Abstract
Background and Aims
The FDA-approved multitarget stool-DNA [mt-sDNA] test is a successful colorectal cancer [CRC] screening tool in average-risk individuals but is not indicated for patients with inflammatory bowel disease [IBD]. We determined the performance of the mt-sDNA assay without the haemoglobin component [mt-sDNAHgb-] in patients with IBD, while measuring sensitivity for colorectal cancer and advanced colorectal neoplasia [ACRN].
Methods
This was a multi-centre, proof-of-concept investigation in persons aged 18–84 years with a diagnosis of IBD, or primary sclerosing cholangitis [PSC] with IBD. Enrolment occurred between March 2013 and May 2016. Stool was tested with the mt-sDNA molecular markers only, minus the immunochemical haemoglobin component.
Results
The analysis set contained 355 samples. The median age was 52 [range 39–62] years, 45.6% were female and 93% were White. Two-thirds [63%] had ulcerative colitis [UC] and 10.1% had PSC/IBD. Colonoscopy revealed cancer in 8.5% [N = 30], advanced precancerous lesions [APLs] in 9.3% [N = 33] and non-advanced precancerous lesions in 7.6% [N = 27], and three-quarters [74.7%, N = 265] had negative findings. mt-sDNAHgb- sensitivity was 73.3% for any stage cancers, and 76.2% for ACRN. Sensitivity was highest for IBD-associated high-grade dysplasia at 100% and 84.6% for IBD-associated low-grade dysplasia ≥1 cm. The test showed higher sensitivity and lower specificity in UC than in Crohn’s disease. Increasing inflammation score was associated with a significant decrease in mt-sDNAHgb- test score [ = 0.028] amongst neoplasia-negative individuals, but not in patients with ACRN.
Conclusions
These data highlight the potential of multitarget stool-DNA marker testing as an important addition to colorectal cancer surveillance by complementing colonoscopic evaluations in IBD patients.
Keywords: Colorectal neoplasms/prevention and control, inflammatory bowel diseases, stool/liquid biopsy
Graphical Abstract
Graphical Abstract.

1. Introduction
There are some patients with inflammatory bowel disease [IBD], including ulcerative colitis [UC] and Crohn’s disease [CD] of the colon, who are at increased risk of developing colorectal cancer.1,2 Clinical guidelines recommend that screening colonoscopy begin 8–10 years following IBD diagnosis, with regular surveillance colonoscopy subsequently performed every 1–5 years—the interval varying based on the findings of each exam, and also by different gastrointestinal societies.3–6 Those with comorbid primary sclerosing cholangitis [PSC] should undergo surveillance annually beginning when PSC is diagnosed.3–6 This imposes a high lifetime burden of colonoscopies on these often-young individuals. In addition, IBD carries the unique challenge of having difficult to detect dysplasia, which can be obscured by active inflammation and is challenging to visualize, even with modern endoscopic imaging techniques.6 Identification of colorectal cancer [CRC]-prone dysplasia has long depended on biopsies of visible lesions, as well as numerous random biopsy specimens from all colon segments to detect invisible dysplasia.3,7–9 Thus, confidence that surveillance colonoscopy can successfully find and remove dysplastic lesions, thereby reducing CRC incidence in IBD, is often undermined by the elusive nature of dysplastic lesions and operator endoscopist’s experience. Tests that are complementary to colonoscopy could theoretically improve the detection of IBD-associated dysplasia and offer greater prevention against CRC. Indeed, recent studies indicate that many patients with long-standing IBD would prefer a theoretical highly sensitive blood, stool or imaging test over surveillance colonoscopy, even if the latter is done every 5 years.10
Molecular markers have demonstrated effectiveness in screening for sporadic colorectal neoplasia, yet their application to IBD-associated neoplasia remains under-explored.11,12 The multitarget stool DNA test [mt-sDNA; Cologuard®; Exact Sciences Corp.] is an FDA-approved, non-invasive option for use in screening average-risk persons 45 years or older for CRC. mt-sDNA is highly sensitive [92.3–100%] for CRC.13,14 It consists of molecular assays for aberrantly methylated BMP3 and NDRG4 promoter regions, seven point mutations on KRAS [codons 12 and 13], and quantitative DNA (ACTB [β-actin] – human DNA quantity reference gene), along with a faecal immunochemical assay for human haemoglobin.14,15 A logistic-regression algorithm incorporates the measurements of each marker, and a composite score above a pre-defined threshold value indicates a positive test result.14 Moreover, mt-sDNA specificity for CRC has been reported as 87–95%.14,16 In its current formulation, the mt-sDNA test is not approved for use in patients with IBD.17 This is because intestinal bleeding would contribute to the haemoglobin detection component of the mt-sDNA assay, resulting in many false-positives.
Research suggests that there is potential in utilizing the mt-sDNA CRC DNA biomarker panel without the haemoglobin component, to develop a clinically useful IBD-advanced colorectal neoplasia [ACRN] surveillance test, herein referred to as mt-sDNAHgb-. A case control study in an IBD cohort of patients [19 with colorectal neoplasia, 35 without] revealed that methylated tumour suppressor genes BMP3 and NDRG4—the two methylated tumour suppressor genes included in the mt-sDNA test—were highly discriminant for CRC individually, and detected 100% of IBD-associated CRC [9/9] at 89% specificity.12 More recently, a study of stool samples from 332 patients with IBD revealed a similarly strong association between methylation levels of the BMP3 and VAV3 genes with the detection of CRC and high-grade dysplasia [HGD].11 The use of such a test in the high-risk IBD population may help alleviate the burden of CRC in patients with IBD, for example when combined with surveillance colonoscopy to enhance dysplasia detection.
In this study, we explored the performance of the nucleic acid mt-sDNA molecular markers in patients with IBD. Our primary objective was to determine the sensitivity and specificity of the mt-sDNAHgb- test for CRC in patients with IBD, while secondarily measuring the sensitivity of the test to detect CRC plus advanced precancerous lesions. Colonoscopy and histopathology were used as confirmatory standards.
2. Methodology
2.1. Design
This study [Clinical Trials Registration Number: NCT01819766] was designed as a case-controlled, cross-sectional, multi-centre investigation to determine the sensitivity and specificity of the mt-sDNAHgb- test for detecting CRC alone, or in combination with HGD and/or low-grade dysplasia [LGD]. By design, all stool samples were tested using the approved mt-sDNA assay molecular markers, but the immunochemical haemoglobin test tube was not collected.
As with the FDA-approved test, no restrictions were imposed regarding concomitant medications during the stool collection. Enrolment occurred between March 2013 and May 2016, and data were analysed in December 2020. Institutional Review Board [IRB] approval was obtained from Copernicus Group IRB, and all participants provided written informed consent prior to any study-related procedures.
2.2. Sample collection and study population
Eligible individuals included persons aged 18–84 years who had a diagnosis of IBD, or a diagnosis of PSC with IBD prior to enrolment date. Along with being capable of providing informed consent, patients also had to be candidates for surveillance colonoscopy [with the intention of CRC/dysplasia surveillance]; those without CRC/dysplasia comprised the control group. Cases included surveillance colonoscopy-detected CRC/dysplasia or were referred for surgical intervention based on prior histological confirmation of CRC/dysplasia. The latter exclusion criteria included: a history of aerodigestive tract cancer, prior colorectal resection [except ileocolic resection in CD patients], IBD limited only to the rectum and without a concurrent PSC diagnosis, the subject participated in any clinical study within the previous 30 days wherein an investigational compound or device was introduced into the subject, or if the investigator deemed the patient’s condition unfit for participation.
Enrolled patients provided a single stool sample within 30 days of enrolment [prior to bowel prep for post enrolment colonoscopy], at least 7 days after their most recent pre-enrolment colonoscopy, or prior to any surgical intervention. No subject underwent a second colonoscopy after sample collection for the purpose of the study. All stool samples were shipped to Exact Sciences for processing.
2.3. Colonoscopy and histopathological procedures
Participants underwent a colonoscopy procedure no more than 120 days after the date of enrolment [unless the subject was enrolled with a recent dysplasia or CRC diagnosis and surgical intervention was indicated]. Bowel preparation procedures and post-enrolment colonoscopy were performed according to the established standard of practice at each clinical site. Results of each colonoscopy examination were collected, with personnel performing the colonoscopy blinded to the outcome of the mt-sDNAHgb- screening test. Histopathological diagnosis was obtained for all tissue removed based on the interpretation of the local pathologist with expertise in gastrointestinal [GI] pathology. For subjects in whom surgical intervention was indicated, the procedure had to occur no more than 120 days after the date of study enrolment and histopathological information was collected for all tissue removed during surgery. All subjects were considered to have completed the study when they produced and sent in an evaluable stool sample and completed the post-enrolment colonoscopy or surgical intervention. Sites were responsible for acquiring and maintaining histopathological interpretation of endoscopic biopsies, polypectomy specimens and excisional surgical pathology specimens for all subjects with tissue excised at colonoscopy.
To ensure high-quality surveillance and follow-up colonoscopies in this study, all medical personnel conducting colonoscopies were required to be experienced in performing IBD-related surveillance colonoscopies or be under the supervision of a board-certified gastroenterologist with this expertise. Unless a neoplastic mass prevented a full colonoscopy, a complete colonoscopy required photo-documentation of the caecum. Index lesion types were categorized, as in Table 1. The hierarchy of clinical significance for index lesions is defined in Appendix A.
Table 1.
Participant demographics, disease history and sample pathology.
| Sensitivity and specificity analysis population [N = 355] | |||
|---|---|---|---|
| Total cohort | Cases [N = 63] | Controls [N = 292] | |
| Age [years], n [IQR] | |||
| Median | 52 [39–62] | 55 [47–67] | 51 [38–62] |
| Sex, n [%] | |||
| Female | 162 [45.6] | 19 [30.2] | 143 [49.0] |
| Male | 193 [54.4] | 44 [69.8] | 149 [51.0] |
| Race, n [%] | |||
| White | 330 [93] | 58 [92.1] | 272 [93.2] |
| Black or African American | 14 [3.9] | 1 [1.6] | 13 [4.5] |
| Asian | 5 [1.4] | 2 [3.2] | 3 [1.0] |
| Other | 6 [1.7] | 2 [3.2] | 4 [1.4] |
| Ethnicity, n [%] | |||
| Hispanic or Latino | 18 [5.1] | 4 [6.4] | 14 [4.8] |
| Not Hispanic or Latino | 337 [94.9] | 59 [93.7] | 278 [95.2] |
| Disease status, n [%] | |||
| Crohn’s | 128 [36.1] | 21 [33.3] | 107 [36.6] |
| Ulcerative colitis | 222 [62.5] | 40 [63.5] | 182 [62.3] |
| Indeterminate colitis | 5 [1.4] | 2 [3.2] | 3 [1.0] |
| Primary sclerosing cholangitis [PSC], n [%] | |||
| Yes | 36 [10.1] | 50 [79.4] | 269 [92.1] |
| No | 319 [89.9] | 13 [20.6] | 23 [7.9] |
| IBD duration [years], n [IQR] | |||
| Median | 16 [11–25] | 18 [11–30] | 16 [11–24] |
| History of dysplasia [≥ 1 year prior to enrolment], n [%] | |||
| Yes | 11 [3.1] | 9 [14.3] | 2 [0.7] |
| No | 344 [96.9] | 54 [85.7] | 290 [99.3] |
| Case pathologies | |||
| Cancer [Category 1], n [%] | 30 [8.5] | ||
| Left-sided | Right-sided | ||
| Stage I | 2 | 0 | |
| Stage II | 4 | 4 | |
| Stage III | 6 | 6 | |
| Stage IV | 1 | 1 | |
| Stage unknown | 3 | 3 | |
| APL [Category 2], n [%] | 33 [9.3] | ||
| Category 2.1—Adenoma, HGD | 1 | ||
| Category 2.2—Adenoma, villous growth | 0 | ||
| Category 2.3—Adenoma ≥ 1 cm | 1 | ||
| Category 2.4—Sessile serrated polyp, serrated lesion ≥ 1 cm | 7 | ||
| Category 2.5—IBD-HGD, any size | 11 | ||
| Category 2.6—IBD-LGD, ≥ 1 cm | 13 | ||
| Non-advanced precancerous lesions [Categories 3–5], n [%] | 27 [7.6] | ||
| Category 3—1 or 2 adenomas > 0.5 cm, <1 cm, IBD-LGD < 1 cm | 13 | ||
| Category 4—≥3 adenomas, < 1 cm | 1 | ||
| Category 5—1 or 2 adenomas, ≤ 0.5 cm | 13 | ||
| Negative [Category 6]a, n [%] | 265 [74.7] | ||
IQR: interquartile range; APL: advanced precancerous lesions; HGD: high-grade dysplasia; LGD: low-grade dysplasia; IBD-HGD/LGD: inflammatory bowel disease-associated dysplasia [high- or low-grade].
aIncludes hyperplastic polyps <1 cm and no findings.
For this study, the term advanced precancerous lesions [APL] refers to: advanced adenomas [HGD or with ≥25% villous histological features, or measuring ≥1 cm]; sessile serrated polyps measuring ≥1 cm; IBD-associated HGD; and IBD-associated LGD measuring ≥1 cm. Most [73%] of the lesions in this category were IBD-associated dysplasia, with others falling within conventional definitions of advanced adenomas and advanced sessile serrated polyps. Non-advanced precancerous lesions [non-APLs] were included among Category 3–5 lesions. Histologic Activity Index was recorded for each colonic segment and ranged from 0 to 3: inactive/quiescent colitis [0], mildly active inflammation [1], moderately active inflammation [2] and severely active inflammation [3] [Appendix B]. Based on this, an Inflammation Score [IS] was calculated as the average score across the total number of colonic segments evaluated per colonoscopy.
2.4. Statistical analysis
The co-primary endpoints for this study were the sensitivity and specificity of the mt-sDNAHgb- test for CRC. This was based on the histopathological diagnosis of all lesions discovered at colonoscopy which were either biopsied or removed during colonoscopy, or subsequently removed after colonoscopy. The largest of the non-APLs was considered the index lesion for each case where CRC or APLs were not detected. For each sample, the algorithm for the mt-sDNA test was applied using a value of zero for the Hgb component, and the resulting score was compared to the established clinical decision point to generate a positive or negative call. The algorithm effectively normalizes the signal from the methylation and mutation markers to the reference gene.
Targeted enrolment was ~440 subjects at 20 sites to achieve a minimum number of enrollees in the following categories: 50 positive cases [35 CRC and 15 HGD] and 315 negative controls.
Primary goals were to establish a clinically meaningful sensitivity estimate for the mt-sDNAHgb- test for CRC according to a one-sided 95% lower bound, as well as a clinically meaningful specificity estimate for the test according to a one-sided 95% lower bound. Likewise, the secondary goal was to estimate sensitivity of the test for IBD-associated CRC or HGD.
Sample size calculations for the mt-sDNAHgb- test were set according to the one-sided 95% lower bounds [Table 2] around pre-specified, clinically meaningful performance targets of 85% CRC sensitivity, 55% HGD sensitivity and CRC + HGD sensitivity of 76% (based on a 7:3 CRC to HGD sample size ratio [CRC + HGD combined sensitivity = 0.7 × CRC sensitivity + 0.3 × HGD sensitivity]). Control patient sample size was determined by the 95% confidence interval [CI] lower bound for a target specificity of 90%.
Table 2.
Sensitivity and specificity analyses by pathology.
| Pathology | N | Specificity, % [95% CI] | Target values | |
|---|---|---|---|---|
| Specificity | Lower bound | |||
| Negative and non-APL | 292 | 80.8 [75.9–84.9] | 90% | 86% |
| Negative | 265 | 83.4 [78.4–87.4] | NA | NA |
| Pathology | N | Sensitivity, % [95% CI] | Target values | |
| Sensitivity | Lower bound | |||
| CRC | 30 | 73.3 [55.6–85.8] | 85% | 66% |
| CRC & HGD | 42 | 78.6 [64.1–88.3] | 76% | 60% |
| CRC & APL | 63 | 76.2 [64.4–85.0] | NA | NA |
| APL | 33 | 78.8 [62.2–89.3] | NA | NA |
| IBD-HGD | 11 | 100 [74.1–100] | NA | NA |
| IBD-LGD ≥ 1 cm | 13 | 84.6 [57.8–95.7] | NA | NA |
APL: advanced precancerous lesions [includes serrated lesions]; CRC: colorectal cancer; HGD: high-grade dysplasia; IBD-HGD/LGD: inflammatory bowel disease-associated dysplasia [high- or low-grade]; non-APL: non-advanced precancerous lesions; NA: not applicable.
For the primary endpoint of sensitivity and specificity for CRC, subjects with negative colonoscopic findings [‘negative’ population] were included in the specificity analysis as negative outcomes. The ‘negative’ population included those subjects with a colonoscopy showing no findings, small [<1 cm] conventional adenomatous polyps, small [<1 cm] IBD-associated LGD, non-neoplastic disease, and/or hyperplastic polyps <1 cm in diameter. Subjects with a clinical history of IBD-associated dysplasia [Categories 2.5, 2.6], or subjects who were referred to the study with findings of IBD-associated HGD [Category 2.5] or IBD-associated LGD [Category 2.6] and were subsequently determined to have a negative colonoscopy or surgical intervention, were not included in the specificity analysis. Subjects with colonoscopy/histopathological findings that confirmed CRC were considered as positive outcomes and included in the sensitivity calculations for CRC. Those subjects with colonoscopic/histopathological index lesion findings of IBD-associated dysplasia and APLs were not included in the primary sensitivity analysis.
The secondary endpoint of sensitivity and specificity included subjects with colonoscopic findings of CRC and/or HGD as positive for the composite CRC/HGD sensitivity calculations. Subjects with negative colonoscopic findings were considered negative for the specificity calculation. The ‘negative’ population included those subjects with a colonoscopy showing no findings, small [<1 cm] conventional adenomatous polyps, non-neoplastic disease and/or hyperplastic polyps <1 cm in diameter. Subjects with IBD-associated LGD and LGD associated with APLs were not included in this analysis. In another secondary analysis to determine sensitivity and specificity of the test for histopathologically confirmed ACRN, subjects with colonoscopic findings of CRC, IBD-associated HGD, IBD-associated LGD ≥ 1 cm or APLs were considered as positive for composite ACRN sensitivity calculations.
Spearman’s correlation was used to assess correlation of the IBD inflammation score [IS] and the mt-sDNAHgb- score for both negative samples and ACRN. Spearman’s correlation was also used to assess correlation of the IS and reference marker levels for the negative samples.
3. Results
From 20 sites, 495 participants were enrolled after providing informed consent. Prior to testing, 110 participants were initially excluded due to protocol violations, study incompletion or categorization issues. An additional 30 participants were excluded from the specificity analyses for sample invalidity, a clinical history of IBD-associated LGD/HGD, or a negative colonoscopy result after being referred for dysplasia. A total of 355 participants were included in the sensitivity and specificity analysis [Figure 1].
Figure 1.

Patient consort diagram.
The evaluable cohort had a median age of 52 (interquartile range [IQR], 39–62) years and was 45.6% female. Most participants were White [93%] and had a diagnosis of UC [62.5%]. PSC was present in 10.1% of the population. There were no appreciable differences in demographics or disease state between cases and controls [Table 1].
Pathology based on colonoscopy results revealed CRC in 8.5% [N = 30], APLs in 9.3% [N = 33] and non-APLs in 7.6% [N = 27]; while nearly three-quarters [74.7%, N = 265] of the cohort yielded negative findings including hyperplastic polyps <1 cm. Amongst those with histopathologically confirmed APLs, 24/33 [73%] lesions were considered colitis-associated dysplasias [Table 1].
3.1. mt-sDNAHgb- [IBD-ACRN] surveillance test performance
Among the 355 participants included in the sensitivity and specificity analyses, the mt-sDNAHgb- test sensitivity was 73.3% [CI, 55.6–85.8%] for cancers of any stage, and 76.2% [CI, 64.4–85.0%] for all ACRN [includes CRC and the entire Category 2 spectrum of APLs]. Sensitivity was highest for IBD-associated HGD at 100% [CI, 74.1–100%], and 84.6% [CI, 57.8–95.7%] for IBD-associated LGD ≥ 1 cm. When considering the 280 participants with negative findings [no neoplasia or only non-APLs], specificity was 80.8% [CI, 75.9–84.9%]; specificity was 83.4% [CI, 78.4–87.4%] for subjects with no neoplasia alone [Table 2]. Receiver operating characteristic [ROC] analysis of cases [CRC and ACRN] and controls [non-APL and negatives] revealed an area undre the curve [AUC] of 0.840. mt-sDNA test sensitivity was shown to increase with lesion size, although with wide CIs [Table 3]. Sensitivity and specificity by IBD type revealed that the mt-sDNAHgb- test measured higher sensitivity and lower specificity in UC than in CD [although differences were not statistically significant] [Table 4].
Table 3.
Sensitivity and specificity by lesion size.
| Sensitivity, % [95% CI] | Specificity, % [95% CI] | |||||||
|---|---|---|---|---|---|---|---|---|
| Lesion size [cm] | Total | Combined CRC + APL | N | CRC | N | APL | N | Negative, non-APL |
| <0.5 | 3 | 66.7% [20.8–93.9%] | 1 | 0% | 2 | 100% [34.2–100%] | 271 | 81.6% [76.5–85.7%] |
| 0.5–0.9 | 2 | 100% [34.2–100%] | 1 | 100% [20.7–100%] | 1 | 100% [20.7–100%] | 21 | 71.4% [50.0–86.2%] |
| 1–1.9 | 17 | 70.6% [46.9–86.7%] | 3 | 100% [43.9–100%] | 14 | 64.3% [38.8–83.7%] | 0 | NA |
| 2–2.9 | 20 | 75.0% [53.1–88.8%] | 9 | 66.7% [35.4–87.9%] | 11 | 81.8% [52.3–94.9%] | 0 | NA |
| ≥3 | 21 | 81.0% [60.0–92.3%] | 16 | 75.0% [50.5–89.8%] | 5 | 100% [56.6–100%] | 0 | NA |
APL: advanced precancerous lesions [all categories]; CRC: colorectal cancer [all stages]; non-AA: non-advanced precancerous lesions; NA: not applicable.
Table 4.
Sensitivity and specificity by IBD type
| Crohn’s disease | Ulcerative colitis | p-value | |||
|---|---|---|---|---|---|
| Estimate [CI] | N | Estimate [CI] | N | ||
| CRC sensitivity | 55.6% [26.7–81.1%] | 9 | 78.9% [56.7–91.5%] | 19 | 0.3715 |
| APL sensitivity | 58.3% [32.0–80.7%] | 12 | 90.5% [71.1–97.3%] | 21 | 0.0709 |
| Specificity [negative, non-AA] | 86.9% [79.2–92.0%] | 107 | 77.5% [70.9–82.9%] | 182 | 0.0620 |
APL: advanced precancerous lesions [includes serrated lesions]; CRC: colorectal cancer; non-APL: non-advanced precancerous lesions.
In the negative population, recovery of the ACTB control marker was higher as the IS increased [ρ = 0.246, p = 0.006] [Figure 2]. As such, increasing IS was associated with a significant decrease in mt-sDNAHgb- test score [ρ = −0.176, p = 0.049] [Figure 3]. In patients with CRC and APLs, there was no association between IS and mt-sDNAHgb- score [p = 0.732].
Figure 2.
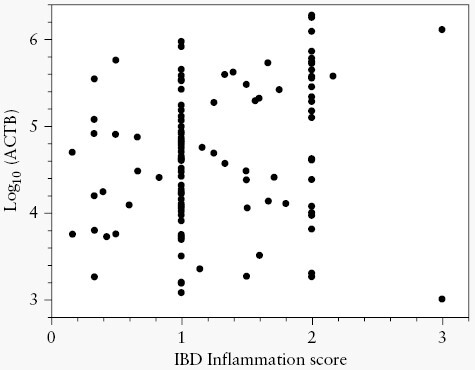
Inflammatory bowel disease inflammation score vs log10ACTB recovery for negatives and non-advanced precancerous lesions.
Figure 3.

Analysis of test score by inflammatory bowel disease inflammation score for negatives and non-advanced precancerous lesions.
4. Discussion
CRC is an established, potentially life-threatening complication of chronic IBD. As such, subsets of patients are encouraged to participate in a lifelong programmem of regular colonoscopic surveillance to find precancerous [dysplastic] or early cancerous lesions, thereby reducing morbidity and mortality.18 There are several important challenges to the implementation of this recommendation. Dysplasia in patients with IBD is often more difficult to detect by colonoscopy and is more likely to be multifocal than sporadic dysplasia.9 Furthermore, adherence to recommended colonoscopy intervals is often suboptimal.19 Even for patients who adhere to regular colonoscopic exams, post-colonoscopy CRCs [so-called interval cancers] are still rather common, seen in as many as 29% of patients.20 The advent of technology to detect molecular alterations in stool has strong biological rationale and now plays an important role in screening for sporadic CRC.14,21 It has been anticipated that molecular testing by stool assays in patients with IBD could overcome some of the aforementioned challenges.
Prior research has demonstrated the potential for using stool molecular markers to assist in the detection of neoplastic lesions in patients with IBD. An earlier case-control study in a cohort of IBD patients revealed that the same methylated tumour suppressor genes [BMP3 and NDRG4] that are included in the FDA-approved mt-sDNA test were highly discriminant for CRC individually, and detected 100% of IBD-associated CRC at 89% specificity.12 This served as the incentive for the present multi-centre, prospective case-control study designed to see whether DNA markers incorporated in the mt-sDNA test—yet without the haemoglobin component—can be useful for IBD neoplasia detection.
We observed that the mt-sDNA molecular markers provided a sensitivity of 73.3% for CRC, and 78.6% for CRC and HGD, with specificity of 80.8%. Sensitivity was 100% for lesions with HGD, and 84.6% for IBD-LGD ≥ 1 cm. For APLs ≥2 cm, sensitivity was 81.8–100%. Our findings also suggest that ~19% of patients with long-standing IBD who are found to have no neoplasia on colonoscopy may have a false-positive stool DNA test or subtle lesions not detected by colonoscopy, a rate slightly higher than the general population [DeeP-C study].14
It is worth noting that endoscopists in this study were blinded to the results of the stool DNA test. As shown by others, when providers are aware of a positive mt-sDNA test, they tend to have higher adenoma detection rates presumably due to more careful colonoscopies.21 In the IBD patient population, a stool-based DNA test might be positioned as an interval test between surveillance colonoscopies. The reason as to why patients with UC had higher mt-sDNAHgb- scores than patients with CD is not clear but could perhaps relate to the extent of mucosal involvement.
In the neoplasia-negative population, there was a significant decrease in mt-sDNAHgb- test score [p = 0.028] as IBD IS increased. Because the reference standard [ACTB] is a measure of total human DNA and can be found in white blood cells, the concomitant increase in ACTB with increasing IS suggests that some of the changes observed coincided with those of the reference standard. In a previous study of patients with IBD, Kisiel et al. revealed that methylated BMP3 [as used in this study] and methylated VAV demonstrated a sensitivity of 92% and specificity of 90%, using ZDHHC1—which is methylated in human colonic epithelial cell DNA, but not white blood cells, as the reference standard.14 That study demonstrated that ZDHHC1-corrected markers are less affected by inflammation than ACTB. Inflammatory activity is also a barrier to the performance of surveillance colonoscopy, which should ideally be performed when patients are in clinical remission.9
Strengths of this study are a well-characterized, prospectively enrolled patient population with detailed pathology related to neoplasia and inflammation, and the use of approved technology for collecting, extracting and analysaing stool DNA. There are also important limitations. First, the study did not meet the enrolment targets for CRC and HGD lesions in IBD patients, providing lesser precision than anticipated in our reported assay performance estimates. Additionally, there were 86 patients whose index lesion categorization was based on clinical data rather than confirmation by a research pathology review. This is mitigated, in part, by the study environment; patients were enrolled from expert referral centres for IBDs. Nonetheless, the data herein contribute to the proof-of-concept regarding the potential of mt-sDNA markers to inform CRC surveillance.
Stool-based molecular testing combined with surveillance colonoscopy offers theoretical advantages over the current standard of care, including patient factors [ease of access, patient convenience, no bowel purgation, non-invasiveness, no missed time from work or other activities, lower cost] and biological factors [entire colorectum can be surveyed as the exfoliated markers from the whole colon surface are present in the stool, objective report on molecular markers]. With refinements in molecular markers, such as the next generation of mt-sDNA tests currently being developed to screen average-risk individuals,22,23 stool-based molecular tests in the high-risk IBD population could become an important addition to cancer surveillance.
Supplementary Material
Acknowledgments
Medical writing and editorial support were provided by William K. Johnson, PhD, an employee of Exact Sciences. The authors would also like to thank Katie Draheim for study planning and execution, and Martin Krockenberger for statistical analysis.
Contributor Information
Steven Itzkowitz, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Francis A Farraye, Mayo Clinic, Rochester, MN, USA.
Paul J Limburg, Exact Sciences Corporation, Madison, WI, USA.
Zubin Gagrat, Exact Sciences Corporation, Madison, WI, USA.
Marilyn C Olson, Exact Sciences Corporation, Madison, WI, USA.
Julia Zella, Exact Sciences Corporation, Madison, WI, USA.
John B Kisiel, Mayo Clinic, Rochester, MN, USA.
Funding
This work was supported by Exact Sciences Corporation, LLC [Madison, WI, USA].
Conflict of Interest
Employment and disclosures: SI has received consulting fees and research support from Exact Sciences Corporation, research support from Freenome, and consulting fees from Geneoscopy. Mayo Clinic and Exact Sciences own intellectual property under which JBK is listed as an inventor and may receive royalties in accordance with Mayo Clinic policy. ZG, MO, JZ and MK are employees and shareholders of Exact Sciences. FAF has no relevant conflicts. PJL serves as Chief Medical Officer for Exact Sciences, initially through a contracted services agreement with Mayo Clinic [under which he and Mayo Clinic had contractual rights to receive royalties] and later as an Exact Sciences employee. The authors have no other relevant financial or non-financial interests to disclose.
Author Contributions
Conceptualization—SI, FF, JBK, PJL, MO; Data curation—ZG, JZ; Formal analysis—ZG; Investigation—SI, FF, JBK, PJL, ZG; Methodology—ZG; Project administration—JZ, MO; Resources—MO; Supervision—MO, PJL; Validation—ZG; Visualization—SI, PJL, JBK, ZG; Writing [original draft]—All authors; Writing [review & editing]—All authors.
Data Availability
Upon request with a clearly stated purpose, data will be made available following approval from Exact Sciences.
References
- 1. Jess T, Rungoe C, Peyrin-Biroulet L.. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol 2012;10:639–45. doi: 10.1016/J.CGH.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 2. Clarke WT, Feuerstein JD.. Colorectal cancer surveillance in inflammatory bowel disease: Practice guidelines and recent developments. World J Gastroenterol 2019;25:4148–57. doi: 10.3748/WJG.V25.I30.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laine L, Kaltenbach T, Barkun A, McQuaid KR, Subramanian V, Soetikno R; SCENIC Guideline Development Panel. SCENIC Guideline Development Panel. SCENIC international consensus statement on surveillance and management of dysplasia in inflammatory bowel disease. Gastrointest Endosc 2015;81:489–501.e26. doi: 10.1016/J.GIE.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 4. Torres J, Bonovas S, Doherty G, et al. ECCO guidelines on therapeutics in Crohn’s disease: medical treatment. J Crohns Colitis 2020;14:4–22. doi: 10.1093/ECCO-JCC/JJZ180. [DOI] [PubMed] [Google Scholar]
- 5. Lamb CA, Kennedy NA, Raine T, et al. ; IBD guidelines eDelphi consensus group. British Society of Gastroenterology consensus guidelines on the management of inflammatory bowel disease in adults. Gut 2019;68:s1–s106. doi: 10.1136/GUTJNL-2019-318484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shah SC, Itzkowitz SH.. Colorectal cancer in inflammatory bowel disease: mechanisms and management. Gastroenterology Published online October 2021. doi: 10.1053/J.GASTRO.2021.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farraye FA, Odze RD, Eaden J, Itzkowitz SH.. AGA technical review on the diagnosis and management of colorectal neoplasia in inflammatory bowel disease. Gastroenterology 2010;138:746–774.e4. doi: 10.1053/J.GASTRO.2009.12.035. [DOI] [PubMed] [Google Scholar]
- 8. Itzkowitz SH, Present DH; Crohn's and Colitis Foundation of America Colon Cancer in IBD Study Group. Consensus conference: Colorectal cancer screening and surveillance in inflammatory bowel disease. Inflamm Bowel Dis 2005;11:314–21. doi: 10.1097/01.MIB.0000160811.76729.D5. [DOI] [PubMed] [Google Scholar]
- 9. Murthy SK, Feuerstein JD, Nguyen GC, Velayos FS.. AGA clinical practice update on endoscopic surveillance and management of colorectal dysplasia in inflammatory bowel diseases: expert review. Gastroenterology 2021;161:1043–1051.e4. doi: 10.1053/J.GASTRO.2021.05.063. [DOI] [PubMed] [Google Scholar]
- 10. Braithwaite E, Carbonell J, Kane J, Gracie D, Selinger C.. Patients’ perception of colonoscopy and acceptance of colonoscopy based IBD related colorectal cancer surveillance. Expert Rev Gastroenterol Hepatol 2021;15:211–6. doi: 10.1080/17474124.2021.1829971. [DOI] [PubMed] [Google Scholar]
- 11. Kisiel JB, Klepp P, Allawi HT, et al. Analysis of DNA methylation at specific loci in stool samples detects colorectal cancer and high-grade dysplasia in patients with inflammatory bowel disease. Clin Gastroenterol Hepatol 2019;17:914914.–921.e5. doi: 10.1016/J.CGH.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kisiel JB, Yab TC, Hussain FTN, et al. Stool DNA testing for the detection of colorectal neoplasia in patients with inflammatory bowel disease. Aliment Pharmacol Ther 2013;37:546–54. doi: 10.1111/APT.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Redwood DG, Asay ED, Blake ID, et al. Stool DNA testing for screening detection of colorectal neoplasia in Alaska Native People. Mayo Clin Proc 2016;91:61–70. doi: 10.1016/j.mayocp.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 14. Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;370:1287–97. doi: 10.1056/nejmoa1311194. [DOI] [PubMed] [Google Scholar]
- 15. Centers for Medicare & Medicaid Services. National Coverage Analysis - Screening for Colorectal Cancer - Stool DNA Testing (CAG-00440N) - Decision Memo. CMS.gov Website. Published October 9, 2014. Accessed October 6, 2021. https://www.cms.gov/medicare-coverage-database/view/ncacal-decision-memo.aspx?proposed=N&NCAId=277
- 16. Imperiale TF, Kisiel JB, Itzkowitz SH, et al. Specificity of the multi-target stool DNA test for colorectal cancer screening in average-risk 45-49 year-olds: a cross-sectional study A C. Cancer Prev Res (Phil). 2021;14:489–496. doi: 10.1158/1940-6207.CAPR-20-0294 [DOI] [PubMed] [Google Scholar]
- 17. Exact Sciences Corporation LLC. Cologuard ® Physician Brochure. Accessed December 1, 2020. https://www.cologuardtest.com/hs-fs/hub/377740/file-1412311339.pdf
- 18. Ananthakrishnan AN, Cagan A, Cai T, et al. Colonoscopy is associated with a reduced risk for colon cancer and mortality in patients with inflammatory bowel diseases. Clin Gastroenterol Hepatol 2015;13:322–329.e1. doi: 10.1016/J.CGH.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gordon C, Chee D, Hamilton B, et al. Root-cause analyses of missed opportunities for the diagnosis of colorectal cancer in patients with inflammatory bowel disease. Aliment Pharmacol Ther 2021;53:291–301. doi: 10.1111/apt.16155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Troelsen FS, Sørensen HT, Crockett SD, Pedersen L, Erichsen R.. Characteristics and survival of patients with inflammatory bowel disease and postcolonoscopy colorectal cancers. Clin Gastroenterol Hepatol 2022;20:e984–e1005. doi: 10.1016/J.CGH.2021.05.039. [DOI] [PubMed] [Google Scholar]
- 21. Johnson DH, Kisiel JB, Burger KN, et al. Multitarget stool DNA test: clinical performance and impact on yield and quality of colonoscopy for colorectal cancer screening. Gastrointest Endosc 2017;85:657–665.e1. doi: 10.1016/J.GIE.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Domanico M, Kisiel J, Gagrat Z, et al. Novel multi-target stool DNA marker panel yields highly accurate detection of colorectal cancer and premalignant neoplasia. Am J Gastroenterol 2019;114:S191–S191. doi: 10.14309/01.AJG.0000590832.76462.30. [DOI] [Google Scholar]
- 23. Exact Sciences Corporation. Clinical Validation of An Optimized Multi-Target Stool DNA (Mt-sDNA 2.0) Test, for Colorectal Cancer Screening ‘BLUE-C’ - Full Text View - ClinicalTrials.gov. Accessed June 8, 2022. https://clinicaltrials.gov/ct2/show/NCT04144738
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Upon request with a clearly stated purpose, data will be made available following approval from Exact Sciences.


