Abstract
YPEL5 is a member of the Yippee-like (YPEL) gene family that is evolutionarily conserved in eukaryotic species. To date, the physiological function of YPEL5 has not been assessed due to a paucity of genetic animal models. Here, using CRISPR/Cas9-mediated genome editing, we generated a stable ypel5−/− mutant zebrafish line. Disruption of ypel5 expression leads to liver enlargement associated with hepatic cell proliferation. Meanwhile, hepatic metabolism and function are dysregulated in ypel5−/− mutant zebrafish, as revealed by metabolomic and transcriptomic analyses. Mechanistically, Hnf4a is identified as a crucial downstream mediator that is positively regulated by Ypel5. Zebrafish hnf4a overexpression could largely rescue ypel5 deficiency-induced hepatic defects. Furthermore, PPARα signaling mediates the regulation of Hnf4a by Ypel5 through directly binding to the transcriptional enhancer of the Hnf4a gene. Herein, this work demonstrates an essential role of Ypel5 in hepatocyte proliferation and function and provides the first in vivo evidence for a physiological role of the ypel5 gene in vertebrates.
Keywords: YPEL5, hepatomegaly, hepatic function, HNF4A, PPARα signaling
Introduction
YPEL5 belongs to a novel gene family consisting of five members (YPEL1–YPEL5), which is highly conserved among eukaryotic organisms, ranging from fungi to humans (Hosono et al., 2004). Molecular phylogenetic analysis of 100 Yippee-like (YPEL) family members from 68 species reveals that YPEL5 is evolutionarily distinct from the other four members and suggests that YPEL5 is an ortholog of Drosophila Yippee (Hosono et al., 2004, 2010). Drosophila Yippee was initially identified as an intracellular protein that specifically interacted with the blood protein Hemolin of an insect (moth Hyalophora cecropia) (Roxstrom-Lindquist and Faye, 2001). In budding yeast Saccharomyces cerevisiae, deletion of MOH1, the ortholog of Drosophila yippee, enhanced cell viability under various lethal apoptogenic conditions, indicating a pro-apoptotic role of Moh1 protein (Lee et al., 2017). In African green monkey kidney fibroblast COS-7 cells, YPEL5 displays different subcellular localizations during cell cycle progression and positively regulates cell proliferation and growth (Hosono et al., 2010). More recently, it was shown that the Ypel5 gene is located in a locus linked to a network of interferon-stimulated genes in mice. Further examination shows that YPEL5 is a negative regulator of interferon-β1 production and interacts physically with TBK1/IKBKE kinases in cultured cells (Jeidane et al., 2016). In addition, studies using human colorectal cancer cell lines demonstrate that YPEL5 expression is epigenetically suppressed by the METTL3/YTHDF2 m6A axis (Zhou et al., 2021). Despite accumulating in vitro evidence suggests that YPEL5 is multifunctional, its in vivo function and mechanism of action are still poorly understood, mainly due to the lack of a vertebrate knockout animal model.
Over the last decades, zebrafish (Danio rerio) has emerged as a valuable vertebrate animal model for developmental biology. Compared with other experimental animals, the zebrafish model possesses some unique advantages, including low cost of housing and maintenance, high fecundity, external development, optically transparent embryos, and easy genetic manipulation and screening. Additionally, the major organs and tissues of zebrafish share similar molecular, morphological, and physiological features with their mammalian counterparts. Moreover, the zebrafish and human genomes are well conserved, with ∼70% of human genes having at least one zebrafish ortholog (Howe et al., 2013).
In the present study, we characterize the function of ypel5 during embryonic development in zebrafish. By loss-of-function assay using a generated CRISPR mutant, we show that ypel5 deficiency leads to enlarged liver size accompanied by hepatocyte hyperproliferation without significantly increasing cell size. Metabolomic and transcriptomic analyses reveal that hepatic metabolism and function are dysregulated in ypel5−/− mutant zebrafish. Furthermore, Hnf4a is identified as a crucial downstream mediator of Ypel5 signaling. The hepatic defects induced by ypel5 deficiency could be largely rescued by hnf4a overexpression. Moreover, peroxisome proliferator-activated receptor α (PPARα) signaling is involved in the regulation of Hnf4a by Ypel5. Together, our findings reveal for the first time that Ypel5 is required for normal embryonic liver development and function in vertebrates.
Results
Generation of ypel5−/− mutant zebrafish via the CRISPR/Cas9 system
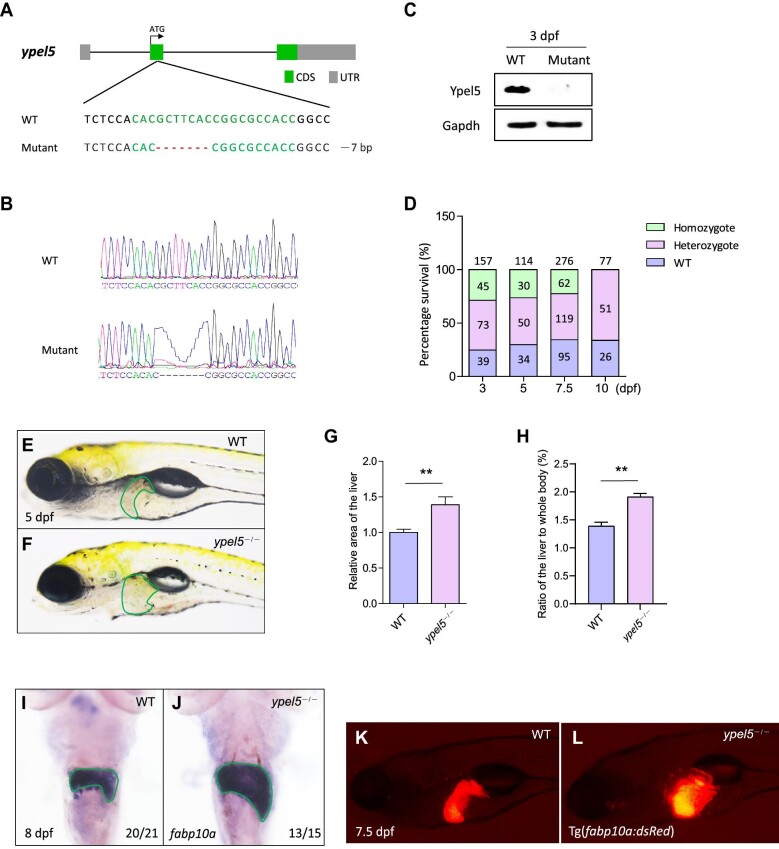
To investigate the physiological role of ypel5, we generated a ypel5-null allele, which harbored a 7-bp deletion in exon 2 (Figure 1A and B). The mutation created a premature stop codon, resulting in the synthesis of a truncated Ypel5 protein lacking 75 C-terminal amino acids. Western blot analysis with the protein extracts from embryos further confirmed the loss of Ypel5 protein in the homozygous mutants (Figure 1C). The ypel5 homozygous mutants did not exhibit gross morphological defects or developmental delays compared with control siblings during early development (data not shown). However, the ypel5−/− larvae could not survive beyond 10 days post fertilization (dpf), as no homozygous mutants were identified at this time point (Figure 1D).
Figure 1.
Disruption of the ypel5 gene causes liver enlargement. (A) Genetic inactivation of zebrafish ypel5 gene based on CRISPR/Cas9. Schematic representation of CRISPR/Cas9 target site at exon 2 as used in this study. The gRNA target site is highlighted in green, and the indel is indicated by a red dash. (B) Genotyping of the ypel5−/− mutant by Sanger sequencing. (C) Western blot analysis of Ypel5 using lysates from wild-type (WT) and ypel5−/− mutant embryos at 3 dpf. Gapdh was used as the loading control. (D) The survival of zebrafish larvae at different developmental stages. No homozygotes were identified at 10 dpf. (E–H) Brightfield images (E and F), relative liver size (G), and the ratio of the liver to whole body (H) of WT and ypel5−/− mutant zebrafish at 5 dpf. The livers are outlined with green lines. Data shown are mean ± SEM. Student's t-test. **P < 0.01. (I and J) WISH assay of fabp10a at 8 dpf. Green lines circle the boundary of the liver. The ratio of embryos with the representative expression pattern is indicated at the right bottom. (K and L) Representative images of WT and ypel5−/−/Tg(fabp10a:dsRed) zabrafish at 7.5 dpf. The liver was marked by red fluorescence.
ypel5 deficiency leads to hepatomegaly in zebrafish
Although ypel5−/− mutants did not display obvious developmental defects in body shape or growth retardation, they exhibited enlarged livers (Figure 1E–H). This prompted us to hypothesize that disruption of ypel5 expression may affect liver development. We then examined the expression of fabp10a, a liver-specific marker, by whole-mount in situ hybridization (WISH) assay. The liver size of ypel5−/− mutants was significantly increased at 5 dpf (Figure 1E–H), and the enlargement was even more pronounced by 8 dpf (Figure 1I and J). This phenotype was further confirmed on ypel5−/−/Tg(fabp10a:dsRed) larvae where the liver was marked by red fluorescence (Figure 1K and L). However, the bile duct development appeared to be unaffected, as revealed by the Tg(Tp1bglob:GFP) line (Supplementary Figure S1A and B). Additionally, the morphologies of other endoderm-derived tissues such as the endocrine pancreas, exocrine pancreas, and intestine were not obviously affected, as determined by the expression of insulin, trypsin, and fabp2, respectively (Supplementary Figure S1C–H). Together, these results suggest that the loss of Ypel5 affects liver development in zebrafish.
ypel5 depletion promotes hepatic cell proliferation
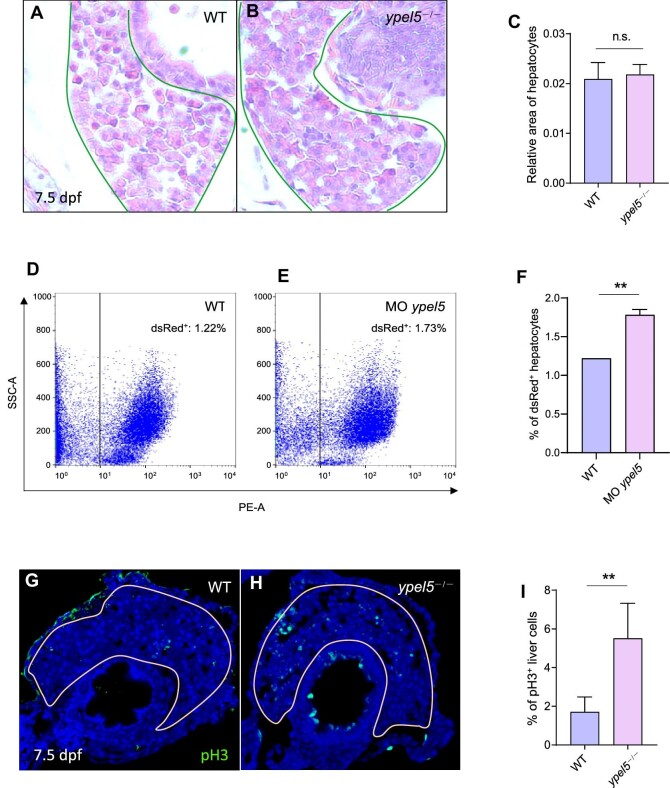
Hepatomegaly in ypel5−/− mutants might result from hepatocyte enlargement and/or hepatic cell hyperproliferation. Hematoxylin and eosin (H&E) histological staining of liver sections did not show significant differences in hepatocyte size between control siblings and ypel5-deficient larvae (Figure 2A–C), indicating that ypel5 deletion has little effect on hepatic cell morphology. In contrast, the number of hepatocytes was significantly increased in the ypel5 morpholino-injected Tg(fabp10a:dsRed) larvae compared with control siblings, as revealed by fluorescence-activated cell sorting analysis (FACS) analysis (Figure 2D–F), suggesting that ypel5 deletion increased hepatic cell number. To further determine whether hepatocyte proliferation was involved in ypel5 deficiency-induced hepatomegaly, phosphorylated histone 3 (pH3), a marker of cell proliferation, was examined in liver samples by immunohistochemistry. The percentage of pH3-positive cells was remarkably increased in ypel5−/− mutants (Figure 2G–I), suggesting that ypel5 depletion enhanced hepatocyte proliferation, which may contribute to liver enlargement.
Figure 2.
ypel5 deficiency results in enhanced hepatic cell proliferation. (A–C) Representative images of H&E staining (A and B) and relative hepatocyte size (C) of WT and ypel5−/− mutant zebrafish at 7.5 dpf. The livers are outlined with green lines. (D–F) FACS analysis (D and E) and quantification (F) of dsRed-positive hepatocytes from WT and Tg(fabp10a:dsRed) larvae at 7.5 dpf. (G–I) Representative images of pH3 staining (green) to label hepatic cell proliferation (G and H) and quantification of pH3-positive liver cells (I) in WT and ypel5−/− mutant zebrafish at 7.5 dpf. The sections were counterstained with DAPI to label the nucleus (blue). White lines circle the boundary of the liver. Data shown are mean ± SEM. Student's t-test. n.s., not significant; **P < 0.01.
Hepatic metabolism and function are dysregulated in ypel5−/− mutant zebrafish
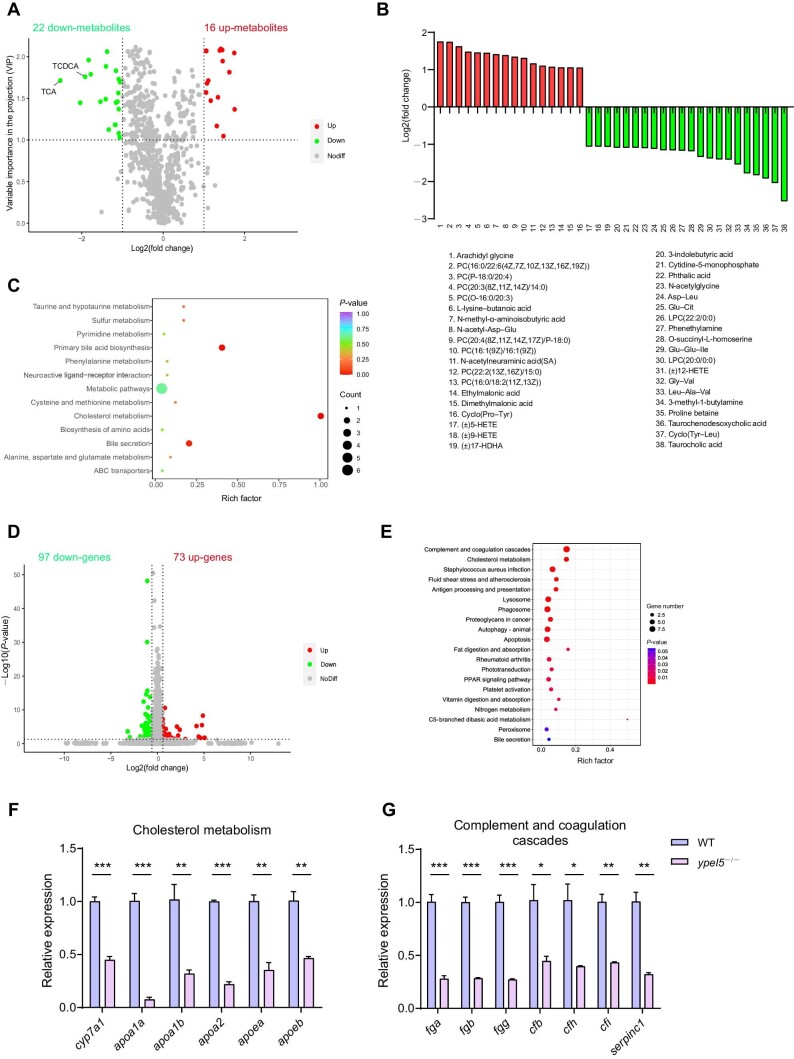
The liver is a central metabolic organ. To determine whether ypel5 deficiency had an impact on hepatic metabolism, a ultrahigh-performance liquid chromatography–electrospray ionization–tandem mass spectrometry (UPLC–ESI–MS/MS)-based, widely targeted metabolomic analysis was applied to reveal the metabolomic profile differences between ypel5−/− mutants and control siblings. A total of 818 metabolites were detected in zebrafish larvae extracts at 7.5 dpf. Among them, 22 metabolites were downregulated and 16 metabolites were upregulated in ypel5−/− mutants, with variable importance in the projection (VIP) ≥1 and fold change ≥2 (Figure 3A and B; Supplementary Table S1). Of note, taurocholic acid (TCA) and taurochenodesoxycholic acid (TCDCA), which are derived from the catabolism of cholesterol in hepatocytes (de Aguiar et al., 2013), were the most downregulated metabolites. To better understand the biological functions of these differential metabolites, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was employed. Multiple lipid metabolism pathways, such as cholesterol metabolism, primary bile acid biosynthesis, and bile secretion, were affected, and a series of amino acid metabolic pathways, including cysteine and methionine metabolism, were also dysregulated (Figure 3C). Since the liver is a central organ for lipid and amino acid homeostasis, the dysfunction could be a consequence of abnormal hepatic metabolism.
Figure 3.
ypel5 depletion leads to impaired hepatic metabolism and function. (A–C) Volcano plot (A) and KEGG enrichment (C) of differential metabolites between control siblings and ypel5−/− mutants at 7.5 dpf. (B) The 16 upregulated and 22 downregulated metabolites. (D and E) Volcano plot (D) and KEGG enrichment (E) of differentially expressed genes between control siblings and ypel5−/− mutants at 3 dpf. (F and G) qPCR analysis of genes related to cholesterol metabolism and complement and coagulation cascades. Data shown are mean ± SEM. Student's t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
In order to unravel the mechanism behind the altered hepatic metabolism in ypel5−/− mutant zebrafish, we conducted RNA sequencing (RNA-seq) with zebrafish larvae at 3 dpf to capture some of the earliest transcriptional changes leading to liver dysfunction. DE-Seq2 was used to determine differentially expressed genes (≥1.5-fold change, P < 0.05) between control siblings and ypel5−/− mutants. Of the nearly 22693 genes analyzed, 97 genes were downregulated and 73 genes were upregulated in ypel5−/− mutants (Figure 3D; Supplementary Table S2). Then, KEGG pathway enrichment analysis was performed to gain insight into the affected biological pathways. Genes associated with cholesterol metabolism were among the top enriched gene sets (Figure 3E), in line with the metabolomic profile of ypel5−/− mutants. Additionally, genes associated with complement and coagulation cascades were also highly enriched (Figure 3E). Because the liver is a major organ for the synthesis of coagulation factors and complement, dysregulation of this gene set may contribute to liver dysfunction in ypel5−/− mutants.
To further confirm the gene expression differences revealed by RNA-seq, quantitative real-time polymerase chain reaction (qPCR) was performed. The transcriptional level of cyp7a1, a rate-limiting enzyme involved in cholesterol metabolism into bile acid, was significantly decreased in ypel5−/− mutants, and several genes related to cholesterol transport and metabolism were also downregulated in ypel5−/− mutants, including apoa1a, apoa1b, apoa2, apoea, and apoeb (Figure 3F). Meanwhile, the expression levels of the complement and coagulation cascade genes, fga, fgb, fgg, cfb, cfh, cfi, and serpinc1, were remarkably reduced in ypel5−/− mutants compared to control siblings (Figure 3G). To demonstrate these gene expression changes in the liver, we microinjected ypel5 morpholino into Tg(fabp10a:dsRed) embryos and then sorted the dsRed-positive hepatic cells for qPCR analysis. Consistently, the expression levels of fga, fgb, fgg, cfb, and cfi were sharply reduced in ypel5 morphants compared with control embryos (Supplementary Figure S2A). The expression levels of cell cycle-related genes, such as tp53 and wee1, were also decreased in ypel5 morphants (Supplementary Figure S2B), confirming that ypel5 deficiency leads to aberrant hepatocyte proliferation. Additionally, we examined the expression of hnf4a, apoa1b, and cfb by WISH. As expected, their expression was restricted to the liver and downregulated in the mutants (Supplementary Figure S2C).
Taken together, the metabolomic and transcriptomic data suggest that, in addition to controlling hepatocyte proliferation, Ypel5 is also involved in physiological regulation of liver function.
Hnf4a is a crucial downstream mediator of Ypel5
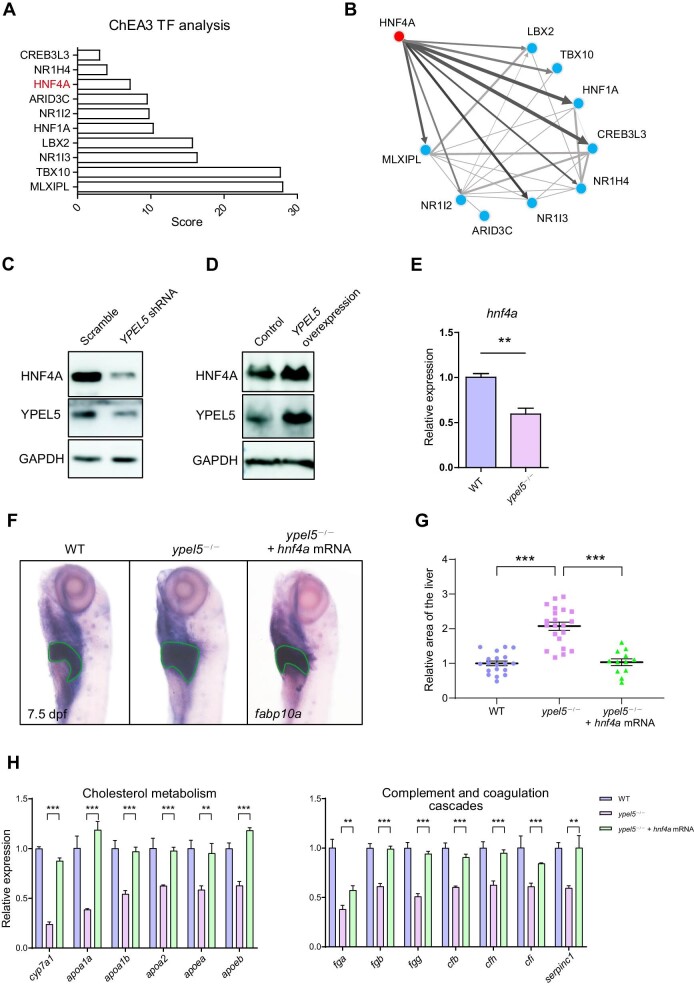
In order to identify potential regulators of the ypel5 deficiency-induced gene expression changes, we performed a transcription factor enrichment analysis, ChEA3 (Keenan et al., 2019). HNF4A was among the top predicted regulators and able to directly regulate other transcription factors (Figure 4A and B). HNF4A is a liver-enriched master regulator that plays a key role in liver development and metabolism (Dubois et al., 2020). As shown in Figure 4C, the protein level of HNF4A was sharply decreased when YPEL5 was knocked down by using short-hairpin RNA (shRNA) in Huh-7 cells. Reciprocally, ectopic expression of YPEL5 induced HNF4A protein level (Figure 4D). The hnf4a mRNA level was also significantly reduced in ypel5−/− mutants compared with control siblings (Figure 4E; Supplementary Figure S2C). These data suggest that Hnf4a is a downstream effector of Ypel5 and its expression level is positively regulated by Ypel5. To further confirm that Hnf4a mediates the hepatic defects induced by ypel5 deficiency, we performed a rescue experiment by ectopically expressing hnf4a in ypel5−/− mutant zebrafish. The hepatomegaly in ypel5−/− mutants could be largely ameliorated by hnf4a overexpression (Figure 4F and G). Furthermore, the decreased expression levels of genes related to cholesterol metabolism and complement and coagulation cascades were also restored (Figure 4H). Collectively, these findings suggest that Hnf4a is a crucial mediator of ypel5 deficiency-induced hepatic defects.
Figure 4.
Hnf4a acts as a crucial downstream effector of Ypel5. (A and B) ChEA3 transcription factor (TF) analysis was performed on differentially expressed genes. The top 10 proteins and their co-regulatory networks are shown. (C and D) Western blot analysis of HNF4A in Huh-7 cells. (E) qPCR analysis of hnf4a expression in control siblings and ypel5−/− mutants at 3 dpf. (F and G) WISH assay of fabp10a (F) and relative liver size (G) of zebrafish larvae at 7.5 dpf. Green lines circle the boundary of the liver. (H) qPCR analysis of genes related to cholesterol metabolism and complement and coagulation cascades. Data shown are mean ± SEM. Student's t-test. **P < 0.01; ***P < 0.001.
Ypel5 regulates Hnf4a expression via the PPARα signaling pathway
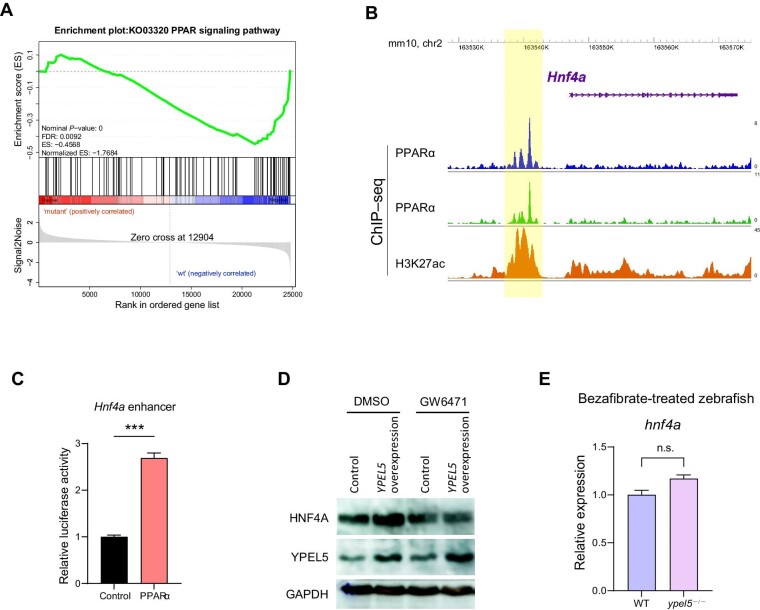
Gene set enrichment analysis (GSEA) of the RNA-seq data revealed that the PPAR signaling pathway was significantly enriched and downregulated in ypel5−/− mutants (Figure 5A). PPARs are lipid-activated transcription factors that belong to the nuclear receptor superfamily. There are three members of this family: PPARα regulates lipid metabolism in the liver, PPARβ/δ promotes fatty acid β-oxidation largely in extrahepatic organs, and PPARγ stores triacylglycerol in adipocytes (Wang et al., 2020). It has been shown that PPARα via HNF4A participated in maintaining metabolic homeostasis in the liver (Contreras et al., 2015). To determine whether PPARα signaling was involved in the regulation of Hnf4a expression by Ypel5, we first examined the genome-wide PPARα protein binding sites by applying Cistrome algorithm (Mei et al., 2017) on two PPARα chromatin immunoprecipitation sequencing (ChIP–seq) data sets in hepatic cells (Boergesen et al., 2012; Soltis et al., 2017). There was an intragenic PPARα-bound region ∼4 kb upstream of the Hnf4a transcription start site (Figure 5B). This PPARα binding site correlated with histone modification H3K27ac, which is a specific marker for the active enhancers (Creyghton et al., 2010). Then, we cloned this regulatory region into pGL3 luciferase reporter vector and performed transient transfection in Huh-7 cells. As shown in Figure 5C, PPARα overexpression significantly increased luciferase activity. These data suggest that Hnf4a may be a direct transcriptional target of PPARα. Furthermore, the elevated HNF4A protein level induced by YPEL5 overexpression was recovered by GW6471 (a potent antagonist of PPARα) treatment in Huh-7 cells (Figure 5D). Bezafibrate, an agonist of PPARα, could fully restore the reduced hnf4a expression in ypel5−/− mutants (Figure 5E). Taken together, our findings indicate that PPARα signaling mediates the regulation of Hnf4a by Ypel5.
Figure 5.
PPARα signaling mediates the regulation of Hnf4a by Ypel5. (A) GSEA showing the enrichment of the PPAR signaling pathway in ypel5−/− mutants. (B) Genomic browser view showing ChIP–seq signals of PPARα (GSM864671, GSM1163175) and H3K27ac (GSM2540257) at the Hnf4a gene locus in hepatocytes. The peaks are highlighted in yellow. (C) Luciferase activity assay in Huh-7 cells transfected with control vectors or PPARα and reporter constructs. The Renilla plasmid was used as an internal control. (D) Western blot analysis of Huh-7 cells treated for 48 h with control vehicle or GW6471. (E) qPCR analysis of hnf4a expression between control siblings and ypel5−/− mutants treated for 48 h with bezafibrate. Data shown are mean ± SEM. Student's t-test. n.s., not significant; ***P < 0.001.
Discussion
YPEL5 is an evolutionarily conserved protein present in a wide variety of eukaryotic organisms, ranging from fungi to vertebrates. To date, its physiological function remains largely unknown. In the current study, we generated a stable ypel5−/− mutant zebrafish line by using CRISPR/Cas9 technology and found that the ypel5 homozygous mutants could not survive beyond 10 dpf. Interestingly, when raised in the sterile egg water supplemented with appropriate antibiotics (Shan et al., 2015), a very small number of ypel5−/− larvae could live longer (Supplementary Figure S3A), implying that the compromised immunity might be a reason for the early lethality. Furthermore, ypel5 depletion led to hepatomegaly accompanied by hepatocyte hyperproliferation without significantly increasing cell size. Consistent with a previous report (Hosono et al., 2004), ypel5 was ubiquitously expressed during zebrafish embryonic development, including in the liver (Supplementary Figure S4A and B). It is unclear why the ypel5 depletion-caused predominant tissue-specific phenotypes were generally restricted to the liver. One possible explanation is that the Ypel5 function is compensated by its paralogs in other organs but not in the liver. Supporting this idea, a single-cell transcriptome atlas in developing zebrafish revealed abundant expression of ypel5 but low or not detectable expression of its paralogs in the liver (Farnsworth et al., 2020; Supplementary Figure S4C). However, we cannot rule out a functional role of Ypel5 in other tissues and organs.
Combined metabolomic and transcriptomic analyses revealed that hepatic metabolism and function were dysregulated in ypel5−/− mutant zebrafish. Based on KEGG pathway enrichment analyses of differentially expressed genes and differential metabolites, cholesterol metabolism, one of the important functions of the liver, was identified as the most significantly enriched pathway. TCA and TCDCA are the end-products of cholesterol metabolism in the liver and involved in bile acid biosynthesis. Their levels were sharply reduced in ypel5−/− mutants, indicating that cholesterol metabolism, as well as bile acid metabolism, is significantly affected. Accordantly, the transcription level of cyp7a1, which catalyzes the first, rate-limiting step in the neutral pathway of bile acid synthesis, was strikingly decreased in ypel5−/− mutants. In addition, the mRNA expression levels of genes related to complement and coagulation cascades, which are primarily synthesized in the liver, were also reduced. To overcome the limitations of this study, e.g. not using ypel5-depleted hepatocytes for metabolomic and transcriptomic analyses, a mouse line carrying a conditionally null allele of the Ypel5 gene should be developed and used in future studies.
Zebrafish ypel5 depletion led to the hepatic phenotype somewhat reminiscent of that seen in Hnf4a knockout mice. HNF4A is a principal transcription factor required for liver development and function. It regulates genes specifically implicated in lipid metabolism, glucose metabolism, amino acid metabolism, and blood coagulation (Hayhurst et al., 2001; Inoue et al., 2006). In addition, HNF4A is critical for hepatocyte proliferation. The hepatocyte-specific Hnf4a knockout mice displayed severe hepatomegaly and increased hepatocyte proliferation (Hayhurst et al., 2001; Bonzo et al., 2012; Walesky et al., 2013). Here, by performing ChEA3 transcription factor enrichment analysis, we identified Hnf4a as a potential transcription factor that was likely to regulate differentially expressed genes in ypel5−/− mutants. We further demonstrated that Hnf4a expression was positively regulated by Ypel5, and overexpression of hnf4a could largely rescue the hepatic defects triggered by ypel5 deficiency. These data indicate that Hnf4a is a key mediator of Ypel5 signaling. Previous reports suggest that YPEL5 is one of the peripheral components of the carboxy-terminal to LisH complex, which has E3 ubiquitin ligase activity (Huffman et al., 2019). However, the regulation of HNF4A by YPEL5 seems not driven by a proteasome-dependent degradation process, because the addition of a proteasome inhibitor did not rescue HNF4A protein levels (Supplementary Figure S5A). Actually, the hnf4a mRNA expression level was remarkably decreased in ypel5−/− mutants, indicating that the regulation of hnf4a expression occurred at the transcriptional level. GSEA of the RNA-seq data revealed that the PPAR signaling pathway was significantly changed in ypel5−/− mutants. Further analysis demonstrated that PPARα signaling mediated the regulation of Hnf4a by Ypel5 through directly binding to the transcriptional enhancer of the Hnf4a gene. It will be of interest to determine how YPEL5 regulates the PPARα signaling pathway during liver development in future studies.
In summary, our results provide the first genetic evidence that Ypel5 is essential for liver development and function, expanding our knowledge to understand the biological functions of the YPEL family and contributing to the development of new targets for the treatment of liver diseases in the future.
Materials and methods
Zebrafish
Zebrafish maintenance and staging were performed as described previously (Kimmel et al., 1995). The transgenic lines Tg(lfabf:dsRed;elaA:EGFP) (hereafter referred to as Tg(fabp10a:dsRed); Korzh et al., 2008) and Tg(Tp1bglob:GFP) (Parsons et al., 2009) were used. Relative liver size of zebrafish larvae was measured using ImageJ software. Zebrafish embryos at 24 h post fertilization were treated with 20 μM bezafibrate (Selleck, S4159) or DMSO for 48 h, and then collected for qPCR analysis. The zebrafish facility and study were approved by the Ethics Committee of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, and the methods were carried out in accordance with the approved guidelines.
CRISPR/Cas9 mutagenesis
Zebrafish ypel5 deletion was generated by CRISPR/Cas9-mediated genome editing as described before (Chang et al., 2013). Briefly, guide RNA (gRNA) was designed using an online tool, ZiFiT Targeter software (http://zifit.partners.org/ZiFiT), synthesized in vitro with a PCR product-based approach, and purified using mirVana miRNA Isolation Kit (Ambion). Cas9 mRNA was synthesized by in vitro transcription using SP6 mMESSAGE mMACHINE Kit (Ambion). Approximately 200 pg of Cas9 mRNA and 50 pg of gRNA were co-injected into 1-cell-stage zebrafish embryos to generate F0 line with mosaic mutation. Further outcrossing of F0 line with wild-type line was performed to generate F1 heterozygous lines. Finally, F2 mutant line with the homozygous mutation was obtained via incrossing of F1 heterozygotes. The ypel5−/− mutant line was genotyped by Sanger sequencing of PCR fragments covering the gRNA target site.
WISH
WISH was performed as described previously (Thisse and Thisse, 2008). Digoxigenin-labeled RNA probes were transcribed with T7, T3, or SP6 polymerase (Ambion). The probes were detected using alkaline phosphatase-coupled anti-digoxigenin Fab fragment antibodies (Roche) with BCIP/NBT staining (Vector Laboratories). The stained embryos were then photographed using a stereomicroscope (Nikon) equipped with a digital camera.
Histology and pH3 immunostaining
Zebrafish larvae at 7.5 dpf were fixed in 4% paraformaldehyde, dehydrated, embedded in paraffin, cut into 4 μm-thick sections, and then stained with H&E solution according to standard protocols. Relative hepatocyte area was measured using the ImageJ software package.
For pH3 immunostaining, zebrafish larvae at 7.5 dpf were fixed in 4% paraformaldehyde at 4°C overnight, embedded in OCT compound (SAKURA), and cryosectioned into 4-μm slices. After blocked with 10% fetal bovine serum (FBS) for 1 h at room temperature, the sections were incubated with rabbit anti-pH3 antibody (1:100 dilution, Santa Cruz) at 4°C overnight and then with Alexa Fluor 488-conjugated anti-rabbit IgG (Invitrogen) for 1 h at room temperature. The sections were counterstained with DAPI (Vector Labs) to label cell nuclei.
FACS
Tg(fabp10a:dsRed) transgenic larvae at 5 and 7.5 dpf were dissociated into single cells using 0.25% trypsin (Sigma-Aldrich). Single-cell suspension was obtained by centrifugation at 400× g for 5 min, washing twice with phosphate-buffered saline, and passing through a 40-μm nylon mesh filter. FACS was performed with FACSAria II (BD Biosciences) to collect homogenous dsRed-positive cells. Data were analyzed with FlowJo 10.8.1 software (FlowJo LLC).
Targeted metabolomic analysis
Metabolomic analysis was carried out by Wuhan Metware Biotechnology Co., Ltd. Briefly, zebrafish larvae at 7.5 dpf were anesthetized for collection and washed twice with cold HPLC-grade water. The zebrafish were immediately snap-frozen and lysed in 50 μl of pre-cooled 70% methanol/water mixture on ice. Following homogenization, additional 150 μl of pre-cooled 70% methanol/water mixture was supplemented to each tube, followed by incubation on ice for 15 min and centrifugation at 12000 rpm for 10 min. Then, the supernatant was transferred into a new centrifuge tube. After incubating at −20°C for 30 min, the supernatant was centrifuged at 12000 rpm for 15 min at 4°C. The sample extracts were analyzed using an LC–ESI–MS/MS system (UPLC, ExionLC AD; MS, QTRAP® System). The analytical conditions for UPLC were as follows. Column: Waters ACQUITY UPLC HSS T3 C18 (1.8 μm, 2.1 mm × 100 mm); column temperature: 40°C; flow rate: 0.4 ml/min; injection volume: 2 μl; solvent system: water (0.1% formic acid):acetonitrile (0.1% formic acid); gradient program: 95:5 (v/v) at 0 min, 10:90 (v/v) at 11.0 min, 10:90 (v/v) at 12.0 min, 95:5 (v/v) at 12.1 min, and 95:5 (v/v) at 14.0 min. The metabolites were identified using the Metware database (MWDB). The metabolite abundances were quantified according to their peak areas. Metabolites were considered to be differentially accumulated when VIP ≥ 1 and |log2(fold change)| ≥ 1.
RNA-seq and data analysis
RNA-seq was carried out by Suzhou Transcriptome Biotechnology Co., Ltd. Briefly, zebrafish larvae at 3 dpf were harvested and lysed in 500 μl TRIzol (Invitrogen), and RNA was extracted following the manufacturer's standard procedures. Libraries were prepared with Illumina TruSeq Stranded mRNA kit and quantified with a Qubit 2.0 Fluorometer (dsDNA HS kit; ThermoFisher), and the size distribution was determined with the Agilent 2100 Bioanalyzer System prior to pooling. RNA-seq was performed on an Illumina HiSeq2000 platform. Sequence reads were aligned to the zebrafish GRCz11 genome assembly with HISAT2 (Kim et al., 2015). Cufflinks v2.2 was used to generate FPKM values, and the differentially expressed genes were determined by R package DE-Seq2 (v 1.34.0) (Trapnell et al., 2013). KEGG pathway enrichment analysis with differentially expressed genes was conducted by R package ClusterProfiler (v 4.2.2). ChEA3 transcription factor enrichment analysis was performed as previously described (Keenan et al., 2019). GSEA was performed by clusterProfiler v3.14.3, using the curated gene set C2 of Molecular Signature Database.
qPCR
Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's recommendations. cDNA was synthesized with the ReverTra Ace-α-™ kit (TOYOBO). qPCR was performed by a LightCycler 480 System (Roche) following the manufacturer's protocols. Relative expression was quantitated using the ΔΔCt method. The primers for qPCR are shown in Supplementary Table S3.
Plasmid construction and mRNA synthesis
Zebrafish hnf4a, human YPEL5, and mouse PPARα were cloned into pCS2+ vector. Mouse Hnf4a enhancer was cloned into pGL3 promoter vector. The indicated primers for construction are shown in Supplementary Table S1. For shRNA-mediated knockdown, the shRNA (5′-CCGGGTTCGAGATGTGAGCTGCAAACTCGAGTTTGCAGCTCACATCTCGAACTTTTTG-3′) targeting human YPEL5 mRNA was acquired from MISSION shRNA (Sigma) and cloned into pLKO.1-GFP vector. A scrambled shRNA was used as a control.
Capped mRNAs were synthesized from linearized plasmids using the mMessage mMachine SP6 kit (Invitrogen) and diluted to 60 ng/μl for microinjection.
Cell culture and transfection
Huh-7 cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Gibco) at 37°C in a humidified atmosphere with 5% CO2 and transfected with plasmids using Effectene Transfection Reagent (QIAGEN) according to the manufacturer's protocols. In some experiments, Huh-7 cells were incubated with 20 μM MG132 (MCE, HY-13259) or 10 μM GW6471 (Selleck, S2798) for 24 h or 48 h, respectively, and subjected to western blot analysis.
Western blot analysis
Western blot analysis was performed using standard methodology with the following antibodies: YPEL5 (Eterlife, EL801477), HNF4A (Cell Signaling Technology, 3113S), and GAPDH (Proteintech, 60004-1).
ChIP–seq data analysis
PPARα and H3K27ac ChIP–seq data sets in hepatic cells were reanalyzed from previously published work (Boergesen et al., 2012; Goldstein et al., 2017; Soltis et al., 2017). ChIP–seq data were processed by the Cistrome analysis pipeline and loaded into WashU genome browsers for visualization.
Luciferase activity assay
Huh-7 cells were transfected with the indicated plasmids using Effectene Transfection Reagent (QIAGEN) and harvested after 72 h. Luciferase activities were analyzed using the Dual Luciferase Reporter Assay Kit (Promega) according to the manufacturer's protocols. Luciferase activity was normalized to Renilla activity.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software. All values are expressed as mean ± SEM except where noted. Differences between two groups were examined using the Student's t-test. P < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgements
We thank Dr Jun Chen (Zhejiang University) for providing transgenic zebrafish lines, as well as Xiangwei Wu (Suzhou Transcriptome Biotechnology Co., Ltd) and Shanhe Yu (Ruijin Hospital) for their excellent technical support and helpful discussion.
Contributor Information
Yun Deng, Shanghai Institute of Hematology, State Key Laboratory of Medical Genomics, National Research Center for Translational Medicine at Shanghai, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; CNRS-LIA Hematology and Cancer, Sino-French Research Center for Life Sciences and Genomics, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Xiao Han, Shanghai Institute of Hematology, State Key Laboratory of Medical Genomics, National Research Center for Translational Medicine at Shanghai, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; CNRS-LIA Hematology and Cancer, Sino-French Research Center for Life Sciences and Genomics, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Huiqiao Chen, Department of Hematology, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou 310011, China.
Chaoxian Zhao, Shanghai Institute of Hematology, State Key Laboratory of Medical Genomics, National Research Center for Translational Medicine at Shanghai, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; CNRS-LIA Hematology and Cancer, Sino-French Research Center for Life Sciences and Genomics, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Yi Chen, Shanghai Institute of Hematology, State Key Laboratory of Medical Genomics, National Research Center for Translational Medicine at Shanghai, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Jun Zhou, Shanghai Institute of Hematology, State Key Laboratory of Medical Genomics, National Research Center for Translational Medicine at Shanghai, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; CNRS-LIA Hematology and Cancer, Sino-French Research Center for Life Sciences and Genomics, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Hugues de The, CNRS-LIA Hematology and Cancer, Sino-French Research Center for Life Sciences and Genomics, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; Université de Paris 7/INSERM/CNRS UMR 944/7212, Equipe Labellisée Ligue Nationale Contre le Cancer, Hôpital St. Louis, Paris 75010, France.
Jun Zhu, CNRS-LIA Hematology and Cancer, Sino-French Research Center for Life Sciences and Genomics, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; Université de Paris 7/INSERM/CNRS UMR 944/7212, Equipe Labellisée Ligue Nationale Contre le Cancer, Hôpital St. Louis, Paris 75010, France.
Hao Yuan, Shanghai Institute of Hematology, State Key Laboratory of Medical Genomics, National Research Center for Translational Medicine at Shanghai, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; CNRS-LIA Hematology and Cancer, Sino-French Research Center for Life Sciences and Genomics, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Funding
This work was supported by the National Natural Science Foundation of China (31371479).
Conflict of interest: none declared.
Author contributions: Y.D., X.H., H.C., C.Z., and Y.C. performed experiments and collected data; J. Zhou and H.d.T. provided suggestions on experimental design and data presentation; J. Zhu provided suggestions on experimental design and analyzed data; and H.Y. designed the research, performed experiments, analyzed data, and wrote the manuscript.
References
- Boergesen M., Pedersen T.A., Gross B.et al. (2012). Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol. Cell. Biol. 32, 852–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonzo J.A., Ferry C.H., Matsubara T.et al. (2012). Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4α in adult mice. J. Biol. Chem. 287, 7345–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N., Sun C., Gao L.et al. (2013). Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras A.V., Rangel-Escareno C., Torres N.et al. (2015). PPARα via HNF4α regulates the expression of genes encoding hepatic amino acid catabolizing enzymes to maintain metabolic homeostasis. Genes Nutr. 10, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M.P., Cheng A.W., Welstead G.G.et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aguiar V.T., Tarling E.J., Edwards P.A. (2013). Pleiotropic roles of bile acids in metabolism. Cell Metab. 17, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois V., Staels B., Lefebvre P.et al. (2020). Control of cell identity by the nuclear receptor HNF4 in organ pathophysiology. Cells 9, 2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth D.R., Saunders L.M., Miller A.C. (2020). A single-cell transcriptome atlas for zebrafish development. Dev. Biol. 459, 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein I., Paakinaho V., Baek S.et al. (2017). Synergistic gene expression during the acute phase response is characterized by transcription factor assisted loading. Nat. Commun. 8, 1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst G.P., Lee Y.H., Lambert G.et al. (2001). Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 21, 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono K., Noda S., Shimizu A.et al. (2010). YPEL5 protein of the YPEL gene family is involved in the cell cycle progression by interacting with two distinct proteins RanBPM and RanBP10. Genomics 96, 102–111. [DOI] [PubMed] [Google Scholar]
- Hosono K., Sasaki T., Minoshima S.et al. (2004). Identification and characterization of a novel gene family YPEL in a wide spectrum of eukaryotic species. Gene 340, 31–43. [DOI] [PubMed] [Google Scholar]
- Howe K., Clark M.D., Torroja C.F.et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman N., Palmieri D., Coppola V., (2019). The CTLH complex in cancer cell plasticity. J. Oncol. 2019, 4216750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Peters L.L., Yim S.H.et al. (2006). Role of hepatocyte nuclear factor 4α in control of blood coagulation factor gene expression. J. Mol. Med. 84, 334–344. [DOI] [PubMed] [Google Scholar]
- Jeidane S., Scott-Boyer M.P., Tremblay N.et al. (2016). Association of a network of interferon-stimulated genes with a locus encoding a negative regulator of non-conventional IKK kinases and IFNB1. Cell Rep. 17, 425–435. [DOI] [PubMed] [Google Scholar]
- Keenan A.B., Torre D., Lachmann A.et al. (2019). ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 47, W212–W224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L., (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R.et al. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. [DOI] [PubMed] [Google Scholar]
- Korzh S., Pan X., Garcia-Lecea M.et al. (2008). Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev. Biol. 8, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Jun D.Y., Park J.E.et al. (2017). Pro-apoptotic role of the human YPEL5 gene identified by functional complementation of a yeast moh1Δ mutation. J. Microbiol. Biotechnol. 27, 633–643. [DOI] [PubMed] [Google Scholar]
- Mei S., Qin Q., Wu Q.et al. (2017). Cistrome data browser: a data portal for ChIP–seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 45, D658–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M.J., Pisharath H., Yusuff S.et al. (2009). Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 126, 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxstrom-Lindquist K., Faye I. (2001). The Drosophila gene Yippee reveals a novel family of putative zinc binding proteins highly conserved among eukaryotes. Insect Mol. Biol. 10, 77–86. [DOI] [PubMed] [Google Scholar]
- Shan Y., Fang C., Cheng C.et al. (2015). Immersion infection of germ-free zebrafish with Listeria monocytogenes induces transient expression of innate immune response genes. Front. Microbiol. 6, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis A.R., Motola S., Vernia S.et al. (2017). Hyper- and hypo-nutrition studies of the hepatic transcriptome and epigenome suggest that PPARα regulates anaerobic glycolysis. Sci. Rep. 7, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M.et al. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walesky C., Gunewardena S., Terwilliger E.F.et al. (2013). Hepatocyte-specific deletion of hepatocyte nuclear factor-4α in adult mice results in increased hepatocyte proliferation. Am. J. Physiol. Gastrointest. Liver Physiol. 304, G26–G37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Nakajima T., Gonzalez F.J.et al. (2020). PPARs as metabolic regulators in the liver: lessons from liver-specific PPAR-null mice. Int. J. Mol. Sci. 21, 2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Tang W., Xu Y.et al. (2021). METTL3/YTHDF2 m6A axis accelerates colorectal carcinogenesis through epigenetically suppressing YPEL5. Mol. Oncol. 15, 2172–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.