Abstract
Mitochondria are essential organelles found in eukaryotic cells that play a crucial role in ATP production through oxidative phosphorylation (OXPHOS). Mitochondrial DNA depletion syndrome (MTDPS) is a group of genetic disorders characterized by the reduction of mtDNA copy number, leading to deficiencies in OXPHOS and mitochondrial functions. Mutations in FBXL4, a substrate-binding adaptor of Cullin 1-RING ubiquitin ligase complex (CRL1), are associated with MTDPS, type 13 (MTDPS13). Here, we demonstrate that, FBXL4 directly interacts with the mitophagy cargo receptors BNIP3 and BNIP3L, promoting their degradation through the ubiquitin-proteasome pathway via the assembly of an active CRL1FBXL4 complex. However, MTDPS13-associated FBXL4 mutations impair the assembly of an active CRL1FBXL4 complex. This results in a notable accumulation of BNIP3/3L proteins and robust mitophagy even at basal levels. Excessive mitophagy was observed in Knockin (KI) mice carrying a patient-derived FBXL4 mutation and cortical neurons (CNs)-induced from MTDPS13 patient human induced pluripotent stem cells (hiPSCs). In summary, our findings suggest that abnormal activation of BNIP3/BNIP3L-dependent mitophagy impairs mitochondrial homeostasis and underlies FBXL4-mutated MTDPS13.
Subject terms: Ubiquitin ligases, Disease genetics
Introduction
Mitophagy is a highly regulated process whereby damaged or dysfunctional mitochondria are selectively eliminated through autophagy [1–3]. The efficient clearance of defective mitochondria is crucial for cellular homeostasis, and dysregulation of mitophagy has been implicated in the pathogenesis of numerous neurodegenerative disorders, including Alzheimer’s disease (AD) and Parkinson’s disease (PD) [4]. Mitochondrial quality control is regulated by two proteins associated with PD: the mitochondrial kinase PINK1 and E3-ubiquitin (Ub) ligase, PARKIN. Together, they play a central role in the process of mitophagy. When defective mitochondria are present, PINK1 accumulates on their surface, leading to the translocation of PARKIN from the cytosol to initiate the clearance of these damaged mitochondria via autophagy [5, 6]. Despite extensive investigation of the Parkin-PINK1 pathway, mito-QC reporter mice lacking PINK1 display no obvious loss of basal mitophagy across a broad range of tissues of high metabolic demand, including neurons and cardiomyocytes [7].
In addition to the Parkin/PINK1-dependent pathway, mitophagy can also be initiated by various mitochondria cargo receptors. These receptors are transmembrane proteins located on the outer mitochondrial membrane that bind to autophagosome membranes and facilitate the recruitment of mitochondria into autophagosomes [8]. Mitophagy receptors include but are not limited to BNIP3, BNIP3L/NIX, FUNDC1, and BCL2L13, which are involved in various cellular processes such as development, differentiation, and apoptosis. The activation of these receptors results in the formation of mitophagosomes around damaged or dysfunctional mitochondria and their subsequent delivery to lysosomes for degradation [9].
FBXL4 mutations are one of the causes of MTDPS [10, 11]. This disorder is part of a group of rare genetic disorders characterized by a decreased amount of mtDNA in affected tissues [12]. The clinical presentation of FBXL4-related MTDPS varies widely, with symptoms ranging from severe multisystem involvement in infancy to late-onset neurodegenerative disorders. The most common clinical features include hypotonia, developmental delay, muscle weakness, seizures, neuropathy, and optic atrophy. The severity and progression of symptoms depend on the extent and distribution of mtDNA depletion in affected tissues, with highly metabolic tissues such as muscle and brain being the most affected [10, 11]. Treatment options for FBXL4-related mitochondrial DNA depletion syndromes are limited and mainly supportive [13]. Studies conducted with patient-derived fibroblasts, as well as using FBXL4 knockout mice, have demonstrated that FBXL4 deficiency contributes to excessive mitophagy activity, leading to the depletion of mtDNA in patients [14]. FBXL4 belongs to the F-BOX and leucine-rich repeat protein family and is believed to function as an adaptor protein by binding to substrate proteins and facilitating their recognition by the CUL1-RBX1-SKP1 E3 ubiquitin ligase complex [15]. However, the physiological substrates regulated by CRL1FBXL4 complex and the pathogenic mechanisms underlying FBXL4 mutation-driven MTDPS13 are still poorly understood.
In this study, we demonstrate that BNIP3 and BNIP3L are proteolytic substrates for CRL1FBXL4 complex. Loss-of-function mutations of FBXL4 in human cells and mouse models stimulates robust basal mitophagy and mitochondrial dysfunction, which is dependent on BNIP3/3 L accumulation. Furthermore, we observed abnormal activation of mitophagy in cortical neurons induced from hiPSCs derived from MTDPS13 patients. Therefore, our findings support the model that CRL1FBXL4-mediated degradation of BNIP3/BNIP3L tightly suppresses basal mitophagy in multiple organs, and its deregulation leads to severe symptoms of MTDPS.
Results
Identification of BNIP3 and BNIP3L family proteins as FBXL4 interacting proteins
As FBXL4 is a substrate-binding adaptor of the CRL1 E3 ubiquitin ligase complex, it is plausible that FBXL4 targets specific mitophagy-related proteins for ubiquitination and degradation. To determine the molecular mediators of FBXL4-mediated mitophagy regulation, we utilized affinity purification coupled with mass spectrometry (AP-MS) methods to identify FBXL4-interacting proteins. Our AP-MS results showed that, besides the core components of the CRL1 E3 (CUL1 and RBX1), FBXL4 existed in a complex with BNIP3 and BNIP3L, which are well-known mitophagy cargo receptors (Fig. 1A, Supplementary Table 1). We suspect that FBXL4 may regulate mitophagy through its interaction with BNIP3 and BNIP3L.
Fig. 1. Identification of BNIP3 and BNIP3L proteins as FBXL4 interacting proteins.
A Tandem affinity purification of FBXL4-containing protein complexes was conducted from 293 T cells stably overexpressing FLAG-FBXL4. The number of total/unique peptides identified by mass spectrometry analysis are shown in the Table. B, C 293 T cells were transfected with the indicated plasmids. The whole cell lysates (WCL) were prepared and subjected to co-IP with anti-FLAG antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies. D Recombinant expressed GST-FBXL4 protein or GST bound to glutathione-Sepharose beads and incubated with recombinant expressed His-BNIP3 or BNIP3L proteins. Bound His-BNIP3 or BNIP3L proteins were detected by WB with anti-His antibody. E Schematic representation of FBXL4 deletion mutants. F 293 T cells were transfected with the indicated plasmids. The WCL were prepared and subjected to co-IP with anti-FLAG antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies.
To validate our hypothesis, we first conducted co-immunoprecipitation (co-IP) assays and demonstrated that ectopically overexpressed FBXL4 interacts with both BNIP3 and BNIP3L (Fig. 1B). Additionally, FLAG-FBXL4 could immunoprecipitate endogenous BNIP3/BNIP3L (Fig. 1C). To determine the direct interaction between FBXL4 and BNIP3/3L, we conducted in vitro experiments using purified recombinant proteins. Specifically, we expressed and purified recombinant GST-FBXL4 and His-BNIP3/3L from bacteria. As shown in Fig. 1D, GST-FBXL4 protein, but not GST alone, exhibited binding with His-BNIP3 or BNIP3L, as indicated by the results of GST-pulldown assays. This confirms the physical interaction between FBXL4 and BNIP3/3L. Besides, FBXL4 did not interact with other mitophagy cargo receptors like PHB2, NIPSNAP1, and FUNDC1, which underscores the highly specific interaction between FBXL4 and BNIP3/BNIP3L (Fig. 1B). Furthermore, we found that the leucine-rich repeat domain of FBXL4 is responsible for its interaction with both BNIP3/BNIP3L (Fig. 1E, F).
Thus, these data provide evidence supporting the notion that FBXL4 specifically interacts with BNIP3/BNIP3L in cells.
FBXL4 mediates ubiquitination and degradation of BNIP3/BNIP3L
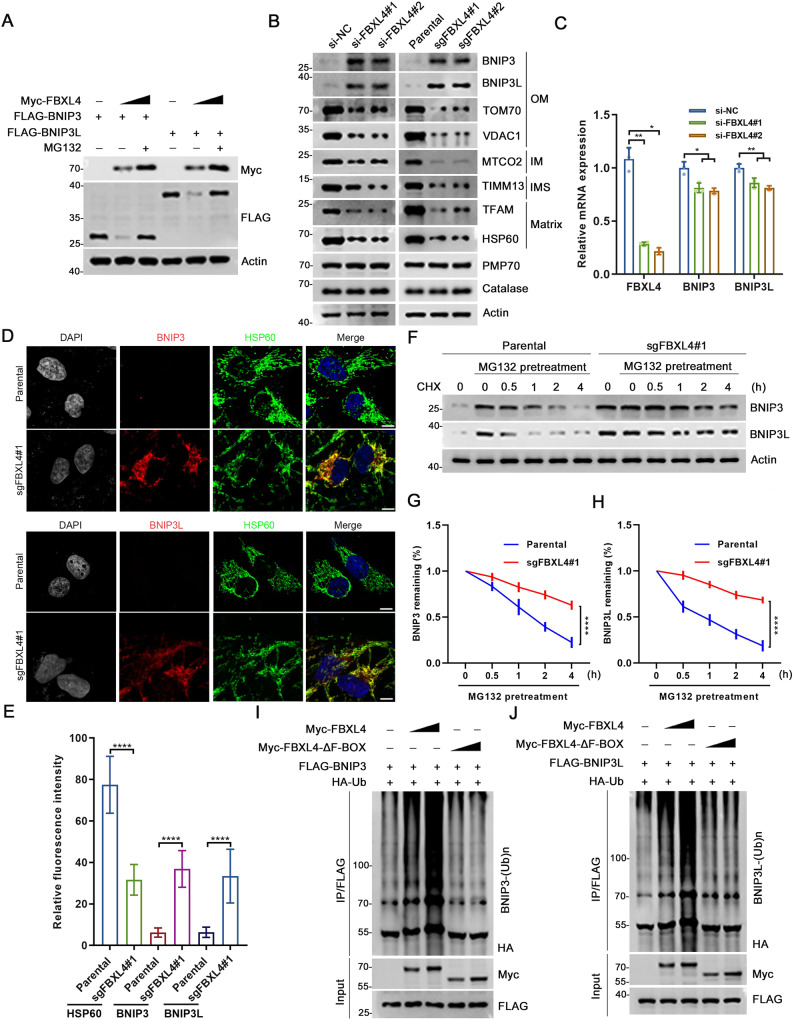
To investigate the potential roles of FBXL4 in promoting the ubiquitination and subsequent degradation of BNIP3/BNIP3L, we performed ectopic overexpression experiments. Our results showed that the wild-type FBXL4 markedly decreased the protein levels of BNIP3 or BNIP3L in a dose-dependent manner, while the ΔF-BOX mutant of FBXL4 had no such effect (Supplementary Fig. 1A, B). Notably, the proteasome inhibitor MG132 completely reversed the effect of FBXL4 on BNIP3/BNIP3L protein levels (Fig. 2A). Furthermore, we observed that depletion of FBXL4 through siRNA-mediated knockdown (KD) or CRISPR/Cas9-mediated knockout (KO) in HeLa and H1299 cells led to a marked increase in the steady-state levels of endogenous BNIP3/BNIP3L (Fig. 2B, Supplementary Fig. 1C, D). Notably, we observed that the mRNA levels of BNIP3/BNIP3L were even reduced in FBXL4-depleted cells compared to control cells (Fig. 2C), probably to buffer the notable accumulation of BNIP3/3 L protein,indicating that FBXL4 plays a role in the post-transcriptional regulation of BNIP3/BNIP3L. This reduction in mRNA levels may potentially serve as a compensatory mechanism to counterbalance the strong accumulation of BNIP3/3 L protein. Immunofluorescence (IF) analysis further revealed that the intensity of BNIP3/BNIP3L was markedly upregulated in FBXL4-KO cells (Fig. 2D, E). Moreover, the half-life of BNIP3/BNIP3L was remarkably prolonged in FBXL4-KO cells (Fig. 2F–H). Importantly, our data also demonstrated that the impact of FBXL4 on BNIP3/BNIP3L is independent of macroautophagy, as seen in ATG7-KO cells that FBXL4 KD still markedly upregulated BNIP3/BNIP3L protein levels (Supplementary Fig. 1E). It is worth mentioning that despite extensive efforts made to optimize antibody usage, none of the commercial antibodies were successful in yielding specific signals for FBXL4.
Fig. 2. FBXL4 mediates ubiquitination and degradation of BNIP3 and BNIP3L.
A 293 T cells were transfected with FLAG-BNIP3, FLAG-BNIP3L and increasing amount of Myc-FBXL4. The WCL were prepared for WB with the indicated antibodies. B WCL from HeLa cells transfected with FBXL4-specific siRNA or negative control (siNC) were prepared for WB with the indicated antibodies. FBXL4 KO cell lines were generated through LentiCRISPRv2 methods. The WCL from parental and FBXL4 KO HeLa cells were prepared for WB with the indicated antibodies. C RT-qPCR measurement of FBXL4, BNIP3 and BNIP3L mRNA expression in HeLa cells transfected with FBXL4-specific siRNA or siNC, Data are shown as means ± SD (n = 3). P values are calculated by the Two-way ANOVA test. *p < 0.05, **p < 0.01. D, E Representative IF images from parental and FBXL4 KO HeLa cells, stained with BNIP3 (or BNIP3L), HSP60 and DAPI. Scale bar, 10 μm. The relative intensity of HSP60, BNIP3 (or BNIP3L) and BNIP3L were quantified and shown in E. Data were shown as means ± SD (n = 50). P values are calculated by the Two-way ANOVA test. ****p < 0.0001. F–H WB analysis of the indicated proteins in the WCL of parental and FBXL4 KO HeLa cells pretreated with DMSO or MG132 (20 µM) for 5 h and then treated with cycloheximide (CHX, 50 μg/ml) and harvested at different time points. At each time point, the intensity of BNIP3 (G) and BNIP3L (H) was normalized to the intensity of Actin and then to the value at 0 h. P values are calculated by the Two-way ANOVA test. ****p < 0.0001. I, J WB analysis of the indicated proteins in vivo ubiquitination assays performed products and WCL from 293 T cells transfected with the indicated plasmids.
We then investigated whether other subunits of the CRL1FBXL4 complex were indispensable for BNIP3/BNIP3L destabilization. To do so, we depleted RBX1 or each Cullin (CUL1, 2, 3, 4A, 4B, or 5) in HeLa cells, and observed a marked increase in protein levels of BNIP3/BNIP3L upon depletion of RBX1, CUL1, or CUL2 (Supplementary Fig. 1F). Additionally, overexpression of the dominant-negative fragment of CUL1 or CUL2 (CUL1-DN, CUL2-DN) also resulted in elevated protein levels of BNIP3/BNIP3L, while such an impact was not observed when overexpressing other CULs-DN (Supplementary Fig. 1G). It is not surprising that CUL2 depletion also upregulates BNIP3/BNIP3L, as the CUL2VHL E3 ubiquitin complex is known to regulate protein stability of HIF1/2α, which in turn mediates transcriptional regulation of BNIP3/ BNIP3L [16].
We found that FBXL4-WT, but not the ΔF-BOX mutant, can promote polyubiquitination of BNIP3/BNIP3L (Fig. 2I, J). We utilized a panel of linkage-specific K27/K29/K48/K63-Ub antibodies to demonstrate that ubiquitinated BNIP3/BNIP3L mainly contain K48-Ub linkages, which is not unexpected since K48-Ub linkage is the canonical signal for protein degradation by the proteasome (Supplementary Fig. 2A, B). Ubiquitinome datasets deposited in the Phosphosite database (www.phosphosite.org) indicate that ubiquitination occurs at six lysine residues in BNIP3 and three lysine residues in BNIP3L, on their cytosolic sides (Supplementary Fig. 2C). By mutating these lysine residues to arginine, we found that FBXL4-mediated BNIP3/BNIP3L ubiquitination was completely abolished (Supplementary Fig. 2D, E), resulting in a significant extension of the protein half-life for both BNIP3/BNIP3L (Supplementary Fig. 2F–I). These results indicate that at least some of these lysine residues serve as ubiquitin attachment sites targeted by CRL1FBXL4 complex.
Having established the involvement of the ubiquitin-proteasome pathway in regulating BNIP3/3 L protein levels, it becomes intriguing to investigate whether this process is influenced by upstream signals or physiological conditions. Iron loss and hypoxia treatments are well-known inducers of mitophagy, leading to a notable increase in BNIP3/3 L protein levels. Nevertheless, it is currently believed that this upregulation occurs through HIFα-mediated transcription of BNIP3/3 L mRNAs. A question arises regarding whether these conditions also enhance the protein stability of BNIP3/3 L. However, our investigation revealed that the protein levels of exogenously overexpressed BNIP3/3L remained unchanged upon treatment with the iron chelator deferiprone (DFP) or exposure to hypoxia (2% O2), despite observing strong stabilization of HIF1α (Supplementary Fig. 3A–D). These findings indicate that, at least in our experimental conditions, iron loss or hypoxia did not affect the protein stability of BNIP3/3L.
Collectively, these data suggest that CRL1FBXL4 complex facilitates the ubiquitination and subsequent degradation of both BNIP3/BNIP3L. This degradation mechanism is reliant on the ubiquitin-proteasome pathway, but not macroautophagy.
MTDPS13 patient-associated FBXL4 mutants are defective in promoting ubiquitination and degradation of BNIP3/BNIP3L
In previous studies, more than 60 MTDPS13-associated FBXL4 mutations were reported (Supplementary Table 2). Approximately half of these mutations are frameshift or nonsense mutations, which typically predict loss of expression. Thus, we focused our analysis on the functional impact of missense mutations on FBXL4 function. We found that the protein levels of ectopically-overexpressed FBXL4 mutants are generally lower compared to FBXL4-WT. However, this distinction can be mitigated by treatment with MG132, suggesting that at least some, if not all, MTDPS13-associated FBXL4 mutations are destabilizing (Fig. 3A–C). Given that the majority of missense mutations of FBXL4 are located in the leucine-rich repeat domain responsible for binding BNIP3/BNIP3L, we also hypothesized that these mutations may alter the binding affinity between FBXL4 and BNIP3 or BNIP3L. We expressed wild-type or MTDPS13-associated FBXL4 mutants at similar levels and conducted semi-endogenous co-IP assays. Surprisingly, all tested FBXL4 mutants showed similar binding capacity to BNIP3 or BNIP3L as FBXL4-WT. However, these mutants exhibited reduced binding capacity to RBX1, SKP1, and CUL1, indicating that MTDPS13-associated FBXL4 mutants are generally defective in recruiting other subunits to assemble a CRL1FBXL4 complex (Fig. 3D). Furthermore, FBXL4-mediated ubiquitination of BNIP3 or BNIP3L was abolished or markedly attenuated in these mutants (Fig. 3E, F). In contrast to FBXL4-WT, reintroduction of the MTDPS13-associated FBXL4 mutants into FBXL4-KO cells cannot reverse the FBXL4 deficiency–caused accumulation of BNIP3/BNIP3L proteins (Fig. 3G).
Fig. 3. MTDPS13 patient-associated FBXL4 mutants are defective in promoting ubiquitination and degradation of BNIP3/BNIP3L.
A Schematic representation of six MTDPS13-associated FBXL4 mutations. B WB analysis of the indicated proteins in WCL from 293 T cells transfected with the indicated plasmids and treated with DMSO or MG132 (20 μM) for 5 h. C Quantification of the intensity of the indicated protein in B. (n = 3). The intensity of each band was normalized to the intensity of Actin, and One-way ANOVA test in A. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. D 293 T cells were transfected with the indicated plasmids. The WCL were prepared and subjected to co-IP with anti-FLAG antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies. E, F WB analysis of the indicated proteins in vivo ubiquitination assays performed products and WCL from 293 T cells transfected with the indicated plasmids. G WB analysis of the indicated proteins in WCL from parental and FBXL4 KO H1299 cells stably overexpressing EV, FLAG-FBXL4-WT, -I205T or –S410P mutant.
Collectively, these data suggest the ability of MTDPS13-associated FBXL4 mutants to promote BNIP3/BNIP3L degradation is severely compromised, resulting in BNIP3/BNIP3L accumulation in cells.
FBXL4 deficiency activates mitophagy, which is dependent on BNIP3/BNIP3L
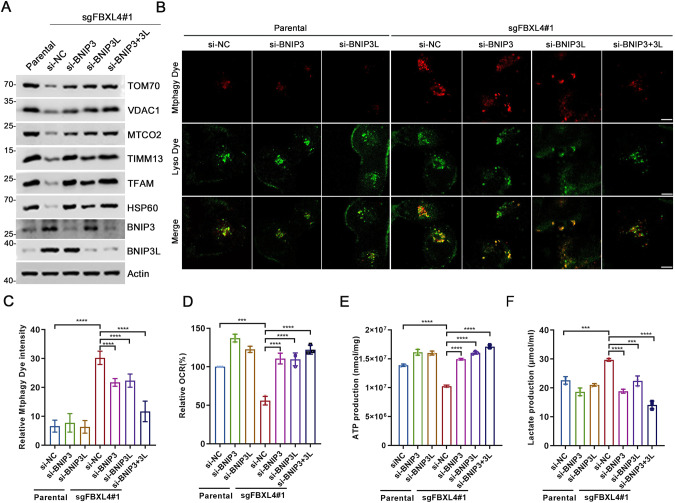
In line with previous research stating that mitophagy is hyperactive in FBXL4-KO mouse model [14], we found that KD or KO of FBXL4 in HeLa or H1299 cells led to a marked decrease in mitochondrial marker proteins across four sub-compartments (outer membrane, inner membrane, intermembrane space, and matrix) (Fig. 2B, Supplementary Fig. 1C). To investigate whether BNIP3/BNIP3L mediate the hyperactivation of mitophagy resulting from FBXL4 deficiency, we carried out individual or combined KD of BNIP3 and BNIP3L in FBXL4 KO cells. Our results indicated that KD of either BNIP3 or BNIP3L partially rescued the FBXL4 KO-induced downregulation of mitochondrial proteins, while combined KD of BNIP3/BNIP3L completely rescued it (Fig. 4A). In ATG7 KO HeLa cells, we observed that FBXL4 KD notably increased BNIP3/3L protein levels, while the levels of mitochondrial marker proteins remained unchanged. This result suggests that FBXL4 plays a crucial role in regulating mitochondrial contents by utilizing the canonical autophagy pathway (Supplementary Fig. 1E).
Fig. 4. FBXL4 deficiency activates mitophagy in a BNIP3/BNIP3L-dependent manner.
A WB analysis of the indicated proteins in the WCL from parental or FBXL4 KO HeLa cells transfected with indicated siRNAs. B, C Representative IF images from parental and FBXL4 KO HeLa cells transfected with indicated siRNAs, stained with Mtphagy Dye and Lyso Dye. Scale bar, 10 μm. The relative intensity of Mtphagy Dye was quantified and shown in C. Data were shown as means ± SD. (n = 50). D The OCR of parental and FBXL4 KO HeLa cells transfected with the indicated siRNA or siNC were measured using OCR assay Kit. Data are shown as means ± SD (n = 3). E The intracellular ATP production of parental and FBXL4 KO HeLa cells transfected with indicated siRNAs were measured using ATP production assay Kit. Data are shown as means ± SD (n = 3). F The intracellular lactate levels of parental and FBXL4 KO HeLa cells transfected with indicated siRNAs were measured using Lactate Assay Kit. Data are shown as means ± SD (n = 3). P values are calculated by the One-way ANOVA test and the Two-way ANOVA test in C–F. ***p < 0.001, ****p < 0.0001.
To monitor mitophagy, we utilized Mthagy Dye, which accumulates in intact mitochondria and displays weak fluorescence under normal conditions. Upon induction of mitophagy, damaged mitochondria fuse with lysosomes, and Mthagy Dye then emits strong fluorescence [17]. Our analysis showed that FBXL4 KO dramatically augmented mitophagy signals. However, KD of BNIP3 or BNIP3L alone partially blocked the activation of mitophagy, while combined KD of BNIP3/BNIP3L totally blocked it (Fig. 4B, C). We also assessed several mitochondrial metabolic indexes to evaluate mitochondrial function. FBXL4 KO resulted in decreased oxygen consumption rates (OCR) and ATP production, coupled with an increase in lactate production. Notably, the KD of BNIP3/BNIP3L substantially reversed these effects (Fig. 4D–F).
Collectively, these data suggest that FBXL4 deficiency-induced hyperactivation of mitophagy occurs due to accumulation of BNIP3 and BNIP3L.
FBXL4 deficiency activates BNIP3/BNIP3L-dependent mitophagy in Fbxl4 mutation knockin (KI) mouse model
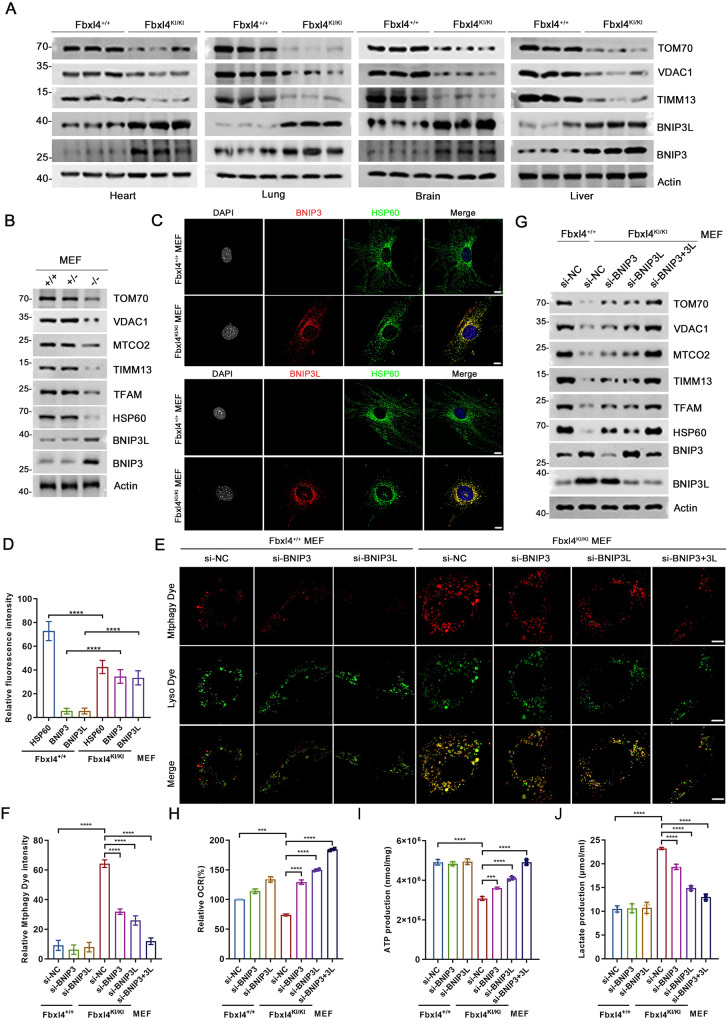
Previously, we reported a homozygous variant, c.993insA, in FBXL4, which causes a frameshift mutation resulting in a p.L332Tfs*3 variant in an affected patient. Clinical examination of a 1-year-old girl with this mutation revealed typical MTDPS13 features, including severe encephalopathy, early-onset lactic acidosis, hypotonia, developmental delay, and facial deformity [18, 19]. To better understand the pathophysiological impact of FBXL4 mutations on BNIP3/BNIP3L proteins and mitophagy in vivo, we generated a corresponding Fbxl4 c.994insA (p.L332Tfs*20) KI mouse model (Supplementary Fig. 4A). Among 224 offspring mice, only 2 homozygous KI mice were recovered at weaning, indicating most Fbxl4 KI pups die in the neonatal period (Supplementary Fig. 4B, C). This result is largely consistent with what was previously described in an Fbxl4 KO mouse model [14]. We collected multiple tissues, including heart, lung, and brain, from P0 mice. WB analysis indicated that the tissues of Fbxl4KI mice exhibited varying degrees of upregulation in BNIP3/BNIP3L proteins, compared to those of wild-type mice. Additionally, mitochondrial proteins were inevitably downregulated across all the examined tissues (Fig. 5A). Interestingly, a previous proteomic analysis revealed that BNIP3 was the sole mitochondrial protein that showed an upregulation in the liver tissues of Fbxl4-KO mice as compared to wild-type mice (Supplementary Fig. 4D) [14]. We then prepared mouse embryonic fibroblasts (MEFs). BNIP3/BNIP3L protein levels were markedly upregulated in homozygous Fbxl4KI/KI MEFs, compared to wild-type or heterogenous Fbxl4KI/+ MEFs (Fig. 5B–D). Similar to the observation in human cells, Fbxl4KI/KI MEFs showed a high level of basal mitophagy, which was nearly undetectable in wild-type MEFs. Importantly, KD of BNIP3 or BNIP3 alone partially blocked, while simultaneous KD of BNIP3/BNIP3L totally blocked Fbxl4 KI-induced mitophagy activation (Fig. 5E–G). Fbxl4KI/KI MEFs showed reduced OCR and ATP production, as well as increased in lactate production, compared to wild-type MEFs. Importantly, these effects were largely reversed by KD of BNIP3/BNIP3L (Fig. 5H–J).
Fig. 5. BNIP3/BNIP3L-dependent mitophagy was abnormally activated in Fbxl4 mutation KI mouse model.
A WB analysis of the indicated proteins in WCL from the indicated tissues of Fbxl4+/+ and Fbxl4KI/KI mice. B WB analysis of the indicated proteins in WCL from Fbxl4+/+, Fbxl4+/KI and Fbxl4KI/KI MEFs. C, D Representative IF images from Fbxl4+/+and Fbxl4KI/KI MEFs, stained with BNIP3 (or BNIP3L), HSP60 and DAPI. Scale bar, 10 μm. The relative intensity of HSP60, BNIP3 (or BNIP3L) and BNIP3L were quantified and shown in D. Data were shown as means ± SD (n = 50). P values are calculated by the Two-way ANOVA test. ****p < 0.0001. E, F Representative IF images from Fbxl4+/+and Fbxl4KI/KI MEFs transfected with the indicated siRNAs, stained with Mtphagy Dye and Lyso Dye. Scale bar, 10 μm. The relative intensity of Mtphagy Dye was quantified and shown in D. Data were shown as means ± SD (n = 50). G WB analysis of the indicated proteins in WCL from Fbxl4+/+and Fbxl4KI/KI MEFs transfected with the indicated siRNAs. H The OCR of Fbxl4+/+ and Fbxl4KI/KI MEFs transfected with indicated siRNAs were measured using OCR assay Kit. Data are shown as means ± SD (n = 3). I The intracellular ATP production of Fbxl4+/+ and Fbxl4KI/KI MEFs transfected with the indicated siRNA or siNC were measured using ATP production assay Kit. Data are shown as means ± SD (n = 3). J The intracellular lactate levels of Fbxl4+/+ and Fbxl4KI/KI MEFs transfected with the indicated siRNAs were measured using Lactate Assay Kit. Data are shown as means ± SD (n = 3). P values are calculated by the One-way ANOVA test and the Two-way ANOVA test in F–J. ***p < 0.001, ****p < 0.0001.
Collectively, these data verified that Fbxl4 deficiency-induced mitophagy hyperactivation is due to BNIP3/BNIP3L accumulation in Fbxl4 mutation KI mouse model.
Mitophagy is abnormally activated in neural progenitor cells (NPCs) and cortical neurons (CNs) induced from MTDPS13 patient hiPSCs
Mitophagy dysregulation is an early indicator of neuronal dysfunction, which is commonly observed in both neurodevelopmental and age-related neurodegenerative disorders [4]. To investigate whether FBXL4 mutations cause hyperactive mitophagy in neurons, we modeled the disease in a set of neural induction experiments, using a hiPSC clone (FBXL4MUT) generated by reprograming of human peripheral blood mononuclear cells from our MTDPS13 patient [18]. Human embryonic stem cells H9 (hESCs) were used as a healthy control. To model MTDPS13, we induced CNs from hiPSCs or H9 hESCs up to the stage of neurospheres on Day 30 (Supplementary Fig. 5A–C). Western blot (WB) analyses revealed that FBXL4MUT hiPSCs had higher levels of BNIP3/BNIP3L proteins but lower levels of mitochondrial marker proteins compared to control H9 ESCs. Similar trends were observed in CNs-induced from both hiPSCs and hESCs (Fig. 6A). We then performed IF analysis and labeled NPCs with SOX2 and postmitotic CNs with Doublecortin (DCX). Both H9 hESCs and FBXL4MUT hiPSCs had similar neurosphere-forming capacities. However, the IF intensity of BNIP3/BNIP3L proteins was much higher, while that of mitochondrial marker protein HSP60 was much lower in FBXL4MUT hiPSC-induced NPCs or CNs than in H9 ESC-induced NPCs or CNs (Fig. 6B, C, Supplementary Fig. 6A, B). DFP induces mitophagy via HIF1α-dependent transcriptional upregulation of BNIP3/BNIP3L [20]. We observed significantly higher basal mitophagy in FBXL4MUT hiPSC-induced NPCs or CNs when compared to H9 ESC-induced NPCs or CNs. Moreover, DFP-induced mitophagy was further exacerbated in FBXL4MUT hiPSC-induced NPCs or CNs compared to H9 ESC-induced NPCs or CNs (Fig. 6D–F, Supplementary Fig. 6C–E).
Fig. 6. Mitophagy is abnormally activated in CNs induced from MTDPS13 patient hiPSCs.
A WB analysis of the indicated proteins in PSC (H9 ESC and FBXL4MUT iPSC) and CNs derived from H9 ESC and FBXL4MUT iPSC (on Day 30). PSC: Pluripotent stem cells; ESC: Embryonic stem cells; iPSC: Induced pluripotent stem cells; CNs: cortical neurons. Representative images (B) of DCX, HSP60, BNIP3 and BNIP3L antibody immunostaining in 500 µM DFP treated H9/FBXL4MUT CNs (right) with untreated H9/FBXL4MUT CNs (left). Scar bar, 5 µm. Comparing the HSP60, BNIP3 (or BNIP3L) immunostaining intensity of DCX positive cells (C). Data were shown as means ± SD (n = 50). P values are calculated by the Two-way ANOVA test. **P < 0.005; ****P < 0.0001. Representative live cell imaging (D) of 500 µM DFP treated H9/FBXL4MUT CNs with untreated H9/FBXL4MUT CNs, stained with Mtphagy Dye and Lyso Dye, and quantitative analysis (E) of mitophagy levels. Scar bar, 10 µm. Data were shown as means ± SD (n = 50). P values are calculated by the Two-way ANOVA test. ****P < 0.0001. F WB analysis of the indicated proteins in DFP (500 µM)-treated H9/FBXL4MUT CNs with untreated H9/FBXL4MUT CNs (left) on Day 30.
Collectively, these data suggest that FBXL4 mutations lead to excessive mitophagy in NPCs or CNs, which may result in neuronal dysfunction in individuals with MTDPS13.
Discussion
The identification of new genes associated with MTDPS offers valuable insights into the molecular pathways underlying mitochondrial dysfunction and potential avenues for targeted therapies. However, effective treatments for MTDPS remain elusive due to multi-organ involvement, and a more comprehensive understanding of the molecular mechanisms underlying the disease is necessary [12]. Our study reveals that FBXL4, a gene linked to MTDPS13, plays a critical role in mitophagy by assembling an active CRL1FBXL4 complex to target BNIP3/BNIP3L for proteasomal degradation. Notably, MTDPS13-associated FBXL4 mutants are generally unstable and exhibit reduced capacity to assemble an active CRL1FBXL4 complex, resulting in overactivation of BNIP3/3L-dependent mitophagy. The excessive mitophagy observed in this scenario can lead to the degradation of healthy mitochondria, ultimately resulting in organ failure in tissues with high metabolic demand (such as the brain, heart, and skeletal muscle) and the onset of MTDPS13 (Fig. 7).
Fig. 7. A working model proposed in this study.
A schematic diagram is illustrated to depict a model in which FBXL4 mutations lead to the overactivation of BNIP3/BNIP3L-dependent mitophagy in MTDPS13 patients.
Mitophagy has been observed to occur even under apparently normal conditions, a phenomenon referred to as “basal” mitophagy [2]. Despite not yet fully understanding the precise function or functions of basal mitophagy, researchers have utilized fluorescent mitophagy reporters such as mt-Keima and mito-QC to demonstrate that basal mitophagy is prevalent across tissues in both mice and flies [7, 21]. Our study has shown evidence of robust mitophagy in FBXL4-null human and mouse cells, even without any overt external stresses. These findings support the notion that basal mitophagy is a general housekeeping mechanism for maintaining mitochondrial homeostasis. it is widely understood that hypoxia and iron chelation are known to trigger mitophagy by increasing BNIP3/BNIP3L transcription in a manner dependent on the HIF1/2α pathway [16, 22]. Our results have shown that the protein half-life of BNIP3/BNIP3L is relatively short under normal conditions. However, in FBXL4-deficient cells, BNIP3/BNIP3L is strongly stabilized. It is possible that the ubiquitin-proteasome pathway tightly regulates BNIP3/BNIPL protein levels, enabling quick accumulation of BNIP3/NIP3L post-translationally in response to specific stresses, ultimately initiating mitophagy. Yet, the precise triggers for this pathway remain unknown and warrant further investigation. In this study, we made an intriguing observation regarding the decrease in BNIP3/3 L mRNAs following FBXL4 depletion. This finding suggests the presence of a cellular mechanism that transcriptionally suppresses BNIP3/3L expression in order to prevent excessive mitophagy, which could potentially impair cell fitness.
In addition to mitochondria, BNIP3/BNIP3L are also localized to peroxisomes to promote pexophagy, which is a selective autophagy that targets peroxisomes and is essential for the maintenance of homeostasis of peroxisomes [23, 24]. iron chelator DFP can induce both mitophagy and pexophagy in a BNIP3/BNIP3L-dependent manner [20]. it is intriguing to note that the protein levels of peroxisomal markers (Catalase and PMP70) were not altered in FBXL4-KO cells (Fig. 2B, Supplementary Fig. 1C), indicating that FBXL4 does not regulate pexophagy. FBXL4 is primarily localized in the mitochondria and may only regulate mitochondrial pools of BNIP3/BNIP3L, thus playing a specific role in mitophagy. Besides their role in mitophagy, BNIP3/BNIP3L are also important regulators of other forms of programmed cell death, including apoptosis, necrosis, necroptosis, pyroptosis, and ferroptosis, which are essential for maintaining tissue homeostasis and responding to cellular stress and damage [25]. Future studies could explore whether dysregulation of BNIP3/BNIP3L in these pathways can contribute to FBXL4 mutation-induced MTDPS13, providing further insight into the full molecular mechanisms underlying this disease.
MTDPS is a group of genetic disorders that are caused by mutations, resulting in the reduction of mtDNA copy number in affected tissues, impaired mitochondrial function and multi-organ dysfunction [12]. Today, a dozen of gene mutations has been characterized to cause MTDPS. The genetic heterogeneity of MTDPS highlights the complexity of mtDNA maintenance mechanisms. Almost all these genes are directly involved in mtDNA synthesis, replication, maintenance, and stability [26]. Our findings indicate that MTDPS13 may represent a unique subtype, as the mutations in the FBXL4 gene lead to mitochondrial content depletion, resulting in a proportionate decrease in mtDNA rather than regulating mtDNA synthesis or replication. Until now, treatment options for FBXL4-related MTDPS13 are still limited and mainly supportive. Based on our findings, we proposed that design and discovery of small-molecule PROTAC BNIP3/BNIP3L degraders can be pursued as a new therapeutic strategy to overcome BNIP3/BNIP3L accumulation in MTDPS13 patients. However, finding a small molecule that can bind highly specifically to BNIP3/BNIP3L is a big challenge. mDivi-1 is a small molecule that targets dynamin-related protein, a crucial regulator of mitochondrial division, and has demonstrated inhibitory effects on mitochondrial fission. Mitochondrial fragmentation and mitophagy inhibition are among its beneficial effects through its ability to inhibit Drp1 [27]. It would be worthwhile to investigate whether mitochondrial fission inhibitors, such as mDivi-1, can counteract the excessive mitophagy triggered by FBXL4 deficiency, restore mitochondrial dynamics, and ultimately alleviate the clinical symptoms of MTDPS13 patients. This area of research warrants further investigation.
Materials and methods
Cell lines, lentiviral infection and transfection
HeLa, H1299, and 293 T cells were acquired from American Type Culture Collection (ATCC). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C with 5% CO2. We routinely perform DNA fingerprinting and PCR to verify the authenticity of the cell lines and to ensure they are free of mycoplasma infection. We conducted transient transfection using EZ Trans (Shanghai Life-iLab Biotech). For lentiviral transfection, we transfected pLKO shRNA KD or pCDH overexpression plasmids and virus-packing constructs into 293 T cells. The viral supernatant was collected after 48 h. The cells were then infected with the viral supernatant in the presence of polybrene (8 µg/ml) and selected in growth media containing puromycin (1.5 μg/ml). The gene-specific shRNA or siRNA sequences can be found in Supplementary Table 3.
Antibodies, recombinant proteins, and chemicals
The information of antibodies, recombinant proteins, and chemicals used in this study is listed in Supplementary Tables 4, 5.
GST pull-down assays
GST fusion proteins were immobilized on glutathione-Sepharose beads (GE Healthcare). The beads were subsequently washed with pull-down buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40, 1 mM DTT, 10% glycerol, 1 mM EDTA, 2.5 mM MgCl2, and 1 µg/ml leupeptin. Afterward, the beads were incubated with the recombinant His-tagged proteins for 2 h. The beads were then washed 5 times with binding buffer and resuspended in sample buffer. Finally, the bound proteins were subjected to SDS–PAGE and WB analysis.
Gene KO cell line generation
The sgRNAs targeting the FBXL4 gene were designed using an online CRISPR design tool (http://crispr.mit.edu) and subcloned into the LentiCRISPRv2 vector from Dr. Feng Zhang’s lab. HeLa and H1299 cells were plated and transfected with LentiCRISPRv2 and virus-packing constructs. The viral supernatant was collected after 48 h. The cells were then infected with the viral supernatant in the presence of polybrene (8 µg/ml). HeLa and H1299 cells were selected with 1.5 μg/ml puromycin to generate the stable cell lines. Supplementary Table 3 lists the sequences of gene-specific sgRNAs.
RNA isolation and quantitative real-time reverse transcription PCR (qRT-qPCR)
Total RNAs were isolated from cells using the TRIzol reagent (Thermo) following the manufacturer’s instructions. Concentrations and purity of RNAs were determined by measuring the absorption of ultra-violet lights using a NanoDrop spectrophotometer (Thermo). cDNAs were reversed-transcribed using a HiScript III 1st Strand cDNA Synthesis Kit (Vazyme), followed by amplification of cDNA using ChamQ SYBR qPCR Master Mix (Vazyme). The relative mRNA levels of genes were quantified using the 2-ΔΔCt method, with normalization to Actin. The primer sequences are listed in Supplementary Table 3.
IF and confocal microscopy
HeLa, H9 hESCs, FBXL4MUT hiPSCs, NPCs, or CNs were seeded on glass coverslips in 24-well plates and harvested at 80% confluence. The cells were washed with PBS and fixed with 4% paraformaldehyde in PBS. After permeabilization with 0.4% Triton X-100 for 10 min and then in the blocking solution (PBS plus 5% donkey serum), for 30 min at room temperature (RT). The cells were then incubated with primary antibodies at 4 °C overnight. After washing with PBST buffer, fluorescence-labelled secondary antibodies were applied. DAPI was utilized to stain nuclei. The glass coverslips were mounted on slides and imaged using a confocal microscope (LSM880, Zeiss) with a 63*/1.4NA Oil PSF Objective. Quantitative analyses were performed using ImageJ software.
Oxygen consumption assays
OCR was measured under basal conditions in the presence of the mitochondrial inhibitors oligomycin (0.25 μmol/L, Calbiochem). Experiments were performed at 37 °C. OCR was calculated by the oligomycin-induced changes in comparison to basal rates. H1299 cells were seeded at a density of 20,000 cells in the cell culture microplate. The total protein of each well was determined by Bradford assay and used as the reference to normalize the OCR.
ATP measurement assays
H1299 and MEF cells were seeded in 6-well plates at 2 × 106 per well and were transfected with indicated siRNAs. After 48 h of transfection, the cells were collected for determination of ATP production by using the ATP assay kit (Beyotime).
Lactate measurement assays
HeLa and MEF Cells were seeded into six-well plates at a density of 1 × 105 cells in 2 ml DMEM medium per well and cultured overnight. According to the manual of the Lactate Acid Assay Kit (Solarbio), The resulting color was assayed was measured at 570 nm using a microplate absorbance reader.
Generation and breeding of fbxl4 c.994insA KI mice
Mice with c.993-994insA mutation of murine Fbxl4 were designed and generated from Shanghai Model Organisms Center, Inc. (Shanghai, China). In brief, Cas9 mRNA was in vitro transcribed with mMESSAGE T7 Ultra Kit (Ambion) according to the manufacturer’s instructions, and subsequently purified using the MEGAclearTM Kit (Thermo) Cas9 targeted guide RNA (sgRNA) was in vitro transcribed using the MEGAshortscript Kit (Thermo) and subsequently purified using MEGAclearTM Kit. The transcribed Cas9 mRNA and sgRNA as well as a 200 base pairs single-stranded oligo deoxy nucleotide (ssODN) were co-injected into zygotes of C57BL/6 J mouse. Obtained F0 mice were validated by PCR and sequencing. The F0 mice with expected point mutation were chosen and crossed with C57BL/6 J mice to produce F1 mice. The genotype of F1 mice was identified by PCR and confirmed by sequencing. The sequences used for CRISPR/Cas9 editing and the primers used for genotyping of the mutant site (c.993dupA) are listed in Supplementary Table 3.
Mice were maintained under a 12 h/12 h light/dark cycle at 22–25 °C and 40–50% humidity with standard food and water available ad libitum. Studies were performed with E13 mouse pups. The ethical approval for the study was obtained from the Regional Committee for Medical and Health Research Ethics, Children’s Hospital of Shanghai. All procedures for animal care and animal experiments were carried out in accordance with the guidelines of the Care and Use of Laboratory Animals proposed by Children’s Hospital of Shanghai and Shanghai Municipality, PR China. The protocol was approved by the Science and Technology Commission of Shanghai Municipality (Permit Number: SYXK (hu) 2020-0032).
MEFs generation and immortalization
Timed pregnant female mice at embryonic day 12.5 to 14.5 were sacrificed, and the embryos were carefully dissected to remove the cerebrum, internal organs, and limbs. The remaining tissues were cut into small pieces and treated with trypsin-EDTA (0.25%) for 10 min at 37 °C. The trypsin was neutralized with DMEM, a complete medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. The culture media were changed every 2-3 days until the cells reached confluence. To immortalize MEFs, they were passaged up to approximately 10 times before infection with lentiviral vectors expressing the SV40 large T-antigen. Stable transduction was achieved with puromycin selection. The successful integration of the immortalizing gene was confirmed through Sanger sequencing and WB analysis.
Maintenance of human pluripotent stem cells
All procedures were approved by the Ethics Committee of the Ethics Committees at Shanghai Children’s Hospital (2023RY041-E01). hiPSCs from a one-year-old Chinese girl with MTDPS13 were established and reported previously [18]. The whole exome sequencing showed a homologous variant c.993insA(p.L332Tfs*3) in the FBXL4 gene. FBXL4MUT hiPSCs and human embryonic stem cell line H9 ESCs were cultured and maintained in feeder-free conditions (Matrigel, (Corning) using E8 medium (Gibco) at 37 °C with 5% CO2. Half of the medium was exchanged with fresh E8 medium daily, and the cells were passaged every 5–7 days when they reached 80% confluence using EDTA (0.5 mM). On day 7, stem cells were dissociated with Accutase (Gibco) at 37 °C for 2 min and placed onto poly-ornithine/laminin-coated coverslips. Then these cells were cultured in the E8 medium for use.
Differentiation of cortical neurons
hESCs and hiPSCs were detached with Accutase and then allowed to form embryoid bodies (EBs) the next day according to the methods of previous reports [28, 29]. A total of 9,000 dissociated cells were plated into each well of a V-bottom ultra-low attachment 96-well plate (Sumitomo Bakelite, MS-9096VZ). EBs were cultured in half E8 medium and half neural induction medium (NIM; DMEM/F-12, 1% (v/v) N2 supplement (Gibco) MEM-NEAA (Gibco), 1% (v/v) GlutaMAX (Gibco) with 0.3 μm LDN-193189 (STEMGENT4) and 2 μm SB431542 (Ametek Scientific). On Day 6, the medium was replaced with the neural induction medium. On day 10, the EBs were attached to the plastic or laminin-coated substrate in six-well plates and cultured in the neural induction medium and neural tube-like rosettes were observed starting on days 10–11. On Day 15, the rosette clones were gently blown off using a 1-ml pipette. The cells were then continuously floated in the same medium until day 30. From Day 30, neuroepithelial spheres were dissociated with Accutase at 37 °C for 2–3 min and placed onto poly-ornithine/laminin-coated coverslips in the neural induction medium.
Statistical analysis
Band intensities of WB results were calculated by ImageJ in accordance with the manufacturer’s instructions. Statistical analysis was performed using GraphPad Prism (GraphPad Software), and the differences between the two groups were analyzed using one-way analysis of variance (ANOVA) or two-way analysis of variance ANOVA. All data were displayed as means ± S.D. values for experiments conducted with at least three replicates. * represents p < 0.05; ** represents p < 0.01; *** represents p < 0.001, **** represents p < 0.0001.
Supplementary information
Acknowledgements
The image in graphical model is produced by online program FigDraw (https://www.figdraw.com).
Author contributions
CW conceived the study. YC, DJ, YL, XX, YW, and XL performed the experiments and data analyses. CW, LM, SZ, YC, YL, HS, and KG analyzed and interpreted the data. CW wrote the manuscript.
Funding
This work was in part supported by the National Natural Science Foundation of China (No. 91957125, 81972396 to CW; 31821002, 31930062 to SZ; 82071269 to LM; 82272992, 91954106 to KG), the State Key Development Programs of China (No. 2022YFA1104200 to CW; 2018YFA0800300 to SZ; 2021YFA1101302 to LM), the Natural Science Foundation of Shanghai (No. 22ZR1406600 to CW; 22ZR1449200 to KG), and the Open Research Fund of State Key Laboratory of Genetic Engineering, Fudan University (No. SKLGE-2111 to KG). Science and Technology Research Program of Shanghai (No. 9DZ2282100), Shanghai key clinical specialty project (No. shslczdzk05705 to YC), National Fund Cultivation Special Project of Shanghai Children’s Hospital (No. 2021YGZQ05 to XL).
Data availability
The mass spectrometry proteomics data have been deposited to the via the PRIDE partner repository with the dataset identifier PXD040601. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yingji Chen, Dongyue Jiao, Yang Liu.
Contributor Information
Yucai Chen, Email: chenyc@shchildren.com.cn.
Shi-Min Zhao, Email: zhaosm@fudan.edu.cn.
Lixiang Ma, Email: lxma@fudan.edu.cn.
Chenji Wang, Email: chenjiwang@fudan.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01205-1.
References
- 1.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–59. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montava-Garriga L, Ganley IG. Outstanding Questions in Mitophagy: What We Do and Do Not Know. J Mol Biol. 2020;432:206–30. doi: 10.1016/j.jmb.2019.06.032. [DOI] [PubMed] [Google Scholar]
- 3.Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20:1013–22. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 4.Misgeld T, Schwarz TL. Mitostasis in Neurons: Maintaining Mitochondria in an Extended Cellular Architecture. Neuron. 2017;96:651–66. doi: 10.1016/j.neuron.2017.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newman LE, Shadel GS. Pink1/Parkin link inflammation, mitochondrial stress, and neurodegeneration. J Cell Biol. 2018;217:3327–9. doi: 10.1083/jcb.201808118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harper JW, Ordureau A, Heo JM. Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol. 2018;19:93–108. doi: 10.1038/nrm.2017.129. [DOI] [PubMed] [Google Scholar]
- 7.McWilliams TG, Prescott AR, Montava-Garriga L, Ball G, Singh F, Barini E, et al. Basal Mitophagy Occurs Independently of PINK1 in Mouse Tissues of High Metabolic Demand. Cell Metab. 2018;27:439–49 e435. doi: 10.1016/j.cmet.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20:233–42. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Killackey SA, Philpott DJ, Girardin SE. Mitophagy pathways in health and disease. J Cell Biol. 2020;11:219. [DOI] [PMC free article] [PubMed]
- 10.Gai X, Ghezzi D, Johnson MA, Biagosch CA, Shamseldin HE, Haack TB, et al. Mutations in FBXL4, encoding a mitochondrial protein, cause early-onset mitochondrial encephalomyopathy. Am J Hum Genet. 2013;93:482–95. doi: 10.1016/j.ajhg.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnen PE, Yarham JW, Besse A, Wu P, Faqeih EA, Al-Asmari AM, et al. Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am J Hum Genet. 2013;93:471–81. doi: 10.1016/j.ajhg.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Hattab AW, Scaglia F. Mitochondrial DNA depletion syndromes: review and updates of genetic basis, manifestations, and therapeutic options. Neurotherapeutics. 2013;10:186–98. doi: 10.1007/s13311-013-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavorato M, Nakamaru-Ogiso E, Mathew ND, Herman E, Shah N, Haroon S, et al. Dichloroacetate improves mitochondrial function, physiology, and morphology in FBXL4 disease models. JCI Insight. 2022;7:e156346. [DOI] [PMC free article] [PubMed]
- 14.Alsina D, Lytovchenko O, Schab A, Atanassov I, Schober FA, Jiang M, et al. FBXL4 deficiency increases mitochondrial removal by autophagy. EMBO Mol Med. 2020;12:e11659. doi: 10.15252/emmm.201911659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–80. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee P, Chandel NS, Simon MC. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat Rev Mol Cell Biol. 2020;21:268–83. doi: 10.1038/s41580-020-0227-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwashita H, Torii S, Nagahora N, Ishiyama M, Shioji K, Sasamoto K, et al. Live Cell Imaging of Mitochondrial Autophagy with a Novel Fluorescent Small Molecule. ACS Chem Biol. 2017;12:2546–51. doi: 10.1021/acschembio.7b00647. [DOI] [PubMed] [Google Scholar]
- 18.Yuan F, Wang C, Xi J, Wang S, Lin L, Wang Y, et al. Generation of an induced pluripotent stem cell line SHCDNi001-A from a one-year-old Chinese girl with mitochondrial DNA depletion syndrome 13. Stem Cell Res. 2020;45:101832. doi: 10.1016/j.scr.2020.101832. [DOI] [PubMed] [Google Scholar]
- 19.Wang S, Lin L, Wang Y, Wang A, Liu Z, Wu S, et al. Novel homozygous mutation in the FBXL4 gene is associated with mitochondria DNA depletion syndrome-13. J Neurol Sci. 2020;416:116948. doi: 10.1016/j.jns.2020.116948. [DOI] [PubMed] [Google Scholar]
- 20.Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–35. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McWilliams TG, Prescott AR, Allen GF, Tamjar J, Munson MJ, Thomson C, et al. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol. 2016;214:333–45. doi: 10.1083/jcb.201603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao JF, Rodger CE, Allen GFG, Weidlich S, Ganley IG. HIF1alpha-dependent mitophagy facilitates cardiomyoblast differentiation. Cell Stress. 2020;4:99–113. doi: 10.15698/cst2020.05.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilhelm LP, Zapata-Munoz J, Villarejo-Zori B, Pellegrin S, Freire CM, Toye AM, et al. BNIP3L/NIX regulates both mitophagy and pexophagy. EMBO J. 2022;41:e111115. doi: 10.15252/embj.2022111115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn WA, Jr., Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, et al. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–46. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Hattab AW, Craigen WJ, Scaglia F. Mitochondrial DNA maintenance defects. Biochim Biophys Acta Mol Basis Dis. 2017;1863:1539–55. doi: 10.1016/j.bbadis.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 27.Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma L, Hu B, Liu Y, Vermilyea SC, Liu H, Gao L, et al. Human embryonic stem cell-derived GABA neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell. 2012;10:455–64. doi: 10.1016/j.stem.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Saiyin H, Liu Y, Wang Y, Li X, Ji R, et al. Human striatal organoids derived from pluripotent stem cells recapitulate striatal development and compartments. PLoS Biol. 2022;20:e3001868. doi: 10.1371/journal.pbio.3001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the via the PRIDE partner repository with the dataset identifier PXD040601. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.