Abstract
The treatment options for advanced papillary thyroid cancer (PTC) and anaplastic thyroid cancer (ATC) refractory to standard therapies are limited. Although anti-PD-1 therapy has a manageable safety profile and has been effective in a small percentage of patients with advanced PTC and refractory ATC, the majority of the patients either do not respond or develop resistance to anti-PD-1 therapy. N6-methyladenosine (m6A) modification is a critical determinant of the complexity of the tumor microenvironment (TME). However, it is unclear whether and how m6A modification in tumor cells shapes the immune landscape of PTC and ATC. In this study, we performed bulk and single cell RNA sequencing analysis of PTC and ATC tissues, and found that low METTL3 expression not only correlated to poor response to immune checkpoint blockade (ICB) but was also associated with increased TNF family-related ligand-receptor interactions in the immunosuppressive Tregs and exhausted T cells. Furthermore, overexpression of METTL3 in PTC and ATC cells enhanced the efficacy of anti-PD-1 therapy in a peripheral blood mononuclear cell humanized NCG (huPBMC-NCG) mouse model. Mechanistically, M2 macrophage-derived extracellular vesicles (M2 EVs) inhibited METTL3 expression in PTC and ATC cells via miR-21-5p. Downregulation of METTL3 promoted demethylation of CD70 mRNA, which prevented YTHDF2-mediated degradation of the transcripts. The stabilization of CD70 mRNA, and the subsequent upregulation in CD70 protein levels increased the abundance of the immunosuppressive Tregs and terminally exhausted T cells, thereby inducing resistance to anti-PD-1 therapy. Furthermore, blocking CD70 using cusatuzumab, a high-affinity monoclonal antibody, reversed the anti-PD-1 therapy resistance induced by M2 EVs in vivo. Finally, we demonstrated that METTL3 expression negatively correlated with CD70 expression and M2 macrophages and Tregs infiltration in PTC and ATC tissues. Our findings provide new insights into developing novel therapies for advanced PTC and ATC.
Subject terms: Cancer microenvironment, Immune evasion, Epigenetics
Introduction
Thyroid cancer is one of the most common malignancies of the endocrine system, and ranked 9th in terms of incidence in 2020 with approximately 586,000 cases worldwide [1]. Although most patients with papillary thyroid cancer (PTC) can benefit from surgical resection, the majority develop recurrent invasive disease and distant metastases. Anaplastic thyroid cancer (ATC) is a rare, aggressive form of thyroid cancer with nearly 100% disease-specific mortality and no standard therapeutic approach [2, 3]. Newer molecular targeted therapies such as sorafenib and lenvatinib have been approved for the treatment of thyroid cancers. However, their efficacy is limited due to poor cytotoxicity, rapid development of drug resistance, and considerable side effects. Therefore, there is an urgent need to develop new treatments for these aggressive thyroid cancers [4].
Over the past decade, immune checkpoint blockade (ICB) therapies have consistently improved the overall survival of cancer patients [5]. Blockers targeting the programmed cell death protein 1 (PD-1) and programmed death-ligand 1 (PD-L1) are increasingly being considered for treating thyroid cancer [6] since both PD-1 and PD-L1 are upregulated in PTC and ATC cells, as well as the tumor-infiltrating lymphocytes in patients with advanced cancer [7]. Nevertheless, PD-1/PD-L1 blockade monotherapies have been effective in only about 9% of the patients with advanced PTC, and 19% of ATC patients [3, 8]. Therefore, it is necessary to elucidate the mechanisms underlying the relationship between the tumor cells and immune system in thyroid cancer.
Thyroid cancer genesis involves a series of genetic, epigenetic and epitranscriptomic alterations. N6-methyladenosine (m6A) is the most common epigenetic modification in RNA, and occurs at the N6 position of adenine. Methyltransferases, also known as m6A “writers”, include METTL3/14, WTAP and RBM15, among others, and mediate methylation. On the other hand, the demethylases or m6A “erasers” such as FTO and ALKBH5 reverse the modification. The m6A “readers” are RNA-binding proteins, including YTHDF1/2, YTHDC1/2/3 and HNRNPC, that recognize the methylated residues on RNA, and mediate the translation and degradation of the transcripts. A growing number of studies show that aberrant m6A modification plays a crucial role in the development of various cancers [9]. In particular, METTL3-mediated m6A modification is a key regulator of thyroid cancer development [10, 11]. However, it is unclear whether and how METTL3 deregulation in thyroid tumor cells dictates the immune landscape.
Herein, we revealed that low METTL3 expression correlated with poor response to ICB, and overexpressing METTL3 in tumor cells could enhance the efficacy of anti-PD-1 therapy in PTC and ATC. At the molecular level, M2 macrophage downregulated METTL3 expression in tumor cells through secreting extracellular vesicles (EVs) containing miR-21-5p, resulting in demethylation and stabilization of CD70 mRNA in a YTHDF2-dependent manner. The subsequent upregulation in CD70 protein levels increased the abundance of the immunosuppressive Tregs and terminally exhausted T cells, thereby inducing resistance to anti-PD-1 therapy. Blocking CD70 with cusatuzumab could reverse the anti-PD-1 therapy resistance induced by M2 EVs, which provided a novel strategy for the treatment of recalcitrant thyroid cancer.
Materials and methods
Bioinformatics analysis
The bulk RNA sequencing (RNA-seq) data (FPKM value) and clinical-related information of 503 PTC tissues were collected from The Cancer Genome Atlas (TCGA) dataset obtained from the online data portal THCA Xena (https://xenabrowser.net/datapages/). Bulk RNA-seq data of 20 ATC tissues GSE76039 was downloaded from the Gene Expression Omnibus (GEO). According to the median expression level of METTL3, the patients were divided into the METTL3high (above median value) and METTL3low (below median level) groups. CD70high and CD70low groups were defined in the same way. ImmuCellAI (Immune Cell Abundance Identifier (http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/analysis) was used to estimate the immune cell infiltration and predict patients’ response to immune checkpoint blockade therapy from gene expression dataset [12]. The biological processes were identified by Gene Ontology (GO) enrichment analysis (https://david.ncifcrf.gov/) [13]. CIBERSORT analysis of tumor-infiltrating immune cells was performed by TIMER2.0 (http://timer.cistrome.org/) [14].
The single cell RNA sequencing (scRNA-seq) data of 7 PTC and 5 ATC tissues were collected from the GEO datasets (GSE184362 for PTC and GSE148673 for ATC). The cells were visualized by using Uniform Manifold Approximation and Projection (UMAP) embedding. Differentially expressed genes between every pair of clusters were identified by using FindMarkers [15]. The cancer cell clusters in PTC and ATC tissues were classified as METTL3+ or METTL3− depending on whether METTL3 expression was detected (value > 0) or undetected (value = 0) respectively. CellPhoneDB (https://github.com/Teichlab/cellphonedb), a public repository of ligands, receptors, and their interactions, was used to perform the cell-cell communication analysis [16].
Cell culture, generation of the stable cell line
One thyroid follicular epithelial cell line (Nthy-ori 3-1), two PTC cell lines (BCPAP, K1) and two ATC cell lines (C643, BHT101) were included in this research. Nthy-ori 3–1 cell line was purchased from the American Type Culture Collection (USA). BCPAP, K1, and BHT101 cell lines were purchased from Guangzhou Cellcook Biotech Co. (Guangzhou, P.R. China). C643 cell line was purchased from the Chinese Academy of Science (Shanghai, P.R. China). BCPAP, K1, and C643 were cultured in 1640 medium (Gibco) supplemented with 10% FBS, and BHT-101 was cultured in DMEM medium (Gibco) supplemented with 10% FBS (Biological industries, Israel). Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2 37 °C was used to incubate the cells in a humidified 5% CO2 incubator. All cell lines were identified by short tandem repeat (STR) analysis before using in the experiments [17].
Cells were infected by lentiviral particles carrying shRNA plasmids or containing METTL3 cDNA for generating stable METTL3 knockdown cell lines or stable METTL3 overexpression cell lines (shMETTL3-1: GGAACATTATGATCCAGAAAC, shMETTL3-3: GGAACTGGATCCTACCTATGT) as previously described [18]. The expression of CD70 and YTHDF2 in cell lines were suppressed by small interfering RNAs (siRNAs): CD70: 5‘- CUGCGUCUCAGCUUCCACCAA-3’, YTHDF2: 5‘- GGUGAAGCUGCUUGGUCUA-3’.
Co-culture experiments and flow cytometry analysis
Ficoll lymphocyte separation solution was used to isolate PBMCs from the peripheral venous blood collected from healthy volunteers by gradient density centrifugation. The thyroid cancer cells were supplemented with fresh medium and co-cultured with PBMCs in the presence of IL-2 (1,000 U/mL) for 72 h in a ratio of 1:5 (cancer cell: PBMCs). After incubation, the cells were collected for flow cytometry analysis. For intracellular staining of TNF-α (tumor necrosis factor -α) and IFN-γ, the cells were treated with a leukocyte activation cocktail with Brefeldin A (423304, Biolegend) in vitro. Cell surface antigen staining was performed before the cells were fixed and permeabilized with the eBioscience™ Foxp3 /Transcription Factor Staining Buffer Set (00-5523-00, Thermo Fisher) for subsequent intracellular staining. Fluorochrome-conjugated mAbs to the following human antigens were used: CD8 (344714), TNF-α (502909), IFN-γ (502524), GzmB (515406), Perforin (353310), PD-1 (329931), Tim-3 (345034), CD45 (368504), CD4 (357408), Foxp3 (357408) and Zombie NIR™ (423105) from Biolegend. Flow cytometry data were analyzed with FlowJo software.
M6A-methylated RNA immunoprecipitation sequencing (MeRIP-Seq)
MeRIP-sequencing and analysis was carried out by LC-BIO Biotech (Hangzhou, China) as previously described [19]. Briefly, the quality and quantity of total RNA were evaluated, and Poly (A) mRNA was purified and fragmented into oligonucleotides of length ~100 nt. Then, the cleaved RNA fragments were incubated with an m6A-specific Ab in IP buffer supplemented with bovine serum albumin. The mixture was incubated with protein-A beads and eluted with elution buffer. Eluted m6A-containing fragments and untreated input control fragments were converted to a final cDNA library. The paired-end 2 × 150-bp libraries were sequenced on an Illumina Novaseq 6000 platform at LC-BIO Biotech Limited following their recommended protocol. Sequencing data were deposited in the Gene Expression Omnibus (GSE218627).
RNA-seq analysis
Total RNA was extracted with TRIzol reagent (Invitrogen, USA). RNA purification, reverse transcription, library construction and sequencing were performed at Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China) according to the manufacturer’s instructions (Illumina, San Diego, CA). Briefly, cDNA libraries were generated and sequenced on an Illumina NovaSeq 6000 platform. The raw paired end reads were trimmed and quality controlled by fastp (https://github.com/OpenGene/fastp) [20] with default parameters. Then clean reads were separately aligned to reference genome with orientation mode using HISAT2 (http://ccb.jhu.edu/software/hisat2/index.shtml) software [21]. The mapped reads of each sample were assembled by StringTie (https://ccb.jhu.edu/software/stringtie/) in a reference-based approach [22]. Sequencing data were deposited in the Gene Expression Omnibus (GSE217851).
MeRIP- real-time quantitative PCR (RT-qPCR)
Briefly, total RNA was fragmented by RNA Fragmentation Buffer (100 mM Tris-HCl, 100 mM ZnCl2 in nuclease free H2O,10X), protein-A magnetic beads (10002D, Thermo Fisher Scientific), and protein-G magnetic beads (88847, Thermo Fisher Scientific) were tumbled with anti-m6A antibody (ABE572, Millipore) at 4 °C for at least 6 h. The antibody-bead mixture was resuspended in the IP reaction mixture containing fragmented total RNA, IP buffer(150 mM NaCl, 10 mM Tris-HCl, pH 7.5, 0.1% IGEPAL CA-630 in nuclease free H2O), and RNasin Plus RNase Inhibitor (N2611, Promega, Madison, WI), and incubated for 2 h at 4 °C. The RNA reaction mixture was then washed twice in IP buffer, twice in low-salt IP buffer (50 mM NaCl, 10 mM Tris-HCl, pH 7.5, 0.1% IGEPAL CA-630 in nuclease free H2O), and twice in high-salt IP buffer (500 mM NaCl, 10 mM Tris-HCl, pH 7.5, 0.1% IGEPAL CA-630 in nuclease free H2O) for 10 min each at 4 °C. After extensive washing, the m6A enriched fragmented RNA was eluted from the beads in RLT buffer supplied in RNeasy Mini Kit (74106, QIAGEN, Germany) for 2 min at room temperature and was detected of CD70 expression by RT-qPCR. All m6A sites of CD70 were predicted using SRAMP (http://www.cuilab.cn/sramp). CD70 was amplified using primers listed below: forward primer 8334: CCACCACTTACATCAGCGTGACC; reverse primer 8334: ATTTGGGTTGGTTCCAAG-TCTTTGC; forward primer 9303 and 9310: GGGACGTGCCTTGTCTCAGATG; reverse primer 9303 and 9310: TGTCCTCAAACTAGTCATGCCTTCC.
RNA extraction, RT-qPCR, and mRNA stability assays
We performed RNA extraction, RT-qPCR, and mRNA stability assays as previously described [11]. RT-qPCR was performed using SYBR Premix Ex Taq Kit (Takara), according to the manufacturer’s instructions. Data were collected and analyzed using the Light Cycler 480 instrument (Roche). MiRNA was reverse transcribed and relatively quantitated using the TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific, USA). Primers were listed below: CD70 forward primer: ATGGCATCTACATGGTACACAT; CD70 reverse primer: CCCCAGTTCCAAAAT-CCAAAAA; METTL3 forward primer: CTTCAGCAGTTCCTGAATTAGC; METTL3 reverse primer: ATGTTAAGGCCAGATCAGAGAG; TaqMan probes were purchased from Thermo Fisher Scientific. The internal reference gene for METTL3 and CD70 was β-actin, and that for miRNA was U6. The relative expression levels for each sample were calculated based on the 2 − ΔΔCt method. To evaluate the mRNA stability of PTC and ATC cells, actinomycin D (Sigma, USA) at 5 μg/mL was added. After incubation for the indicated times, cells were harvested, and total RNA was isolated for RT-qPCR to detect stability.
Western blot analysis
Western blotting was performed as previously described [23]. Briefly, we generated total protein from each group by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred the proteins to polyvinylidene fluoride membranes. The membranes were incubated with the primary antibody METTL3 (1:1000, ab195352, Abcam), CD70 (PA5-102557, Invitrogen, 1:1000), and GAPDH (sc-47724, Santa Cruz Biotechnology, 1:2000) overnight at 4 °C and then incubated with secondary antibodies specific to the mouse-, rabbit-primary antibodies (1:4,000; Santa Cruz Biotechnology) at room temperature for 1 h. Afterward, the membranes were exposed using an enhanced chemiluminescence reagent (Chemicon International, USA). Full-length original western blots in this article are provided in Supplementary File.
Luciferase reporter assay of CD70 m6A sites
We performed Luciferase reporter assay as previously described [11]. Fragments of the CD70-3’UTR containing the wild-type m6A motifs and mutant m6A motifs were constructed by (Hanbio, China) and fused into the pmirGLO dual-luciferase expression vector (Promega, Madison, WI, USA). The relative level of firefly luciferase activity was normalized to the Renilla luciferase activity levels and represents the effect of m6A modification on CD70 expression.
Induction of M2 macrophage and isolation of M2 EVs
The human leukemia monocytic cell line THP-1 was purchased from Guangzhou Cellcook Biotech Co. (Guangzhou, P.R. China). THP-1 cells were incubated with 50 ng/ml PMA for 24 h to make cells adhere to the wall, then the cells were treated with 20 ng/mL IL-4 and 20 ng/mL IL-13 for 24 h to induct M2 macrophage. EV experiment was performed as previously described [23]. The culture medium was collected, centrifuged at 3,000 rpm for 10 min at 4 °C, and then centrifuged at 10,000 × g for 30 min at 4 °C to remove cellular debris. Next, the supernatant was filtered using a 0.22-μm filter. EVs were obtained by ultracentrifugation at 100,000 × g for 90 min and then collected and diluted in PBS. To fluorescently label EVs, resuspended EVs were incubated with 20 µM CFSE (5(6)-carboxyfluorescein diacetate N-succinimidyl ester; Sigma-Aldrich) at 37 °C for 2 h before the PBS washing and then used to treat cells.
Overexpression and inhibition of miR-21-5p
The cells were grown to 70-80% confluency and then transfected with miR-21-5p mimic/inhibitor or the respective controls (Hanbio, Shanghai, China) using Lipofectamine RNAiMAX (Invitrogen, USA) according to the manufacturer’s instructions. The sequence of the miR-21-5p mimic was 5’-ACCUUGGCUCUAGACUGCUUACU-3’ and that of the miR-21-5p inhibitor was 5’-AGUAAGCAGUCUAGAGCCAAGGU-3’.
Luciferase reporter assay of miR-21-5p and METTL3
The WT 3’UTR and mutant-type (Mut) luciferase reporter vectors of METTL3 were constructed and cloned into the XhoI and NotI sites of the psiCHECK-2 vector (Hanbio, China). Then, 293 T cells were co-transfected with 0.8 mg 30 UTR WT/Mut luciferase reporter plasmid and 5 pmol mmu-miRNA mimics/negative control (Hanbio, China) using transfection reagents (Lipofectamine 2000 Reagent; Invitrogen, USA), according to the manufacturer’s instructions. After 48 h, the lysates of the 293 T cells were harvested, and the luciferase activities were measured using a dual-luciferase reporter assay system (Promega, USA). The data were normalized for transfection efficiency by dividing the RLuc activity by the corresponding FLuc activity.
HuPBMC-NCG xenograft mice model
NCG mice (female, 4-5 weeks old, purchased from Jiangsu GemPharmatech) were acclimated for 1 week and were selected randomly to subcutaneously (s.c.) injected with 100 μl suspensions of 1 × 106 thyroid cancer cell lines. All mice were housed in animal facility under specific pathogen-free conditions. After 7 day, 5 × 106 huPBMCs were intravenously (i.v.) inoculated into mice. Mice were treated intravenously with anti-PD-1 mAb (nivolumab, 200 μg per injection), Combination (nivolumab + anti-CD70 mAb cusatuzumab, 200 μg per injection each), or PBS once a week. The treatment with M2-TAMs EVs or PBS continued in mice at 2ug EVs per injection at the tumor site twice a week [24]. The body weight of mice and the length (mm) and width (mm) of tumors were monitored every 3 or 4 days. Tumor volume (mm3) was calculated as Length × Width2 × 1/2. Tumor growth analyses were limited to 3 to 4 weeks because following this period mice started to show signs of xenograft-versus-host disease (xGVHD). Therefore, after 14 days or 21 days of PBMCs implantation, the mice were euthanized [25, 26]. In accordance with the ethical guidelines, the tumor volume did not reach 1500 mm3 or ulcers happened. Each tumor tissue was divided evenly into two parts, one for flow cytometry analysis and the other for immunofluorescence (IF) staining. Lung tissues were processed for haematoxylin-eosin (HE) staining.
All animal experiments complied with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines, and were performed in accordance with the National Institutes of Health guidelines for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978). The protocols were approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (AE-2022032). Furthermore, there were no blinding study for different animal groups.
Quantification of metastatic nodules
The lung tissues were removed from the tumor-brearing mice, fixed in paraformaldehyde, embedded in paraffin, and cut into 5-μm sections. As previously described [27], one consecutive section was taken and then a number of consecutive sections were discarded (typically 20–40 depending on the size of the tumor nodule) before collecting another one consecutive section. This procedure was repeated and applied to the entire lung. Metastatic nodules were counted on each HE section under a microscope and the sum was taken as the final number of lung metastatic nodules.
Immune cell infiltration analysis of xenografts
Tumor tissue preparation and flow cytometry were performed as previously described [28]. Tumor tissues were prepared into single-cell suspension by enzymatic digestion (collagenase IV 1 mg/ml and Dnase I 50 μg/ml). All single-cell suspension was passed through a 70-μm cell strainer (BD Falcon, New Jersey, USA) to remove impurities before erythrocyte lysis was performed. Live/Death cells were selected/excluded based on Zombie Violet™ Fixable Viability Kit (423113, Biolegend). Fluorochrome-conjugated mAbs to the following human antigens were used: CD3 (300316/317306), CD8 (344714), TNF-α (502909), IFN-γ (502524), Granzyme B (515406/372203), Perforin (353310), PD-1 (329931), Tim-3 (345034), CD45 (368504), CD4 (357408), Foxp3 (357408) from Biolegend. IF staining was performed as previously described [23], tumor tissues were incubated with anti-CD8 (1:1000, 66868-1-Ig, Proteintech). CD8+ T cells were counted for positively stained CD8 cells at the most densely infiltrated area inside tumors per 400 × magnification.
Human clinical specimens
A total of 100 patients with PTC and 22 patients with ATC were enrolled from Tianjin Medical University Cancer Institute and Hospital. Paired tumor and adjacent normal tissues were collected from 29 PTC patients for RNA isolation and expression analysis. The tissues from the remaining 71 PTC patients and the 22 ATC patients were embedded in paraffin for immunohistochemistry (IHC) staining. Human samples were collected following approval by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (EK2021149), and informed consent was obtained from all patients. The experiments were performed in accordance with the principles of the Declaration of Helsinki.
IHC staining assay
We performed IHC staining using the avidin-biotin-peroxidase complex method, as previously described [23]. All samples were incubated overnight at 4 °C with anti-METTL3 (1:500, ab195352, Abcam), anti-CD70 (1:200, ab195352, Abcam), anti-CD163 (1:200, ZM-0428, ZSGB-BIO) and anti-Foxp3(1:200, ab20034, Abcam). The staining scores were analyzed from three independent, random fields and were evaluated by two experienced pathologists. A standard semiquantitative double-scoring system was applied to evaluate the intensity and proportion of positive cells stained at 200 × magnification for METTL3 and CD70. The staining intensity was graded as 0 (no staining), 1 (slightly brown), 2 (moderately brown) and 3 (dark brown). The percentage of positive cells was scored as 0 (<25%), 1 (25-50%), 2 (50–75%), and 3 (>75%). The total staining score for each sample was calculated as the sum of the staining intensity and percentage of positively stained cells. Samples with final score ≥4 were classified as high expression, and those with final score <4 as low expression [29]. Regarding CD163, positively stained cells were counted at 200 × magnification. Regarding Foxp3, positively stained cells were counted at 400 × magnification.
Transwell migration assay
The culture supernatants of PTC or ATC cells were harvested after cells were seeded for 72 h. 4.0um transwell inserts were placed onto a 24-well plate. A total of 500 μl of supernatant was applied to the bottom of the well and 200 μl of 5 × 104 THP1-derived M2 macrophages on the top of the inserts. After 24 h incubation at 37 °C in a CO2 incubator, the inserts were collected and stained with crystal violet as previously described [30].
Statistical analysis
GraphPad Prism 9.0 (GraphPad Software, La Jolla, CA, USA) was applied to analyze the data. Unless otherwise stated, the data are presented as the mean ± SD of at least three independent experiments. We used Student’s t test or ANOVA to compare quantitative data between groups and the chi-square test to compare categorical variables. Pearson’s correlation coefficient was used to assess the relationship between METTL3 and CD70 expression in clinical samples. P value < 0.05 was considered significant.
Results
Low METTL3 expression in PTC and ATC leads to an immunosuppressive TME and resistance to ICB therapy
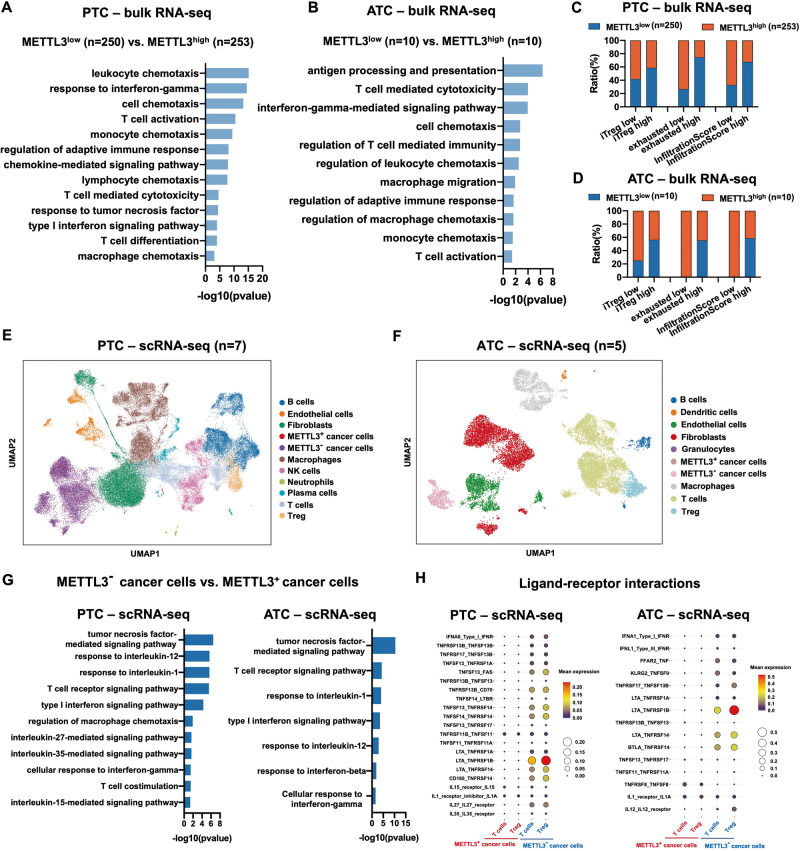
To explore the mechanisms underlying the resistance of PTC and ATC cells to ICB therapy, we analyzed the bulk RNA-seq data of 503 PTC tissues and 20 ATC tissues from previously published datasets and used the ImmuCellAI web tool to predict the ICB response. GO enrichment analysis revealed that pathways involved in mRNA regulation were significantly enriched in the responsive versus the non-responsive PTC and ATC patients (Fig. S1A). Since m6A is the most abundant mRNA modification [31], we also compared the ICB response rate of patients with different expression levels of the m6A regulatory genes. In the PTC tissues, low expression of METTL3 or high expression of METTL14, ZC3H13, FTO and YTHDF3 were associated with ICB resistance. In contrast, ICB resistance in ATC was associated with low expression of METTL3, IGF2BP1, IGF2BP2 and YTHDF1. Furthermore, PTC and ATC patients with low METTL3 expression were less responsive to ICB compared to those expressing high levels of METTL3, indicating that the latter has an impact on the efficacy of ICB against thyroid cancer (Fig. S1B). Low METTL3 expression also correlated with the enrichment of GO terms related to inflammatory cell chemotaxis and T cell-mediated immunity pathways (Fig. 1A, B), along with higher immune cell infiltration scores and greater infiltration of induced Tregs (iTreg) and exhausted T cell infiltration as per ImmuCellAI analysis (Fig. 1C, D). These results suggested that PTC and ATC tissues expressing low levels of METTL3 are immunosuppressive and, therefore, resistant to ICB therapy.
Fig. 1. Low METTL3 expression in PTC and ATC leads to an immunosuppressive TME and resistance to ICB therapy.
A, B GO analysis of the bulk RNA-seq data showing enriched pathways and (C, D) ImmuCellAI analysis showing immune cell infiltration in the METTL3low and METTL3high PTC and ATC tissues (PTC: METTL3low group n = 250, METTL3high group n = 253; ATC: METTL3low group n = 10, and METTL3high group n = 10). E, F Identification of infiltrating cell types in PTC (n = 7) and ATC (n = 5) tissues using scRNA-seq data. G GO analysis of enriched pathways and (H) bubble plots showing TNF, IFN, IL-1 and IL-12-related ligand-receptor interactions in T cells and Tregs in the METTL3+ and METTL3− PTC and ATC tissues.
To further explore the role of METTL3 in the immune landscape of thyroid tumors, we retrieved the scRNA-seq of 7 PTC tissues and 5 ATC tissues from previously published datasets. The cells from PTC and ATC tissues were robustly grouped into 10 and 9 clusters respectively, including cancer cells, B cells, endothelial cells, fibroblasts, macrophages, T cells, Tregs, etc. As shown in Fig. 1E, F, most cancer cells were METTL3 negative. Moreover, METTL3 mRNA was significantly downregulated in the tumor tissues of 29 PTC patients compared to the paired normal adjacent tissues (Fig. S1C). Compared to the thyroid follicular epithelial cell line, thyroid cancer cell lines expressed lower levels of METTL3 (Fig. S1D). These results indicated that low intrinsic expression of METTL3 in thyroid tumors may contribute to their malignant transformation. Furthermore, the differentially expressed genes between the METTL3+ and METTL3− cancer cells showed significant enrichment of pathways involved in regulating immune cell cytotoxicity, including TNF-mediated signaling pathway, T cell receptor signaling pathway, type I interferon signaling pathway, cellular response to interferon-gamma, etc. (Fig. 1G). In particular, CellPhoneDB analysis predicted significant receptor-ligand interactions involving TNF-family proteins between METTL3− cancer cells and exhausted T cells or Tregs (Fig. 1H). Taken together, low METTL3 expression in thyroid tumors may increase the abundance of exhausted T cells and Tregs via TNF signaling, which results in an immunosuppressive milieu that is resistant to ICB therapy.
METTL3 overexpression in the PTC and ATC cells enhanced the efficacy of anti-PD-1 therapy
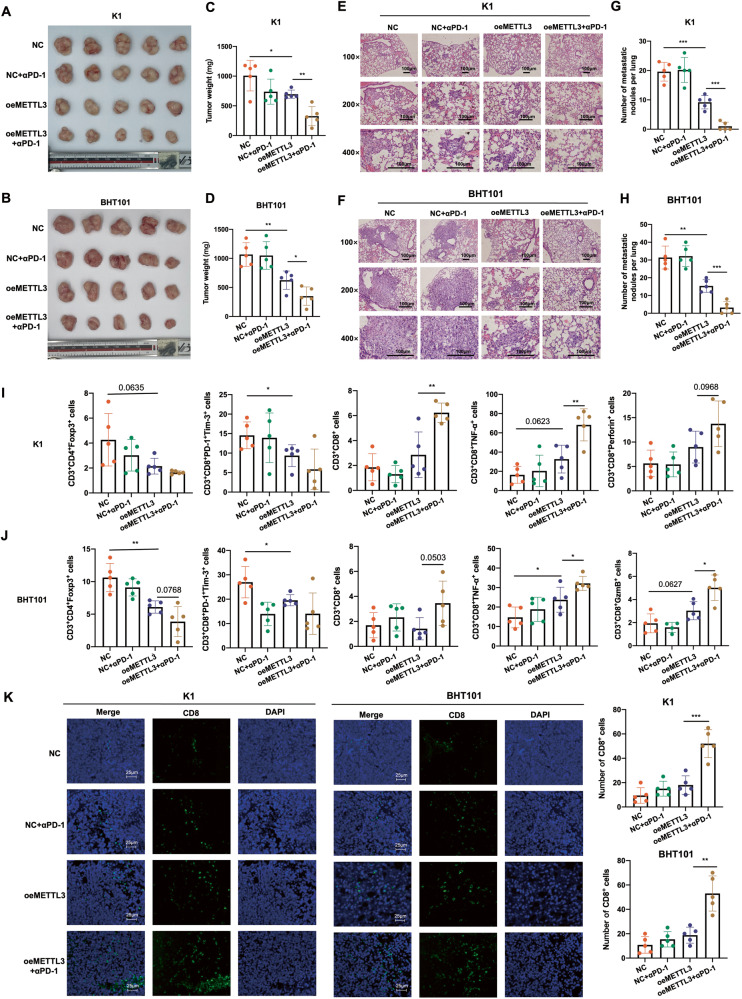
To determine whether METTL3 is a determinant of the response of thyroid tumors to ICB, we overexpressed METTL3 (oeMETTL3) in the K1 and BHT101 cell lines that have relatively low intrinsic METTL3 expression (Fig. S1E), and subcutaneously transplanted the negative control (NC) and oeMETTL3 cells into huPBMC-NCG mice. The tumor-bearing mice were treated with anti-PD-1 (αPD-1) mAb as shown in the experimental outline in Fig. S2A. There was no significant difference between the body weights of αPD-1 mAb-treated mice and untreated control mice, indicating lack of treatment toxicity (Fig. S2B). Furthermore, the K1oeMETTL3 and BHT101oeMETTL3 cells formed smaller primary tumors and fewer lung metastatic nodules compared to their NC counterparts expressing normal levels of METTL3. As expected, αPD-1 mAb treatment significantly decreased the tumor volume, tumor weight and the number of metastatic nodules in the oeMETTL3 group but had no effect on the tumor growth and metastasis in the NC group (Fig. 2A–H and Fig. S2B-C). Moreover, the oeMETTL3 xenografts had significantly lower abundance of the immunosuppressive CD3+CD4+Foxp3+ Tregs and CD3+CD8+PD-1+Tim-3+ terminally exhausted T cells compared to that in NC tumors, which corresponded to higher infiltration of the cytotoxic CD3+CD8+ T cells expressing TNF-α, GzmB or perforin after αPD-1 therapy in the former (Fig. 2I, J and Fig. S2D-E). IF staining of the xenografts further confirmed the significant increase in CD8+ T cells in the oeMETTL3 group after αPD-1 therapy (Fig. 2K). Taken together, METTL3 overexpression can reverse the immunosuppressive microenvironment of thyroid tumors by decreasing the infiltration of Tregs and terminally exhausted T cells, and increasing the abundance of effector cytotoxic T cells after PD-1 blockade.
Fig. 2. METTL3 overexpression in the PTC and ATC cells enhanced the efficacy of anti-PD-1 therapy.
A, B General view of tumor mass, C, D tumor weights, E, F representative images of HE-stained pulmonary metastatic nodules (scale bars, 100 μm) and G, H quantification of metastatic nodules from mice bearing K1NC, K1oeMETTL3, BHT101NC and BHT101oeMETTL3 xenografts with or without αPD-1 mAb treatment. I, J Percentage of CD3+CD4+Foxp3+ Tregs, CD3+CD8+PD-1+Tim-3+ terminally exhausted T cells, CD3+CD8+ T cell, CD3+CD8+ TNF-α+ T cells and CD3+CD8+ perforin+ T cells in the xenografts of above groups. K Representative IF staining images showing CD8+ T cells in the above groups (scale bars, 25 μm). Data are presented as the mean ± SD of 5 biologically independent animals. The p-values in panel C, D, G, H, I, J and K were calculated by Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
METTL3 silencing in thyroid cancer cells promoted an immunosuppressive phenotype via CD70 demethylation
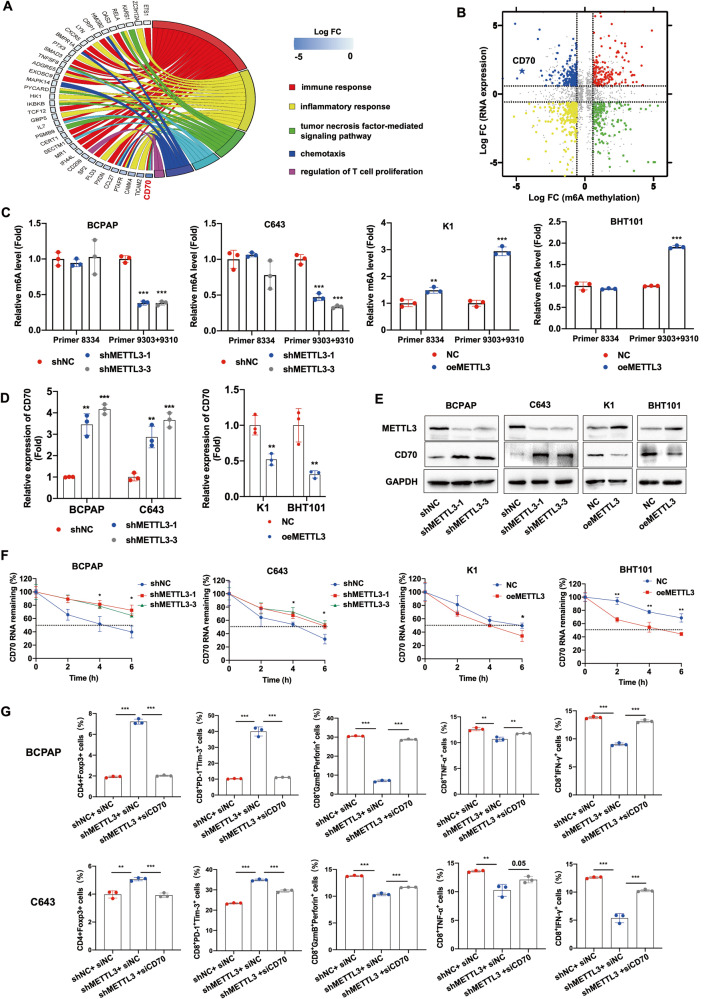
To investigate the possible mechanism underlying the immunosuppressive effect of METTL3 downregulation in thyroid cancer, we knocked down the METTL3 gene in the BCPAP and C643 cells lines that have relatively high intrinsic METTL3 expression, and RNA-seq analysis was performed in BCPAP cells with stable transfection of shMETTL3 and shNC. GO analysis of the differentially expressed genes revealed significant enrichment of the signaling pathways regulating T cell functions in shMETTL3 cells (Fig. S3A). Furthermore, MeRIP-seq was also performed to detect the m6A transcripts in control and shMETTL3 cells. As shown in Fig. S3B-C, m6A signals were abundant in the coding sequence (CDS), stop codon and 3’ Un-Translated region (UTR), and the consensus motif GGAC was highly enriched in the control and METTL3-knockdown groups. Since METTL3 is a methyltransferase, we analyzed the genes with m6A downregulation in the shMETTL3 cells, and found that CD70, a member of the TNF superfamily that is involved in TNF-mediated signaling and regulation of T cell proliferation (Fig. 3A), showed the most obvious demethylation following METTL3 knockdown. As shown in Fig. 3B, the m6A peak of CD70 decreased by 23.9-fold and its transcript level increased by 3.18-fold in the shMETTL3 cells compared to the control. We also confirmed the demethylation of CD70 mRNA in METTL3-knockdown cells based on the MeRIP-seq data (Fig. S3D).
Fig. 3. METTL3 silencing in thyroid cancer cells promoted an immunosuppressive phenotype via CD70 demethylation.
A GO enrichment analysis of pathways in m6A-downregulated genes. B Star plot showing distribution of genes with differential methylation (X axis; |fold change | ≥1.5) and expression levels (Y axis; |fold change | ≥1.5) in shMETTL3 and shNC BCPAP cells. C CD70 RNA m6A levels, D CD70 mRNA levels and E CD70 protein levels in the METTL3-knockdown and METTL3-overexpressing PTC and ATC cells. F The half-life of CD70 mRNA in the METTL3-knockdown and METTL3-overexpressing PTC and ATC cells after treatment with actinomycin D. G Percentage of CD4+Foxp3+ Tregs, CD8+PD-1+Tim-3+ terminally exhausted T cells, CD8+GzmB+perforin+ T cells, CD8+TNF-α+ T cells and CD8+IFN-γ+ T cells in PBMCs co-cultured with METTL3-knockdown BCPAP and C643 cells transfected with CD70-specific siRNA. Data of are presented as the mean ± SD of three independent experiments. The p-values in panel C, D, F and G were calculated by Student’s t-test. *p < 0.05; **p < 0.01; ***p < 0.001.
To further identify the specific m6A site affected by METTL3, we predicted the mRNA sequence of CD70 using the SRAMP website, which revealed 3 potential m6A modification sites with very high confidence in the 3’UTR (Fig. S3E). Furthermore, MeRIP-RT-qPCR indicated significant reduction in the m6A levels at positions 9303 and 9310 in the METTL3-silenced cells, and an increase in METTL3-overexpressing cells (Fig. 3C). In line with this, METTL3 knockdown significantly increased CD70 mRNA and protein levels in BCPAP and C643 cell lines, while the overexpression of METTL3 in K1 and BHT101 had the opposite effects (Fig. 3D, E). Likewise, METTL3 knockdown and overexpression prolonged and reduced the half-life of CD70 mRNA, respectively (Fig. 3F). These results indicated that low levels of METTL3 stabilize CD70 mRNA via demethylation.
To determine whether CD70 demethylation mediates the immunosuppressive effect of METTL3 in thyroid cancer, we silenced CD70 expression in the METTL3-knockdown BCPAP and C643 cells, and co-cultured PBMCs with shMETTL3 and shMETTL3/si-CD70 cells. While METTL3 knockdown significantly increased the percentage of CD4+Foxp3+ Tregs and CD8+PD-1+Tim-3+ terminally exhausted T cells, and decreased that of the cytotoxic CD8+GzmB+perforim+ T cells, CD8+TNF-α+ T cells, and CD8+IFN-γ+ T cells, CD70 silencing reversed this immunosuppressive phenotype (Fig. 3G and Fig. S3F). Taken together, the low expression of METTL3 in PTC and ATC tissues promotes an immunosuppressive landscape via demethylation and stabilization of CD70 mRNA.
METTL3-mediated regulation of CD70 is dependent on YTHDF2
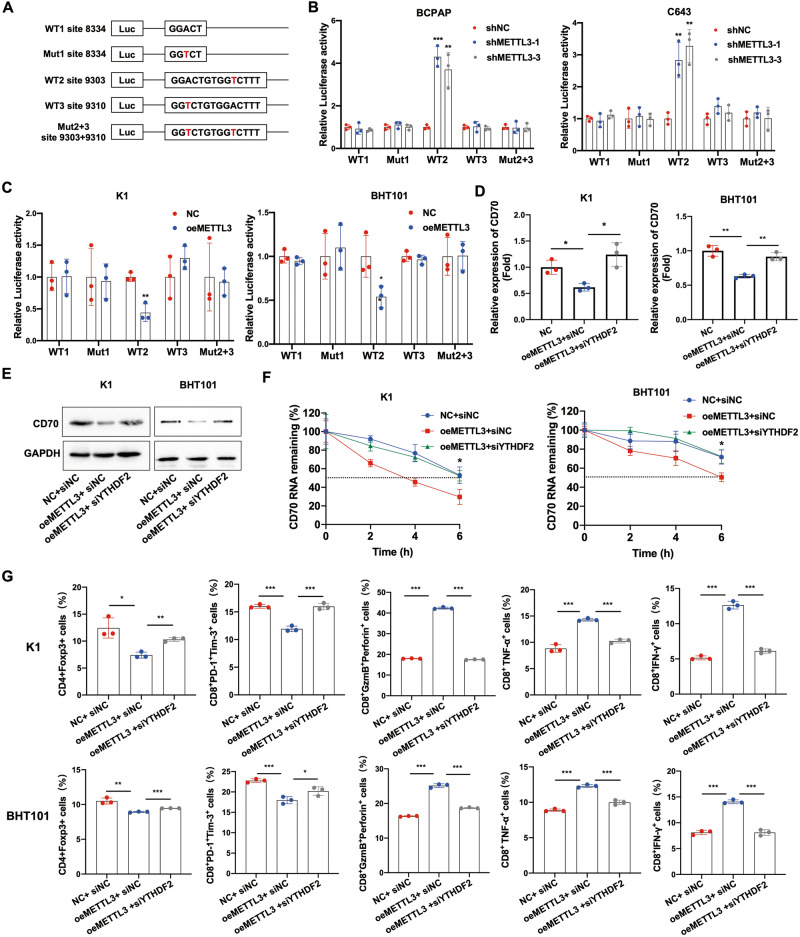
To further understand the mechanism underlying METTL3-mediated regulation of CD70, we respectively cloned three synonymous mutations at the putative m6A sites in CD70 3’UTR into the pmirGlo dual-luciferase reporter. The relative luciferase activity of the reporter carrying CD70 with m6A-modified residue 9303 reduced significantly in the METTL3-knockdown BCPAP and C643 cells. However, a mutation at that residue nullified the effect of METTL3 silencing (Fig. 4A, B). The opposite results were observed in METTL3-overexpressing K1 and BHT101 cells (Fig. 4C). These results suggested that the GGAC sequence of 9303 in the 3’UTR of CD70 is essential for METTL3-mediated methylation. Since CD70 expression is negatively regulated by m6A modification, we hypothesized that the m6A reader YTHDF2 recognizes the methylated sites of CD70 mRNA and facilitates its degradation. Indeed, YTHDF2 knockdown reversed the downregulation of CD70 mRNA and protein in METTL3-overexpressing cells (Fig. 4D, E), and also extended the half-life of CD70 mRNA after treatment with actinomycin D (Fig. 4F). To further explore whether the immunomodulatory function of METTL3 is also dependent on YTHDF2, we analyzed the percentage of different T cell populations in PBMCs co-cultured in the presence of METTL3-overexpressing thyroid cancer cells with or without YTHDF2 knockdown. While METTL3 overexpression significantly decreased the percentage of Tregs and terminally exhausted T cells, and increased that of cytotoxic CD8+ T cells, YTHDF2 silencing reversed these effects (Fig. 4G and Fig. S4). In summary, the immunoregulatory function of METTL3 in thyroid cancer is dependent on YTHDF2-mediated recognition of the m6A sites in CD70 mRNA and subsequent degradation of the transcripts.
Fig. 4. METTL3-mediated regulation of CD70 is dependent on YTHDF2.
A Putative m6A modification sites in the CD70 mRNA 3’UTR. B, C The relative luciferase activity of the CD70 3’UTR reporter construct with wild type or synonymous mutant m6A sites in METTL3-knockdown and METTL3-overexpressing PTC and ATC cells are shown. D CD70 mRNA levels, E CD70 protein levels and F half-life of CD70 mRNA after actinomycin D treatment in METTL3-overexpressing K1 and BHT101 cells transfected with si-YTHDF2. G Percentage of CD4+Foxp3+ Tregs, CD8+PD-1+Tim-3+ terminally exhausted T cells, CD8+GzmB+perforin+ T cells, CD8+TNF-α+ T cells and CD8+IFN-γ+ T cells in PBMCs co-cultured in the presence of METTL3-overexpressing K1 and BHT101 cells transfected with si-YTHDF2. Data of are presented as the mean ± SD of three independent experiments. The p-values in (B), (C), (D), (F) and (G) were calculated by Student’s t-test. *p < 0.05; **p < 0.01; ***p < 0.001.
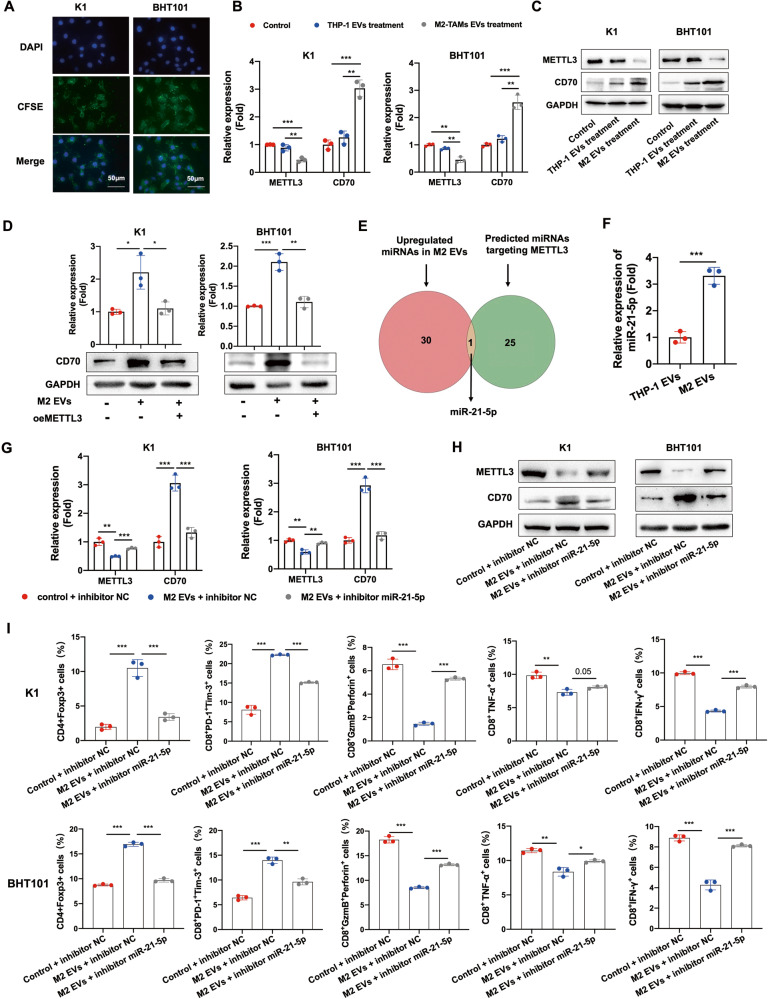
M2 macrophage-derived EVs downregulated METTL3 in PTC and ATC cells through miR-21-5p
GO analysis of the bulk RNA-seq data of PTC and ATC tissues revealed that pathways involved in the regulation of EVs were significantly enriched in the METTL3low group compared to the METTL3high group (Fig. S5A-B), which suggested that the expression of METTL3 in thyroid tumors may be regulated by EVs. To explore the possible source of EVs, we analyzed the components of the TME using the CIBERSORT algorithm, and found that macrophages were the most abundant immune cells in the tumors. Furthermore, macrophage infiltration was particularly high in the METTL3low and CD70high tissues (Fig. S5C-D). M2 macrophages are key immunosuppressive cells in the TME, and can transfer multiple bioactive molecules to tumor cells in order to modulate the biological functions of the latter [32]. In a previous study, we found that M2 macrophages can transport non-coding RNAs (ncRNAs) to cancer cells through EVs [23]. Therefore, we hypothesized that M2 macrophages may regulate the expression of METTL3 in thyroid tumor tissues via EVs.
To confirm our hypothesis, we incubated KI and BHT101 cells with carboxyfluorescein succinimidyl ester (CFSE)-labeled M2 EVs. The CSFE-labeled EVs were detected in the K1 and BHT101 cells, which indicated that the vesicles were phagocytosed (Fig. 5A). Furthermore, METTL3 was downregulated and CD70 was upregulated in cells co-cultured with M2 EVs compared to the controls or cells co-cultured with THP-1 cells (Fig. 5B, C). Overexpressing METTL3 reversed the M2 EVs-induced upregulation of CD70, indicating that M2 EVs increased CD70 expression by downregulating METTL3 (Fig. 5D).
Fig. 5. M2 macrophage-derived EVs downregulated METTL3 in PTC and ATC cells through miR-21-5p.
A Representative images of K1 and BHT101 cells with phagocytosed CSFE-labeled M2 EVs (scale bars, 50 μm). The cell nucleus were stained with DAPI. B, C METTL3 and CD70 mRNA and protein levels in K1 and BHT101 cells co-cultured with M2 EVs, co-cultured with THP-1 cells and cultured alone. D CD70 mRNA and protein levels in K1oeMETTL3 and BHT101 oeMETTL3 cells co-cultured with M2 EVs. E Overlap between upregulated miRNAs in M2 EVs and predicted miRNAs targeting METTL3. F The expression of miR-21-5p in THP-1 EVs and M2 EVs. G, H METTL3 and CD70 mRNA and protein levels in K1 and BHT101 cells co-cultured with M2 EVs in the presence or absence of the miR-21-5p inhibitor. I Percentage of CD4+Foxp3+ Tregs, CD8+PD-1+Tim-3+ terminally exhausted T cells, CD8+GzmB+perforin+ T cells, CD8+TNF-α+ T cells and CD8+IFN-γ+ T cells in PBMCs co-cultured with M2 EVs-treated K1 and BHT101 cells expressing the miR-21-5p inhibitor. Data of are presented as the mean ± SD of three independent experiments. The p-values in (B), (D), (F), (G), (H) and (I) were calculated by Student’s t-test. *p < 0.05; **p < 0.01; ***p < 0.001.
To explore the mechanisms underlying the regulatory effect of M2 EVs on METTL3, we used TargetScan algorithm [33] to predict miRNAs targeting METTL3, and screened the GSE97467 dataset for differentially expressed miRNAs between the exosomes secreted by monocytes and M2 macrophages. we identified miR-21-5p as a putative regulator of METTL3, and detected high expression levels of miR-21-5p in the M2 EVs compared to THP-1 EVs (Fig. 5E), which was also confirmed by RT-qPCR (Fig. 5F). Moreover, TCGA data indicated that miR-21-5p was significantly upregulated in PTC patients with METTL3low and CD70high tissues compared to the other groups (Fig. S5E). These results suggested that M2 macrophages may downregulate METTL3 and upregulate CD70 in thyroid cancer cells by transmitting EVs containing miR-21-5p. Consistent with this hypothesis, miR-21-5p significantly repressed the luciferase activity of the METTL3-WT construct whereas miR-NC had no effect (Fig. S5F). In addition, miR-21-5p decreased METTL3 expression and increased that of CD70 (Fig. S5G-H), and inhibition of miR-21-5p reversed the effect of M2 EVs on METTL3 and CD70 mRNA and protein levels (Fig. 5G, H). Interestingly, K1 and BHT101 cells incubated with M2 EVs significantly increased the percentage of Tregs and terminally exhausted T cells, and decreased that of cytotoxic CD8+ T cells among the co-cultured PBMCs. However, inhibition of miR-21-5p reversed the immunosuppressive effect of M2 EVs (Fig. 5I and Fig. S6A). Taken together, the M2 EVs downregulate METTL3 in PTC and ATC cells via miR-21-5p, thus promoting CD70-induced immunosuppression.
Since high CD70 expression in the tumor cells promotes infiltration of M2 macrophages in glioblastomas [30], we hypothesized that the METTL3/CD70 axis may affect chemotaxis of M2 macrophages in thyroid tumors as well. Indeed, conditioned media from METTL3-knockdown PTC and ATC cells attracted more M2 macrophages compared to that from control cells, and this effect was reversed by silencing CD70 in the tumor cells (Fig. S6B). These results suggested that M2 macrophages and the METTL3/CD70 axis may form a positive feedback loop in PTC and ATC cells to promote immunosuppression and resistance to PD-1 blockade.
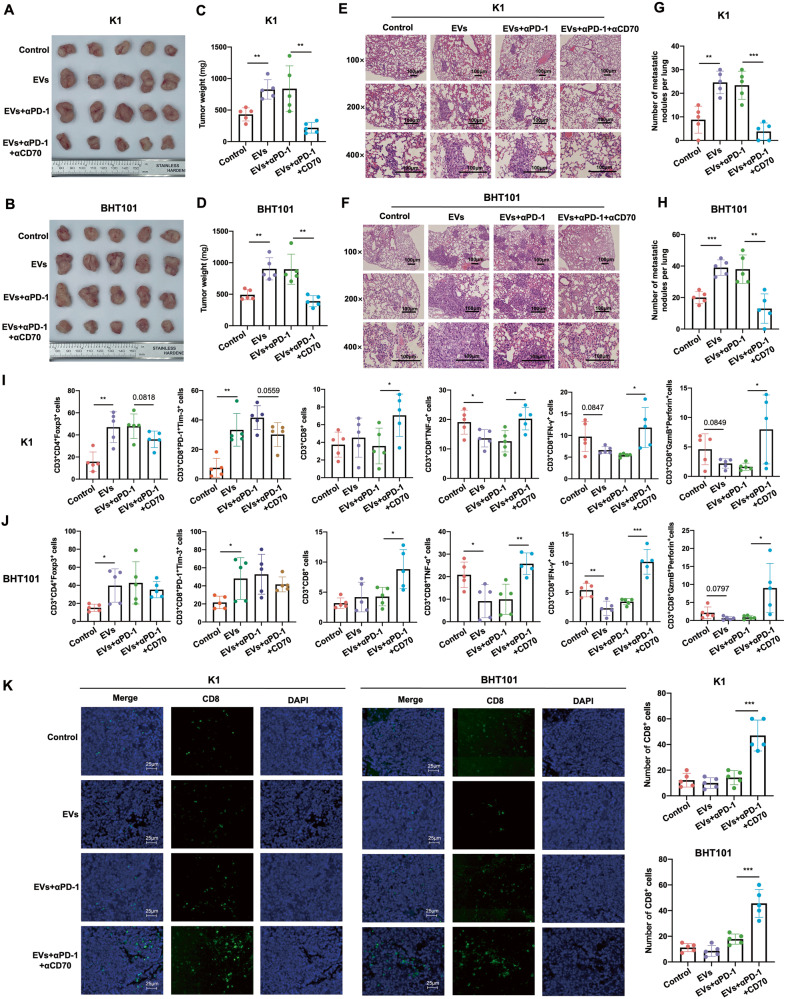
CD70 blockade reversed αPD-1 resistance induced by M2 EVs in PTC and ATC
To explore the possibility of targeting CD70 in order to improve the outcomes of αPD-1 therapy in vivo, we transplanted K1 and BHT101 cells into the huPBMC-NCG mice, and treated the tumor-bearing mice with M2 EVs, followed by αPD-1 mAb with or without αCD70 mAb (cusatuzumab) (Fig. S7A-B). The presence of M2 EVs significantly increased the volume and weight of the primary tumors, as well as the number of pulmonary metastatic nodules formed by K1 and BHT101 cells compared to that in the control group. Interestingly, while αPD-1 mAb did not exert any obvious anti-tumor effect in the M2 EVs-treated mice, the combination of αPD-1 mAb and αCD70 mAb significantly decreased the tumor volume, tumor weight and number of metastatic nodules (Fig. 6A–H and Fig. S7C). Furthermore, the M2 EVs increased the percentage of immunosuppressive Tregs and terminally exhausted T cells, and decreased that of cytotoxic CD3+CD8+ T cells producing TNF-α, IFN-γ, GzmB and perforin in the K1 and BHT101 xenografts. On the other hand, the combination of αPD-1 mAb and αCD70 mAb significantly increased the infiltration of CD3+CD8+ T cells and CD3+CD8+ T cells producing TNF-α, IFN-γ, GzmB and perforin, and the effect was stronger compared to that of αPD-1 mAb monotherapy (Fig. 6I, J and Fig. S7D). IF staining of the xenografts further confirmed a significant increase in CD8+ T cells after the combination treatment compared to αPD-1 mAb monotherapy (Fig. 6K). Taken together, CD70 blockade can effectively reverse the immunosuppressive phenotype induced by M2 EVs and therefore improve the therapeutic outcomes of anti-PD-1 therapy in PTC and ATC.
Fig. 6. CD70 blockade reversed αPD-1 resistance induced by M2 EVs in PTC and ATC.
A, B General view of tumor mass, C, D tumor weights, E, F representative images of HE-stained pulmonary metastatic nodules (scale bars, 100 μm) and G, H quantification of metastatic nodules from mice bearing K1and BHT101xenografts treated with placebo, M2 EVs, M2 EVs + αPD-1 mAb, and M2 EVs + αPD-1 mAb and CD70 mAb. I, J Percentage of CD3+CD4+Foxp3+ Tregs, CD3+CD8+PD-1+Tim-3+ terminally exhausted T cells, CD3+CD8+ T cells, CD3+CD8+ TNF-α+ T cells, CD3+CD8+ IFN-γ+ T cells and CD3+CD8+GzmB+perforin+ T cells in the above groups. K Representative IF staining images showing CD8+ T cells in the above groups (scale bars, 25 μm). Data are presented as the mean ± SD of 5 biologically independent animals. The p-values in (C), (D), (G), (H), (I), (J) and (K) were calculated by Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
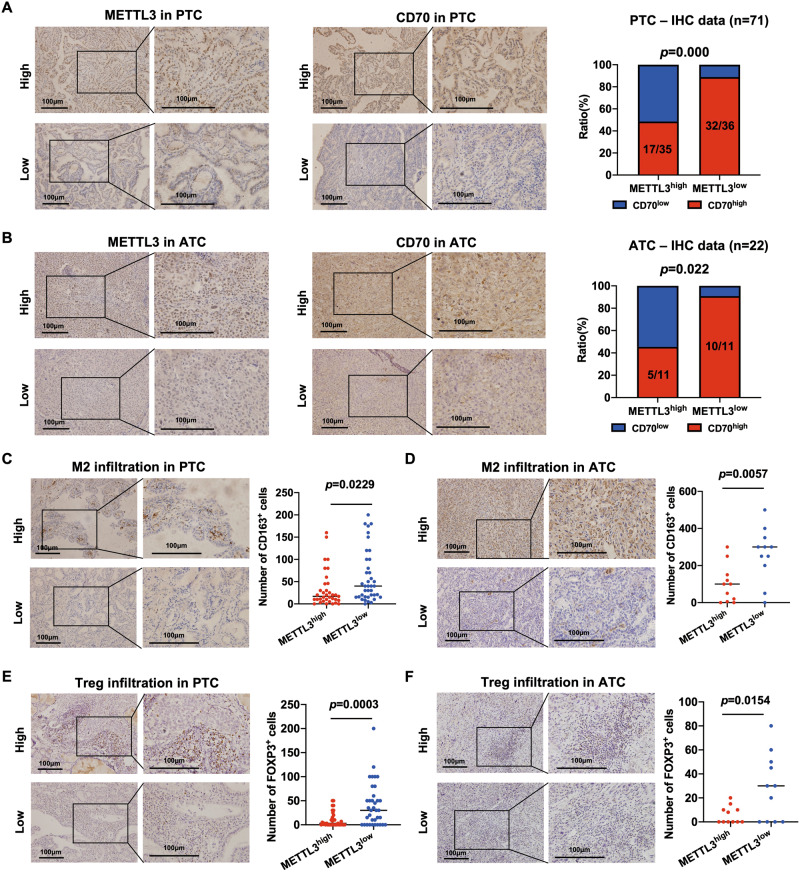
METTL3 expression negatively correlates with CD70 expression and M2 macrophages and Tregs infiltration in PTC and ATC tissues
To determine the clinical correlation between METLL3 and CD70, we analyzed their expression levels in PTC and ATC tissues using bulk RNA-seq data, RT-qPCR data and IHC staining data. As shown in Fig. S8A-C, CD70 mRNA expression was negatively correlated with that of METTL3 mRNA as per the bulk RNA-seq data of 503 PTC and 22 ATC tissues, as well as the RT-qPCR data of 29 PTC tissues (Fig. S8A-C). Moreover, low METTL3 mRNA expression and high CD70 mRNA expression in PTC were associated with larger tumors, higher lymph node metastasis rate, and higher TNM stage compared to the other groups (Table S1). We validated the correlation between METTL3 and CD70 proteins using IHC staining data of 71 PTC and 22 ATC tissue specimens, and the results were consistent (Fig. 7A, B). In addition, low METTL3 protein expression and high CD70 protein expression also correlated to poor prognoses in the PTC patients (Table S2).
Fig. 7. METTL3 expression negatively correlates with CD70 expression and M2 macrophages and Tregs infiltration in PTC and ATC tissues.
(A-B) Representative IHC images showing in situ METTL3 and CD70 protein expression in PTC (n = 71) and ATC (n = 22) tissues. The chi-square test was used to analyze the correlation between METTL3 and CD70 expression. (C-D) Representative IHC images showing CD163+ M2 macrophages in PTC and ATC tissues. (E-F) Representative IHC images showing Foxp3+ Tregs in PTC and ATC tissues. Scale bars, 100 μm. The p-values in panel C, D, E and F were calculated by Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
The correlation between macrophages or Tregs infiltration and METTL3 expression was analyzed using the TISIDB database (http://cis.hku.hk/TISIDB/index.php) [34]. METTL3 mRNA expression was negatively correlated with the infiltration of macrophages and Tregs in thyroid tumors (Fig. S8D-E). Furthermore, IHC staining of the PTC and ATC tissues for CD163 and Foxp3 indicated higher infiltration of M2 macrophages (Fig. 7C, D) and Tregs (Fig. 7E, F) respectively in samples with low METTL3 expression compared with those with high METTL3 expression.
Discussion
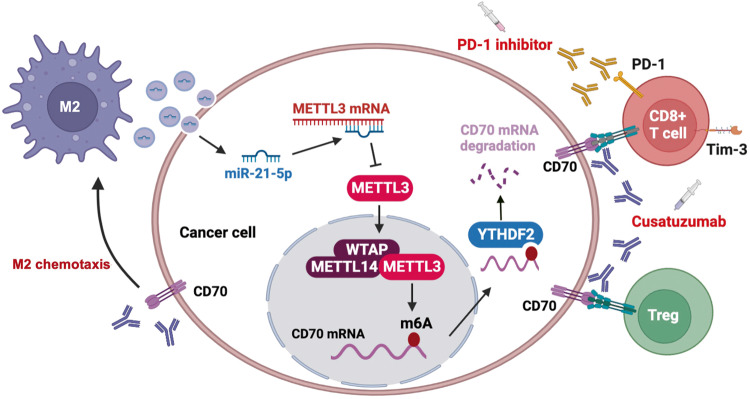
In this study, we found that M2 macrophages can downregulate METTL3 expression in tumor cells by secreting EVs that contain miR-21-5p, resulting in demethylation and stabilization of CD70 mRNA in a YTHDF2-dependent manner. The subsequent upregulation in CD70 protein levels increases the abundance of the immunosuppressive Tregs and terminally exhausted T cells, and promotes M2 macrophage chemotaxis, thereby forming a positive feedback loop of immunosuppression and resistance to anti-PD1 therapy. Targeting CD70 with cusatuzumab can disrupt this loop and reverse anti-PD-1 resistance, thus offering a novel strategy for the treatment of recalcitrant thyroid cancer (Fig. 8).
Fig. 8. The proposed model illustrates the mechanism by which M2 EVs induced METTL3 inhibition to drives anti-PD-1 therapy resistance via m6A-CD70-mediated immune suppression in thyroid cancer.
M2 macrophages in the thyroid TME secrete EVs containing miR-21-5p, which downregulates METTL3 in the tumor cells, resulting in demethylation and stabilization of CD70 mRNA in a YTHDF2-dependent manner, eventually leading to increased CD70 protein levels. CD70 increases the proportion of Tregs and terminally exhausted T cells, and promotes M2 macrophage chemotaxis, thus forming a positive feedback loop of immunosuppression and anti-PD-1 therapy resistance. Targeting CD70 using cusatuzumab can break this loop and reverse anti-PD-1 resistance.
M6A modification plays a significant role in shaping the TME, and the m6A patterns of tumor cells can predict the immune landscape of the TME and guide immunotherapy [35]. In addition, m6A modification in tumor cells can aid in immune evasion by promoting the secretion of chemokines with oncogenic effects and suppressing those involved in anti-tumor immunity [36]. METTL3 is an established oncogene [37] that inhibits immune responses in most cancer types, such as breast cancer [38], lung cancer [39], colorectal cancer [40, 41] and bladder cancer [42]. Inhibition of METTL3 in colorectal cancer and melanoma cells significantly enhanced the response to anti-PD-1 therapy [43]. Interestingly, low METTL3 expression was associated with ICB resistance in thyroid cancer tissues, and overexpressing METTL3 in tumor cells reversed the immunosuppressive microenvironment and boosted the efficacy of anti-PD-1 therapy, which contradicts the observations with other cancers. Nevertheless, our findings are consistent with previous reports stating that METTL3 is downregulated in thyroid cancer and inhibits cancer progression [10, 11]. Therefore, METTL3 activators are a promising approach for the treatment of thyroid cancer.
The mechanism underlying the immunosuppressive effect of METTL3 downregulation was further explored, which revealed that the immune checkpoint molecule CD70 was regulated by METTL3-mediated m6A modification. Previous studies have shown that CD70 is aberrantly expressed in many hematological and solid malignancies and is associated with tumor progression and immunosuppression [44]. Consistently, high expression of CD70 in the cancer cells can facilitate immune evasion via three mechanisms: increased survival and proliferation of Tregs, induction of T cell apoptosis, and skewing T cells towards the exhausted phenotype [45]. However, the role of CD70 in thyroid cancer has not been reported so far. In our study, we observed that knocking down CD70 in PTC and ATC cells significantly decreased the percentage of immunosuppressive Tregs and terminally exhausted T cells, and increased that of cytotoxic T cells among the co-cultured PBMCs. However, CD70 knockdown had no effect on the proportion of CD8+ T cells (data not shown), suggesting that CD70 does not regulate T cell apoptosis in thyroid cancer. Both mono- and combination therapies of anti-CD70 antibody have been explored in acute myeloid leukemia (AML) [44], although it is unclear whether patients with thyroid cancer can benefit from this approach. We found that 69% of PTC tissues and 68% of ATC tissues expressed high levels of CD70, which strongly indicates the potential therapeutic value of targeting CD70 in these tumors. Cusatuzumab, a human anti-CD70 monoclonal antibody with enhanced cytotoxicity, can reportedly eliminate AML stem cells in patients treated with hypomethylating agents [46]. In our study as well, the combination of cusatuzumab and the anti-PD-1 antibody nivolumab significantly decreased the growth and metastases of PTC and ATC xenografts in mice. Thus, the combination of cusatuzumab and nivolumab should be considered for CD70+ PTC and ATC patients.
Tumor-associated macrophages (TAMs) mainly exhibit the M2 phenotype to induce an immunosuppressive milieu and can limit the efficacy of anti–PD-1 therapy in many tumors [47, 48]. A previous study showed that approximately 50% of PTC tissues and most ATC tissues have extensive infiltration of TAMs, and high infiltration of M2-TAMs supports the immunosuppressive TME and promotes thyroid cancer progression [49]. However, the underlying molecular mechanisms have not yet been elucidated. Our study found that M2-TAMs downregulated METTL3 in the thyroid cancer cells and promoted immunosuppressive TME through transmiting EVs containing miR-21-5p and inhibition of miR-21-5p could reverse the immunosuppressive effect of M2-TAMs. This finding indicated that targeting miR-21-5p may improve M2-TAMs-mediated anti-PD1 therapy resistance.
Despite these significant findings, our study does have some limitations. First, the ICB response analysis in PTC and ATC patients was based on the prediction software due to lack of clinical data. Thus, clinical trials will be required to validate whether METTL3 and CD70 can predict ICB response for thyroid cancer. In addition, given the heterogeneity and complexity of the thyroid TME, spontaneous tumor-developing transgenic mice are needed to explore the role of METTL3 in other immune cells and validate METTL3 and CD70 as therapeutic targets in thyroid cancer. Moreover, other m6A regulatory genes, such as METTL14, ZC3H13, FTO, YTHDF3, IGF2BP2 and YTHDF1, were also differently expressed in the PTC or ATC tissues, and their impact on the immune microenvironment and ICB response of thyroid cancer also need to be elucidated.
In summary, our study reveals a novel regulatory mechanism between tumor cells and TME that mediates anti-PD-1 therapy resistance in PTC and ATC. The immunosuppressive effect of the M2 EVs via the METTL3/CD70 axis can be potentially targeted to enhance sensitivity of recalcitrant thyroid cancer to anti-PD-1 therapy.
Supplementary information
Author contributions
MG, XZ and XR performed study concept, design and supervision; JN, XH and JH designed and performed most of the experiments, analyzed the data, and created the figures. WZ and YH participated in some experiments. JN and XH wrote and revised the manuscript. YS helped to revise the manuscript. MG, XZ, XR, and NJ provided financial support. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82203668, 82172821, 82103386, 81872169), Tianjin Municipal Science and Technology Project (19JCYBJC27400, 21JCZDJC00360), Beijing-Tianjin-Hebei Basic Research Cooperation Project (20JCZXJC00120), the Science &Technology Development Fund of Tianjin Education Commission for Higher Education (2021ZD033), Tianjin Medical Key Discipline (Specialty) Construction Project (TJYXZDXK-058B), Tianjin Health Research Project (TJWJ2022XK024).
Data availability
The data supporting the present study are available from the corresponding author upon reasonable request.
Code availability
Detailed code is available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (No. EK2021149 for human cancer specimens and NO. AE-2022032 for in vivo mouse experiments). Written informed consent was obtained from all subjects. All experiments were performed in accordance with the principles of the Declaration of Helsinki.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Junya Ning, Xiukun Hou, Jie Hao.
Contributor Information
Xianhui Ruan, Email: tjruanxianhui@163.com.
Xiangqian Zheng, Email: xzheng05@tmu.edu.cn.
Ming Gao, Email: headandneck2008@126.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01217-x.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.French JD, Bible K, Spitzweg C, Haugen BR, Ryder M. Leveraging the immune system to treat advanced thyroid cancers. Lancet Diabetes Endocrinol. 2017;5:469–81. doi: 10.1016/S2213-8587(16)30277-7. [DOI] [PubMed] [Google Scholar]
- 3.Capdevila J, Wirth LJ, Ernst T, Ponce Aix S, Lin CC, Ramlau R, et al. PD-1 blockade in anaplastic thyroid carcinoma. J Clin Oncol. 2020;38:2620–7. doi: 10.1200/JCO.19.02727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giannini R, Moretti S, Ugolini C, Macerola E, Menicali E, Nucci N, et al. Immune profiling of thyroid carcinomas suggests the existence of two major phenotypes: An ATC-Like and a PDTC-Like. J Clin Endocrinol Metab. 2019;104:3557–75. doi: 10.1210/jc.2018-01167. [DOI] [PubMed] [Google Scholar]
- 5.Lei Y, Li X, Huang Q, Zheng X, Liu M. Progress and challenges of predictive biomarkers for immune checkpoint blockade. Front Oncol. 2021;11:617335. doi: 10.3389/fonc.2021.617335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray KD, McCloskey JE, Vedvyas Y, Kalloo OR, Eshaky SE, Yang Y, et al. PD1 blockade enhances ICAM1-directed CAR T therapeutic efficacy in advanced thyroid cancer. Clin Cancer Res. 2020;26:6003–16. doi: 10.1158/1078-0432.CCR-20-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bastman JJ, Serracino HS, Zhu Y, Koenig MR, Mateescu V, Sams SB, et al. Tumor-infiltrating T Cells and the PD-1 checkpoint pathway in advanced differentiated and anaplastic thyroid cancer. J Clin Endocrinol Metab. 2016;101:2863–73. doi: 10.1210/jc.2015-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehnert JM, Varga A, Brose MS, Aggarwal RR, Lin CC, Prawira A, et al. Safety and antitumor activity of the anti-PD-1 antibody pembrolizumab in patients with advanced, PD-L1-positive papillary or follicular thyroid cancer. BMC Cancer. 2019;19:196. doi: 10.1186/s12885-019-5380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu ZH, Feng ST, Zhang D, Cao XC, Yu Y, Wang X. The functions and prognostic values of m6A RNA methylation regulators in thyroid carcinoma. Cancer Cell Int. 2021;21:385. doi: 10.1186/s12935-021-02090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Y, Peng X, Zhou Q, Tan L, Zhang C, Lin S, et al. METTL3-mediated m6A modification of STEAP2 mRNA inhibits papillary thyroid cancer progress by blocking the Hedgehog signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis. 2022;13:358. doi: 10.1038/s41419-022-04817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He J, Zhou M, Yin J, Wan J, Chu J, Jia J, et al. METTL3 restrains papillary thyroid cancer progression via m(6)A/c-Rel/IL-8-mediated neutrophil infiltration. Mol Ther. 2021;29:1821–37. doi: 10.1016/j.ymthe.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miao YR, Zhang Q, Lei Q, Luo M, Xie GY, Wang H, et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinh) 2020;7:1902880. doi: 10.1002/advs.201902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou X, Shi X, Zhang W, Li D, Hu L, Yang J, et al. LDHA induces EMT gene transcription and regulates autophagy to promote the metastasis and tumorigenesis of papillary thyroid carcinoma. Cell Death Dis. 2021;12:347. doi: 10.1038/s41419-021-03641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–w514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen H, Shen X, Feng M, Wu D, Zhang C, Yang Y, et al. A universal approach for integrating super large-scale single-cell transcriptomes by exploring gene rankings. Brief Bioinform. 2022;23:bbab573. doi: 10.1093/bib/bbab573. [DOI] [PubMed] [Google Scholar]
- 16.Luo H, Xia X, Kim GD, Liu Y, Xue Z, Zhang L, et al. Characterizing dedifferentiation of thyroid cancer by integrated analysis. Sci Adv. 2021;7:eabf3657. doi: 10.1126/sciadv.abf3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao X, Dang L, Zheng X, Lu Y, Lu Y, Ji R, et al. Targeting super-enhancer-driven oncogenic transcription by CDK7 inhibition in anaplastic thyroid carcinoma. Thyroid. 2019;29:809–23. doi: 10.1089/thy.2018.0550. [DOI] [PubMed] [Google Scholar]
- 18.Ruan X, Tian M, Kang N, Ma W, Zeng Y, Zhuang G, et al. Genome-wide identification of m6A-associated functional SNPs as potential functional variants for thyroid cancer. Am J Cancer Res. 2021;11:5402–14. [PMC free article] [PubMed] [Google Scholar]
- 19.Song H, Song J, Cheng M, Zheng M, Wang T, Tian S, et al. METTL3-mediated m(6)A RNA methylation promotes the anti-tumour immunity of natural killer cells. Nat Commun. 2021;12:5522. doi: 10.1038/s41467-021-25803-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–5. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ning J, Ye Y, Bu D, Zhao G, Song T, Liu P, et al. Imbalance of TGF-β1/BMP-7 pathways induced by M2-polarized macrophages promotes hepatocellular carcinoma aggressiveness. Mol Ther. 2021;29:2067–87. doi: 10.1016/j.ymthe.2021.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan W, Wu X, Zhou W, Fong MY, Cao M, Liu J, et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol. 2018;20:597–609. doi: 10.1038/s41556-018-0083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanmamed MF, Rodriguez I, Schalper KA, Onate C, Azpilikueta A, Rodriguez-Ruiz ME, et al. Nivolumab and urelumab enhance antitumor activity of human T lymphocytes engrafted in Rag2-/-IL2Rgammanull immunodeficient mice. Cancer Res. 2015;75:3466–78. doi: 10.1158/0008-5472.CAN-14-3510. [DOI] [PubMed] [Google Scholar]
- 26.Feng Y, Xie K, Yin Y, Li B, Pi C, Xu X, et al. A Novel Anti-B7-H3 x Anti-CD3 bispecific antibody with potent antitumor activity. Life (Basel) 2022;12:157. doi: 10.3390/life12020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li R, Huang Y, Lin J. Distinct effects of general anesthetics on lung metastasis mediated by IL-6/JAK/STAT3 pathway in mouse models. Nature. Communications. 2020;11:642. doi: 10.1038/s41467-019-14065-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, Meng Y, Liu L, Lv Y, Yu W, Liu T, et al. CD4(+) T cells are required to improve the efficacy of CIK therapy in non-small cell lung cancer. Cell Death Dis. 2022;13:441. doi: 10.1038/s41419-022-04882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao P, Long X, Zhang L, Ye Y, Guo J, Liu P, et al. Neurotensin/IL-8 pathway orchestrates local inflammatory response and tumor invasion by inducing M2 polarization of Tumor-Associated macrophages and epithelial-mesenchymal transition of hepatocellular carcinoma cells. Oncoimmunology. 2018;7:e1440166. doi: 10.1080/2162402X.2018.1440166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge H, Mu L, Jin L, Yang C, Chang YE, Long Y, et al. Tumor associated CD70 expression is involved in promoting tumor migration and macrophage infiltration in GBM. Int J Cancer. 2017;141:1434–44. doi: 10.1002/ijc.30830. [DOI] [PubMed] [Google Scholar]
- 31.Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608–24. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Zhao M, Liu S, Guo J, Lu Y, Cheng J, et al. Macrophage-derived extracellular vesicles: diverse mediators of pathology and therapeutics in multiple diseases. Cell Death Dis. 2020;11:924. doi: 10.1038/s41419-020-03127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–2. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 35.Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19:53. doi: 10.1186/s12943-020-01170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lou X, Wang JJ, Wei YQ, Sun JJ. Emerging role of RNA modification N6-methyladenosine in immune evasion. Cell Death Dis. 2021;12:300. doi: 10.1038/s41419-021-03585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. 2020;13:117. doi: 10.1186/s13045-020-00951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W, et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. 2022;21:60. doi: 10.1186/s12943-021-01447-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z, Wang T, She Y, Wu K, Gu S, Li L, et al. N(6)-methyladenosine-modified circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer. 2021;20:105. doi: 10.1186/s12943-021-01398-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, Pan Y, Zhou Q, Liang C, Wong CC, Zhou Y, et al. METTL3 Inhibits Antitumor Immunity by Targeting m(6)A-BHLHE41-CXCL1/CXCR2 Axis to Promote Colorectal Cancer. Gastroenterology. 2022;163:891–907. doi: 10.1053/j.gastro.2022.06.024. [DOI] [PubMed] [Google Scholar]
- 41.Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, et al. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell. 2022;82:1660–1677.e1610. doi: 10.1016/j.molcel.2022.02.033. [DOI] [PubMed] [Google Scholar]
- 42.Ni Z, Sun P, Zheng J, Wu M, Yang C, Cheng M, et al. JNK signaling promotes bladder cancer immune escape by regulating METTL3-Mediated m6A Modification of PD-L1 mRNA. Cancer Res. 2022;82:1789–802. doi: 10.1158/0008-5472.CAN-21-1323. [DOI] [PubMed] [Google Scholar]
- 43.Wang L, Hui H, Agrawal K, Kang Y, Li N, Tang R, et al. m(6) A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. Embo j. 2020;39:e104514. doi: 10.15252/embj.2020104514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flieswasser T, Van den Eynde A, Van Audenaerde J, De Waele J, Lardon F, Riether C, et al. The CD70-CD27 axis in oncology: the new kids on the block. J Exp Clin Cancer Res. 2022;41:12. doi: 10.1186/s13046-021-02215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacobs J, Deschoolmeester V, Zwaenepoel K, Rolfo C, Silence K, Rottey S, et al. CD70: An emerging target in cancer immunotherapy. Pharmacol Ther. 2015;155:1–10. doi: 10.1016/j.pharmthera.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Riether C, Pabst T, Höpner S, Bacher U, Hinterbrandner M, Banz Y, et al. Targeting CD70 with cusatuzumab eliminates acute myeloid leukemia stem cells in patients treated with hypomethylating agents. Nat Med. 2020;26:1459–67. doi: 10.1038/s41591-020-0910-8. [DOI] [PubMed] [Google Scholar]
- 47.Duan Z, Luo Y. Targeting macrophages in cancer immunotherapy. Signal Transduct Target Ther. 2021;6:127. doi: 10.1038/s41392-021-00506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhulai G, Oleinik E. Targeting regulatory T cells in anti-PD-1/PD-L1 cancer immunotherapy. Scand J Immunol. 2022;95:e13129. doi: 10.1111/sji.13129. [DOI] [PubMed] [Google Scholar]
- 49.Liu Q, Sun W, Zhang H. Roles and new insights of macrophages in the tumor microenvironment of thyroid cancer. Front Pharmacol. 2022;13:875384. doi: 10.3389/fphar.2022.875384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the present study are available from the corresponding author upon reasonable request.
Detailed code is available from the corresponding author upon request.