Abstract
Prostate cancer progression is connected to the activity of conventional oncogenes and tumour suppressors and driven by circulating steroid hormones. A key issue has been how to identify and care for aggressively developing prostate tumours. Here we discuss how expression of the splicing regulators ESRP1 and ESRP2, and how their role as “masterminds” of epithelial splicing patterns, have been identified as markers of aggressively proliferating prostate primary tumours. We suggest that the origin of prostate cancer within epithelial cells, and the subsequent association of ESRP1 and ESRP2 expression with more aggressive disease progression, identify ESRP1 and ESRP2 as lineage survival oncogenes. To move this field on in the future it will be important to identify the gene expression targets controlled by ESRP1/2 that regulate prostate cancer proliferation. Potential future therapies could be designed to target ESRP1 and ESRP2 protein activity or their regulated splice isoforms in aggressive prostate tumours. Design of these therapies is potentially complicated by the risk of producing a more mesenchymal splicing environment that might promote tumour metastasis.
Subject terms: RNA splicing, Oncogenes
Introduction
The prostate gland is located underneath the bladder in men and is the origin of the most frequent male-specific cancer [1, 2]. Prostate cancers can have variable disease progression: while some prostate tumours are relatively indolent and require monitoring alone, others can progress more aggressively and need to be treated with either radiotherapy or surgery. A key research priority is to identify the disease mechanisms that cause more aggressive cancers to develop, so that these can be detected and treated.
The molecular mechanisms driving prostate cancer have been intensively investigated, and involve hormone-driven signalling pathways, “classical” oncogenes including ERG and MYC, and tumour suppressors like PTEN [3, 4]. Since the 1940s it has been known that circulating levels of male steroid hormones called androgens (including testosterone and dihydroxy-testosterone) are critical drivers for prostate cancer progression [5]. Androgens exert their biological functions by interacting with a transcription factor called the androgen receptor (abbreviated AR). The AR protein is cytoplasmic in the absence of circulating androgens, but forms dimers in response to androgen exposure, and moves into the nucleus to control transcription of target genes important in cancer development. Approximately 50% of prostate cancers contain a genetic fusion between the androgen-responsive TMPRSS2 gene and the ERG transcription factor gene. This genetic fusion places ERG (and therefore ERG-regulated downstream target genes that control prostate cancer cell proliferation, differentiation and apoptosis) under androgen control—thus converting ERG into a hormone-responsive oncogene [6, 7].
Many genes that are controlled by androgens have downstream effects on prostate cancer proliferation. For this reason, both locally advanced and metastatic prostate cancer are often treated with Androgen Deprivation Therapy (ADT) [3, 8], a treatment that reduces the production of androgens and promotes tumour regression. Currently used drugs used for ADT include luteinizing hormone releasing hormone (LHRH) antagonists that block the hypothalamic-pituitary gonadal axis of testosterone production. Other drugs include abiraterone, which interferes with enzymes required for androgen biosynthesis [9]; and bicalutamide (brand name Casodex) and enzalutamide (brand name Xtandi), which bind to the AR and blocks its ligand-dependent activity. Although ADT initially promotes prostate tumour shrinkage, cancers can relapse over time and become unresponsive to ADT—a condition described as castration-resistant prostate carcinoma (CRPC) [3].
Most human protein-coding genes are split into introns and exons, and are transcribed as precursor messenger RNAs (pre-mRNAs). Exons are spliced together to create mature mRNAs by a nuclear complex called the spliceosome [10]. Most human protein coding genes can be alternatively spliced to produce multiple different mRNAs. Alternative splicing plays a key role in prostate cancer progression, providing a possible angle of treatment, and this has been the topic of excellent recent reviews [11–13]. Much of the focus of alternative splicing research in prostate cancer has been on investigating alternatively spliced isoforms encoded by the androgen receptor gene (AR) itself. This is because alternative splicing can create AR splice variants that encode variant AR proteins that are constitutively active without androgens, and that become important later in disease progression as castrate resistant prostate cancer (CRPC) develops [11]. These include the splice variant ARv7 that includes a cryptic terminal exon, causing it to encode a variant androgen receptor containing a DNA binding domain but lacking a ligand binding domain. Expression of splice variants such as ARv7 contribute to androgen hormone-independent patterns of gene expression in patients with advanced prostate cancer and in response to ADT. How these AR splice variants are regulated has been the focus of intensive investigation [11]. Recent evidence indicates that regulation of epithelial splicing patterns is also important early in the disease progression of aggressive prostate cancer, and this is the topic of this review.
Amplification of the ESRP1 gene in early onset and highly aggressive prostate cancer
The most significant risk factor for a man to develop prostate cancer is age. The average age for prostate cancer diagnosis is 66 years, and prostate cancer is rarely found in men below the age of 40 years (https://www.cancer.org/cancer/prostate-cancer/about/key-statistics.html). Early onset aggressive prostate cancer (EOPC) develops in men below the age of 55 years, and this is likely to be the result of active oncogenic pathways driving tumour development. Identification and understanding of these oncogenic pathways is therefore of key importance. Genome analyses of prostate tumours have revealed amplification of the ESRP1 gene (encoding the Epithelial Splicing Regulator Protein 1) to be associated with early onset aggressive prostate cancer [14]. Although the ESRP1 gene is located on chromosome 8 close to the MYC oncogene (an important oncogene that is also frequently amplified in prostate cancer [15]), ESRP1 amplification can be observed in prostate tumours independently of MYC alterations, indicating that it is not a bystander event [14]. Indeed, MYC-independent ESRP1 amplification in early onset prostate cancer correlates with more advanced cancer histology and high mitotic activity of tumour cells, and is also predictive of a shorter time to disease recurrence [14]. Clonal analyses of tumour cells also show that ESRP1 gene amplification occurs step-wise in the development of aggressive early onset prostate cancer, following genetic changes in tumour-suppressor genes such as KLF5, BRCA1, RB1, PTEN and TP53 [14].
ESRP1 and ESRP2 are “splicing masterminds” within epithelial cells
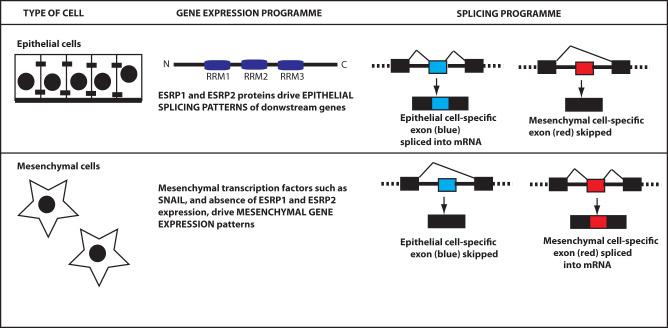
ESRP1 and its closely related paralog ESRP2 are known as “splicing masterminds” of epithelial cells [16]. Epithelial cells are tightly connected to adjacent cells and to basement membranes [17], and are anchored within monolayer sheets where they often function in secretion and boundary formation (Fig. 1) [18]. Epithelial cells typically grow in sheets within tissues, in close connection to other cells and often anchored to basement membranes. In contrast, mesenchymal cells usually exist as individual cells, and are able to move (Fig. 1). Human and mouse genomes each contain ESRP1 and ESRP2 proteins expressed by distinct paralog genes (with ESRP1 on human chromosome 8, and ESRP2 on human chromosome 16). Transcriptome analyses show that mouse organs and tissues containing a high proportion of epithelial cells also express high levels of both ESRP1 and ESRP2 (these high expressing tissues include the normal prostate) [19]. A major difference between ESRP1 and ESRP2 is at the level of overall expression. For example, the mouse Esrp1 gene is expressed at higher levels than Esrp2, and genetic knockout of mouse Esrp1 causes postnatal death due to cleft lip and palate [19]. In contrast, mice deleted for Esrp2 are viable, while double knockout of Esrp1 and Esrp2 prevents many organs from properly forming [19], indicating ESRP1 and ESRP2 proteins can compensate for each other during normal development. Although ESRP1/ESRP2 proteins have mainly been investigated as splicing regulators within the nucleus, the human ESRP1 gene can also produce an alternatively spliced transcript that encodes a cytoplasmic protein isoform that may regulate cytoplasmic aspects of epithelial function [20, 21].
Fig. 1. ESRP1 and ESRP2 are splicing masterminds of epithelial cells.
Epithelial cells grow together in sheets of physically connected cells. Epithelial cells express high levels of ESRP1 and ESRP2 proteins that direct epithelial patterns of gene splicing. ESRP1 and ESRP2 contain three RNA Recognition Motif (RRM) domains that bind to target RNAs. Mesenchymal cells have a different morphology, tend to be mobile and have different patterns of gene expression from epithelial cells.
Some key genes express alternative epithelial and mesenchymal mRNA splice isoforms that are specialised to function within their respective cell types. This means that the same gene will be made into a slightly different mRNA, often encoding a different protein isoform, depending on whether it is expressed in an epithelial or mesenchymal cell environment. ESRP1 and ESRP2 proteins regulate production of epithelial-specific alternative mRNA isoforms—explaining why they are known as “splicing masterminds” of epithelial cells. Human ESRP1 and ESRP2 proteins share an overall 60% amino acid identity. ESRP1 and ESRP2 proteins contain three domains called RRMs (RNA Recognition Motifs) that enable them to bind to pre-mRNA to control splicing patterns (Fig. 1) [22–24]. The RRM protein domains of ESRP1 and ESRP2 share higher levels of amino acid sequence conservation compared to their total protein sequences (RRM1: 88.5% identity; RRM2: 71% identity; RRM3: 80% identity). ESRP1 protein binds to a UG-rich RNA target sequence within introns and exons [25]. Binding of ESRP proteins to RNA within or immediately upstream of exons generally causes exon skipping, while ESRP binding within intron sequences downstream of regulated exons promotes splicing inclusion [18, 26]. Mesenchymal cells do not express ESRP1 or ESRP2, but do express specific transcription factors such as SNAIL (Fig. 1) that establish mesenchymal patterns of transcription.
High expression of epithelial-specific splicing regulator proteins in primary prostate tumours correlates with a more aggressive disease trajectory
The study of EOPC provides a window into the development of aggressive prostate cancer, but EOPC only affects a small population of men. However, there is also data indicating that high levels of ESRP1 and ESRP2 gene expression more generally contribute to aggressive prostate primary tumour development and correlate with poorer patient prognosis. RNAseq data from general (i.e. not EOPC) primary prostate adenocarcinomas available in The Cancer Genome Atlas (TCGA) [27] show that high ESRP1 (but not ESRP2) gene expression levels correlate with poorer patient prognosis [28–30]. Similar data have been obtained at the protein level by analysing tissue microarrays stained for ESRP1 and ESRP2. Immunohistochemistry of ~19,000 prostate tumours correlated high levels of ESRP1 and ESRP2 protein expression with more rapid biochemical tumour recurrence, higher Gleason scores, higher tumour stages and increased numbers of lymph node metastases [28, 29, 31].
Androgens control ESRP1 and ESRP2 expression and their splicing targets in prostate cancer cells
Recent data have identified an important mechanism controlling ESRP1 and ESRP2 expression within prostate cancer cells that is linked to disease progression. Although androgens have been long known to control transcription patterns, the latest data show androgens also control alternative splicing [30, 32–34]. Two recent studies, one from our group (Munkley et al. [30]) and one by Shah et al. [32], identified a novel gene expression axis controlling epithelial splicing patterns in prostate cancer cells: androgen levels control expression of ESRP1 and ESRP2 proteins, which then control downstream epithelial splicing patterns [30, 32]. RNA-seq data from prostate tumours show that ADT reduces expression levels of both ESRP1 and ESRP2 genes [30, 32]. Furthermore, ESRP2 expression can be directly induced within prostate cancer cell lines by exposure to androgens [30]. ESRP1 protein levels detected on tissue microarrays also directly correlate with pre-operative PSA levels, further linking androgen-driven gene expression with ESRP1 [14].
Importantly, many ESRP-regulated exons in prostate cancer cells have reciprocal splicing patterns when prostate cancer cells were stimulated with androgens versus treatment with a drug called Casodex that is used to antagonise the androgen receptor [30]. Expression levels of ESRP1 and ESRP2 genes also decrease in response to enzalutamide, a drug blocking AR function in prostate cancer cells [32]. As a consequence, treatment with enzalutamide also changed ESRP-regulated splicing patterns [32]. Treatment of cultured prostate cancer cells with Casodex and enzalutamide model the molecular changes that occur during ADT, and has established a clear link between ESRP1/ESRP2 and androgens.
The mechanisms through which ESRP1 and ESRP2 are controlled by androgen exposure have not been fully elucidated. The simplest model would be if ESRP1 and ESRP2 are direct transcriptional activated targets of the AR protein, and genomic ChIP (chromatin immunoprecipitation) identified AR binding sites within the vicinity of the ESRP2 gene [30]. Analysis of ESRP1/ESRP2 protein staining in thousands of prostate tumours show that levels of ESRP1/ESRP2 proteins are highest in prostate cancers that are also positive for the TMPRS22-ERG genetic fusion [28]. Thus, ESRP1 and ESRP2 expression could also be indirectly activated via androgen-regulation of the ERG transcription factor in prostate tumours containing the TMPRS22-ERG genetic fusion. However, arguing against this indirect model, other immunohistochemical analyses have shown that higher levels of ESRP1 protein correlate with a shorter time to prostate cancer biochemical recurrence independent of ERG translocation status [14].
Each of the above bits of evidence indicate that increased expression levels of ESRP1 and ESRP2 in primary prostate tumours are associated with a more aggressive disease trajectory. This means that immunohistochemical analyses linking ESRP1 and ESRP2 expression with more aggressive disease progression might also be useful as prostate cancer biomarkers to apply in parallel to Gleason grading [28]. Such independent markers are needed since although Gleason grades are a good prognostic marker, the same tumours can be frequently assessed differently by different histopathologists [28].
High levels of ESRP1 and ESRP2 in primary prostate tumours correlate with a more proliferative gene expression profile
An intriguing possibility is that the epithelial splicing patterns established by increased levels of ESRP proteins in response to circulating levels of androgens in prostate cancer patients could be identical to those splicing patterns established by ESRP1 gene amplification in early onset prostate cancer. Thus, upregulation of ESRP1/ESRP2 gene expression in aggressive tumours, by either gene amplification or in response to androgens could be different means to the same end, and drive the progression of aggressive prostate cancer in a similar way. This means that identifying the precise targets and functional consequences of the AR-ESRP gene expression axis could make a big contribution to understanding the biological basis of aggressive prostate tumour development.
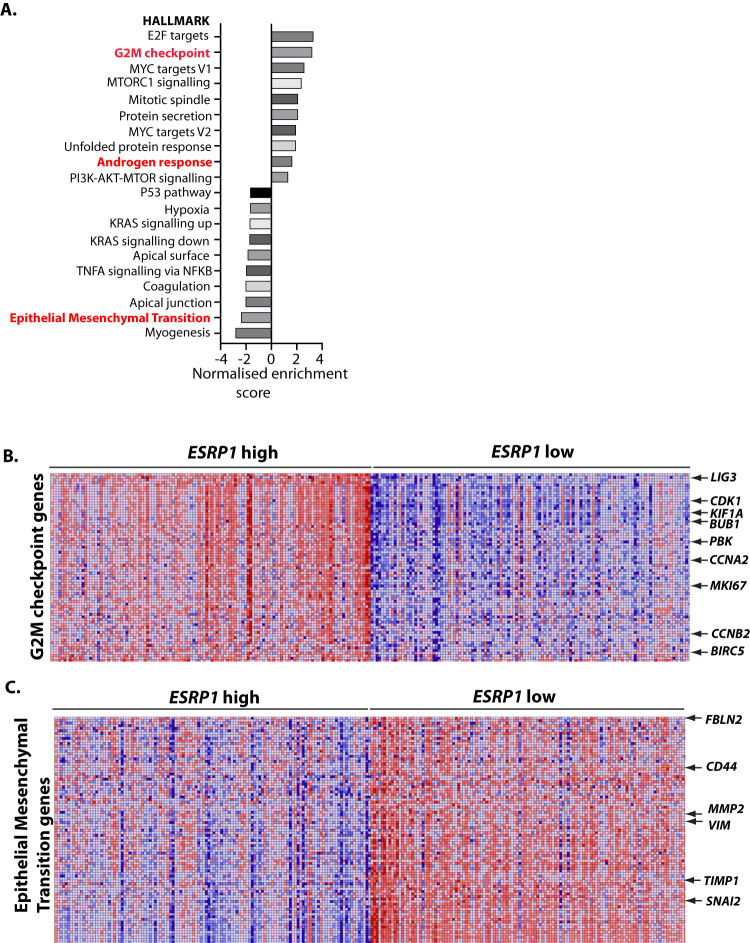
What could be the selective advantage for more aggressive prostate tumours to express higher levels of ESRP1 and ESRP2? Increased expression of ESRP1 and ESRP2 direct epithelial splicing patterns, and epithelial cells have been reported to replicate more rapidly than mesenchymal cells [35, 36]. Tissue microarray data show that high levels of ESRP1 correlate with an increased cellular proliferation rate [14]. Hence, by expressing higher levels of ESRP1 and ESRP2, more aggressive primary prostate tumours may maintain an epithelial gene expression environment to result in a growth advantage. Supporting this, Gene Set Enrichment Analysis (GSEA) of gene expression patterns in 250 primary prostate tumours with high levels of ESRP1 in the TCGA PRAD cohort [27] revealed positive enrichment for gene sets associated with DNA replication and mitotic activity (Fig. 2A). Consistent with the above studies linking ESRP1/ESRP2 gene expression with testosterone, the gene expression sets associated with high levels of ESRP1 or ESRP2 gene expression include the androgen response (Fig. 2A).
Fig. 2. High levels of ESRP1 and ESRP2 in primary prostate tumours correlate with a more proliferative gene expression profile.
A Analysis of the transcriptomes of 250 primary prostate tumours in the cancer genome atlas (TCGA) PRAD cohort identified enriched gene expression sets that correlate with ESRP1 levels. The bar chart shows positively and negatively enriched sets of gene expression in primary prostate tumours stratified by ESRP1 gene expression levels (highest and lowest 25%). B Heat map showing higher expression of genes associated with the G2M transition in ESRP1 high versus ESRP1 low tumours. C Heat map showing reduced expression of EMT-associated genes in ESRP1-high tumours versus ESRP1-low tumours.
Distinct patterns of gene expression within tumours with elevated or reduced levels of ESRP1 expression are illustrated by the heat maps in Fig. 2B, C. These heat maps show individual levels of gene expression activity. Tumours expressing elevated levels of ESRP1 also have higher activity of genes associated with G2M checkpoints, compared to tumours expressing lower levels of ESRP1 (high gene expression is indicated by red, and lower gene expression levels in blue in Fig. 2B, C). The genes with increased expression levels in ESRP1 high tumours include MKI67 (encoding the proliferation marker Mki67 [37]); the cell cycle regulator encoding genes CDK1, CCNA2, and CCNB2; and BIRC5 that inhibits apoptosis.
The possession of epithelial and mesenchymal states are somewhat plastic, and are controlled by expression levels of ESRP1 and ESRP2 and the levels of mesenchymal transcription factors [38, 39]. Metastasis (the movement of cells from the primary tumour to other anatomic locations) involves a switch in cell morphology from epithelial to mesenchymal cells, called an Epithelial to Mesenchymal Transition or EMT. The biological changes that occur during metastasis mirror similar changes in cell properties that happen during normal development and wound healing (reviewed by [40]). ESRP1 and ESRP2 are key players in maintaining the epithelial gene expression environment. Experiments in human non-small cell lung cancer cells have shown that many (but not all) of the splice changes involved in EMT are directly controlled by changing concentrations of ESRP1 and ESRP2 [26].
Further meta-analysis of prostate cancer TCGA data show down-regulation of EMT pathways in primary prostate tumours expressing high levels of ESRP1 (Fig. 2A). In contrast, lower levels of ESRP1 correlate with patterns of gene expression associated with mesenchymal patterns of gene expression, including the mesenchymal transcription factors VIM and SNAI2 (Fig. 2C). Interestingly, established metastases can de-differentiate to regain epithelial properties [39]. Hence re-expression of ESRP1 and ESRP2 proteins could in principle be part of a gene expression programme enabling successful metastatic growth once secondary tumours have seeded. Experimental analysis using cells and tissues from outside of the prostate show that ESRP1 expression is transcriptionally controlled during metastasis. The mesenchymal transcription factor SNAIL binds within the ESRP1 promoter to repress its transcription, and induce EMT of human mammary epithelial cells [41]. There are also changes in the chromatin architecture of the ESRP1 gene following ectopic expression of the mesenchymal transcription factor Twist1 in mammary epithelial cells [42]. Inhibition of Twist1 promotes successful growth of mouse squamous cell carcinomas once metastases are established [36]. Such dynamic cell status switches between epithelial and mesenchymal cell gene expression programmes could be important within metastases, since epithelial cells proliferate more rapidly than mesenchymal cells, and mesenchymal transcription factors can inhibit genes important for cell growth [39]. Conversely the mesenchymal state is more compatible with increased cell mobility, drug efflux and immune evasion [39].
ESRP1 and ESRP2 as lineage-survival oncogenes in early aggressive prostate cancer
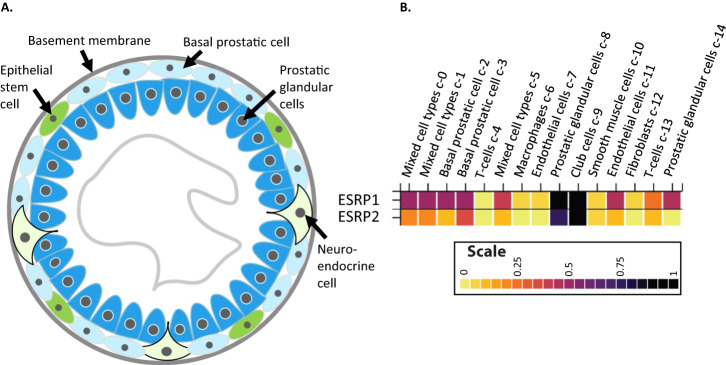
Why might primary prostate tumours express high levels of ESRP1 and ESRP2 as a strategy to increase tumour growth? The answer to this question might be found in the normal cell biology of the prostate gland. The healthy prostate gland cell population is dominated by epithelial cells surrounding a central lumen (Fig. 3A). These epithelial cells include basal cells on the glandular periphery that also contain a stem cell population, and luminal cells which secrete fluid into the central lumen that nourishes sperm and helps their mobility and survival.
Fig. 3. ESRP1 and ESRP2 as lineage survival oncogenes.
A The cellular organisation of the prostate gland. B Cell type expression profiles of ESRP1 and ESRP2 from single cell RNAseq data downloaded from the Human Protein Atlas. Epithelial Cells (high levels of ESRP1/2 expression): Luminal Secretory cells, Basal cells, Neuroendocrine cells, Club cells, and Hillock cells. Stromal cells (low levels of ESRP expression): Smooth muscle cells, T cells and fibroblasts.
Prostate cancers usually initiate as a prostatic intraepithelial neoplasia, which then forms a localized prostate carcinoma. Most prostate tumours originate within the prostatic glandular cells and basal cells of the prostate gland, that have been identified from single RNAseq data to have high levels of ESRP1 gene expression (Fig. 3B) [3, 43]. Highly aggressive hormone resistant prostate cancers have overall gene expression profiles that resemble basal rather than luminal cell signatures, and prostate cancer cells can also trans-differentiate into neuroendocrine cells in response to androgen deprivation [44, 45]. However, whether prostate cancer cells derive from luminal, basal or trans-differentiated neuroendocrine cells, importantly an epithelial cell type of origin is still involved, and so this originating cell would be expected to have an epithelial splicing programme directed by ESRP expression (Fig. 3).
The fact that prostate tumours originate from epithelial cells that already express ESRP1 and ESRP2, and then the ESRP1 and ESRP2 genes subsequently become important within a disease context, fits with the definition of a specific group of oncogenes called lineage-survival oncogenes [46]. By definition, lineage-survival oncogenes are important originally for the operation of healthy cell lineages but then become oncogenes during cancer initiation. Other known lineage-specific oncogenes include SOX2 in lung and oesophageal squamous cell carcinomas, and MITF (microphthalmia-associated transcription factor) in melanoma [46, 47].
Implications of ESRP1 and ESRP2 expression patterns for treating aggressive prostate tumours
Global RNAseq studies have identified multiple splicing targets for ESRP1 and ESRP2 in prostate cancer cells (Table 1) [30, 32]. Analysis of RNA expression data from prostate cancer patients RNA available in The Cancer Genome Atlas shows that expression of some of these ESRP-regulated splice isoforms correlates with a less favourable patient outcome [30, 32]. ESRP1 and ESRP2 both activate splicing of NUMB exon 3, and splicing of this exon in prostate tumours correlates with a reduced time until biochemical tumour recurrence in patients [30]. NUMB encodes a tumour suppressor protein that binds to MDM2 protein to prevent degradation of p53 and also inhibits Notch signalling [30, 48]. Other examples of ESRP-regulated splicing events that correlate with poorer patient prognosis include repression of exon 23 of the MYO1B gene [30]. The MYO1B gene encodes for an actin filament motor protein involved in intra-cellular vesicle transport. Exon skipping of MYO1B exon 23 correlates with a shorter time to prostate tumour biochemical recurrence [30].
Table 1.
Some ESRP1/ESRP2 regulated splice events detected in prostate cancer cells.
| GeneGene | Protein Function | ESRP regulation | Clinical relevance |
|---|---|---|---|
| ADAM15 | Transmembrane glycoprotein involved in cell adhesion | Activates inclusion of exon 20 | ESRP controls poorer prognosis isoform |
| MINK1 | Serine/threonine kinase | Activates inclusion of exon 18 | ESRP controls poorer prognosis isoform |
| MYO1B | Motor protein | Represses exon 23 | ESRP controls poorer prognosis isoform |
| NUMB | Tumour suppressor | Activates inclusion of exon 6 | NUMB exon 6 shows more skipping in primary prostate tumours |
| MLPH | Rab effector protein | Represses exon 9 | ESRP controls poorer prognosis isoform |
| MYH10 | Non-muscle actin dependent myosin | Represses exon 6 | ESRP controls poorer prognosis isoform |
| RPS24 | needed for cell proliferation | Represses exon 5 | ESRP controls poorer prognosis isoform |
| TUFT1 | adaptation to hypoxia, mesenchymal stem cell function, and neuronal differentiation | Activates inclusion of exon 2 | ESRP controls poorer prognosis isoform |
| DOCK7 | guanine nucleotide exchange factor | Represses inclusion of exon 23 | DOCK7 exon 23 shows more skipping in primary prostate tumours |
| ARFGAP2 | ADP Ribosylation Factor GTPase Activating Protein 2 | Activates inclusion of exon 8 | ARFGAP2 exon 8 shows more inclusion in primary prostate tumours |
| ENAH | cell motility and adhesion | Activates inclusion of exon 11A | ENAH exon 11 A shows more inclusion in primary prostate tumours |
| FN1 | cell adhesion and maintenance of cell shape | Represses exon 25 | |
| FLNB | Connects cell membrane to actin cytoskeleton | Activates inclusion of exon 30 | Exon 30 included form in primary prostate tumours correlates with better prognosis |
| MAP3K7 | controls transcription and apoptosis | Activates inclusion of exon 12 | MAP3K7 genetic deletion leads to poor clinical prognosis |
This selection of regulated splice events are taken from Munkley et al. 2019 [30] to illustrate how ESRP1/ESRP2 expression levels could control aspects of prostate cancer biology.
Since ESRP1 and ESRP2 are over-expressed in aggressive prostate tumours, could they also be targeted to reduce the growth of primary prostate tumours? Modulating expression of ESRP1 and ESRP2 in aggressive tumours could inhibit cell proliferation, but also have unintended side effects. The splicing patterns of NUMB exon 3, MYO1B exon 23 and other ESRP-target exons can be experimentally modulated either by depleting ESRP1 and ESRP2 protein levels in prostate cancer cells, or by using drugs like Casodex to interfere with AR signalling [30]. However, a complication of this possible approach is that any treatment that decreases activity of ESRP1 and ESRP2 may also dampen epithelial splicing patterns. This in turn could generate a prostate cancer cell that could metastasise more easily. Supporting this scenario in prostate cancer, ADT treatment has been linked to increased chance of metastasis in prostate cancer [49, 50]. This increased metastatic potential could be related to splice isoform production. For example, a splice isoform is produced from the MAP3K7 gene (that encodes a mitogen activated protein kinase enzyme) that skips exon 12 in response to ESRP1/ESRP2 depletion in prostate cancer cells. This splice isoform, called MAP3K7Δexon12, is normally expressed in highly metastatic cancer cell lines [30]. The FLNB gene (encoding an actin binding protein) makes two different mRNA isoforms, one including exon 30 (made in epithelial cells) and one skipping exon 30 (made in mesenchymal cells). Increased skipping of FLNB exon 30 (an exon normally activated by the AR-ESRP axis in prostate cancer), is tightly linked to the development of metastases in breast cancer [51]. CRISPR-mediated deletion of FLNB exon 30 from the genome of breast cancer cells is sufficient to convert breast cancer cells from an epithelial to a mesenchymal phenotype [51].
Changes in ESRP1 and ESRP2 expression are part of more global changes in the splicing environment during prostate cancer progression
Almost 68% of splicing regulator protein genes are differentially expressed over prostate cancer development, and there are also parallel changes in splicing patterns [52]. Hence expression changes in ESRP1 and ESRP2 in clinical prostate cancer disease progression, and downstream modulation of mRNA splicing patterns, most likely occur within the broader context of global changes taking place in the splicing environment [11, 53].
Could ESRP1 and ESRP2 be lineage survival oncogenes in other kinds of tumour?
Could ESRP1 and ESRP2 also operate as lineage-survival oncogenes during the development of other tumours, many of which also develop from epithelial tissues? Supporting this, several studies indicate that ESRP1 overexpression plays a crucial role in various cancers where they also correlate with poorer survival statistics [24]. Higher levels of ESRP1 expression have been detected in breast cancer, and have been associated with poor prognosis in oestrogen receptor positive breast cancer cells [54]. ESRP1 is controlled by the steroid hormone receptor ERα in breast cancer cells, and there are ERα binding sites within ESRP1 and ESRP2 promoters [55]. ESRP1 is also controlled by the transcription factor E2F1 in breast cancer cells and sensitive to oxygen levels [56]. ESRP1 protein regulates splicing of the CD44 cell surface receptor mRNA, that encodes a protein which interacts with tyrosine kinases in breast cancer cells [57]. The CD44 gene contains group of variable exons which are repressed by ESRP1, to produce a splice variant called CD44s which positively correlates with co-expression of genes involved in cell proliferation [57]. Loss of ESRP1 expression leads to inclusion of these CD44 variable exons, leading to production of variant CD44v protein isoform that is able to activate the PDGFRβ/Stat3 intracellular signalling pathway to promote the formation of a “cancer stem cell” population with mesenchymal properties [57]. Higher expression of ESRP1 also results in poor survival in non-small-cell lung cancer [58] and in colorectal carcinoma [59]. ESRP1 is overexpressed in early stage colon cancer, and ESRP1 downregulation leads to colon cancer cell death [60].
Conclusions
In summary, prostate cancer initiates within epithelial cell types. While androgen hormone signalling and classical oncogenes play a key role in prostate cancer development, recent data link the development of more aggressive primary prostate cancers with high levels of epithelial splicing regulator protein expression. ESRP1 and ESRP2 are encoded by genes that are important within the epithelial cells from which prostate tumours are derived, and then play an important role in subsequent tumour development. This gene expression pattern fits within the definition of a group of oncogenes called lineage-survival oncogenes. Future work will be needed to more precisely identify the genes controlled by ESRP1 and ESRP2 that are important in aggressive prostate cancer, how they result in tumour growth, and how expression of these genes are modulated in response to ADT and during tumour metastasis. Important questions for the future include whether the specific splice isoforms that cause aggressive prostate cancer properties can be identified and then therapeutically targeted.
Acknowledgements
We thank Prostate Cancer UK (grants PG12-34 and RIA16-ST2-011), and the BBSRC (grants BB/W002019/1 and BB/S008039/1) for funding this work. We thank Dr. Ralf Kist for helpful discussion of lineage survival oncogenes.
Author contributions
Conceptualisation: DJE, JM and RS; Funding acquisition: DJE; Writing -original draft: DJE and RS; Investigation: DJE, ES, SL, CD. Writing –review and editing: JM, SL, ES and JW.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Romer, AS, Parsons TS. The Vertebrate Body 5th ed. Saunders: Philadelphia, 1977.
- 2.Aaron L, Franco O, Hayward SW. Review of prostate anatomy and embryology and the etiology of BPH. Urol Clin North Am. 2016;43:279–88. doi: 10.1016/j.ucl.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang G, Zhao D, Spring DJ, Depinho RA. Genetics and biology of prostate cancer. Genes Dev. 2018;32:1105–40. doi: 10.1101/gad.315739.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies AH, Zoubeidi A. Targeting androgen receptor signaling: a historical perspective. Endocr Relat Cancer. 2021;28:T11–T18. doi: 10.1530/ERC-21-0116. [DOI] [PubMed] [Google Scholar]
- 6.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X-W, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 7.Adamo P, Ladomery MR. The oncogene ERG: a key factor in prostate cancer. Oncogene. 2016;35:403–14. doi: 10.1038/onc.2015.109. [DOI] [PubMed] [Google Scholar]
- 8.Huggins C, Hodges CV. Studies on prostatic cancer i. the effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. Cancer Res. 1941;1:293–7. [Google Scholar]
- 9.Livermore KE, Munkley J, Elliott J. D. Androgen receptor and prostate cancer. AIMS Mol Sci. 2016;3:280–99. doi: 10.3934/molsci.2016.2.280. [DOI] [Google Scholar]
- 10.Papasaikas P, Valcárcel J. The spliceosome: the ultimate RNA Chaperone and sculptor. Trends Biochem Sci. 2016;41:33–45. doi: 10.1016/j.tibs.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Paschalis A, Sharp A, Welti JC, Neeb A, Raj GV, Luo J, et al. Alternative splicing in prostate cancer. Nat Rev Clin Oncol. 2018;15:663–75. doi: 10.1038/s41571-018-0085-0. [DOI] [PubMed] [Google Scholar]
- 12.Mehterov N, Kazakova M, Sbirkov Y, Vladimirov B, Belev N, Yaneva G, et al. Alternative RNA splicing-the trojan horse of cancer cells in chemotherapy. Genes (Basel) 2021;12. 10.3390/genes12071085. [DOI] [PMC free article] [PubMed]
- 13.Zou C, Zhang D The spliceosome as a new therapeutic vulnerability in aggressive prostate cancer. Mol Cell Oncol. 2020;7. 10.1080/23723556.2020.1778420. [DOI] [PMC free article] [PubMed]
- 14.Gerhauser C, Favero F, Risch T, Simon R, Feuerbach L, Assenov Y, et al. Molecular evolution of early-onset prostate cancer identifies molecular risk markers and clinical trajectories. Cancer Cell. 2018;34:996–1011.e8. doi: 10.1016/j.ccell.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebello RJ, Oing C, Knudsen KE, Loeb S, Johnson DC, Reiter RE, et al. Prostate cancer. Nat Rev Dis Prim. 2021;7:9. doi: 10.1038/s41572-020-00243-0. [DOI] [PubMed] [Google Scholar]
- 16.Tavanez JP, Valcárcel J. A splicing mastermind for EMT. EMBO J. 2010;29:3217–8. doi: 10.1038/emboj.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pradella D, Naro C, Sette C, Ghigna C. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer. 2017;16:8. doi: 10.1186/s12943-016-0579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallinjoud P, Villemin JP, Mortada H, Espinoza MP, Desmet FO, Samaan S, et al. Endothelial, epithelial, and fibroblast cells exhibit specific splicing programs independently of their tissue of origin. Genome Res. 2014;24:511–21. doi: 10.1101/gr.162933.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bebee TW, Park JW, Sheridan KI, Warzecha CC, Cieply BW, Rohacek AM, et al. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. Elife. 2015;4. 10.7554/eLife.08954. [DOI] [PMC free article] [PubMed]
- 20.Yang Y, Carstens RP. Alternative splicing regulates distinct subcellular localization of Epithelial splicing regulatory protein 1 (Esrp1) isoforms. Sci Rep. 2017;7:3848. doi: 10.1038/s41598-017-03180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fagoonee S, Bearzi C, Di Cunto F, Clohessy JG, Rizzi R, Reschke M, et al. The RNA binding protein ESRP1 fine-tunes the expression of pluripotency-related factors in mouse embryonic stem cells. PLoS One. 2013;8:e72300. doi: 10.1371/journal.pone.0072300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023; 51:D523–D531. [DOI] [PMC free article] [PubMed]
- 23.Liu Y, Li Y, Du C, Kuang S, Zhou X, Zhang J, et al. Underlying mechanisms of epithelial splicing regulatory proteins in cancer progression. J Mol Med (Berl). 2022. 10.1007/s00109-022-02257-5. [DOI] [PubMed]
- 24.Hayakawa A, Saitoh M, Miyazawa K, Dual roles for epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) in cancer progression. In: Advances in Experimental Medicine and Biology. 2017, pp 33–40. [DOI] [PubMed]
- 25.Peart NJ, Hwang JY, Quesnel-Vallières M, Sears MJ, Yang Y, Stoilov P, et al. The global Protein-RNA interaction map of ESRP1 defines a post-transcriptional program that is essential for epithelial cell function. iScience. 2022;25:105205. doi: 10.1016/j.isci.2022.105205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Park JW, Bebee TW, Warzecha CC, Guo Y, Shang X, et al. Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the epithelial-to-mesenchymal transition. Mol Cell Biol. 2016;36:1704–19. doi: 10.1128/MCB.00019-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304.e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freytag M, Kluth M, Bady E, Hube-Magg C, Makrypidi-Fraune G, Heinzer H, et al. Epithelial splicing regulatory protein 1 and 2 (ESRP1 and ESRP2) upregulation predicts poor prognosis in prostate cancer. BMC Cancer. 2020;20:1220. doi: 10.1186/s12885-020-07682-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee HH, Lee AJ, Park WS, Lee J, Park J, Park B, et al. Epithelial splicing regulatory protein (ESPR1) expression in an unfavorable prognostic factor in prostate cancer patients. Front Oncol. 2020;10:556650. doi: 10.3389/fonc.2020.556650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munkley J, Ling L, Krishnan SRG, Hysenaj G, Scott E, Dalgliesh C, et al. Androgen-regulated transcription of ESRP2 drives alternative splicing patterns in prostate cancer. Elife. 2019;8. 10.7554/elife.47678. [DOI] [PMC free article] [PubMed]
- 31.Stinnesbeck M, Kristiansen A, Ellinger J, Hauser S, Egevad L, Tolkach Y, et al. Prognostic role of TSPAN1, KIAA1324 and ESRP1 in prostate cancer. APMIS. 2021;129:204–12. doi: 10.1111/apm.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah K, Gagliano T, Garland L, O’Hanlon T, Bortolotti D, Gentili V, et al. Androgen receptor signaling regulates the transcriptome of prostate cancer cells by modulating global alternative splicing. Oncogene. 2020;39:6172–89. doi: 10.1038/s41388-020-01429-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajan P, Dalgliesh C, Carling PJ, Buist T, Zhang C, Grellscheid SN, et al. Identification of novel androgen-regulated pathways and mRNA isoforms through genome-wide exon-specific profiling of the LNCaP transcriptome. PLoS One. 2011;6:e29088. doi: 10.1371/journal.pone.0029088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munkley J, Maia TM, Ibarluzea N, Livermore KE, Vodak D, Ehrmann I, et al. Androgen-dependent alternative mRNA isoform expression in prostate cancer cells. F1000Research. 2018;7:1189. doi: 10.12688/f1000research.15604.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vega S, Morales AV, Ocaña OH, Valdés F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–43. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–36. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammarsten P, Josefsson A, Thysell E, Lundholm M, Hägglöf C, Iglesias-Gato D, et al. Immunoreactivity for prostate specific antigen and Ki67 differentiates subgroups of prostate cancer related to outcome. Mod Pathol J U S Can Acad Pathol Inc. 2019;32:1310–9. doi: 10.1038/s41379-019-0260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Göttgens E-L, Span PN, Zegers MM. Roles and regulation of epithelial splicing regulatory proteins 1 and 2 in epithelial-mesenchymal transition. Int Rev Cell Mol Biol. 2016;327:163–94. doi: 10.1016/bs.ircmb.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 39.Brabletz S, Schuhwerk H, Brabletz T, Stemmler MP. Dynamic EMT: a multi-tool for tumor progression. EMBO J. 2021;40:e108647. doi: 10.15252/embj.2021108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2020;21:341–52. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinke LM, Xu Y, Cheng C. Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial-mesenchymal transition. J Biol Chem. 2012;287:36435–42. doi: 10.1074/jbc.M112.397125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malouf GG, Taube JH, Lu Y, Roysarkar T, Panjarian S, Estecio MR, et al. Architecture of epigenetic reprogramming following Twist1-mediated epithelial-mesenchymal transition. Genome Biol. 2013;14:R144. doi: 10.1186/gb-2013-14-12-r144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang D, Zhao S, Li X, Kirk JS, Tang DG. Prostate luminal progenitor cells in development and cancer. Trends cancer. 2018;4:769–83. doi: 10.1016/j.trecan.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang D, Park D, Zhong Y, Lu Y, Rycaj K, Gong S, et al. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat Commun. 2016;7:10798. doi: 10.1038/ncomms10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith BA, Sokolov A, Uzunangelov V, Baertsch R, Newton Y, Graim K, et al. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci USA. 2015;112:E6544–52. doi: 10.1073/pnas.1518007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 47.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–42. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colaluca IN, Basile A, Freiburger L, D’Uva V, Disalvatore D, Vecchi M, et al. A Numb-Mdm2 fuzzy complex reveals an isoform-specific involvement of Numb in breast cancer. J Cell Biol. 2018;217:745–62. doi: 10.1083/jcb.201709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Y, Wang B-E, Leong KG, Yue P, Li L, Jhunjhunwala S, et al. Androgen deprivation causes epithelial–mesenchymal transition in the prostate: implications for androgen-deprivation therapy. Cancer Res. 2012;72:527–36. doi: 10.1158/0008-5472.CAN-11-3004. [DOI] [PubMed] [Google Scholar]
- 50.Zhifang M, Liang W, Wei Z, Bin H, Rui T, Nan W, et al. The androgen receptor plays a suppressive role in epithelial- mesenchymal transition of human prostate cancer stem progenitor cells. BMC Biochem. 2015;16:13. doi: 10.1186/s12858-015-0042-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Choi PS, Chaffer CL, Labella K, Hwang JH, Giacomelli AO, et al. An alternative splicing switch in FLNB promotes the mesenchymal cell state in human breast cancer. Elife. 2018;7. 10.7554/eLife.37184. [DOI] [PMC free article] [PubMed]
- 52.Zhang D, Hu Q, Liu X, Ji Y, Chao HP, Liu Y, et al. Intron retention is a hallmark and spliceosome represents a therapeutic vulnerability in aggressive prostate cancer. Nat Commun. 2020;11. 10.1038/s41467-020-15815-7. [DOI] [PMC free article] [PubMed]
- 53.Munkley J, Livermore K, Rajan P, Elliott DJ. RNA splicing and splicing regulator changes in prostate cancer pathology. Hum Genet. 2017;136:1143–54. doi: 10.1007/s00439-017-1792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gökmen-Polar Y, Neelamraju Y, Goswami CP, Gu Y, Gu X, Nallamothu G, et al. Splicing factor ESRP1 controls ER-positive breast cancer by altering metabolic pathways. EMBO Rep. 2019;20:e46078. doi: 10.15252/embr.201846078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elhasnaoui J, Ferrero G, Miano V, Franchitti L, Tarulli I, Coscujuela Tarrero L, et al. A regulatory axis between epithelial splicing regulatory proteins and estrogen receptor α modulates the alternative transcriptome of luminal breast cancer. Int J Mol Sci. 2022;23. 10.3390/ijms23147835. [DOI] [PMC free article] [PubMed]
- 56.Ashok C, Ahuja N, Natua S, Mishra J, Samaiya A, Shukla S. E2F1 and epigenetic modifiers orchestrate breast cancer progression by regulating oxygen-dependent ESRP1 expression. Oncogenesis. 2021;10:58. doi: 10.1038/s41389-021-00347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Brown RL, Wei Y, Zhao P, Liu S, Liu X, et al. CD44 splice isoform switching determines breast cancer stem cell state. Genes Dev. 2019;33:166–79. doi: 10.1101/gad.319889.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui J, Ren P, Li Y, Ma Y, Wang J, Lin C, et al. ESRP1 as a prognostic factor of non-small-cell lung cancer is related to the EMT transcription factor of Twist. Thorac Cancer. 2021;12:2449–57. doi: 10.1111/1759-7714.14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fagoonee S, Picco G, Orso F, Arrigoni A, Longo DL, Forni M, et al. The RNA-binding protein ESRP1 promotes human colorectal cancer progression. Oncotarget. 2017;8:10007. doi: 10.18632/oncotarget.14318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vadlamudi Y, Kang SC. Silencing ESRP1 expression promotes caspase-independent cell death via nuclear translocation of AIF in colon cancer cells. Cell Signal. 2022;91:110237. doi: 10.1016/j.cellsig.2021.110237. [DOI] [PubMed] [Google Scholar]