATF (R = 0.79), suggesting that many of the anergy-accessible peaks contain AICE sites. On the other hand, JUN proteins and NR4A1 showed less bias toward binding within anergy-associated chromatin peaks (Fig. 2, C and D; and Fig. S1 B). Distinct patterns of TF binding were also observed when ATAC-seq accessibility peaks were selected for analysis based on strong variance in PCA dimension 1 (PC1). NFATc1, BATF, and IRF4 binding were essentially restricted to anergy-associated cluster PC(1−) DNA coordinates whereas DNA binding by two other TFs (TCF1, CBFb) was limited to cluster PC(1+) gene loci that are inaccessible within anergic cells (Fig. 2, F and G).
As one additional test of the role of BATF in the epigenetic regulation of T cell tolerance, we made use of a publicly available ATAC-seq dataset obtained from normal and BATF-deficient polyclonal CD4+ T cells stimulated in vitro for 72 h with a combination of mAbs directed to CD3 and CD28 (Karwacz et al., 2017). WT “Th0” T cells stimulated under these conditions acquired an epigenetic landscape more similar (R = 0.40) to bona fide anergic T cells than to any other cell group when interrogated using our own 9,365 differentially accessible open chromatin regions (Fig 2, C and E). Like anergic cells, these chronically stimulated T cells also demonstrated chromatin accessibility peaks at loci that accommodate DNA-binding for both BATF and NFATc1 (R = 0.47 and 0.31, respectively) (Fig. 2, C–H). Interestingly, genetic deletion of Batf resulted in only a small change in the overall pattern of chromatin accessibility across the 9,365 peaks (R = 0.81), yet PC(1−) selected chromatin accessibility peaks enriched in anergic cells and containing BATF binding sites appeared to be preferentially lost from the Batf−/− T cells (Fig. 2, E and H). Thus, BATF protein induced during chronic TCR stimulation can influence chromatin structure at DNA loci that become accessible specifically in anergic T cells and that contain a BATF consensus binding motif. The finding aligns with previous reports of BATF’s capacity to enact initial chromatin remodeling and recruitment as a “pioneer” factor (Ciofani et al., 2012; Pham et al., 2019). Taken together, this multiomic analysis suggests that anergic T cells utilize a distinct TF program driven by a BATF-containing AP-1 protein complex cooperating with IRF4 and NFAT.
BATF is necessary for the normal induction and maintenance of anergic T cells
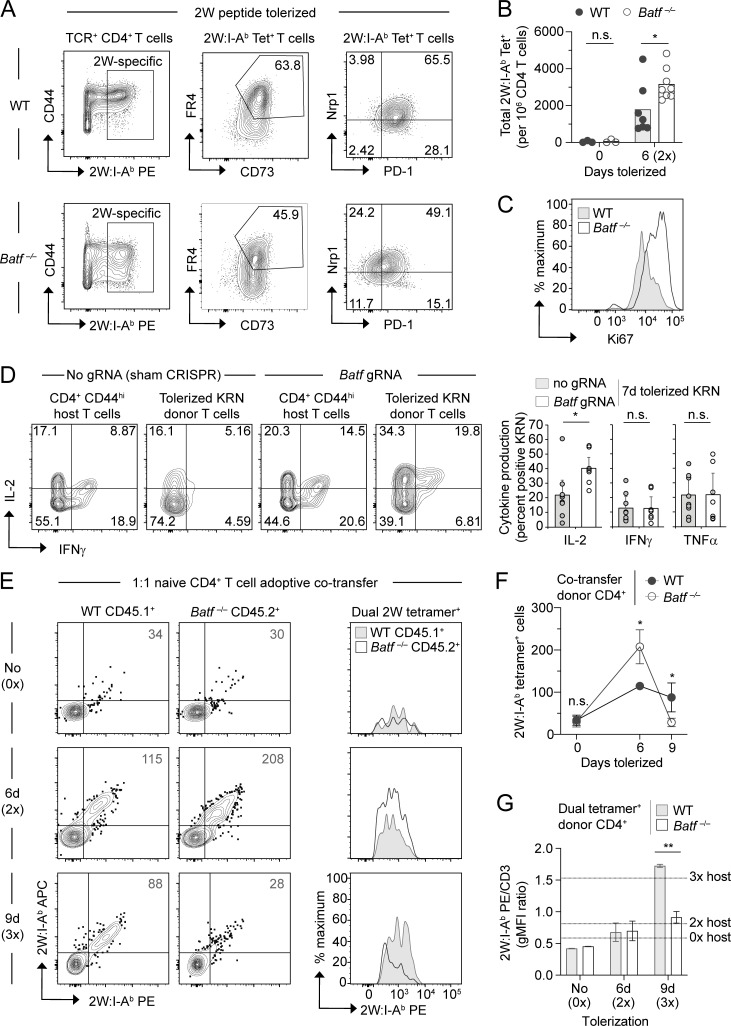
In mice, CD4+ T cell anergy can be induced in vivo following repeated exposure to a soluble peptide antigen in the absence of adjuvant or infection (Kearney et al., 1994; Pape et al., 1998). To formally test if BATF plays a role in the anergy program, we employed a similar tolerization strategy wherein multiple injections of the model peptide antigen 2W (Rees et al., 1999) were delivered intravenously in PBS to wildtype (WT) versus Batf−/− B6 mice. Polyclonal antigen-specific CD4+ T cells were then tracked using 2W peptide-loaded MHC class II (2W:I-Ab) tetramers (Moon et al., 2007). Whereas comparable numbers of WT and Batf−/− 2W-specific T cells were detected in peptide-naïve animals, Batf−/− cells displayed signs of anergy resistance with approximately twofold greater clonal expansion during the first 6 days of 2W peptide tolerization (Fig. 3, A and B). Reduced expression of the checkpoint inhibitor protein PD-1 and increased expression of the proliferation marker Ki67 correspondingly suggested that tolerized 2W-specific T cells failed to undergo their typical cell cycle arrest in the absence of BATF (Fig. 3, A and C). Despite the delay in proliferative arrest shown here, T cells undergoing anergy induction in the absence of BATF demonstrated only subtle differences in the phenotypic expressions of FR4, CD73, and NRP1 (Fig. 3 A).
Figure 3.
Deletion compensates for defective anergy induction in BATF-deficient T cells. (A–C) CD4+ T cells from the SLOs of WT and Batf−/− mice either untreated or tolerized with two i.v. injections (6 d, 2×) of 2W peptide on days 0 and 3 and analyzed on day 6. (A) Representative flow cytometry plots for 2W:I-Ab tetramer-binding CD4 T cells (left) with expression of FR4, CD73 (middle), and Nrp1 and PD-1 (right). (B) Total 2W:I-Ab tetramer-binding cell numbers per million total CD4+ T cells. (C) Representative Ki67 staining of 2W:I-Ab tetramer-binding T cells following 6-day (2×) peptide tolerization. n = 3–8 mice per group, three independent experiments. *P < 0.05 by unpaired t test; n.s. = not significant. (D) Intracellular cytokine staining (left, representative flow cytometry plots; right, percent cytokine positive) for tolerized KRN T cells following in vitro stimulation. CRISPR-Cas9 treated (using either Batf gRNA or no gRNA as a control) KRN TCR transgenic T cells were transferred into B6.I-Ab/g7 host mice for in vivo tolerization, recovered from the SLOs on day 7, and stimulated for 4 h with PMA and ionomycin. Normal host CD44hi conventional CD4+ T cells are also shown left for reference. Bars represent the mean ± SEM of IL-2, IFNγ, and TNFα percent positive KRN T cells. n = 8–9 mice per sample group, three independent experiments. *P < 0.05 by unpaired t test; n.s. = not significant. (E–G) Bulk polyclonal naïve CD4+ T cells from congenically marked WT and Batf−/− mice were mixed equally, transferred into B6 CD90.1 recipient mice, and then tolerized with i.v. 2W peptide injections for the indicated durations beginning 1 day after co-transfer (n = 2–3 mice per time point, two independent experiments). (E) Gated donor CD4+ T cells stained with a combination of PE- and allophycocyanin (APC)-conjugated 2W:I-Ab tetramers. Histogram overlays of the dual tetramer-positive populations are displayed on the right. Numbers in the flow cytometry plots are the mean dual tetramer-positive cell number. (F) Mean numbers ± SD of dual 2W:I-Ab tetramer-positive cells (two-way ANOVA with Sidak’s correction; *P < 0.05, n.s. = not significant). (G) CD3-normalized geometric mean fluorescence intensity (gMFI) of PE-conjugated 2W:I-Ab tetramer within the dual 2W:I-Ab tetramer-positive cell population. Dashed lines indicate the mean value for host dual tetramer-positive cells at the corresponding time points. Bars represent mean ± SEM (paired t test; **P < 0.01).
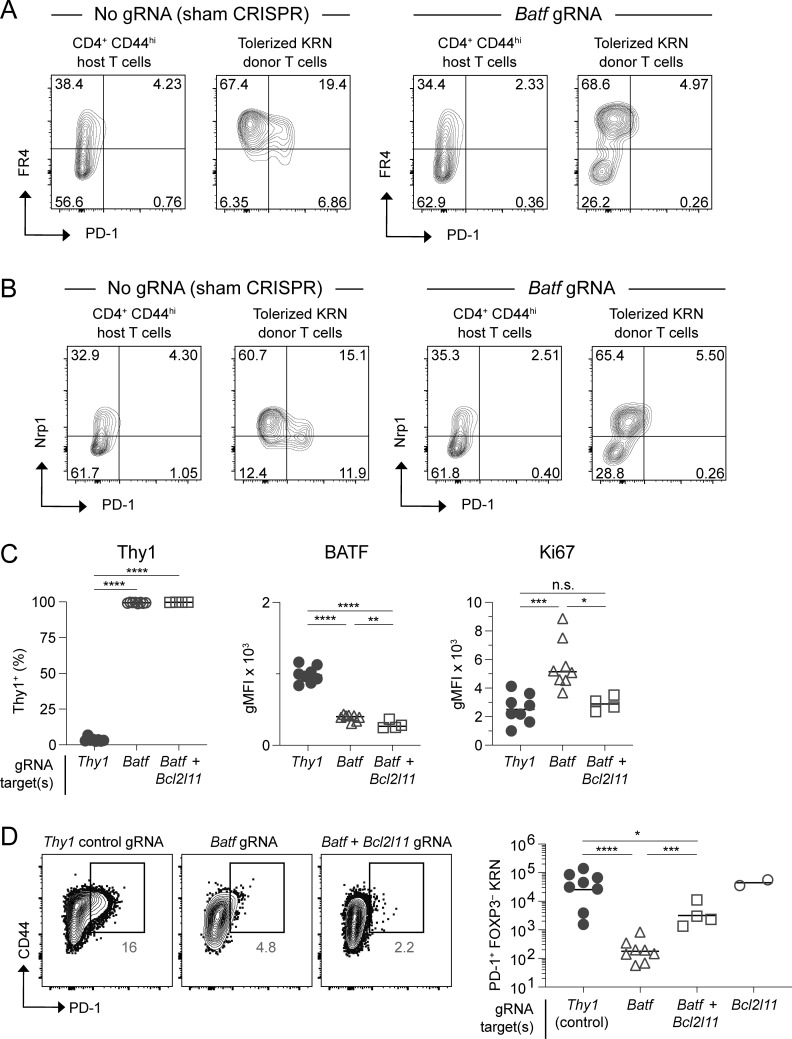
To validate this finding and further probe for a functional anergy defect, we employed a second strategy for generating T cell anergy in vivo—namely, the transfer of monoclonal self-reactive T cells into a setting with ubiquitous self-antigen expression. KRN CD4+ T cells express a TCR transgene that reacts with a self peptide from glucose-6-phosphate isomerase when presented on the MHC class II molecule I-Ag7 (Matsumoto et al., 1999). Previous work has shown that naïve KRN T cells undergo abortive clonal expansion and robust anergy induction (including defective IL-2, IFNγ, and TNFα production) within 1 wk from transfer into normal I-Ag7-expressing mice (Martinez et al., 2012). Here, we utilized the CRISPR-Cas9 nuclease to knock down Batf in primary naïve KRN T cells prior to adoptive transfer. Importantly, Batf knockdown led to increased IL-2 growth factor inducibility following this tolerance regimen (Fig. 3 D). On the other hand, the ability of tolerized KRN T cells to produce IFNγ and TNFα upon restimulation was not significantly improved by Batf knockdown. Consistent with a selective regulation of anergy by BATF, tolerant CRISPR/Batf-modified KRN T cells also demonstrated a reduction in PD-1, yet the expression of the anergy markers FR4 and NRP1 remained largely unaffected (Fig. S2, A and B). Therefore, BATF appears to mediate certain key features of anergy induction—such as defects in autocrine growth factor production, proliferation, and the upregulation of PD-1—whereas other characteristics of anergy develop independently of BATF.
Figure S2.
BATF drives expression of PD-1 but is dispensable for FR4 and NRP1 upregulation during tolerance induction. (A and B) Naïve KRN T cells were modified by CRISPR-Cas9 in the presence or absence of Batf gRNA (as indicated) and then subjected to tolerization for 7 days by adoptive transfer into B6.I-Ab/g7 hosts. Shown are the day 7 FR4 × PD-1 (A) and NRP1 × PD-1 (B) flow cytometry profiles for tolerized KRN T cells. Normal host polyclonal CD44hi CD4+ T cells are also displayed for reference. Mice as in Fig. 3 D. (C and D) KRN T cells were modified by CRISPR-Cas9 in the presence of Thy1, Batf, or Batf plus Bcl2l11 gRNAs and then subjected to tolerization for 7 days. (C) Expression of Thy1, BATF, and Ki67 in modified T cells. (D) PD-1 expression in tolerized KRN T cells with representative flow plots shown (left) and conventional PD-1+ KRN T cell numbers quantitated (right). Unpaired t test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. = not significant. Source data as in Fig. 6.
To confirm the T-intrinsic effects of BATF, we tested 2W peptide-specific Batf−/− T cells in a transfer setting over an extended tolerization period. Pooled polyclonal CD4+ T cells from untreated WT CD45.1+ and Batf−/− CD45.2+ mice were mixed at equal numbers (20 × 106 for each cell genotype) and then transferred into congenically mismatched peptide-naïve hosts (Fig. 3, E–G). Consistent with the early anergy breach seen within intact Batf-/- mice, transferred 6-day-tolerized 2W-specific Batf−/− cells were twice as proliferative as WT cells. However, within just three additional days of tolerization, this competitive advantage reversed and the majority of the Batf−/− population was lost (Fig. 3, E and F). This disappearance of Batf−/− T cells indicated that, in addition to its role in anergy development, BATF may also control deletional tolerance. Clonal deletion typically results in reduced tetramer binding intensity due to the preferential removal of high-affinity TCR-bearing T cell clones (Moon et al., 2011). In keeping with the kinetics of the observed Batf−/− cell loss, a reduction in the relative 2W:I-Ab tetramer binding strength of Batf−/− cells was uniquely detected on day 9 of tolerization (Fig. 3, E and G). This result confirmed that, without BATF, high-affinity T cell clones that would have otherwise been selected for survival were instead being removed from the repertoire. Thus, clonal deletion compensates for defective anergy induction in peptide-tolerized Batf−/− T cells.
BATF activates anergy genes and represses an apoptosis gene
Having observed a sudden switch from anergy to deletional tolerance after 1 wk of T cell tolerization in the absence of BATF, we aimed to find a mechanistic explanation for BATF pleiotropically regulating these two modes of peripheral tolerance. A bioinformatics approach was applied to determine whether the clonal deletion outcome could have been predicted a priori based on the specific gene targets of BATF in these ATAC-seq peaks. In this analysis, we integrated the synchronous ATAC-seq and RNA-seq datasets—resulting in positive or negative gene-regulation information for 3,746 peaks—and then annotated the presence or absence of BATF-binding DNA consensus sequences within each peak. Pathway analysis on the genes that were in the vicinity of called peaks revealed enrichment for apoptosis-related pathways only among the set that contained BATF sites (Fig. 4 A). Similarly, ATAC-seq accessibility peaks assigned to PCA clusters PC(1−) and PC(1+) and ranked according to PC1 loading value were found to be highly associated with genes important for the “Cell survival” functional annotation (n = 46 molecules, P value = 1.53e−9, bias-corrected activation Z-score = 2.18) (Table S1).
Figure 4.
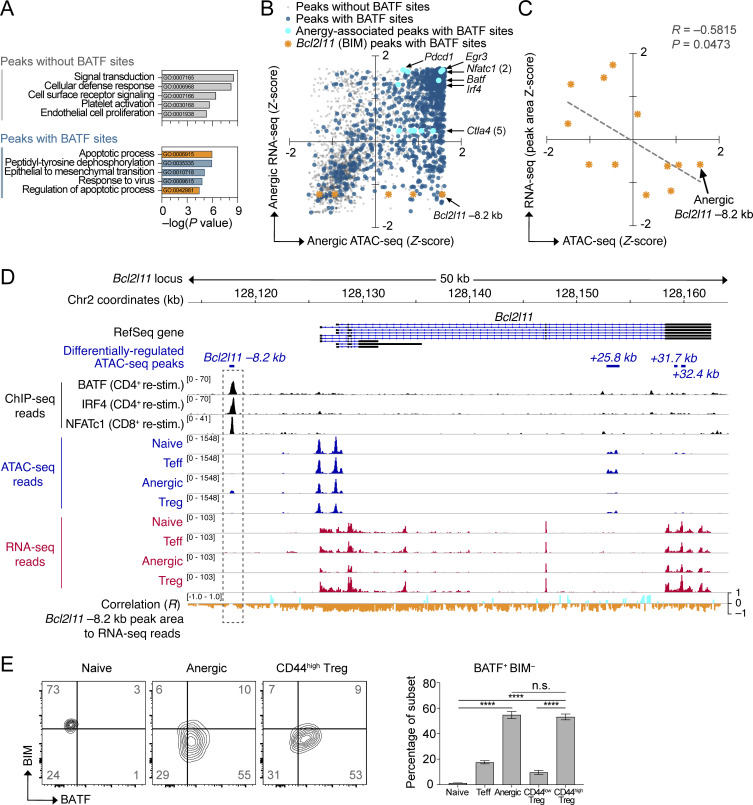
Negative association between BATF and BIM (Bcl2l11) expression. (A) Pathway enrichment of genes near differentially regulated ATAC-seq peaks either lacking de novo BATF motifs (top) or containing BATF motifs (bottom). Analysis was performed on 3,746 unique peaks corresponding to a differentially expressed gene. The top five significant pathways for each group are displayed (database: GO Biological Process 2017). Pathways related to programmed cell death are shown in orange. (B) Integrated ATAC-seq and nearest neighbor RNA-seq mRNA expression data (3,746 total chromatin peaks) were plotted according to Z-scores within the anergic sample group. Peaks without BATF motifs (transparent gray; 995 peaks), peaks with BATF motifs (dark blue; 2,751 peaks), anergy-associated gene peaks containing BATF motifs (aqua highlight with annotation), and Bcl2l11 peaks with BATF motifs (orange highlight with annotation of predicted Bcl2l11 −8.2 kb repressor site). (C) Linear regression analysis of ATAC-seq read Z-score at 3 BATF-containing Bcl2l11 peaks versus the Z-scores for RNA-seq reads across the identical DNA peak coordinates. Data shown are pooled for all four sample groups at each of the three loci (naïve, Teff, anergic, Treg; n = 12 total points). Note that the three ATAC-seq peaks selected for analysis contained both a BATF consensus binding motif and also demonstrated increased chromatin accessibility in the Anergic sample group (lower right quadrant in B). (D) Stacked ChIP-seq, ATAC-seq, and RNA-seq tracks at the Bcl2l11 gene locus with the calculated correlation of transcriptional activity to the pattern of Bcl2l11 −8.2 kb peak reads (dashed rectangle highlight). Data tracks are as indicated (top to bottom): condensed RefSeq gene open-reading frames; differentially regulated ATAC-seq peak coordinates (relative to the putative transcriptional start site); BATF, IRF4, and NFATc1 ChIP-seq reads from published datasets of primed T cells receiving secondary stimulation (black; Iwata et al., 2017; Klein-Hessling et al., 2017); ATAC-seq mean read densities for each experimental group (blue); RNA-seq mean read densities for each experimental group (red); multivariate correlation plot between the Bcl2l11 −8.2 kb peak ATAC-seq reads versus RNA-seq reads at given coordinates across the locus (positive correlation = aqua, negative correlation = orange). (E) Flow cytometry plots of BATF versus BIM intracellular protein staining in WT polyclonal CD4+ T cell subsets (left; two concatenated samples per subset shown) and quantification (right; n = 18 mice, two independent experiments) of BATF+ BIM− cell percentages in the lower-right quadrant. The represented CD4+ T cell subsets were pre-gated using the antibody markers defined in Fig. 1. Numbers in the quadrants and right panel are mean percentages ± SD (one-way ANOVA with Tukey’s correction, ****P < 0.0001, n.s. = not significant). Source ATAC-seq and RNA-seq data for A–D as in Fig. 1.
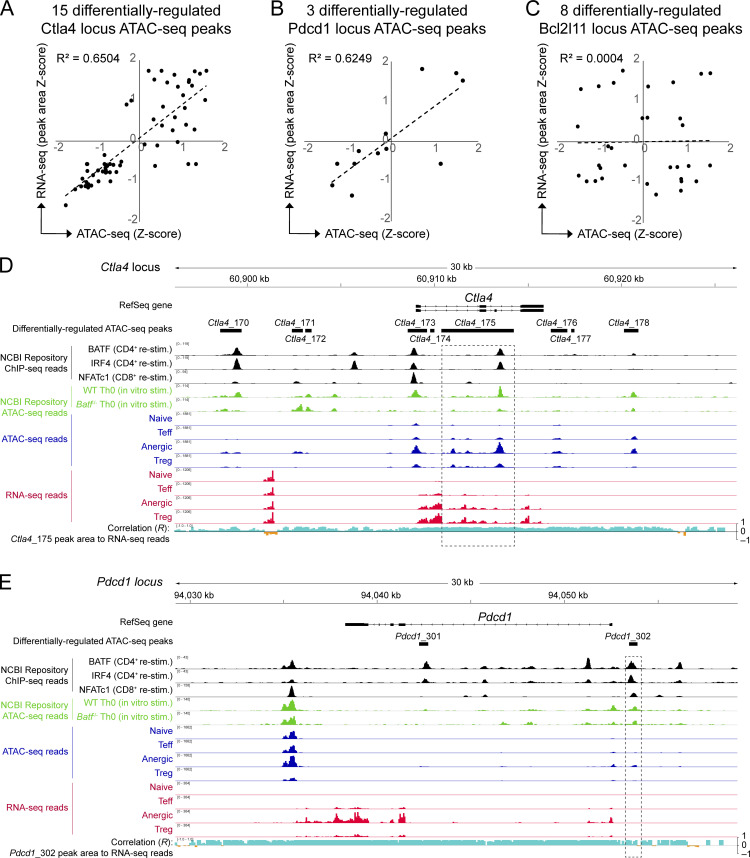
As expected for an AP-1 family TF and consistent with past reports, most open chromatin peaks that contained a BATF consensus binding motif in anergic T cells were associated with upregulated mRNA transcription at the adjacent gene (Fig. 4 B, upper right quadrant). Peaks corresponding to several well-known anergy-associated genes (e.g., Pdcd1 and Ctla4) exhibited this direct transcriptional activator pattern—consistent with the actions of DNA promoters and enhancers within these peaks (Fig. 4 B and Fig. S3). Nevertheless, a small set of BATF site-containing peaks showed a different gene regulatory pattern that was more indicative of repression (Fig. 4 B, lower right quadrant). Most notably, this included three distinct peaks within the locus for the apoptosis-associated gene Bcl2l11 (BIM). Compared with the other T cell subpopulations, anergic T cells expressed little mRNA for Bcl2l11 (anergic Z-score = −1.23). Remarkably, one particular area of BATF-associated chromatin residing 8.2 kb upstream of the Bcl2l11 transcriptional start site (Bcl2l11 −8.2 kb) was highly accessible in only anergic cells (Fig. 4, B–D). Bcl2l11 −8.2 kb (identified as peak Bcl2l11_5390 in Table S1) contained overlapping BATF, IRF4, and NFATc1 ChIP-seq binding sites and assumed a repressor-like activity based on negatively correlating RNA reads across the entire locus (Fig. 4, C and D). Direct intracellular staining of polyclonal CD4+ T cell subsets confirmed that BATF and BIM protein expression are inversely correlated in the steady state (Fig. 4 E). A BATF+ and BIM− state was found to be especially prevalent for both anergic T cells and CD44high Treg cells, perhaps reflecting the recency or chronicity of antigen exposure within these subpopulations (Kalekar et al., 2016; Rudensky et al., 2006). Taken together, these results supported the hypothesis that TCR-mediated repression of Bcl2l11 gene expression facilitates the continued survival of anergic T cells and suggested that BATF may be important to this gene repression.
Figure S3.
Tn5 transposase chromatin accessibility directly correlates with adjacent RNA transcriptional activity at the Ctla4 and Pdcd1 loci, but not at Bcl2l11. (A–C) Naïve, Teff, anergic, and Treg sample group Z-scores for both ATAC-seq chromatin accessibility and adjacent RNA-seq reads were calculated for the same ATAC-seq peak sequence coordinates and compared, with correlation coefficients (R2) as shown. Correlation is shown for (A) 15 differentially regulated Ctla4 locus peaks (Ctla4_166 through Ctla4_180), (B) three differentially regulated Pdcd1 locus peaks (Pdcd1_301 through Pdcd1_303), and (C) eight differentially regulated Bcl2l11 locus peaks (Bcl2l11_5387 through Bcl2l11_5394). Note that the Bcl2l11_5390 ATAC-seq peak is referred to as “Bcl2l11 −8.2 kb” in Figs. 4 and 5 as well as in the text. (D and E) Ctla4 (D) and Pdcd1 (E) loci are displayed using the Integrated Genomics Viewer. Data tracks are as indicated (top to bottom): Condensed Refseq gene open-reading frames; differentially regulated ATAC-seq peak coordinates; BATF, IRF4, and NFATc1 ChIP-seq DNA-binding from published datasets (black); published ATAC-seq mean read densities for in vitro stimulated WT and Batf−/− Th0 cells (green); ATAC-seq mean read densities for each experimental group (blue); RNA-seq mean read densities for each experimental group (red); plot of multivariate correlation observed between locus RNA-seq reads and one nearby BATF-binding ATAC-seq chromatin accessibility peak (Ctla4_175 in D; Pdcd1_302 in E; intervals highlighted in dashed rectangles) with positive correlation shown in aqua and negative correlation in orange. Source ATAC-seq and RNA-seq data as in Fig. 1.
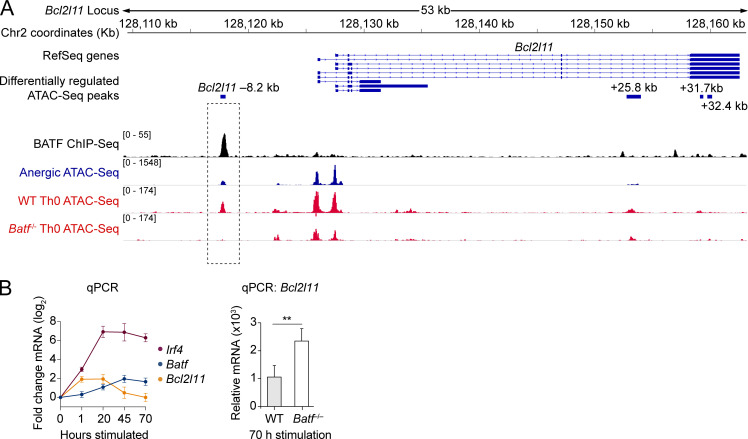
Given the known pioneering capacity of BATF in chronically stimulated Th0 cells (Fig. 2 H), we questioned whether BATF might directly orchestrate chromatin accessibility at the anergy-associated Bcl2l11 −8.2 kb peak. 72 h in vitro stimulated WT Th0 T cells shared an identical BATF-binding Bcl2l11 −8.2 kb chromatin accessibility peak with anergic T cells (Fig. 5 A). Genetic ablation of Batf in the Th0 cells, however, led to greatly reduced accessibility at this chromatin peak. Similar in vitro stimulation of WT polyclonal CD4+ T cells confirmed an upregulation of Batf and Irf4 and yet downregulation of Bcl2l11 mRNA under these conditions (Fig. 5 B). Finally, the genetic deletion of Batf led to an enhanced and sustained induction of Bcl2l11 mRNA. Taken together, these analyses suggest that BATF—acting in a manner distinct from its usual transcriptional-activating function—may counteract clonal deletion in anergic cells by pioneering and recruiting a repressor complex to the Bcl2l11 −8.2 kb locus.
Figure 5.
BATF-dependent chromatin remodeling at a putative DNA repressor element 8.2 kb upstream of the Bcl2l11 promoter. (A) Stacked ChIP-seq and ATAC-seq tracks at the Bcl2l11 gene locus with focus on the Bcl2l11 −8.2 kb peak (dashed rectangle highlight). Data tracks are as indicated (top to bottom): condensed RefSeq gene open-reading frames; differentially regulated ATAC-seq peak coordinates (relative to the putative transcriptional start site); BATF ChIP-seq reads (black; Iwata et al., 2017; Klein-Hessling et al., 2017); anergic group ATAC-seq reads (blue); ATAC-seq reads from 72 h in vitro CD3/CD28 mAb stimulated WT and Batf−/− CD4 T cells (red; Karwacz et al., 2017). Source ATAC-seq track data as in Fig. 1. (B) Relative mRNA expression measured by qPCR following in vitro stimulation of naïve CD4+ T cells with anti-CD3 and anti-CD28 mAbs. Left: Time course for Irf4, Batf, and Bcl2l11 relative mRNA levels in WT T cells. Data shown are the mean fold-change (log2) ± SD (n = 3 unique cell samples at each time point, two independent experiments). Right: Bcl2l11 mRNA observed in WT versus Batf−/− T cells after 70 h of stimulation. Data shown are the mean ± SEM (n = 6–7 samples in two independent experiments; unpaired t test; **P < 0.01).
The BATF–BIM axis controls peripheral deletion
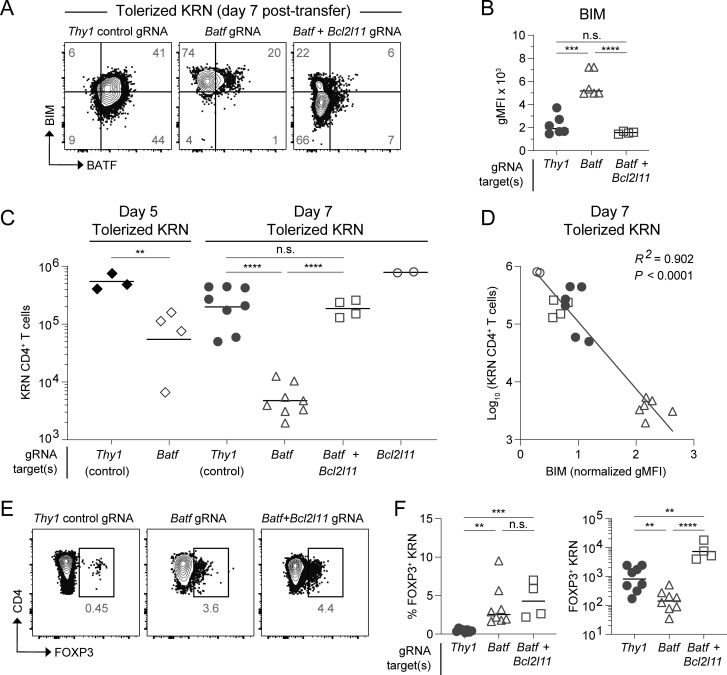
Given the molecular relationships gleaned from bioinformatics and the inverse correlations observed between BATF and BIM levels, we next sought to directly test the hypothesis that BATF restrains BIM expression in anergic T cells. If true, this could account for the compensatory clonal deletion observed in Batf−/− T cells following tolerization. On day 7 after transfer into normal B6.I-Ab/g7 hosts, control self-reactive KRN T cells (with a knockdown of the irrelevant Thy1 gene) displayed clear BATF upregulation and BIM reduction (Fig. 6 A). In contrast, Batf knockdown caused an increase in the level of BIM (Fig. 6, A and B). Coordinately, extensive clonal deletion was observed in the KRN T cells following Batf knockdown; however, this cell loss was completely abrogated when both Batf and Bcl2l11 genes were simultaneously targeted (Fig. 6 C). The degree of clonal deletion seen across all experimental groups proved highly dependent on the amount of BIM protein present (R2 = 0.90; Fig. 6 D). Thus, anergic T cells upregulate BATF to inhibit BIM expression and avert apoptosis.
Figure 6.
BIM-dependent apoptosis of BATF-deficient T cells. CRISPR-Cas9 modified KRN transgenic T cells transferred into B6.I-Ab/g7 host mice and analyzed from the SLOs. Knockdown target(s) are indicated by the gRNA label. (A) BATF versus BIM intracellular protein staining 7 days after transfer. Numbers in the quadrants are mean percentages. Gating was set based on BATF and BIM expression in host-derived irrelevant naïve CD4 T cells (two concatenated samples shown). (B) gMFI of BIM protein in day 7 tolerized KRN T cells. Lines indicate the mean. (C) Total tolerized KRN T cell numbers recovered from B6.I-Ab/g7 host SLOs on days 5 and 7. Lines indicate the geometric mean. (D) Linear regression analysis of log-transformed total day 7 tolerized KRN T cell numbers versus sample-normalized BIM gMFI. Symbols correspond to day 7 experimental groups as in C (n = 18 total points from two independent experiments). (E) FOXP3 intracellular protein staining (two concatenated samples shown) 7 days after adoptive transfer. Numbers in the flow cytometry plots are mean percentages for the group among all samples. (F) Percentage (left) and total recovered cell numbers (right) of FOXP3+ KRN T cells 7 days after adoptive transfer. Lines indicate the mean (left) cell percentage or geometric mean (right) cell number. n = 2–8 mice per experimental group, four independent experiments (unpaired t test; **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. = not significant).
BATF maintains the survival of anergy-derived Treg cells
Having established several mechanisms by which BATF can regulate tolerant T cell fates, we next asked whether sustained BATF expression influences the differentiation plasticity of anergic cells. Molecular analysis suggested that BATF could be a positive regulator of anergy target genes such as Pdcd1 and Ctla4 (Fig. 4 B and Fig. S3). Supporting this notion, loss of BATF prevented KRN T cell differentiation to a PD-1+ phenotype during tolerance induction and this PD-1 expression was only minimally restored by simultaneous Bcl2l11 knockdown (Fig. S2). In contrast, we found no evidence that BATF could directly regulate the Foxp3 gene locus and, in fact, complete restoration of FOXP3+ KRN Treg cell numbers by Batf plus Bcl2l11 double knockdown was observed (Fig. 6, E and F). This suggested that BATF primarily regulates FOXP3+ Treg cell expansion by preventing BIM-dependent deletion rather than by modulating the Foxp3-mediated differentiation program.
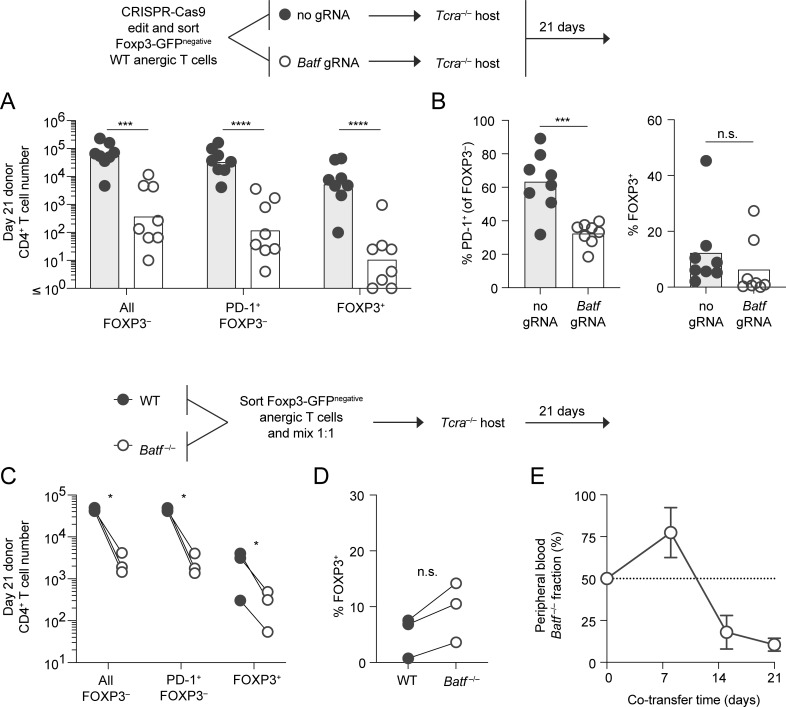
The anergic polyclonal CD4+ T cell population—as defined by the markers used in this study—contains progenitors for FOXP3+ peripheral Treg cell differentiation. Their differentiation potential is most pronounced upon adoptive transfer into lymphopenic TCR α-chain–deficient (Tcra−/−) hosts. We therefore also reconstituted Tcra−/− mice with sorted polyclonal anergic T cells to evaluate how CRISPR-Cas9–mediated Batf gene deletion could affect this late-stage tolerance program. Batf knockdown led to >100-fold reduction in all the anergy-derived T cell subpopulations that could be recovered on day 21 after transfer, and no specific block in FOXP3+ Treg cell generation was observed (Fig. 7, A and B). In contrast, the interruption of Batf expression led to a significant reduction in the generation of PD-1+ conventional CD4 T cells from the anergic donor cells (Fig. 7 B).
Figure 7.
Both conventional T cells and anergy-derived Treg cells rely on BATF for long-term survival following anergy reversal. (A) CD44high Foxp3(GFP)− CD73high FR4high (anergic) polyclonal CD4+ T cells were modified with CRISPR-Cas9 (with gRNA as indicated) and transferred into separate Tcra−/− recipient mice to facilitate anergy reversal. Recipient SLOs were then analyzed on day 21 after transfer for resulting conventional and Treg cell subpopulations as indicated (n = 8 mice, four independent experiments). Bars represent the geometric mean for the indicated cell phenotype (unpaired t test; ***P < 0.001, ****P < 0.0001). (B) Mean percentage of PD-1–expressing FOXP3− conventional (left) and FOXP3+ Treg (right) polyclonal donor T cells recovered from day 21 host mice following CRISPR-Cas9 treatment in the presence of Batf gRNA or no gRNA (sham control) as in A (unpaired t test; ***P < 0.001, n.s = not significant). n = 8 mice, in four independent experiments. (C–E) Anergic polyclonal CD4+ T cells were sorted from congenically marked WT and Batf−/− mice, mixed 1:1, and co-transferred into shared Tcra−/− recipients (n = 3 mice per group, one experiment). Donor-derived cell numbers based on phenotype (C) and (D) percentage of WT and Batf−/− donor-derived FOXP3+ Treg cells recovered from recipient SLOs on day 21 (paired t test; *P < 0.05, n.s. = not significant). (E) Fraction of donor CD4+ T cells in the peripheral blood that were Batf-deficient at the indicated time points (mean ± SD).
Using a complementary approach, WT versus Batf−/− anergic polyclonal T cell populations were sorted separately, mixed at equal numbers, and transferred to a shared Tcra−/− host (Fig. 7, C and D). Kinetic tracking of donor cells in the peripheral blood demonstrated an early but transient competitive advantage for the Batf−/− fraction (Fig. 7 E). This result was reminiscent of the failure to undergo proliferative arrest early during tolerance induction in the absence of Batf (shown in Fig. 3) and again may have related to an inability of Batf−/− T cells to upregulate the checkpoint inhibitor protein PD-1. Nevertheless, BATF was eventually necessary to maintain the survival of both conventional cells and FOXP3+ Treg cells at late times following anergy reversal (Fig. 7, C–E). Collectively, these results demonstrate that sustained BATF expression is required for the long-term survival of Treg cells arising from anergic progenitors, but BATF plays no role in promoting their differentiation to the Treg fate.
The BATF–BIM axis shapes several antigen-experienced T cell populations
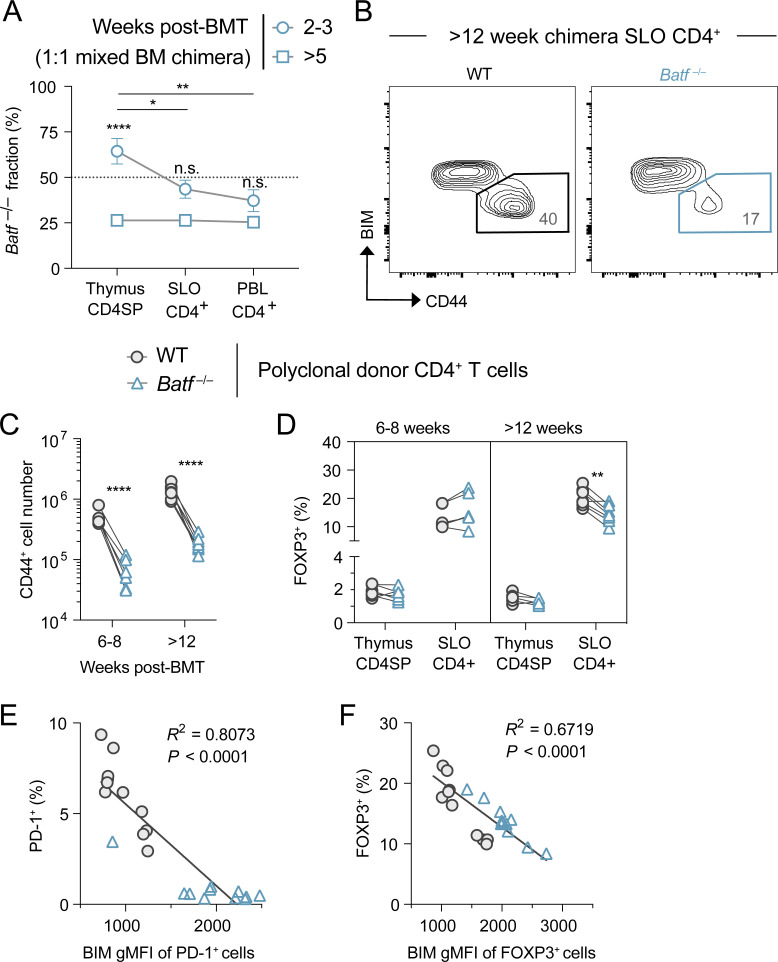
In a final series of experiments, we aimed to broaden our investigation of the BATF–BIM regulatory axis by assessing its impact on the homeostasis of the polyclonal peripheral CD4+ T cell compartment at large. To explore the T cell–intrinsic effects of BATF under steady-state conditions, we established bone marrow (BM) chimera mice with an equal mixture of congenically mismatched WT and Batf−/− BM cells. As early as 6 wk after lethal irradiation and BM reconstitution, Batf−/− T cells interestingly displayed a prethymic developmental disadvantage that persisted into the periphery (Fig. S4 A). This observation aligned well with a previously reported survival defect in the common lymphoid progenitor hematopoietic population following Batf knockdown (Wang et al., 2012). Notwithstanding this developmental defect, Batf−/− polyclonal T cells in the periphery of stable chimeric mice (≥8 wk after BM transfer) also demonstrated a selective reduction in CD44+ antigen-experienced cells and FOXP3+ Treg cells (Fig. S4, B–D). We speculated that this difference was due to disturbed BIM repression in the absence of BATF that led to their peripheral deletion. In support of this notion, measured BIM levels were higher in the post-thymic Batf−/− T cell compartment (Fig. 8 A). An inverse correlation between the frequency of CD44+ cells within each donor group and their measured BIM content supported the hypothesis that BIM directly antagonizes the survival of antigen-experienced T cells (Fig. 8 B). Similar inverse correlations were observed for gated PD-1–expressing conventional CD4+ T cells and FOXP3+ Treg cells (Fig. S4, E and F). All of these findings supported the model that BATF-dependent repression of BIM shapes antigen-experienced subpopulations in the peripheral CD4+ T cell repertoire.
Figure S4.
Expression of BIM, CD44, FOXP3, and PD-1 is dysregulated in the Batf−/− donor-derived polyclonal CD4+ T cells observed in 1:1 WT and Batf−/− mixed BM chimeric mice. (A) Kinetics of relative T cell reconstitution in thymus, SLOs, and blood of mixed BM chimeric mice. Data represent the mean ± SD percentage of Batf−/− donor-derived T cells in samples, with significant differences between time points or tissue source as indicated (one-way ANOVA; *P < 0.05, **P < 0.01, ****P < 0.0001, n.s. = not significant). (B) Representative CD44+ BIMlo polyclonal CD4 T cell SLO accumulation in week 12 mixed BM chimeras. (C and D) Quantitation of CD44+ conventional (C) and FOXP3+ Treg (D) cell generation over time. Lines connect donor cell groups of individual mixed BM chimeric mice, with significant differences between WT (circles) and Batf−/− (triangles) as indicated (paired t test; **P < 0.01, ****P < 0.0001). (E and F) Correlation observed between BIM expression and the frequency of PD-1+ conventional (E) or FOXP3+ Treg (F) cells in both WT (circles) and Batf−/− (triangles) donor-derived populations. Source data as in Fig. 8, A and B.
Figure 8.
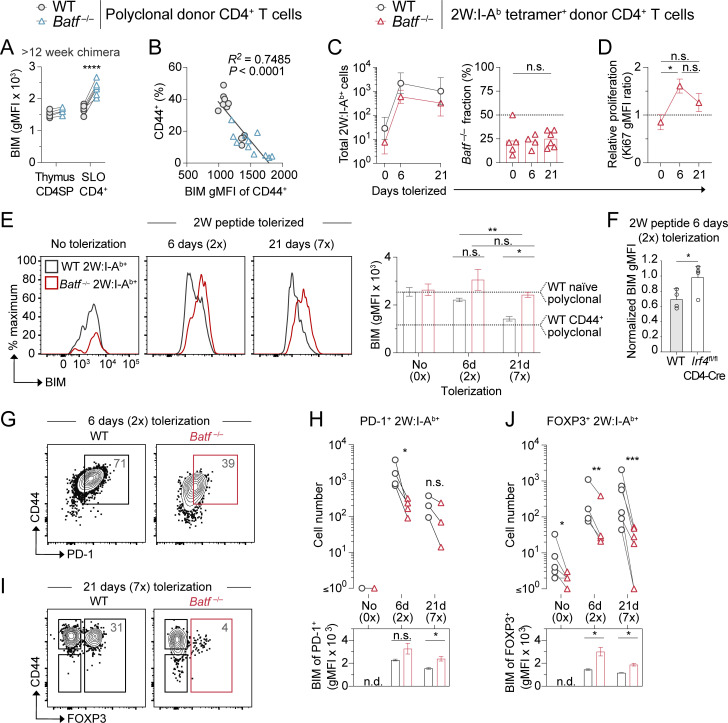
BATF-mediated BIM repression retains antigen-experienced T cells. (A) BIM protein expression measured in total polyclonal CD4+ T cells from chimeric mice lethally irradiated and reconstituted with 50% WT CD45.1+ and 50% Batf−/− CD45.2+ BM. Chimeras were analyzed >12 wk after BM transfer. Lines connect donor cell groups of individual mixed BM chimeric mice (n = 5–8 mice per organ group, one experiment; unpaired t test, ****P < 0.0001). (B) Linear regression analysis of BIM protein expression within CD44+ T cells versus the percent CD44+ T cells in mixed BM chimera SLOs as in A. Analyzed ≥ 8 wk after BM transfer (n = 22 total points, two independent experiments). (C) 2W:I-Ab tetramer-positive T cell numbers (left, mean ± SEM) and Batf−/− fraction (right, mean percentage) of mixed BM chimera SLOs. Mice were left untreated (0 days, 0×) or tolerized with two (6 days, 2×) or seven (21 days, 7×) i.v. injections of 2W peptide (one-way ANOVA, n.s. = not significant). n = 4–6 mice per group, in three independent experiments. (D) Ki67 protein expression within Batf−/− donor BM-derived 2W:I-Ab+ T cells divided by WT T cell expression, as measured in SLOs from 2W peptide-tolerized mixed BM chimeric mice (mice as in C; one-way ANOVA, *P < 0.05). (E) Histogram overlays (left) and quantification (right) of BIM expression measured in mixed BM chimera 2W:I-Ab tetramer+ T cells as in C (n = 3–4, two independent experiments). Bars in the graph represent mean ± SEM (paired t test comparing groups at single time point and unpaired t test comparing same group between time points; *P < 0.05, **P < 0.01, n.s. = not significant). Dashed lines indicate the mean expression measured in naïve or CD44+ polyclonal WT CD4+ T cells within the same chimera mice. Mice as in C. (F) Normalized BIM expression detected by flow cytometry in 2W:I-Ab tetramer-binding CD4 T cells from the SLOs of WT and Irf4-deficient mice tolerized with two i.v. injections (2×) of 2W peptide on days 0 and 3 and analyzed on day 6. Data shown are the mean ± SEM ratio of BIM gMFI in tolerized tetramer-binding T cells to BIM gMFI in tetramer-negative naïve polyclonal CD4 T cells from the same mouse (n = 4 mice per group, two independent experiments; unpaired t test; *P < 0.05). (G and H) PD-1 protein staining on day 6 (G), cell numbers (H, top), and subpopulation BIM expression (H, bottom) of mixed BM chimera 2W:I-Ab+ T cells following 2W peptide tolerization. Numbers in plots are mean percentages (paired t test; *P < 0.05). Mice as in C. (I and J) FOXP3 protein staining on day 21 (I), cell numbers (J, top), and subpopulation BIM expression (J, bottom) of mixed BM chimera 2W:I-Ab+ T cells following 2W peptide tolerization. Numbers in plots are mean percentages (paired t test; *P < 0.05, **P < 0.01, ***P < 0.001). Mice as in C.
This observed BIM upregulation and apparent deletion within bulk polyclonal subsets represented an average effect with different CD4+ T cell specificities experiencing TCR engagement at different times. To synchronize antigen exposure and better understand the timing of BIM repression by BATF, we returned to a 2W peptide tolerization approach and used 2W:I-Ab tetramers to monitor tolerized T cells over 21 days. In mixed BM chimeras undergoing tolerization to repeated i.v. challenge with 2W peptide, Batf−/− 2W-specific polyclonal CD4+ T cells displayed expansion kinetics similar to WT cells, with Batf−/− cells representing ∼25% of the total 2W-specific responders at all time points (Fig. 8 C). This similar expansion kinetic was observed even though the Batf−/− fraction expressed less PD-1 and their rate of cell-cycle progression (based on Ki67 expression) was increased (Fig. 8, D and G). As expected, BIM levels gradually declined in 2W-specific WT T cells during tolerization to the level of polyclonal CD44+ WT cells (Fig. 8 E). In contrast, the Batf−/− fraction failed to suppress BIM expression during 2W tolerization. A similar failure to suppress BIM expression was observed with T-intrinsic deficiency of Irf4 (Fig. 8 F). Consistent with BIM-dependent deletion of 2W peptide responder T cells in the absence of BATF, tetramer-binding PD-1–expressing conventional CD4+ T cells and FOXP3+ Treg cells were selectively lost (Fig. 8, G–J). Thus, a BATF–BIM regulatory axis appears to control antigen-experienced CD4+ T cell homoeostasis in the periphery.
Discussion
Here we have identified and investigated a previously unknown AP-1 gene regulatory program in T cell peripheral tolerance. This program was revealed through a chromatin accessibility analysis of naturally arising polyclonal anergic CD4+ T cells and is now shown to be driven by the non-classical AP-1 family member BATF. Our findings indicate that BATF holds dual roles in the development of peripheral immune tolerance by both promoting anergy and preventing clonal deletion. At a molecular level, BATF appears to execute these functions in opposing ways. In most cases, BATF appears to act like a traditional transactivator, binding at anergy-associated gene DNA promoters/enhancers (e.g., Pdcd1, Ctla4) to increase the expression of co-inhibitory proteins during tolerization and promote cell cycle arrest. Nonetheless, our data also indicate that BATF binding near select genes (e.g., −8.2 kb upstream of Bcl2l11) alternatively leads to repression of mRNA transcription. For the case of Bcl2l11, repression of BIM protein expression by BATF appears to be necessary to maintain the survival of chronically stimulated CD4+ T cells.
While BATF’s gene transactivating functionality has been well documented (Schraml et al., 2009; Glasmacher et al., 2012; Kurachi et al., 2014; Murphy et al., 2013), our study now provides evidence that it may also act as a transcriptional repressor at Bcl2l11 −8.2 kb. A major question that follows is what biochemical context allows for BATF to exert repressor function at certain genetic loci. At an epigenetic level, this could be dictated by distinct chromatin accessibility or cooperation of BATF with different TFs. An elegant study recently dissected some of these complex epigenetic cues in effector CD8+ T cells and also speculated that BATF may play a role as a repressor within the Bcl2l11 locus (Tsao et al., 2022). A study of Junb-deficient T cells further demonstrated that JUNB (a frequent binding partner for BATF within AP-1 heterodimers) represses BIM expression and promotes T cell survival following immunization, and this effect also appeared to map to a region of IRF4 DNA binding just upstream of the Bcl2l11 transcriptional start site (Hsieh et al., 2022). We now provide evidence for the involvement of both BATF and IRF4 in anergic T cell Bcl2l11 gene repression. Furthermore, BATF’s role in inhibiting Bcl2l11 gene transcription includes an ability to pioneer chromatin remodeling and accessibility at the Bcl2l11 −8.2 putative repressor element. Two versions of the AICE motif (AICE1 and AICE2) have been reported and result from different binding conformations of the same BATF–JUN–IRF complex (Glasmacher et al., 2012; Li et al., 2012; Iwata et al., 2017). It is intriguing to speculate that the less common AICE2 motif, or else a third unknown sequence, might preferentially recruit a repressor complex to Bcl2l11 −8.2 kb.
The anergy epigenetic landscape presented in this study proved to be largely consistent with that previously reported for in vivo tolerized CD4+ T cells assessed for regions of open chromatin using a DNase hypersensitivity analysis (Bevington et al., 2020). This previous study likewise indicated regions of uniquely accessible chromatin in tolerant T cells enriched for both NFAT and AP-1 consensus binding motifs. However, their footprint analysis suggested reduced AP-1 binding in tolerant T cells compared to naïve cells after activation. The authors suggested that, although accessible, tolerized cells fail to productively occupy many of these AP-1 sites due to TCR signaling disruptions and poor accumulation of traditional AP-1 family members. Nevertheless, their study also indicated that increased factor binding to a composite AP-1/IRF motif might occur in tolerant T cells. We find that the prevailing anergic T cell epigenetic signature is indeed driven by a non-classical BATF-containing AP-1 complex acting in concert with IRF4. The extended AICE site seen bound in both studies of tolerized T cells now implies that a unique AP-1 program is utilized during tolerance induction and maintenance. Discovery of such an alternative AP-1 site occupant furthermore reconciles these data with the historical notion that T cell anergy lacks AP-1 recruitment (Mondino et al., 1996; Macián et al., 2002; Macián, 2005; Fathman and Lineberry, 2007; Martinez et al., 2015), as most earlier studies would not have directly probed for BATF.
In a model where anergy and exhaustion are related cellular states (Trefzer et al., 2021; Pereira et al., 2017; ElTanbouly and Noelle, 2021), the identification of a novel BATF–BIM axis helps clarify BATF’s role in the context of chronic antigen stimulation. Similar to anergic T cells, BATF is highly expressed in exhausted pathogen-specific T cells and appears sufficient to prevent effector-like proliferation (Man et al., 2017; Quigley et al., 2010). However, other recent studies suggest that BATF counteracts the exhaustion program to generate more potent effectors against cancer (Topchyan et al., 2021) and chronic infection (Chen et al., 2021). In one study, BATF was overexpressed in tumor-specific chimeric antigen receptor (CAR) T cells, which led to improved CAR T cell survival and tumor control (Seo et al., 2021). An earlier study also showed that non-specific BATF overexpression during T cell development caused peripheral T cell accumulation due to an apoptotic defect (Logan et al., 2012). These outcomes would be predicted based on BATF’s repression of the Bcl2l11 gene as shown here. In more physiological circumstances, BIM counter-regulation may also explain why BATF is required to generate TH17 cells (Schraml et al., 2009; Ciofani et al., 2012; Yosef et al., 2013), non-lymphoid tissue Treg cells (Delacher et al., 2020, 2021), T follicular helper cells, and germinal center B cells (Ise et al., 2011; Betz et al., 2010), despite it activating only a portion of their respective differentiation programs. Future studies will need to determine if simultaneous BATF and BIM knockdown allows for differentiation to these various cell fates.
The data presented in this study demonstrate that distinct peripheral tolerance outcomes—anergy, deletion, and later Treg cell maintenance—are mechanistically linked by a central BATF–BIM axis. BATF deficiency in T cells undergoing anergy induction here did not prevent peripheral tolerance, as this AP-1 TF’s layered functionality ensures that these chronically stimulated T cells are instead deleted through BIM upregulation. Future investigations will determine when the continued presence of anergic T cells and sustained self-antigen recognition are advantageous. The ability of anergic T cells to use ongoing TCR signaling to both maintain functional unresponsiveness and yet sustain their viability has previously been reported (Pape et al., 1998; Stritesky et al., 2013). This can importantly retain a population of self-tolerant T cells that are ideal progenitors for peripheral FOXP3+ Treg cells with demonstrated protective function (Kalekar et al., 2016). Nevertheless, one can also imagine how this axis might equally constrain immunity against host threats including cancer by promoting tolerance. It is also possible that a pathogen or malignancy could exploit such a mechanism to the detriment of the host. Emerging evidence now suggests that the co-opting of a similar BATF–BIM regulatory pathway in fact occurs in human B cell malignancies (Ma et al., 2017; Di Bernardo et al., 2008; Fedele et al., 2021). Further molecular investigation of BATF and BIM in diverse cellular contexts will improve our understanding of peripheral lymphocyte differentiation and self-tolerance to guide the development of targeted interventions.
Materials and methods
Study design
The aim of this study was to identify TFs underlying T cell anergy and investigate a mechanism for BATF control of T cell peripheral tolerance fates. Bioinformatic analyses indicated unique BATF functionality at the Bcl2l11 locus. Mechanistic details were determined with antigen-specific tolerization systems incorporating germline knockout mice and CRISPR-Cas9 gene editing of primary T cells.
Mice
WT C57BL/6 (B6), congenic B6 CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ), congenic B6 CD90.1 (B6.PL-Thy1a/CyJ), Tcra−/− (B6.129S2-Tcratm1Mom/J), CD4-Cre (B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ), Irf4fl/fl (B6.129S1-Irf4tm1Rdf/J; Klein et al., 2006), and Batf−/− (B6.129S-Batftm1.1Kmm/J; Schraml et al., 2009) mice were purchased from the Jackson Laboratory or Charles River Breeding Laboratories. Tcra−/− CD45.1, Foxp3GFP congenic, Foxp3DTR/GFP (Kim et al., 2007) congenic, Batf−/− Foxp3GFP congenic, KRN transgenic (Kouskoff et al., 1996), CD4-cre Irf4fl/fl, and B6.I-Ag7 (Kouskoff et al., 1996) mice were bred and maintained in-house. For tolerance induction, KRN adoptive transfer experiments were performed in B6 × B6.I-Ag7 F1 cross (B6.I-Ab/g7) recipients. Mixed BM chimeras were generated from lethally irradiated B6 CD90.1 mice reconstituted with an equal mixture of B6 CD45.1 and B6 Batf−/− (CD45.2) total BM cells (20 × 106 of each donor). For chimera tolerization experiments, host mice were allowed at least 8 wk to reconstitute. All mice were housed in a specific pathogen–free facility at the University of Minnesota—Twin Cities. All experiments were performed in accordance with institutional (University of Minnesota Institutional Animal Care and Use Committee, approval 2207-40250A) and federal guidelines.
Cell isolation
Single-cell suspensions from pooled lymph nodes and spleen were prepared in sort buffer (PBS + 2% fetal bovine serum ± 0.1% sodium azide). Samples were treated with ACK lysis buffer (Gibco) and anti-CD16/CD32 antibody Fc block (Invitrogen). CD4+ T cell populations of interest were magnetically enriched by either negative isolation or 2W:I-Ab tetramer staining for 1 h at room temperature followed by phycoerythrin (PE) and/or APC positive selection. All enrichment procedures were performed with EasySep immunomagnetic beads and magnets (STEMCELL Technologies). BM was isolated by rapid centrifugation of femoral and tibial shafts.
Antigen tolerization
Mice were injected intravenously with 100 μg of 2W peptide (EAWGALANWAVDSA; Rees et al., 1999) every 3 days for variable durations. Cells were harvested and analyzed 3 days after the last injection, which is the time point indicated by “days tolerized.”
Fluorescence-activated cell sorting (FACS)
Physical cell sorting was performed for bulk ATAC-seq, bulk RNA-seq, and adoptive transfer of the anergic CD4+ T cell population. Following the isolation of CD4+ T cells, samples from FOXP3-GFP reporter mice were surface stained for the following markers: CD4, CD44, CD73, FR4, and a single-fluorescence cocktail to exclude non-CD4+ T cells (B220, CD8a, CD11b, CD11c, F4/80) and dead cells (Tonbo Ghost Dye Red 780). For the sequencing experiments, antibodies against Nrp1, CD25, and CXCR5 were also included. Cells were physically sorted under aseptic conditions at the University of Minnesota Flow Cytometry Core Facility on a FACSAria (BD Biosciences). For adoptive transfer experiments, the purified CD4+ T cells were resuspended in sterile PBS prior to intravenous transfer (5 × 104 to 2 × 105 cells) into congenic WT or Tcra−/− B6 recipients.
Phenotypic flow cytometry and cell counts
T cells were isolated and enriched as described above. Samples were then stained for 30 min at 4°C with the fluorophore-conjugated antibodies described in the “FACS” section, as well as antibodies for the following targets: PD-1, Nrp1, and in some cases CD45.1, CD45.2, CD90.1, and CD90.2. Cells were then treated with FOXP3/Transcription Factor Staining Buffer solutions (Tonbo) and stained overnight at 4°C with fluorophore-conjugated antibodies specific for BATF, BIM, FOXP3, and Ki67. Samples were acquired with Fortessa (BD) flow cytometers and analyzed with FlowJo software. Cell counts were determined from calculations relative to AccuCheck Counting Beads (Invitrogen) included in each sample tube. Only cells without Ghost Dye Red 780 (Tonbo) reactivity were considered during analysis. In tetramer enrichment experiments, only 2W:I-Ab tetramer-positive events identified in the magnet-bound sample fraction were included in enumerations.
ATAC-seq
Chromatin accessibility in sorted CD4+ T cell populations was determined using the ATAC-seq method as previously described (Buenrostro et al., 2015). Briefly, 5 × 104 captured cells from each of the indicated groups (n = 2 or 3) were pelleted, washed with PBS, resuspended in cold sterile-filtered lysis buffer (10 mM Tris-Cl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% Igepal CA-630), and immediately centrifuged for 10 min (500 × g, 4°C). Pellets were then resuspended in the transposition reaction mix (Nextera TD buffer, Nextera TDE1 transposase, and nuclease-free H2O) and incubated for 30 min at 37°C. After transposition, DNA was purified with the MinElute PCR Purification kit (QIAGEN) according to the manufacturer’s instructions. Library amplification (Nextera dual-indexed), quality control (agilent sizing and PicoGreen quantification), and sequencing (Illumina NovaSeq SP flow cell 2 × 50-bp paired-end run) were carried out at the University of Minnesota Genomics Center.
ATAC-seq data processing and analysis
Trimmed ATAC-seq reads (Trimmomatic 0.38.1; Bolger et al., 2014) were aligned to the mm10 mouse genome assembly using Bowtie2 2.3.3.1 (Langmead and Salzberg, 2012) with the “--very-sensitive” preset, then sorted and converted in SAMtools 1.13 (Li et al., 2009). Peak-calling with integrated PCR duplicate removal and Tn5 transposase cut site interval adjustment was performed on individual samples with Genrich 0.5 (https://github.com/jsh58/Genrich) in ATAC-seq mode. Reads mapping to mitochondrial DNA and ENCODE blacklist regions (Amemiya et al., 2019) were excluded from the analysis. From the individual peak-called samples, a total of 38,509 consensus peak sites were determined by DiffBind 2.10.0 (https://bioconductor.org/packages/release/bioc/html/DiffBind.html), and the normalized peak accessibility matrix was exported for custom differential comparisons in R. Differentially accessible peaks (9,365 genomic intervals; Table S1) were defined as having a Bonferroni FDR-corrected ANOVA P value <0.01 and an absolute log2 fold change >2.5 in at least one pairwise comparison. Peaks were assigned to the gene with the nearest transcription start site using the annotatePeaks program in HOMER v4.11.1 (Heinz et al., 2010) with reference to the mm10 genome assembly and RefSeq annotation coordinates. Enriched de novo motifs within an entire peak region were identified and enumerated by the findMotifsGenome program in HOMER. Global footprinting analysis was performed using TOBIAS 0.13.2 (Bentsen et al., 2020) while visualization of cut-site probability around specific TFs was performed separately on fixed and de-duplicated read pairs (SAMtools) for 9,365 differentially accessible chromatin peaks using a combination of MotifDb 1.34.0 and ATACseqQC 1.16.0 in R (Ou et al., 2018). Differentially regulated ATAC-seq peak reads were also pooled from sample replicates using the bigwigCompare tool in deepTools 3.5.1.0.1 (Ramírez et al., 2016), and then rank-ordered peak intervals were visualized simultaneously using the combination of computeMatrix and plotHeatmap. For select gene loci, pooled ATAC-seq sample bigwig files were displayed as read density tracks using Integrated Genomics Viewer 2.8.2 (Robinson et al., 2011).
Bioinformatic analysis of public ATAC-seq and ChIP-seq datasets
Publicly available ATAC-seq SRA datafiles processed as fastq reads were aligned to mm10_canonical using Bowtie2 and further analyzed as bigwig files. Publicly available ChIP-seq SRA datafiles processed as fastq reads were aligned to mm10_canonical using Bowtie2 and then peak discovery was performed using MACS 1.0.1 (Zhang et al., 2008). ATAC-seq and ChIP-seq bigwig tracks were subsequently interrogated with the differentially regulated ATAC-seq peak coordinates and displayed using computeMatrix and plotHeatmap. ATAC-seq and ChIP-seq sample bigwig files were also visualized as tracks at select gene loci using the Integrated Genomics Viewer.
National Center for Biotechnology Information (NCBI) public ATAC-seq datasets interrogated are as follows:
GSM2441752: Th0_72h_1; ATAC-seq (SRR5134239)
GSM2441752: Th0_72h_1; ATAC-seq (SRR5134240)
GSM2441753: Th0_72h_2; ATAC-seq (SRR5134241)
GSM2441753: Th0_72h_2; ATAC-seq (SRR5134242)
GSM2441770: BATFko_72hrs_Th0_1; ATAC-seq (SRR5134275)
GSM2441770: BATFko_72hrs_Th0_1; ATAC-seq (SRR5134276)
GSM2441771: BATFko_72hrs_Th0_2; ATAC-seq (SRR5134277)
GSM2441771: BATFko_72hrs_Th0_2; ATAC-seq (SRR5134278).
NCBI public ChIP-seq datasets interrogated are as follows:
GSM2607978: Nfatc1/A-Bio.Rosa26BirA; ChIP-Seq (SRR5515786)
GSM1570761: CA-RIT-RV NFAT1; ChIP-Seq (SRR1731137)
GSM2259177: BATF_WT; ChIP-Seq (SRR3997695)
GSM2259178: IRF4_WT; ChIP-Seq (SRR3997696)
GSM3383358: Tcf1 ChIP-Seq; ChIP-Seq (SRR7816507)
GSM3744545: CBFb; ChIP-Seq (SRR9001316)
GSM2735960: HA-NUR77 rep 2; ChIP-Seq (SRR5914768).
RNA isolation and quantitative PCR (qPCR)
Total RNA was extracted from physically sorted (for RNA-seq) or column-isolated and then cultured (for qPCR) CD4+ T cell populations using a QIAGEN RNeasy Kit according to the manufacturer’s instructions. For qPCR, cDNA was generated using the qScript cDNA Synthesis Kit (Quantabio). qPCR primer sets (Integrated DNA Technologies) used are given in Table 1.
Table 1.
qPCR primer sets
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Batf | 5′-ACAGAGACAGACACAGAAAGC-3′ | 5′-CGGTGAGCTGTTTGATCTCTT-3′ |
| Irf4 | 5′-CTCTTTGACACAGCAGTTTC-3′ | 5′-TCACCAAAGCACAGAGTCAC-3′ |
| Fos | 5′-ACAGCCTTTCCTACTACCATTC-3′ | 5′-GGCACTAGAGACGGACAGAT-3′ |
| Jun | 5′-GCATGGACCTAACATTCGATCT-3′ | 5′-GAGCACTACAGAAGCAATCTACA-3′ |
| Bcl2l11 | 5′-TGTCTGACTCTGATTCTCGGA-3′ | 5′-TGCAATTGTCCACCTTCTCTG-3′ |
| 18s | 5′-CCCCAAAATGGTTAAGGTTGC-3′ | 5′-AACAAAGTCTGGCCTGTATCC-3′ |
RNA-seq analysis
Purified RNA quality control (Agilent sizing and RiboGreen quantification), library creation (Takara SMARTer Stranded Total RNA-seq Kit v2—Pico Input Mammalian), and sequencing (Illumina NovaSeq SP flow cell 2 × 50-bp paired-end run) were carried out at the University of Minnesota Genomics Center. RNA-seq reads were processed using an established pipeline (https://github.com/msi-ris/CHURP/wiki/PURR-Manual-Page). This pipeline incorporates the following software components: Trimmomatic (Bolger et al., 2014), HISAT2 (Kim et al., 2019), SAMtools (Li et al., 2009), featureCounts (Liao et al., 2014), and edgeR (Robinson et al., 2010). Differentially expressed genes (4,349 genes; Table S1) were defined as having a Benjamini-Hochberg FDR-corrected P value <0.05 and an absolute fold change >2 in any pairwise comparison. For visualization at select gene loci, RNA-seq reads were pooled from sample replicates using the bigwigCompare tool and then displayed using Integrated Genomics Viewer.
Integrated ATAC-seq, ChIP-seq, and RNA-seq analyses
Differentially accessible ATAC-seq peaks that also corresponded to a differentially expressed gene were extracted for integrative analysis based on assigned gene names using the dplyr package (https://github.com/tidyverse/dplyr) in R. Predicted BATF binding sites were annotated by HOMER (Heinz et al., 2010) using the software’s default known motif for BATF (derived from a ChIP-seq experiment in GSE39756). Pathway analyses were performed on this dataset using Enrichr (Kuleshov et al., 2016) with reference to the GO Biological Process 2017 database. Correlations between pooled Tn5 transposase chromatin accessibility (ATAC-seq) and candidate nuclear factor DNA-binding (ChIP-seq) within differentially regulated ATAC-seq peak intervals were determined using the multiBigWigSummary and plotCorrelation programs in deepTools. To quantitate and visualize correlations between the pattern of Tn5 transposase chromatin accessibility in a select ATAC-seq peak and nearby transcriptional activity, pooled sample ATAC-seq peak areas were correlated with adjacent (±50,000 kbp) RNA-seq read densities (bin size = 100 bp) for each sample group using the multivariate analysis tool in JMP Pro 15.1.0 (https://www.jmp.com). Correlation matrix R values for each bin were then plotted as wig data across individual gene loci using Integrated Genomics Viewer. Alternatively, sample RNA-seq reads identified within ATAC-seq chromatin accessibility peak coordinates were expressed as a Z-score and plotted as a function of the ATAC-seq read density Z-score within the same sample peak, and the correlation coefficient for the entire dataset was then calculated.
PCA
Both ATAC-seq differentially regulated chromatin accessibility peaks and RNA-seq differentially regulated mRNAs datasets were subjected to a PCA using pcaExplorer 2.14.2 in R. Additionally, ATAC-seq peaks demonstrating high variance in principal components (PC) 1 (61.5% of variance), PC2 (27.7% of variance), or PC3 (8.2% of variance; Z-score ≥ 2 or else less than or equal to −2; 1,046 selected peaks) were assigned to PCA clusters based on a t-distributed stochastic neighbor embedding (TSNE) approach using the Rtsne 0.16 tool in R. A total of 245 differentially regulated ATAC-seq peaks were assigned to the PCA clusters PC(1−) (n = 186) or PC(1+) (n = 59). PC(1−) and PC(1+) cluster gene assignments were subsequently pooled and subjected to functional pathways analysis using the Ingenuity Pathways Analysis tool (QIAGEN Sciences).
CRISPR-Cas9 gene editing
CRISPR-Cas9–mediated gene knockout in primary T cells was performed with some modifications to a previous protocol (Seki and Rutz, 2018; Oh et al., 2019). Briefly, naïve T cells from KRN transgenic mice were purified by negative isolation and assessed for counts and purity. Each sgRNA (Synthego, sequences listed below) was incubated with Alt-R S.p. Cas9 Nuclease V3 (Integrated DNA Technologies) for 10 min at room temperature to form a separate RNP complex, then combined into a single tube for each nucleofection condition. Cells were resuspended in P3 Primary Cell Nucleofector Solution with Supplement 1 (Lonza) plus Alt-R Cas9 Electroporation Enhancer (Integrated DNA Technologies) and immediately aliquoted to the tubes with combined RNP complexes. Mixed cells and sgRNA-Cas9 complexes were transferred to nucleocuvette wells (Lonza) and pulsed with the DN100 program on a Lonza 4D-Nucleofector. Electroporated cells were resuspended and washed in complete media, then assessed again for viable cell counts. KRN T cells were resuspended in sterile PBS prior to intravenous transfer (2 × 105 cells) into B6.I-Ab/g7 recipients. CRISPR sgRNA sequences used are given in Table 2.
Table 2.
CRISPR sgRNA sequences
| Gene target | Position | sgRNA sequence |
|---|---|---|
| Thy1 | chr9:44046628 | 5′-CAGUCUUGCAGGUGUCCCGA-3′ |
| Thy1 | chr9:44046707 | 5′-CCGCCAUGAGAAUAACACCA-3′ |
| Batf | chr12:85689295 | 5′-AUGAUGUGAGGAAAGUUCAG-3′ |
| Batf | chr12:85689368 | 5′-GUGGGUACUCACCAGGUGAA-3′ |
| Bcl2l11 | chr2:128147103 | 5′-UUCCAUACGACAGUCUCAGG-3′ |
| Bcl2l11 | chr2:128147186 | 5′-AGUUCAACGAAACUUACACA-3′ |
Statistical analysis
Numerical data were analyzed in JMP Pro, Prism, R, and Excel software. The applied statistical tests are indicated in the figure legends.
Online supplemental material
Fig. S1 shows that ATAC-seq chromatin accessibility peaks are bound by BATF, IRF4, and NFAT proteins in anergic CD4+ T cells. Fig. S2 shows that BATF drives the expression of PD-1 but is dispensable for FR4 and NRP1 upregulation during tolerance induction. Fig. S3 shows that Tn5 transposase chromatin accessibility directly correlates with adjacent RNA transcriptional activity at the Ctla4 and Pdcd1 loci, but not at Bcl2l11. Fig. S4 shows that expression of BIM, CD44, FOXP3, and PD-1 are dysregulated in the Batf−/− donor-derived polyclonal CD4+ T cells observed in 1:1 WT and Batf−/− mixed BM chimeric mice. Table S1 contains 9,365 differentially regulated chromatin accessibility peaks by ATAC-seq (tab A), 4,349 differentially regulated genes by RNA-seq (tab B), and the top ingenuity pathways analysis disease or function annotations for 245 combined PCA clusters PC(1+) and PC(1−) ATAC-seq chromatin accessibility peaks (tab C).
Supplementary Material
shows 9,365 differentially regulated chromatin accessibility peaks by ATAC-seq (tab A), 4,349 differentially regulated genes by RNA-seq (tab B), and top Ingenuity Pathways Analysis disease or function annotations for 245 combined PCA clusters PC(1+) and PC(1−) ATAC-seq chromatin accessibility peaks (tab C).
Acknowledgments
We wish to thank Drs. Marc Jenkins, Kristin Hogquist, and Peter Igarashi for critical reading of this manuscript prior to publication.
This work was supported by National Institutes of Health grant P01 AI35296 (D.L. Mueller), National Institutes of Health grant T32 AI007313 (K.A. Hogquist), and the Rheumatology Research Foundation Rheumatology Future Physician Scientist Award (P.J. Titcombe).
Author contributions: Conceptualization: P.J. Titcombe and D.L. Mueller. Methodology: P.J. Titcombe, M. Silva Morales, and N. Zhang. Investigation: P.J. Titcombe, M. Silva Morales, N. Zhang, and D.L. Mueller. Visualization: P.J. Titcombe and D.L. Mueller. Funding acquisition: D.L. Mueller. Supervision: D.L. Mueller. Writing—original draft: P.J. Titcombe. Writing—review & editing: D.L. Mueller.
Data availability
NCBI Gene Expression Omnibus series records GSE244276 and GSE244278 provide access to all primary sequencing data generated in this study.
References
- Amemiya, H.M., Kundaje A., and Boyle A.P.. 2019. The ENCODE blacklist: Identification of problematic regions of the genome. Sci. Rep. 9:9354. 10.1038/s41598-019-45839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron, L., Knoechel B., Lohr J., and Abbas A.K.. 2008. Cutting edge: Contributions of apoptosis and anergy to systemic T cell tolerance. J. Immunol. 180:2762–2766. 10.4049/jimmunol.180.5.2762 [DOI] [PubMed] [Google Scholar]
- Bentsen, M., Goymann P., Schultheis H., Klee K., Petrova A., Wiegandt R., Fust A., Preussner J., Kuenne C., Braun T., et al. 2020. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat. Commun. 11:4267. 10.1038/s41467-020-18035-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz, B.C., Jordan-Williams K.L., Wang C., Kang S.G., Liao J., Logan M.R., Kim C.H., and Taparowsky E.J.. 2010. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 207:933–942. 10.1084/jem.20091548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington, S.L., Ng S.T.H., Britton G.J., Keane P., Wraith D.C., and Cockerill P.N.. 2020. Chromatin priming renders T cell tolerance-associated genes sensitive to activation below the Signaling threshold for immune response genes. Cell Rep. 31:107748. 10.1016/j.celrep.2020.107748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A.M., Lohse M., and Usadel B.. 2014. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics. 30:2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro, J.D., Wu B., Chang H.Y., and Greenleaf W.J.. 2015. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109:21.29.1–21.29.9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappert, P., and Schwartz R.H.. 2010. Induction of T cell anergy: Integration of environmental cues and infectious tolerance. Curr. Opin. Immunol. 22:552–559. 10.1016/j.coi.2010.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Zander R.A., Wu X., Schauder D.M., Kasmani M.Y., Shen J., Zheng S., Burns R., Taparowsky E.J., and Cui W.. 2021. BATF regulates progenitor to cytolytic effector CD8+ T cell transition during chronic viral infection. Nat. Immunol. 22:996–1007. 10.1038/s41590-021-00965-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani, M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkhurst C.N., Muratet M., et al. 2012. A validated regulatory network for Th17 cell specification. Cell. 151:289–303. 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo, J., Sun H., Welling T.H., Tian Z., and Zou W.. 2013. T cell anergy, exhaustion, senescence, and stemness in the tumor microenvironment. Curr. Opin. Immunol. 25:214–221. 10.1016/j.coi.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacher, M., Imbusch C.D., Hotz-Wagenblatt A., Mallm J.-P., Bauer K., Simon M., Riegel D., Rendeiro A.F., Bittner S., Sanderink L., et al. 2020. Precursors for nonlymphoid-tissue Treg cells reside in secondary lymphoid organs and are programmed by the transcription factor BATF. Immunity. 52:295–312.e11. 10.1016/j.immuni.2019.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacher, M., Simon M., Sanderink L., Hotz-Wagenblatt A., Wuttke M., Schambeck K., Schmidleithner L., Bittner S., Pant A., Ritter U., et al. 2021. Single-cell chromatin accessibility landscape identifies tissue repair program in human regulatory T cells. Immunity. 54:702–720.e17. 10.1016/j.immuni.2021.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bernardo, M.C., Crowther-Swanepoel D., Broderick P., Webb E., Sellick G., Wild R., Sullivan K., Vijayakrishnan J., Wang Y., Pittman A.M., et al. 2008. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat. Genet. 40:1204–1210. 10.1038/ng.219 [DOI] [PubMed] [Google Scholar]
- ElTanbouly, M.A., and Noelle R.J.. 2021. Rethinking peripheral T cell tolerance: Checkpoints across a T cell’s journey. Nat. Rev. Immunol. 21:257–267. 10.1038/s41577-020-00454-2 [DOI] [PubMed] [Google Scholar]
- Fathman, C.G., and Lineberry N.B.. 2007. Molecular mechanisms of CD4+ T-cell anergy. Nat. Rev. Immunol. 7:599–609. 10.1038/nri2131 [DOI] [PubMed] [Google Scholar]
- Fedele, P.L., Liao Y., Gong J.-N., Yao Y., van Delft M.F., Low M.S.Y., Tai L., Herold M.J., Jackson J.T., Teh C.E., et al. 2021. The transcription factor IRF4 represses proapoptotic BMF and BIM to licence multiple myeloma survival. Leukemia. 35:2114–2118. 10.1038/s41375-020-01078-0 [DOI] [PubMed] [Google Scholar]
- Garces de Los Fayos Alonso, I., Liang H.-C., Turner S.D., Lagger S., Merkel O., and Kenner L.. 2018. The role of activator protein-1 (AP-1) family members in CD30-positive lymphomas. Cancers. 10:93. 10.3390/cancers10040093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasmacher, E., Agrawal S., Chang A.B., Murphy T.L., Zeng W., Vander Lugt B., Khan A.A., Ciofani M., Spooner C.J., Rutz S., et al. 2012. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 338:975–980. 10.1126/science.1228309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist, K.A., Baldwin T.A., and Jameson S.C.. 2005. Central tolerance: Learning self-control in the thymus. Nat. Rev. Immunol. 5:772–782. 10.1038/nri1707 [DOI] [PubMed] [Google Scholar]
- Hsieh, T., Sasaki D., Taira N., Chien H., Sarkar S., Seto Y., Miyagi M., and Ishikawa H.. 2022. JunB is critical for survival of T helper cells. Front. Immunol. 13:901030. 10.3389/fimmu.2022.901030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise, W., Kohyama M., Schraml B.U., Zhang T., Schwer B., Basu U., Alt F.W., Tang J., Oltz E.M., Murphy T.L., and Murphy K.M.. 2011. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 12:536–543. 10.1038/ni.2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, A., Durai V., Tussiwand R., Briseño C.G., Wu X., Grajales-Reyes G.E., Egawa T., Murphy T.L., and Murphy K.M.. 2017. Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF-IRF4 transcription factor complex. Nat. Immunol. 18:563–572. 10.1038/ni.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, M.K., and Schwartz R.H.. 1987. Antigen presentation by chemically modified splenocytes induces antigen-specific T cell unresponsiveness in vitro and in vivo. J. Exp. Med. 165:302–319. 10.1084/jem.165.2.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalekar, L.A., and Mueller D.L.. 2017. Relationship between CD4 regulatory T cells and anergy in vivo. J. Immunol. 198:2527–2533. 10.4049/jimmunol.1602031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalekar, L.A., Schmiel S.E., Nandiwada S.L., Lam W.Y., Barsness L.O., Zhang N., Stritesky G.L., Malhotra D., Pauken K.E., Linehan J.L., et al. 2016. CD4(+) T cell anergy prevents autoimmunity and generates regulatory T cell precursors. Nat. Immunol. 17:304–314. 10.1038/ni.3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin, M., Liu Z.g., and Zandi E.. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240–246. 10.1016/s0955-0674(97)80068-3 [DOI] [PubMed] [Google Scholar]
- Karwacz, K., Miraldi E.R., Pokrovskii M., Madi A., Yosef N., Wortman I., Chen X., Watters A., Carriero N., Awasthi A., et al. 2017. Critical role of IRF1 and BATF in forming chromatin landscape during type 1 regulatory cell differentiation. Nat. Immunol. 18:412–421. 10.1038/ni.3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney, E.R., Pape K.A., Loh D.Y., and Jenkins M.K.. 1994. Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity. 1:327–339. 10.1016/1074-7613(94)90084-1 [DOI] [PubMed] [Google Scholar]
- Kim, D., Paggi J.M., Park C., Bennett C., and Salzberg S.L.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37:907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.M., Rasmussen J.P., and Rudensky A.Y.. 2007. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8:191–197. 10.1038/ni1428 [DOI] [PubMed] [Google Scholar]
- Klein, U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., and Dalla-Favera R.. 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7:773–782. 10.1038/ni1357 [DOI] [PubMed] [Google Scholar]
- Klein-Hessling, S., Muhammad K., Klein M., Pusch T., Rudolf R., Flöter J., Qureischi M., Beilhack A., Vaeth M., Kummerow C., et al. 2017. NFATc1 controls the cytotoxicity of CD8+ T cells. Nat. Commun. 8:511. 10.1038/s41467-017-00612-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskoff, V., Korganow A.S., Duchatelle V., Degott C., Benoist C., and Mathis D.. 1996. Organ-specific disease provoked by systemic autoimmunity. Cell. 87:811–822. 10.1016/s0092-8674(00)81989-3 [DOI] [PubMed] [Google Scholar]
- Kuczma, M.P., Szurek E.A., Cebula A., Ngo V.L., Pietrzak M., Kraj P., Denning T.L., and Ignatowicz L.. 2021. Self and microbiota-derived epitopes induce CD4+ T cell anergy and conversion into CD4+Foxp3+ regulatory cells. Mucosal Immunol. 14:443–454. 10.1038/s41385-020-00349-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov, M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. 2016. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44:W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurachi, M., Barnitz R.A., Yosef N., Odorizzi P.M., DiIorio M.A., Lemieux M.E., Yates K., Godec J., Klatt M.G., Regev A., et al. 2014. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 15:373–383. 10.1038/ni.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., Spolski R., Liao W., Wang L., Murphy T.L., Murphy K.M., and Leonard W.J.. 2012. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 490:543–546. 10.1038/nature11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2014. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Logan, M.R., Jordan-Williams K.L., Poston S., Liao J., and Taparowsky E.J.. 2012. Overexpression of Batf induces an apoptotic defect and an associated lymphoproliferative disorder in mice. Cell Death Dis. 3:e310. 10.1038/cddis.2012.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y., Walsh M.J., Bernhardt K., Ashbaugh C.W., Trudeau S.J., Ashbaugh I.Y., Jiang S., Jiang C., Zhao B., Root D.E., et al. 2017. CRISPR/Cas9 screens reveal epstein-barr virus-transformed B cell host dependency factors. Cell Host Microbe. 21:580–591.e7. 10.1016/j.chom.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macián, F. 2005. NFAT proteins: Key regulators of T-cell development and function. Nat. Rev. Immunol. 5:472–484. 10.1038/nri1632 [DOI] [PubMed] [Google Scholar]
- Macián, F., García-Rodríguez C., and Rao A.. 2000. Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J. 19:4783–4795. 10.1093/emboj/19.17.4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macián, F., García-Cózar F., Im S.-H., Horton H.F., Byrne M.C., and Rao A.. 2002. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 109:719–731. 10.1016/s0092-8674(02)00767-5 [DOI] [PubMed] [Google Scholar]
- Man, K., Gabriel S.S., Liao Y., Gloury R., Preston S., Henstridge D.C., Pellegrini M., Zehn D., Berberich-Siebelt F., Febbraio M.A., et al. 2017. Transcription factor IRF4 promotes CD8+ T cell exhaustion and limits the development of memory-like T cells during chronic infection. Infect. Immun. 47:1129–1141.e5. 10.1016/j.immuni.2017.11.021 [DOI] [PubMed] [Google Scholar]
- Martinez, G.J., Pereira R.M., Äijö T., Kim E.Y., Marangoni F., Pipkin M.E., Togher S., Heissmeyer V., Zhang Y.C., Crotty S., et al. 2015. The transcription factor NFAT promotes exhaustion of activated CD8⁺ T cells. Immunity. 42:265–278. 10.1016/j.immuni.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, R.J., Zhang N., Thomas S.R., Nandiwada S.L., Jenkins M.K., Binstadt B.A., and Mueller D.L.. 2012. Arthritogenic self-reactive CD4+ T cells acquire an FR4hiCD73hi anergic state in the presence of Foxp3+ regulatory T cells. J. Immunol. 188:170–181. 10.4049/jimmunol.1101311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, I., Staub A., Benoist C., and Mathis D.. 1999. Arthritis provoked by linked T and B cell recognition of a glycolytic enzyme. Science. 286:1732–1735. 10.1126/science.286.5445.1732 [DOI] [PubMed] [Google Scholar]
- Mondino, A., Whaley C.D., DeSilva D.R., Li W., Jenkins M.K., and Mueller D.L.. 1996. Defective transcription of the IL-2 gene is associated with impaired expression of c-Fos, FosB, and JunB in anergic T helper 1 cells. J. Immunol. 157:2048–2057. 10.4049/jimmunol.157.5.2048 [DOI] [PubMed] [Google Scholar]
- Moon, J.J., Chu H.H., Pepper M., McSorley S.J., Jameson S.C., Kedl R.M., and Jenkins M.K.. 2007. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 27:203–213. 10.1016/j.immuni.2007.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, J.J., Dash P., Oguin T.H. III, McClaren J.L., Chu H.H., Thomas P.G., and Jenkins M.K.. 2011. Quantitative impact of thymic selection on Foxp3+ and Foxp3− subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc. Natl. Acad. Sci. USA. 108:14602–14607. 10.1073/pnas.1109806108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, D.L. 2010. Mechanisms maintaining peripheral tolerance. Nat. Immunol. 11:21–27. 10.1038/ni.1817 [DOI] [PubMed] [Google Scholar]
- Murphy, T.L., Tussiwand R., and Murphy K.M.. 2013. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 13:499–509. 10.1038/nri3470 [DOI] [PubMed] [Google Scholar]
- Nurieva, R.I., Liu X., and Dong C.. 2011. Molecular mechanisms of T-cell tolerance. Immunol. Rev. 241:133–144. 10.1111/j.1600-065X.2011.01012.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S.A., Seki A., and Rutz S.. 2019. Ribonucleoprotein transfection for CRISPR/Cas9-Mediated gene knockout in primary T cells. Curr. Protoc. Immunol. 124:e69. 10.1002/cpim.69 [DOI] [PubMed] [Google Scholar]
- Ohkura, N., Kitagawa Y., and Sakaguchi S.. 2013. Development and maintenance of regulatory T cells. Immunity. 38:414–423. 10.1016/j.immuni.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Ou, J., Liu H., Yu J., Kelliher M.A., Castilla L.H., Lawson N.D., and Zhu L.J.. 2018. ATACseqQC: A bioconductor package for post-alignment quality assessment of ATAC-seq data. BMC Genomics. 19:169. 10.1186/s12864-018-4559-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape, K.A., Merica R., Mondino A., Khoruts A., and Jenkins M.K.. 1998. Direct evidence that functionally impaired CD4+ T cells persist in vivo following induction of peripheral tolerance. J. Immunol. 160:4719–4729. 10.4049/jimmunol.160.10.4719 [DOI] [PubMed] [Google Scholar]
- Pereira, R.M., Hogan P.G., Rao A., and Martinez G.J.. 2017. Transcriptional and epigenetic regulation of T cell hyporesponsiveness. J. Leukoc. Biol. 102:601–615. 10.1189/jlb.2RI0317-097R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, D., Moseley C.E., Gao M., Savic D., Winstead C.J., Sun M., Kee B.L., Myers R.M., Weaver C.T., and Hatton R.D.. 2019. Batf pioneers the reorganization of chromatin in developing effector T cells via ets1-dependent recruitment of Ctcf. Cell Rep. 29:1203–1220.e7. 10.1016/j.celrep.2019.09.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley, M., Pereyra F., Nilsson B., Porichis F., Fonseca C., Eichbaum Q., Julg B., Jesneck J.L., Brosnahan K., Imam S., et al. 2010. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat. Med. 16:1147–1151. 10.1038/nm.2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., and Manke T.. 2016. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44:W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees, W., Bender J., Teague T.K., Kedl R.M., Crawford F., Marrack P., and Kappler J.. 1999. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 96:9781–9786. 10.1073/pnas.96.17.9781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan, J., Mowen K.A., McBride K.D., Smith E.D., Singh H., and Glimcher L.H.. 2002. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 195:1003–1012. 10.1084/jem.20011128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudensky, A.Y., Gavin M., and Zheng Y.. 2006. FOXP3 and NFAT: Partners in tolerance. Cell. 126:253–256. 10.1016/j.cell.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Safford, M., Collins S., Lutz M.A., Allen A., Huang C.-T., Kowalski J., Blackford A., Horton M.R., Drake C., Schwartz R.H., and Powell J.D.. 2005. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat. Immunol. 6:472–480. 10.1038/ni1193 [DOI] [PubMed] [Google Scholar]
- Schraml, B.U., Hildner K., Ise W., Lee W.-L., Smith W.A.-E., Solomon B., Sahota G., Sim J., Mukasa R., Cemerski S., et al. 2009. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 460:405–409. 10.1038/nature08114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, R.H. 2003. T cell anergy. Annu. Rev. Immunol. 21:305–334. 10.1146/annurev.immunol.21.120601.141110 [DOI] [PubMed] [Google Scholar]
- Seki, A., and Rutz S.. 2018. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J. Exp. Med. 215:985–997. 10.1084/jem.20171626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H., González-Avalos E., Zhang W., Ramchandani P., Yang C., Lio C.W.J., Rao A., and Hogan P.G.. 2021. BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat. Immunol. 22:983–995. 10.1038/s41590-021-00964-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritesky, G.L., Xing Y., Erickson J.R., Kalekar L.A., Wang X., Mueller D.L., Jameson S.C., and Hogquist K.A.. 2013. Murine thymic selection quantified using a unique method to capture deleted T cells. Proc. Natl. Acad. Sci. USA. 110:4679–4684. 10.1073/pnas.1217532110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topchyan, P., Xin G., Chen Y., Zheng S., Burns R., Shen J., Kasmani M.Y., Kudek M., Yang N., and Cui W.. 2021. Harnessing the IL-21-BATF pathway in the CD8+ T cell anti-tumor response. Cancers. 13:1263. 10.3390/cancers13061263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trefzer, A., Kadam P., Wang S.-H., Pennavaria S., Lober B., Akçabozan B., Kranich J., Brocker T., Nakano N., Irmler M., et al. 2021. Dynamic adoption of anergy by antigen-exhausted CD4+ T cells. Cell Rep. 34:108748. 10.1016/j.celrep.2021.108748 [DOI] [PubMed] [Google Scholar]
- Tsao, H.-W., Kaminski J., Kurachi M., Barnitz R.A., DiIorio M.A., LaFleur M.W., Ise W., Kurosaki T., Wherry E.J., Haining W.N., and Yosef N.. 2022. Batf-mediated epigenetic control of effector CD8+ T cell differentiation. Sci. Immunol. 7:eabi4919. 10.1126/sciimmunol.abi4919 [DOI] [PubMed] [Google Scholar]
- Tuncel, J., Benoist C., and Mathis D.. 2019. T cell anergy in perinatal mice is promoted by T reg cells and prevented by IL-33. J. Exp. Med. 216:1328–1344. 10.1084/jem.20182002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Sun Q., Morita Y., Jiang H., Gross A., Lechel A., Hildner K., Guachalla L.M., Gompf A., Hartmann D., et al. 2012. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 148:1001–1014. 10.1016/j.cell.2012.01.040 [DOI] [PubMed] [Google Scholar]
- Wing, K., and Sakaguchi S.. 2010. Regulatory T cells exert checks and balances on self tolerance and autoimmunity. Nat. Immunol. 11:7–13. 10.1038/ni.1818 [DOI] [PubMed] [Google Scholar]
- Xing, Y., and Hogquist K.A.. 2012. T-Cell tolerance: Central and peripheral. Cold Spring Harb. Perspect. Biol. 4:a006957. 10.1101/cshperspect.a006957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef, N., Shalek A.K., Gaublomme J.T., Jin H., Lee Y., Awasthi A., Wu C., Karwacz K., Xiao S., Jorgolli M., et al. 2013. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 496:461–468. 10.1038/nature11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows 9,365 differentially regulated chromatin accessibility peaks by ATAC-seq (tab A), 4,349 differentially regulated genes by RNA-seq (tab B), and top Ingenuity Pathways Analysis disease or function annotations for 245 combined PCA clusters PC(1+) and PC(1−) ATAC-seq chromatin accessibility peaks (tab C).
Data Availability Statement
NCBI Gene Expression Omnibus series records GSE244276 and GSE244278 provide access to all primary sequencing data generated in this study.