Abstract
Pyroptosis is a new type of programmed cell death involved in all stages of tumorigenesis. Herein, a comprehensive study was conducted to evaluate the prognostic significance of pyroptosis-related lncRNAs in bladder cancer. Consensus clustering analysis was performed to identify the subclusters of bladder cancer. The prognostic pyroptosis-related lncRNA signature was constructed using LASSO Cox regression analysis. Consensus clustering identified 2 clusters of bladder cancer. Interestingly, significant differences in the ESTIMAE score, immune cell infiltration and immune checkpoint expression were obtained between the 2 clusters. A signature consisting of 11 pyroptosis-related lncRNAs was established and it had a good performance in predicting the overall survival rate of bladder cancer, with an AUC of 0.713. Moreover, pyroptosis-related lncRNA signature acted as a risk factor in bladder cancer. Bladder cancer patients with high-risk score had a higher tumor grade and higher clinical stage. A significant correlation was obtained between the risk score and immune cell infiltration. The expression of most checkpoints was higher in bladder cancer patients with high-risk score. A novel pyroptosis-related lncRNA signature was identified with prognostic value for bladder cancer patients. Pyroptosis-related lncRNAs have a potential role in cancer immunology and may serve as prognostic or therapeutic targets in bladder cancer.
Keywords: bladder cancer, immunotherapy target, lncRNA, prognostic signature, pyroptosis
1. Introduction
Bladder cancer ranks among the top ten most prevalent malignancies globally, accounting for 3.0% of all new cases of malignancies and 2.1% of all malignancy-related deaths.[1] It is estimated that there are over 550,000 new cases of bladder cancer worldwide every.[1] The therapy of bladder cancer has been restricted to surgery and immunotherapy or chemotherapy until now.[2] Once a patient is diagnosed with bladder cancer, the patient needs to be followed up for a lifetime, causing great financial and mental burden.[3] Prognosis varies widely among patients with bladder cancer, with a 5-year overall survival (OS) rate ranging from 5% to 90%.[4] The heterogeneity of bladder cancer calls for further studies of its molecular properties to identify new prognostic and therapeutic markers.
Pyroptosis is a new type of programmed cell death initiated by inflammasomes that differs from other forms of cell death.[5] Accumulating studies have suggested the vital role of pyroptosis in cancer due to its effect on all stages of tumorigenesis.[6] Regulated by noncoding RNAs, pyroptosis can regulate tumor cell proliferation, invasion and metastasis, thus affecting the prognosis of patients.[7] It was recommended that pyroptosis could serve as a prognostic marker for cancer.[8] Accumulating studies have identified that pyroptosis-related signatures could serve as prognostic biomarkers in many types of cancers.[9,10] Wan Li et al suggested that the pyroptosis-related signature could serve as a prognostic biomarker for lung adenocarcinoma.[10] Another pyroptosis-related signature could predict the prognosis and tumor infiltration in colon adenocarcinoma.[11] Moreover, the pyroptosis-related signature was correlated with the prognosis and immune microenvironment in gastric cancer.[12]
Long noncoding RNAs (lncRNAs), a category of RNAs with transcript lengths >200 nucleotides, regulate 70% of gene expression in mammals by interacting with DNA, RNA, and proteins.[13] Accumulating studies have demonstrated that lncRNAs are prognostic markers in bladder cancer and are associated with biological progression regulation. For example, a meta-analysis indicated lncRNA as a diagnostic and prognostic marker for bladder cancer.[14] Another study demonstrated lncRNA PCAT6 as a diagnostic and prognostic marker in bladder cancer.[15] This evidence suggests that pyroptosis-related lncRNAs may also play a vital role in bladder cancer.
In our study, we systematically analyzed the prognostic value of pyroptosis-related lncRNAs in bladder cancer. Using LASSO Cox analysis, we constructed a pyroptosis-related lncRNA signature. Moreover, the correlation between the risk score and immune infiltration in bladder cancer was also evaluated.
2. Materials and methods
2.1. Data sources
The flow diagram of the current study is shown in Supplementary Figure 1, http://links.lww.com/MD/K357. The RNA sequencing data of bladder cancer patients corresponding to clinical information were downloaded from TCGA (https://portal.gdc.cancer.gov/) on March 15, 2021. The immunophenoscore (IPS) of bladder cancer patients was downloaded from the TCIA (https://www.tcia.at) on March 15, 2021. A total of 412 bladder cancer cases (Supplementary Table 1, http://links.lww.com/MD/K358) and 19 normal cases were obtained.
2.2. Pyroptosis-related lncRNA screening
Based on previous publications[16–19] on March 15, 2021, we acquired 33 pyroptosis genes, which are shown in Supplementary Table 2, http://links.lww.com/MD/K359. The lncRNA annotation file of humans was downloaded from GENCODE. LncRNAs were defined as the long noncoding RNA annotation file of the GENCODE website, and 7 types of transcripts were obtained, including lincRNA, antisense, processed transcript, sense intronic, prime overlapping ncRNA, sense overlapping, and macro lncRNA. We obtained 14087 lncRNAs in the TCGA bladder cancer cohort according to the Ensemble IDs. For further analysis, the expression of pyroptosis genes and lncRNAs was then normalized to fragment per kilobase million values. Using the “igraph” packages in the R project, we then constructed a network revealing the correlation between pyroptosis genes and lncRNAs with a coefficient >0.3 and P < .01. Pyroptosis-related lncRNAs were defined as lncRNAs significantly correlated with pyroptosis genes. This was followed by prognostic pyroptosis-related lncRNA screening using the “survival” package (P < .001). By performing univariate Cox analysis, we drew a prognostic forest map of pyroptosis-related lncRNAs. Moreover, we then explored the levels of these lncRNAs in tumor and normal tissues with the “limma” package.
2.3. Consensus cluster analysis
Consensus cluster analysis was performed using the “ConsensusClusterPlus” package, and the threshold value was as follows: iteration = 100 and resample rate = 80%.[20–22] The difference in each cluster in bladder cancer was evaluated with the “survival” package. The differences in clinical characteristics between the 2 clusters of bladder cancer were evaluated with the “pheatmap” package. Using the ESTIMATE algorithm, we then compared the immune score, stroma score, ESTIMATE score and abundance of immune cells.[23,24] The results were visualized with the “vioplot” or “ggpubr” package. Moreover, immune checkpoint expression and IPS score in each cluster were visualized with the “ggpubr” package.
2.4. Prognostic signature construction
LASSO Cox regression analysis (“glmnet” package) was conducted to construct a prognostic signature using the prognostic pyroptosis-related lncRNAs screened above.[25,26] In this analysis, the entire TCGA BLCA dataset involving 412 patients was randomized to a training cohort (n = 206) and a testing cohort (n = 206) to enhance the dependability of the prediction model through the R “caret” package. The penalty parameter (λ) was determined with the minimum criteria, and pyroptosis-related lncRNAs were retained in this prognostic module. Bladder cancer patients in the TCGA cohort were separated into 2 subgroups (low- and high-risk) based on the risk score. The survival curve of the 2 subgroups in OS time was drawn with the “survival” package, and the ROC curve was drawn with the “timeROC” package. Kaplan–Meier survival analysis and time-dependent ROC analysis were applied to draw Kaplan–Meier curves and ROC curves. To further identify prognostic factors, univariate and multivariable Cox regression analyses were conducted considering age, sex, clinical stage, TNM stage and risk score. Univariate and multivariable Cox regression analyses were performed using the “survival” package with “P < .001” as the threshold value. The log-rank test was performed to calculate the p values, hazard ratio and 95% confidence interval (CI). Moreover, we also evaluated the correlation between the risk score and clinical characteristics and the abundance of 22 immune cell types.
2.5. Statistical analysis
All analyses were conducted with R version 4.0.1. Gene expression in cancer tissues versus normal tissues was performed with the Mann–Whitney U test. The difference between 2 subgroups was compared with Student t test. The chi-square test was performed to compare categorical variables. Survival curves were drawn with the Kaplan–Meier method, and the P value was calculated by the log-rank test. Univariate and multivariate analyses were performed to identify the independent prognostic value of the risk score integrated with other clinical features.
3. Results
3.1. Defining pyroptosis-related lncRNAs in bladder cancer
The coexpression network of pyroptosis genes and lncRNAs is shown in Figure 1A, and a total of 722 pyroptosis-related lncRNAs were obtained. Figure 1B shows the results of univariate Cox regression analysis, indicating that 23 pyroptosis-related lncRNAs were significantly correlated with the OS rate in bladder cancer (P < .001, Supplementary Table 3, http://links.lww.com/MD/K360). Further expression analysis suggested that most pyroptosis-related lncRNAs were upregulated, while the expression of AC008050.1, AC104825.1, AL133415.1, RBMS3-AS3, and LINC02762 was decreased in bladder cancer (Fig. 1C).
Figure 1.
Pyroptosis-related lncRNA screening in bladder cancer. (A) The network revealed the lncRNAs correlated with pyroptosis genes in bladder cancer with a coefficient >0.3 and P < .01. The orange dots represent pyroptosis genes, and the orange dots represent pyroptosis-related lncRNAs. (B) Forest map showing 23 pyroptosis-related lncRNAs with significant prognostic value. (C) The expression landscape of 23 pyroptosis-related lncRNAs in bladder cancer. *P < .05, **P < .01, and ***P < .001. lncRNAs = long noncoding RNAs.
3.2. Consensus clustering categorized bladder cancer patients
We used consensus clustering to categorize bladder cancer patients using the above prognostic lncRNAs. As a result, k = 2 was considered the optimal clustering stability from k = 2 to 9 according to the similarity displayed by lncRNA expression and the proportion of ambiguous clustering measures (Fig. 2A). Figure 2B and D shows the cumulative distribution function, increment in the AUC, and tracking plot of subgroups for k = 2 to k = 9, respectively. The survival curve revealed a worse OS rate in cluster 1 versus cluster 2 in bladder cancer (Fig. 2E, P = .003). However, there was no significant difference in age, sex, tumor grade, clinical stage or TNM stage (Fig. 2F, P > .05). Interestingly, the levels of most pyroptosis-related lncRNAs in cluster 2 were increased versus those in cluster 1 (Fig. 2F).
Figure 2.
Consensus clustering analysis. (A) Consensus clustering matrix for k = 2. The consensus clustering CDF (B), relative change in area under CDF curve (C) and tracking plot (D) for k = 2–9. (E) OS curve suggested poor prognosis in cluster 1 versus cluster 2. (F) Heatmap revealing the differences in clinicopathologic features and lncRNA expression between cluster 1 and cluster 2. CDF = cumulative distribution function, OS = overall survival.
3.3. Clusters correlated with different immune infiltration
To clarify the difference between the 2 clusters in the tumor environment in bladder cancer, we then analyzed the correlation between cluster and immune infiltration. As shown in Figure 3A and 3C, the immune score (P = 7.7e−11), stroma score (P = 6e−9) and ESTIMATE score (P = 1e−10) in cluster 1 were higher than those in cluster 2. Moreover, we also found a remarkable difference in the abundance of certain immune cells (Fig. 3D). Specifically, cluster 1 was correlated with a low abundance of plasma cells (P = .038) and regulatory T cells (P = .008) and a high abundance of M1 macrophages (P = .022) and M2 macrophages (P = .038) (Fig. 3E and H). Furthermore, the results suggested an upregulation of PD-L1, CTLA4, HAVCR2, IDO1, LAG3, PDCD1, and PDCD1LG2 and downregulation of SIGLEC15 in cluster 1 compared with cluster 2 (Fig. 4, P < .001). We then compared the IPS in these 2 clusters, which revealed a higher IPS in cluster 2 in the CTLA4_neg_PD1_neg group (P = 8.6e-7, Fig. 5A) and CTLA4_pos_PD1_neg group (P = 5.5e-5, Fig. 5C) but not in the CTLA4_neg_PD1_pos (P = .25, Fig. 5B) group than in cluster 1. The results also suggested a lower IPS in cluster 2 in the CTLA4_pos_PD1_pos group than in cluster 1 (P = .035, Fig. 5D).
Figure 3.
Clusters correlated with distinct immune cell infiltration in bladder cancer. Cluster 1 had a high StromaScore (A), ImmuneScore (B), and ESTIMATEScore (C) versus cluster 2 in bladder cancer. (D) The infiltrating landscape of 22 immune cells in 2 clusters. (E–H) The abundance of plasma cells, regulatory T cells, M1 macrophages and M2 macrophages in the 2 clusters.
Figure 4.
Immune checkpoint expression in different clusters of bladder cancer. The expression of PD-L1 (A), CTLA4 (B), HAVCR2 (C), IDO1 (D), LAG3 (E), PDCD1 (F), PDCD1LG2 (G), and SIGLEC15 (H) in cluster 1 and cluster 2 of bladder cancer. ***P < .001.
Figure 5.
The immunophenoscore in different clusters of bladder cancer. A higher immunophenoscore was observed in cluster 2 in the CTLA4_neg_PD1_neg group (A), CTLA4_pos_PD1_neg group (C), and CTLA4_pos_PD1_pos group (D) but not in the CTLA4_neg_PD1_pos (B) group than in cluster 1.
3.4. Construction of the pyroptosis-related lncRNA prognostic signature in bladder cancer
To better predict the prognosis of bladder cancer patients, we then constructed a pyroptosis-related lncRNA prognostic signature based on 11 lncRNAs using LASSO Cox analysis. Figure 6A and B presents the coefficient and partial likelihood deviance of the prognostic signature. According to the coefficient of each gene, the risk score of each patient was calculated as follows: risk score = EHMT2-AS1 expression × (−0.146) + STAG3L5P-PVRIG2P-PILRB expression × (−0.037) + AC068196.1 expression × (−0.224) + AC008050.1 expression × (0.117) + AL136295.2 expression × (−0.087) + AC093788.1 expression × (−0.201) + AC005840.4 expression × (−0.195) + AC021321.1 expression × (−0.111) + AL133415.1 expression × (0.533) + RBMS3-AS3 expression × (0.089) + LINC02762 expression × (0.008). Bladder cancer cases were separated into a high-risk group and a low-risk group. Further OS analysis demonstrated a worse OS rate in the high-risk group than in the low-risk group in the training cohort (P = .002), test cohort (P < .001), and all bladder cancer cohorts (P < .001), with AUCs of 0.713, 0.661 and 0.684 in the training cohort, test cohort, and all bladder cancer cohorts, respectively (Fig. 6C and E). Figure 6F and H presents the risk score distribution, survival status of patients and lncRNA expression in the training cohort, test cohort, and all bladder cancer cohorts. To further explore the factors affecting the prognosis of bladder cancer patients, we also constructed univariate and multivariate Cox regression analyses. Interestingly, the data indicated that the risk score was an independent factor affecting the prognosis of bladder cancer patients in the training cohort (Fig. 7A), test cohort (Fig. 7B), and all bladder cancer cohort (Fig. 7C).
Figure 6.
Construction of a pyroptosis-related lncRNA prognostic signature in bladder cancer. (A, B) The coefficient and partial likelihood deviance of the prognostic signature. (C–E) OS curve in the high/low risk group and the ROC curve for prognosis prediction in the training cohort, test cohort and bladder cancer cohort. (F–H) Risk score distribution and survival status of patients and the expression of lncRNAs in the training cohort, test cohort and all bladder cancer cohorts. lncRNAs = long noncoding RNAs, OS = overall survival.
Figure 7.
Univariate and multivariate Cox hazard ratio analysis considering risk score and age, sex, clinical stage, and TNM stage in the training cohort (A), test cohort (B), and bladder cancer cohort (C).
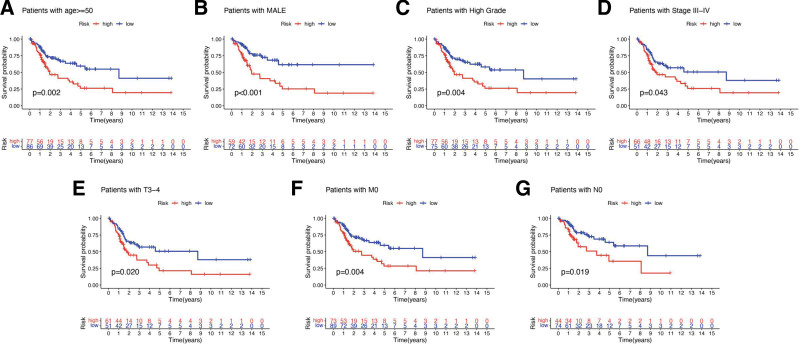
We also verified whether this prognostic was suitable for all BLCA patients. However, the results suggested a poor OS rate in the high-risk group versus the low-risk group in bladder cancer patients in the age >= 50 subgroup (P = .002), MALE subgroup (P < .001), high-grade subgroup (P = .004), stage III-IV subgroup (P = .043), pT3-4 subgroup (P = .020), pM0 subgroup (P = .004) and pN0 subgroup (P = .019) (Fig. 8A and G).
Figure 8.
The OS curve in different subgroups of bladder cancer. The survival curve suggested a poor OS rate in the high-risk group versus the low-risk group in bladder cancer patients in the age >= 50 subgroup (A), MALE subgroup (B), high-grade subgroup (C), stage III to IV subgroup (D), pT3-4 subgroup (E), pM0 subgroup (F), and pN0 subgroup (G). OS = overall survival.
3.5. Risk score correlated with different clinical characteristics and immune infiltration in bladder cancer
Figure 9A shows the expression of lncRNAs and the difference in clinical characteristics in the high- and low-risk groups in bladder cancer. The results indicated that bladder cancer patients with a high tumor grade had a higher risk score than those with a low tumor grade (P = 9.6e-5, Fig. 9B). Moreover, the risk score increased as clinical stage (P = .00034), pT stage (P = .0065), and pN stage increased (P = .00045) (Fig. 9C and E). A significantly different risk score was revealed in different immuneScore groups (P = 1.9e-9) and different cluster groups (P = 5e-13) of bladder cancer patients (Fig. 9F and G).
Figure 9.
Risk score correlated with clinicopathological features in bladder cancer. (A) The heatmap revealed the difference in clinical characteristics between the high- and low-risk groups in bladder cancer. (B–G) The risk score in different tumor grades (B), clinical stages (C), pT stages (D), pN stages (E), ImmuneScore (F), and clusters (G) of bladder cancer patients. *P < .05, **P < .01, and ***P < .001.
We also obtained a significant positive correlation between the risk score and infiltration levels of eosinophils (R = 0.18), M0 macrophages (R = 0.16), M2 macrophages (R = 0.28), activated mast cells (R = 0.23) and neutrophils (R = 0.18) in bladder cancer (Fig. 10A, P < .05). However, the infiltration levels of plasma cells (R = −0.3), CD8+ T cells (R = −0.16), and follicular helper T cells (R = −0.26) decreased as the risk score increased (Fig. 10A, P < .05). Figure 10B shows the correlation between the risk score and immune checkpoints, which revealed higher expression of PD-L1, CTLA4, HAVCR2, IDO1, LAG3, PDCD1, and PDCD1LG2 in the high-risk group than in the low-risk group in bladder cancer (Fig. 10B, all P < .05). SIGLEC15 expression decreased as the risk score increased (Fig. 10B, P = 4.5e-13).
Figure 10.
Risk score correlated distinct immune infiltration. (A) The risk score showed a significant correlation with the abundance of immune cells. (B) The expression of immune checkpoints in the high- and low-risk groups of bladder cancer patients.
4. Discussion
Pyroptosis, first reported by Zychlinsky in 1992, is inflammasome-induced programmed cell death mediated by gasdermins.[27] Accumulating studies have highlighted the vital role of pyroptosis in cancer since it may affect all stages of tumorigenesis and progression.[28] It is well known that lncRNAs may play tumor-suppressive and tumor-oncogenic roles in cancer and that lncRNA expression may accelerate or inhibit tumorigenesis and metastasis.[13] The lncRNA signature could serve as a prognostic biomarker and showed a correlation with immune infiltration in many cancers, including breast cancer and gastric cancer.[29,30] In our study, we aimed to explore the role of pyroptosis-related lncRNAs in the prognosis and tumor microenvironment (TME) of bladder cancer.
We first constructed a coexpression network and identified 722 pyroptosis-related lncRNAs. A univariate Cox regression analysis was performed, and 23 pyroptosis-related prognostic lncRNAs were obtained in the bladder cancer cohort. Based on these 23 lncRNAs, consensus clustering was conducted to separate the bladder cancer cohort into cluster 1 and cluster 2. Interestingly, these 2 clusters showed conspicuous differences in prognosis and TME characterization. Cluster 1 was associated with a high immune score, stroma score and ESTIMATE score and abundant immune cell infiltration, indicating hot tumors.[31] Cluster 2 was an immune-desert phenotype and associated with immune suppression.[32,33] The immune-desert phenotype is a major treatment challenge for cancer immunotherapy.[32] We also found that cluster 1 had a higher IPS than cluster 2. A previous study revealed that subgroup analysis and identification of different subgroups with distinctive molecular characteristics could provide potential treatment options for patients.[34] Another study suggested that the distinct immune orientations of the colorectal cancer molecular subtypes pave the way for tailored immunotherapies.[35] Moreover, the tumor subtyping method can serve as a tumor-agnostic biomarker for immune checkpoint inhibitor response prediction and will improve decision making in cancer treatment.[36] Thus, molecular subtype analysis and identification of subtypes of patients with different molecular characteristics would be of great significance for precision immunotherapy.
Using LASSO Cox analysis, we constructed a pyroptosis-related lncRNA signature, which showed a favorable discrimination performance for predicting bladder cancer patient prognosis. Several lncRNA-related signatures have been established in bladder cancer. A prognostic signature based on 8 lncRNAs served as a prognostic marker for bladder cancer.[37] Another bioinformatics study identified an immune-related lncRNA prognostic module in bladder cancer, which had good performance in the prediction of prognosis and immunotherapeutic response.[38] Another immune-related lncRNA signature was suggested as a prognostic marker for bladder cancer.[39] Our study first constructed a pyroptosis-related lncRNA prognostic signature for bladder cancer, which had never been studied before. Further analysis also suggested the risk score as an independent prognostic factor of bladder cancer.
The results also demonstrated a significant positive correlation between the risk score and infiltration of some immune cells, such as neutrophils. Interestingly, a previous study demonstrated that neutrophils correlated with high recurrence risk and OS rate.[40] Our study suggested poor overall survival in bladder cancer patients with a high-risk score, which was consistent with the results of a previous study. Immune-cell infiltration exerts a vital function in cancer progression and metastasis and affects the prognosis of cancer patients.[41,42]
Some limitations could be found in our study. GEO cohort should be used to verify the molecular subtypes and prognostic signature. Furthermore, it would be better to verify the expression and prognosis of lncRNAs using clinical tissues. Moreover, further study should be performed to explore the potential mechanism of the screened biomarkers.
5. Conclusions
Overall, this study constructed a pyroptosis-related lncRNA signature and clarified its correlation with immune infiltration, suggesting the vital role of pyroptosis-related lncRNAs in the prognosis and TME in bladder cancer. Further study should be conducted to verify our results.
Author contributions
Formal analysis: Jianzhong Ye.
Investigation: Tao Zeng.
Methodology: Jianzhong Ye, Heng Wang.
Project administration: Wen Tian.
Software: Heng Wang.
Validation: Wen Tian.
Visualization: Jianzhong Ye, Heng Wang.
Writing – original draft: Tao Zeng, Wen Tian.
Supplementary Material
Abbreviations:
- CI
- confidence interval
- IPS
- immunophenoscore
- lncRNAs
- long noncoding RNAs
- OS
- overall survival
- TIDE
- tumor immune dysfunction and exclusion
- TME
- tumor microenvironment
TZ and JY contributed equally to this work.
The current study was supported by Science and Technology Research Project of Hubei Educational Committee(B2021265) and Scientific Research Program of Hubei Provincial Department of Education(B2022247).
All methods were carried out in accordance with relevant guidelines and regulations.
The authors have no conflicts of interest to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Supplemental Digital Content is available for this article.
How to cite this article: Zeng T, Ye J, Wang H, Tian W. Identification of pyroptosis-related lncRNA subtype and signature predicts the prognosis in bladder cancer. Medicine 2023;102:42(e35195).
References
- [1].Richters A, Aben KKH, Kiemeney LALM. The global burden of urinary bladder cancer: an update. World J Urol. 2020;38:1895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Siracusano S, Rizzetto R, Porcaro AB. Bladder cancer genomics. Urologia. 2020;87:49–56. [DOI] [PubMed] [Google Scholar]
- [3].Oeyen E, Hoekx L, De Wachter S, et al. Bladder cancer diagnosis and follow-up: the current status and possible role of extracellular vesicles. Int J Mol Sci. 2019;20:821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J Clin. 2020;70:404–23. [DOI] [PubMed] [Google Scholar]
- [5].Guo H, Xie M, Zhou C, et al. The relevance of pyroptosis in the pathogenesis of liver diseases. Life Sci. 2019;223:69–73. [DOI] [PubMed] [Google Scholar]
- [6].Ruan J, Wang S, Wang J. Mechanism and regulation of pyroptosis-mediated in cancer cell death. Chem Biol Interact. 2020;323:109052. [DOI] [PubMed] [Google Scholar]
- [7].Fang Y, Tian S, Pan Y, et al. Pyroptosis: a new frontier in cancer. Biomed Pharmacother. 2020;121:109595. [DOI] [PubMed] [Google Scholar]
- [8].Ye Y, Dai Q, Qi H. A novel defined pyroptosis-related gene signature for predicting the prognosis of ovarian cancer. Cell Death Discov. 2021;7:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li X, He J. A novel pyroptosis-related gene signature for early-stage lung squamous cell carcinoma. Int J Gen Med. 2021;14:6439–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lin W, Chen Y, Wu B, et al. Identification of the pyroptosis-related prognostic gene signature and the associated regulation axis in lung adenocarcinoma. Cell Death Discov. 2021;7:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu J, Hong M, Li Y, et al. Programmed cell death tunes tumor immunity. Front Immunol. 2022;13:847345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shao W, Yang Z, Fu Y, et al. The pyroptosis-related signature predicts prognosis and indicates immune microenvironment infiltration in gastric cancer. Front Cell Dev Biol. 2021;9:676485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Quan J, Pan X, Zhao L, et al. LncRNA as a diagnostic and prognostic biomarker in bladder cancer: a systematic review and meta-analysis. Onco Targets Ther. 2018;11:6415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang D, Du D, Yi S, et al. LncRNA PCAT6: a potential biomarker for diagnosis and prognosis of bladder cancer. Ann Diagn Pathol. 2020;49:151642. [DOI] [PubMed] [Google Scholar]
- [16].Karki R, Kanneganti TD. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat Rev Cancer. 2019;19:197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xia X, Wang X, Cheng Z, et al. The role of pyroptosis in cancer: pro-cancer or pro-“host?”. Cell Death Dis. 2019;10:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang B, Yin Q. AIM2 inflammasome activation and regulation: a structural perspective. J Struct Biol. 2017;200:279–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu Z, Wang L, Liu L, et al. The identification and validation of two heterogenous subtypes and a risk signature based on ferroptosis in hepatocellular carcinoma. Front Oncol. 2021;11:619242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang L, Liu Z, Zhu R, et al. Multiomics landscape and clinical significance of a SMAD4-driven immune signature: implications for risk stratification and frontline therapies in pancreatic cancer. Comput Struct Biotechnol J. 2022;20:1154–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu Z, Guo C, Dang Q, et al. Integrative analysis from multicenter studies identities a consensus machine learning-derived lncRNA signature for stage II/III colorectal cancer. EBioMedicine. 2022;75:103750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu Z, Guo C, Li J, et al. Somatic mutations in homologous recombination pathway predict favorable prognosis after immunotherapy across multiple cancer types. Clin Transl Med. 2021;11:e619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liu Z, Lu T, Li J, et al. Development and clinical validation of a novel six-gene signature for accurately predicting the recurrence risk of patients with stage II/III colorectal cancer. Cancer Cell Int. 2021;21:359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu Z, Weng S, Xu H, et al. Computational recognition and clinical verification of TGF-β-derived miRNA signature with potential implications in prognosis and immunotherapy of intrahepatic cholangiocarcinoma. Front Oncol. 2021;11:757919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zychlinsky A, Prevost MC, Sansonetti PJ. Shigella flexneri induces apoptosis in infected macrophages. Nature. 1992;358:167–9. [DOI] [PubMed] [Google Scholar]
- [28].Liu L, Bai X, Wang J, et al. Combination of TMB and CNA stratifies prognostic and predictive responses to immunotherapy across metastatic cancer. Clin Cancer Res. 2019;25:7413–23. [DOI] [PubMed] [Google Scholar]
- [29].Liu Z, Mi M, Li X, et al. A lncRNA prognostic signature associated with immune infiltration and tumor mutation burden in breast cancer. J Cell Mol Med. 2020;24:12444–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ma B, Li Y, Ren Y. Identification of a 6-lncRNA prognostic signature based on microarray reannotation in gastric cancer. Cancer Med. 2020;9:335–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang B, Wu Q, Li B, et al. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bonaventura P, Shekarian T, Alcazer V, et al. Cold tumors: a therapeutic challenge for immunotherapy. Front Immunol. 2019;10:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kim JM, Chen DS. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann Oncol. 2016;27:1492–504. [DOI] [PubMed] [Google Scholar]
- [34].Kang H, Seo MK, Park B, et al. Characterizing intrinsic molecular features of the immune subtypes of salivary mucoepidermoid carcinoma. Transl Oncol. 2022;24:101496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Becht E, de Reyniès A, Giraldo NA, et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res. 2016;22:4057–66. [DOI] [PubMed] [Google Scholar]
- [36].Takamatsu S, Hamanishi J, Brown JB, et al. Mutation burden-orthogonal tumor genomic subtypes delineate responses to immune checkpoint therapy. J ImmunoTher Cancer. 2022;10:e004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lian P, Wang Q, Zhao Y, et al. An eight-long noncoding RNA signature as a candidate prognostic biomarker for bladder cancer. Aging (Albany NY). 2019;11:6930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wu Y, Zhang L, He S, et al. Identification of immune-related LncRNA for predicting prognosis and immunotherapeutic response in bladder cancer. Aging (Albany NY). 2020;12:23306–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang J, Shen C, Dong D, et al. Identification and verification of an immune-related lncRNA signature for predicting the prognosis of patients with bladder cancer. Int Immunopharmacol. 2021;90:107146. [DOI] [PubMed] [Google Scholar]
- [40].Xu Q, Wang C, Yuan X, et al. Prognostic value of tumor-infiltrating lymphocytes for patients with head and neck squamous cell carcinoma. Transl Oncol. 2017;10:10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yang S, Liu T, Nan H, et al. Comprehensive analysis of prognostic immune-related genes in the tumor microenvironment of cutaneous melanoma. J Cell Physiol. 2019;235:1025–35. [DOI] [PubMed] [Google Scholar]
- [42].Bremnes RM, Busund LT, Kilvaer TL, et al. The role of tumor-infiltrating lymphocytes in development, progression, and prognosis of non-small cell lung cancer. J Thorac Oncol. 2016;11:789–800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.