Abstract
The coding sequences of developmental genes are expected to be deeply conserved, with cis-regulatory change driving the modulation of gene function. In contrast, proteins with roles in defense are expected to evolve rapidly, in molecular arms races with pathogens. However, some gene families include both developmental and defense genes. In these families, does the tempo and mode of evolution differ between genes with divergent functions, despite shared ancestry and structure? The leucine-rich repeat receptor-like kinase (LRR-RLKs) protein family includes members with roles in plant development and defense, thus providing an ideal system for answering this question. LRR-RLKs are receptors that traverse plasma membranes. LRR domains bind extracellular ligands; RLK domains initiate intracellular signaling cascades in response to ligand binding. In LRR-RLKs with roles in defense, LRR domains evolve faster than RLK domains. To determine whether this asymmetry extends to LRR-RLKs that function primarily in development, we assessed evolutionary rates and tested for selection acting on 11 subfamilies of LRR-RLKs, using deeply sampled protein trees. To assess functional evolution, we performed heterologous complementation assays in Arabidopsis thaliana (Arabidopsis). We found that the LRR domains of all tested LRR-RLK proteins evolved faster than their cognate RLK domains. All tested subfamilies of LRR-RLKs had strikingly similar patterns of molecular evolution, despite divergent functions. Heterologous transformation experiments revealed that multiple mechanisms likely contribute to the evolution of LRR-RLK function, including escape from adaptive conflict. Our results indicate specific and distinct evolutionary pressures acting on LRR versus RLK domains, despite diverse organismal roles for LRR-RLK proteins.
Keywords: HAESA, CLAVATA1, plant evolution, domain evolution, protein evolution
Introduction
Signaling from outside a cell to direct cellular behavior is critical in both response to pathogens and development. In defense signaling, receptors directly bind pathogen proteins extracellularly to trigger intracellular defense processes. In development, extracellular signals also bind receptors to trigger intracellular cell identity and physiological responses. Receptor proteins are often structurally similar, but for those with roles in defense, signal-binding ectodomains often evolve more rapidly and with greater rates of positive selection (X.S. Zhang et al. 2006; Fischer et al. 2016; Ahmad et al. 2021; Ghosh et al. 2022). Rapid evolution in defense ectodomains may be a response to rapidly evolving pathogen proteins in a molecular arms race, while intracellular domains still trigger conserved host reactions and may evolve slower (Lehti-Shiu et al. 2012; Parys et al. 2021; Zhang et al. 2021). In contrast, for developmental genes, coding sequence changes that modulate protein structure and function are hypothesized to be less important than regulatory changes (King and Wilson 1975; Atchley and Hall 1991; Carroll 2008; Wittkopp and Kalay 2011; Long et al. 2016; Marand et al. 2023). Proteins in the leucine-rich repeat receptor-like kinase (LRR-RLK) family have roles in defense and development and a conserved domain structure. The LRR domain is the ligand-binding ectodomain at the N terminus, and the RLK domain is an intracellular kinase domain, with the 2 domains separated by a single-strand transmembrane α-helix (Shiu and Bleecker 2001; Liu et al. 2017). Here, we investigate whether asymmetric evolution of extracellular signal perception and intracellular signal response domains is limited to LRR-RLKs with roles in defense, or if the trend also applies to LRR-RLKs with roles in development.
There is suggestive evidence that sequences encoding LRR and RLK domains are evolving asymmetrically. First, although encoded by single genes, LRR and RLK domains are physically separated by the plasma membrane and have distinct molecular functions (Santiago et al. 2016; Hohmann et al. 2018). Second, different subfamilies of developmental LRR-RLKs respond to distinct ligands, but activate similar intracellular signaling cascades, implying asymmetric functional shifts (Hohmann et al. 2018; Zheng et al. 2019; Zhu et al. 2019; Liu et al. 2022). Third, the coordination between cognate LRR and RLK domains is interchangeable: functional output by RLK domains can be activated by alternate LRR domains in chimeric proteins (Osakabe et al. 2005; Diévart et al. 2006; Brutus et al. 2010; Zheng et al. 2019; Hohmann et al. 2020). This uncoupled domain function may permit independent evolutionary trajectories (Bhattacharyya et al. 2006; Di Roberto and Peisajovich 2014; Sato et al. 2014). Fourth, over deep time, truncations or fusions with unrelated domains are more common in LRR domains than in RLK domains, and more LRR-only genes are conserved and expressed (Man et al. 2020). Taken together, these data suggest that LRR and RLK domains may have distinct evolutionary trajectories, regardless of their diverse organismal functions in things like development, defense, and physiology.
To determine whether LRR and RLK domains are evolving asymmetrically, we leveraged the One Thousand Plant Transcriptomes database (One Thousand Plant Transcriptomes Initiative 2019) and inferred deeply sampled peptide trees for 11 subfamilies of LRR-RLK proteins (Shiu and Bleecker 2001; Dufayard et al. 2017; Man et al. 2020). We used these trees to test for distinct evolutionary forces acting on LRR versus RLK domains. We also used heterologous transformation experiments to test for divergent functional evolution of LRR versus RLK domains. We discovered a clear signal of asymmetric evolution between LRR and RLK domains encoded by the same gene, but our heterologous transformation experiments revealed more complexity. Our work highlights the multiple interacting mechanisms that drive the evolution of gene and protein function.
Results
LRR Domains Are Evolving Faster Than RLK Domains
To determine the evolutionary trajectories of individual protein domains, we needed deeply sampled trees that included multiple LRR-RLK subfamilies with diverse functions. Therefore, we selected 9 LRR-RLK subfamilies in clade XI with roles in plant development and physiology and 2 LRR-RLK subfamilies with roles in defense (Table 1). We also included CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1), a defense receptor LysM-RLK that binds chitin derivatives (Chinchilla et al. 2006; Gimenez-Ibanez et al. 2009; Zhang et al. 2015; Table 1). CERK1 binds pathogen-associated molecular patterns, is a target of bacterial effectors, and is expected to be under intense positive selection pressure (Gimenez-Ibanez et al. 2009; Hogenhout et al. 2009; Lohmann et al. 2010; De Mita et al. 2014; Wang et al. 2018).
Table 1.
Protein subfamilies included in gene tree inference
| Name (A. thaliana) | Gene ID | Biological function | Cladea | Genesb | References |
|---|---|---|---|---|---|
| BARELY ANY MERISTEM (BAM1) | AT5G65700 | Meristem and tissue specification | XI | 447 | DeYoung et al. (2006) |
| C-TERMINALLY ENCODED PEPTIDE RECEPTOR 2 (CEPR2) | AT1G72180 | Nitrate uptake | XI | 358 | Tabata et al. (2014) |
| CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1) | AT3G21630 | Microbe detection | NAc | 367 | Miya et al. (2007) |
| CLAVATA1 (CLV1) | AT1G75820 | Shoot meristem size | XI | 156 | Clark et al. (1993) |
| FLAGELLIN-SENSITIVE 2 (FLS2) | AT5G46330 | Microbe detection | XII | 60 | Gómez-Gómez and Boller (2000) |
| GASSHO2 (GSO2) | AT5G44700 | Epidermis development | XI | 17 | Tsuwamoto et al. (2008) |
| HAESA (HAE) | AT4G28490 | Abscission and cell separation | XI | 409 | Jinn et al. (2000) |
| HAESA-LIKE3 (HSL3) | AT5G25930 | Stomatal closure | XI | 208 | Liu et al. (2020), Liu et al. (2022) |
| HAIKU2 (IKU2) | AT3G19700 | Seed and embryo development | XI | 354 | Luo et al. (2005) |
| PLANT ELICITOR PEPTIDE1 RECEPTOR1 (PEPR1) | AT1G73080 | Innate immunity | XI | 41 | Liu et al. (2013) |
| PHLOEM INTERCALATED WITH XYLEM (PXY) | AT5G61480 | Vascular development | XI | 137 | Hirakawa et al. (2008) |
| ROOT MERISTEM GROWTH FACTOR1 INSENSITIVE5 (RGI5) | AT1G34110 | Root meristem size | XI | 8 | Ou et al. (2016) |
aSubfamily assignments from (Man et al. 2020).
bNumber of genes recovered in each subfamily.
cCERK1 is a LysM-RLK.
Gene trees with deep taxonomic sampling are needed to detect signatures of selection and variation in evolutionary rate. However, fully assembled plant genomes are sparse given the immense diversity of multicellular land plants and are not phylogenetically evenly distributed. This limits bioinformatic resolution and skews statistical inference toward overrepresented clades, like the grasses (Kress et al. 2022). To compensate for this sampling bias, we leveraged the One Thousand Plant Transcriptomes project (1KP), which includes phylogenetically informed sampling from across multicellular land plants (One Thousand Plant Transcriptomes Initiative 2019). Using the 1KP data and an iterative search and tree inference algorithm we developed (Man et al. 2020), we inferred deeply sampled peptide trees for all 11 LRR-RLK subfamilies, each of which had between 8 and 447 members (Table 1). This wide discrepancy in the number of genes recovered may be due to the 1KP sampling strategy, which focused preferentially on above-ground tissue, biasing against root-expressed genes like RGI5 (Ou et al. 2016; One Thousand Plant Transcriptomes Initiative 2019).
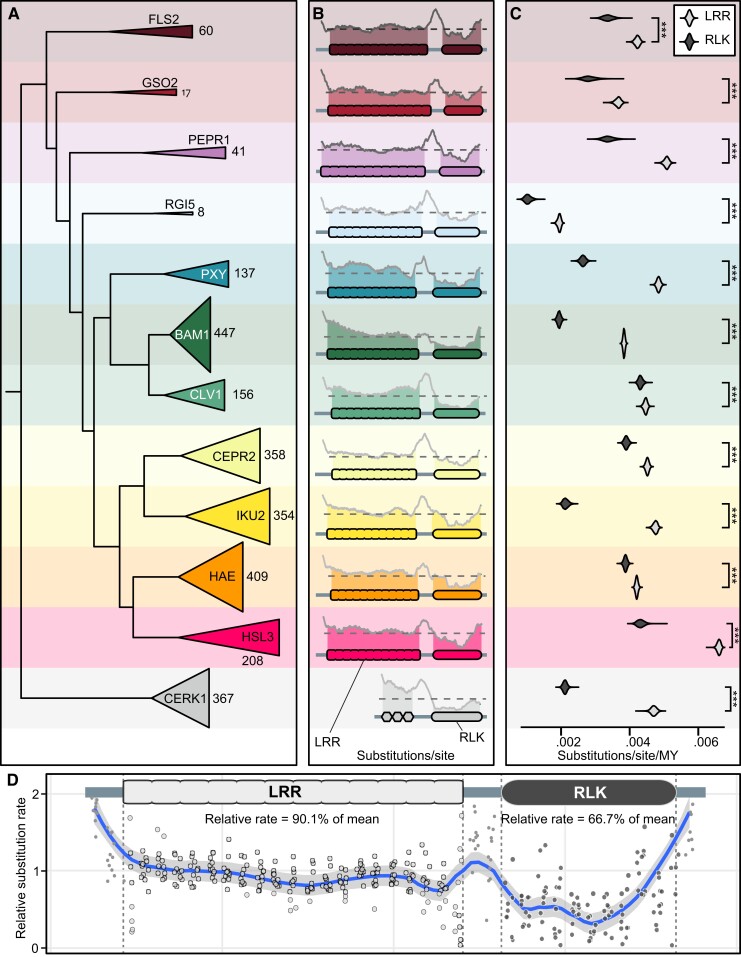
To ask how the tempo of evolution might differ between LRR and RLK domains, we used our trees to estimate evolutionary rates of amino acid substitutions in angiosperm LRR-RLK sequences using multiple methods. First, we used our trees to infer site-specific relative substitution rates using an empirical Bayesian approach implemented in IQ-TREE (Fig. 1; Nguyen et al. 2015). This analysis permitted the comparison of amino acid site substitution rates in various regions of each protein, relative to overall mean substitution rates. For example, in the CLAVATA1 (CLV1) subfamily, substitution rates were lowest in the region of the RLK domain (34.2% of the overall mean rate), substantially higher rates in the LRR domain (95.7% of the overall mean rate), and the fastest rates were in interdomain sequences. Despite the different functional roles of proteins in each subfamily (Table 1), these analyses revealed similar patterns of site substitution rates between subfamilies (Fig. 1b and c). Averaged over all 11 subfamilies, sequences of LRR domains evolve at 90.1% of the subfamily's overall mean substitution rate, while sequences of RLK domains evolve slower at 66.7% of the mean (Fig. 1d). We next employed a tangential approach, a Markov chain Monte Carlo (MCMC) Bayesian analysis implemented in BEAST, to infer the substitution rate for each domain that includes probability estimates of the entire tree structure (Bouckaert et al. 2014). This analysis revealed that LRR domains were evolving significantly faster than RLK domains in each subfamily (unpaired Wilcoxon tests, P-value <0.001) (Fig. 1d). Notably, while the relative rates of ectodomain evolution were substantially higher for CERK1 than for the 9 developmental proteins we analyzed, this was not the case for FLS2 or PEPR1, which are middle of the pack with respect to asymmetric evolution rate. Therefore, the differences in evolutionary rates between LRR and RLK domains are not solely a function of protein function. Independent evolutionary rates for LRR and RLK domains may be consistent across LRR-RLK proteins, regardless of their functional roles.
Fig. 1.
Evolutionary rates differ between LRR and RLK domains. a) Relationships of 11 LRR-RLK clades and CERK1. Triangle height proportional to clade size. b) Substitution rate (relative to mean) at each amino acid residue for each clade, 75-residue sliding mean (gray trace). Dashed lines, mean rates for both domains. c) Bayesian posterior rate probability for LRR and RLK domains of each clade. ***P-value <0.001 (unpaired Wilcoxon). d) Relative evolution rate for each LRR-RLK clade, 20 LRR bins and 10 RLK bins, fitted with a local polynomial regression.
Positive Selection Is More Common in LRR Domains Than in RLK Domains
The divergent evolutionary rates we detected between LRRs and RLK domains in developmental proteins suggest divergent selective pressures acting on LRR versus RLK domains, despite their close juxtaposition in the same protein. To determine whether LRR and RLK domains are under different selective regimes, we used our deeply sampled trees and 2 codon-based tests for selection implemented in the HyPhy software suite: Mixed Effects Model of Evolution (MEME) to detect episodic diversifying (positive) selection and Fast, Unconstrained Bayesian AppRoximation (FUBAR) to detect signatures of negative selection (Kosakovsky Pond and Frost 2005; Murrell et al. 2013). We performed these tests on our 11 LRR-RLK subfamilies as well as on our outgroup subfamily CERK1.
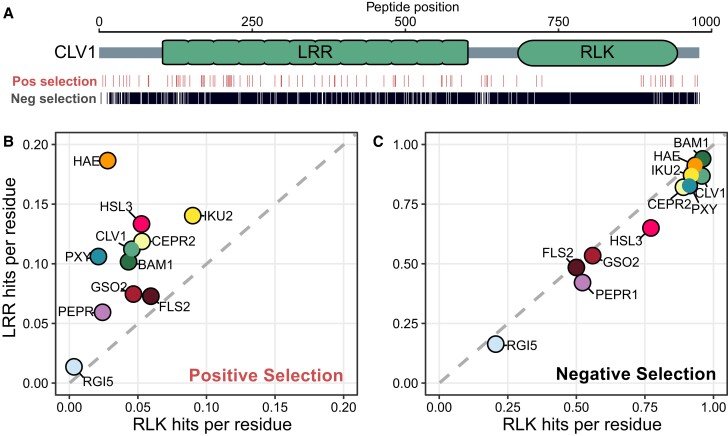
MEME detected much higher rates of positive selection acting on all sampled LRR domains. Both MEME and FUBAR are sensitive to the number of genes included, with fewer opportunities to detect selection in subfamilies with fewer members (Poon et al. 2009). Therefore, to normalize for different numbers of genes in each subfamily, we represented our results as ratios of selection pressure between each subfamily's LRR and RLK domains, rather than as absolute values (Fig. 2b and c). As expected (Bishop et al. 2000; X.S. Zhang et al. 2006), the CERK1 subfamily showed a strong bias for positive selection in sequences encoding extracellular (LysM) domains (∼41% of LysM domain residues vs. ∼6% of RLK residues, ∼6.8-fold difference) (supplementary fig. S1A, Supplementary Material online). Similarly, across all 11 LRR-RLK subfamilies, positive selection occurred more frequently in gene regions encoding LRR domains (∼10% of residues) compared with RLK domains (4% of residues) (χ2 test, P-value < 0.001) (Fig. 2b). The sensitivity of MEME and FUBAR to sequence number means that the absolute rates of positive selection cannot be compared between subfamilies. However, a bias toward higher positive selection rates in LRR domains was true within every subfamily, including the smallest we sampled (Fig. 2b). For example, 11.2% of LRR domain residues and ∼4.6% of RLK residues in CLV1 were under positive selection, a ∼2.4-fold difference (Fig. 2a). Interestingly, the defense proteins FLS2 and PEPR1 did not show the strong bias for positive selection acting on the LRR domain that was characteristic of CERK1. Instead, FLS2 and PEPR1 were very similar to the developmental LRR-RLKs in this assay (Fig. 2b, supplementary fig. S1, Supplementary Material online). This suggests similar evolutionary pressures acting on developmental and defense LRR-RLKs.
Fig. 2.
Signatures of selection are unequally distributed between LRR and RLK domains. a) CLV1, as an example, with the locations of sites predicted to be under positive or negative selection shown. b) Rates per residue of signatures of positive selection in RLK (x-axis) versus LRR (y-axis) domains in each clade. c) Rates per residue of signatures of negative selection in RLK (x-axis) versus LRR (y-axis) domains in each clade. Gray dashed line defines the region of no ratiometric bias between domains.
The higher positive selection acting on LRR versus RLK domains occurred against a background of pervasive negative selection. For example, in CLV1, nearly all residues (∼87% of LRR residues and ∼96% of RLK residues, a ∼1.1-fold difference) were under negative selection (Fig. 2a). This was expected, as clv1 mutants have severe cell proliferation phenotypes in diverse species, consistent with CLV1 being a conserved developmental gene, and thus under strong purifying selection (Clark et al. 1993; Bommert et al. 2005; Xu et al. 2015; Whitewoods et al. 2018). This pattern extended to every subfamily of LRR-RLKs that we analyzed (Fig. 2c). Negative selection was detected at most residues in both domains (∼74% of RLK residues overall vs. ∼66% of LRR residues overall), and this difference was not significant for any single subfamily (χ2 test, P-value >0.05) (Fig. 2c). Pervasive negative selection indicates that the structures of LRR domains are under strong evolutionary constraint, even while they undergo positive selection at certain sites. These features of LRR-RLK evolution appear to be independent of proteins’ diverse roles in development or defense. Thus, differences in domain structural constraints or defense arms races are unlikely to explain asymmetry in LRR versus RLK domain evolution.
Transcriptome Sequencing Bias, LRR Motif Misalignment, and Saturation Cannot Explain Asymmetric Evolutionary Dynamics
The higher positive selection acting on LRR versus RLK domains may have been an analytical artifact, caused by factors independent of evolutionary pressures. We first reasoned that transcriptome sequencing bias might explain our results. Most genes in our analyses are from transcriptomes sequenced in the 3′ to 5′ direction (One Thousand Plant Transcriptomes Initiative 2019). LRR domains are encoded in the 5′ portions of each transcript; therefore, we reasoned that sequencing errors, which preferentially cluster in 5′ sequence, could explain asymmetric positive selection. To check that sequencing error bias was not responsible for the elevated rate of positive selection detection in LRR domains, we performed twin analyses of the PHLOEM INTERCALATED WITH XYLEM (PXY) subfamily using 42 sequences each: one set translated from 1KP transcriptome sequences, and a second translated from whole genome assemblies that are not directionally biased. These analyses found an identical number of positive selection hits in both domains (25 in LRRs vs. 2 in RLKs) (supplementary fig. S1, Supplementary Material online). This indicates that transcriptome sequencing bias likely does not explain the consistently elevated ratio of positive selection detected in LRR domains relative to RLK domains.
We next reasoned that variability in LRR motif number could cause misalignments, incorrect homology assessments, and biased selection estimates. Three lines of evidence indicated that this was not the case for our analyses. First, we extracted individual LRR motifs from Amborella trichopoda BAM1 and realigned each to an alignment of full-length BAM1 sequences without A. trichopoda BAM1. Each of the 21 LRRs realigned at its original position, despite the lack of positional context from proximal sequence (supplementary fig. S2, Supplementary Material online). This indicates that, at least for BAM1, there is significant phylogenetic information in individual LRR motifs to prevent misalignment. Second, we reasoned that if variability in LRR motif number caused misalignment and biased selection estimates, then variation in LRR number should correlate with the number of positively selected sites in each subfamily. To evaluate this prediction, we used an LRR prediction tool optimized for plant genomes to identify LRR motifs (Chen 2021) and assessed variability in LRR motif number in each sequence in each subfamily (supplementary fig. S3A, Supplementary Material online). We tested for correlations between variance in LRR motif number and the number of positively selected residues in LRR domains and found no evidence that these variables were related (supplementary fig. S3B, Supplementary Material online). Third, we would have expected significantly lower negative selection acting on LRR domains if misalignment of the highly variable LRR domains could explain our results. Instead, negative selection rates were similar between LRR and RLK domains (Fig. 2c). Thus, variability in LRR motif number and misalignment likely cannot explain our results.
Last, we thought that high substitution rates in LRR domains might have led to saturation over deep divergence times and caused artifactually high positive selection estimates in LRR domains. To account for this, we repeated our tests for positive selection on our largest subfamilies: BAM1, CEPR2, CERK1, and HAE, using only sequences from Papaveraceae species. This subsetted analysis traverses ∼88 My of evolutionary divergence between species, rather than 140 to 270 My in the full analysis of all angiosperms (Kumar et al. 2017; Sauquet et al. 2022). If saturation had led to higher levels of positive selection in LRR domains in the full analysis, then we expected this signal to disappear in the subsetted analysis. This is not what we found. Positive selection was still more common in LRR domains in the subsetted analysis, and negative selection was similar between LRR and RLK domains, with a slight bias toward RLK domains, as in the full analysis (supplementary table S1, Supplementary Material online). Taken together, these analyses indicate that higher positive selection rates in LRR domains are not due to sequencing bias, misalignment, or saturation but rather due to differing evolutionary pressures acting on LRR versus RLK domains.
Changes in Domain Function Have Likely Contributed to Functional Divergence of LRR-RLKs
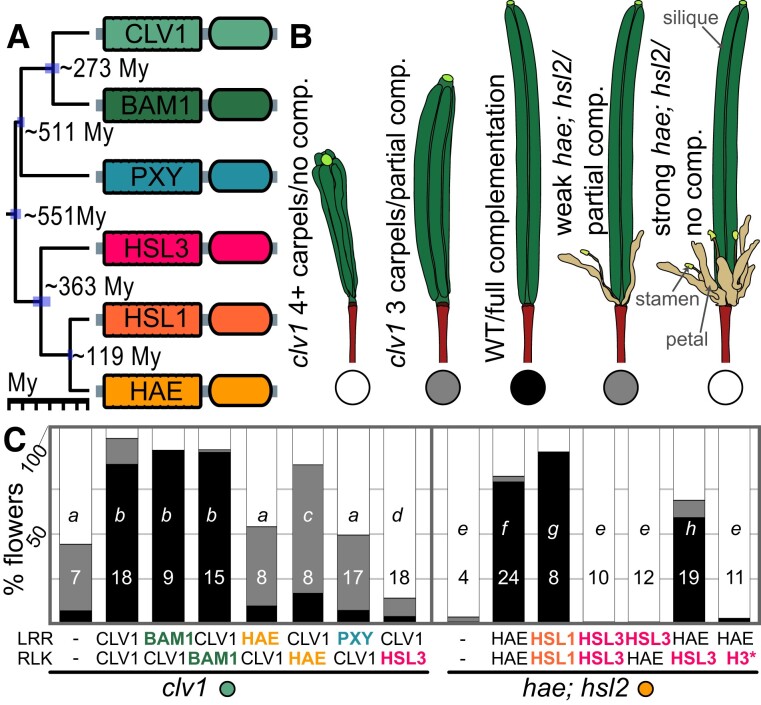
Divergent evolutionary pressures acting on LRR and RLK domains suggest that the 2 domains have independently diverged in function. To evaluate this prediction, we next designed in planta experiments to assess functional conservation of divergent LRR-RLK domains. We transformed chimeric LRR-RLKs with swapped domains into Arabidopsis clv1 single mutants and hae;hsl2 double mutants (supplementary fig. S4, Supplementary Material online; Scholl and Anderson 1994; Nimchuk et al. 2011). HAE and HSL2 are close paralogs and are functionally redundant with respect to floral organ abscission (Cho et al. 2008). Therefore, we will refer to these genes as HAE/HSL2 from here on. CLV1 and HAE/HSL2 are distantly related LRR-RLK genes within clade XI (Man et al. 2020), separated from each other by ∼551 My of evolution (Fig. 3a; supplementary fig. S5, Supplementary Material online). Over this deep time, CLV1 and HAE/HSL2 have diverged considerably in function. HAE and HSL2 regulate cell separation at the abscission zone of sepals, petals, and stamens. hae;hsl2 double mutants fail to shed these floral organs, which remain attached to pedicels (Fig. 3b; Jinn et al. 2000; Kumpf et al. 2013). CLV1 controls proliferation of stem cells in meristems, and clv1 mutants produce fasciated siliques with extra carpels (Fig. 3b; Clark et al. 1993, 1997). The mutant alleles we used (clv1-15, hae-3/hsl2-3) are recessive and produce clear, quantifiable phenotypes (Clark et al. 1993; Jinn et al. 2000), allowing us to ask whether divergent molecular evolution was mirrored by divergent functional evolution of LRR and RLK domains. We took quantitative measures of transgene complementation, when compared with positive (CLV1 or HAE coding sequence under their native promoters) and negative (untransformed mutant) controls. We binned these quantitative data into “no complementation,” “partial complementation,” and “full complementation,” with stringent limits on the “no complementation” and “full complementation” bin boundaries, so that we could detect partial complementation by deeply divergent genes with the gross phenotypic measures we used (supplementary fig. S6, Supplementary Material online). Binning our complementation data allowed us to compare 2 very different sets of mutants and phenotypic assays.
Fig. 3.
RLK and LRR domain functions have distinct evolutionary trajectories. a) Time-calibrated chronogram of 6 A. thaliana LRR-RLK genes showing approximate divergence times between paralogs. b) Illustrations of representative Arabidopsis siliques from mutants transformed with (chimeric) transgenes. c) Percentage of flowers from clv1 or hae/hsl2 mutants complemented by various paralogous domains. Numbers in bars indicate independent events (5 to 10 flowers per event). Letters indicate significantly different groups (analysis of variance, Tukey's post hoc). H3*, catalytically dead HSL3 variant.
We first assessed the conservation of function between CLV1, HAE/HSL2, and their closest respective paralogs. CLV1 is most closely related to the BAM genes, which are in a subfamily separated from CLV1 by ∼273 My of evolution (Fig. 3a; supplementary fig. S5, Supplementary Material online; Man et al. 2020). Despite this deep divergence time, BAM genes also function in stem cell proliferation and can partially compensate for the loss of clv1 ortholog function through transcriptional regulation (DeYoung et al. 2006; Nimchuk et al. 2015; Nimchuk 2017; Rodriguez-Leal et al. 2019). Consistent with this, CLV1pro::BAM1LRR:CLV1RLK usually fully complemented the clv1 phenotype, as did the reciprocal swap (CLV1pro::CLV1LRR:BAM1RLK, Fig. 3c). Therefore, the LRR and RLK domains encoded by CLV1 and BAM1 have conserved functions. HAE/HSL2 and their closest paralog HSL1 are separated by only 119 My of evolution (Fig. 3a; supplementary fig. S5, Supplementary Material online) but regulate distinct processes: HSL1 regulates stomatal development, and native HSL1 is neither necessary nor sufficient to drive hae;hsl2-regulated floral organ abscission (Cho et al. 2008; Qian et al. 2018). Despite this divergence in HSL1 function, and similar to our result for BAM1 and CLV1, the full HSL1 coding sequence driven under the HAE promoter can complement hae;hsl2 double mutants (Fig. 3c), consistent with what others have found (Roman et al. 2022). These data indicate that the failure of the endogenous HSL1 locus to trigger abscission in hae;hsl2 mutants is likely due to HSL1's divergent expression pattern, not the functional evolution of the HSL1 protein (Cho et al. 2008; Stenvik et al. 2008; Roman et al. 2022). Our results indicate that functional differences between CLV1 and BAM1, and between HAE/HSL2 and HSL1, are not due to changes in protein domain function, despite large differences in the divergence times among CLV1, HAE/HSL2, and their closest paralogs.
We next assessed complementation of the hae/hsl2 double mutant with domains encoded by HSL3, a gene separated from hae/hsl2 by ∼363 My of evolution. The full HSL3 coding sequence, expressed under the HAE promoter, failed to complement the hae;hsl2 phenotype, as did the chimeric transgene that had the LRR domain encoded by HSL3 and the RLK domain encoded by HAE (Fig. 3c). However, the reciprocal domain swap with the LRR domain encoded by HAE and the RLK domain encoded by HSL3 (HAEpro::HAELRR:HSL3RLK) partially complemented the hae;hsl2 phenotype, suggesting substantial functional conservation of the HAE/HSL2 and HSL3 RLK domains (Fig. 3c). To verify that catalytic activity of the RLK domain encoded by HSL3 was required for complementation, we created a catalytically dead variant by introducing a single amino acid change in the ATP binding pocket (K714E) (Taylor et al. 2016). This variant (HAEpro::HAELRR:HSL3RLK(K714E)) failed to complement the hae;hsl2 phenotype (Fig. 3c), indicating that the conserved catalytic activity of the HSL3 RLK domain is necessary for its partial replacement of HAE/HSL2. Taken together, these data are consistent with our bioinformatic findings (Figs. 1 and 2) and indicate that the LRR and RLK domains encoded by HAE/HSL2 and HSL3 have had different trajectories of functional divergence. Their LRR domains have diverged considerably and can no longer replace each other, while their RLK domains retain some conserved functionality.
Last, we assessed the ability of LRR and RLK domains encoded by deeply divergent paralogs to complement our focal mutants. We assessed the complementation of clv1 with domains encoded by PXY, HAE, and HAESA-LIKE3 (HSL3), representing ∼511 to 551 My of divergence (Fisher and Turner 2007). Chimeric constructs encoding LRR domains from HAE and PXY or with the RLK domain from HSL3 failed to complement the clv1 mutant. In contrast, the RLK domain encoded by HAE, together with the reciprocal CLV1 domain, partially complemented the clv1 phenotype (CLV1pro::CLV1LRR:HAERLK, Fig. 3c). Together, these data suggest that the HAE and CLV1 RLK domains retain vestiges of conserved function over deep time, while their LRR domains have completely diverged in function, mirroring their asymmetric rates of evolution.
Discussion
Sequences encoding the LRR domains of defense LRR-RLKs evolve under accelerated evolution with increased directional selection relative to their cognate RLK domains (Tang et al. 2010; Fischer et al. 2014, 2016; Dufayard et al. 2017; Parys et al. 2021; Zhang et al. 2021). Here, we show that this disjunct molecular evolution between LRR and RLK domains extends to subfamilies of clade XI LRR-RLKs with divergent functions in development, physiology, and defense. Sequences encoding LRR domains evolve consistently faster than their RLK counterparts (Fig. 1). This accelerated rate was likely not due to less stringent purifying selection: rates of purifying selection were similar in both domains (Fig. 2c). In contrast, rates of positive selection were higher in LRR domains (Fig. 2b). In heterologous assays of gene function, LRR domains encoded by HAE and HSL3 could not substitute for LRR domains encoded by CLV1 or HAE, respectively, while their RLK domains could partially substitute (Fig. 3c). Thus, the asymmetric molecular evolution of LRR and RLK domains is mirrored somewhat by asymmetric functional evolution.
Asymmetric domain evolution is a widespread feature of LRR-RLKs and plant signaling proteins. Some LRR-RLKs have dual roles in defense and development, and there may be undiscovered crosstalk driving some of the asymmetry in evolutionary rates between LRR and RLK domains in the “developmental” proteins we assayed (Lal et al. 2018; Rosas-Diaz et al. 2018; Verma et al. 2022). However, the disjunct evolution of LRR versus RLK domains that we detected across many clade XI subfamilies (Figs. 1 and 2) extends across clades of LRR-RLKs (X.S. Zhang et al. 2006; Fischer et al. 2016; Dufayard et al. 2017; Hosseini et al. 2020). More broadly, positive selection hits are more prevalent in the ectodomains of receptor-like kinase proteins from Charophyte algae, proteins with many diverse signaling roles (Gong and Han 2021). LRR domains are often found in association with nucleotide-binding domains in NLR proteins, which are critical regulators of plant defense (Baggs et al. 2017). Positive selection is higher in the LRR domains of NLR proteins (Mondragón-Palomino et al. 2002), although LRR-RLK evolution may be more constrained than NLR evolution on a genome-wide scale (Baggs et al. 2017; Bailey et al. 2018; Pruitt et al. 2021; Kileeg et al. 2023). Taken together, these data indicate that signaling and response domains in plant receptor proteins are subject to distinct evolutionary forces and constraints and are evolving somewhat independently.
Escape from adaptive conflict in LRR domains may be contributing to the functional evolution of LRR-RLKs. Cis-regulatory evolution can deploy conserved regulatory modules to additional, new contexts (Carroll 2008; Kramer and Li 2017). However, the dual use of a single gene in multiple contexts can incur adaptive conflict, in which genetic adaptations beneficial in one context are detrimental to another, constraining evolution (Hittinger and Carroll 2007; Flagel and Wendel 2009). Escape from adaptive conflict is the release of this constraint through gene duplications, followed by coding sequence evolution of one or both genes, allowing each gene to specialize in its respective function (Ohno 1970; Hughes 1994; Vogel and Chothia 2006). LRR-RLKs bear the hallmarks of a gene family in which escape from adaptive conflict drove functional evolution: LRR-RLKs regulate diverse developmental processes, recurrent gene duplication drove expansion of the gene family, and as we show here, they contain signatures of adaptive evolution, even in genes with deeply conserved developmental roles (Lehti-Shiu et al. 2012; Fischer et al. 2016; Dufayard et al. 2017; Man et al. 2020). In addition, divergent LRR-RLKs can share conserved outputs but have divergent inputs (Zheng et al. 2019; Zhu et al. 2019; Liu et al. 2022). Prior to gene duplication, there may be high adaptive conflict in LRR domains but low adaptive conflict in RLK domains. Gene duplication may allow for accelerated optimization of formerly conflicted LRR domains, leaving RLK domains largely conserved in function.
Results from our domain swap experiments with HAE/HSL2 and HSL3 are consistent with functional divergence through escape from adaptive conflict in single (LRR) domains. HAE/HSL2 and HSL3 are paralogs that control unrelated developmental functions—HAE and HSL2 regulate floral organ abscission (Jinn et al. 2000), while HSL3 regulates stomatal closure (Liu et al. 2020, 2022). The LRR domains of HAE and HSL2 bind IDA peptides, but the LRR domain of HSL3 binds structurally distinct, nonhomologous peptides called CTNIPs (Kumpf et al. 2013; Rhodes et al. 2022). In contrast to this divergent LRR binding, the RLK domains of HAE/HSL2 and HSL3 phosphorylate conserved downstream targets such as MPK3 and MPK6 (Zhu et al. 2019; Liu et al. 2022; Rhodes et al. 2022). The HSL3 LRR domain, in combination with the HAE RLK domain, failed to complement the hae;hsl2 mutant, but the HSL3 RLK domain did complement hae;hsl2 in the reciprocal domain swap (Fig. 3c). Together, these data indicate that the outputs of the HAE/HSL2 and HSL3 cytoplasmic signaling modules have not appreciably diverged in function since their duplication ∼363 Ma, but that their inputs have. Lower positive selection acting on the RLK domains of HAE/HSL2 and HSL3 conserved downstream targets, alongside some functional conservation (Fig. 3c), suggests low adaptive conflict in the output (RLK) domain of the last common ancestor of these proteins. In contrast, high levels of positive selection acting on their input (LRR) domains, nonconserved upstream triggers, and nonconservation of function indicate that diversification of the input was beneficial to fitness, perhaps because of escape from adaptive conflict.
Many mechanisms in addition to escape from adaptive conflict may contribute to evolutionary rate variation between LRR and RLK domains (Wolf et al. 2008, 2010; Zhang and Yang 2015; Echave et al. 2016; Roberts and Josephs 2023). Intracellular and extracellular domains of transmembrane proteins are folded in different cellular environments (the lumen of the endoplasmic reticulum vs. the cytosol, respectively), which may release extracellular (LRR) domains from evolutionary constraints related to protein misfolding (Pál et al. 2001; Drummond and Wilke 2008; Feyertag et al. 2017; Sarkar and Alvarez-Ponce 2022). Buried amino acid residues evolve slower than solvent-exposed residues (Lin et al. 2007; Franzosa and Xia 2009). RLK and LRR domains may have different ratios of buried versus solvent-exposed residues, thus contributing to evolutionary rate variation. Exon edge conservation can constrain sequence divergence (Bush et al. 2015). LRR domains are encoded by single exons, but RLK domain sequences span introns (Zan et al. 2013; Man et al. 2020); thus, exon edge conservation may be constraining RLK evolution. Recombination errors may drive variation in LRR repeat number within LRR domains (Kuang et al. 2004; Wicker et al. 2007), thus increasing the rate of LRR domain evolution (although we found no relationship between variation in LRR number and positive selection [supplementary fig. S3, Supplementary Material online]). Proteins with many interactors evolve slower than those with fewer interactors (Alvarez-Ponce et al. 2017). Both LRR and RLK domains mediate protein–protein interactions (Hohmann et al. 2017), but perhaps RLK domains have a richer set of interactors than LRR domains and thus evolve slower. Any number of these and other mechanisms may contribute to the faster divergence of LRR versus RLK domains, in addition to divergence in domain functions in signal perception and signal response.
Higher evolutionary rates and higher levels of positive selection suggest that LRR function is changing faster than RLK function both between and within subfamilies of LRR-RLKs. Within subfamilies, the evolution of LRR domains may be subtly modulating protein function over deep time. Although ligand–receptor pairs have not been confirmed beyond Arabidopsis for most LRR-RLKs, receptor–ligand relationships are deeply conserved in at least the CLV1 and HAE subfamilies (Clark et al. 1993; Jinn et al. 2000; Bommert et al. 2005; Santiago et al. 2016; Whitewoods et al. 2018; Wang et al. 2019). In addition, key residues that mediate receptor–ligand interactions are deeply conserved within LRR domains (Santiago et al. 2016; Hohmann et al. 2018; Rhodes et al. 2022; Snoeck et al. 2022). However, receptor–ligand relationships are rarely one-to-one (Deyoung and Clark 2008; Müller et al. 2008; Ou et al. 2016; Je et al. 2018; Qian et al. 2018; Rodriguez-Leal et al. 2019). The changes to LRR domains we detected may modulate which particular suite of ligands bind a particular LRR domain (e.g. which specific CLE paralogs bind which LRR domain) or modulate ligand-binding kinetics. In addition, interactions with coreceptors, often mediated by LRR domains (Cui et al. 2018; Hohmann et al. 2018), can modulate receptor trafficking to and from the membrane, and the outcomes of ligand–receptor binding (Nimchuk et al. 2011; Je et al. 2018; Qi et al. 2020). Future dissections of coevolving, positively selected residues between LRR-RLKs, and between LRR-RLKs and their ligands, may be useful in resolving complex interactions among receptors, coreceptors, and ligands. Thus, while the function of particular LRR-RLKs may be broadly conserved over deep time, rapid evolutionary change and positive selection may indicate incremental and subtle change to protein (and LRR domain) function.
Our work has implications for agriculture and crop engineering, where the subtle modulation of plant traits was critical in domestication, and is becoming useful in crop improvement using genome engineering. This subtle modulation is often accomplished through cis-regulatory change (Meyer and Purugganan 2013; Rodríguez-Leal et al. 2017; Stitzer and Ross-Ibarra 2018; Bartlett et al. 2022). However, coding changes have been important in plant evolution and development (Bartlett and Whipple 2013; Bartlett 2019; Bartlett et al. 2022). Coding sequence change could be more easily implemented and may have more predictable outcomes than cis-regulatory sequences, which are incompletely understood, especially in plants (Rodríguez-Leal et al. 2017; Marand et al. 2023). Importantly, the LRR-RLK protein family regulates many agronomically important aspects of plant development and has already been targeted in crop improvement efforts (Bommert et al. 2005; Xu et al. 2015; Je et al. 2016; Yang et al. 2018; Shao et al. 2019). More extensive or fine-scaled phenotypic analyses than our coarse endpoint phenotyping (e.g. measuring meristem or abscission zone size) may reveal subtle differences in function between LRR-RLK orthologs from diverse species. Detailed phenotypic and molecular dissections of how residues under selection impact protein function over deep time could provide novel solutions in crop improvement.
Materials and Methods
Sequence Retrieval and Curation
For each subfamily, we collected gene locus identifiers (Man et al. 2020), and peptide sequences were obtained from the primary transcript annotation databases for Arabidopsis thaliana, A. trichopoda, Brachypodium distachyon, Oryza sativa (rice), Solanum lycopersicum (tomato), Populus trichocarpa (poplar), Selaginella moellendorffii, and Physcomitrella patens, and Zea mays (maize) from Phytozome ver. 12 (Goodstein et al. 2012). We aligned these sequences using MAFFT v. 7.313 (Katoh and Standley 2013) and built a Hidden Markov Model (HMM) profile using HMMER v. 3.1b2 (Altschul et al. 1990; Eddy 2011). We used this HMM profile to scan every peptide database from the One Thousand Plants Transcriptome Initiative (1KP), and the top 2 scoring hits from each genome were collected (One Thousand Plant Transcriptomes Initiative 2019). We then scanned hits for protein domains using HMMER v3.1b2 and the Pfam protein profile HMM database v31.0 using the “trusted cutoff” bit score gathering threshold (Eddy 2011; El-Gebali et al. 2019). Only hits with detected LRR and RLK domains and with a length ≥85% of the Arabidopsis anchor gene were included. We aligned hits using the L-INS-i iterative refinement algorithm implemented in MAFFT v. 7.313 (Katoh and Standley 2013), filtered for homoplastic positions by Noisy v1.5.12 (Dress et al. 2008), and inferred a gene tree using IQtree v1.6.3 (Nguyen et al. 2015). We interpreted and visualized the tree using package ggtree v1.10.0 in R v3.4.3 (R Core Team 2017; Yu et al. 2017), and the 1KP genes falling into each target subfamily were iteratively used as seed sequences in another round of BLAST and HMM profiling until stable protein trees for each subfamily were inferred (Table 1).
Maximum Likelihood Subfamily Gene Tree Inference
For each subfamily, we built alignments using search hits and ERECTA (AT2G26330) as the outgroup using the L-INS-i iterative refinement algorithm implemented in MAFFT v. 7.313 (Katoh and Standley 2013). Next, we filtered for homoplastic positions using Noisy v1.5.12 (Dress et al. 2008) and used the alignment to determine the best-fitting model of protein evolution and infer a gene tree using maximum likelihood analysis with 1,000 bootstrap replicates using IQtree v1.6.3 (Nguyen et al. 2015).
Rate of Evolution by Site
Because the 1KP dataset was enriched for angiosperms and because we wanted densely sampled protein trees, we trimmed each alignment and tree to include only angiosperm peptide sequences, with A. trichopoda genes positioned as sister to all other sequences in each analysis (Mathews and Donoghue 1999; Soltis et al. 2008). For each subfamily, we aligned peptide sequences from each subfamily using the L-INS-i iterative refinement algorithm implemented in MAFFT v. 7.313 (Katoh and Standley 2013) and used this to infer the posterior mean site evolution rate with a Yule speciation prior and a random starting tree using the -wsr function in IQtree v1.6.3 (Nguyen et al. 2015). To prevent alignment artifacts from distorting the mean rate calculations, we removed positions from the alignment not present in the Arabidopsis gene from which the subfamily is named (anchor gene). We calculated and visualized the sliding window mean rate trace for each residue of the Arabidopsis anchor gene using a custom R script utilizing the packages Biostrings v.2.54.0 (Pagès et al. 2017), ggplot2 v3.3.0 (Wickham et al. 2019), and zoo v1.8-7 (Zeileis and Grothendieck 2005). LRR and RLK domain coordinates for the Arabidopsis anchor gene were taken from UniProt definitions (UniProt Consortium 2015). To calculate mean evolution rates across all subfamilies, we binned rates for each subfamily into 3 N-terminal, 20 LRR, 3 transmembrane, 10 RLK, and 2 C-terminal bins, shown fitted with a local polynomial regression with an α = 0.1.
Rate of Evolution by Domain
For each subfamily alignment, LRR and RLK partitions were defined based on the UniProt domain annotations of the subfamily's Arabidopsis anchor gene (UniProt Consortium 2015). We performed Bayesian analyses using BEAST v2.6.1 (Bouckaert et al. 2019) on the CIPRES Science Gateway (Miller et al. 2010). In these analyses, the peptide sequence alignment is partitioned into LRR and RLK domains, and the tree and partition substitution rates are coestimated and then weighted over tree-space posterior probabilities to give a rate distribution estimate per domain. To infer relative domain rates, we used the following parameters: tree and clock models, but not site models, were linked during the analysis, to provide independent site rate estimates for the partitions. A gamma site model with 6 discrete estimated rate categories was set for each partition to ensure appropriate gamma rate heterogeneity, using a JTT amino acid substitution matrix which was found to best fit LRR-RLKs (Susko et al. 2002; Man et al. 2020). Strict and relaxed molecular clock models were compared using a nested sampling approach to generate marginal likelihoods for Bayes Factor estimation and a likelihood ratio test (Kass and Raftery 1995; Brown and Yang 2011; Russel et al. 2019). Based on these tests, we proceeded using a relaxed clock model, which was subsequently used for all further analyses. Subfamily trees were then inferred under a Calibrated Yule model and a birth–death model and had statistically identical Maximum Clade Credibility trees as calculated by Tree Annotator v2.6.2 (Bouckaert et al. 2019); therefore, Birth–death models were used in all subfamilies as in (Magallón et al. 2015). The node containing all major angiosperm subfamilies other than A. trichopoda was calibrated to 135 to 137 Ma (lognormal prior distribution, 95% posterior density credibility interval; Magallón et al. 2015). MCMC chains were run for sufficient length to ensure complete mixing and convergence of molecular clock rate estimate and other parameters were used, thresholded by Effective Sample Sizes (ESS) >100, measured using Tracer v1.6 (Rambaut et al. 2018). Within each subfamily, rate estimate distributions did not have equivalent variances (F-test, P < 0.001 for all subfamilies) and were therefore compared using nonparametric unpaired Wilcoxon tests.
Selection Tests
We performed tests looking for signatures of selection using the nucleotide sequences corresponding to the peptide MSAs used to infer evolutionary rates. We aligned nucleotide sequences by their codon translations using the BLOSUM45 scoring matrix in MAFFT v. 7.313 (Katoh and Standley 2013) as implemented in Geneious v10.0.8 (Kearse et al. 2012). These MSAs were used to run tests in the HyPhy suite (Pond et al. 2005): FUBAR (Murrell et al. 2013) and MEME (Murrell et al. 2012), as implemented on the Datamonkey web server (Weaver et al. 2018). MEME hits were thresholded at the default P-value of 0.1, and FUBAR hits were thresholded at a more stringent threshold of 0.999 posterior probability. Site-by-site selection evidence from these tests was visualized using a custom R script utilizing ggplot2 v3.3.0 (R Core Team 2017; Wickham et al. 2019). The alternate GSO2 selection tests utilized only CDS sequences from genomic assemblies available from homologs detected from Ensembl Plants (Kersey et al. 2018).
Chronogram for Inference of Gene Subfamily Divergence Times and Tree Topology
Four genes from each subfamily were extracted: the A. trichopoda gene, and a representative from rosids, asterids, and monocots, generally A. thaliana, tomato, and rice, and well-supported S. moellendorffii and P. patens genes that are sister to these subfamilies. We aligned these using the L-INS-i iterative refinement algorithm implemented in MAFFT v. 7.313 (Katoh and Standley 2013). We used this alignment to perform Bayesian analyses using BEAST v2.6.1 (Bouckaert et al. 2019) on the CIPRES Science Gateway (Miller et al. 2010). This analysis was run as a single partition, using the JTT substitution matrix, and a relaxed clock model. Priors included a Yule tree model and Most Recent Common Ancestor (MRCA) nodes: eudicot MRCA was uniform 111 to 131 My, angiosperm MRCA was uniform 173 to 199 My, tracheophyte MRCA was uniform 410 to 468 My, and embryophyte MRCA was uniform 465 to 533 My (Magallón et al. 2015; Kumar et al. 2017). MCMC chains were run for sufficient length to ensure complete mixing and convergence of molecular clock rate estimate and other parameters were used, thresholded by ESS > 100, measured using Tracer v1.6 (Rambaut et al. 2018). The final tree was visualized using FigTree v1.4.3 (Bouckaert et al. 2019; supplementary fig. S2, Supplementary Material online) or ggtree v1.10.0 in R v3.4.3 (R Core Team 2017; Yu et al. 2017) (tree backbone Fig. 1).
Construction of Chimeric Transgenes
We utilized the MoClo system of Golden-Gate molecular assembly to make full-length domain swaps constructs using sequences encoding promoters, LRR domains, RLK domains, and terminators (Engler et al. 2014). For CLV1 promoter constructs, we replicated the promoter/5′UTR and terminator/3′UTR coordinates published previously (Nimchuk et al. 2011). For HAE promoter constructs, we determined the promoter/5′UTR by conservation with other species using Vista plots in Phytozome's JBrowse (Goodstein et al. 2012) to be 1,609 bp upstream the gene's start codon and utilized the terminator from the nos gene in Agrobacterium tumefaciens (Engler et al. 2014). The sequence encoding protein domains was amplified using PCR with Col-0 gDNA template or synthesized commercially. We assembled all transgenic constructs into either the pICH86966 T-DNA plant transformation vector from the MoClo kit (Engler et al. 2014) or a plasmid derived from pH7m24GW (O’Connor et al. 2017) and verified by Sanger sequencing or whole plasmid sequencing. To standardize domain coordinates across genes used for domain swaps, we made a translation alignment between all genes and engineered a synthetic 4-bp Golden-Gate assembly joining overlap within the transmembrane domain that introduced only synonymous coding alterations in all of the genes used in our assay.
Plant Growth Conditions and Plant Transformation
Seeds were sown on the surface of soilless medium in 3″ pots, watered, and stratified covered in dark at 4°C for 3 d and then transferred to a growing rig (photosynthetic active radiation ∼100 μmol/m2/s), with a 16 h light and 8 h dark cycle; constant temperature ∼20°C. clv1-15 (WiscDsLox489-492B1) was kindly provided by Zachary Nimchuck (Nimchuk et al. 2011). hsl2-3/hae-3 (CS69822) seed was obtained from the Arabidopsis Biological Resource Center (Scholl and Anderson 1994). Both mutants are in the Columbia background. Into these plant lines, we transfected LRR-RLK constructs using A. tumefaciens strain GV3101 with the floral dip method (X. Zhang et al. 2006). Transgenic T1 seeds were selected according to (Davis et al. 2009) and transplanted from selection plates to soilless medium in pots at 2 wk of age and grown to maturity.
Phenotypic Scoring
All T1 plants were phenotyped as soon as the oldest 10 flowers on the primary inflorescence could be scored for abscission of floral organs (for hae;hsl2 rescue constructs) or carpel number (for clv1 rescue constructs). Events from HAE constructs were scored by lightly brushing the inflorescence twice, with a 90° turn between, through a pair of soft bristle paint brushes affixed with bristles oriented toward each other. After brushing, we assessed hae;hsl2 double mutant complementation by binning each flower into 1 of 3 categories: fully complemented (all floral organs abscised), partially complemented (some abscised, some remain attached to pedicel, complementation strength of 1), or not complemented (no floral organs abscised).
CLV1 construct transformants were scored by counting carpels in each of up to 10 siliques from each mature transformant (8 to 24 independent T1 lines) using a dissecting microscope. Flowers were binned into 3 categories: fully complemented (two carpels per silique), partially complemented (three carpels per silique), or not complemented (four or more carpels per silique).
Supplementary material
Supplementary material is available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank Courtney Babbitt, Remco Bouckaert, Heather Bracken-Grissom, Joseph Gallagher, Jason Kamilar, Zachary Nimchuk, and Devin O’Connor, for technical support, helpful discussion, reagents, and seed. Amber de Neve, Joseph Gallagher, Erin Patterson, and 2 anonymous reviewers provided helpful comments on an earlier version of the manuscript.
Contributor Information
Jarrett Man, Department of Biology, University of Massachusetts Amherst, Amherst, MA 01002, USA.
T A Harrington, Department of Biology, University of Massachusetts Amherst, Amherst, MA 01002, USA.
Kyra Lally, Department of Biology, University of Massachusetts Amherst, Amherst, MA 01002, USA.
Madelaine E Bartlett, Department of Biology, University of Massachusetts Amherst, Amherst, MA 01002, USA.
Funding
This work was supported by the National Science Foundation (IOS-2129189 and IOS-1546837 to M.E.B.), a Lotta Crabtree fellowship (J.M.), a UMass Biotechnology Training Program fellowship (T.A.H.), and the UMass Center for Agriculture, Food, and the Environment.
Data Availability
All alignments, trees, rate test results, selection test results, and phenotyping data are available at dryad (doi:10.5061/dryad.jm63xsjg5). Code for analyses is available on the Bartlett lab's GitHub (https://github.com/BartlettLab/Domain_evolution).
References
- Ahmad HI, Afzal G, Iqbal MN, Iqbal MA, Shokrollahi B, Mansoor MK, Chen J. Positive selection drives the adaptive evolution of mitochondrial antiviral signaling (MAVS) proteins-mediating innate immunity in mammals. Front Vet Sci. 2021:8:814765. 10.3389/fvets.2021.814765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990:215(3):403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alvarez-Ponce D, Feyertag F, Chakraborty S. Position matters: network centrality considerably impacts rates of protein evolution in the human protein-protein interaction network. Genome Biol Evol. 2017:9(6):1742–1756. 10.1093/gbe/evx117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Hall BK. A model for development and evolution of complex morphological structures. Biol Rev Camb Philos Soc. 1991:66(2):101–157. 10.1111/j.1469-185X.1991.tb01138.x. [DOI] [PubMed] [Google Scholar]
- Baggs E, Dagdas G, Krasileva KV. NLR diversity, helpers and integrated domains: making sense of the NLR IDentity. Curr Opin Plant Biol. 2017:38:59–67. 10.1016/j.pbi.2017.04.012. [DOI] [PubMed] [Google Scholar]
- Bailey PC, Schudoma C, Jackson W, Baggs E, Dagdas G, Haerty W, Moscou M, Krasileva KV. Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 2018:19(1):23. 10.1186/s13059-018-1392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett M. Looking back to look forward: protein–protein interactions and the evolution of development. New Phytol. 2019:225(3):1127–1133. 10.1111/nph.16179. [DOI] [PubMed] [Google Scholar]
- Bartlett ME, Moyers BT, Man J, Subramaniam B, Makunga NP. The power and perils of de novo domestication using genome editing. Annu Rev Plant Biol. 2022:74(1):727–750. 10.1146/annurev-arplant-053122-030653. [DOI] [PubMed] [Google Scholar]
- Bartlett ME, Whipple CJ. Protein change in plant evolution: tracing one thread connecting molecular and phenotypic diversity. Front Plant Sci. 2013:4:382. 10.3389/fpls.2013.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya RP, Reményi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006:75(1):655–680. 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- Bishop JG, Dean AM, Mitchell-Olds T. Rapid evolution in plant chitinases: molecular targets of selection in plant-pathogen coevolution. Proc Natl Acad Sci U S A. 2000:97(10):5322–5327. 10.1073/pnas.97.10.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, Hake S, Werr W. Thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development. 2005:132(6):1235–1245. 10.1242/dev.01671. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014:10(4):e1003537. 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, et al. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2019:15(4):e1006650. 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RP, Yang Z. Rate variation and estimation of divergence times using strict and relaxed clocks. BMC Evol Biol. 2011:11(1):271. 10.1186/1471-2148-11-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci U S A. 2010:107(20):9452–9457. 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush SJ, Kover PX, Urrutia AO. Lineage-specific sequence evolution and exon edge conservation partially explain the relationship between evolutionary rate and expression level in A. thaliana. Mol Ecol. 2015:24(12):3093–3106. 10.1111/mec.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008:134(1):25–36. 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Chen T. Identification and characterization of the LRR repeats in plant LRR-RLKs. BMC Mol Cell Biol. 2021:22(1):9. 10.1186/s12860-021-00344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006:18(2):465–476. 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn T-L, Zhang S, Walker JC. Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008:105(40):15629–15634. 10.1073/pnas.0805539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993:119(2):397–418. 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997:89(4):575–585. 10.1016/S0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Cui Y, Hu C, Zhu Y, Cheng K, Li X, Wei Z, Xue L, Lin F, Shi H, Yi J, et al. CIK receptor kinases determine cell fate specification during early anther development in Arabidopsis. Plant Cell. 2018:30(10):2383–2401. 10.1105/tpc.17.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AM, Hall A, Millar AJ, Darrah C, Davis SJ. Protocol: streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods. 2009:5(1):3. 10.1186/1746-4811-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mita S, Streng A, Bisseling T, Geurts R. Evolution of a symbiotic receptor through gene duplications in the legume–rhizobium mutualism. New Phytol. 2014:201(3):961–972. 10.1111/nph.12549. [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006:45(1):1–16. 10.1111/j.1365-313X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- Deyoung BJ, Clark SE. BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics. 2008:180(2):895–904. 10.1534/genetics.108.091108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diévart A, Hymes MJ, Li J, Clark SE. Brassinosteroid-independent function of BRI1/CLV1 chimeric receptors. Funct Plant Biol. 2006:33(8):723–730. 10.1071/FP06080. [DOI] [PubMed] [Google Scholar]
- Di Roberto RB, Peisajovich SG. The role of domain shuffling in the evolution of signaling networks. J Exp Zool B Mol Dev Evol. 2014:322(2):65–72. 10.1002/jez.b.22551. [DOI] [PubMed] [Google Scholar]
- Dress AWM, Flamm C, Fritzsch G, Grünewald S, Kruspe M, Prohaska SJ, Stadler PF. Noisy: identification of problematic columns in multiple sequence alignments. Algorithms Mol Biol. 2008:3(1):7. 10.1186/1748-7188-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008:134(2):341–352. 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufayard J-F, Bettembourg M, Fischer I, Droc G, Guiderdoni E, Périn C, Chantret N, Diévart A. New insights on leucine-rich repeats receptor-like kinase orthologous relationships in angiosperms. Front Plant Sci. 2017:8:381. 10.3389/fpls.2017.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echave J, Spielman SJ, Wilke CO. Causes of evolutionary rate variation among protein sites. Nat Rev Genet. 2016:17(2):109–121. 10.1038/nrg.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011:7(10):e1002195. 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019:47(D1):D427–D432. 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert T-M, Werner S, Jones JDG, Patron NJ, Marillonnet S. A golden gate modular cloning toolbox for plants. ACS Synth Biol. 2014:3(11):839–843. 10.1021/sb4001504. [DOI] [PubMed] [Google Scholar]
- Feyertag F, Berninsone PM, Alvarez-Ponce D. Secreted proteins defy the expression level-evolutionary rate anticorrelation. Mol Biol Evol. 2017:34(3):692–706. 10.1093/molbev/msw268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer I, Dainat J, Ranwez V, Glémin S, Dufayard J-F, Chantret N. Impact of recurrent gene duplication on adaptation of plant genomes. BMC Plant Biol. 2014:14(1):151. 10.1186/1471-2229-14-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer I, Diévart A, Droc G, Dufayard J-F, Chantret N. Evolutionary dynamics of the leucine-rich repeat receptor-like kinase (LRR-RLK) subfamily in angiosperms. Plant Physiol. 2016:170(3):1595–1610. 10.1104/pp.15.01470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher K, Turner S. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr Biol. 2007:17(12):1061–1066. 10.1016/j.cub.2007.05.049. [DOI] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009:183(3):557–564. 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- Franzosa EA, Xia Y. Structural determinants of protein evolution are context-sensitive at the residue level. Mol Biol Evol. 2009:26(10):2387–2395. 10.1093/molbev/msp146. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Basak S, Dutta S. Natural selection shaped the evolution of amino acid usage in mammalian toll like receptor genes. Comput Biol Chem. 2022:97:107637. 10.1016/j.compbiolchem.2022.107637. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009:19(5):423–429. 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000:5(6):1003–1011. 10.1016/S1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Gong Z, Han G-Z. Flourishing in water: the early evolution and diversification of plant receptor-like kinases. Plant J. 2021:106(1):174–184. 10.1111/tpj.15157. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012:40(D1):D1178–D1186. 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci U S A. 2008:105(39):15208–15213. 10.1073/pnas.0808444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007:449(7163):677–681. 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- Hogenhout SA, Van der Hoorn RAL, Terauchi R, Kamoun S. Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact. 2009:22(2):115–122. 10.1094/MPMI-22-2-0115. [DOI] [PubMed] [Google Scholar]
- Hohmann U, Lau K, Hothorn M. The structural basis of ligand perception and signal activation by receptor kinases. Annu Rev Plant Biol. 2017:68(1):109–137. 10.1146/annurev-arplant-042916-040957. [DOI] [PubMed] [Google Scholar]
- Hohmann U, Ramakrishna P, Wang K, Lorenzo-Orts L, Nicolet J, Henschen A, Barberon M, Bayer M, Hothorn M. Constitutive activation of leucine-rich repeat receptor kinase signaling pathways by BAK1-interacting receptor-like kinase 3 chimera. Plant Cell. 2020:32(10):3311–3323. 10.1105/tpc.20.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann U, Santiago J, Nicolet J, Olsson V, Spiga FM, Hothorn LA, Butenko MA, Hothorn M. Mechanistic basis for the activation of plant membrane receptor kinases by SERK-family coreceptors. Proc Natl Acad Sci U S A. 2018:115(13):3488–3493. 10.1073/pnas.1714972115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini S, Schmidt EDL, Bakker FT. Leucine-rich repeat receptor-like kinase II phylogenetics reveals five main clades throughout the plant kingdom. Plant J. 2020:103(2):547–560. 10.1111/tpj.14749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc Biol Sci. 1994:256(1346):119–124. 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Je BI, Gruel J, Lee YK, Bommert P, Arevalo ED, Eveland AL, Wu Q, Goldshmidt A, Meeley R, Bartlett M, et al. Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat Genet. 2016:48(7):785–791. 10.1038/ng.3567. [DOI] [PubMed] [Google Scholar]
- Je BI, Xu F, Wu Q, Liu L, Meeley R, Gallagher JP, Corcilius L, Payne RJ, Bartlett ME, Jackson D. The CLAVATA receptor FASCIATED EAR2 responds to distinct CLE peptides by signaling through two downstream effectors. Elife. 2018:7:e35673. 10.7554/eLife.35673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000:14(1):108–117. 10.1101/gad.14.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995:90(430):773–795. 10.1080/01621459.1995.10476572. [DOI] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013:30(4):772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012:28(12):1647–1649. 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Allen JE, Allot A, Barba M, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Grabmueller C, et al. Ensembl genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018:46(D1):D802–D808. 10.1093/nar/gkx1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kileeg Z, Haldar A, Khan H, Qamar A, Adam Mott G. Differential expansion and retention patterns of LRR-RLK genes across plant evolution. bioRxiv 549740. 10.1101/2023.07.26.549740v1. 26 July 2023, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975:188(4184):107–116. 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005:22(5):1208–1222. 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Li W. A transcriptomics and comparative genomics analysis reveals gene families with a role in body plan complexity. Front Plant Sci. 2017:8:869. 10.3389/fpls.2017.00869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Soltis DE, Kersey PJ, Wegrzyn JL, Leebens-Mack JH, Gostel MR, Liu X, Soltis PS. Green plant genomes: what we know in an era of rapidly expanding opportunities. Proc Natl Acad Sci U S A . 2022:119(4):e2115640118. 10.1073/pnas.2115640118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang H, Woo S-S, Meyers BC, Nevo E, Michelmore RW. Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell. 2004:16(11):2870–2894. 10.1105/tpc.104.025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 2017:34(7):1812–1819. 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Kumpf RP, Shi C-L, Larrieu A, Stø IM, Butenko MA, Péret B, Riiser ES, Bennett MJ, Aalen RB. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc Natl Acad Sci U S A. 2013:110(13):5235–5240. 10.1073/pnas.1210835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal NK, Nagalakshmi U, Hurlburt NK, Flores R, Bak A, Sone P, Ma X, Song G, Walley J, Shan L, et al. The receptor-like cytoplasmic kinase BIK1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host Microbe. 2018:23:485–497.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Zou C, Shiu S-H. Origin, diversity, expansion history, and functional evolution of the plant receptor-like kinase/pelle family. In: Tax F, Kemmerling B, editors. Receptor-like kinases in plants: from development to defense. Berlin, Heidelberg: Springer; 2012. p. 1–22. [Google Scholar]
- Lin Y-S, Hsu W-L, Hwang J-K, Li W-H. Proportion of solvent-exposed amino acids in a protein and rate of protein evolution. Mol Biol Evol. 2007:24(4):1005–1011. 10.1093/molbev/msm019. [DOI] [PubMed] [Google Scholar]
- Liu P-L, Du L, Huang Y, Gao S-M, Yu M. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol Biol. 2017:17(1):47. 10.1186/s12862-017-0891-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Hou S, Rodrigues O, Wang P, Luo D, Munemasa S, Lei J, Liu J, Ortiz-Morea FA, Wang X, et al. Phytocytokine signalling reopens stomata in plant immunity and water loss. Nature. 2022:605(7909):332–339. 10.1038/s41586-022-04684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X-S, Liang C-C, Hou S-G, Wang X, Chen D-H, Shen J-L, Zhang W, Wang M. The LRR-RLK protein HSL3 regulates stomatal closure and the drought stress response by modulating hydrogen peroxide homeostasis. Front Plant Sci. 2020:11:548034. 10.3389/fpls.2020.548034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou J-M. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci U S A. 2013:110(15):6205–6210. 10.1073/pnas.1215543110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann GV, Shimoda Y, Nielsen MW, Jørgensen FG, Grossmann C, Sandal N, Sørensen K, Thirup S, Madsen LH, Tabata S, et al. Evolution and regulation of the Lotus japonicus LysM receptor gene family. Mol Plant Microbe Interact. 2010:23(4):510–521. 10.1094/MPMI-23-4-0510. [DOI] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016:167(5):1170–1187. 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci U S A. 2005:102(48):17531–17536. 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 2015:207(2):437–453. 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- Man J, Gallagher JP, Bartlett M. Structural evolution drives diversification of the large LRR-RLK gene family. New Phytol. 2020:226(5):1492–1505. 10.1111/nph.16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marand AP, Eveland AL, Kaufmann K, Springer NM. cis-Regulatory elements in plant development, adaptation, and evolution. Annu Rev Plant Biol. 2023:74(1):111–137. 10.1146/annurev-arplant-070122-030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S, Donoghue MJ. The root of angiosperm phylogeny inferred from duplicate phytochrome genes. Science. 1999:286(5441):947–950. 10.1126/science.286.5441.947. [DOI] [PubMed] [Google Scholar]
- Meyer RS, Purugganan MD. Evolution of crop species: genetics of domestication and diversification. Nat Rev Genet. 2013:14(12):840–852. 10.1038/nrg3605. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop. New Orleans, LA; 2010. p. 1–8. 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2007:104(49):19613–19618. 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón-Palomino M, Meyers BC, Michelmore RW, Gaut BS. Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 2002:12(9):1305–1315. 10.1101/gr.159402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008:20(4):934–946. 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. FUBAR: a fast, unconstrained Bayesian approximation for inferring selection. Mol Biol Evol. 2013:30(5):1196–1205. 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012:8(7):e1002764. 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015:32(1):268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk ZL. CLAVATA1 controls distinct signaling outputs that buffer shoot stem cell proliferation through a two-step transcriptional compensation loop. PLoS Genet. 2017:13(3):e1006681. 10.1371/journal.pgen.1006681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr Biol. 2011:21(5):345–352. 10.1016/j.cub.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk ZL, Zhou Y, Tarr PT, Peterson BA, Meyerowitz EM. Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development. 2015:142(6):1043–1049. 10.1242/dev.119677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor DL, Elton S, Ticchiarelli F, Hsia MM, Vogel JP, Leyser O. Cross-species functional diversity within the PIN auxin efflux protein family. Elife. 2017:6:e31804. 10.7554/eLife.31804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- One Thousand Plant Transcriptomes Initiative . One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019:574(7780):679–685. 10.1038/s41586-019-1693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell. 2005:17(4):1105–1119. 10.1105/tpc.104.027474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Lu X, Zi Q, Xun Q, Zhang J, Wu Y, Shi H, Wei Z, Zhao B, Zhang X, et al. RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana. Cell Res. 2016:26(6):686–698. 10.1038/cr.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès H, Aboyoun P, Gentleman R, DebRoy S. 2017. Biostrings: efficient manipulation of biological strings. R package version 2.
- Pál C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001:158(2):927–931. 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parys K, Colaianni NR, Lee H-S, Hohmann U, Edelbacher N, Trgovcevic A, Blahovska Z, Lee D, Mechtler A, Muhari-Portik Z, et al. Signatures of antagonistic pleiotropy in a bacterial flagellin epitope. Cell Host Microbe. 2021:29(4):620–634.e9. 10.1016/j.chom.2021.02.008. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005:21(5):676–679. 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Poon AFY, Frost SDW, Pond SLK. Detecting signatures of selection from DNA sequences using Datamonkey. Methods Mol Biol. 2009:537:163–183. 10.1007/978-1-59745-251-9_8. [DOI] [PubMed] [Google Scholar]
- Pruitt RN, Locci F, Wanke F, Zhang L, Saile SC, Joe A, Karelina D, Hua C, Fröhlich K, Wan W-L, et al. The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature. 2021:598(7881):495–499. 10.1038/s41586-021-03829-0. [DOI] [PubMed] [Google Scholar]
- Qi X, Yoshinari A, Bai P, Maes M, Zeng SM, Torii KU. The manifold actions of signaling peptides on subcellular dynamics of a receptor specify stomatal cell fate. Elife. 2020:9:e58097. 10.7554/eLife.58097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian P, Song W, Yokoo T, Minobe A, Wang G, Ishida T, Sawa S, Chai J, Kakimoto T. The CLE9/10 secretory peptide regulates stomatal and vascular development through distinct receptors. Nat Plants. 2018:4(12):1071–1081. 10.1038/s41477-018-0317-4. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018:67(5):901–904. 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing. Vienna: (Austria: ): R Foundation for Statistical Computing; 2017. [Google Scholar]
- Rhodes J, Roman A-O, Bjornson M, Brandt B, Derbyshire P, Wyler M, Schmid MW, Menke FLH, Santiago J, Zipfel C. Perception of a conserved family of plant signalling peptides by the receptor kinase HSL3. Elife. 2022:11:e74687. 10.7554/eLife.74687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M, Josephs EB. Weaker selection on genes with treatment-specific expression consistent with a limit on plasticity evolution in Arabidopsis thaliana. Genetics. 2023:224(2):iyad074. 10.1093/genetics/iyad074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017:171(2):470–480. 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Xu C, Kwon C-T, Soyars C, Demesa-Arevalo E, Man J, Liu L, Lemmon ZH, Jones DS, Van Eck J, et al. Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat Genet. 2019:51(5):786–792. 10.1038/s41588-019-0389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman A-O, Jimenez-Sandoval P, Augustin S, Broyart C, Hothorn LA, Santiago J. HSL1 and BAM1/2 impact epidermal cell development by sensing distinct signaling peptides. Nat Commun. 2022:13(1):876. 10.1038/s41467-022-28558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Diaz T, Zhang D, Fan P, Wang L, Ding X, Jiang Y, Jimenez-Gongora T, Medina-Puche L, Zhao X, Feng Z, et al. A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. Proc Natl Acad Sci U S A. 2018:115:1388–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel PM, Brewer BJ, Klaere S, Bouckaert RR. Model selection and parameter inference in phylogenetics using nested sampling. Syst Biol. 2019:68(2):219–233. 10.1093/sysbio/syy050. [DOI] [PubMed] [Google Scholar]
- Santiago J, Brandt B, Wildhagen M, Hohmann U, Hothorn LA, Butenko MA, Hothorn M. Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. Elife. 2016:5:e15075. 10.7554/eLife.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar C, Alvarez-Ponce D. Extracellular domains of transmembrane proteins defy the expression level-evolutionary rate anticorrelation. Genome Biol Evol. 2022:14(1):evab235. 10.1093/gbe/evab235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato PM, Yoganathan K, Jung JH, Peisajovich SG. The robustness of a signaling complex to domain rearrangements facilitates network evolution. PLoS Biol. 2014:12(12):e1002012. 10.1371/journal.pbio.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauquet H, Ramírez-Barahona S, Magallón S. What is the age of flowering plants? J Exp Bot. 2022:73(12):3840–3853. 10.1093/jxb/erac130. [DOI] [PubMed] [Google Scholar]
- Scholl R, Anderson M. Arabidopsis biological resource center. Plant Mol Biol Rep. 1994:12(3):242–244. 10.1007/BF02668747. [DOI] [Google Scholar]
- Shao G, Lu Z, Xiong J, Wang B, Jing Y, Meng X, Liu G, Ma H, Liang Y, Chen F, et al. Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in rice. Mol Plant. 2019:12(8):1090–1102. 10.1016/j.molp.2019.04.008. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE. 2001:2001(113):re22. 10.1126/stke.2001.113.re22. [DOI] [PubMed] [Google Scholar]
- Snoeck S, Abramson BW, Garcia AGK, Egan AN, Michael TP, Steinbrenner AD. Evolutionary gain and loss of a plant pattern-recognition receptor for HAMP recognition. Elife. 2022:11:e81050. 10.7554/eLife.81050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Bell CD, Kim S, Soltis PS. Origin and early evolution of angiosperms. Ann N Y Acad Sci. 2008:1133(1):3–25. 10.1196/annals.1438.005. [DOI] [PubMed] [Google Scholar]
- Stenvik G-E, Tandstad NM, Guo Y, Shi C-L, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell. 2008:20(7):1805–1817. 10.1105/tpc.108.059139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzer MC, Ross-Ibarra J. Maize domestication and gene interaction. New Phytol. 2018:220(2):395–408. 10.1111/nph.15350. [DOI] [PubMed] [Google Scholar]
- Susko E, Inagaki Y, Field C, Holder ME, Roger AJ. Testing for differences in rates-across-sites distributions in phylogenetic subtrees. Mol Biol Evol. 2002:19(9):1514–1523. 10.1093/oxfordjournals.molbev.a004214. [DOI] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science. 2014:346(6207):343–346. 10.1126/science.1257800. [DOI] [PubMed] [Google Scholar]
- Tang P, Zhang Y, Sun X, Tian D, Yang S, Ding J. Disease resistance signature of the leucine-rich repeat receptor-like kinase genes in four plant species. Plant Sci. 2010:179(4):399–406. 10.1016/j.plantsci.2010.06.017. [DOI] [Google Scholar]
- Taylor I, Wang Y, Seitz K, Baer J, Bennewitz S, Mooney BP, Walker JC. Analysis of phosphorylation of the receptor-like protein kinase HAESA during Arabidopsis floral abscission. PLoS One. 2016:11(1):e0147203. 10.1371/journal.pone.0147203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuwamoto R, Fukuoka H, Takahata Y. GASSHO1 and GASSHO2 encoding a putative leucine-rich repeat transmembrane-type receptor kinase are essential for the normal development of the epidermal surface in Arabidopsis embryos. Plant J. 2008:54(1):30–42. 10.1111/j.1365-313X.2007.03395.x. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium . UniProt: a hub for protein information. Nucleic Acids Res. 2015:43(D1):D204–D212. 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A, Lin M, Smith D, Walker JC, Hewezi T, Davis EL, Hussey RS, Baum TJ, Mitchum MG. A novel sugar beet cyst nematode effector 2D01 targets the Arabidopsis HAESA receptor-like kinase. Mol Plant Pathol. 2022:23:1765–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Chothia C. Protein family expansions and biological complexity. PLoS Comput Biol. 2006:2(5):e48. 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Sha Y, Hu J, Yang T, Piao X, Zhang X. Genetic signatures of plant resistance genes with known function within and between species. Genetica. 2018:146(6):517–528. 10.1007/s10709-018-0044-9. [DOI] [PubMed] [Google Scholar]
- Wang F, Zheng Z, Yuan Y, Li J, Zhao M. Identification and characterization of HAESA-like genes involved in the fruitlet abscission in litchi. Int J Mol Sci. 2019:20(23):5945. 10.3390/ijms20235945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S, Shank SD, Spielman SJ, Li M, Muse SV, Kosakovsky Pond SL. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol Biol Evol. 2018:35(3):773–777. 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitewoods CD, Cammarata J, Nemec Venza Z, Sang S, Crook AD, Aoyama T, Wang XY, Waller M, Kamisugi Y, Cuming AC, et al. CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Curr Biol. 2018:28(15):2365–2376.e5. 10.1016/j.cub.2018.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Yahiaoui N, Keller B. Illegitimate recombination is a major evolutionary mechanism for initiating size variation in plant resistance genes. Plant J. 2007:51(4):631–641. 10.1111/j.1365-313X.2007.03164.x. [DOI] [PubMed] [Google Scholar]
- Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, et al. Welcome to the Tidyverse. J Open Source Softw. 2019:4(43):1686. 10.21105/joss.01686. [DOI] [Google Scholar]
- Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2011:13(1):59–69. 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Gopich IV, Lipman DJ, Koonin EV. Relative contributions of intrinsic structural-functional constraints and translation rate to the evolution of protein-coding genes. Genome Biol Evol. 2010:2:190–199. 10.1093/gbe/evq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf MY, Wolf YI, Koonin EV. Comparable contributions of structural-functional constraints and expression level to the rate of protein sequence evolution. Biol Direct. 2008:3(1):40. 10.1186/1745-6150-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Liberatore KL, MacAlister CA, Huang Z, Chu Y-H, Jiang K, Brooks C, Ogawa-Ohnishi M, Xiong G, Pauly M, et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat Genet. 2015:47(7):784–792. 10.1038/ng.3309. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhu K, Li H, Han S, Meng Q, Khan SU, Fan C, Xie K, Zhou Y. Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol J. 2018:16(7):1322–1335. 10.1111/pbi.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017:8(1):28–36. 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- Zan Y, Ji Y, Zhang Y, Yang S, Song Y, Wang J. Genome-wide identification, characterization and expression analysis of populus leucine-rich repeat receptor-like protein kinase genes. BMC Genomics. 2013:14(1):318. 10.1186/1471-2164-14-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeileis A, Grothendieck G. 2005. zoo: S3 infrastructure for regular and irregular time series. arXiv, arXiv:0505527, preprint: not peer reviewed.
- Zhang XS, Choi JH, Heinz J, Chetty CS. Domain-specific positive selection contributes to the evolution of Arabidopsis leucine-rich repeat receptor-like kinase (LRR RLK) genes. J Mol Evol. 2006:63(5):612–621. 10.1007/s00239-005-0187-z. [DOI] [PubMed] [Google Scholar]
- Zhang X, Dong W, Sun J, Feng F, Deng Y, He Z, Oldroyd GED, Wang E. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 2015:81(2):258–267. 10.1111/tpj.12723. [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin S-S, Niu Q-W, Chua N-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006:1(2):641–646. 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- Zhang L, Hua C, Pruitt RN, Qin S, Wang L, Albert I, Albert M, van Kan JAL, Nürnberger T. Distinct immune sensor systems for fungal endopolygalacturonases in closely related Brassicaceae. Nat Plants. 2021:7(9):1254–1263. 10.1038/s41477-021-00982-2. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang J-R. Determinants of the rate of protein sequence evolution. Nat Rev Genet. 2015:16(7):409–420. 10.1038/nrg3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Bai Q, Wu L, Liu H, Liu Y, Xu W, Li G, Ren H, She X, Wu G. EMS1 And BRI1 control separate biological processes via extracellular domain diversity and intracellular domain conservation. Nat Commun. 2019:10(1):4165. 10.1038/s41467-019-12112-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Shao Y, Ge S, Zhang M, Zhang T, Hu X, Liu Y, Walker J, Zhang S, Xu J. A MAPK cascade downstream of IDA-HAE/HSL2 ligand-receptor pair in lateral root emergence. Nat Plants. 2019:5(4):414–423. 10.1038/s41477-019-0396-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All alignments, trees, rate test results, selection test results, and phenotyping data are available at dryad (doi:10.5061/dryad.jm63xsjg5). Code for analyses is available on the Bartlett lab's GitHub (https://github.com/BartlettLab/Domain_evolution).