Abstract
The COVID-19 pandemic, caused by the β-coronavirus (β-CoV) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), continues to cause significant global morbidity and mortality. While vaccines have reduced the overall number of severe infections, there remains an incomplete understanding of viral entry and innate immune activation, which can drive pathology. Innate immune responses characterized by positive feedback between cell death and cytokine release can amplify the inflammatory cytokine storm during β-CoV–mediated infection to drive pathology. Therefore, there remains an unmet need to understand innate immune processes in response to β-CoV infections to identify therapeutic strategies. To address this gap, here we used an MHV model and developed a whole genome CRISPR-Cas9 screening approach to elucidate host molecules required for β-CoV infection and inflammatory cell death, PANoptosis, in macrophages, a sentinel innate immune cell. Our screen was validated through the identification of the known MHV receptor Ceacam1 as the top hit, and its deletion significantly reduced viral replication due to loss of viral entry, resulting in a downstream reduction in MHV-induced cell death. Moreover, this screen identified several other host factors required for MHV infection-induced macrophage cell death. Overall, these findings demonstrate the feasibility and power of using genome-wide PANoptosis screens in macrophage cell lines to accelerate the discovery of key host factors in innate immune processes and suggest new targets for therapeutic development to prevent β-CoV-induced pathology.
Subject terms: Cell biology, Microbiology
A CRISPR screen identified host factors that promote viral entry and inflammatory cell death, PANoptosis, upon infection with the murine hepatitis virus, a type of beta-coronavirus, suggesting potential therapeutic targets for coronavirus infections.
Introduction
Coronavirus infections are among the current leading causes of global morbidity and mortality. The global burden of confirmed COVID-19 cases has surpassed 775 million, resulting in more than 6.9 million deaths as of May 22, 20231. COVID-19 is caused by the β-coronavirus (β-CoV) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)2. β-CoVs are positive sense single-stranded RNA (ssRNA) viruses that carry a relatively large RNA genome of about 30 kb3,4. These viruses cause respiratory infections that result in severe inflammatory immune pathology that drives morbidity and mortality5. Newly developed vaccines have significantly reduced symptomatic COVID-19 infections and mortality6. However, the progress in developing therapeutics to treat these infections is lagging behind, and there remains an urgent unmet need to provide treatment for patients and prevent severe disease manifestations.
Identifying new treatment strategies requires a mechanistic understanding of viral entry and innate immune activation to identify therapeutic targets. Innate immunity generally provides the first line of defense against infection and disease, but in the case of β-CoV infections, excess innate immune activation has been associated with the release of high levels of pro-inflammatory cytokines and chemokines, leading to a cytokine storm and pathogenesis7–12. Exacerbated cytokine release and organ failure upon infection with respiratory RNA viruses can be caused by aberrant innate immune-mediated regulated cell death13–18. Cell death pathways can be characterized as non-lytic (e.g., apoptosis) and lytic (e.g., pyroptosis and necroptosis). Mechanistically, apoptosis is initiated by caspase-8 and -9 and executed by caspase-3 and -719; pyroptosis is induced by inflammatory caspases, caspase-1 and caspase-11 (mouse)/caspase-4 and -5 (human), and executed by gasdermin family members20–24; and caspase-8 inhibition leads to necroptosis via the RIPK3/MLKL pathway25–29. Growing evidence has shown extensive crosstalk among these cell death pathways, leading to the identification of PANoptosis. PANoptosis is a unique innate immune lytic, inflammatory cell death pathway that is driven by caspases and RIPKs and regulated by multiprotein PANoptosome complexes. PANoptosomes form upon cytosolic pattern recognition receptor sensing of pathogens, pathogen-associated molecular patterns (PAMPs), damage-associated molecular patterns (DAMPs), or the cytokines produced downstream30–35.
PANoptosis has been extensively implicated in β-CoV pathology. For instance, mouse hepatitis virus (MHV), a prototypical β-CoV that mimics many of the key aspects of human β-CoV biology, induces NLRP3 inflammasome activation and PANoptosis in murine bone marrow-derived macrophages (BMDMs)36. Additionally, SARS-CoV-2 and MHV induce PANoptosis in response to IFN therapy in both human and mouse cells, and this cell death drives an inflammatory immune response and lethality in murine models15. Furthermore, TNF and IFN-γ released during SARS-CoV-2 infection induce robust PANoptosis that drives inflammation, cytokine storm, and morbidity and mortality in murine models16. Acute respiratory distress syndrome (ARDS), a determining feature of COVID-19 disease severity, is promoted by myeloid cell death in patients with COVID-19 and in the MHV mouse model10,15.
Given the clear connections between innate immune inflammatory cell death, PANoptosis, and pathology in β-CoV infections, it is critical to understand the molecular mechanisms involved in this pathway to identify new therapeutic targets. We therefore utilized a whole genome CRISPR-based knockout screen to identify the key host factors driving cell death in response to MHV infection. We identified Ceacam1 as a critical molecule that regulates viral infection and acts as an important node in inflammatory cell death. Our CRISPR screen also identified several other molecules that are likely to have critical roles in β-CoV-mediated cell death that require further evaluation in future studies. Blocking these mechanisms presents a promising strategy to inhibit inflammatory cell death and reduce pathology in β-CoV infections.
Results
Genome-wide CRISPR screen identified host factors required for MHV-induced cell death
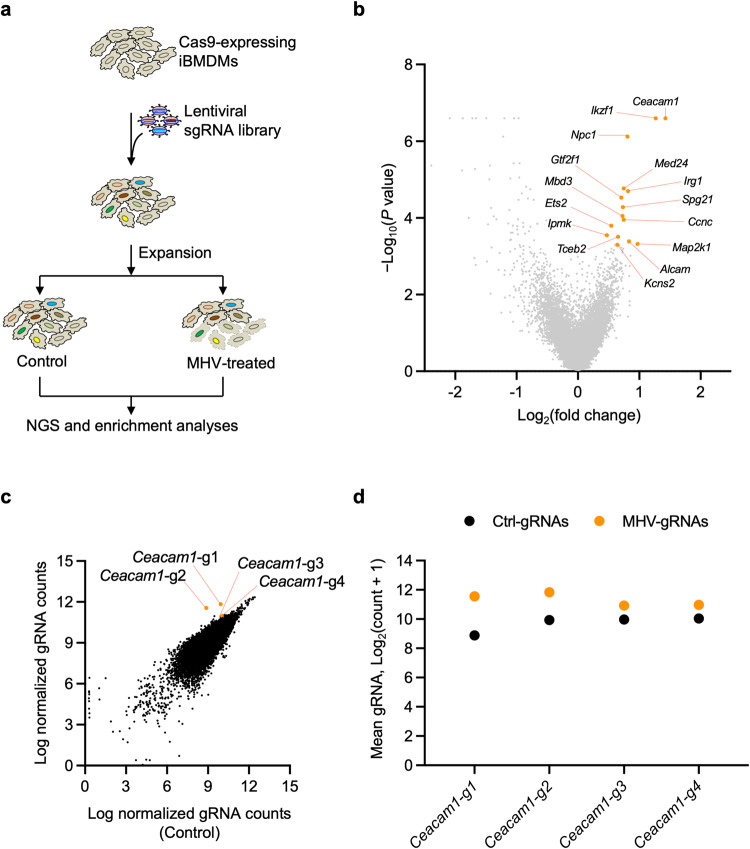
While extensive connections have been found between innate immune inflammatory cell death, PANoptosis, and pathology in β-CoV infections15,16,36, a need for a comprehensive understanding of the host cellular pathways involved in β-CoV infections remains to identify additional therapeutic targets. Therefore, to identify the host factors that are essential for β-CoV infection-induced cell death, we performed a whole genome CRISPR screen using the prototypical β-CoV MHV as a model. To develop the screen, we first prepared iBMDMs from Cas9-transgenic mice. These iBMDMs have several advantages, including their ease of use and nearly unlimited supply, and they represent a highly susceptible cell type with intact protein machinery to induce innate immune and cell death pathways.
To perform the CRISPR screen, we used the whole genome mouse pooled Brie library (Addgene, 73633), containing 78,637 individual guides targeting a total of 19,674 genes, with an average of 4 guides for each gene, as well as 1000 control non-targeting gRNAs37. We generated a pool of iBMDMs with individual genes deleted by CRISPR and infected these with MHV (MOI = 0.1) for 24 h. The surviving cells were analyzed to identify gRNAs that were enriched, suggesting that these genes have a role in inducing cell death in response to MHV infection (Fig. 1a). The screen identified several host genes that are potentially involved in MHV-induced cell death (pro-death host factors) (Fig. 1b). Analysis of the enriched genes indicated that gene sets corresponding to multiple biological processes and cellular components were significantly enriched among the pro-death host genes (Supplementary Fig. 1a, b). Several of the enriched gene sets encompass the host molecular machinery required for RNA transcription, amino acid metabolism, and the antiviral interferon signaling cascades (Supplementary Fig. 1a). Moreover, GO term analyses focused on the cellular components identified that most of the enriched gRNAs were regulatory genes of the endo-lysosomal compartment and mediators of RNA polymerase-II-dependent transcription (Supplementary Fig. 1b); these are also known to be important for both viral replication and intracellular maturation of the β-CoV particles38–42.
Fig. 1. CRISPR screen identifies host factors required for β-CoV-induced cell death.
a Schematic of the CRISPR screen workflow in mouse hepatitis virus (MHV)-infected immortalized bone marrow-derived macrophages (iBMDMs). b Representative volcano plot showing the log2 mean fold change for the gRNAs in the CRISPR screen following infection of iBMDMs carrying gRNAs with MHV (MOI 0.1) for 24 h. The top 15 CRISPR screen hits are labeled. c Scatter plot highlighting the enrichment of all four gRNAs targeting Ceacam1 in the pool of iBMDMs carrying gRNAs from the whole genome CRISPR screen following infection with MHV (MOI 0.1) for 24 h. d Scatter plot depicting the distribution of the normalized gRNA counts in log scale for the individual Ceacam1 gRNAs, represented in panels (b) and (c). Ctrl-gRNAs denotes the count in the control pool of cells that were not infected; MHV-gRNAs denotes the count in the pool of cells following infection with MHV (MOI 0.1) for 24 h.
We next focused on the most highly enriched individual gRNAs and found that these corresponded to the MHV viral receptor carcinoembryonic antigen-related cell adhesion molecule 1 (Ceacam1) (Fig. 1b). Ceacam1 is known to be a critical host factor that dictates the cell tropism of MHV43, providing validation for the specificity of the findings from the CRISPR screen. Moreover, individual gRNA analyses showed uniform enrichment of all four unique gRNAs targeting Ceacam1, confirming that gRNAs for Ceacam1 are among the most positively enriched genes in the MHV CRISPR screen (Fig. 1c, d). Together, these findings suggest that a whole genome CRISPR screen can identify host molecules that are essential for β-CoV–mediated cellular effects.
MHV infection induces Ceacam1-dependent inflammatory cell death, PANoptosis, and cytokine release in iBMDMs
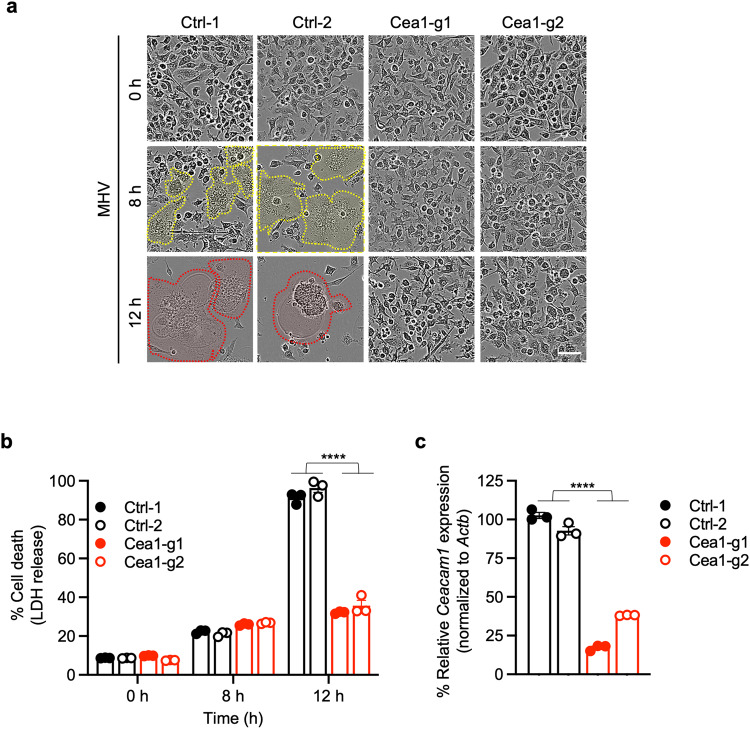
We next sought to validate the CRISPR screen findings using Ceacam1 as the lead candidate gene. To this end, we first assessed the kinetics of cell death in MHV-infected iBMDMs. We observed robust cell death in MHV-infected wild-type (WT) iBMDMs that increased over time (Fig. 2a, b), while gRNA-mediated deletion of Ceacam1 significantly protected the iBMDMs from cell death (Fig. 2a–c). Moreover, the characteristic syncytia formation mediated by the virus spike protein during MHV infection was observed in WT iBMDMs (Fig. 2a), similar to other cell types44,45, but not in cells with gRNA-mediated deletion of Ceacam1 (Fig. 2a, c).
Fig. 2. Ceacam1 is required for MHV-induced cell death.
a Cell death analysis in mouse hepatitis virus (MHV; MOI 0.1)-infected immortalized bone marrow-derived macrophages (iBMDMs) with and without Ceacam1 gRNA treatment with two different guides (Cea1-g1 and Cea1-g2). The yellow dotted lines denote syncytia, and the red dotted lines denote ballooning and dying syncytia. b Percentage of cell death at indicated time points after MHV infection in iBMDMs with and without Ceacam1 gRNA treatment in terms of LDH release. c Knockdown efficiency of the two different Caecam1 gRNAs, Cea1-g1 and Cea1-g2, employed in this study. Actb was used to normalize the Caecam1 expression. The data presented are representative of three independent experiments (a–c). Data are shown as mean ± SEM (b, c). Analysis was performed using the Student’s t-test; ****P < 0.0001. Ctrl: Control with no gRNA. The scale bar is representative of 50 μm.
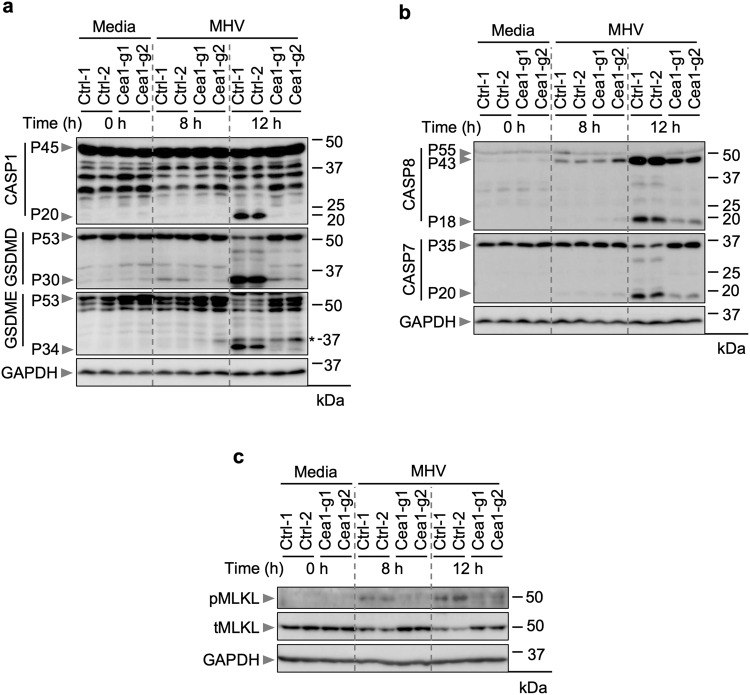
Previous studies showed that MHV infection in macrophages induces PANoptosis, characterized by the activation of multiple caspases and cell death markers15,36. These include caspase-1 to promote the processing of GSDMD to its P30 N-terminal fragment (GSDMD-N)15,36, which forms pores in the cell membrane to trigger cell death20–22,46,47. We therefore sought to determine whether the same is true in iBMDMs and understand how the loss of Ceacam1 affects these molecular processes. We observed the cleaved, active form of caspase-1 in WT iBMDMs after MHV infection (Fig. 3a), indicating the occurrence of inflammasome activation. Additionally, we observed the P30 fragment of GSDMD and the P34 fragment of GSDME (Fig. 3a). Consistent with the cell death data (Fig. 2a, b), gRNA-based deletion of Ceacam1 markedly reduced the activation of caspase-1 and GSDMD (Fig. 3a). Also, in line with previous reports in other cell types48–51, we observed activation of caspases-8 and -7 in WT iBMDMs in response to MHV infection (Fig. 3b). However, the gRNA-based deletion of Ceacam1 reduced the cleavage and activation of these caspases (Fig. 3b). In addition, in WT iBMDMs, we detected phospho-MLKL (Fig. 3c), another pore-forming molecule known to induce cell death, and phospho-MLKL was markedly reduced in Caecam1-deficient iBMDMs (Fig. 3c). Furthermore, gRNA-mediated knockdown of Ceacam1 reduced the release of pro-inflammatory cytokines IL-1β and KC at 12 h post-MHV infection (Supplementary Fig. 2a–c). Together, these observations confirm Ceacam1’s positive role in MHV-induced PANoptotic cell death and subsequent inflammatory cytokine release.
Fig. 3. Ceacam1 modulates MHV-induced PANoptosis.
a–c Immunoblot analysis of pro- (P45) and cleaved caspase-1 (P20; CASP1), pro- (P53) and activated (P30) gasdermin D (GSDMD), and pro- (P53) and activated (P34) gasdermin E (GSDME) (a); pro- (P55) and cleaved caspase-8 (P43, P18; CASP8) and pro- (P35) and cleaved caspase-7 (P20; CASP7) (b); and phospho- (pMLKL) and total MLKL (tMLKL) (c) from mouse hepatitis virus (MHV; MOI 0.1)-infected immortalized bone marrow-derived macrophages (iBMDMs) with and without Ceacam1 gRNA treatment with two different guides (Cea1-g1 and Cea1-g2) at the indicated time points. Asterisk denotes a non-specific band. Blots were reprobed for GAPDH to serve as the internal control. The data presented are representative of three independent experiments (a–c). Ctrl: Control with no gRNA.
Ceacam1 is required for MHV viral replication
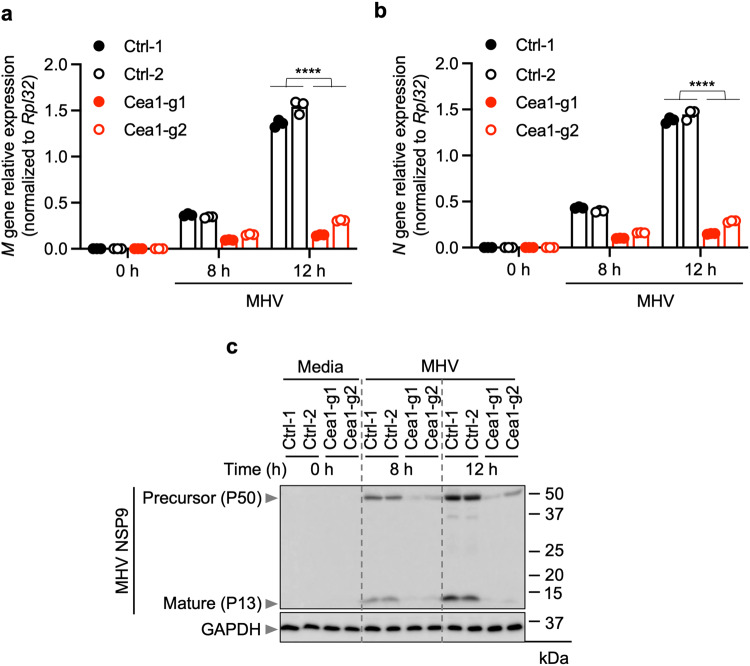
During infection, inflammatory cell death is activated downstream of innate immune sensing of viral PAMPs and DAMPs52. Inhibition of viral replication may reduce the amounts of viral PAMPs and DAMPs present, thereby reducing cell death. Given the known function of Ceacam1 in viral entry53,54, we hypothesized that infectivity may be the mechanism by which loss of Ceacam1 inhibits cell death. To test this hypothesis, we examined intracellular levels of viral nucleic acids and proteins from MHV as a measure of viral replication and viral PAMP abundance. We observed a substantial increase in the expression of MHV mRNA for the structural proteins M and N in WT iBMDMs, but not in Ceacam1-deficient cells (Fig. 4a, b). Moreover, we also observed the time-dependent production of the MHV non-structural protein NSP9 in both its pro- and cleaved/matured forms in WT iBMDMs, but not in Ceacam1 gRNA-treated iBMDMs (Fig. 4c). Treatment of iBMDMs with GC376, an inhibitor of viral replication that blocks the activity of the β-CoV main protease (Mpro), substantially reduced the MHV-infected cell death, PANoptosis (Supplementary Fig. 3a–c). Together, these data establish the Ceacam1 receptor as an important viral entry point for MHV, allowing the virus to enter the cytosol to replicate and induce subsequent inflammatory cell death in iBMDMs.
Fig. 4. Loss of Ceacam1 reduces MHV replication.
a, b Quantification of mRNA levels of the mouse hepatitis virus (MHV) M (a) and N (b) structural proteins at the indicated time points after MHV infection (MOI 0.1) in immortalized bone marrow-derived macrophages (iBMDMs) with and without Ceacam1 gRNA treatment with two different guides (Cea1-g1 and Cea1-g2). c Immunoblot analysis of non-structural protein 9 (NSP9) precursor (P50 and P37) and mature forms (P13) in iBMDMs following the indicated treatment. Blots were reprobed for GAPDH to serve as the internal control. The data presented are representative of three independent experiments (a–c). Data are shown as mean ± SEM (a, b). Analysis was performed using the Student’s t-test; ****P < 0.0001. Ctrl: Control with no gRNA.
Discussion
β-CoV-induced cell death pathways have been widely implicated in disease pathogenesis5,6,8–10,15,16. Our findings here show that the MHV-induced, Ceacam1-dependent cell death in myeloid iBMDMs is characterized by activation of caspases-1, -8, and -7, GSDMD, GSDME, and MLKL. These results suggest that Ceacam1 acts upstream of multiple cell death molecules by promoting the intracellular entry of MHV, which is required for replication of the virus to produce viral PAMPs and DAMPs that induce cell death activation. Moreover, we found that GC376 also inhibits MHV-induced inflammatory cell death, PANoptosis. GC376 is a highly potent dipeptidyl aldehyde bisulfite adduct inhibitor that acts as a prodrug and releases active GC373 inside the cells. Once in the cell, GC373 specifically binds to the active site cysteines of the Mpro of positive sense ssRNA viruses, such as β-CoVs, and blocks the proteolytic processing of the viral polyprotein to prevent viral replication55,56. Together, these observations establish a link between viral replication and the induction of inflammatory cell death in myeloid cells. While several studies have demonstrated that the replication of β-CoVs in myeloid cells in vivo is poor, this replication is also not essential for pathogenesis. Myeloid cells can phagocytose or sense the DAMPs and PAMPs from infected and dying epithelial and other non-immune cells around them and further amplify the inflammatory mechanisms of cell death and pathophysiology16,57–59, highlighting the importance of understanding the induction of cell death in this population. Furthermore, our study also emphasizes the utility of using whole genome CRISPR screening technology to identify new cellular processes in innate immune myeloid cellular models of viral infections. Using this technology enables the evaluation of both known and unknown cell death mediators in parallel to discover new regulatory mechanisms. Our CRISPR screen identified several hits that are likely to be involved in regulating β-CoV-induced cell death, and these hits can be further assessed in future studies to characterize therapeutic targets.
MHV is known to mimic many aspects of the pathology and pathogenesis of the human β-CoVs, including SARS-CoV-254,60. For example, MHV can induce cytokine storm and syncytia formation, which are important predictive features of mortality in cases of SARS-CoV-2 infection61–63. Syncytia formation is thought to promote viral spread to neighboring cells, which can drive cell death, and syncytia are frequently observed in patients with severe COVID-1964,65. However, MHV does not bind to human CEACAM1 and hence cannot infect human cells53,54. Nonetheless, the many similarities between MHV and SARS-CoV-2 viral replication, pathogenic mechanisms, and the availability of well-established mouse models have helped the field substantially further our understanding of SARS-CoV-2-related pathologies, highlighting the continued utility of this model.
Overall, our study leveraged a whole genome CRISPR screen approach to identify the host cellular receptor Ceacam1 as a key regulator of MHV-mediated cell death, demonstrating the feasibility and power of using genome-wide screens in macrophage cell lines to accelerate the discovery of key host factors in innate immune processes. Moreover, our study also supports the potential of using blocking antibodies or targeted degradation of receptors such as Ceacam1 to block viral entry and the associated pathologies. Indeed, comparable approaches to block viral entry and replication are being employed and tested successfully in preclinical and clinical studies in a variety of viral infections, both in animal models and in clinical trials66,67. Understanding the molecular mechanisms of cell death in β-CoV infections provides new insights for the development of novel therapeutic and prophylactic strategies to counteract current and emerging pathogens and prevent pathology.
Methods
Generation of Brie lentiviral library
The Mouse Brie CRISPR KO library was a gift from David Root and John Doench (Addgene #73632 and #73633). The plasmid library was amplified and validated in the Center for Advanced Genome Engineering at St. Jude Children’s Research Hospital (St. Jude) as described in the Broad GPP protocol, with the only exception being the use of Endura DUOs electrocompetent cells. The St. Jude Hartwell Center Genome Sequencing Facility provided all NGS sequencing. Single-end 100 cycle sequencing was performed on a NovaSeq 6000 (Illumina). Validation to check gRNA presence and representation was performed using calc_auc_v1.1.py (https://github.com/mhegde/) and count_spacers.py68. Viral particles were produced by the St. Jude Vector Development and Production Laboratory. CRISPR KO screens were analyzed using Mageck-Vispr v0.5.769.
Mouse hepatitis virus (MHV) culture
The mouse hepatitis virus (A59 strain) was propagated in 17Cl-1 cells as previously described70. The virus titer was measured by plaque assay in 17Cl-1 cells.
CRISPR screen
Cas9-expressing immortalized bone marrow-derived macrophages (iBMDMs) were generated from Cas9-GFP knock-in mice (Jackson Laboratories, 026179). All work with animals was reviewed and approved by the St. Jude Children’s Research Hospital IACUC. The iBMDMs were infected with lentiviral particles at an MOI of 0.3 to ensure that the majority of the iBMDMs received a single gRNA per cell to generate a pool of individual knockout cells. Two replicates of an adequate number of cells were used as controls to obtain a representation (screen depth) of >500 cells for each sgRNA in the library, and a similar number of cells from the same batch of virus preparation were infected with MHV at a multiplicity of infection (MOI) of 0.1 for 24 h. The uninfected control cell population and the surviving cells from the MHV-infected samples were subjected to CRISPR screen enrichment analysis. Total genomic DNA was isolated using NucleoSpin® Blood kits (Takara Bio Inc., USA; 740954 and 740950), and the concentrations of the isolated gDNA samples were measured using NanoDrop (Thermo Fisher Scientific, USA).
The MAGeCK pipeline was used to estimate the log2 (fold change) with significance levels for each gene in the CRISPR screen. Genes with positive fold change were expected to be required for cell death. The top gene hits along with their significance from the CRISPR screen were visualized using a volcano plot and an RRA score plot using MAGeCKFlute v2.0.071. The expression profiles of individual gRNAs for control and treatment samples were visualized using a scatter plot. The gene ontology (GO)-term analyses of biological processes and cellular components of the significantly enriched genes from the upregulated list of gRNAs in the MHV whole genome CRISPR screen were analyzed using the widely used web-based application package, Enrichr72–74, which integrates gene-set libraries and ranks the gene sets based on the enrichment scores of the given set of genes (https://maayanlab.cloud/Enrichr/).
Cas9-iBMDM culture and infection
Cas9-expressing iBMDMs were electroporated separately with gRNAs for Ceacam1 (sequences: Ceacam1.gRNA1 (Cea1-g1): 5’-ATAGTAATATGAATTTCACG-3’; Ceacam1.gRNA2 (Cea1-g2): 5’-TTGTTGTCTTCAGCAACCTG-3’). The gRNA electroporated cells were allowed to rest for 5 days to achieve CRISPR-based deletion of the targeted genes. The cells were then expanded and seeded in 12- and 24-well plates at a seeding density of 1 × 106 cells/mL and 0.5 × 106 cells/mL, respectively, in complete DMEM (DMEM supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS; S1620, Biowest) and 1% penicillin-streptomycin (15070-063, Thermo Fisher Scientific)), and rested for about 24 h before the experiments. The cells were then washed with PBS and infected with MHV at an MOI of 0.1 in DMEM plain media (Sigma, D6171) with 1% penicillin/streptomycin (Thermo Fisher Scientific, 15070-063). After 1 h incubation with MHV, cells were supplemented with FBS to a final concentration of 10% and used for IncuCyte imaging or incubated for the indicated times before collecting samples for downstream analyses. Where indicated, the iBMDMs were treated with GC376 (Cayman Chemical, #31469), starting at 1 h post-infection with MHV, using the indicated concentrations.
Cell death analysis
The IncuCyte® S3 and/or SX5 Live-Cell Analysis System was used to image and analyze cell death in real-time, as previously described75. In brief, Cas9-iBMDMs electroporated with non-targeting and Ceacam1 gRNA were seeded in 24-well plates (0.5 × 106 cells/well) and infected with MHV as described above. After 1 h of incubation, propidium iodide (PI; Life Technologies, P3566) was added to the cells together with FBS. The plate was scanned for phase-contrast images (4 image fields/well) in real-time for 24 h at an interval of 1 h. Cell death was quantified in terms of percentage LDH release and assessed using the Cyto Tox 96® Non-Radioactive Cytotoxicity assay kit (Promega G1782) according to the manufacturer’s instructions.
ELISA
Cytokines IL-1β, KC, and IL-6 were quantified according to the manufacturer’s instructions using the MILLIPLEX® MAP Mouse cytokine/chemokine bead panel (Millipore, MCYTOMAG-70K). The samples were analyzed using the MILLIPLEX Analyzer with the xPONENT software.
Western blotting
To blot for caspases, Cas9-iBMDMs were lysed along with the supernatant using 125 μL 4× SDS loading buffer, containing 50 μL of caspase lysis buffer (1× protease inhibitors, 1× phosphatase inhibitors, 10% NP-40, and 25 mM DTT). To detect other proteins in the cell lysates alone, the supernatants were aspirated, and the cells were lysed using RIPA buffer. Caspase and RIPA lysates were separated on 10–12% polyacrylamide gels, then transferred onto PVDF membranes. After blocking the membranes with 5% skim milk for 1 h at room temperature, the blots were incubated with their respective primary antibodies overnight at 4 °C. Subsequently, the blots were washed in 1× TBST and incubated with secondary antibodies conjugated with HRP for 1 h at room temperature. The GE Amersham Imager 600 was used to develop the blots. The antibodies used were as follows: anti-caspase-1 (AdipoGen, AG-20B-0042, 1:1000), anti-caspase-7 (CST, #9492, 1:1000), anti-cleaved caspase-7 (CST, #9491, 1:1000), anti-caspase-8 (CST, #4927, 1:1000), anti-cleaved caspase-8 (CST, #8592, 1:1000), anti-GSDMD (Abcam, ab209845, 1:1000), anti-GSDME (Abcam, AB215191, 1:1000), anti-pMLKL (CST, #37333, 1:1000), anti-tMLKL (Abcepta, AP14272B, 1:1000), GAPDH (CST, #5174S, 1:5000), NSP9 of MHV (Rockland Immunochemicals, 200-301-A56, 1:1000), and HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, anti-rabbit [111-035-047], 1:5000; anti-mouse [315-035-047], 1:5000).
Quantitative PCR
Total RNA was isolated using TRIzol® reagent (Ambion, #15596018). A total of 500 ng of RNA was reverse transcribed to cDNA using the High-capacity cDNA synthesis reverse transcription kit (Applied Biosystems, #4368813), as per the manufacturer’s instructions. The cDNA was used for measurement of Ceacam1, viral M gene, and viral N gene expression in MHV-infected Cas9-expressing iBMDMs with or without Ceacam1 depletion by gRNA treatment (described above). mRNA levels were quantified in real time using Sybr® Green (Applied Biosystems, #4367659) and the Quant Studio™ 7 Flex Real-Time PCR System (Applied Biosystems). The expression levels were normalized to Rpl32 in the case of the viral genes and Actb for the host cellular Ceacam1 gene expression and presented as fold change. Forward and reverse primer sequences used in the study are as follows: M gene, Forward: 5-GGAACTTCTCGTTGGGCATTATACT-3, Reverse: 5-ACCACAAGATTATCATTTTCACAACATA-3; N gene, Forward: 5-CAGATCCTTGATGATGGC GTAGT-3, Reverse: 5- AGAGTGTCCTATCCCGACTTTCTC-3; Rpl32, Forward: 5-AAGCGAAA CTGGCGGAAAC-3, Reverse: 5-TAACCGATGTTGGGCATCAG-3; Ceacam1, Forward: 5-ATTT CACGGGGCAAGCATACA-3, Reverse: 5-GTCACCCTCCACGGGATTG-3; and Actb, Forward: 5-CAGCTTCTTTGCAGCTCCTT-3, Reverse: 5-CACGATGGAGGGGAATACAG-3.
Statistics and reproducibility
Data analysis was performed using GraphPad Prism 7.0 software. Data are presented as mean ± SEM from three independent repeats. The student’s t test was used to determine the statistical significance. P values ≤0.05 were considered significant, where **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank all the members of the Kanneganti laboratory for their comments and suggestions during the development of this manuscript. We thank R. Tweedell, PhD, and A. Sharma, PhD, for scientific editing and writing support. Work from our laboratory is supported by the US National Institutes of Health (AI101935, AI124346, AI160179, AR056296, and CA253095 to T.-D.K.) and the American Lebanese Syrian Associated Charities (to T.-D.K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
R.K.S.M., R.R.B., and T.-D.K. designed the study. R.K.S.M., R.R.B., R.M., and J.P.C. performed experiments. R.K.S.M., R.R.B., R.M., J.P.C., S.M.P.-M., and T.-D.K. analyzed the data. R.K.S.M. and R.R.B. wrote the manuscript with input from all authors. T.-D.K. oversaw the project.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Si Ming Man and Joao Valente.
Data availability
Next-generation sequencing results from the CRISPR screen are deposited in BioProject: PRJNA1009133. All other datasets are included in the published article and the supplementary information. The uncropped western blots are included in supplementary information (Supplementary Figs. 4–6). The numerical data points for the graphs are provided in Supplementary Data 1. All other data are available from the corresponding authors upon reasonable request.
Code availability
This paper does not report original code.
Competing interests
T.-D.K. was a consultant for Pfizer. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: R. K. Subbarao Malireddi, Ratnakar R. Bynigeri.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-023-05414-9.
References
- 1.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan W, et al. A novel coronavirus genome identified in a cluster of pneumonia cases - Wuhan, China 2019–2020. China CDC Wkly. 2020;2:61–62. doi: 10.46234/ccdcw2020.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartenian E, et al. The molecular virology of coronaviruses. J. Biol. Chem. 2020;295:12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watson OJ, et al. Global impact of the first year of COVID-19 vaccination: a mathematical modelling study. Lancet Infect. Dis. 2022;22:1293–1302. doi: 10.1016/S1473-3099(22)00320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco-Melo D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadjadj J, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karki R, Kanneganti TD. The ‘cytokine storm’: molecular mechanisms and therapeutic prospects. Trends Immunol. 2021;42:681–705. doi: 10.1016/j.it.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore JB, June CH. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 2020;146:119–127.e114. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujikura, D. & Miyazaki, T. Programmed Cell Death in the Pathogenesis of Influenza. Int. J. Mol. Sci.19, 10.3390/ijms19072065 (2018). [DOI] [PMC free article] [PubMed]
- 14.Herold S, Becker C, Ridge KM, Budinger GR. Influenza virus-induced lung injury: pathogenesis and implications for treatment. Eur. Respir. J. 2015;45:1463–1478. doi: 10.1183/09031936.00186214. [DOI] [PubMed] [Google Scholar]
- 15.Karki R, et al. ZBP1-dependent inflammatory cell death, PANoptosis, and cytokine storm disrupt IFN therapeutic efficacy during coronavirus infection. Sci. Immunol. 2022;7:eabo6294. doi: 10.1126/sciimmunol.abo6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karki R, et al. Synergism of TNF-alpha and IFN-gamma triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184:149–168.e117. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeung ML, et al. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat. Microbiol. 2016;1:16004. doi: 10.1038/nmicrobiol.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue Y, et al. SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9:904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kesavardhana S, Malireddi RKS, Kanneganti TD. Caspases in cell death, inflammation, and pyroptosis. Annu. Rev. Immunol. 2020;38:567–595. doi: 10.1146/annurev-immunol-073119-095439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He WT, et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 2015;25:1285–1298. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kayagaki N, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 22.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 24.Zhou, Z. et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science368, 10.1126/science.aaz7548 (2020). [DOI] [PubMed]
- 25.Hitomi J, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser WJ, et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang TB, et al. Caspase-8 serves both apoptotic and nonapoptotic roles. J. Immunol. 2004;173:2976–2984. doi: 10.4049/jimmunol.173.5.2976. [DOI] [PubMed] [Google Scholar]
- 28.Oberst A, et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, et al. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature. 2011;471:373–376. doi: 10.1038/nature09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christgen S, et al. Identification of the PANoptosome: a molecular platform triggering pyroptosis, apoptosis, and necroptosis (PANoptosis) Front. Cell Infect. Microbiol. 2020;10:237. doi: 10.3389/fcimb.2020.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malireddi, R. K. S. et al. Innate immune priming in the absence of TAK1 drives RIPK1 kinase activity-independent pyroptosis, apoptosis, necroptosis, and inflammatory disease. J. Exp. Med.217, 10.1084/jem.20191644 (2020). [DOI] [PMC free article] [PubMed]
- 32.Samir P, Malireddi RKS, Kanneganti TD. The PANoptosome: a deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis) Front Cell Infect. Microbiol. 2020;10:238. doi: 10.3389/fcimb.2020.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng M, Karki R, Vogel P, Kanneganti TD. Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense. Cell. 2020;181:674–687 e613. doi: 10.1016/j.cell.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, S. et al. AIM2 forms a complex with pyrin and ZBP1 to drive PANoptosis and host defence Nature597, 415-419 (2021). [DOI] [PMC free article] [PubMed]
- 35.Sundaram, B. et al. NLRP12-PANoptosome activates PANoptosis and pathology in response to heme and PAMPs. Cell186, 2783-2801.e20 (2023). [DOI] [PMC free article] [PubMed]
- 36.Zheng M, et al. Impaired NLRP3 inflammasome activation/pyroptosis leads to robust inflammatory cell death via caspase-8/RIPK3 during coronavirus infection. J. Biol. Chem. 2020;295:14040–14052. doi: 10.1074/jbc.RA120.015036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burkard C, et al. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang H, Luo D, Liao H, Li S. Coronavirus usurps the autophagy-lysosome pathway and induces membranes rearrangement for infection and pathogenesis. Front. Microbiol. 2022;13:846543. doi: 10.3389/fmicb.2022.846543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao G, et al. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell. 2021;56:427–442 e425. doi: 10.1016/j.devcel.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rovnak J, Quackenbush SL. Exploitation of the Mediator complex by viruses. PLoS Pathog. 2022;18:e1010422. doi: 10.1371/journal.ppat.1010422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, et al. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korner, R. W., Majjouti, M., Alcazar, M. A. A. & Mahabir, E. Of mice and men: the coronavirus MHV and mouse models as a translational approach to understand SARS-CoV-2. Viruses12, 10.3390/v12080880 (2020). [DOI] [PMC free article] [PubMed]
- 44.Lavi E, Wang Q, Weiss SR, Gonatas NK. Syncytia formation induced by coronavirus infection is associated with fragmentation and rearrangement of the Golgi apparatus. Virology. 1996;221:325–334. doi: 10.1006/viro.1996.0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ou X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding J, et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 47.Liu X, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belyavsky M, Belyavskaya E, Levy GA, Leibowitz JL. Coronavirus MHV-3-induced apoptosis in macrophages. Virology. 1998;250:41–49. doi: 10.1006/viro.1998.9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen, K. W. et al. Extrinsic and intrinsic apoptosis activate pannexin-1 to drive NLRP3 inflammasome assembly. EMBO J.38, 10.15252/embj.2019101638 (2019). [DOI] [PMC free article] [PubMed]
- 50.Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taabazuing CY, Okondo MC, Bachovchin DA. Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem. Biol. 2017;24:507–514 e504. doi: 10.1016/j.chembiol.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanneganti TD. Intracellular innate immune receptors: life inside the cell. Immunol. Rev. 2020;297:5–12. doi: 10.1111/imr.12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dveksler GS, et al. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 1991;65:6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams RK, Jiang GS, Holmes KV. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl Acad. Sci. USA. 1991;88:5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim Y, Mandadapu SR, Groutas WC, Chang KO. Potent inhibition of feline coronaviruses with peptidyl compounds targeting coronavirus 3C-like protease. Antivir. Res. 2013;97:161–168. doi: 10.1016/j.antiviral.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim Y, et al. Broad-spectrum inhibitors against 3C-like proteases of feline coronaviruses and feline caliciviruses. J. Virol. 2015;89:4942–4950. doi: 10.1128/JVI.03688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S, et al. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target Ther. 2020;5:235. doi: 10.1038/s41392-020-00334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu Q, et al. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity. 2021;54:1304–1319.e1309. doi: 10.1016/j.immuni.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sefik E, et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature. 2022;606:585–593. doi: 10.1038/s41586-022-04802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo S, et al. The NLRP3 inflammasome and IL-1beta accelerate immunologically mediated pathology in experimental viral fulminant hepatitis. PLoS Pathog. 2015;11:e1005155. doi: 10.1371/journal.ppat.1005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang KJ, et al. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perlman S, Dandekar AA. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Braga L, et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature. 2021;594:88–93. doi: 10.1038/s41586-021-03491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z, et al. SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination. Cell Death Differ. 2021;28:2765–2777. doi: 10.1038/s41418-021-00782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mazzon, M. & Marsh, M. Targeting viral entry as a strategy for broad-spectrum antivirals. F1000Res8, 10.12688/f1000research.19694.1 (2019). [DOI] [PMC free article] [PubMed]
- 68.Joung J, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 2017;12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li W, et al. Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR. Genome Biol. 2015;16:281. doi: 10.1186/s13059-015-0843-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schickli JH, Zelus BD, Wentworth DE, Sawicki SG, Holmes KV. The murine coronavirus mouse hepatitis virus strain A59 from persistently infected murine cells exhibits an extended host range. J. Virol. 1997;71:9499–9507. doi: 10.1128/jvi.71.12.9499-9507.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang B, et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat. Protoc. 2019;14:756–780. doi: 10.1038/s41596-018-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie Z, et al. Gene set knowledge discovery with enrichr. Curr. Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malireddi RKS, et al. TAK1 restricts spontaneous NLRP3 activation and cell death to control myeloid proliferation. J. Exp. Med. 2018;215:1023–1034. doi: 10.1084/jem.20171922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Next-generation sequencing results from the CRISPR screen are deposited in BioProject: PRJNA1009133. All other datasets are included in the published article and the supplementary information. The uncropped western blots are included in supplementary information (Supplementary Figs. 4–6). The numerical data points for the graphs are provided in Supplementary Data 1. All other data are available from the corresponding authors upon reasonable request.
This paper does not report original code.